Abstract
In mouse models of biliary tract diseases, macrophages are recruited to the periductal milieu and promote injury and cholestasis. Although cell necrosis with release of biomolecules termed damage-associated molecular patterns (DAMPs) promotes recruitment and activation of macrophages, necrosis was not observed in these studies. Because extracellular vesicles (EVs) are important in cell-to-cell communication, we postulated that activated cholangiocytes may release EVs containing DAMPs as cargo. Both the human (NHC) and mouse cholangiocyte (603B) cell lines display constitutive activation with mRNA expression of chemokines. Proteomic analysis revealed that EVs from both cell lines contained the DAMP S100A11, a ligand for the receptor for advanced glycation end products (RAGE). Bone marrow-derived macrophages (BMDM) incubated with EVs derived from the mouse 603B cell line increased mRNA expression of proinflammatory cytokines. Genetic or pharmacologic inhibition of RAGE reduced BMDM expression of proinflammatory cytokines treated with EVs. RAGE signaling resulted in activation of the canonical NF-κB pathway, and consistently, proinflammatory cytokine expression was blunted by the IKKα/β inhibitor TPCA-1 in BMDM incubated with EVs. We also demonstrated that primary mouse cholangiocyte-derived organoids express chemokines indicating cholangiocyte activation, release EVs containing S100A11, and stimulate proinflammatory cytokine expression in BMDM by a RAGE-dependent pathway. In conclusion, these observations identify a non-cell death mechanism for cellular release of DAMPs by activated cholangiocytes, namely by releasing DAMPs as EV cargo. These data also suggest RAGE inhibitors may be salutary in macrophage-associated inflammatory diseases of the bile ducts.
Keywords: cholestatic liver injury, DAMPs, extracellular vesicles, macrophages, sclerosing cholangitis
INTRODUCTION
Cholangiocytes are polarized epithelial cells that line the intra- and extrahepatic bile ducts. Major physiologic functions of cholangiocytes include modification of the bile, the formation of a barrier between biliary constituents in the bile duct lumen and the tissue surrounding the bile duct, and providing access to the immune and vascular systems in the peribiliary space. Unfortunately, cholangiocytes are affected by inflammation and fibrosis in a variety of diseases termed the cholangiopathies (18). These cholangiopathies not only impair bile formation causing cholestasis but may also lead to extensive hepatic fibrosis, cirrhosis, and end-stage liver disease with its sequela. Treatment options for the cholangiopathies are limited, and collectively, the cholangiopathies cause considerable morbidity and mortality. For example, the cholangiopathy primary sclerosing cholangitis is the indication for 6% of all liver transplants performed in the United States (19). The inflammatory processes mediating the cholangiopathies are incompletely understood, and more information is necessary to help guide the development of mechanism-based therapies.
Evolving information suggests that cholangiocytes may actively participate in the cholangiopathies by inciting recruitment to and activation of inflammatory cells within the periductal milieu. These “activated cholangiocytes” are characterized by secretion of chemokines and cytokines, especially IL-8 (1, 24, 25, 34, 38). For example, in mouse models of cholestatic liver injury, monocyte-derived macrophages are recruited to the periductal microenvironment due to cholangiocyte secretion of the chemoattractants CCL2 and IL-8 (8, 24, 25). This observation is notable as it implicates the innate immune system and sterile inflammatory processes in the pathogenesis of cholangiopathies. However, additional mechanisms also promote sterile inflammation in pathogenic conditions. Cell death by necrosis with release of intracellular damage-associated molecular pattern (DAMP) biomolecules into the extracellular space can initiate and perpetuate sterile inflammatory responses, especially those processes mediated by the innate immune system such as macrophages (32). However, the cholangiopathies are not characterized by cholangiocyte necrosis suggesting that perhaps another mechanism for cholangiocyte release of DAMPs may exist.
Cells communicate not only via direct contact and soluble factors but also by membrane-derived nanometer-sized vesicles termed extracellular vesicles (EVs). Emerging studies have identified important roles for EVs in liver diseases (10). EVs are classified into three groups based on their cellular biogenesis: exosomes, microvesicles, and apoptotic bodies. EVs derived from multivesicular bodies are termed exosomes, while EVs termed microvesicles bud directly from the plasma membrane. However, there are no widely accepted markers for these subtypes of EVs (23), and therefore, EVs is an all-encompassing term. Exosomes and microvesicles are similar in size, mostly ranging from ~40 nm up to 200 nm, unlike apoptotic bodies, which are considerably larger (consistently >500 nm) and are generated in the late stage of apoptosis as a consequence of cell fragmentation (28). We have recently identified DAMPs as cargo originating from lipotoxic hepatocyte-derived EVs by mass spectrometry assessment (14). This observation suggests that other liver cell types, such as activated cholangiocytes, may also release DAMP containing EVs as a mechanism for promoting macrophage activation and sterile inflammation.
In the current study, we employed mouse and human cholangiocyte cell lines and primary mouse cholangiocyte organoids to examine if released EVs contain DAMPs, which may activate bone marrow-derived macrophages (BMDM). We observed that indeed these cholangiocyte models release EVs containing the established DAMP S100A11 (37), which activates BMDM by binding the receptor for advanced glycation end products (RAGE) (20). These observations identify a non-cell death mechanism for cellular release of DAMPs and have implications for the potential treatment of cholangiopathies, namely the use of pharmacologic RAGE inhibitors.
MATERIALS AND METHODS
Antibodies and reagents.
The following primary antibodies were used: for Western blot, anti-TSG101 [no. ab125011, Research Resource Identifier (RRID): AB_10974262], anti-RAGE (no. ab37647, RRID: AB_777613), and anti-phospho IκB-α (ab133462, RRID: AB_2801653) from Abcam (Cambridge, MA); anti-β-actin (no. sc-8432, RRID: AB_626630) and anti-CD81 (no. sc-9158, RRID: AB_638255) from Santa Cruz Biotechnology (Santa Cruz, CA); and anti-IκB-α (no. 4814, RRID: AB_390781) and anti-Alix (no. 2171, RRID: AB_2299455) from Cell Signaling Technology (Beverly, MA). Anti-S100A11 (10237-1-AP, RRID: AB_2183478; Proteintech Group, