Summary
Protein complexes built of Structural Maintenance of Chromosomes (SMC) and kleisin subunits, including cohesin, condensin and the Smc5/6 complex, are master organizers of genome architecture in all kingdoms of life. How these large ring-shaped molecular machines use the energy of ATP hydrolysis to change the topology of chromatin fibers has remained a central unresolved question of chromosome biology. A currently emerging concept suggests that the common principle that underlies the essential functions of SMC protein complexes in the control of gene expression, chromosome segregation or DNA damage repair is their ability to expand DNA into large loop structures. Here, we review the current knowledge about the biochemical and structural properties of SMC proteins complexes that might allow them to accomplish DNA loop extrusion and compare their action to other motor proteins and nucleic acid translocases. We evaluate the currently predominant models of active loop extrusion and propose a detailed version of a ‘scrunching’ model, which reconciles much of the available mechanistic data and provides an elegant explanation for how SMC protein complexes fulfill an array of seemingly diverse tasks during the organization of genomes.
Introduction: SMC protein complexes and the DNA loop extrusion hypothesis
The sheer length of the DNA molecule in each of our chromosomes poses unique challenges to DNA transactions that need to take place within the confines of the nuclear or cellular space. A family of large protein complexes present in all domains of life has evolved to deal with the large-scale organization of our genetic material [1]. These complexes are characterized by long coiled-coil protein subunits named Structural Maintenance of Chromosomes (SMC), which use the energy of adenosine triphosphate (ATP) hydrolysis to drive conformational changes in DNA topology. In prokaryotes, SMC or SMC-related homodimers assemble with additional subunits to form SMC–ScpAB, MukBEF or MksBEF complexes that play key roles in the successful segregation of the bacterial nucleoid (Figure 1A) [2]. Eukaryotic SMC complexes are, in contrast, built from heterodimers of different SMC subunits that form the basis for cohesin, condensin and Smc5/6 complexes [3,4]. Each complex has distinct functions: Cohesin holds together sister chromatids and is responsible for the dynamic compartmentalization of chromosomes during interphase into Topologically Associating Domains (TADs). Condensin plays a key role in the compaction and individualization of chromatids in preparation for their segregation by spindle microtubules during cell divisions. Smc5/6 complexes have been implicated in the repair of DNA damage by homologous recombination and the resolution of DNA supercoiling that results from DNA replication.
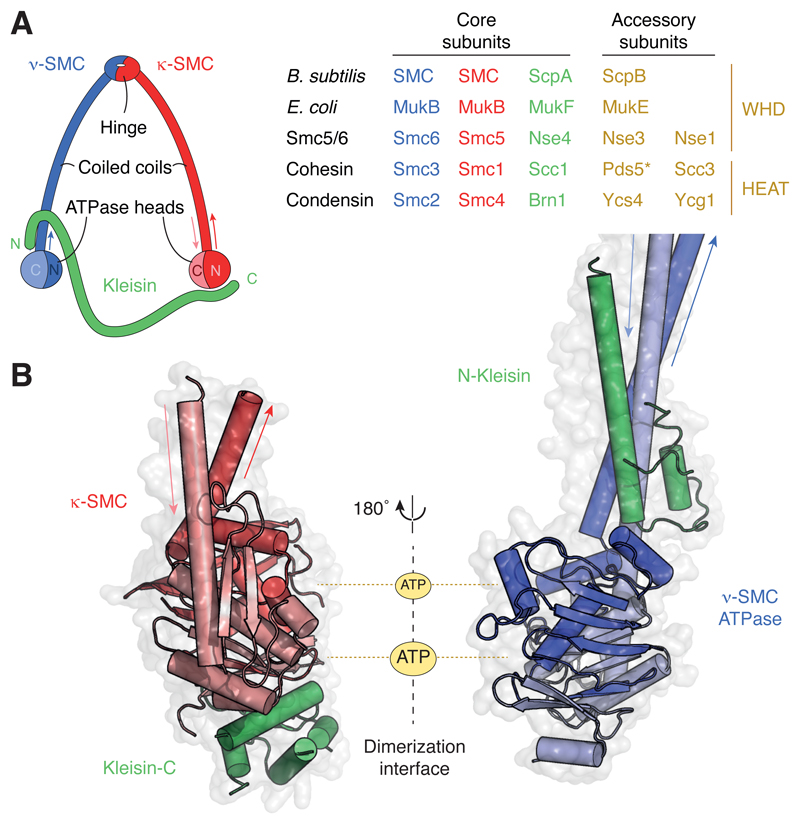
Figure 1. Architecture and composition of SMC complexes.
(A) Ring-like architecture created by the association of the SMC and kleisin subunits and subunit composition of common prokaryotic (top two) and eukaryotic (bottom three; S. cerevisiae protein names) SMC complexes. *Note that Pds5 associates with the other cohesin subunits transiently and might hence not be a stoichiometric subunit of cohesin complexes. (B) Structure models of a κ-SMC ATPase head domain bound to the WHD located at the C terminus of the kleisin subunit (pdb: 1w1w; S. cerevisiae cohesin Smc1–Scc1) and of a ν-SMC ATPase head domain bound to the helical domain located at the N terminus of the kleisin subunit (pdb: 4ux3; S. cerevisiae cohesin Smc3–Scc1).
SMC proteins fold around a globular ‘hinge’ domain, situated in the middle of their peptide chain, into anti-parallel coiled coils of varying average lengths (Figure 1A), ranging between ~270 amino acid (aa) residues for the coiled coils of the Smc5–Smc6 to ~350 aa for the coiled coils of Smc1–Smc3 and Smc2–Smc4 heterodimers of cohesin and condensin, respectively [5]. The combined N and C termini at the end of the coiled coil create a globular ‘head’ domain that contains ATP binding and hydrolysis motifs of the ATP-Binding Cassette (ABC) transporter family. In addition to the constitutive dimerization via the association of their hinge domains, two SMC molecules transiently interact when their head domains sandwich a pair of ATP molecules, which creates two hydrolysis-competent catalytic pockets, similar to other ABC ATPases. The ATPase head domains are connected by a so-called ‘kleisin’ subunit, which binds the coiled-coil base region immediately adjacent to one SMC (designated as ν-SMC) head domain via an α-helix located at its N terminus [6–8] and to the distal side of the other SMC (designated as κ-SMC) head domain via a Winged-Helix Domain (WHD) at its C terminus (Figure 1B) [6,9]. This arrangement imposes overall asymmetry even upon prokaryotic complexes and results in a closed ring structure, which is able to encircle DNA within its circumference [7,10–12]. In addition, all kleisin proteins recruit supplementary protein subunits that are either composed of α-helical repeat motifs found in Huntingtin, Elongation factor 3, protein phosphatase 2A and TOR1 kinase (HEAT) proteins, as in the case of cohesin and condensin, or tandem WHDs, as in the case of Smc5–Smc6 and prokaryotic SMC complexes (Figure 1A) [13,14]. Several other proteins, some with distinct enzymatic activities, have been found to also associate with different SMC complexes (for an overview, see [15]).
How do SMC protein complexes organize chromosome architecture? An increasing amount of evidence supports the notion that the common molecular mechanism that underlies the action of SMC complexes is their ability to create and progressively enlarge loops of DNA in a process termed loop extrusion (Figure 2A) [16,17]. This simple mechanism can bring two distant elements on the same DNA molecule into close physical proximity. For example, enhancer sequences that can be hundreds of kilobase pairs (kbp) away can be targeted to the promoter sequences they regulate for the activation of gene expression. Loop extrusion furthermore provides an elegant method for folding the chromatin fiber into rod-shaped mitotic chromosomes. In computational simulations that use polymer physics, loop extrusion can indeed account for the dynamic sub-compartmentalization of the genome into TADs [18,19] as well as mitotic chromosome formation and sister chromatid individualization [20,21]. By encircling and linking together DNA helices [10,11], SMC protein complexes seem to be excellently suited to direct the formation of DNA loops in the genome. Whether they fulfill the requirements that are imposed onto loop-extruding factors in theoretical modeling approaches remains, however, unclear. In this review, we discuss possible mechanisms for SMC-mediated DNA loop extrusion based on the biochemical and structural properties of these large ring-shaped protein complexes.
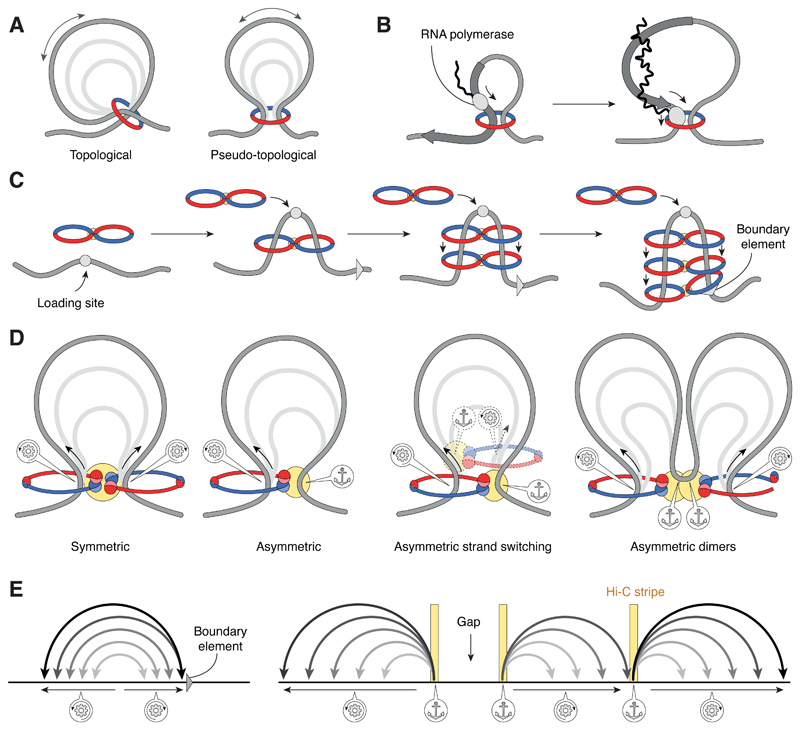
Figure 2. Principles of DNA loop extrusion.
(A) SMC rings might capture the bases of DNA loops either in a topological manner (which requires ring opening for DNA entry or exit) or a pseudo-topological manner (which does not require ring opening for DNA entry). (B) Loop Extrusion by external motors. The transcription machinery might push loops to the 3’ end of actively transcribed genes, thereby enlarging loops. (C) Motor-free models of loop extrusion. The continuous loading of SMC complexes at a specific site creates osmotic pressure that pushes already loaded rings along the DNA until they encounter a barrier (boundary element), where they accumulate and dissociate. (D) SMC complexes as DNA motors. Loop Extrusion could be driven by SMC dimers (handcuffs) that reel in DNA symmetrically from both sides or by individual SMC complexes that anchor DNA at one side and reel in DNA from the other. Symmetric loop extrusion could alternatively be achieved if individual complexes repeatedly switch strands or if two complexes that each anchor DNA assemble in a head-to-head orientation, producing two loops as they reel in DNA from opposite sides. (E) Symmetric loop extrusion until halted by the encounter of boundary elements reproduces TAD formation. Asymmetric loop extrusion is expected to produce ‘stripes’ in Hi-C contact maps and might lead to gaps that are not folded into loops.
