Abstract
Background
The present study analyzed the relationship between clinical features and the T790M mutation in non-small cell lung cancer (NSCLC) patients resistant to epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) treatment.
Methods
NSCLC patients with resistance to first-generation EGFR-TKIs in which the disease control time was more than 6 months after initial TKI treatment were enrolled. T790M mutation analysis was performed using one of the following methods according to each manufacturer’s protocols: Cobas EGFR mutation test (41/105, 39.0%), digital PCR (42/105, 40.0%) or Scorpion amplification refractory mutation system (ARMS) (22/105, 21.0%). Sample type of T790M was from tissue only (53/105, 50.5%), plasma only (46/105, 43.8%), tissue and plasma (6/105, 5.7%).
Results
Of 105 patients, 57 were T790M-positive and 48 were T790M-negative. T790M-positive patients had longer progression-free survival (PFS) after initial EGFR-TKI treatment (p = 0.019). T790M positivity was more frequent in patients treated with gefitinib than in those treated with icotinib (65% vs 40.54%, p = 0.018). The rate of T790M positivity was lower in patients with EGFR L858R (44.44%, 12/27) before TKI treatment than in those with EGFR 19del (72.0%, 36/50, p = 0.036). Patients who achieved PR after initial EGFR-TKI treatment had a higher rate of T790M positivity than those with SD (75.76% vs 50%, p = 0.023). There was no relationship between T790M status and age, gender, primary site, metastasis site, or treatment before TKI.
Conclusion
Progression-free survival (PFS), drug type, response to initial EGFR-TKI treatment, and EGFR status before initial EGFR treatment were associated with the frequency of T790M mutation.
Keywords: lung cancer, EGFR, T790M, gefitinib, icotinib
Introduction
In East Asia, the most common driver mutation in lung adenocarcinoma is the EGFR mutation, which accounts for approximately 50% of cases.1 First- and second-generation epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) such as gefitinib, erlotinib, icotinib, and afatinib show good responses in non-small cell lung cancer (NSCLC) patients, especially those with exon 19 deletions (Del 19) and exon 21 substitution (L858R).2–5 However, after 12 months of treatment, most patients progress.6,7 The resistance mechanisms to first- and second-generation EGFR-TKIs include threonine at position 790 (T790M) in EGFR, small cell lung cancer transformation, mutation of phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha, mesenchymal–epithelial transition, amplification, and epithelial–mesenchymal transition. Among these resistance mechanisms, T790M in EGFR is the most common, accounting for 50–60% of cases.6,8
Third-generation EGFR-TKIs such as osimertinib show good efficacy in patients with T790M-positive NSCLC who acquire resistance to prior EGFR-TKI treatment. Compared with pemetrexed plus platinum therapy, patients treated with osimertinib show longer progression-free survival (PFS) (10.1 vs 4.4 months; hazard ratio, 0.30; and 95% confidence interval, 0.23–0.41).9–11 Recently, in the FLAURA trial, osimertinib was compared with first-generation EGFR-TKIs in the treatment of EGFR mutation-naive patients with advanced NSCLC. Median PFS was superior with ostimertinib (18.9 vs 10.2 months)12, median OS was also longer in patients with ostimertinib (38.6 vs 31.8 months; HR=0.799, P=0.0462).12,13 Based on this, in 2018, the FDA approved osimertinib as the first-line treatment of NSCLC patient with EGFR mutant. However, after the failure of first-line treatment with ostimertinib, there will be no TKI available. The only option is chemotherapy. Chemotherapy has a significant impact on the quality of life of patients. Maximizing the duration of chemotherapy-free treatment is important for many patients, so how to maximize survival benefit and the most effective TKI treatment sequence is still the biggest problem we are facing.
Previous studies reported that longer PFS in patients treated with EGFR-TKIs and 19 del in EGFR is associated with higher rates of EGFR T790M positivity, although the number of cases in these studies were small.14,15 In the present study, we investigated the relationship between clinical characteristics and the EGFR T790M mutation in 105 NSCLC patients with resistance to prior EGFR-TKI treatment. We also showed that patients treated with gefitinib had a higher frequency of the T790M mutation than those treated with icotinib.
Methods
Patient Samples
This was a retrospective observational study that included patients enrolled from Jiangsu Province Hospital in China between January 2017 and October 2018. This study was approved by the ethical committee of the Jiangsu Province Hospital Medical Ethics Committee. The study was performed in accordance with the approved guidelines, and written informed consents were obtained from all participants included in the study. The inclusion criteria were as follows: lung adenocarcinoma, presence of resistance to treatment with first-generation EGFR-TKIs, the disease control time was more than 6 months after initial TKI treatment. Clinical data included age, gender, tumor location, treatment with first EGFR-TKIs, presence of EGFR mutations before and after EGFR-TKI treatment, PFS, effectiveness of previous EGFR-TKI treatment, metastatic sites, and presence of T790M after previous EGFR-TKI treatment.
