Abstract
Background
Mechanisms of neuropathic pain are not fully understood. Molecular changes in spinal dorsal horn take part in the initiation and development of neuropathic pain.
Methods
To detect the transcriptome changes in the dorsal spinal cord of neuropathic pain rat, sciatic nerve chronic constriction injury (CCI) rats were used. Then, the CCI ipsilateral dorsal spinal cords of lumbar L3-L5 segments were collected at 14th day post-CCI and subjected to microRNA and long non-coding RNA (lncRNA)/mRNA microarray. To evaluate functions of differential mRNAs, bioinformatics methods including gene ontology (GO) and KEGG pathway analysis were conducted for significantly up- and downregulated mRNAs.
Results
MicroRNA microarrays showed that 13 microRNAs were differently expressed between CCI and sham-operated rats (fold change ≥ 2.0). Six of them were upregulated, and the other seven were downregulated in CCI group. MicroRNA-1b overexpressed 18.7 times after CCI. LncRNA/mRNA microarray detected 876 lncRNAs with significant differential expression (fold change ≥ 2.0). Among them, 339 were significantly upregulated, and 537 were downregulated in CCI group. Sixteen of them differentially expressed more than 10 times and the lncRNA XR_356687 overexpressed as high as 53 times. In addition, 950 mRNAs were differentially expressed (fold change ≥ 2.0), including 405 upregulated and 545 downregulated in CCI group. Ten of these mRNAs with changed expressions of more than 10 times. The Hspa1b (encodes heat shock protein 70) overexpressed 24 times in CCI rats. Gene ontology analysis revealed that hundreds of differentially expressed mRNAs involved in the biological processes, cellular component, and molecular function. In addition, these genes significantly enriched into 32 KEGG pathways, including the TNF, FoxO, cytokine–cytokine receptor interaction, and calcium signaling pathways.
Conclusion
Neuropathic pain induced comprehensive changes of transcription profile in the dorsal spinal cord. These differentially expressed transcripts in spinal cord could be potential targets in defeating neuropathic pain.
Keywords: neuropathic pain, peripheral nerve injury, microRNA, long non-coding RNA, mRNA, pathways, spinal dorsal horn, chronic constriction injury
Introduction
Neuropathic pain is still a major health and economic burden worldwide.1,2 However, the mechanisms of neuropathic pain are not fully understood, and the elucidation of molecular changes during neuropathic pain is fundamental for the development of mechanism-oriented treatments.3
Spinal dorsal horn is the docking site of primary and secondary sensory neurons. It also harbors the descending terminals from supraspinal structures, such as locus coeruleus and periaqueductal grey.4–6 Evidence shows that spinal dorsal horn plays important roles in the generation and maintain of neuropathic pain, because it is one of the key areas where central sensitization take place in neuropathic pain and other kinds of chronic pain situation.7–10 Therefore, transcriptomic changes of functional molecules such as mRNAs and non-coding RNAs in the spinal dorsal horn influence the development and outcome of pathological pain.11,12
MicroRNAs are small non-coding molecules, which post-transcriptionally modulate many kinds of pathological processes including neuropathic pain.13,14 Abnormal expression of microRNA in spinal cord has been reported in animal models of neuropathic pain.15 More than 100 microRNAs were detected abnormally expressed in the dorsal horn of CCI rats with TaqMan® Low Density Array.16 By using sequencing analysis,17 12 differential microRNAs were detected in spinal cord of spared nerve injury (SNI) rats. It is also reported that miR-203,18 miR-155,19 and miR-38120 showed abnormal expression in the dorsal spinal cord of CCI rats. Microarray analysis of microglia derived from dorsal spinal cord showed that miR-29c may be involved in the occurrence and maintenance of neuropathic pain in L4 spinal nerve transected mice.21
Long non-coding RNAs (lncRNAs) are more than 200 nucleotides in length. Although without protein-coding potential, lncRNAs regulate gene expression and they are characterized as key modulators of neuronal functions.22 Emerging evidence showed that lncRNAs were differentially expressed and play pivotal roles in the situation of neuropathic pain.23 For example, lncRNA CRNDE upregulated in the dorsal spinal cord enhanced neuropathic pain via modulating miR-136/IL6R axis and neuroinflammation in CCI rats.24 LncRNA XIST in the spinal cord accelerated neuropathic pain progression through regulation of miR-150 and ZEB1 in CCI rat models.25 An lncRNA named as Kcna2 antisense RNA facilitates neuropathic pain by silencing Kcna2 in primary afferent neurons.26 LncRNA uc.48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root ganglia.27 LncRNA BC168687 in the dorsal root ganglion is involved in TRPV1-mediated diabetic neuropathic pain in rats.28
mRNA and lncRNA microarrays of spinal cord tissue identified a total of 511 lncRNAs and 493 mRNAs with significant differential expression in spinal nerve ligation (SNL) mice.29 By using whole transcriptome shotgun sequencing, 134 lncRNA and 1066 mRNAs were found to be significantly deregulated on day 14 post-surgery in SNI rats.17 A total of 1200 lncRNAs and 739 mRNAs were differentially expressed in the ipsilateral spinal cord of SNI mice.30 However, when neuropathic pain occurs, the expression profile and specific neuropathic pain regulation mechanisms of lncRNA in the dorsal horn of spinal cord are largely unknown.
In view of the inconsistent changes of transcripts in spinal cord in neuropathic pain models, more transcriptomics studies are needed to focus on molecular changes in the dorsal spinal cord. To detect transcriptomic changes in the dorsal spinal cord during neuropathic pain and to explore the expression profiles of microRNA, lncRNA, and mRNAs, we established the widely used CCI neuropathic pain model and detected the differentially expressed transcripts with microRNA and lncRNA/mRNA microarrays. Differentially expressed mRNAs were subjected to Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses to evaluate their functions and possible mechanisms.
Methods
Animals
Experimental protocols were approved by the Experimental Animal Care and Use Committee of Zunyi Medical University. All experimental processes were carried out in accordance with the Guide for the Care and Use of Laboratory Animals.31 Sprague–Dawley rats (male, 7–8 weeks, 250 to 300 g) were housed under approved conditions with 12-/12-hr light/dark cycles with food and water provided ad libitum. Twelve rats were used in this study.
