Abstract
Heart failure (HF) is a dominant cause of morbidity and mortality in the developed world, with available pharmacotherapies limited by high rates of residual mortality and a failure to directly target the changes in cell state that drive adverse cardiac remodeling. Pathologic cardiac remodeling is driven by stress-activated cardiac signaling cascades that converge on defined components of the chromatin regulatory apparatus in the nucleus, triggering broad shifts in transcription and cell state. Thus, studies focusing on how cytosolic signaling pathways couple to the nuclear gene control machinery has been an area of therapeutic interest in HF. In this review, we discuss current concepts pertaining to the role of chromatin regulators in HF pathogenesis, with a focus on specific proteins and RNA-containing macromolecular complexes that have shown promise as druggable targets in the experimental setting.
Heart failure (HF) is a global epidemic and represents a leading cause of morbidity and mortality in the developed world [1–5]. Lifetime risk for developing HF has been estimated to be as high as 20%, with the prevalence projected to increase over the next two decades. This increased prevalence is not only the result of our success in treating patients with myocardial infarction (MI) and our growing ability to stabilize acute cardiovascular events [5–7], but is also caused by an aging populace and rising rates of comorbidities including obesity, hypertension, and diabetes [8,9]. Currently available therapeutic modalities to treat HF, which mostly focus on blockade of circulating neurohormone activity, are inadequate as reflected by high rates of residual mortality in patients adhering to guideline directed medical therapy. Furthermore, neurohormonal antagonism does not directly alter root-cause defects in cardiac tissue and often only slows disease progression rather than preventing or reversing it. The fact that nearly half of those who develop HF die within 5 years of diagnosis highlights the urgent need to identify completely new axes of disease pathogenesis and leverage this knowledge toward the development of novel therapies [4,10].
Abnormalities in cardiac gene regulation represent a new axis of HF pathogenesis and emerging research implicates the transcriptional apparatus as a novel therapeutic target. The last decades have seen major advances in our understanding of how stress- or injury-induced cardiac signaling cascades converge on the nucleus to trigger global shifts in gene expression that contribute to adverse cardiac remodeling and impaired cardiac function [11,12]. Importantly, a host of studies using genetic gain- and loss-of-function approaches have highlighted the functions of a set of core transcription factors (TFs), such as NFAT, MEF2, NF-κB, GATA4 and C-MYC, in sustaining and amplifying the gene regulatory networks (GRNs) critical for pathological cardiac remodeling in vivo [12]. These stress-induced gene programs drive pathologic processes including cardiomyocyte (CM) hypertrophy, altered substrate metabolism and energetics, myofibroblast (myoFB) activation, and innate inflammatory responses, all of which collectively fuel a vicious cycle that culminates in cardiac structural changes and progressive contractile dysfunction. Current pharmacological therapies generally target very proximal steps in stress-dependent cardiac signaling (e.g., antagonists of the ßl adrenergic receptor and blockade of renin-angiotensin signaling) [5,13]. These stress-induced pathways ultimately converge on TFs and the chromatin regulatory apparatus in the nucleus, which transduce these broad upstream signals into changes in gene expression and cell identity. For these reasons, the study of how cytosolic signaling pathways couple to the nuclear gene control machinery has been an area of intense scientific and therapeutic interest. In this review, we provide an overview of current concepts pertaining to the role of chromatin regulators in HF, with a particular focus on protein and RNA-containing macromolecular complexes that have been shown to have translational potential in proof-of-concept experimental studies.
1. Epigenetic regulation of gene expression
How cells within the human body, all of which share the same DNA sequence, differentiate into the myriad of distinct cell types with highly specialized functions remains one of the most fascinating questions in biology. This remarkable process is achieved, in a large part, through epigenetic control of gene expression, which orchestrates strict spatio-temporal control of ceil state-defining gene programs. The term epigenetics [14]. refers to the layer of chemical modifications that exists above (‘epi’) the DNA sequence (‘genetic’) and allows the genome to function distinctively in different cell types. The epigenome comprises all of the processes that dynamically shape chromatin to modulate cell-state specific gene expression, including methylation of DNA and post-translational modification of histone tails [15, 16]. Active transcription of genes is influenced by the activity of DNA regulatory elements called enhancers, defined as cis-acting DNA modules that are needed to activate and sustain transcription at their target promoters. Enhancer activity is dynamic and can be restricted to a particular tissue or cell type, specific time point, or a precise physiologic, pathologic, or environmental condition [17]. Collectively, these features allow for the temporal, spatial, and quantitative control of gene expression. Enhancers are the key information processing units that regulate the expression of cell-state defining gene regulatory networks (GRNs) and are characterized by enrichment of regulatory transcription factor (TF) binding sites, active chromatin marks (e.g., acetylation of histone H3 lysine 27, H3K27ac), and binding of co-activator proteins (e.g., BRD4, MED1) [18]. Cell state is dictated and maintained by the binding of core TFs to their cognate DNA motifs on target enhancers, which ultimately results in remodeling of regional chromatin and assembly of the transcriptional machinery. Several studies have defined how changes in the enhancer landscape dictates cardiac cell fate specification and differentiation [19–21]. In an analogous manner, the activation of pathologic transcriptional programs in the stressed heart (e.g., transformation of a healthy CM into one that is hypertrophic and dysfunctional) is triggered by pathologic rewiring of the enhancer and transcriptional landscape [22,23]. In mouse models of pressure overload cardiac hypertrophy, chromatin immunoprecipitation coupled with massively parallel sequencing (ChIP-seq) has been used to map global deposition of several histone modifications associated with the activation of a defined epigenomic landscape [24]. In particular, the regulation of histone methylation has been shown to play a key role in the maintenance of cardiac homeostasis and hypertrophy [24–26]. Recently, the laboratory of Gianluigi Condorelli has characterized the dual function of the histone methyl-transferase G9a in maintaining correct gene expression programs in normal CMs as well as in driving the gene expression changes underlying cardiac hypertrophy [27]. G9a exerts this double function by its ability to repress, via dimethylation of lysine 9 at histone 3 (H3K9me2), different sets of genes depending upon the functional status of the heart. Interestingly, G9a interacts with MEF2C in CMs, suggesting that broadly expressed histone methyltransferase might achieve cell- and locus-specific activity by interacting with cardiac TFs [27].