EGFR Mutation Test
T790M mutation analysis was performed using one of the following methods according to each manufacturer’s protocols: Cobas EGFR mutation test (the cobas 4800 FFPET test utilized as per manufacturer’s instructions (Roche Molecular Systems Inc.) and the cobas 4800 blood test (Roche Molecular Systems Inc.)), digital PCR (ddPCRTM., Bio-Rad/MolecularMD, Hercules, CA, USA) or Scorpion amplification refractory mutation system (ARMS) (QIAGEN, Venlo, The Netherlands), or analysis of biopsy specimens and plasma specimens.
Statistical Analysis
The relationship between the frequency of T790M mutation and clinicopathologic parameters (Table 1) was assessed using the chi-square test, Fisher’s exact test and log rank test (Table 1 and Table 6). P values ≤0.05 were statistically significant. All statistical tests were performed using SPSS ver. 24.0 (IBM Corp., Armonk, NY, USA).
Table 4.
Univariate Analysis Of The Association Between Patients’ Age
| T790M Status | 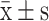 |
t-Value | P-value | |
|---|---|---|---|---|
| Age | Negative | 60.92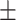 9.86 9.86 |
−0.486 | 0.628 |
| Positive | 60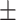 9.41 9.41 |
Table 1.
Patients’ Characteristics And Demographic Data
| Characteristics | No (%) n=105 |
|---|---|
| Age, median (range, yr) | 60 (37–85) |
| Gender | |
| Male | 39 (37.1) |
| Female | 66 (62.9) |
| Primary site | |
| Left lung | 44 (41.9) |
| Right lung | 60 (57.1) |
| Unknown | 1 (1.0) |
| Performance status | |
| ECOG < 2 | 96 (91.4) |
| ECOG ≥ 2 | 9 (8.6) |
| Initial EGFR-TKI | |
| Gefitinib | 60 (57.1) |
| Icotinib | 37 (35.2) |
| Other | 8 (7.6) |
| EGFR mutation status before TKI | |
| Exon 19 deletions | 50 (47.6) |
| Exon 21 L858R | 27 (25.7) |
| Other mutation | 3 (2.9) |
| Unknown | 25(23.8) |
| EGFR mutation status after TKI | |
| Exon 19 deletions | 37 (35.2) |
| Exon 21 L858R | 21 (20.0) |
| No common mutation (Exon 19 deletion and Exon 21 L858R) | 22 (21.0) |
| Unknown | 25 (23.8) |
| PFS of initial TKI, median (range, month) | 17 (1–63) |
| Response of initial TKI | |
| PR | 33 (31.4) |
| SD | 42 (40.0) |
| Unknown | 30 (28.6) |
| Metastasis site | |
| Lung | |
| Yes | 92 (87.6) |
| No | 11 (10.5) |
| Unknown | 2 (1.9) |
| Bone | |
| Yes | 50 (47.6) |
| No | 53 (50.5) |
| Unknown | 2 (1.9) |
| Brain | |
| Yes | 32 (30.5) |
| No | 71 (67.6) |
| Unknown | 2 (1.9) |
| Other | |
| Yes | 19 (18.1) |
| No | 84 (80.0) |
| Unknown | 2 (1.9) |
| Treatment before TKI | |
| Chemotherapy | |
| Yes | 50 (47.6) |
| No | 46 (43.8) |
| Unknown | 9 (8.6) |
| Operation | |
| Yes | 17 (16.2) |
| No | 84 (80.0) |
| Unknown | 4 (3.8) |
Table 6.
Patients’ Characteristics And Demographic Data Between Gefitinib And Ecotinib
| Characteristics | Gefitinib | Ecotinib | p-Value | ||
|---|---|---|---|---|---|
| Age, median (range, yr) | 61(44–85) | 60 (37–76) | 0.265 | ||
| Gender | Male | 26(76.47%) | 8(23.53%) | 0.029 | |
| Female | 34(53.97%) | 29(46.03%) | |||
| Primary site | Left lung | 29(67.44%) | 14(32.56%) | 0.281 | |
| Right lung | 31(58.49%) | 22(41.51%) | |||
| Unknown | 0(0.00%) | 1(100.00%) | |||
| Performance status | ECOG <2 | 56(62.92%) | 33(37.08%) | 0.476 | |
| ECOG ≥ 2 | 4(50.00%) | 4(50.00%) | |||
| EGFR mutation status before TKI | Exon 19 deletions | 29(60.42%) | 19(39.58%) | 0.460 | |
| Exon 21 L858R | 19(70.37%) | 8(29.63%) | |||
| Other mutation | 0(0.00%) | 1(100.00%) | |||
| Unknown | 12(57.14%) | 9 (42.86%) | |||
| EGFR mutation status after TKI | Exon 19 deletion | 18(50.00%) | 18(50.00%) | 0.248 | |
| Exon 21 L858R | 11 (61.11%) | 7 (38.89%) | |||
| No common mutation (Exon 19 deletion and Exon 21 L858R) | 14 (70.00%) | 6 (30.00%) | |||
| Unknown | 17 (73.91%) | 6 (26.09%) | |||
| PFS of initial TKI, median (range, months) | 18(1–56) | 16 (3–63) | 0.204 | ||
| Response of initial TKI | PR | 19 (57.58%) | 14 (42.42%) | 0.643 | |
| SD | 23 (60.53%) | 15 (39.47%) | |||
| Unknown | 18(69.23%) | 8(30.77%) | |||
| Metastasis site | Lung | Yes | 52(60.47%) | 34(39.53%) | 0.837 |
| No | 7(70.00%) | 3(30.00%) | |||
| Unknown | 1(100.00%) | 0(0.00%) | |||
| Bone | Yes | 30(63.83%) | 17(36.17%) | 0.802 | |
| No | 29(59.18%) | 20(40.82%) | |||
| Unknown | 1(100.00%) | 0(0.00%) | |||
| Brain | Yes | 22(73.33%) | 8(26.67%) | 0.141 | |
| No | 37(56.06%) | 29(43.94%) | |||
| Unknown | 1(100.00%) | 0(0.00%) | |||
| Other | Yes | 11(68.75%) | 5(31.25%) | 0.743 | |
| No | 48(60.00%) | 32(40.00%) | |||
| Unknown | 1(100.00%) | 0(0.00%) | |||
| Treatment before TKI | Chemotherapy | Yes | 29(59.18%) | 20(40.82%) | 0.852 |
| No | 26(63.41%) | 15(36.59%) | |||
| Unknown | 5(71.43%) | 2(28.57%) | |||
| Operation | Yes | 8(50.00%) | 8(50.00%) | 0.289 | |
| No | 49(62.82%) | 29(37.18%) | |||
| Unknown | 3(100.00%) | 0(0.00%) | |||
| Sample type of T790M | Tissue only | 29 (59.18%) | 20 (40.82%) | 0.633 | |
| Plasma only | 28 (66.67%) | 14 (33.33%) | |||
| Tissue and Plasma | 3 (50.00%) | 3 (50.00%) | |||
Results
Patient Characteristics
The study enrolled 105 patients with advanced NSCLC with resistance to treatment with initial EGFR-TKIs. The patients’ characteristics and demographic data are shown in Table 1. The median age was 60 years (range, 37–85 years), and there were 39 men and 66 women. The primary site was the left lung in 44 patients and the right lung in 60 patients. Most patients (n = 60) received gefitinib as initial EGFR-TKI treatment, and 37 patients were treated with icotinib. Before TKI treatment, 50 patients had exon 19 deletions, and exon 21 L858R occurred in 27 patients. After TKI treatment, 22 patients had neither EGFR 19 deletion nor 21 L858R mutation, 37 patients harbored exon 19 deletions, and 21 patients harbored the exon 21 L858R mutation. After initial EGFR-TKI treatment, the median PFS was 17 months (range, 1–63 months). Response to initial EGFR-TKI treatment was PR in 33 patients and SD in 42 patients. After EGFR-TKI treatment, 92 patients had a new metastasis site in the lung, 50 in bone, 32 in the brain, and 19 in other sites. Before TKI treatment, 50 patients received chemotherapy and 17 underwent surgery.
T790M Mutation Status
T790M mutation status and baseline EGFR mutation status are shown in Table 2. Fifty-seven patients were T790M-positive, and 48 were negative. Forty-one patients were analyzed by the Cobas EGFR mutation test, 42 by Digital PCR, and 22 by Scorpion ARMS. Samples used for T790M analysis were tissues only in 53 patients, plasma only in 46 patients, and both tissue and plasma in six patients.
Table 2.
T790M Mutation Status
| T790M Status After TKI | |
| Positive | 57 (54.3) |
| Negative | 48 (45.7) |
| T790M analysis | |
| Digital PCR | 42 (40.0) |
| Cobas EGFR Mutation Test | 41 (39.0) |
| Scorpion ARMS | 22 (21.0) |
| Sample type of T790M | |
| Tissue only | 53 (50.5) |
| Plasma only | 46 (43.8) |
| Tissue and plasma | 6 (5.7) |
Analysis Of The Association Between Patient Characteristics And T790M Status
The relationship between patient characteristics and T790M status is shown in Tables 3–5. T790M-positive patients had longer PFS after initial EGFR-TKI treatment. Patients treated with gefitinib had a higher frequency of T790M positivity than those treated with icotinib. Patients harboring EGFR L858R before TKI treatment had a lower T790M positivity rate than those with EGFR 19del. Patients who achieved PR after initial EGFR-TKI treatment had a higher rate of T790M positivity than those who showed SD. There was no relationship between T790M status and age, gender, primary site, metastasis site, and treatment before TKI.