Neuropathic Pain Model
The left sciatic nerve underwent CCI surgeries to induce neuropathic pain. Animals were randomly allocated to sham operation (Sham) or CCI group (n=6 in each group). Neuropathic pain was induced with the CCI as previously described by Bennett and Xie32 with a 5–0 chromic gut suture.33 In brief, after sodium pentobarbital anesthesia (35 mg/kg, i.p.), after the sciatic nerve of the mid-thigh level on the left side was exposed, four snug ligatures of chromic gut suture were loosely tied around the nerve with about 1-mm space between the knots, so as not to compromise the vascular supply. Sham rats underwent the same anesthesia and surgical procedures but the sciatic nerves were not ligated.
Mechanical Pain Threshold Testing
Baseline mechanical pain thresholds were detected for the aforementioned 12 rats before CCI surgery. Mechanical pain intensity was monitored at 2, 6, 10, and 14 days post-surgery. Mechanical hypersensitivity was determined using the electronic von Frey plantar aesthesiometer (model 2390, IITC, Wood Dale, IL, USA) as we previously reported.34 Before tests, rats were given 15 min to settle down and habituate to the test environment. A rigid polypropylene tip was applied against the mid-plantar surface of the left hind paw. The paw withdrawal threshold was automatically recorded by the device and the cut-off was set at 50 g. The rigid tip was presented perpendicularly to the plantar surface, and brisk withdrawal or paw flinching was considered as positive responses; the digital number presented on the monitor was recorded as the paw mechanical withdrawal threshold (MWT). Three successive stimuli were applied and data were represented by mean values.
Thermal Pain Threshold Testing
Baseline thermal pain thresholds were detected for the aforementioned 12 rats before CCI surgery. To assess nociceptive responses to thermal stimuli, rats were unrestrained and acclimatized in acrylic cubicle (10 × 20 ×12 cm) on a uniform glass surface up to 60 mins before testing. Radiant heat was applied to the plantar surface of the test paw and a cut-off time of 20 s was used to prevent tissue damage. The thermal withdrawal latency from the radiant heat was recorded with a plantar test (Hargreaves’ method35) analgesia meter (model 390, IITC Life Science, Woodland Hills, CA, USA). Abrupt paw withdrawal, licking, and shaking were taken to be positive responses.36
Ipsilateral Dorsal Spinal Cord Samples Collection
Two weeks post-surgery, rats were deeply anesthetized with isoflurane and decapitated. The lumbar enlargement segments (L3-L5) of spinal cords were transversely sectioned and hemi-dissected along the midline. Only the surgery ipsilateral dorsal half of the lumbar enlargement was collected under a microscope and stored at −80°C.37
RNA Isolation, Purification, And Hybridization
Samples of Sham and CCI group were collected by pooling 6 ipsilateral L3-L5 dorsal spinal cords. Total RNA from each sample was quantified using the NanoDrop (ND-1000, NanoDrop, USA) and RNA integrity was determined by gel electrophoresis.
MicroRNA labeling and array hybridization were according to Exiqon’s manual. After stopping the labeling procedure, the Hy3™-labeled samples were hybridized on the miRCURYTM LNA Array (v.19.0) (Exiqon) according to array manual. Then, the slides were scanned using the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA).
For lncRNA microarray, sample labeling and array hybridization were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology) with minor modifications. Briefly, mRNA was purified and each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 3ʹ bias utilizing a random priming method (Arraystar Flash RNA Labeling Kit, Arraystar). The labeled cRNAs were purified by RNeasy Mini Kit (Qiagen). The concentration and specific activity of the labeled cRNAs were measured by NanoDrop and fragmented, and then the hybridization solution was dispensed into the gasket slide and assembled to the LncRNA expression microarray slide. The slides were incubated for 17 hrs at 65°C in an Agilent Hybridization Oven. The hybridized arrays were washed, fixed, and scanned using the Agilent DNA Microarray Scanner (part number G2505C).
Microarray Data Analysis
MicroRNA array images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. MiRNAs that with intensities≥30 in all samples were chosen for calculating normalization factor.
Agilent Feature Extraction software (version 11.0) was used to analyze acquired lncRNA array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX v12.1 software package (Agilent Technologies). After quantile normalization of the raw data, LncRNAs and mRNAs that have flags in Present or Marginal (“All Targets Value”) were chosen for further data analysis.
Differentially expressed microRNAs, lncRNAs, and mRNAs between two samples were identified through fold change filtering. RNAs having fold changes ≥2 were set as significantly differentially expressed. Hierarchical clustering was performed to show the distinguishable expression pattern of microRNAs, lncRNAs, and mRNAs between Sham and CCI group.
Bioinformatics Analysis
Gene Ontology (GO) analyses were performed to explore the function of differential mRNAs in terms of biological processes, cellular components, and molecular functions. Pathways defined by Kyoto Encyclopedia of Genes and Genomes (KEGG), Biocarta and Reactome (http://www.genome.jp/kegg/) were identified by Database for Annotation, Visualization and Integrated Discovery (DAVID; http://www.david.abcc.ncifcrf.gov/).
Statistical Analysis
The results were reported as mean ± SD. Statistically significant differences of mechanical and thermal pain thresholds between Sham and CCI groups were estimated by the repeated measures two-way Analysis of Variance (ANOVA) followed by Sidak’s multiple comparisons tests with GraphPad Prism (v7.0, GraphPad Software, USA). P < 0.05 was considered as being statistically significant.
Results
Neuropathic Pain Model Confirmation With Mechanical Hypersensitivity After CCI Surgery
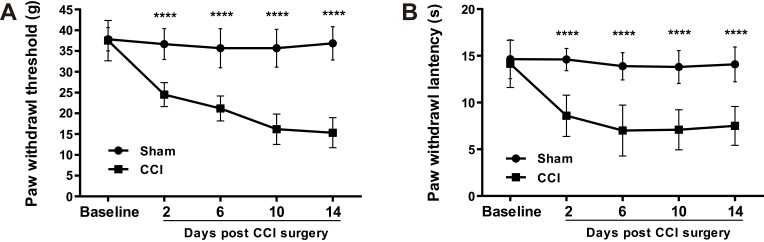
Compared with control rats (Sham group), rats with neuropathic pain (CCI group) showed significantly increased mechanical (Figure 1A) and thermal hypersensitivity (Figure 1B) on the ipsilateral hind paw in the next 2 weeks after CCI surgery.