Previous studies have highlighted the dynamic interplay between histone modification and CpG site methylation (mCpG) during cardiac development and CM maturation. Interestingly, mCpG of coding and cis-regulatory regions is remarkably stable in the setting of HF [28]. While bisulfite sequencing can resolve DNA methylation patterns at the single nucleotide level, it does not discriminate between 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) [29]. Recent evidence suggests that the dynamic modulation of hydroxymethylation at gene bodies is indeed an epigenetic feature that accompanies pathologic transcriptional changes in CMs during hypertrophy [30]. Genome-wide distribution of 5hmC positively correlates with gene expression and, following pathological hypertrophy, the 5hmC landscape of CMs is shifted toward a neonatal-like pattern. 5hmC also seems to define a small subset of enhancer regions that dynamically respond to pro-hypertrophic cardiac stress [30]. 5hmC at these enhancer regions correlates with gene expression pattern following cardiac hypertrophy, suggesting the possibility that this subset of hydroxymethylated cis-regulatory elements may play a role in sustaining the pathological response in CMs. Mechanistically, depletion of 5hmC via downregulation of Tet methylcytosine dioxygenase 2 (Tet2) affects enrichment of the active chromatin mark H3K27ac within enhancer regions, with consequent decreased expression of local genes [30]. Recent studies have also shown that DNA hydroxymethylation is positively correlated with enhancer activity and chromatin accessibility, and is enriched at the boundaries of low methylated regions (LMRs) [31]. Importantly, LMRs with enhancer signatures in CMs are enriched for single nucleotide polymorphisms (SNPs) linked to coronary heart disease and cardiac arrhythmias [28]. The pathologic relevance of cardiac cis-acting regulatory elements is further supported by the observation that cardiac disease-associated SNPs are enriched in heart-specific super-enhancers (SEs), regions of the mammalian genome comprising of multiple enhancer clusters that are disproportionately enriched with regulatory TF binding sites, active chromatin marks (e.g., H3K27ac), and coactivator protein binding [32,33]. These data support the contention that aberrant enhancer-mediated signaling plays a critical role in pathogenesis of human heart disease.
2. Interdicting signaling from enhancers to RNA Pol II-dependent transcription as a therapeutic strategy for HF
In response to cardiac stress, a defined set of coordinately activated TFs bind specific enhancers in a combinatorial manner. These enhancer-bound TFs then recruit diverse cofactors that alter local chromatin structure and/or signal to the transcription machinery. Two major classes of cofactors are those that mobilize the nucleosomes (e.g. the ATP-dependent chromatin remodeling complexes) and those that add or remove post-translational modifications (PTMs) to histone proteins. Of the PTMs, reversible acetylation of histone tail lysine residues has been the most studied in the context of HF pathogenesis [16,34,35]. Dynamic lysine acetylation is coordinately regulated by histone acetyltransferases (HATs) and histone deacetylascs (HDACs), also known as epigenetic “writers” and “erasers”, respectively. These marks are recognized and “read” by proteins that specifically bind these post-translational marks. Emerging studies have highlighted the importance of epigenetic “readers” as important new players in cardiac biology [16]. These proteins harbor recognition motifs for histone modifications and also contain domains that scaffold the formation of protein and RNA complexes that mediate downstream transcriptional signaling.
Given the known importance of lysine acetylation in cardiac biology, a class of epigenetic readers that contain an evolutionary conserved acetyl-lysine recognition module, or bromodomain (BD), have drawn particular attention [36,37]. Of these BD-containing reader proteins, the BET (Bromodomain and Extra-Terminal) family, which comprise the ubiquitously expressed BRD2, BRD3, BRD4 and testis-specific BRDT, has become the subject of intense study given their role in a variety of cancers and the advent of potent and specific small molecule inhibitors against this family [38]. BET proteins are characterized by two tandem N-terminal BDs which are each 110 amino acids in length [36,37]. These BDs are comprised of four alpha-helices (Z, A, B, C) and two α-helix-connecting loops (ZA, BC) that provide a deep hydrophobic pocket for the specific recognition of acetylated lysine. Of the BET family, BRD4 is the most extensively characterized given its known role in transcriptional elongation [39,40] and its active development as a drug target in cancer. BRD4 was originally identified as a mitotic chromosome-binding protein that binds acetylated chromatin throughout the cell cycle, thus providing “epigenetic memory” for post mitotic transcription [41]. BRD4 is localized genome-wide to both actively transcribed promoters and enhancers, with a disproportionate enrichment at SEs [32,42–46]. A direct role of BRD4 in co-activating transcription is evidenced by its ability to associate with and allosterically activate CDK9, the core kinase in the positive transcription elongation factor b (P-TEFb) complex [39,40]. P-TEFb is a critical effector of Pol II transcriptional elongation via its ability to phosphorylate and inhibit pausing factors such as the DS1F/NELF complex as well as its ability to phosphorylate key serine residues on C-terminal heptapeptide repeats of Pol II [47].
Important roles for BRD4 have recently been described in a host of cell state transitions, including oncogenesis, where BRD4’s function is associated with epithelial-to-mesenchymal transition (EMT), stem cell-like conversion, and other broad transcriptional rewiring events that characterize tumor evolution [44,48–53]. Murine loss-of-function alleles for Brd2 and Brd4 in the germline demonstrate critical developmental roles for these proteins with homozygous mutant animals demonstrating early embryonic lethality [54,55]. The recent development of potent, specific, and reversible BET bromodomain inhibitors, such as the first-in-class tri-azolo-thienodiazepinc small-molecule JQ1, has significantly accelerated the therapeutic interest in the BET family [52,56]. JQ1 binds the acetyl-lysine binding pocket of BRD2, 3, and 4 with exquisite shape complementarity, high specificity, and nanomolar affinity, competitively displacing BET proteins from their endogenous acetylated interaction partners [52,56]. Pharmacological inhibition of BET proteins with JQ1 is therefore a reversible and dose-titratable tool for understanding the gene regulatory function of BRD4 as molecular amplifier of enhancer-to-promoter signaling. Importantly, drug derivatives of the tool compound JQ1 are now progressing in early phase cancer trials, providing a runway for considering BET inhibition in other disease settings [57]. Mice harboring conditionally targeted Brd4 alleles have recently been developed, allowing for the study of allele-specific and cell-restricted gene deletion in vivo [58]. While JQ1 binds all BET proteins and widely distributes to many tissues in vivo, it should be emphasized that these tool compounds allow functional probing of BET proteins in a manner that is wholly different from that which can be achieved by gene deletion/knockdown or bulk protein depletion (as occurs with heterobifunctional molecules that target BET protein degradation by bringing them into proximity with cellular E3 ubiquitin ligases) [59]. Importantly, pharmacological inhibition of BET proteins by pulsatile exposure of the small molecule JQ1 is reversible and therefore may overcome the potential toxicity associated with Brd4 gene silencing [60].
Two independent and contemporaneous publications from our groups were the first to establish proof of principle that BET reader proteins are essential co-activators of stress-dependent gene expression programs during HF pathogenesis [42,61] (for review, see Haidar and McKinsey 2014 [62]). BET inhibition using either JQ1 or siRNA- mediated Brd4 knockdown in isolated neonatal rat ventricular myocytes (NRVM) inhibited hallmark features of agonist-induced hypertrophy and suppressed stress-mediated gene induction, supporting a cell-autonomous role of BRD4 in controlling CM cell state during pathological stimulation [42]. To provide human relevance, we subsequently demonstrated that JQ1 could suppress cndothelin-1 induced hypertrophy and global stress-gene transactivation in cultured human iPSC-derived CMs [63]. Importantly, early administration of JQ1 in the murine transverse aortic constriction (TAC) model of LV pressure overload potently prevented LV systolic dysfunction, adverse cardiac remodeling, and cardiomyocyte hypertrophy/fibrosis [42]. In line with its active chromatin-binding activity, BRD4 ChIP-seq in adult mouse heart tissue demonstrated that BRD4 enriched at active cardiac enhancers and promoters. During TAC-mediated stress, Pol II ChIP-seq revealed widespread pause-release of Pol II at stress activated genes, a mechanism attenuated by JQ1 [42]. These data are consistent with the ability of enhancer-bound BRD4 to interact with the P-TEFb complex and promote productive Pol II elongation at stress activated genes. In addition, these findings corroborate previous work from the laboratory of Michael Schneider demonstrating that P-TEFb inhibition attenuates cardiomyocyte hypertrophy in vitro, while transgenic overexpression of cyclin-Tl (the key regulatory cyclin in the P-TEFb complex) in murine cardiomyocytes in vivo is sufficient to drive pathological cardiac remodeling [64]. Together, these findings support a model in which stress-activated signaling pathways deploy a defined set of DNA-binding transcription factors, which then bind specific enhancer elements in the stressed myocardium. In turn, BRD4 is recruited to these enhancer elements, possibly via interactions with acetylated histones and acetylated transcription factors, where it scaffolds transcription complexes such as CDK9/P-TEFb and triggers pause-release of genes that have actively poised Pol II at the transcription start site and are primed for activation [42]. Along this line, recent work in NRVMs has shown that during agonist mediated stress, BRD4 bound at basal enhancers robustly redistributes genome-wide, and becomes asymmetrically reallocated to a defined set of cardiomyocyte SEs [65]. Importantly, activity of these SEs and their target genes appear to be preferentially sensitive to local depletion of BRD4 via small molecule BET inhibition. Furthermore, the gain vs. loss of BRD4 at enhancers tends to correlate with concordant changes in transcriptional activity at the respective target genes, suggesting that reallocation of rate-limiting quantities of BRD4 directly affects gene transactivation. These findings are consistent with previous work in cultured endothelial cells during innate-inflammatory activation [46]. In addition to the BRD4/PTEFb complex, recent studies have also demonstrated that the general transcription factor TFIIB may also play a role in aberrant cardiac gene regulation during HF pathogenesis, raising the possibility that stress-dependent regulation of Pol II initiation at a subset of transcription start sites may also promote adverse cardiac remodeling [66,67].
While these index studies provided the initial insights into the role of BET protein function in the heart, they relied on early administration of JQ1 in the prevention of adverse cardiac remodeling and HF pathogenesis. More recently, our group has provided proof of principle for the therapeutic efficacy of BET inhibition in both the murine pressure overload and myocardial infarction (MI) models [63]. In the setting of prolonged pressure overload or a large anterior wall MI, late administration of JQ1 was able to treat pre-established HF, attenuating multiple hallmark features of HF progression in vivo, including LV dysfunction, LV cavity dilation, cardiomegaly, CM hypertrophy and left ventricle (LV) fibrosis [63], Using RNA-seq from whole LV tissue across both the murine TAC and MI treatment models, we defined a shared set of JQ1-suppressed genes that represent a common program mediating the observed therapeutic effects. This common gene signature was strongly enriched for transcripts involved in pro-fibrotic and innate inflammatory processes, with representation of genes that were expressed in several cell types that populate the stressed myocardium. Gene set enrichment and ingenuity pathway analyses revealed that JQ1 had a strong preference for suppressing TGFβ- and NFκB-driven transcriptional programs [63]. We found that the effect of JQ1 on differential gene expression was much more prominent in the stressed heart than in the baseline state (i.e. sham-treated), in line with our observation that JQ1 does not profoundly alter baseline cardiac structure or function at the doses used in our study. This is consistent with the observation that stress-dependent gene transactivation requires massive flux of BRD4 to new SE loci, an event likely to be preferentially sensitive to JQ1 [42,46]. Consistent with the lack of a striking effect on the unstressed heart, JQ1 treatment at the same doses used in our disease models did not attenuate exercise-induced cardiac growth in mice, a remodeling process that does not feature robust activation of fibrosis and inflammation [63,68,69]. These observations are consistent with a strong predilection for inhibiting pro-fibrotic and pro-inflammatory cell states during cardiac stress/injury.
While it is clear that JQ1 exerts potent on-target bioactivity in the heart and strongly attenuates TGFβ- and NFκB-dependent transcriptional programs, the precise identity of cell-types that mediate these therapeutic effects remains a major unanswered question with important therapeutic implications. Pro-fibrotic and inflammatoty gene transactivation can emanate from multiple cell types in the adult heart. As our initial transcriptional analyses were performed on whole heart tissue, further dissections of cell-intrinsic effects are an important area of ongoing investigation. We postulate that the therapeutic efficacy of JQ1 in these models may be via effects on several cell types including cardiomyocytes, fibroblasts, endothelial cells, and immune cell subsets. The bulk left ventricular (LV) tissue transcriptomic analyses indicate that the activated cardiac myoFBs, a key player in wound healing responses, may serve as a particularly important therapeutic target of BET inhibitors in vivo [63]. During cardiac stress, resident cardiac fibroblasts transition to the activated myoFB cell state, a process that features massive transcriptional rewiring allowing the capacity to proliferate, migrate, secrete cytokines, elaborate extracellular matrix, contract, and recruit immune cells [70]. Similar to observations in hepatic stellate cells [71], our data suggest that the cell state transition from a resident/quiescent cardiac fibroblast to an activated myoFB is highly sensitive to BET inhibition. Finally, we note that there are numerous other co-regulatory molecules that signal from enhancers to the transcription machinery, raising the possibility that other proteins may also serve as novel epigenetic targets in HF.
3. IncRNAs: gene regulatory switches that may be therapeutically manipulated in HF
In addition to proteins, several species of non-coding RNAs have been shown to play critical roles in chromatin regulation and broad control of gene expression programs. In this section, we turn attention to a subset of long noncoding RNAs QncRNAs) that have emerged as potentially druggable epigenetic targets in preclinical models of HF.