Table 3.
Univariate Analysis Of The Association Between Patients’ Characteristics And T790M Status
| Characteristics | T790M Status | p-Value | |||
|---|---|---|---|---|---|
| Positive | Negative | ||||
| Gender | Male | 20(51.28%) | 19(48.72%) | 0.635 | |
| Female | 37(56.06%) | 29(43.94%) | |||
| Primary site | Left lung | 25(56.82%) | 19(43.18%) | 0.724 | |
| Right lung | 32(53.33%) | 28(46.67%) | |||
| Initial EGFR-TKI | Gefitinib | 39(65.0%) | 21(35.0%) | 0.018 | |
| Icotinib | 15(40.54%) | 22(59.46%) | |||
| EGFR mutation status before TKI | Exon 19 deletions | 36(72.0%) | 14(28.0%) | 0.036 | |
| Exon 21 L858R | 12(44.44%) | 15(55.56%) | |||
| Other mutation | 1(33.33%) | 2(66.67%) | |||
| EGFR mutation status after TKI | Exon 19 deletion | 24(64.86%) | 13(35.14%) | =0.001 | |
| Exon 21 L858R | 13(61.9%) | 8(38.1%) | |||
| No common mutation (Exon 19 deletion and Exon 21 L858R) | 4(18.18%) | 19(81.82%) | |||
| Response of initial TKI | PR | 25(75.76%) | 8(24.24%) | 0.023 | |
| SD | 21(50%) | 21(50%) | |||
| Metastasis site | Lung | Yes | 50(54.35%) | 42(45.65%) | 0.990 |
| No | 6(54.55%) | 5(45.45%) | |||
| Bone | Yes | 29(58%) | 21(42%) | 0.472 | |
| No | 27(50.94%) | 26(49.06%) | |||
| Brain | Yes | 20(62.5%) | 12(37.5%) | 0.266 | |
| No | 36(50.7%) | 35(49.3%) | |||
| Other | Yes | 7(36.84%) | 12(63.16%) | 0.089 | |
| No | 49(58.33%) | 35(41.67%) | |||
| Treatment before TKI | Operation | Yes | 7(41.18%) | 10(58.82%) | 0.228 |
| No | 48(57.14%) | 36(42.86%) | |||
| Chemotherapy | Yes | 26(52%) | 24(48%) | 0.274 | |
| No | 29(63.04%) | 17(36.96%) | |||
Table 5.
Multivariate Analysis Of The Association Between Patients’ Characteristics And T790M Status
| Characteristics | OR (95% CI) | p-Value |
|---|---|---|
| Initial EGFR-TKI icotinib (vs gefitinib) | 0.05(0.003–0.803) | 0.034 |
| EGFR mutation status before TKI | ||
| Exon 19 deletions (vs Exon 21 L858R) | 0.081(0.008–0.793) | 0.031 |
| Exon 19 deletions (vs other mutations) | 0.005(0.000–0.143) | 0.002 |
| Response of initial TKI PR (vs SD) | 21.569(1.791–259.808) | 0.016 |
Abbreviations: OR, odds ratio; CI, confidence interval.
Relationship Between T790M Status And EGFR Mutation
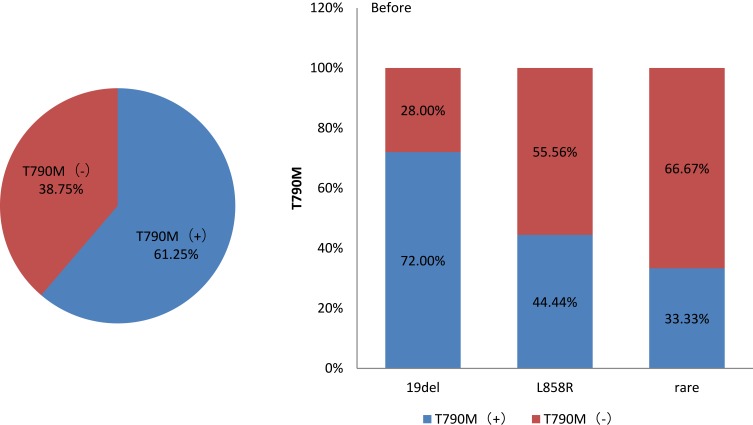
After initial first-generation EGFR TKIs treatment, of 50 patients with EGFR 19 deletion mutation before TKI treatment, 36 harbored the T790M mutation. Of 27 patients with EGFR 21 L858R mutation, 12 had the T790M mutation (p = 0.036; Figure 1).