Figure 1.
CCI of sciatic nerve induces mechanical and thermal hypersensitivity. Paw mechanical withdrawal thresholds (A) and thermal withdrawal latency (B) were assessed with electronic Von Frey and plantar test analgesia meter, respectively, on ipsilateral hind paws of CCI and sham-operated rats. n = 6 in both groups. All data were represented as mean ± SD. Statistical analyses consisted of repeated measures two-way ANOVA tests followed by Sidak’s multiple comparisons tests. ****P < 0.0001 between Sham and CCI groups.
MicroRNA Profiles
The miRCURY™ LNA Array (v7.0, Exiqon) detected 705 rat microRNAs (Supplementary Excel 1). Hierarchical clustering and scatter plot visualization showed the microRNA expression levels were different. Overall, 13 of them were calculated as differential microRNAs (fold change ≥ 2.0) between the CCI and Sham groups. Six of them were significantly upregulated, and the other seven were downregulated in CCI group. microRNA-1b overexpressed 18.7 times (intensity 2282 vs 113) after CCI insult. The 13 differential microRNAs are listed in Table 1.
Table 1.
Differentially Expressed microRNAs Between Sham-Operated And CCI Rats
| microRNA | Fold Change | Intensity (CCI) | Intensity (Sham) |
|---|---|---|---|
| Upregulated in CCI | |||
| rno-miR-1b | 18.7 | 2282 | 113 |
| rno-miR-98-5p | 3.7 | 784 | 195 |
| rno-miR-31a-3p | 2.6 | 281 | 101 |
| rno-miR-376b-5p | 2.2 | 529 | 218 |
| rno-miR-1-3p | 2.2 | 57 | 24 |
| rno-miR-214-3p | 2.1 | 37 | 16 |
| Downregulated in CCI | |||
| rno-miR-329-5p | −3.7 | 226 | 781 |
| rno-miR-675-5p | −2.9 | 60 | 163 |
| rno-miR-342-5p | −2.7 | 34 | 84 |
| rno-miR-203a-3p | −2.6 | 28 | 66 |
| rno-let-7d-5p | −2.5 | 121 | 279 |
| rno-miR-542-5p | −2.4 | 19 | 41 |
| rno-miR-672-5p | −2.2 | 16 | 32 |
LncRNA Profiles
By using the Arraystar Rat LncRNA/mRNA chips (v2.0, 4 × 44k, Arraystar), 10,430 rat lncRNAs were detected (Supplementary Excel 2). Hierarchical clustering and scatter plot visualization showed the lncRNAs expression levels were different. Overall, 876 of them were calculated as differential lncRNAs (fold change ≥ 2.0) between the CCI and Sham groups. Among them, 339 were significantly upregulated, and the other 537 were downregulated in CCI group (Supplementary Excel 3). Sixteen of them differentially expressed more than 10 times and the lncRNA XR_356687 (sequence: GTTCATTATCGGAATTAACCAGACATATCGTTCCACCAACTAAGAACGGCCATGCACCAC) overexpressed 53 times (raw intensity: 19,188 vs 364). The top 20 significantly up- and downregulated lncRNAs are listed, respectively, in Table 2.
Table 2.
Differentially Expressed lncRNAs Between Sham-Operated And CCI Rats
| lncRNA (Seq Name) | Gene Symbol | Fold Change | Raw Intensity (CCI) | Raw Intensity (Sham) |
|---|---|---|---|---|
| Top 20 upregulated in CCI | ||||
| XR_356687 | LOC102552325 | 52.5 | 19,188 | 364 |
| XR_592109 | LOC102546410 | 22.3 | 107 | 5 |
| XR_598798 | LOC102549116 | 15.2 | 73 | 5 |
| XR_347276 | Fancm | 13.9 | 67 | 5 |
| XR_347276 | Fancm | 13.9 | 67 | 5 |
| XR_592810 | LOC102551503 | 13.0 | 63 | 5 |
| XR_593301 | LOC100912242 | 12.7 | 61 | 5 |
| XR_347392 | Lin52 | 8.8 | 58 | 7 |
| XR_593865 | LOC102550632 | 8.3 | 40 | 5 |
| XR_601438 | LOC103692636 | 7.8 | 40 | 5 |
| XR_595336 | LOC103693645 | 6.7 | 32 | 5 |
| XR_346855 | LOC102548844 | 5.9 | 57 | 10 |
| XR_357290 | LOC102553550 | 5.6 | 38 | 7 |
| XR_590881 | LOC103691449 | 5.6 | 27 | 5 |
| XR_340868 | LOC102550064 | 5.6 | 95 | 18 |
| XR_591045 | LOC103691528 | 5.5 | 27 | 5 |
| XR_592900 | LOC103692568 | 5.4 | 76 | 15 |
| XR_339110 | LOC102548550 | 5.4 | 27 | 5 |
| XR_594520 | LOC103693250 | 5.3 | 37 | 7 |
| XR_340350 | LOC102553293 | 5.2 | 30 | 6 |
| Top 20 downregulated in CCI | ||||
| ENSRNOT00000034363 | AABR06081233.1 | −19.4 | 183 | 3600 |
| XR_338868 | LOC102546527 | −10.9 | 5 | 57 |
| NR_046246 | Rn28s | −10.9 | 11,145 | 113,335 |
| NR_046246 | Rn28s | −10.9 | 11,145 | 113,335 |
| NR_046246 | Rn28s | −10.9 | 11,145 | 113,335 |
| NR_046246 | Rn28s | −10.9 | 11,145 | 113,335 |