The mammalian genome is predominantly composed of non-protein coding sequence, raising the possibility for diverse regulatory functions of these regions. Deep RNA-sequencing approaches have revealed that the majority of the non-coding genome is actively transcribed, generating thousands of non-coding RNAs IncRNAs) [72–75]. While GRN activity has been long known to be controlled by TF proteins binding to their cognate DNA regulatory elements, emerging evidence strongly supports a similarly critical role for networks of ncRNAs [76,77]. The best characterized ncRNAs are microRNAs (miRNAs), small single-stranded ribonucleotides that lead to post-transcriptional silencing by targeting messenger RNAs for degradation or via translational suppression. Deep genome-wide transcriptomic profiling has identified other functional classes of transcripts with potentially important regulatory functions. Long non-coding RNAs (IncRNAs) are operationally defined as ncRNAs that are longer than 200 nucleotides in length and exhibit minimal protein coding potential. Currently, the best characterized IncRNAs are processed transcripts that are Pol II transcribed, multi-exonic, alternatively spliced, and poly-adenylated [78]. While IncRNAs are typically expressed at a lower level than mRNAs, they often feature much greater tissue and cell-type specificity [19,73]. LncRNAs have been shown to regulate diverse biological processes and are emerging as critical regulators of nearly every aspect of GRN activity, including transcriptional control, post-transcriptional processing, and chromatin remodeling [79]. LncRNAs are unique in their ability to spatially amplify regulatory information encoded by their underlying DNA where they can act in both cis and trans. Recently, IncRNAs have been shown to fine-tune GRNs controlling cardiac lineage specification [19,80,81]. Progress in transcription sequencing technologies have revealed that many functional enhancers are themselves transcription units that generate ncRNAs [18,82,83]. These exist as two distinct classes: bidirectional non-poly-adenylated transcripts (enhancer RNAs or cRNAs); and unidirectional, multi-exonic, spliced, poly-adenylated processed transcripts (enhancer-associated IncRNAs or clncRNAs). clncRNAs are important for concentrating TFs to specific DNA regions and modifying the local chromatin environment and have emerged as potential regulators of nuclear three-dimensional topologies [84–86]. Notably, elncRNAs expression is more cell and tissue specific than ncRNAs transcribed from non-enhancer regions [19]. Furthermore, SEs tend to produce more eRNAs than typical enhancers [18,84,87] with several IncRNAs associated with SEs now emerging as important modulators of cell fate determination and maintenance of cell identity in the heart [88,89].
4. Targeting IncRNAs in HF
Mhrt, the first IncRNA implicated in pathological hypertrophy, is located in the intergenic region between Myh6 and Myh7 in mice, and transcribed in an antisense orientation to Myh7 [90]. In the setting of pressure overload, Mhrt expression is reduced, an event which appears to be permissive for adverse cardiac remodeling. Transgenic overexpression of Mhrt in mice using a cardiomyocyte-specific promoter is sufficient to protect the heart from stress-induced cardiomyocyte hypertrophy and HF progression. Mechanistic studies demonstrate that Mhrt antagonizes the activity of BRG1, a chromatin remodeling factor previously implicated in the control of pathological cardiovascular GRNs. Mhrt acts as a molecular decoy, binding to the helicase domain of BRG1 and titrating it away from its DNA targets, thus suppressing expression of pathological gene programs. Importantly, the human MHRT ortholog is depleted in failing human hearts, suggesting a conserved regulatory role of potential translational significance.
In 2016, two other IncRNAs named Chast and Chaer were discovered upon screening for CM-enriched IncRNA species differentially expressed with cardiac stress [91,92], Like Mhrt, both of these IncRNAs have been shown to have important roles in controlling cardiac remodeling and HF pathogenesis. The group led by Thomas Thum characterized Chast, demonstrating its upregulation in murine CMs following TAC, but noted its concomitant expression in non-cardiac tissues [91], Consistent with these findings, the human ortholog of Chast was found to be significantly up-regulated in the hypertrophic hearts of patients with aortic stenosis. Viral overexpression of Chast was sufficient to trigger CM hypertrophy in both in vitro and in vivo systems. Although targeting of IncRNA transcripts can be challenging due to their generally low expression and subcellular localization (where they are enriched in defined chromatin compartments of the nucleus), modified antisense oligonucleotides (ASOs) have shown efficacy in robust and stable depletion of these species in vitro and in vivo. Indeed, ASO-mediated silencing of Chast attenuated TAC-induced pathological remodeling in adult mice in vivo. The IncRNA Chaer (cardiac-hypertrophy-associated epigenetic regulator) was discovered and characterized by Yibin Wang’s group, who showed that it interacts with the polycomb repressor complex 2 (PRC2) via a bi-tetra-loop motif within the 5′-end of Chaer that is both necessary and sufficient for PRC2 binding [92]. Through this interaction, Chaer interferes with PRC2 genomic localization, thereby potentiating pathological gene transactivation by reducing the repressive H3K27 histone methylation mark at the promoters of these genes. Indeed, germline inactivation of Chaer attenuated TAC-induced hypertrophy in mice in vivo. The translational potential of these findings will require future studies to demonstrate that silencing of Chaer at later stages of established HF is associated with improved indices of cardiac remodeling in preclinical models.
The strategy used to discover the aforementioned IncRNAs all selected for CM-enriched transcripts. Recently, there has been growing interest in understanding the role of non-CM cell types in HF pathogenesis with a particular focus on cardiac fibroblasts, given their highly plastic nature [70,93,94]. Upon cardiac stress, these cells undergo a dramatic epigenetically-regulated shift in their cellular identity. While fibrotic responses may acutely serve to stabilize a focal area of myocardial damage, clinical and experimental studies support the contention that excessive, diffuse, or chronic activation of these processes can be deleterious to long-term cardiac function and patient survival. Recent work from Jeff Molkentin’s group has implicated myoFBs (a sub-population of stress-activated fibroblasts that induce the marker gene Periostin) as a cell type that can fuel the vicious cycle of adverse cardiac remodeling in HF [93,94]. As such, understanding the fundamental mechanisms driving cardiac fibroblast cell state transitions during stress and uncovering novel approaches to target this process are of significant scientific and therapeutic interest. Thierry Pedrazzini’s group discovered Wisper (Wisp2 super-enhancer-associated RNA), a cardiac fibroblast-enriched IncRNA that regulates fibrosis in HF [88]. They identified Wisper using an existing database of annotated cardiac-enriched IncRNAs in HF samples [22]. followed by iterative filtering for those that were transcribed from cardiac SEs, had human orthologues, were induced in the setting of an MI, and were enriched in cardiac fibroblasts. In vitro loss-of-function studies demonstrated that Wisper positively regulated myoFB gene expression and function, including proliferation, migration, survival, and ECM deposition. ASO-mediated depletion of Wisper in adult mice post-MI inhibited cardiac fibrosis and significantly improved LV function. Mechanistically, Wisper was shown to bind the RNA splicing factor TIA1 cytotoxic granule associated RNA binding protein-like 1 (TIAR), preventing its nuclear translocation and subsequent processing of pro-fibrotic transcripts. Indeed, TIAR has been previously shown to control the post-transcriptional processing of PLOD2, a profibrotic enzyme. Importantly, Wisper expression is not detected in lung or kidney fibroblasts at baseline and is not induced in models of kidney fibrosis, supporting the notion that Wisper may have relative specificity for controlling cardiac fibrosis. Furthermore, expression levels of the human Wisper ortholog correlate with the degree of cardiac fibrosis in heart tissue isolated from patients with aortic stenosis, underscoring the translational relevance of these findings. We note that the Wisper locus contains both a SE DNA element as well as an encoded enhancer-associated IncRNA, such that transcription at the locus and/or the RNA itself may be involved in remodeling the local chromatin topology and stabilizing chromatin loops between the SE and its target promoters. Taken together, these findings demonstrate that Wisper is a cardiac fibroblast-enriched IncRNA that is conserved in humans, promotes transcriptional activation of the myoFB cell state, and functions as a potentially druggable target in human HF.
Ultimately, targeting IncRNAs for therapeutic purposes will require a deep mechanistic understanding of how these species regulate gene expression at the molecular level as well as determination of the specific cell types in which they function. While systemic delivery of nucleic acid-based therapies to extrahepatic tissues remains in its early days, this foundational knowledge will help improve the efficacy of this therapeutic strategy and increase our understanding of the toxicides that may arise from manipulating these powerful regulators of cell state.
5. Conclusions
The advent of novel sequencing technologies to dissect transcriptomic and epigenomic landscapes has fundamentally advanced our understanding of the gene control mechanisms that govern cell state. The concept that aberrant cell state contributes to disease and can be manipulated to exert therapeutic benefit is well-established in cancer biology, where novel therapies targeting these pathways arc in active clinical development. In contrast, our understanding of the epigenetic mechanisms that contribute to HF pathogenesis is still in its infancy. Here, we have provided examples of three classes of chromatin regulatory macromolecules (chromatin modifying enzymes such as G9a, co-activator proteins such as BRD4, and chromatin regulatory IncRNAs) that have been shown in proof-of-concept preclinical models to be druggable epigenetic targets in HF (Fig. 1). In order to advance the translational potential of these classes of molecules, future efforts must be directed at understanding the relevant cell types in which they are acting and the mechanisms by which they function. Conditional genetic approaches (where possible), single-cell based transcriptomic and epigenomic interrogations, and definition of the interaction partners for these chromatin regulators (including not only proteins but also RNA species) will be critical. Such studies may ultimately inform novel therapeutic strategies that may be directed in a more cell-type restricted manner, allowing for a greatly expanded therapeutic index of approaches that pharmacologically manipulate epigenetic signaling in human HF.
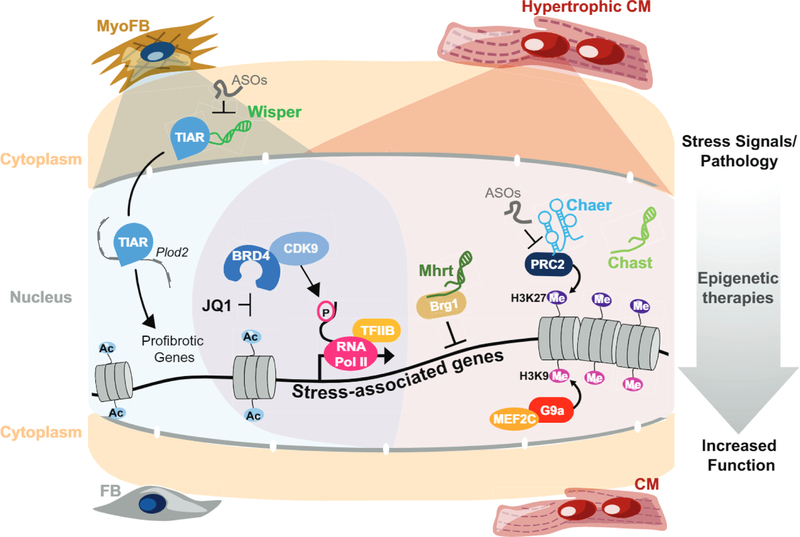
Fig. 1.
Stress-activated cardiac signaling cascades converge on defined components of the chromatin regulatory apparatus. These events trigger activation of stress-associated gene programs, Pol II pause release, cardiomegaly, fibrosis, and ultimately heart failure (HF). Chromatin regulators in HF that can be pharmacologically targeted with demonstration of translational potential include the histone methyltransferase G9a, members of the Bromo- and Extra- Terminal (BET) domain family, and certain long non-coding RNA (IncRNA) molecules, in unstressed CMs, G9a binds to and represses the transcriptional activity of MEF2C. Following stress, BET bromodomain inhibition with JQ1 suppresses stress-dependent transcriptional elongation in the heart and potently blocks pathological remodeling in vivo. CM-enriched IncRNAs such as Mhrt, Chast and Chaer play important roles during maladaptive cardiac remodeling. In contrast, the IncRNA Wisper is a cardiac fibroblast-enriched IncRNA that regulates cardiac fibrosis after injury. FB = Fibroblast. myoFB = Myofibroblast. CM = Cardiomyocyte. ASOs = Antisense Oligonucleotides.
Acknowledgments
We thank Ana Catarina Silva (ana@anasilvaillustrations.com) for drawing the schematic figure.
Sources of funding
M. Alexanian was supported by the Swiss National Science Foundation (project P2LAP3_178056). A. Padmanabhan is supported by the Tobacco-Related Disease Research Program Postdoctoral Fellowship (578649) and A.P. Giannini Foundation Career Development Award (P0527061). S.M. Haldar and T.A. McKinsey were supported by National Institutes of Health R01 HL127240. T.A. McKinsey received additional support from the National Institutes of Health2 (HL116848 and DK119594), as well as the American Heart Association (16SFRN31400013).
Footnotes
Disclosures
S.M. Haldar is an executive and shareholder of Amgen, Inc. and is a shareholder of Tenaya Therapeutics. The other authors report no conflicts.