Figure 1.
Relationship between T790M status and EGFR mutation.
Relationship Between T790M Status And PFS After Initial EGFR-TKI Treatment
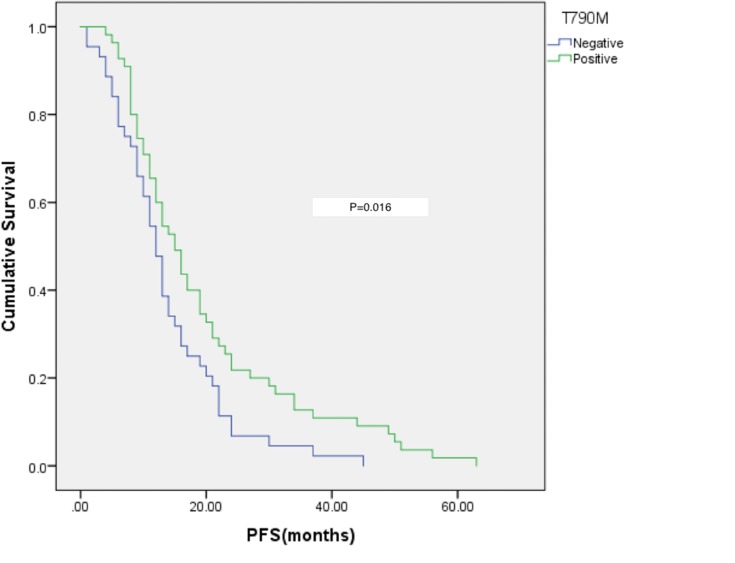
The median PFS of T790M-positive patients was 15 months and that of T790M-negative patients was 12 months (p = 0.019). Patients with a longer PFS after initial EGFR-TKI treatment had a higher rate of T790M positivity (Figure 2).
Figure 2.
Relationship between T790M status and PFS after initial EGFR-TKI treatment.
Relationship Between T790M Status And Response To Initial EGFR-TKI Treatment
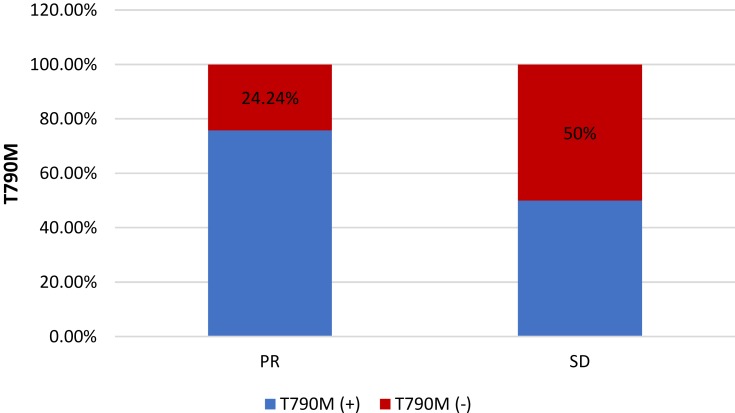
After initial EGFR-TKI treatment, 33 patients achieved PR, and the T790M-positive rate in those patients was 75.76 (25/33); 42 patients showed SD with a T790M-positive rate of 50% (21/42) (p = 0.023; Figure 3).
Figure 3.
Relationship between T790M status and response to initial EGFR-TKI treatment.
Relationship Between T790M Status And Initial EGFR-TKI
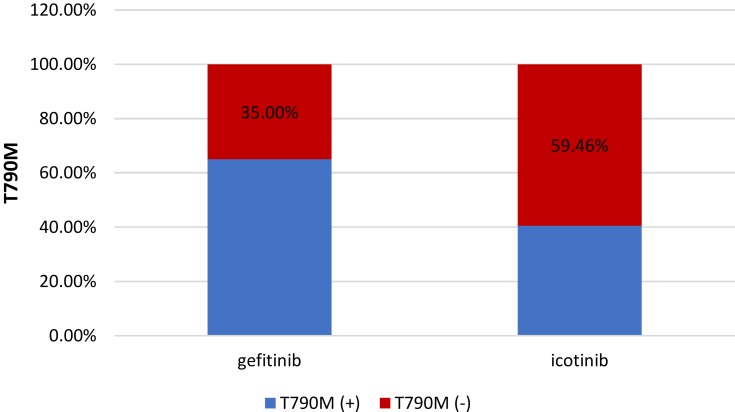
Fifty patients received gefitinib and 37 received icotinib as initial EGFR-TKI treatment, and the rate of T790M-positive cases was 65% (39/50) and 40.54% (15/37), respectively (p = 0.018; Figure 4).
Figure 4.
Relationship between T790M status and initial EGFR-TKI.
Discussion
Third-generation EGFR-TKIs such as osimertinib show good responses in T790M-positive NSCLC patients. The objective response rate is 60–70% and the median PFS was approximately 1 year with minimal adverse effects.9,16 For the detection of T790M mutation, tissue biopsies remain the gold standard,17 although plasma T790M assays are more available, plasma genotyping is associated with an approximately 30% false-negative rate.18 To distinguish false-negative patients from NSCLC patients who can only access liquid biopsy and are not fit for re-biopsy, we analyzed the clinical features of NSCLC patients with T790M-positive mutation.
In the present study, patients with the EGFR exon 19 deletion had higher mutation rates in T790M than those with the L858R mutation, which is consistent with the previous reports.14,15,19–21
The possible mechanisms underlying the differences in T790M mutation status before and after previous EGFR-TKI treatment remains unclear. In a previous study, patients with EGFR 19del had a better response to EGFR-TKI treatment than those with the L858R mutation.22 Bose et al23 reported that EGFR 19del located between strand β3 and helix αC could disrupt the inactive conformation of the EGFR kinase domain and enhance the effectiveness of EGFR-TKIs. Zhang et al24 showed that structural differences played an important role in EGFR kinase domain activation. These intrinsic structural characteristics may underlie the differences in the rates of T790M mutation between patients with the 19del and L858R mutations.
In the present study, we first showed that patients who received gefitinib as a previous EGFR-TKI treatment had a higher rate of T790M mutation than those treated with icotinib. Patients’ characteristics and demographic data between gefitinib and ecotinib are shown in Table 6, and there were no significant differences in other clinical features except gender. As we know, there was no study shown gender influences the positive rate of T790M, it may due to the small sample size of the present study, further research is needed. Mok TS9 found in patients with T790M-positive advanced non-small-cell lung cancer, who had disease progression after first-line EGFR-TKI therapy, that osimertinib had significantly greater efficacy than platinum therapy plus pemetrexed (10.1 months vs 4.4 months; HR: 0.30; 95% CI: 0.23 to 0.41; P<0.001). For these patients, osimertinib is recommended. In Hattori Y’s25 study, in patients with non-squamous non-small-cell lung cancer (NSCLC) who are epidermal growth factor receptor (EGFR) mutation-positive and for whom EGFR-tyrosine kinase inhibitor (TKI) 1st line has failed, patients treated with carboplatin, paclitaxel, and bevacizumab, the median progression-free survival (PFS) was 6.6 months (95% CI; 4.8–12.0 months) and median overall survival was 18.2 months (95% CI; 12.0–23.4 months). For the patients with T790M-negative after EGFR-TKI failure, treatment with carboplatin, paclitaxel, and bevacizumab can be a promising treatment option.
In the study by Wang H,26 the incidence of T790M mutation was 56.1%, which is similar to that observed with other first- and second-generation EGFR TKIs. In Wang H’s study, 41 patients had successfully tissue re-biopsy, the re-biopsy rate was 53.2% and the incidence rate of T790M mutations was 56.1%. In our study, patients with icotinib’s tissue re-biopsy was successful in 23 patients, and the incidence rate of T790M mutations was 56.5% (13/23). In all 37 patients with icotinib, T790M status of 15 patients was positive (40.5%,15/37). This apparent discrepancy can be due to the small sample size of the present study as well as the use of low-sensitivity detection methods (false negative), and further research is needed to confirm the present results. A network meta-analysis27 showed that gefitinib and icotinib had similar effectiveness and adverse effects. However, in a nude mouse model, gefitinib exhibited stronger antitumor activity than icotinib, and gefitinib also inhibited the phosphorylation of EGFR better than icotinib. These differences may be associated with the different stereochemistry between gefitinib and icotinib, although the underlying mechanism remains to be elucidated.
We showed that patients with a partial response to previous EGFR-TKI treatment had a lower rate of T790M mutation than those with EGFR 19 deletion and L858R mutation and that patients with PR had higher T790M mutation rates than those with SD. This result was similar to that reported by Oya et al28 who showed that patients with a better response to EGFR-TKIs had higher mutation rates in EGFR T790M.