| XR_342867 | LOC102555707 | −10.7 | 5 | 55 |
| XR_593141 | LOC102550062 | −10.4 | 5 | 54 |
| XR_591024 | LOC102556203 | −10.3 | 5 | 53 |
| XR_338470 | LOC102553477 | −9.9 | 6 | 60 |
| XR_590615 | LOC102552001 | −9.8 | 6 | 56 |
| XR_590615 | LOC102552001 | −9.8 | 6 | 56 |
| XR_601117 | LOC103695264 | −9.6 | 660 | 6129 |
| XR_352792 | LOC102551164 | −8.5 | 5 | 44 |
| XR_342721 | LOC102550786 | −8.4 | 5 | 44 |
| ENSRNOT00000041435 | LOC367333 | −8.1 | 5 | 42 |
| XR_601956 | LOC103695347 | −7.8 | 5 | 40 |
| XR_589758 | LOC103690882 | −7.4 | 5 | 38 |
| ENSRNOT00000075140 | AABR06019245.1 | −7.1 | 5 | 37 |
| ENSRNOT00000075140 | AABR06019245.1 | −7.1 | 5 | 57 |
mRNA Profiles
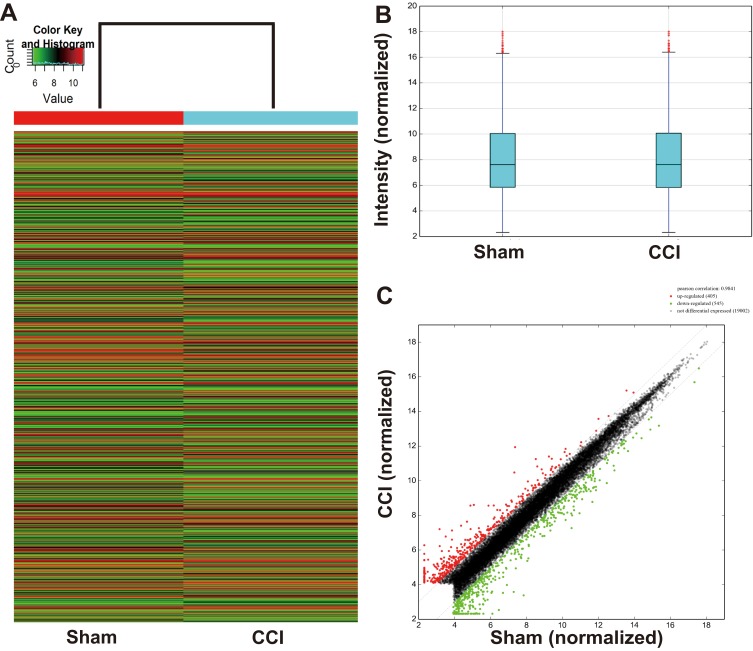
The Arraystar Rat LncRNA/mRNA chips detected 19,952 rat mRNAs (Supplementary Excel 4). Hierarchical clustering and scatter plot visualization showed the mRNA expression levels were different (Figure 2). Overall, 950 of them were calculated as differential mRNAs (fold change ≥ 2.0) between the CCI and Sham groups. Among them, 405 were significantly upregulated, and 545 were downregulated in the CCI group (Supplementary Excel 5). Ten mRNAs changed the expression more than 10 times, and the Hspa1b (encodes heat shock protein 70) overexpressed 24 times in CCI dorsal spinal cord. The top 20 differentially upregulated and downregulated mRNAs are listed, respectively, in Table 3.
Figure 2.
mRNA expression between the CCI and sham-operated rats. (A) Hierarchical clustering shows a differential mRNA expression profiles between two groups. The upregulated mRNAs in the CCI group are shown in red, and the downregulated ones are in green. (B) Box plots show the distribution of mRNAs for the two groups. The distributions (the means, the 25th and 75th percentiles) were nearly the same in the CCI and Sham groups after normalization, indicating that the overall expression of mRNA was uniform between the two groups. (C) Scatter plots assess the mRNA expression differences between CCI and Sham groups. The mRNAs above the top green line (indicated with red dot) and below the bottom green line (indicated with green dot) are differential mRNAs (fold change ≥ 2.0 or ≤ −2.0, respectively), while mRNAs with no expression difference are represented with black dots between the two border lines.
Table 3.
Differentially Expressed mRNAs Between Sham-Operated And CCI Rats
| mRNA (Seq Name) | Gene Symbol | Fold Change | Raw Intensity (CCI) | Raw Intensity (Sham) |
|---|---|---|---|---|
| Top 20 upregulated in CCI | ||||
| NM_212504 | Hspa1b | 23.9 | 4098 | 172 |
| NM_001024740 | Ccdc116 | 16.4 | 141 | 9 |
| NM_001025028 | Fam35a | 15.5 | 74 | 5 |
| NM_001017496 | Cxcl13 | 12.8 | 366 | 30 |
| ENSRNOT00000008897 | Ptprd | 11.6 | 378 | 35 |
| NM_001113390 | Ptprr | 11.0 | 53 | 5 |
| NM_080694 | Cacng6 | 9.6 | 134 | 15 |
| NM_001108873 | Tnfrsf10b | 9.6 | 105 | 11 |
| NM_012912 | Atf3 | 8.9 | 1452 | 168 |
| ENSRNOT00000044252 | Ascl4 | 8.6 | 41 | 5 |
| NM_001000884 | Olr1117 | 8.5 | 41 | 5 |
| ENSRNOT00000026719 | Col6a3 | 8.3 | 72 | 9 |
| ENSRNOT00000018645 | Zc3hav1l | 8.2 | 162 | 21 |
| NM_053551 | Pdk4 | 7.7 | 190 | 27 |
| NM_001031637 | Abca17 | 7.5 | 42 | 6 |
| NM_001145755 | Nlrp1a | 6.7 | 93 | 15 |
| ENSRNOT00000010289 | Apold1 | 6.7 | 2507 | 381 |
| ENSRNOT00000049430 | Rbbp6 | 6.4 | 365 | 62 |
| NM_019216 | Gdf15 | 6.3 | 31 | 5 |
| NM_001145072 | Iqcf5 | 6.2 | 59 | 10 |
| Top 20 downregulated in CCI | ||||
| NM_001108582 | Dapl1 | −22.0 | 11 | 240 |
| ENSRNOT00000032726 | Cylc2 | −15.2 | 6 | 92 |
| NM_133292 | Sval1 | −13.8 | 5 | 72 |
| ENSRNOT00000065875 | AABR06030627.1 | −13.1 | 12 | 163 |
| NM_001108480 | Bcl2l12 | −9.7 | 5 | 50 |
| NM_001305124 | Tmem220 | −9.4 | 147 | 1431 |
| NM_001008931 | Vom1r48 | −9.0 | 12 | 108 |
| ENSRNOT00000046533 | LOC287992 | −8.3 | 5 | 43 |
| NM_001000659 | Olr587 | −7.8 | 5 | 40 |
| ENSRNOT00000038519 | Grhl1 | −7.6 | 16 | 126 |
| NM_001108922 | Stk32c | −7.4 | 424 | 3119 |
| ENSRNOT00000060162 | AABR06088936.1 | −7.3 | 5 | 38 |