References
- [1].Mosterd A, Hoes AW, de Bruyne MC, Deckers JW, Linker DT, Hofman A, Grobbee DE, Prevalence of heart failure and left ventricular dysfunction in the general population; the Rotterdam study, Eur. Heart J 20 (6) (1999) 447–455. [PubMed] [Google Scholar]
- [2].Redfield MM, Jacobsen SJ, Burnett JC, Mahoney DW Jr, Bailey KR, Rodeheffer RJ, Burden of systolic and diastolic ventricular dysfunction in the community: appreciating the scope of the heart failure epidemic, JAMA 289 (2) (2003) 194–202. [DOI] [PubMed] [Google Scholar]
- [3].Bleumink GS, Knetsch AM, Sturkenboom MC, Straus SM, Hofman A, Deckers JW, Witteman JC, Stricker BH, Quantifying the heart failure epidemic: prevalence, incidence rate, lifetime risk and prognosis of heart failure the Rotterdam study, Eur. Heart J 25 (18) (2004) 1614–1619. [DOI] [PubMed] [Google Scholar]
- [4].M. Writing Group, Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Isasi CR, Jimenez MC, Judd SE, Kissela BM, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Magid DJ, McGuire DK, Mohler ER 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Rosamond W, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Woo D, Yeh RW, Turner MB, C. American Heart Association Statistics, S. Stroke Statistics, Heart disease and stroke statistics-2016 update: A report from the American Heart Association, Circulation 133 (4) (2016) e38–360. [DOI] [PubMed] [Google Scholar]
- [5].Metra M, Teerlink JR, Heart failure, Lancet 390 (10106) (2017) 1981–1995. [DOI] [PubMed] [Google Scholar]
- [6].Lloyd-Jones DM, Larson MG, Leip EP, Beiser A, D’Agostino RB, Kannel WB, Murabito JM, Vasan RS, Benjamin EJ, Levy D, Framingham Heart S, Lifetime risk for developing congestive heart failure: the Framingham Heart Study, Circulation 106 (24) (2002) 3068–3072. [DOI] [PubMed] [Google Scholar]
- [7].Heidenreich PA, Albert NM, Allen LA, Bluemke DA, Butler J, Fonarow GC, Ikonomidis JS, Khavjou O, Konstam MA, Maddox TM, Nichol G, Pham M, Pina IL, Trogdon JG, C. American Heart, Association Advocacy Coordinating, T. Council on Arteriosclerosis, B. Vascular, R. Council on Cardiovascular, Intervention, C. Council on Clinical, E. Council on, Prevention, C. Stroke, Forecasting the impact of heart failure in the United States: a policy statement from the American Heart Association, Circ. Heart Fail 6 (3) (2013) 606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ahmad FS, Ning H, Rich JD, Yancy CW, Lloyd-Jones DM, Wilkins JT, Hypertension, obesity, diabetes, and Heart failure-free survival: the cardiovascular disease lifetime risk pooling project, JACC, Heart Fail 4 (12) (2016) 911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lee DS, Massaro JM, Wang TJ, Kannel WB, Benjamin EJ, Kenchaiah S, Levy D, D’Agostino RB Sr., Vasan RS, Antecedent blood pressure, body mass index, and the risk of incident heart failure in later life, Hypertension 50 (5) (2007) 869–876. [DOI] [PubMed] [Google Scholar]
- [10].Taylor CJ, Ryan R, Nichols L, Gale N, Hobbs FR, Marshall T, Survival following a diagnosis of heart failure in primary care, Fam. Pract 34 (2) (2017) 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hill JA, Olson EN, Cardiac plasticity, N. Engl. J. Med 358 (13) (2008) 1370–1380. [DOI] [PubMed] [Google Scholar]
- [12].van Berlo JH, Maillet M, Molkentin JD, Signaling effectors underlying pathologic growth and remodeling of the heart, J. Clin. Invest 123 (1) (2013) 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bristow MR, beta-adrenergic receptor blockade in chronic heart failure, Circulation 101 (5) (2000) 558–569. [DOI] [PubMed] [Google Scholar]
- [14].Waddington CH, The epigenotype. 1942, Int. J. Epidemiol 41 (1) (2012) 10–13. [DOI] [PubMed] [Google Scholar]
- [15].Allis CD, Jenuwein T, The molecular hallmarks of epigenetic control, Nat. Rev. Genet 17 (8) (2016) 487–500. [DOI] [PubMed] [Google Scholar]
- [16].Di Salvo TG, Haldar SM, Epigenetic mechanisms in heart failure pathogenesis, Circ. Heart Fail 7 (5) (2014) 850–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, Haugen E, Heimfeld S, Murry CE, Akey JM, Stamatoyannopoulos JA, Developmental fate and cellular maturity encoded in human regulatory DNA landscapes, Cell 154 (4) (2013) 888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li W, Notani D, Rosenfeld MG, Enhancers as non-coding RNA transcription units: recent insights and future perspectives, Nat. Rev. Genet 17 (4) (2016) 207–223. [DOI] [PubMed] [Google Scholar]
- [19].Alexanian M, Marie D, Jenkinson SP, Mina M, Friedman CE, Ting CC, Micheletti R, Plaisance I, Nemir M, Maison D, Kernen J, Pezzuto I, Villeneuve D, Burdet F, Ibberson M, Leib SL, Palpant NJ, Hernandez N, Ounzain S, Pedrazzini T, A transcribed enhancer dictates mesendoderm specification in pluripotency, Nat. Commun 8 (1) (2017) 1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA, Bruneau BG, Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage, Cell 151 (1) (2012) 206–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dickel DE, Barozzi I, Zhu Y, Fukuda-Yuzawa Y, Osterwalder M, Mannion BJ, May D, Spurrell CH, Plajzer-Frick I, Pickle CS, Lee E, Garvin TH, Kato M, Akiyama JA, Afzal V, Lee AY, Gorkin DU, Ren B, Rubin EM, Visel A, Pennacchio LA, Genome-wide compendium and functional assessment of in vivo heart enhancers, Nat. Commun 7 (2016) 12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, Crippa S, Nemir M, Sarre A, Johnson R, Dauvillier J, Burdet F, Ibberson M, Guigo R, Xenarios I, Heymans S, Pedrazzini T, Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs, Eur. Heart J 36 (6) (2015) 353–68a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].He A, Gu F, Hu Y, Ma Q, Ye LY, Akiyama JA, Visel A, Pennacchio LA, Pu WT, Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease, Nat. Commun 5 (2014) 4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Papait R, Cattaneo P, Kunderfranco P, Greco C, Carullo P, Guffanti A, Vigano V, Stirparo GG, Latronico MV, Hasenfuss G, Chen J, Condorelli G, Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy, Proc. Natl. Acad. Sci. U. S. A 110 (50) (2013) 20164–20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stein AB, Jones TA, Herron TJ, Patel SR, Day SM, Noujaim SF, Milstein ML, Klos M, Furspan PB, Jalife J, Dressler GR, Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes, J. Clin. Invest 121 (7) (2011) 2641–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang QJ, Chen HZ, Wang L, Liu DP, Hill JA, Liu ZP, The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice, J. Clin. Invest 121 (6) (2011) 2447–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Papait R, Serio S, Pagiatakis C, Rusconi F, Carullo P, Mazzola M, Salvarani N, Miragoli M, Condorelli G, Histone methyltransferase G9a is required for cardiomyocyte homeostasis and hypertrophy, Circulation 136 (13) (2017) 1233–1246. [DOI] [PubMed] [Google Scholar]
- [28].Gilsbach R, Schwaderer M, Preissl S, Gruning BA, Kranzhofer D, Schneider P, Nuhrenberg TG, Mulero-Navarro S, Weichenhan D, Braun C, Dressen M, Jacobs AR, Lahm H, Doenst T, Backofen R, Krane M, Gelb BD, Hein L, Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo, Nat. Commun 9 (1) (2018) 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jin SG, Kadam S, Pfeifer GP, Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine, Nucleic Acids Res 38 (11) (2010) el25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Greco CM, Kunderfranco P, Rubino M, Larcher V, Carullo P, Anselmo A, Kurz K, Carell T, Angius A, Latronico MV, Papait R, Condorelli G, DNA hydroxymethylation controls cardiomyocyte gene expression in development and hypertrophy, Nat. Commun 7 (2016) 12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li J, Wu X, Zhou Y, Lee M, Guo L, Han W, Mo W, Cao WM, Sun D, Xie R, Huang Y, Decoding the dynamic DNA methylation and hydroxymethylation landscapes in endodermal lineage intermediates during pancreatic differentiation of hESC, Nucleic Acids Res 46 (6) (2018) 2883–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA, Master transcription factors and mediator establish super-enhancers at key cell identity genes, Cell 153 (2) (2013) 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA, Super-enhancers in the control of cell identity and disease, Cell 155 (4) (2013) 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].McKinsey TA, Olson EN, Toward transcriptional therapies for the failing heart: chemical screens to modulate genes, J. Clin. Invest 115 (3) (2005) 538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].McKinsey TA, Therapeutic potential for HDAC inhibitors in the heart, Annu. Rev. Pharmacol. Toxicol 52 (2012) 303–319. [DOI] [PubMed] [Google Scholar]
- [36].Zeng L, Zhou MM, Bromodomain: an acetyl-lysine binding domain, FEBS Lett 513 (1) (2002) 124–128. [DOI] [PubMed] [Google Scholar]
- [37].Berger SL, The complex language of chromatin regulation during transcription, Nature 447 (7143) (2007) 407–412. [DOI] [PubMed] [Google Scholar]
- [38].Taniguchi Y, The Bromodomain and extra-terminal domain (BET) family: functional anatomy of BET Paralogous proteins, Int. J. Mol. Sci 17 (11) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K, The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription, Mol. Cell 19 (4) (2005) 523–534. [DOI] [PubMed] [Google Scholar]
- [40].Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q, Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4, Mol. Cell 19 (4) (2005) 535–545. [DOI] [PubMed] [Google Scholar]
- [41].Dey A, Ellenberg J, Farina A, Coleman AE, Maruyama T, Sciortino S, Lippincott-Schwartz J, Ozato K, A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition, Mol. Cell. Biol 20 (17) (2000) 6537–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, Alaiti MA, Bullard J, Alazem K, Margulies KB, Cappola TP, Lemieux M, Plutzky J, Bradner JE, Haldar SM, BET bromodomains mediate transcriptional pause release in heart failure, Cell 154 (3) (2013) 569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, Reichert E, Rung AL, Rodig SJ, Young RA, Shipp MA, Bradner JE, Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma, Cancer Cell 24 (6) (2013) 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA, Selective inhibition of tumor oncogenes by disruption of super-enhancers, Cell 153 (2) (2013) 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, N.C.S. Program, Black BL, Visel A, Pennacchio LA, Collins FS, A. National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program, N.C.S.P. Authors, Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants, Proc. Natl. Acad. Sci. U. S. A 110 (44) (2013) 17921–17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Brown JD, Lin CY, Duan Q, Griffin G, Federation A, Paranal RM, Bair S, Newton G, Lichtman A, Kung A, Yang T, Wang H, Luscinskas FW, Croce K, Bradner JE, Plutzky J, NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis, Mol. Cell 56 (2) (2014) 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Adelman K, Lis JT, Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans, Nat. Rev. Genet 13 (10) (2012) 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].French CA, Miyoshi I, Aster JC, Kubonishi I, Kroll TG, Dal Cin P, Vargas SO, Perez-Atayde AR, Fletcher JA, BRD4 bromodomain gene rearrangement in aggressive carcinoma with translocation t(15;19), Am. J. Pathol 159 (6) (2001) 1987–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rodriguez RM, Huidobro C, Urdinguio RG, Mangas C, Soldevilla B, Dominguez G, Bonilla F, Fernandez AF, Fraga MF, Aberrant epigenetic regulation of bromodomain BRD4 in human colon cancer, J. Mol. Med 90 (5) (2012) 587–595. [DOI] [PubMed] [Google Scholar]
- [50].Alluri PG, Asangani IA, Chinnaiyan AM, BETs abet Tam-R in ER-positive breast cancer, Cell Res 24 (8) (2014) 899–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR, BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia, Mol. Cell 58 (6) (2015) 1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE, Selective inhibition of BET bromodomains, Nature 468 (7327) (2010) 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T, Rodig SJ, Kung AL, Bradner JE, Weinstock DM, BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia, Blood 120 (14) (2012) 2843–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shang E, Wang X, Wen D, Greenberg DA, Wolgemuth DJ, Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse, Develop. Dyn 238 (4) (2009) 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS, Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4, Mol. Cell. Biol 22 (11) (2002) 3794–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A, Suppression of inflammation by a synthetic histone mimic, Nature 468 (7327) (2010) 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Andrieu G, Belkina AC, Denis GV, Clinical trials for BET inhibitors run ahead of the science, Drug Discov. Today Technol 19 (2016) 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lee JE, Park YK, Park S, Jang Y, Waring N, Dey A, Ozato K, Lai B, Peng W, Ge K, Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis, Nat. Commun 8 (1) (2017) 2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE, Drug Development, Phthalimide conjugation as a strategy for in vivo target protein degradation, Science 348 (6241) (2015) 1376–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bolden JE, Tasdemir N, Dow LE, van Es JH, Wilkinson JE, Zhao Z, Clevers H, Lowe SW, Inducible in vivo silencing of Brd4 identifies potential toxicities of sustained BET protein inhibition, Cell Rep 8 (6) (2014) 1919–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Spiltoir JI, Stratton MS, Cavasin MA, Demos-Davies K, Reid BG, Qi J, Bradner JE, McKinsey TA, BET acetyl-lysine binding proteins control pathological cardiac hypertrophy, J. Mol. Cell. Cardiol 63 (2013) 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Haldar SM, McKinsey TA, BET-ting on chromatin-based therapeutics for heart failure, J. Mol. Cell. Cardiol 74 (2014) 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Duan Q, McMahon S, Anand P, Shah H, Thomas S, Salunga HT, Huang Y, Zhang R, Sahadevan A, Lemieux ME, Brown JD, Srivastava D, Bradner JE, McKinsey TA, Haidar SM, BET bromodomain inhibition suppresses innate inflammatory and proflbrotic transcriptional networks in heart failure, Sci. Transl. Med 9 (390) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, Michael LH, DeMayo FJ, Schneider MD, Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy, Nat. Med 8 (11) (2002) 1310–1317. [DOI] [PubMed] [Google Scholar]