What drives DNA loop extrusion?
Models that rely on external motors
One key aspect that distinguishes models of SMC-mediated loop extrusion is the energy source that drives the expansion of DNA loops. Despite possessing ATPase domains, there are no clear clues from the architecture of SMC complexes as to how the chemical energy from ATP hydrolysis could be converted into the mechanical motion that would be necessary to actively move DNA. Furthermore, the ATP hydrolysis rates that have so far been measured for different SMC complexes are extremely low – ranging between 0.1 and 2 molecules ATP hydrolyzed per SMC dimer and second – when compared to those of DNA motor proteins, which are capable of hydrolyzing thousands of ATP molecules per second (Table 1) [22,23]. These low ATPase rates make it difficult to imagine that ATP hydrolysis by the SMC subunits could be sufficient for rapid loop extrusion rates, which have been estimated based on Hi-C data to reach speeds of several hundred base pairs (bp) per second in bacteria [41]. It is therefore conceivable that the driving force for loop extrusion might come from external DNA motor proteins that move DNA through the SMC protein ring.
Table 1. ATPase rates of SMC complexes and other motor proteins.
| Protein complex | Max. ATPase rate (s-1) | References | |
|---|---|---|---|
| SMC complexes | B. subtilis SMC | 0.3 | [24–26] |
| B. subtilis SMC–ScpAB | 0.5 | [26] | |
| E. coli MukB | <0.1 | [27,28] | |
| E. coli MukBEF | 0.1 | [28] | |
| S. cerevisiae Smc2–Smc4 | 0.2 | [29] | |
| S. cerevisiae condensin | * 2.0 | [30,31] | |
| X. laevis Smc2–Smc4 | 0.1 | [32] | |
| X. laevis condensin | * 0.9 | [33] | |
| S. pombe condensin | 0.2 | [34] | |
| S. pombe cohesin | ** 0.2 | [35] | |
| ABC transporters | E. coli BtuCD | 2 | [36] |
| S. typhimurium HisP | 8 | [37] | |
| T. maritima TM287/288 | 2 | [38] | |
| Motors | E. coli FtsK translocase | 2,600 | [22] |
| E. coli EcoR124I translocase | 998 | [23] | |
| D. melanogaster kinesin | 25 | [39] | |
| S. cerevisiae dynein | 38 | [40] |
in the presence of DNA
in the presence of the Scc2–Scc4 loader and DNA
The observation cohesin complexes accumulate at sites of convergent gene transcription in budding yeast[42–46] and in mouse embryonic fibroblasts lacking both Wapl and CTCF – two proteins that release cohesin from chromosomes or anchor it to specific sites in the genome, respectively [47] – prompted the idea that RNA polymerases might push cohesin along the chromatin fiber (Figure 2B). In this scenario, cohesin is able to slide along the DNA double helix due to the topological entrapment of DNA within the cohesin ring architecture [48]; a notion that is consistent with the in vitro release of cohesin from circular minichromosomes upon linearization of the DNA double helix [49] and the random translocation of salt-resistant cohesin complexes along immobilized DNA molecules in single molecule imaging experiments [50–52]. The one-dimensional diffusion coefficients measured in the latter studies ranged between 0.2 and 3.8 μm2/s and mobility increased with increasing salt concentrations, which implies that cohesin complexes make electrostatic contacts with the DNA double helix, in addition to encircling it. Whereas cohesin was able to move past DNA-bound obstacles up to the size of nucleosomes (~11 μm diameter), although with much reduced diffusion times, it failed to pass larger objects (20 μm or more), including other cohesin molecules. Remarkably, a bacterial RNA polymerase moved cohesin along the DNA over distances of several kbp [50], as did other DNA motor proteins, including the prokaryotic DNA translocase FtsK [51]. Hence, the combined action of external molecular motors, roadblocks and cohesin rings that diffuse along the DNA helix could result in an expansion of DNA loops – as long as the pushing motor acts within the confines of a pre-formed loop (Figure 2B).
However, a model in which cohesin-dependent loop extrusion is driven by the transcription machinery cannot account for the de novo TAD formation in mouse zygotes before the onset of transcription [53] or in early Drosophila embryos in which transcription elongation has been blocked by small molecule inhibitors [54]. Inhibition of transcription also had no major effects on TAD formation after cohesin depletion and re-induction, or after B cell activation; nor did it massively alter the genomic distribution of cohesin [55]. Pushing of cohesin rings by RNA polymerases hence cannot be the sole explanation for how these complexes move along the chromatin fiber. Moreover, entrapment of DNA by cohesin rings does not seem to be a prerequisite for translocation, since mutations in the SMC hinge domains or in one of the two SMC ATP binding sites of yeast cohesin that are both thought to block stable topological loading onto chromosomes did not interfere with cohesin migration from its loading sites at centromeres up to 30 kbp into the neighboring chromosome regions [56] or affect the diffusive behavior of cohesin in a single molecule assay [52]. It therefore seems unlikely that the driving force behind loop extrusion can be explained by the action of external motors that move passively sliding SMC complexes along the chromatin fiber.
Models of motor-free mechanisms
Does loop extrusion require any motor activity at all? Recent computer simulations suggest that, at least in theory, the processive enlargement of DNA loops can be explained by a combination of local loading of SMC protein complexes coupled to their diffusion from the loading site [57,58]. In these models, the osmotic pressure generated by the loading of new cohesin complexes at a defined site would provide a ‘ratchet’ mechanism to drive cohesins away from the loading site until they reach boundary elements, for example CTCF-bound DNA regions, which confine their diffusion (Figure 2C). If at least some of the cohesin molecules were loaded such that they bridge two DNAs, either by their entrapment within a single ring or through tethering of two rings that each entrap a single DNA, this process would result in the extrusion of a DNA loop that is stabilized once cohesin reaches the boundary elements, thereby reproducing the basic features observed in Hi-C contact maps [57].
Although these models are able to reproduce in silico loop extrusion driven by the energy of localized cohesin loading and the thermal energy of diffusion, they would predict the steady-state enrichment of cohesin around sites occupied by its Scc2–Scc4 loader complex. Whereas such an enrichment is not evident from ChIP-seq data under normal conditions [47], it can be observed after depletion of cellular ATP levels in human cells [55] or when ATP hydrolysis by the SMC subunits is prevented by mutations in yeast cells [59]. ATP hydrolysis is therefore either required to load cohesin complexes onto DNA in a manner that allows their diffusion away from the loading site, or, more likely, for an active movement process along the chromosome. A mechanism that drives loop extrusion by osmotic pressure is furthermore difficult to extend to condensin complexes, which appear to extrude DNA loops at highly variable positions in the genome and hence cannot generate osmotic pressure from clearly defined loading sites [60].
Models of SMC complexes as DNA motors
Since it seems difficult to reconcile available data with passive models for SMC-dependent DNA loop extrusion that rely merely on diffusion or the action of external motors, it is worth considering the possibility that the role of the SMC ATPase cycle goes beyond loading SMC complexes onto DNA. The idea that an SMC complex might not require external motors to fold chromosomes into loops gained strong support from the finding that a mixture of purified condensin I, topoisomerase II, histones and three different histone chaperones was sufficient for the formation of chromatid-like structures in vitro, without the help of additional DNA motor proteins [61]. Direct visual proof for an SMC-powered translocation mechanism came from the observation of purified budding yeast condensin complexes, which actively moved along linear λ-DNA molecules that had been tethered at both ends in a microfluidic setup [30]. On these ‘DNA curtains’, condensin molecules translocated at a moderate velocity (~60 bp/s), yet in a highly processive manner, moving over distances of several kbp without turning around or falling off. Importantly, movement was dependent on the presence of hydrolyzable ATP and was abolished by mutations that prevent ATP binding by the SMC ATPase head domains [30].
Subsequent single-molecule experiments further demonstrated that the linear translocation observed for condensin can be converted into DNA loop extrusion. Addition of purified budding yeast condensin to λ-DNA molecules, which had been tethered under low tension to a passivated surface and then slightly stretched into an arc by buffer flow, allowed the real-time visualization of ATP-dependent formation and gradual expansion of DNA loops of several kbp in size [31]. Loop extrusion rates correlated with the tension in the DNA molecules and ranged from ~100 bp/s for DNA molecules that had been stretched to a degree comparable to the ones in the DNA curtains setup to ~1.5 kbp/s for DNA molecules that had been tethered with more slack. Condensin hence is a highly processive but weak DNA motor that stalls at forces exceeding ~1 pN (Table 2) [31]; a value that is in line with the stalling forces that have been measured for DNA compaction by the same condensin complexes in magnetic tweezers assays [62,63]. In vivo evidence for the notion that SMC protein complexes move actively on DNA was obtained in the bacterium Bacillus subtilis. In this species, a limited number of SMC protein complexes load onto DNA at a specific genomic site in the vicinity of the replication origin (parS) and then zip up the left and right ‘arms’ of the circular chromosome while they translocate along the chromosome [41]. Time-resolved Hi-C and ChIP-seq experiments suggested that these SMC complexes move at a constant rate of ~0.9 kbp/s – a speed that is of the same order of magnitude as the loop extrusion rates measured for yeast condensin [31]. The findings that mutant SMC complexes that are unable to hydrolyze ATP accumulated at parS sites [68] and that inhibition of transcription did not prevent the juxtaposition of the two chromosome arms [41] are difficult to explain by a passive diffusion models and further argue against the notion that the transcription machinery provides the motor force for movement.
Table 2. Translocation rates of SMC complexes and other motor proteins.
| Protein complex | Max. translocation speed (s-1) | Step size (ATP-1) | Stall force (pN) | References | |
|---|---|---|---|---|---|
| SMCs | B. subtilis SMC-ScpAB | * 770 bp | [41] | ||
| G. gallus Condensin | * 170 bp | [60] | |||
| S. cerevisiae Condensin | 1,500 bp | 750 bp | 1 | [31] | |
| Selected motors | E. coli FtsK translocase | 4,800 bp | 2 bp | >60 | [22,64] |
| E. coli EcoR124I translocase | 840 bp | 2 bp | >4 | [23,65] | |
| S. cerevisiae RSC translocase | 25 bp | 2 bp | 30 | [66] | |
| D. melanogaster kinesin | 1,000 nm | 8 nm | 6 | [67] | |
| S. cerevisiae dynein | variable | 8-32 nm | 1 | [67] |
estimates from Hi-C data
Comparison to other translocating motor proteins
To gain insights into the mechanistic basis for the DNA translocation mechanism of condensin, and possibly other SMC protein complexes, we start by considering the working principles of other well-characterized motor proteins, for example the cytoskeletal motor proteins myosin, kinesin or dynein [69]. On a superficial level, these cytoskeletal motors and SMC proteins share the architectural principle of a dimer of ATPase-containing domains that are linked via elongated coiled-coil stalks. This similarity had prompted the hypothesis that SMCs might function analogous to these motor proteins, long before their actual structure had been known [70,71]. Although the speeds by which these motor proteins move (Table 2) are in the range of the condensin-driven DNA loop extrusion rates measured in vitro (~100 nm/s [31]), they do burn significantly more ATP for their movement than SMC protein complexes seem to do. Bulk measurements suggest that a condensin complex can, even when stimulated by the addition of DNA, hydrolyze only very few (up to 2) molecules of ATP per second – an order of magnitude below the ATPase rates of cytoskeletal motor proteins (Table 1). In addition, major differences exist in the functional principles between cytoskeletal and SMC motors. For example, the directionality of movement of cytoskeletal motors is largely determined by the polarity of their substrates actin and tubulin. Since no such polarity is present in the phosphate backbone of the DNA double helix, it remains mysterious what would prevent condensin from constantly changing directions, which is clearly not the case [30]. Moreover, the ATPase heads of cytoskeletal motors complete their reaction cycles sequentially, which is fundamentally different to the concerted ATP binding and hydrolysis steps that are thought to take place at SMC head pairs, based on their homology to ABC transporter ATPases [72].
The movement of ATP-dependent nucleic acid translocases, like, for example, helicases, is generally based on catalytic core domains that are formed by a pair of RecA-like lobes [73]. Through sequential ATP binding and hydrolysis cycles of the two RecA lobes, which always contact the same sugar-phosphate backbone, these enzymes are able to overcome the directionality problem of double-stranded nucleic acids and processively translocate over lengths of several kbp [74]. Notably, the type I restriction enzyme EcoR124I has been demonstrated to employ such a motor activity to reel in DNA adjacent to its recognition site towards itself, essentially extruding a loop [75], before cleaving the DNA helix [65]. Such an ‘inchworming’-type mechanism does, however, only allow for translocation steps of a few bp per ATP hydrolysis cycle. This number is very different to the step sizes of hundreds of bp or more that have been estimated for purified condensin complexes [30,31,62,76]. The DNA-translocation mechanism of SMC complexes therefore must be fundamentally different from those of the well-characterized DNA translocases described above. To understand the translocation and ultimately the DNA loop extrusion mechanism of SMC protein complexes, it will be essential to know (1) how they interact with their chromatin substrates, (2) whether they function as individual complexes or larger molecular assemblies, and (3) how conformational changes in these complexes drive their movement on DNA.
How do SMC protein complexes interact with chromosomes?
Taking into account that the unifying feature of all SMC protein complexes is the presence of a dimer of long coiled-coil ATPases and a kleisin subunit that associate with related, yet heterogeneous sets of additional subunits, it seems reasonable to assume that the basic DNA translocase activity would need to be contained within the SMC–kleisin core. If this hypothesis were true, we would expect that ATPase-cycle-regulated DNA interactions within these subunits, coupled to possible large-scale conformational changes, serve as the motor for directed locomotion. The additional subunits most likely determine the differences in specificity and functionality between the diverse complexes, for example by controlling the speed of the core motor activity, regulating recruitment of SMC complexes to specific chromosomal loci, or anchoring them to chromatin during the movement steps. What is the current evidence that such a concept could represent reality?
Evidence exists that both, the SMC subunits (Box 1) as well as the non-SMC subunits (Box 2) can make stable contacts with the chromatin fiber, although it has remained unclear whether all, some, or none of these potential DNA-binding interfaces are used by SMC protein complexes for DNA loop extrusion. One intriguing possibility is that multiple low-affinity binding sites exist in SMC subunits, to which DNA gains only transient access in a manner that is controlled by the ATPase cycle (see below). Binding to such sites might be considerably augmented by topological DNA confinement. Such a scenario would explain the difficulties in pinpointing the exact locations of DNA binding sites within the SMC subunits.
Box 1. DNA binding sites in the SMC subunits.
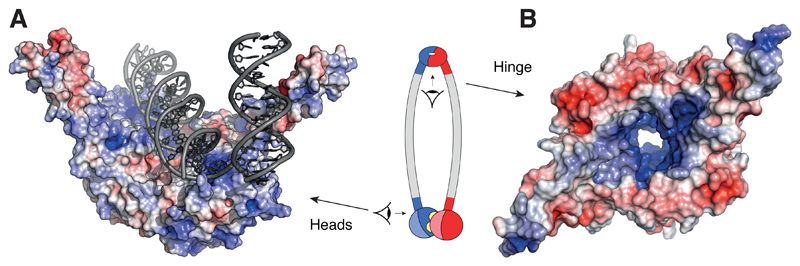
Recent co-crystal structures with double-stranded DNA of Rad50, an SMC-related coiled-coil ATPase with key roles in DNA damage repair, have revealed potential interaction sites either directly at the inner shallow, positively charged cavity of the ATP-dimerized head domains [77,78] or at the head-proximal coiled coils [79] (Figure 3A). The former binding surface is presumably only established upon ATPase head dimerization, which is consistent with the reduced in vitro DNA-binding activity of mutant Rad50 heads that are unable to dimerize in the presence of nucleotide. Remarkably, the crystal structures of engaged prokaryotic SMC [80] and MukB [28] ATPase head domains revealed similar positively charged cavities, which raises the intriguing possibility that the ATPase head domains of (at least prokaryotic) SMCs might possess an ATP-controlled DNA binding site.
A second putative DNA binding site might be located at or near the SMC hinge dimerization domains. In vitro, the purified hinge domains of prokaryotic SMCs [81,82], cohesin [83], condensin [84,85] and Smc5/6 complexes [86] bind to DNA substrates. All but one [83] study reported stronger binding of the SMC hinge dimers to single-stranded than to double-stranded DNA. The functional implications of this substrate preference are currently unclear, but single-stranded DNA binding has recently been linked to the topological loading of a second DNA strand into the cohesin ring for the establishment of sister chromatid cohesion at the DNA replication fork [87]. The reduction in DNA binding upon mutation of positively charged residues suggests that DNA binding might take place at positively charged patches at the inner surface between the stems of the coiled coils [82] or at other surfaces of the hinge structure [81,84,86]. Alternatively, it might be conceivable that the conserved positively charged channel within the torus-shaped hinge domain [88] is, at least in part, responsible for nucleic acid interaction [56,86]; although it would be too small to accommodate a DNA double helix and is absent in the structures of MukB hinge domains [89,90] (Figure 3B).
Box 2. DNA binding sites in the non-SMC subunits.
The Scc2–Scc4 cohesin loader has been shown to bind DNA via its Scc2Nipbl HEAT-repeat subunit in electrophoretic mobility shift assays [35] and the cohesin Scc3SA2 subunit was found to bind and diffuse along DNA until it encounters single-stranded or otherwise structured DNA regions in a microfluidic imaging setup [91]. The presence of an AT-hook motif in the human Scc3SA1 subunit has been implicated in the recruitment of cohesin to telomeric sequences [92,93] and binding of CTCF to the cohesin subunit Scc3SA2 has been suggested to maintain cohesin’s position at promoter and enhancer regions [94]. The Nse4–Nse1–Nse3 kleisin/WHD-domain subunits and the Brn1–Ycs4–Ycg1 kleisin/HEAT-repeat subunits of Smc5/6 or condensin, respectively, were shown to bind short double-stranded DNA templates in vitro [85,95]. A recent crystal structure of the budding yeast Brn1–Ycg1 heterodimer bound to a 18-bp DNA double helix revealed that both subunits contribute to the formation of a positively charged groove within the U-shaped architecture of the Ycg1 HEAT-repeat solenoid, which makes electrostatic contacts with the two DNA helix phosphate backbones [96]. The only moderate affinity to DNA is augmented by the confinement of the bound DNA within a peptide loop of the Brn1 kleisin subunit, which presumably allows the complex to slide along the double helix over short distances until it encounters an obstacle, such as a nucleosome.
The strongest support for a defined role of a DNA binding site exists for the one formed by the condensin Ycg1 HEAT-repeat and Brn1 kleisin subunits [96], which seemingly serves as a DNA anchor site that enables condensin to extrude DNA loops in an asymmetric manner [31]. It is tempting to speculate that the transient DNA-binding activity of the cohesin Scc3 HEAT-repeat subunit [91] could similarly be strengthened upon complex formation with the Scc1 kleisin to provide an equivalent DNA-anchor mechanism in cohesin complexes, which might also be the role of the DNA-binding activity of the Nse3 WHD subunit of Smc5/6 complexes [95].
Do SMC complexes act as individual complexes or higher-order assemblies?
Most theoretical models of SMC-driven loop extrusion are based on the idea that one loop-extruding unit consists of two tethered DNA-binding modules that translocate along the DNA in opposite directions [16,18,20,21]. A bi-directional movement would naturally result in the formation of a symmetric loop centered around its origin, until translocation of one of the two modules would stop, for example when it encounters a boundary element (Figure 2D). What is the evidence that SMC protein complexes function as dimers (or multimers) on chromosomal DNA?
The findings that certain mutations in one allele can genetically suppress the defects caused by different mutations in the second allele of the same cohesin subunit in budding yeast cells [56,97] and that cohesin complexes containing either the SA1 or SA2 HEAT-repeat subunit simultaneously occupy the same binding sites in human cultured cells [98] indeed indicate that two (or more) cohesin complexes might functionally interact on chromosomes. Biochemical evidence for a stable formation of cohesin dimers is, however, less clear, since only one studies indicated oligomerization of human cohesin complexes [99], while other studies did not detect the formation of stable cohesin dimers or multimers by co-immunoprecipitation from yeast or human cell extracts [48,100].
In contrast, there is a solid molecular basis for the existence of dimeric complexes of the Escherichia coli SMC-like complex MukBEF [101], which homodimerize via winged-helix and helical domains at the N termini of their MukF kleisin subunits [28,102]. Although it is conceivable that other SMC complexes could employ similar mechanisms for dimerization, the lack of a WHD at the N termini of other kleisin proteins [26] should caution rash generalization. Nevertheless, the observation that B. subtilis SMC complexes move along the left and right sides of ectopic parS loading sites with different velocities provides an additional argument that the loop-extruding unit needs to consist of two tethered complexes that can translocate at independent rates [41].
Whereas atomic force microscopy imaging, magnetic tweezers and protein cross-linking experiments have provided some indication of condensin multimerization [34,63,103], the discrete one-step bleaching events of fluorescently labeled yeast condensin complexes that localized to the base of emerging DNA loops in single-molecule imaging experiments [31] strongly argue that loop extrusion is achieved by individual condensin complexes. Furthermore, the observation that DNA was only reeled into the loop from one side in a strictly asymmetric manner is difficult to reconcile with models of dimeric motors that reel in DNA from both sides simultaneously as they move into opposite directions. It is instead compatible with a model in which a single condensin complex makes stable contact with the DNA and then uses this contact as an ‘anchor’ while moving along the double helix (Figure 2D). Such a model is also supported by the fact that mutations in the putative DNA anchoring site [96] cause the DNA loop to shift its position, similar to what had been observed in the experimental conditions of the DNA curtains assay [31]. Finally, a careful quantitation of the number of condensin molecules bound to human mitotic chromosomes revealed an average spacing of 60–90 kbp [104], which excellently matches the 60–80 kbp sizes of condensin-mediated loops calculated from Hi-C data of mitotic DT40 cells [60].
Which conformational changes can SMC protein complexes undergo?
SMC coiled-coil gating
The first high-resolution structural model of the entire B. subtilis SMC homodimer, generated from protein cross-linking experiments and crystal structures, revealed a rod-shaped conformation in which the two coiled coils were aligned over much of their lengths [105]. Remarkably, this conformation is incompatible with ATP-dependent head dimerization. Conversion into the canonical ATP-dimerized conformation requires substantial rotational (by 85°) and translational (by 10 Å) movements of the ATPase heads, which results into the tilting of the attached coiled coils into the wide-open configuration seen in crystal structures. SMCs might hence exist in two stable functional states (Figure 4A): In the ATP-bound state, the coiled coils are spread apart and forced into a bent conformation due to their connection at the hinge domains. Upon ATP hydrolysis, tension in the bent coiled coils would be released by straightening, which drives the head domains apart and subsequently allows the coiled coils to ‘zip up’ into a rod-shaped structure. Such a rod-shaped architecture had previously been observed in some EM and AFM images of prokaryotic [106] or eukaryotic [34,48,107–109] SMCs. It is furthermore consistent with cross-linking experiments, crystal structures and small angle x-ray scattering (SAXS) data of constructs that contain hinge and coiled-coil domains of prokaryotic SMCs, condensin or Smc5/6 [86,108], or with cross-linking data of full-length cohesin SMCs [103].
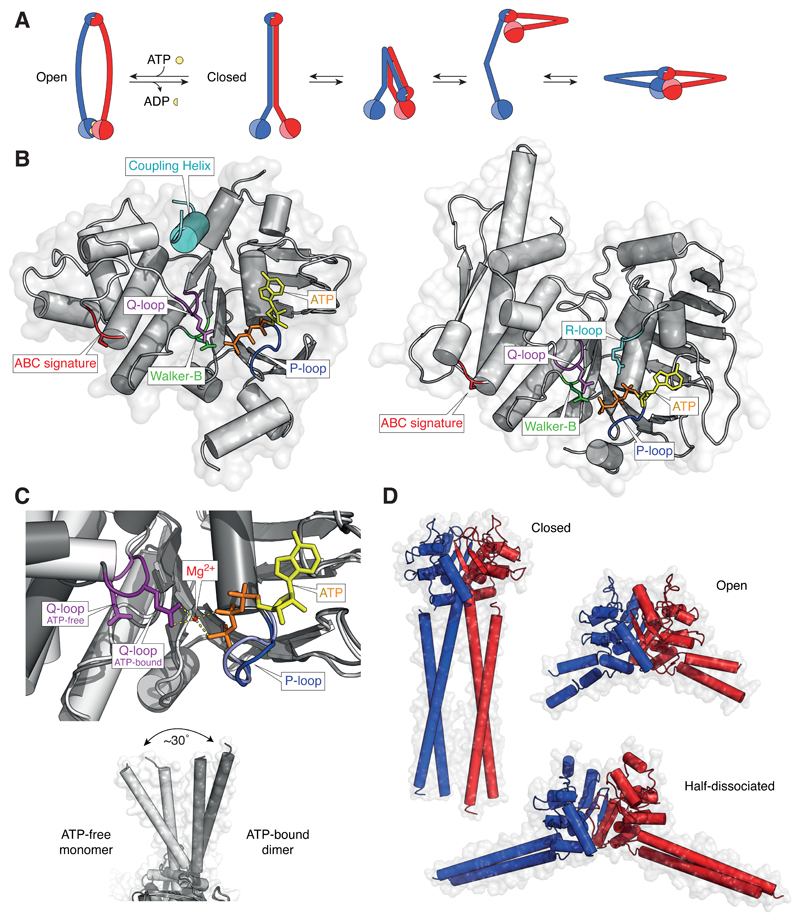
Figure 4. Conformational changes in SMC proteins.
(A) Models of ATP-bound open (ring-shaped) and ATP-free closed (rod-shaped) SMC dimers and conformational transitions of condensin SMC subunits observed in AFM images. (B) Structure models of the ATP-bound NBD of the Staphylococcus aureus Sav1866 ABC transporter ATPase (pdb: 2onj) and of the Geobacillus stearothermophilus SMC ATPase head domain (pdb: 5h68). (C) Close-up view of the Q-loop conformation in structures of the ATP-free monomeric B. subtilis SMC ATPase head domain (light grey; pdb: 5h67) and the ATP-dimerized B. subtilis SMC ATPase head domain (dark grey; pdb: 5xg3) and side-views of the coiled-coil orientations in both structures. (D) Structural models of the Pyrococcus furiosus SMC hinge domain in the closed coiled-coil conformation (pdb: 4rsj), of the T. maritima SMC hinge domain in the open coiled-coil conformation with both hinge interfaces associated (pdb: 1gxl) and of the G. stearothermophilus SMC hinge domain in the open coiled-coil conformation with one hinge interface dissociated (pdb: 5h69).
The idea of strictly stiff coiled coils that store mechanical energy similar to a loaded spring [105] is, however, inconsistent with the EM images of bent or kinked coiled coils that have been recorded for the majority of B. subtilis SMC and E. coli MukB dimers [26,106] or cohesin SMCs [48,107,110]. Even in the rod-shaped conformation, such kinks were frequently identified in the coiled coils of the fission yeast condensin SMCs [34]. It should be noted, however, that these imaging techniques require the deposition of molecules on a solid surface, followed by drying in vacuum, which makes it impossible to exclude that either straight, bent or kinked conformations might be artifacts. High-speed AFM time-lapse recordings of the condensin SMC heterodimer in solution showed a remarkably high degree of flexibility in the coiled-coil arms, which seemed to be able to dynamically explore the entire conformational space available to them, including an arrangement in which one or both of the ATPase head domains bind to the hinge by bending the coil arm by close to 180° (Figure 4A) [109]. Whereas SMCs can assume a stiff rod-shaped conformation, it seems that they can equally well switch to an open conformation that is more flexible than anticipated in the constrained ring hypothesis.
Conformational changes in the SMC ATPase heads
If there were transitions between stiff straight and relaxed circular coiled-coil conformations, then it would be reasonable to assume that these transitions would be driven by the ATPase head domains. How such large-scale changes might initiated, it is worth considering the knowledge gained from the study of the related ABC transmembrane transporters proteins, which catalyze the transfer of small molecule ligands across the lipid bilayer [111]. The nucleotide binding domains (NBDs) of ABC transporters are built from two subdomains. The first subdomain resembles the RecA-like domain found in DNA and RNA translocases and contains the Walker A (P-loop) and B motifs, which are essential for the coordination and hydrolysis of the ATP nucleotide (Figure 4B). The second, initially termed ‘helical’, subdomain contains the so-called ABC signature motif (LSGGQ) and the Q-loop motif (named after a strictly conserved glutamine residue). In all ABC transporters, two complete ATPase sites are only formed upon dimerization of two NBDs in an antiparallel orientation, which brings together Walker A and B motifs of one domain with the signature motif of the other domain [112]. A basic switch mechanism between the ATP-bound and the nucleotide-free conformation is thought to drive ligand translocation through the attached transmembrane domain (TMD) via transmission of the conformational energy through a structurally conserved coupling helix [112,113]. Since there is no evidence for the presence of a coupling helix in SMC proteins, the question arises how the energy from ATP hydrolysis is converted into movement by the SMCs.
The Q-loop might play an essential role in the mechanistic coupling between the two ATPase sites, since it is in a location to link the two subdomains and to position the nucleophilic water molecule for hydrolysis of the ATP phosphodiester bond [72]. Comparison of crystal structures of bacterial SMC ATPase head structures in monomeric nucleotide-free and dimeric ATP-bound states reveal not only a rotation of the helical subdomain and the attached coiled-coil by 30° upon ATP-induced NBD dimerization [114], but also a significant shift in the orientation of the Q-loop (Figure 4C). Movement of the Q-loop therefore has the potential to transmit conformational energy in SMC ATPases without the requirement of a coupling helix. Crystal structures also revealed that ATP binding restructures the flexible R-loop (named after a conserved arginine residue) in the RecA-like subdomain, which contacts the phosphate moieties of the bound nucleotide (Figure 4B) [80,114]. Notably, the R-loop arginine is conserved in SMC and Rad50 proteins, but not in ABC transporters. Although the conformational change in this loop is presumably not necessary to stabilize the bound nucleotide, it might provide a local platform to read out the ATPase state.
Conformational changes in the SMC hinge domains
Crystal structures of various SMC hinge domains suggest that they can assume at least two distinct conformational states (Figure 4D): A ‘closed’ architecture with closely juxtaposed coiled coils [108] or an ‘open’ architecture with outspread coiled coils [48,89,90,114]. Evidence for the existence of either conformation in solution came from SAXS or protein-protein cross-linking experiments [81,84,86,108]. Remarkably, the hinge torus was considerably tilted relative to the coiled-coil axis in the structures of the budding yeast and human condensin [108] and the fission yeast Smc5/6 hinge domain [86] with juxtaposed coiled coils, which suggests that, even when the SMC coiled coils associate into a rod-shaped conformation, their contact point with the hinge domain maintains a high degree of flexibility. If DNA bound to the inner hinge surface between the coiled coils [82], which would obviously only be possible in the open conformation, then an SMC ATPase-mediated switch from closed to open states as put forward in the coiled-coil gating model could regulate access to this binding site [108].
Whereas the hinge domains assembled into a (pseudo-)symmetric torus in most of the available crystal structures, one of the two interfaces between remained dissociated in some crystal forms (Figure 4D) [48,84,114]. If such a half-dissociated state also existed in solution, it would provide an elegant solution for double-stranded DNA helices to gain access to the highly conserved positively charged channel at the center of the hinge torus (Figure 3B). This DNA-binding site might be pivotal for the translocation mechanism during DNA loop extrusion (see below). Alternatively, once DNA has been bound, opening of the second hinge interface and simultaneous re-closure of the first interface would provide a mechanism for the transport of the DNA double helix into the SMC ring structure to yield topological entrapment [115]. These two options might explain why mutation of certain residues in the SMC hinge channel prevent cohesin from encircling chromosomal DNA and from generating sister chromatid cohesion without affecting its translocation on chromosomes [56,88]. If this scenario were true, it is conceivable that topological DNA entrapment resulted as a byproduct during the evolution of SMC proteins whose original function had been to translocate along nucleic acids. How a sequential opening and closing of the SMC hinge might be controlled is still a mystery, but one can imagine that the disengagement of the hinge interfaces might again be controlled by the ATPase head domains, either through a rotation of stiff coiled coils or a direct interaction of head and hinge domains enabled by flexible coiled coils (Figure 4A). However, any model that places emphasis on a defined function of the SMC hinge domains needs to explain how it is possible to replace the torus-shaped SMC hinge by the structurally distinct zinc-hook dimerization domain of Rad50 proteins without eliminating the requirement for SMC complex function during fast cell divisions in B. subtilis [5].
Figure 3. Possible DNA binding sites in SMC ATPase head and hinge domains.
(A) Structure model of DNA bound at the shallow surface of the dimerized Methanocaldococcus jannaschii Rad50 ATPase head domains (pdb: 5f3w) and of DNA bound to the coiled coil of one of the Thermotoga maritima Rad50 ATPase head domains (pdb: 4w9m) projected onto the M. jannaschii structure. (B) Structure model of the dimerized T. maritima SMC hinge domain (pdb: 1gxl). Colors represent positive (blue; +5 keT) or negative (red; –5 keT) electrostatic surface potential values.
Opening and closing the SMC–kleisin ring
Although different studies still disagree whether cohesin complexes open at their SMC hinge or SMC–kleisin interfaces for DNA entry into the ring architecture [56,116], it is generally accepted that DNA exit from cohesin rings is achieved either by the proteolytic cleavage of the kleisin subunit by separase or disengagement of the N terminus of the Scc1 kleisin subunit from the Smc3 ν-SMC coiled coil in a manner that depends on the Pds5 and Wapl subunits [117]. Evidence for the latter pathway comes from the findings that fusion or artificial dimerization of the Smc3 and the Scc1 N terminus prevented cohesin dissociation from chromosomes in yeast [118], fly [119] or human cultured cells [120], and that mutations at this interface interfered with stable binding of cohesin to chromosomes and sister chromatid cohesion in yeast [7] or human cells [110]. Since release of Scc1 from the Smc3 coiled coil by Pds5 could not be observed in the absence of the Smc1 κ-SMC ATPase head [121] and mutation of the SMC ATPases greatly reduced cohesin unloading from plasmid DNA by Pds5 and Wapl [116], it seems reasonable to assume that the ATPase cycle is required to drive the disengagement of Scc1 from the Smc3 coiled coil.
Mutation of the signature motif of the Smc1 ATPase head (but not of the Smc3 ATPase head) strongly reduced the release of cohesin from chromosomes during mitotic prophase [122], which suggests that the two ATPase sites of the cohesin SMCs make different contributions to ring opening. How this is achieved at a mechanistic level is still unknown. One hypothesis suggests that Pds5, Wapl and Scc3 generate a stiff mechanical linker when bound to the Scc1 kleisin subunit [123] that could transduce conformational energy of ATP-dependent SMC head dimerization to the disengagement of Scc1 from Smc3 [116]. It is similarly unclear how ring opening and closing might be coordinated with the transport of DNA out of cohesin rings. Since Pds5, Wapl and Scc3 have some affinity to DNA in vitro, it is possible that these proteins guide the movement of the DNA double helix [123]. Notably, the DNA binding site observed in one crystal structure of Rad50 [79] coincides with the exact same position in the cohesin Smc3 coiled coil where the Scc1 N terminus binds [7], which suggests a possible mechanism to regulate access to a binding site inside the cohesin ring.
Less is known about the possible entry or exit gates of other SMC complexes. It is, however, clear that topological loading of prokaryotic SMC complexes depends on their ATPase activity [12], as it is the case for cohesin [59]. In E. coli, overexpression of the MukF kleisin N terminus results in an efficient displacement of MukBEF from the bacterial chromosome [8], which suggests that disengagement of the kleisin–SMC interface might similarly serve as DNA gate in prokaryotic SMC complexes. Interestingly, binding of the WHD subunit ScpB to the ScpA kleisin subunit displaces it from the B. subtilis SMC neck coiled coil [114] and binding of the WHD subunit MukE to the central region of the MukF kleisin reduces E. coli MukB ATPase activity [8]. The role of the additional WHD subunits might hence be in the control of kleisin–SMC interactions.
Towards a unified model for SMC complexes as DNA-loop-extruding machines
Taking into account the current knowledge of the molecular properties of the different SMC protein complexes, their interaction with DNA and chromatin, as well as the conformational changes these complexes can undergo, is it possible to combine this information into a generally valid model for their anticipated DNA-loop-extrusion activity? It seems that first, a number of challenges need to be overcome before this question can be answered with a definite ‘yes’.
For example, the data obtained from Hi-C experiments hint at a symmetric loop extrusion process by bacterial SMC complexes in vivo [124], whereas single-molecule studies in vitro clearly demonstrated asymmetric loop extrusion by condensin [31]. One caveat of asymmetric loop extrusion is that the DNA region between two complexes that are oriented in a back-to-back configuration would not get folded into a loop, leaving gaps in the compacted chromosome (Figure 2E). Such gaps might, however, not be problematic if condensin complexes frequently dissociated and re-bound in a random orientation – a notion that is compatible with the rapid turnover on chromosomes of most human condensin I complexes within a few minutes [104,125]. A second caveat of anchoring an SMC complex to specific sites in the genome from where it reels in DNA unidirectionally is that this should generate ‘stripes’ in Hi-C contact maps. Stripes were, however, only observed at CTCF-binding sites in activated B cells that coincide with binding sites of the Scc2Nipbl cohesin loader (Figure 2E) [55], but not as a general feature. However, if condensin loaded onto DNA and started extruding loops from random sites in the genome, the absence of stripes in population-averaged Hi-C maps of mitotic chromosomes could be explained [60,126]. Asymmetric could be easily converted into symmetric loop extruders if a single complex repeatedly changed the positions of its anchor and motor sites, or if two complexes always dimerized facing away from each other (Figure 2D), as predicted for cohesin by the handcuff model [97,99]. The latter option would predict, however, that these complexes should still remain bound to their original binding site, which, at least in the case of cohesin complexes that translocate from their chromosomal loading sites [59] or bacterial SMC complexes that move from the replication origin along both arms of the circular bacterial chromosome [41], does not seem to be the case.
Despite these apparent disparities, we will conclude this review by discussing three distinct models for DNA-loop extrusion by SMC protein complexes that have been suggested previously in these or slightly modified versions. All three models are based on ATP-hydrolysis mediated changes in the geometry of the SMC coiled-coil arms, which provide the potential means to overcome the challenges of translocation on chromatin fibers, where nucleosomes and other DNA-bound proteins pose substantial obstacles.
The sequential walking model
In analogy to the translocation mechanism used by cytoskeletal motor proteins (see above), it has been suggested that the two SMC ATPase head domains sequentially bind DNA (Figure 5A): While one head attaches the complex stably to DNA, the other ATPase head is able to reach out to contact another binding site on the same DNA molecule at a distance that is limited either by the combined lengths of the two coiled-coil and hinge domains [30] or by the length of the kleisin linker that connects the two head domains, assuming that the SMC–kleisin ring architecture remains intact [127]. DNA binding of the second ATPase head would then trigger a conformational change that releases the first head to start the next step of the translocation cycle.
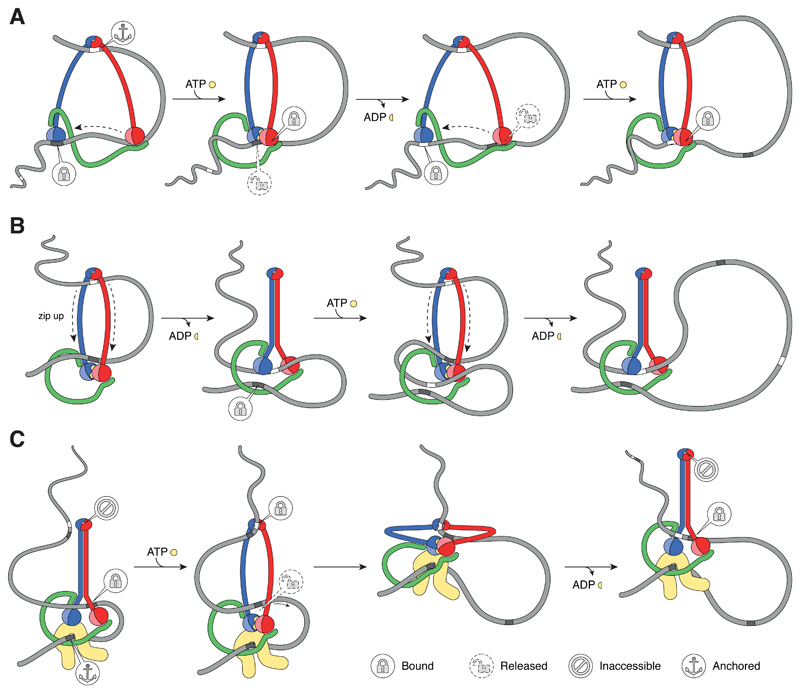
Figure 5. Different models of active SMC-driven Loop Extrusion mechanism.
(A) Sequential walking model. (B) DNA pumping model. (C) Extended scrunching model. See main text for details. Note that the sequential walking model makes no assumptions about the SMC coiled coil conformations, whereas the pumping model assumes stiff coiled coils that are under tension when bent open. The extended scrunching models postulates that SMC coiled coils alternate between stiff and relaxed states.
Such a mechanism could easily account for the 16-nm step sizes that have been calculated for condensin’s movement along DNA [30]. Since translocation does not need to follow the path of the DNA in this model, a single step might cover considerably larger base-pair distances on relaxed DNA [31] or chromatinized substrates. Whereas the mechanical force for translocation could, at least in principle, derive from alternating between opening and closing of the SMC coiled coils [105], the sequential walking model neglects the bi-directionality of the DNA double helix and hence fails to explain how condensin is able to translocate over tens of kbp without changing direction, or how condensin could be prevented from switching to another DNA molecule that comes into bridging distance. Furthermore, the lack of evidence for alternated binding of the monomeric SMC ATPase head domains to DNA make it difficult to reconcile such a model with the available biochemical data.
The DNA pumping model
A very different model has its foundation in the discovery of closely juxtaposed SMC coiled coils in prokaryotic SMC complexes [68,105]. This model relies on pseudo-topological DNA loop capture between open SMC coiled-coils arm (Figure 5B). Following ATP hydrolysis, the coiled coils zip up, starting from the hinge domain, and the base of the entrapped DNA loop is pushed towards and past the now disengaged ATPase heads. Upon exchange of ADP by ATP, the heads re-engage, the SMC coiled coils return to the open conformation and the base of the DNA loop is now entrapped within a second sub-compartment that is confined by the engaged ATPase head domains and the kleisin subunit. In the next cycle, another DNA loop is captured between the coiled coils and pushed into the second sub-compartment, where it merges with the already entrapped DNA loop into an ever-larger loop.
This elegant model raises two key questions. First, how is a pre-formed DNA loop threaded into the lumen between the SMC coiled-coils? It is conceivable that such DNA loops could be generated through ParB-dependent bridging of parS sequences at the SMC loading sites, or through bending of the DNA double helix by other abundant nucleoid-associated proteins [128]. Loops could also be generated through DNA supercoiling, and contacts of the DNA with the SMC hinge and head domains might assist in loop capture. Second, how is directionality of loop extrusion ensured; i.e. what prevents a DNA loop from slipping out of the second sub-compartment when the ATPase heads disengage? It is likely that this would require DNA anchor sites in this sub-compartment, but whether such sites exist has remained unknown.
The extended scrunching model
Initially suggested as an alternative to the sequential walking hypothesis for condensin, the scrunching model anticipates the alternating binding and release of two distinct DNA binding sites, one located at the SMC ATPase head domains and the other located at the hinge domain, for translocation along the chromatin fiber [30]. We here present an extended version of this model, which takes into account the biochemical properties gained from the study of other SMC protein complexes (Figure 5C). We assume that DNA binds to the disengaged ATPase head domains of SMC dimers with juxtaposed coiled coils. Upon ATP-mediated SMC head dimerization, the coiled coils disengage, resulting in a conformational change that provides access to a DNA binding site at the SMC hinge and a concomitant release of the DNA at the head domains. Due to the high flexibility of the disengaged coiled coils, the SMC complex can now (passively) fold into a conformation where the engaged head domains contact the hinge domains. DNA re-binding of the heads and release from the hinge resets the translocation cycle. In this model, the ATPase cycle drives the conformational transitions between coiled-coil opening and closure, analogous to the model put forward for prokaryotic SMCs [68], which are intrinsically coupled to providing DNA access to either of the two DNA binding sites. Stiffening of the SMC coiled-coil arms provides the power stroke that places the hinge at a ~50-nm distance to the head domains. In this model, loop extrusion would be achieved through DNA attachment to an additional, constitutive anchor site, like the one created by the Brn1–Ycg1 subunits of the condensin complex [31,96], which ensures that condensin drags the DNA helix into a loop while it translocates.
Although highly speculative, this model explains several of the features of SMC complexes that we have outlined before: DNA binding at the hinge might take place within the conserved positively charged channel of the torus-shaped domain [88] and alternating access to the channel might be controlled by transitions between the fully associated and half-dissociated conformations observed in crystal structures (Figure 4D). Notably, half-dissociated conformations have only been observed for SMC constructs with open coiled-coils [48,84,114], whereas all crystal structures with juxtaposed coiled-coils displayed fully associated confirmations [108]. This correlation raises the possibility that coiled-coil opening might promote hinge interface disengagement. Control of the DNA binding site at the ATPase head domains would presumably be directly coupled to the head engagement state. For example, the formation of a positively charged DNA-binding groove at the interface between engaged head domains, such as the one observed for Rad50 (Box 1 and Figure 3A), would depend on ATPase head dimerization. Whether such binding sites exist in the SMC hinge and head domains is, however, still unknown. As in the sequential walking model, the power stroke movement does not need to follow the path of the DNA and thereby enables condensin to take steps that are significantly larger than the ~150 bp that would theoretically be possible for a motor that moves 50 nm along a B-type DNA helix. It would also provide the SMC complex with the ability to ‘step over’ nucleosomes and thereby even cover larger base-pair distances. Finally, this model doesn’t rely on pre-formed DNA loops, but instead allows SMC complexes to initiate loop formation de novo.
However, major caveats also remain for the scrunching model. For example, what would prevent condensin from taking a step backwards instead of always stepping into one direction during the power stroke movement? How would the hinge and head domains know that they have come into proximity before handing over DNA binding from the hinge to the head domains? Whereas it is conceivable that such a switch in binding sites could be triggered by a transient association between head and hinge domains, evidence for a direct protein-protein interaction between SMC head and hinge domains is so far only available for cohesin, and even this evidence is limited [35,129]. Finally, the key function of the DNA binding site at the SMC hinge domain put forward by this model makes it difficult to explain how it can be replaced by the Rad50 zinc hook in the B. subtilis SMC complex [5].
Outlook
For two decades, the image of the cohesin complex stably connecting sister chromatids has shaped the perception of SMC protein complexes as static chromatid linker molecules. Yet, a passive structural mode of action has difficulties to explain the emerging evidence for cohesin’s role in transcriptional regulation, or the functions of other SMC complexes. The entrapment of two sister DNA molecules now appears to be a specialized aspect of the family of SMC protein complexes, whose unifying principle most likely can be found in their ability to extrude large DNA loops. How these complexes move along chromatinized DNA and thereby fold genomes into complex structures, like those observed for mitotic chromosomes, has become one of the central questions of chromosome biology.
In this review, we have discussed mechanistic models for the action of SMC protein complexes as DNA-loop-extruding enzymes. Despite their differences, the models underscore the universal requirement for transient DNA binding sites for any processive motor action on DNA helices. Characterization of such sites must take into account the topological restrictions laid out by the unique architecture of SMC–kleisin complexes and the conformational changes that are most likely instructed from distal sites within the complex and controlled by the ATP hydrolysis cycle. Real-time observation of loop extrusion on naked DNA by single condensin molecules has opened up new avenues to systematically test the assumptions made in polymer models for loop extrusion, including the behavior of assemblies that simultaneously act on the same DNA template. What happens when loop-extruding complexes encounter obstacles, such as other loop-extruding enzymes, when they bump into the DNA replication machinery or transcribing RNA polymerases, or when the topology of their DNA substrate changes are exciting questions that will probably be answered soon.
Taking into account that SMC complexes originated in prokaryotes, which lack canonical histones, it is not surprising that many aspects of even eukaryotic SMC complex function can be recapitulated on naked DNA. Yet, the ability to function on chromatinized DNA is one important feature that sets SMC protein complexes apart from other known DNA motor proteins. Remarkably, loop extrusion by condensin – and possibly other SMC complexes – seems to operate in steps that are two orders of magnitude larger than those of conventional DNA motors. Such large steps can be explained if the translocation movement would not need to follow the linear path of the DNA double helix, where histones and a multitude of other proteins might impede processive tracking of the sugar-phosphate backbone. If this were the case, a key prediction of most of the models outlined above is that reducing the length of the SMC coiled coils – a feat that has already been achieved for bacterial SMCs [5] – should shorten step sizes during DNA translocation. Regardless of the outcome of such experiments, it is already clear that whichever mechanism this class of protein complexes uses to form looped chromosomes, it must be something out of the ordinary that has never been seen before.
Supplementary Material
Acknowledgements
We thank all members of the Haering lab and colleagues for stimulating discussions. Work in the Haering lab is supported by the European Molecular Biology Laboratory (EMBL) and the European Research Council (ERC, grant number 681365).
References
- [1].Cobbe N, Heck MMS. The evolution of ATPase activity in SMC proteins. Proteins. 2006;63:685–96. doi: 10.1002/prot.20795. [DOI] [PubMed] [Google Scholar]
- [2].Gruber S. MukBEF on the march: taking over chromosome organization in bacteria? Mol Microbiol. 2011;81:855–859. doi: 10.1111/j.1365-2958.2011.07764.x. [DOI] [PubMed] [Google Scholar]
- [3].Uhlmann F. SMC complexes: from DNA to chromosomes. Nat Rev Mol Cell Biol. 2016;17:399–412. doi: 10.1038/nrm.2016.30. [DOI] [PubMed] [Google Scholar]
- [4].van Ruiten MS, Rowland BD. SMC Complexes: Universal DNA Looping Machines with Distinct Regulators. Trends Genet. 2018;34:477–487. doi: 10.1016/j.tig.2018.03.003. [DOI] [PubMed] [Google Scholar]
- [5].Bürmann F, Basfeld A, Vazquez Nunez R, Diebold-Durand M-L, Wilhelm L, Gruber S. Tuned SMC Arms Drive Chromosomal Loading of Prokaryotic Condensin. Mol Cell. 2017;65:861–869. doi: 10.1016/j.molcel.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bürmann F, Shin H-C, Basquin J, Soh Y-M, Giménez-Oya V, Kim Y-G, et al. An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat Struct Mol Biol. 2013;20:371–379. doi: 10.1038/nsmb.2488. [DOI] [PubMed] [Google Scholar]
- [7].Gligoris TG, Scheinost JC, Bürmann F, Petela N, Chan K-L, Uluocak P, et al. Closing the cohesin ring: structure and function of its Smc3-kleisin interface. Science. 2014;346:963–967. doi: 10.1126/science.1256917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zawadzka K, Zawadzki P, Baker R, Rajasekar KV, Wagner F, Sherratt DJ, et al. MukB ATPases are regulated independently by the N- and C-terminal domains of MukF kleisin. Elife. 2018;7:e31522. doi: 10.7554/eLife.31522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Löwe J. Structure and stability of cohesin's Smc1-kleisin interaction. Mol Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- [10].Haering CH, Farcas A-M, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- [11].Cuylen S, Metz J, Haering CH. Condensin structures chromosomal DNA through topological links. Nat Struct Mol Biol. 2011;18:894–901. doi: 10.1038/nsmb.2087. [DOI] [PubMed] [Google Scholar]
- [12].Wilhelm L, Bürmann F, Minnen A, Shin H-C, Toseland CP, Oh B-H, et al. SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. Elife. 2015;4 doi: 10.7554/eLife.06659. 11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Palecek JJ, Gruber S. Kite Proteins: a Superfamily of SMC/Kleisin Partners Conserved Across Bacteria, Archaea, and Eukaryotes. Structure. 2015;23:2183–2190. doi: 10.1016/j.str.2015.10.004. [DOI] [PubMed] [Google Scholar]
- [14].Wells JN, Gligoris TG, Nasmyth KA, Marsh JA. Evolution of condensin and cohesin complexes driven by replacement of Kite by Hawk proteins. Curr Biol. 2017;27:R17–8. doi: 10.1016/j.cub.2016.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Haering CH, Gruber S. SnapShot: SMC Protein Complexes Part I. Cell. 2016;164:326–326.e1. doi: 10.1016/j.cell.2015.12.026. [DOI] [PubMed] [Google Scholar]
- [16].Alipour E, Marko JF. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–11212. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- [18].Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA. 2015;112:E6456–65. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Goloborodko A, Marko JF, Mirny LA. Chromosome Compaction by Active Loop Extrusion. Biophys J. 2016;110:2162–2168. doi: 10.1016/j.bpj.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Goloborodko A, Imakaev MV, Marko JF, Mirny L. Compaction and segregation of sister chromatids via active loop extrusion. Elife. 2016;5:e14864. doi: 10.7554/eLife.14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Graham JE, Sherratt DJ, Szczelkun MD. Sequence-specific assembly of FtsK hexamers establishes directional translocation on DNA. Proc Natl Acad Sci USA. 2010;107:20263–20268. doi: 10.1073/pnas.1007518107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Seidel R, Bloom JGP, Dekker C, Szczelkun MD. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. Embo J. 2008;27:1388–1398. doi: 10.1038/emboj.2008.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hirano M, Anderson DE, Erickson HP, Hirano T. Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. Embo J. 2001;20:3238–3250. doi: 10.1093/emboj/20.12.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hirano M, Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. Embo J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kamada K, Miyata M, Hirano T. Molecular Basis of SMC ATPase Activation: Role of Internal Structural Changes of the Regulatory Subcomplex ScpAB. Structure. 2013;21:581–594. doi: 10.1016/j.str.2013.02.016. [DOI] [PubMed] [Google Scholar]
- [27].Petrushenko ZM, Lai C-H, Rai R, Rybenkov VV. DNA reshaping by MukB. Right-handed knotting, left-handed supercoiling. J Biol Chem. 2006;281:4606–4615. doi: 10.1074/jbc.M504754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Woo J-S, Lim J-H, Shin H-C, Suh M-K, Ku B, Lee K-H, et al. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell. 2009;136:85–96. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- [29].Stray JE, Lindsley JE. Biochemical analysis of the yeast condensin Smc2/4 complex: an ATPase that promotes knotting of circular DNA. J Biol Chem. 2003;278:26238–26248. doi: 10.1074/jbc.M302699200. [DOI] [PubMed] [Google Scholar]
- [30].Terakawa T, Bisht S, Eeftens JM, Dekker C, Haering CH, Greene EC. The condensin complex is a mechanochemical motor that translocates along DNA. Science. 2017;358:672–676. doi: 10.1126/science.aan6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, et al. Real-time imaging of DNA loop extrusion by condensin. Science. 2018;360:102–105. doi: 10.1126/science.aar7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kimura K, Hirano T. Dual roles of the 11S regulatory subcomplex in condensin functions. Proc Natl Acad Sci USA. 2000;97:11972–11977. doi: 10.1073/pnas.220326097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kimura K, Hirano T. ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- [34].Yoshimura SH, Hizume K, Murakami A, Sutani T, Takeyasu K, Yanagida M. Condensin architecture and interaction with DNA: regulatory non-SMC subunits bind to the head of SMC heterodimer. Current Biology. 2002;12:508–513. doi: 10.1016/s0960-9822(02)00719-4. [DOI] [PubMed] [Google Scholar]
- [35].Murayama Y, Uhlmann F. Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature. 2014;505:367–371. doi: 10.1038/nature12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Borths EL, Poolman B, Hvorup RN, Locher KP, Rees DC. In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporter for vitamin B12 uptake. Biochemistry. 2005;44:16301–16309. doi: 10.1021/bi0513103. [DOI] [PubMed] [Google Scholar]
- [37].Liu CE, Liu PQ, Ames GF. Characterization of the adenosine triphosphatase activity of the periplasmic histidine permease, a traffic ATPase (ABC transporter) J Biol Chem. 1997;272:21883–21891. doi: 10.1074/jbc.272.35.21883. [DOI] [PubMed] [Google Scholar]
- [38].Hohl M, Briand C, Grütter MG, Seeger MA. Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat Struct Mol Biol. 2012;19:395–402. doi: 10.1038/nsmb.2267. [DOI] [PubMed] [Google Scholar]
- [39].Farrell CM, Mackey AT, Klumpp LM, Gilbert SP. The role of ATP hydrolysis for kinesin processivity. J Biol Chem. 2002;277:17079–17087. doi: 10.1074/jbc.M108793200. [DOI] [PubMed] [Google Scholar]
- [40].Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat Struct Mol Biol. 2012;19:492–497–S1. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang X, Brandão HB, Le TBK, Laub MT, Rudner DZ. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science. 2017;355:524–527. doi: 10.1126/science.aai8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Glynn EF, Megee PC, Yu H-G, Mistrot C, Unal E, Koshland DE, et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–95. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- [45].Ocampo-Hafalla M, Muñoz S, Samora CP, Uhlmann F. Evidence for cohesin sliding along budding yeast chromosomes. Open Biology. 2016;6 doi: 10.1098/rsob.150178. 150178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Borrie MS, Campor JS, Joshi H, Gartenberg MR. Binding, sliding, and function of cohesin during transcriptional activation. Proc Natl Acad Sci USA. 2017;114:E1062–1071. doi: 10.1073/pnas.1617309114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Busslinger GA, Stocsits RR, van der Lelij P, Axelsson E, Tedeschi A, Galjart N, et al. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature. 2017;544:503–507. doi: 10.1038/nature22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Haering CH, Löwe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- [49].Ivanov D, Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. doi: 10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- [50].Davidson IF, Goetz D, Zaczek MP, Molodtsov MI, Huis In 't Veld PJ, Weissmann F, et al. Rapid movement and transcriptional re-localization of human cohesin on DNA. Embo J. 2016;35:2671–2685. doi: 10.15252/embj.201695402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Stigler J, Çamdere GÖ, Koshland DE, Greene EC. Single-Molecule Imaging Reveals a Collapsed Conformational State for DNA-Bound Cohesin. Cell Rep. 2016;15:988–998. doi: 10.1016/j.celrep.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kanke M, Tahara E, Huis In't Veld PJ, Nishiyama T. Cohesin acetylation and Wapl-Pds5 oppositely regulate translocation of cohesin along DNA. Embo J. 2016;35:2686–2698. doi: 10.15252/embj.201695756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov SV, Abdennur N, et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544:110–114. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM. Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell. 2017;169:216–219. doi: 10.1016/j.cell.2017.03.024. [DOI] [PubMed] [Google Scholar]
- [55].Vian L, Pekowska A, Rao SSP, Kieffer-Kwon K-R, Jung S, Baranello L, et al. The Energetics and Physiological Impact of Cohesin Extrusion. Cell. 2018;73:1165–1178.e20. doi: 10.1016/j.cell.2018.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Srinivasan M, Scheinost JC, Petela NJ, Gligoris TG, Wissler M, Ogushi S, et al. The Cohesin Ring Uses Its Hinge to Organize DNA Using Non-topological as well as Topological Mechanisms. Cell. 2018;173:1508–1518. doi: 10.1016/j.cell.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Brackley CA, Johnson J, Michieletto D, Morozov AN, Nicodemi M, Cook PR, et al. Extrusion without a motor: a new take on the loop extrusion model of genome organization. Nucleus. 2018;9:95–103. doi: 10.1080/19491034.2017.1421825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yamamoto T, Schiessel H. Osmotic mechanism of the loop extrusion process. Phys Rev E. 2017;96 doi: 10.1103/PhysRevE.96.030402. 030402. [DOI] [PubMed] [Google Scholar]
- [59].Hu B, Itoh T, Mishra A, Katoh Y, Chan K-L, Upcher W, et al. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr Biol. 2011;21:12–24. doi: 10.1016/j.cub.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Nuebler J, et al. A pathway for mitotic chromosome formation. Science. 2018;359:eaao6135. doi: 10.1126/science.aao6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shintomi K, Takahashi TS, Hirano T. Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat Cell Biol. 2015;17:1014–1023. doi: 10.1038/ncb3187. [DOI] [PubMed] [Google Scholar]
- [62].Eeftens JM, Bisht S, Kerssemakers J, Kschonsak M, Haering CH, Dekker C. Real-time detection of condensin-driven DNA compaction reveals a multistep binding mechanism. Embo J. 2017;36:3448–3457. doi: 10.15252/embj.201797596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Keenholtz RA, Dhanaraman T, Palou R, Yu J, D'Amours D, Marko JF. Oligomerization and ATP stimulate condensin-mediated DNA compaction. Sci Rep. 2017;7 doi: 10.1038/s41598-017-14701-5. 14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pease PJ, Levy O, Cost GJ, Gore J, Ptacin JL, Sherratt D, et al. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307:586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- [65].Seidel R, van Noort J, van der Scheer C, Bloom JGP, Dekker NH, Dutta CF, et al. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nat Struct Mol Biol. 2004;11:838–43. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- [66].Sirinakis G, Clapier CR, Gao Y, Viswanathan R, Cairns BR, Zhang Y. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. Embo J. 2011;30:2364–2372. doi: 10.1038/emboj.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Sweeney HL, Holzbaur ELF. Motor Proteins. Cold Spring Harb Perspect Biol. 2018;10:a021931. doi: 10.1101/cshperspect.a021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Minnen A, Bürmann F, Wilhelm L, Anchimiuk A, Diebold-Durand M-L, Gruber S. Control of Smc Coiled Coil Architecture by the ATPase Heads Facilitates Targeting to Chromosomal ParB/parS and Release onto Flanking DNA. Cell Rep. 2016;14:2003–2016. doi: 10.1016/j.celrep.2016.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kato Y, Miyakawa T, Tanokura M. Overview of the mechanism of cytoskeletal motors based on structure. Biophys Rev. 2018;10:571–581. doi: 10.1007/s12551-017-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Guacci V, Yamamoto A, Strunnikov A, Kingsbury J, Hogan E, Meluh P, et al. Structure and function of chromosomes in mitosis of budding yeast. Cold Spring Harb Symp Quant Biol. 1993;58:677–685. doi: 10.1101/sqb.1993.058.01.075. [DOI] [PubMed] [Google Scholar]
- [71].Peterson CL. The SMC family: novel motor proteins for chromosome condensation? Cell. 1994;79:389–392. doi: 10.1016/0092-8674(94)90247-x. [DOI] [PubMed] [Google Scholar]
- [72].Hopfner K-P, Tainer JA. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr Opin Struct Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- [73].Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- [74].Clapier CR, Iwasa J, Cairns BR, Peterson CL. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 2017;18:407–422. doi: 10.1038/nrm.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Riggs AD. DNA methylation and late replication probably aid cell memory, and type I DNA reeling could aid chromosome folding and enhancer function. Philos Trans R Soc Lond, B, Biol Sci. 1990;326:285–297. doi: 10.1098/rstb.1990.0012. [DOI] [PubMed] [Google Scholar]
- [76].Strick TR, Kawaguchi T, Hirano T. Real-time detection of single-molecule DNA compaction by condensin I. 2004;14:874–880. doi: 10.1016/j.cub.2004.04.038. [DOI] [PubMed] [Google Scholar]
- [77].Liu Y, Sung S, Kim Y, Li F, Gwon G, Jo A, et al. ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. Embo J. 2016;35:743–758. doi: 10.15252/embj.201592462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Seifert FU, Lammens K, Stoehr G, Kessler B, Hopfner K-P. Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. Embo J. 2016;35:759–772. doi: 10.15252/embj.201592934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Rojowska A, Lammens K, Seifert FU, Direnberger C, Feldmann H, Hopfner K-P. Structure of the Rad50 DNA double-strand break repair protein in complex with DNA. Embo J. 2014;33:2847–2859. doi: 10.15252/embj.201488889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lammens A, Schele A, Hopfner K-P. Structural Biochemistry of ATP-Driven Dimerization and DNA-Stimulated Activation of SMC ATPases. Current Biology. 2004;14:1778–1782. doi: 10.1016/j.cub.2004.09.044. [DOI] [PubMed] [Google Scholar]
- [81].Griese JJ, Hopfner K-P. Structure and DNA-binding activity of the Pyrococcus furiosus SMC protein hinge domain. Proteins. 2011;79:558–568. doi: 10.1002/prot.22903. [DOI] [PubMed] [Google Scholar]
- [82].Hirano M, Hirano T. Opening closed arms: long-distance activation of SMC ATPase by hinge-DNA interactions. Mol Cell. 2006;21:175–86. doi: 10.1016/j.molcel.2005.11.026. [DOI] [PubMed] [Google Scholar]
- [83].Chiu A, Revenkova E, Jessberger R. DNA interaction and dimerization of eukaryotic SMC hinge domains. J Biol Chem. 2004;279:26233–26242. doi: 10.1074/jbc.M402439200. [DOI] [PubMed] [Google Scholar]
- [84].Griese JJ, Witte G, Hopfner K-P. Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic Acids Res. 2010;38:3454–3465. doi: 10.1093/nar/gkq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Piazza I, Rutkowska A, Ori A, Walczak M, Metz J, Pelechano V, et al. Association of condensin with chromosomes depends on DNA binding by its HEAT-repeat subunits. Nat Struct Mol Biol. 2014;21:560–568. doi: 10.1038/nsmb.2831. [DOI] [PubMed] [Google Scholar]
- [86].Alt A, Dang HQ, Wells OS, Polo LM, Smith MA, McGregor GA, et al. Specialized interfaces of Smc5/6 control hinge stability and DNA association. Nat Commun. 2017;8 doi: 10.1038/ncomms14011. 14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Murayama Y, Samora CP, Kurokawa Y, Iwasaki H, Uhlmann F. Establishment of DNA-DNA Interactions by the Cohesin Ring. Cell. 2018;172:465–477.e15. doi: 10.1016/j.cell.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kurze A, Michie KA, Dixon SE, Mishra A, Itoh T, Khalid S, et al. A positively charged channel within the Smc1/Smc3 hinge required for sister chromatid cohesion. Embo J. 2011;30:364–378. doi: 10.1038/emboj.2010.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Li Y, Schoeffler AJ, Berger JM, Oakley MG. The crystal structure of the hinge domain of the Escherichia coli structural maintenance of chromosomes protein MukB. J Mol Biol. 2010;395:11–19. doi: 10.1016/j.jmb.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ku B, Lim J-H, Shin H-C, Shin S-Y, Oh B-H. Crystal structure of the MukB hinge domain with coiled-coil stretches and its functional implications. Proteins. 2010;78:1483–1490. doi: 10.1002/prot.22664. [DOI] [PubMed] [Google Scholar]
- [91].Countryman P, Fan Y, Gorthi A, Pan H, Strickland J, Kaur P, et al. Cohesin SA2 is a sequence independent DNA binding protein that recognizes DNA replication and repair intermediates. J Biol Chem. 2017;293:1054–1069. doi: 10.1074/jbc.M117.806406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bisht KK, Daniloski Z, Smith S. SA1 binds directly to DNA through its unique AT-hook to promote sister chromatid cohesion at telomeres. J Cell Sci. 2013;126:3493–3503. doi: 10.1242/jcs.130872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lin J, Countryman P, Chen H, Pan H, Fan Y, Jiang Y, et al. Functional interplay between SA1 and TRF1 in telomeric DNA binding and DNA-DNA pairing. Nucleic Acids Res. 2016;44:6363–6376. doi: 10.1093/nar/gkw518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Xiao T, Wallace J, Felsenfeld G. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol. 2011;31:2174–2183. doi: 10.1128/MCB.05093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zabrady K, Adamus M, Vondrova L, Liao C, Skoupilova H, Novakova M, et al. Chromatin association of the SMC5/6 complex is dependent on binding of its NSE3 subunit to DNA. Nucleic Acids Res. 2015;44:1064–1079. doi: 10.1093/nar/gkv1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Kschonsak M, Merkel F, Bisht S, Metz J, Rybin V, Hassler M, et al. Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes. Cell. 2017;171:588–600.e24. doi: 10.1016/j.cell.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Eng T, Guacci V, Koshland D. Interallelic complementation provides functional evidence for cohesin-cohesin interactions on DNA. Mol Biol Cell. 2015;26:4224–4235. doi: 10.1091/mbc.E15-06-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kojic A, Cuadrado A, De Koninck M, Giménez-Llorente D, Rodríguez-Corsino M, Gómez-López G, et al. Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat Struct Mol Biol. 2018;25:496–504. doi: 10.1038/s41594-018-0070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zhang N, Kuznetsov SG, Sharan SK, Li K, Rao PH, Pati D. A handcuff model for the cohesin complex. J Cell Biol. 2008;183:1019–1031. doi: 10.1083/jcb.200801157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters J-M. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Badrinarayanan A, Reyes-Lamothe R, Uphoff S, Leake MC, Sherratt DJ. In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science. 2012;338:528–531. doi: 10.1126/science.1227126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Fennell-Fezzie R, Gradia SD, Akey D, Berger JM. The MukF subunit of Escherichia coli condensin: architecture and functional relationship to kleisins. Embo J. 2005;24:1921–1930. doi: 10.1038/sj.emboj.7600680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Barysz H, Kim JH, Chen ZA, Hudson DF, Rappsilber J, Gerloff DL, et al. Three-dimensional topology of the SMC2/SMC4 subcomplex from chicken condensin I revealed by cross-linking and molecular modelling. Open Biology. 2015;5 doi: 10.1098/rsob.150005. 150005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Walther N, Hossain MJ, Politi AZ, Koch B, Kueblbeck M, Ødegård-Fougner Ø, et al. A quantitative map of human Condensins provides new insights into mitotic chromosome architecture. J Cell Biol. 2018;271:2309–2328. doi: 10.1083/jcb.201801048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Diebold-Durand M-L, Lee H, Ruiz Avila LB, Noh H, Shin H-C, Im H, et al. Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization. Mol Cell. 2017;67:334–347.e5. doi: 10.1016/j.molcel.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Soh Y-M, Bürmann F, Shin H-C, Oda T, Jin KS, Toseland CP, et al. Molecular Basis for SMC Rod Formation and Its Dissolution upon DNA Binding. Mol Cell. 2014;57:290–303. doi: 10.1016/j.molcel.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Eeftens JM, Katan AJ, Kschonsak M, Hassler M, de Wilde L, Dief EM, et al. Condensin Smc2-Smc4 Dimers Are Flexible and Dynamic. Cell Rep. 2016;14:1813–1818. doi: 10.1016/j.celrep.2016.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Huis In 't Veld PJ, Herzog F, Ladurner R, Davidson IF, Piric S, Kreidl E, et al. Characterization of a DNA exit gate in the human cohesin ring. Science. 2014;346:968–972. doi: 10.1126/science.1256904. [DOI] [PubMed] [Google Scholar]
- [111].Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat Rev Mol Cell Biol. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Locher KP. Structure and mechanism of ATP-binding cassette transporters. Philos Trans R Soc Lond, B, Biol Sci. 2009;364:239–245. doi: 10.1098/rstb.2008.0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Linton KJ. Structure and function of ABC transporters. Physiology (Bethesda) 2007;22:122–130. doi: 10.1152/physiol.00046.2006. [DOI] [PubMed] [Google Scholar]
- [114].Kamada K, Su'etsugu M, Takada H, Miyata M, Hirano T. Overall Shapes of the SMC-ScpAB Complex Are Determined by Balance between Constraint and Relaxation of Its Structural Parts. Structure. 2017;25:603–616.e4. doi: 10.1016/j.str.2017.02.008. [DOI] [PubMed] [Google Scholar]
- [115].Gruber S, Arumugam P, Katou Y, Kuglitsch D, Helmhart W, Shirahige K, et al. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- [116].Murayama Y, Uhlmann F. DNA Entry into and Exit out of the Cohesin Ring by an Interlocking Gate Mechanism. Cell. 2015;163:1628–1640. doi: 10.1016/j.cell.2015.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Beckouët F, Srinivasan M, Roig MB, Chan K-L, Scheinost JC, Batty P, et al. Releasing Activity Disengages Cohesin's Smc3/Scc1 Interface in a Process Blocked by Acetylation. Mol Cell. 2016;61:563–574. doi: 10.1016/j.molcel.2016.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Chan K-L, Roig MB, Hu B, Beckouët F, Metson J, Nasmyth K. Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell. 2012;150:961–974. doi: 10.1016/j.cell.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Eichinger CS, Kurze A, Oliveira RA, Nasmyth K. Disengaging the Smc3/kleisin interface releases cohesin from Drosophila chromosomes during interphase and mitosis. Embo J. 2013;32:656–665. doi: 10.1038/emboj.2012.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Buheitel J, Stemmann O. Prophase pathway-dependent removal of cohesin from human chromosomes requires opening of the Smc3-Scc1 gate. Embo J. 2013;32:666–676. doi: 10.1038/emboj.2013.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Ouyang Z, Zheng G, Tomchick DR, Luo X, Yu H. Structural Basis and IP6 Requirement for Pds5-Dependent Cohesin Dynamics. Mol Cell. 2016;62:248–259. doi: 10.1016/j.molcel.2016.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Elbatsh AMO, Haarhuis JHI, Petela N, Chapard C, Fish A, Celie PH, et al. Cohesin Releases DNA through Asymmetric ATPase-Driven Ring Opening. Mol Cell. 2016;61:575–588. doi: 10.1016/j.molcel.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Ouyang Z, Yu H. Releasing the cohesin ring: A rigid scaffold model for opening the DNA exit gate by Pds5 and Wapl. Bioessays. 2017;39 doi: 10.1002/bies.201600207. 1600207. [DOI] [PubMed] [Google Scholar]
- [124].Wang X, Le TBK, Lajoie BR, Dekker J, Laub MT, Rudner DZ. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 2015;29:1661–1675. doi: 10.1101/gad.265876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Gerlich D, Hirota T, Koch B, Peters J-M, Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- [126].Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, et al. Organization of the mitotic chromosome. Science. 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Fudenberg G, Abdennur N, Imakaev M, Goloborodko A, Mirny LA. Emerging Evidence of Chromosome Folding by Loop Extrusion. Cold Spring Harb Symp Quant Biol. 2018;82:45–55. doi: 10.1101/sqb.2017.82.034710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Gruber S. Multilayer chromosome organization through DNA bending, bridging and extrusion. Curr Opin Microbiol. 2014;22:102–210. doi: 10.1016/j.mib.2014.09.018. [DOI] [PubMed] [Google Scholar]
- [129].Mc Intyre J, Muller EGD, Weitzer S, Snydsman BE, Davis TN, Uhlmann F. In vivo analysis of cohesin architecture using FRET in the budding yeast Saccharomyces cerevisiae. Embo J. 2007;26:3783–3793. doi: 10.1038/sj.emboj.7601793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.