| NM_013030 | Slc34a1 | −7.1 | 5 | 37 |
| NM_001137670 | Sept14 | −7.1 | 6 | 45 |
| NM_019622 | Espn | −7.1 | 266 | 1939 |
| NM_001277165 | Ldb3 | −7.0 | 5 | 36 |
| NM_001012140 | Rab34 | −6.7 | 7 | 50 |
| NM_001000809 | Olr1245 | −6.7 | 5 | 34 |
| NM_001244797 | Cidec | −6.5 | 9 | 60 |
| NM_001108926 | Cabp4 | −6.5 | 7 | 49 |
Bioinformatics Analyses Of Differential mRNA
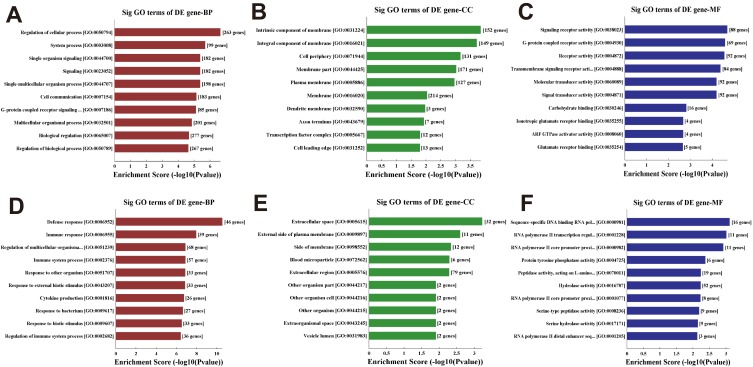
Gene Ontology enrichment analysis of differential mRNAs in CCI rats was conducted and the top 10 significantly enriched (P < 0.05) GO terms for the upregulated and downregulated mRNAs are listed in Figure 3. The upregulated (Figure 3A–C) and downregulated mRNAs (Figure 3D–F) in CCI rats enriched in biological processes, cellular component, and molecular function terms, such as signaling (GO: 0023052, with 182 genes), cell communication (GO: 0007154, with 182 genes), G-protein-coupled activity (GO: 0048856, with 88 genes), cytokine production (GO: 0001816, with 26 genes), etc.
Figure 3.
Gene ontology enrichment analysis. Enriched gene ontology (GO) terms correspond to the 405 differentially upregulated (A-C) mRNAs and 545 downregulated (D-F) mRNAs in CCI group. (A, D) Top 10 significantly (Sig) enriched biological processes (BP) terms for the upregulated and downregulated differentially expressed (DE) mRNAs in CCI group, respectively. (B, E) Top 10 enriched cellular component (CC) terms for the upregulated and downregulated DE mRNAs in CCI group, respectively. (C, F) Top 10 enriched molecular function (MF) terms for the upregulated and downregulated DE mRNAs in CCI group, respectively.
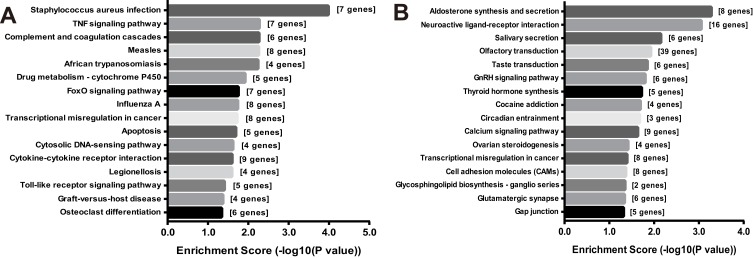
KEGG pathway analysis showed that the differential mRNAs significantly enriched (P < 0.05) in 32 pathways, with the up- and downregulated mRNAs in CCI group enriched to 16 pathways, respectively (Figure 4). The upregulated mRNAs enriched to TNF (7 genes), FoxO (7 genes), apoptosis (5 genes), Cytokine-cytokine receptor interaction (4 genes), Toll-like receptor signaling pathway (5 genes), etc. (Figure 4A), and the downregulated mRNAs enriched to cocaine addiction (4 genes), calcium signaling pathway (9 genes), cell adhesion molecules (CAMs, 8 genes), and glutamatergic synapse (6 genes) pathway (Figure 4B).
Figure 4.
KEGG pathways significantly enriched by differentially expressed mRNAs. (A) Pathways significantly enriched (P < 0.05) by the upregulated mRNAs in CCI group. (B) KEGG pathways significantly enriched (P < 0.05) by the downregulated mRNAs in CCI group.
Discussion
Spinal dorsal horn is an important location for pain central sensatization, pathological pain starting and maintaining, and therefore, neuropathic pain treatment.8 In this study, the expression profiles in neuropathic pain rats’ dorsal spinal cord were detected with different microarrays. Comprehensive expression changes of transcripts including microRNA, lncRNA, and mRNA were found between normal and CCI rats. These results indicate that dorsal spinal cord and these differentially expressed molecules can be targets for neuropathic pain.
In this study, 13 microRNAs were found differentially expressed in neuropathic pain rats, and miR-1b overexpressed 18.7 times in CCI rats, indicating that these differential microRNAs may be involved in the chronic pain process of CCI rats. To date, the expression level of miR-1b in dorsal spinal cord after neuropathic pain is not reported. One study detected the expression of miR-1b in the injured sciatic nerve; interestingly, its expression decreased in the peripheral nerve injury rats. The functional study indicated that miR-1b can negatively regulate nerve regeneration by influencing proliferation, migration, and apoptosis of Schwann cells.38 MicroRNAs are mainly involved in the regulation of biological processes by inhibiting the function of their target genes. To evaluate the function of these microRNAs, prediction of target genes and gain- and/or loss-of-function experiments are needed for subsequent studies.
There are 876 differential lncRNAs between neuropathic pain and normal rats in this study, and 16 of them are differentially expressed more than 10 times. Of note, the lncRNA XR_356687 overexpressed as high as 53 times in CCI rats’ dorsal horn, although its expression level in normal rats is already high (with raw intensity: 19,188 vs 364). This suggests that lncRNAs in the dorsal spinal cord actively participated in the neuropathic pain development. Of course, the biological regulation mechanism of lncRNA is complex, and further studies are needed to confirm these differential lncRNAs’ effects and the specific mechanisms in neuropathic pain environment.
In addition, 405 upregulated and 545 downregulated mRNAs were detected in CCI group. Ten of them show an expression level change of more than 10 times. The Hspa1b mRNA, which encodes heat shock protein 70 (HSP70), overexpressed 24 times in the spinal cord of CCI rats. HSP70 is an abundant and quickly inducible protein, which is constitutively expressed at normal growth temperatures and functions as a molecular chaperone.39 It is reported that HSP70-TLR4 axis attenuated neuroinflammation and ameliorated postoperative pain.40 GO analyses showed they enriched into GO terms such as signaling, cell communication, G-protein-coupled activity, and cytokine production, which are closely related to physical function and in the spinal dorsal horn, the imbalance of their function takes part in neuropathic pain development.10,41–43 KEGG pathway analyses show that these differential mRNAs enriched into 32 KEGG pathways, including TNF, cytokine–cytokine receptor interaction, calcium signaling, and FoxO pathway. The TNF,44,45 cytokine,8,43,46 and calcium-related pathway47,48 contribute to neuropathic pain development in the dorsal spinal cord. Although FoxO has not been confirmed to take part in the neuropathic pain, bioinformatics predicted that FoxO pathway in the dorsal spinal cord37 or dorsal root ganglion49 was related to neuropathic pain.
We previously found that circular RNA (circRNA) expression levels in the dorsal spinal cord of CCI rats were significantly changed.37 A total of 469 circRNAs showed differential expression between CCI and sham-operated rats.37 These data again suggest that neuropathic pain induced comprehensive changes of transcription profile in the dorsal spinal cord.
A growing body of evidence suggests that changes in the transcriptome of the dorsal spinal cord are dynamic in neuropathic pain.16,21,50 However, even in animal models of the same kind of neuropathic pain, such as the peripheral nerve injury induced pain, detected changes in expression profiles and biological pathways are not identical, suggesting that outcomes in dorsal spinal cord molecules may be related to pain duration, pain intensity, and different kinds of species. Besides, recent pieces of evidence revealed that there are reciprocal regulations among the non-coding RNAs and mRNAs. For instance, microRNAs bind to the 3ʹ-untranslated region of their target mRNAs and lead to degradation and translational repression, while lncRNA and circRNAs can function as microRNA sponges51,52 and regulate parent gene expression to affect disease outcomes.52,53 In addition, to further distinguish the cell source of differential gene from the dorsal spinal cord, single-cell separation technique is a feasible method. For example, microglia were collected and subjected to microarray analysis at the spinal dorsal horn; results suggest that differential gene Gria1 is specific for microglia, 56 genes and microRNA-29c may play an important role in the pathogenesis and maintenance of neuropathic pain.21
For the tissue collection, not only the spinal dorsal horn but the whole CCI-ipsilateral dorsal spinal cord was collected, six dorsal spinal cord tissues were pooled as one detection sample; however, repeated microarray detections (e.g., three repeats) and a set P value (e.g., <0.05) are better to detect the differential transcripts. Data in this study are all from male rats; it should be realized that sex difference may influence the detected differential genes.54,55 Expression validation and function-oriented experiments of specific differential microRNA, lncRNA, or mRNA are warranted to clarify their roles in neuropathic pain regulation.
Conclusion
Neuropathic pain induced comprehensive transcriptome changes in the dorsal spinal cord. The differentially expressed transcripts such as microRNA, lncRNA, and mRNA in spinal dorsal horn could be potential targets in defeating neuropathic pain. Strategies targeting these differentially expressed molecules such as microRNA-1b, the lncRNA XR_356687, and HSP70 in spinal dorsal horn may be helpful to relieve neuropathic pain.