In the study of Passiglia F,29 the location of metastatic sites significantly influences the diagnostic accuracy of ctDNA analysis, and the ability to identify resistant T790M mutations in the plasma of NSCLC patients is significantly higher in presence of extrathoracic disease (M1b) than the intrathoracic disease (M1a). In our study, 52 patients used ctDNA analysis in the detection of T790M mutations from plasma. In these patients, 8 were with intrathoracic disease (M1a) and 44 were with extrathoracic disease (M1b) and the T790M-positive rates were 37.5% (3/8) and 50.0% (22/44). The location of metastatic sites may influence the positive rate of T790M, especially in NSCLC patients with M1a. More data are needed to confirm the present results.
The present study had several limitations. First, this was a retrospective study and patient’s backgrounds were diverse. Therefore, the potential bias was greater than that in studies with a prospective design. Second, because of the short period of investigation, survival data were limited. Third, the study was performed in a single center and all patients were Chinese, which may not be representative of other ethnic populations. Fourth, current recommendations are to detect EGFR mutations in fresh, frozen or formalin-fixed paraffin-embedded (FFPE) tissue from a solid biopsy. In the real world, sufficient tumor tissues for multiplex EGFR mutation analysis are difficult to obtain.30 Additionally, because of spatial and temporal heterogeneity, therapeutic guidance throughout the entire process of treatment could not be correctly made by mutation analysis from initial tumor tissue samples at diagnosis, especially after disease progression. For these reasons, new methods such as “liquid biopsies” are being developed. In these new methods, plasma has been shown to be the most promising.31 Compared with the detection of EGFR mutations from tissue samples, ctDNA tests have a sensitivity of 46–82%, a specificity of 90–99% and total coincidence rate of 78–88% in assessing EGFR mutations.32 In study by Buder A,32 T790M testing was performed in 354 plasma samples collected from 129 patients. The concordance rate between ddPCR and Cobas for T790M, sensitivity, and specificity were 86%, 100%, and 85%, respectively. In our study, 46 patients were tested by liquid biopsy testing blood samples which may cause 22 samples to not get common mutation (Exon 19 deletion and Exon 21 L858R) after initial TKI treatment. We still need more data to analyse the relationship between T790M status and EGFR mutation after initial EGFR-TKI treatments. Although this study is the first to show that patients receiving gefitinib had a higher rate of T790M mutation than those treated with icotinib, further research is needed to confirm the present results.
In conclusion, the present study demonstrated that NSCLC patients with EGFR 19del, those treated with gefitinib, and those showing PR after EGFR-TKI treatment had a higher rate of T790M mutation than those without these. These data are potentially important for treatment decision-making after 1st EGFR-TKI progressions, especially in patients with false-negative results that can only access liquid biopsy and are not fit for re-biopsy. Further investigation and basic mechanistic studies are needed to confirm our findings.
Acknowledgments
This study was supported by National Natural Science Foundation of China (grant no.81572262), Natural Science Foundation of Jiangsu Province (grant no.BK20151583), Jiangsu Province’s Key Provincial Talents Program (grant no.ZDRCA2016028), 333 High Class Talented Man Project (grant no.BRA2016516), Six Talents Peak Project of Jiangsu Province (grant no.2013-wsn-032), and The Natural Science Foundation of the Jiangsu Higher Education Institution of China (grant no.18KJB320006).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9(2):154–162. doi: 10.1097/JTO.0000000000000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou C, Wu YL, Chen G, et al. Final overall survival results from a randomised, phase III study of erlotinib versus chemotherapy as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer (OPTIMAL, CTONG-0802). Ann Oncol. 2015;26(9):1877–1883. doi: 10.1093/annonc/mdv276 [DOI] [PubMed] [Google Scholar]
- 3.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–246. doi: 10.1016/S1470-2045(11)70393-X [DOI] [PubMed] [Google Scholar]
- 4.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11(2):121–128. doi: 10.1016/S1470-2045(09)70364-X [DOI] [PubMed] [Google Scholar]
- 5.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 6.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–2247. doi: 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238 [DOI] [PubMed] [Google Scholar]