- [65].Stratton MS, Lin CY, Anand P, Tatman PD, Ferguson BS, Wickers ST, Ambardekar AV, Sucharov CC, Bradner JE, Haldar SM, McKinsey TA, Signal-dependent recruitment of BRD4 to cardiomyocyte super-enhancers is suppressed by a microRNA, Cell Rep 16 (5) (2016) 1366–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sayed D, He M, Yang Z, Lin L, Abdellatif M, Transcriptional regulation patterns revealed by high resolution chromatin immunoprecipitation during cardiac hypertrophy, J. Biol. Chem 288 (4) (2013) 2546–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sayed D, Yang Z, He M, Pfleger JM, Abdellatif M, Acute targeting of general transcription factor UB restricts cardiac hypertrophy via selective inhibition of gene transcription, Circ. Heart Fail 8 (1) (2015) 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bostrom P, Mann N, Wu J, Quintero PA, Plovie ER, Panakova D, Gupta RK, Xiao C, MacRae CA, Rosenzweig A, Spiegelman BM, C/EBPbeta controls exercise-induced cardiac growth and protects against pathological cardiac remodeling, Cell 143 (7) (2010) 1072–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Maillet M, van Berio JH, Molkentin JD, Molecular basis of physiological heart growth: fundamental concepts and new players, Nat. Rev. Mol. Cell Biol 14 (1) (2013) 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Alexanian M, Haidar SM, The cardiac myofibroblast, Circ. Res 123 (12) (2018) 1258–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ding N, Hah N, Yu RT, Sherman MH, Benner C, Leblanc M, He M, Liddle C, Downes M, Evans RM, BRD4 is a novel therapeutic target for liver fibrosis, Proc. Natl. Acad. Sci. U. S. A 112 (51) (2015) 15713–15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta F, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii- Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y, The transcriptional landscape of the mammalian genome, Science 309 (5740) (2005) 1559–1563. [DOI] [PubMed] [Google Scholar]
- [73].Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL, Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses, Genes Dev 25 (18) (2011) 1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES, Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals, Nature 458 (7235) (2009) 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Morris KV, Mattick JS, The rise of regulatory RNA, Nat. Rev. Genet 15 (6) (2014) 423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kaikkonen MU, Lam MT, Glass CK, Non-coding RNAs as regulators of gene expression and epigenetics, Cardiovasc. Res 90 (3) (2011) 430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Mercer TR, Dinger ME, Mattick JS, Long non-coding RNAs: insights into functions, Nat. Rev. Genet 10 (3) (2009) 155–159. [DOI] [PubMed] [Google Scholar]
- [78].Perry RB, Ulitsky I, The functions of long noncoding RNAs in development and stem cells, Development 143 (21) (2016) 3882–3894. [DOI] [PubMed] [Google Scholar]
- [79].Mercer TR, Mattick JS, Structure and function of long noncoding RNAs in epigenetic regulation, Nat. Struct. Mol. Biol 20 (3) (2013) 300–307. [DOI] [PubMed] [Google Scholar]
- [80].Grote P, Wittier L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG, The tissue-specific IncRNA Fendrr is an essential regulator of heart and body wall development in the mouse, Dev. Cell 24 (2) (2013) 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, Abo R, Tabebordbar M, Lee RT, Burge CB, Boyer LA, Braveheart, a long noncoding RNA required for cardiovascular lineage commitment, Cell 152 (3) (2013) 570–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Marques AC, Hughes J, Graham B, Kowalczyk MS, Higgs DR, Ponting CP, Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs, Genome Biol 14 (11) (2013) HI 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ounzain S, Pezzuto I, Micheletti R, Burdet F, Sheta R, Nemir M, Gonzales C, Sarre A, Alexanian M, Blow MJ, May D, Johnson R, Dauvillier J, Pennacchio L, Pedrazzini T, Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease, J. Mol. Cell. Cardiol 76 (2014. November) 55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis AC, Sharp PA, Young RA, Transcription factor trapping by RNA in gene regulatory elements, Science 350 (6263) (2015) 978–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Mele M, Rinn JL, “Cat’s cradling” the 3D genome by the act of LncRNA transcription, Mol. Cell 62 (5) (2016) 657–664. [DOI] [PubMed] [Google Scholar]
- [86].Tan JY, Smith AAT, Ferreira da Silva M, Matthey-Doret C, Rueedi R, Sonmez R, Ding D, Kutalik Z, Bergmann S, Marques AC, Cis-acting complex-trait-associated lincRNA expression correlates with modulation of chromosomal architecture, Cell Rep 18 (9) (2017) 2280–2288. [DOI] [PubMed] [Google Scholar]
- [87].Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, Young RA, Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells, Proc. Natl. Acad. Sci. U. S. A 110 (8) (2013) 2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Micheletti R, Plaisance I, Abraham BJ, Sarre A, Ting CC, Alexanian M, Marie D, Maison D, Nemir M, Young RA, Schroen B, Gonzalez A, Ounzain S, Pedrazzini T, The long noncoding RNA Wisper controls cardiac fibrosis and remodeling, Sci. Transl. Med 9 (395) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ounzain S, Micheletti R, Aman C, Plaisance L, Cecchi D, Schroen B, Reverter F, Alexanian M, Gonzales C, Ng SY, Bussotti G, Pezzuto I, Notredame C, Heymans S, Guigo R, Johnson R, Pedrazzini T, CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis, J. Mol. Cell. Cardiol 89 (2015) 98–112 Pt A. [DOI] [PubMed] [Google Scholar]
- [90].Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien H, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HV, Quertermous T, Chang CP, A long noncoding RNA protects the heart from pathological hypertrophy, Nature 514 (7520) (2014) 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, Dittrich M, Maetzig T, Zimmer K, Remke J, Just A, Fendrich J, Scherf K, Bolesani E, Schambach A, Weidemann F, Zweigerdt R, de Windt LJ, Engelhardt S, Dandekar T, Batkai S, Thum T, Long noncoding RNA Chast promotes cardiac remodeling, Sci. Transl. Med 8 (326) (2016) 326ra22. [DOI] [PubMed] [Google Scholar]
- [92].Wang Z, Zhang XJ, Ji YX, Zhang P, Deng KQ, Gong J, Ren S, Wang X, Chen I, Wang H, Gao C, Yokota T, Ang YS, Li S, Cass A, Vondriska TM, Li G, Deb A, Srivastava D, Yang HT, Xiao X, Li H, Wang Y, The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy, Nat. Med 22 (10) (2016) 1131–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, SC JL, Aronow BJ, Tallquist MD, Molkentin JD, Genetic lineage tracing defines myofibroblast origin and function in the injured heart, Nat. Commun 7 (2016) 12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Khalil H, Kanisicak O, Prasad V, Correll RN, Fu X, Schips T, Vagnozzi RJ, Liu R, Huynh T, Lee SJ, Karch J, Molkentin JD, Fibroblast-specific TGF-beta-Smad2/3 signaling underlies cardiac fibrosis, J. Clin. Invest 127 (10) (2017) 3770–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]