Acknowledgments
The study was supported by a scientific research project from The Science and Technology Department of Guizhou Province (LH[2015]7554) and the National Natural Science Foundation of China (81660201). This work is also supported by the Excellent Young Talents Project (18zy-004) and the Innovative Training Program ([2015]3109) of Zunyi Medical University.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Van HO, Austin SK, Khan RA, Smith BH, Torrance N. Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain. 2014;155(4):654–662. doi: 10.1016/j.pain.2013.11.013 [DOI] [PubMed] [Google Scholar]
- 2.Loeser JD. Economic implications of pain management. Acta Anaesthesiol Scand. 1999;43(9):957–959. doi: 10.1034/j.1399-6576.1999.430914.x [DOI] [PubMed] [Google Scholar]
- 3.Baron R, Binder A, Wasner G. Neuropathic pain: diagnosis, pathophysiological mechanisms, and treatment. Lancet Neurol. 2010;9(8):807–819. doi: 10.1016/S1474-4422(10)70143-5 [DOI] [PubMed] [Google Scholar]
- 4.Pertovaara A. Noradrenergic pain modulation. Prog Neurobiol. 2006;80(2):53–83. doi: 10.1016/j.pneurobio.2006.08.001 [DOI] [PubMed] [Google Scholar]
- 5.Bannister K, Bee LA, Dickenson AH. Preclinical and early clinical investigations related to monoaminergic pain modulation. Neurotherapeutics. 2009;6(4):703–712. doi: 10.1016/j.nurt.2009.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pertovaara A, Kontinen VK, Kalso EA. Chronic spinal nerve ligation induces changes in response characteristics of nociceptive spinal dorsal horn neurons and in their descending regulation originating in the periaqueductal gray in the rat. Exp Neurol. 1997;147(2):428–436. doi: 10.1006/exnr.1997.6555 [DOI] [PubMed] [Google Scholar]
- 7.Ji RR, Chamessian A, Zhang YQ. Pain regulation by non-neuronal cells and inflammation. Science. 2016;354(6312):572–577. doi: 10.1126/science.aaf8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji RR, Xu ZZ, Gao YJ. Emerging targets in neuroinflammation-driven chronic pain. Nat Rev Drug Discov. 2014;13(7):533–548. doi: 10.1038/nrd4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West SJ, Bannister K, Dickenson AH, Bennett DL. Circuitry and plasticity of the dorsal horn – toward a better understanding of neuropathic pain. Neuroscience. 2015;300:254. doi: 10.1016/j.neuroscience.2015.05.020 [DOI] [PubMed] [Google Scholar]
- 10.Todd AJ. Neuronal circuitry for pain processing in the dorsal horn. Nat Rev Neurosci. 2010;11(12):823–836. doi: 10.1038/nrn2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu R, Li L, Li D, et al. Downregulation of Cdh1 signalling in spinal dorsal horn contributes to the maintenance of mechanical allodynia after nerve injury in rats. Mol Pain. 2016;12:174480691664737. doi: 10.1177/1744806916647376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gangadharan V, Kuner R. Pain hypersensitivity mechanisms at a glance. Dis Model Mech. 2013;6(4):889. doi: 10.1242/dmm.011502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andersen HH, Duroux M, Gazerani P. MicroRNAs as modulators and biomarkers of inflammatory and neuropathic pain conditions. Neurobiol Dis. 2014;71(11):159–168. doi: 10.1016/j.nbd.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 14.Jiangpan P, Qingsheng M, Zhiwen Y, Tao Z. Emerging role of microRNA in neuropathic pain. Curr Drug Metab. 2016;17(4):336–344. doi: 10.2174/1389200216666151015113400 [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Gonzalez MJ, Landry M, Favereaux A. MicroRNA and chronic pain: from mechanisms to therapeutic potential. Pharmacol Ther. 2017;180:1–15. doi: 10.1016/j.pharmthera.2017.06.001 [DOI] [PubMed] [Google Scholar]
- 16.Genda Y, Arai M, Ishikawa M, Tanaka S, Okabe T, Sakamoto A. microRNA changes in the dorsal horn of the spinal cord of rats with chronic constriction injury: A TaqMan® low density array study. Int J Mol Med. 2013;31(1):129–137. doi: 10.3892/ijmm.2012.1163 [DOI] [PubMed] [Google Scholar]
- 17.Zhou J, Xiong Q, Chen H, Yang C, Fan Y. Identification of the spinal expression profile of non-coding RNAs involved in neuropathic pain following spared nerve injury by sequence analysis. Front Mol Neurosci. 2017;10:91. doi: 10.3389/fnmol.2017.00091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Huang Y, Ma C, Yu X, Zhang Z, Shen L. MiR-203 involves in neuropathic pain development and represses Rap1a expression in nerve growth factor differentiated neuronal PC12 cells. Clin J Pain. 2015;31(1):36–43. doi: 10.1097/AJP.0000000000000070 [DOI] [PubMed] [Google Scholar]
- 19.Dou L, Lin H, Wang K, et al. Long non-coding RNA CCAT1 modulates neuropathic pain progression through sponging miR-155. Oncotarget. 2017;8(52):89949–89957. doi: 10.18632/oncotarget.v8i52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia LX, Ke C, Lu JM. NEAT1 contributes to neuropathic pain development through targeting miR-381/HMGB1 axis in CCI rat models. J Cell Physiol. 2018;233(9):7103–7111. doi: 10.1002/jcp.26526 [DOI] [PubMed] [Google Scholar]
- 21.Jeong H, Na YJ, Lee K, et al. High-resolution transcriptome analysis reveals neuropathic pain gene-expression signatures in spinal microglia after nerve injury. Pain. 2016;157(4):964–976. doi: 10.1097/j.pain.0000000000000470 [DOI] [PubMed] [Google Scholar]
- 22.Clark BS, Blackshaw S. Long non-coding RNA-dependent transcriptional regulation in neuronal development and disease. Front Genet. 2014;5:164. doi: 10.3389/fgene.2014.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z, Li X, Chen X, et al. Emerging roles of long non-coding RNAs in neuropathic pain. Cell Prolif. 2019;52(1):e12528. doi: 10.1111/cpr.2019.52.issue-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D, Mou JY, Wang F. CRNDE enhances neuropathic pain via modulating miR-136/IL6R axis in CCI rat models. J Cell Physiol. 2019;234(12):22234–22241. doi: 10.1002/jcp.28790 [DOI] [PubMed] [Google Scholar]
- 25.Yan XT, Xu Y, Cheng XL, et al. SP1, MYC, CTNNB1, CREB1, JUN genes as potential therapy targets for neuropathic pain of brain. J Cell Physiol. 2019;234(5):6688–6695. doi: 10.1002/jcp.27413 [DOI] [PubMed] [Google Scholar]
- 26.Zhao X, Tang Z, Zhang H, et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci. 2013;16(8):1024–1031. doi: 10.1038/nn.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Xu H, Zou L, et al. LncRNA uc.48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root ganglia. Purinergic Signal. 2016;12(1):139–148. doi: 10.1007/s11302-015-9488-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C, Li C, Deng Z, Du E, Xu C. Long non-coding RNA BC168687 is involved in TRPV1-mediated diabetic neuropathic pain in rats. Neuroscience. 2018;374:214–222. doi: 10.1016/j.neuroscience.2018.01.049 [DOI] [PubMed] [Google Scholar]
- 29.Jiang BC, Sun WX, He LN, Cao DL, Zhang ZJ, Gao YJ. Identification of lncRNA expression profile in the spinal cord of mice following spinal nerve ligation-induced neuropathic pain. Mol Pain. 2015;11:43. doi: 10.1186/s12990-015-0047-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z, Liang Y, Wang H, et al. LncRNA expression in the spinal cord modulated by minocycline in a mouse model of spared nerve injury. J Pain Res. 2017;10:2503–2514. doi: 10.2147/JPR.S147055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.National Research Council. Guide for the care and use of laboratory animals National Academies Press; 2010. [Google Scholar]
- 32.Bennett GJ, Xie YK. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain. 1988;33(1):87–107. doi: 10.1016/0304-3959(88)90209-6 [DOI] [PubMed] [Google Scholar]
- 33.Parent AJ, Tétreault P, Roux M, et al. Descending nociceptive inhibition is modulated in a time-dependent manner in a double-hit model of chronic/tonic pain. Neuroscience. 2015;315(3):70–78. doi: 10.1016/j.neuroscience.2015.11.065 [DOI] [PubMed] [Google Scholar]
- 34.Cao S, Xiao Z, Sun M, Li Y. D-serine in the midbrain periaqueductal gray contributes to morphine tolerance in rats. Mol Pain. 2016;12:174480691664678. doi: 10.1177/1744806916646786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hargreaves K, Dubner R, Brown F, Flores C, Joris J. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain. 1988;32(1):77–88. doi: 10.1016/0304-3959(88)90026-7 [DOI] [PubMed] [Google Scholar]
- 36.Aman Y, Pitcher T, Simeoli R, Ballard C, Malcangio M. Reduced thermal sensitivity and increased opioidergic tone in the TASTPM mouse model of Alzheimer’s disease. Pain. 2016;157(10):2285–2296. doi: 10.1097/j.pain.0000000000000644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao S, Deng W, Li Y, et al. Chronic constriction injury of sciatic nerve changes circular RNA expression in rat spinal dorsal horn. J Pain Res. 2017;10:1687–1696. doi: 10.2147/JPR.S139592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu YP, Xu P, Guo CX, et al. miR-1b overexpression suppressed proliferation and migration of RSC96 and increased cell apoptosis. Neurosci Lett. 2018;687:137–145. doi: 10.1016/j.neulet.2018.09.041 [DOI] [PubMed] [Google Scholar]
- 39.Hartl FU. Molecular chaperones in cellular protein folding. Nature. 1996;381(6583):571–579. doi: 10.1038/381571a0 [DOI] [PubMed] [Google Scholar]
- 40.Fan YX, Qian C, Liu B, et al. Induction of suppressor of cytokine signaling 3 via HSF-1-HSP70-TLR4 axis attenuates neuroinflammation and ameliorates postoperative pain. Brain Behav Immun. 2018;68:111–122. doi: 10.1016/j.bbi.2017.10.006 [DOI] [PubMed] [Google Scholar]
- 41.Julius D, Basbaum AI. Molecular mechanisms of nociception. Nature. 2001;413(6852):203–210. doi: 10.1038/35093019 [DOI] [PubMed] [Google Scholar]
- 42.Ji RR, Nackley A, Huh Y, Terrando N, Maixner W. Neuroinflammation and central sensitization in chronic and widespread pain. Anesthesiology. 2018;129(2):343–366. doi: 10.1097/ALN.0000000000002130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen G, Zhang YQ, Qadri YJ, Serhan CN, Ji RR. Microglia in pain: detrimental and protective roles in pathogenesis and resolution of pain. Neuron. 2018;100(6):1292–1311. doi: 10.1016/j.neuron.2018.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu D, Fan T. Progressive increase of inflammatory CXCR4 and TNF-Alpha in the dorsal root ganglia and spinal cord maintains peripheral and central sensitization to diabetic neuropathic pain in rats. Mediators Inflamm. 2019;2019:4856156. doi: 10.1155/2019/4856156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W, Bai Y, Qiao Y, et al. 8-O-acetyl shanzhiside methylester from lamiophlomis rotata reduces neuropathic pain by inhibiting the ERK/TNF-alpha pathway in spinal astrocytes. Front Cell Neurosci. 2018;12:54. doi: 10.3389/fncel.2018.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popiolek-Barczyk K, Mika J. Targeting the microglial signaling pathways: new insights in the modulation of neuropathic pain. Curr Med Chem. 2016;23(26):2908–2928. doi: 10.2174/0929867323666160607120124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alles SR, Garcia E, Balasubramanyan S, et al. Peripheral nerve injury increases contribution of L-type calcium channels to synaptic transmission in spinal lamina II: role of alpha2delta-1 subunits. Mol Pain. 2018;14:1744806918765806. doi: 10.1177/1744806918765806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, Li L, Chen SR, et al. The alpha2delta-1-NMDA receptor complex is critically involved in neuropathic pain development and gabapentin therapeutic actions. Cell Rep. 2018;22(9):2307–2321. doi: 10.1016/j.celrep.2018.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen CJ, Liu DZ, Yao WF, et al. Identification of key genes and pathways associated with neuropathic pain in uninjured dorsal root ganglion by using bioinformatic analysis. J Pain Res. 2017;10:2665–2674. doi: 10.2147/JPR.S143431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Liu F, Wei M, et al. Chronic constriction injury-induced microRNA-146a-5p alleviates neuropathic pain through suppression of IRAK1/TRAF6 signaling pathway. J Neuroinflammation. 2018;15(1):179. doi: 10.1186/s12974-018-1215-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272. doi: 10.1038/nrg.2016.20 [DOI] [PubMed] [Google Scholar]
- 52.Zheng J, Liu X, Xue Y, et al. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1beta/Derlin-1 pathway. J Hematol Oncol. 2017;10(1):52. doi: 10.1186/s13045-017-0422-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602. doi: 10.1093/eurheartj/ehv713 [DOI] [PubMed] [Google Scholar]
- 54.Sorge RE, Mapplebeck JC, Rosen S, et al. Different immune cells mediate mechanical pain hypersensitivity in male and female mice. Nat Neurosci. 2015;18(8):1081–1083. doi: 10.1038/nn.4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mapplebeck JC, Beggs S, Salter MW. Sex differences in pain: a tale of two immune cells. Pain. 2016;157(Suppl 1):S2–S6. doi: 10.1097/j.pain.0000000000000389 [DOI] [PubMed] [Google Scholar]