- 9.Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376(7):629–640. doi: 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372(18):1689–1699. doi: 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 11.Cross DA, Ashton SE, Ghiorghiu S, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4(9):1046–1061. doi: 10.1158/2159-8290.CD-14-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378(2):113–125. doi: 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- 13.Ramalingam SS, Gray J, Ohe Y, et al. Osimertinib vs comparator EGFR-TKI as first-line treatment for EGFRm advanced NSCLC (FLAURA): final overall survival analysis. Ann Oncol. 2019;30:v914–v915. doi: 10.1093/annonc/mdz394.076 [DOI] [Google Scholar]
- 14.Joo JW, Hong MH, Shim HS. Clinical characteristics of T790M-positive lung adenocarcinoma after resistance to epidermal growth factor receptor-tyrosine kinase inhibitors with an emphasis on brain metastasis and survival. Lung Cancer. 2018;121:12–17. doi: 10.1016/j.lungcan.2018.04.013 [DOI] [PubMed] [Google Scholar]
- 15.Matsuo N, Azuma K, Sakai K, et al. Association of EGFR exon 19 deletion and EGFR-TKI treatment duration with frequency of T790M mutation in EGFR-mutant lung cancer patients. Sci Rep. 2016;6:36458. doi: 10.1038/srep36458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang JC, Ahn MJ, Kim DW, et al. Osimertinib in pretreated T790M-positive advanced non-small-cell lung cancer: AURA study phase II extension component. J Clin Oncol. 2017;35(12):1288–1296. doi: 10.1200/JCO.2016.70.3223 [DOI] [PubMed] [Google Scholar]
- 17.Zarogoulidis P, Gaga M, Huang H, Darwiche K, Rapti A, Hohenforst-Schmidt W. Tissue is the issue and tissue competition. Re-biopsy for mutation T790: where and why? Clin Transl Med. 2017;6(1):6. doi: 10.1186/s40169-017-0135-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34(28):3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang YH, Hsu KH, Tseng JS, et al. The association of acquired T790M mutation with clinical characteristics after resistance to first-line epidermal growth factor receptor tyrosine kinase inhibitor in lung adenocarcinoma. Cancer Res Treat. 2018. doi: 10.4143/crt.2017.512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaburagi T, Kiyoshima M, Nawa T, et al. Acquired EGFR T790M mutation after relapse following EGFR-TKI therapy: a population-based multi-institutional study. Anticancer Res. 2018;38(5):3145–3150. doi: 10.21873/anticanres.12577 [DOI] [PubMed] [Google Scholar]
- 21.Ke EE, Zhou Q, Zhang QY, et al. A higher proportion of the EGFR T790M mutation may contribute to the better survival of patients with exon 19 deletions compared with those with L858R. J Thorac Oncol. 2017;12(9):1368–1375. doi: 10.1016/j.jtho.2017.05.018 [DOI] [PubMed] [Google Scholar]
- 22.Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015;16(2):141–151. doi: 10.1016/j.cell.2006.05.013 [DOI] [PubMed] [Google Scholar]
- 23.Bose R, Zhang X. The ErbB kinase domain: structural perspectives into kinase activation and inhibition. Exp Cell Res. 2009;315(4):649–658. doi: 10.1016/j.yexcr.2008.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125(6):1137–1149. [DOI] [PubMed] [Google Scholar]
- 25.Hattori Y, Satouchi M, Shimada T, et al. A phase 2 study of bevacizumab in combination with carboplatin and paclitaxel in patients with non-squamous non-small-cell lung cancer harboring mutations of epidermal growth factor receptor (EGFR) after failing first-line EGFR-tyrosine kinase inhibitors (HANSHIN Oncology Group 0109). Lung Cancer. 2015;87(2):136–140. doi: 10.1016/j.lungcan.2014.12.007 [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Zhang L, Si X, Zhang X, Wang M. Re-biopsy status among Chinese non-small-cell lung cancer patients who progressed after icotinib therapy. Onco Targets Ther. 2018;11:7513–7519. doi: 10.2147/OTT [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Zhang Y, Feng G, et al. Comparison of effectiveness and adverse effects of gefitinib, erlotinib and icotinib among patients with non-small cell lung cancer: a network meta-analysis. Exp Ther Med. 2017;14(5):4017–4032. doi: 10.3892/etm.2017.5094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oya Y, Yoshida T, Kuroda H, et al. Association between EGFR T790M status and progression patterns during initial EGFR-TKI treatment in patients harboring EGFR mutation. Clin Lung Cancer. 2017;18(6):698–705 e692. doi: 10.1016/j.cllc.2017.05.004 [DOI] [PubMed] [Google Scholar]
- 29.Passiglia F, Rizzo S, Rolfo C, et al. Metastatic site location influences the diagnostic accuracy of ctDNA EGFR- mutation testing in NSCLC patients: a pooled analysis. Curr Cancer Drug Targets. 2018. doi: 10.2174/1568009618666180308125110 [DOI] [PubMed] [Google Scholar]
- 30.Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol. 2012;7(1):115–121. doi: 10.1097/JTO.0b013e3182307f98 [DOI] [PubMed] [Google Scholar]
- 31.Yung TK, Chan KC, Mok TS, Tong J, To KF, Lo YM. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res. 2009;15(6):2076–2084. doi: 10.1158/1078-0432.CCR-08-2622 [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Zhuo M, Ye X, et al. Quantification of mutant alleles in circulating tumor DNA can predict survival in lung cancer. Oncotarget. 2016;7(15):20810–20824. doi: 10.18632/oncotarget.8021 [DOI] [PMC free article] [PubMed] [Google Scholar]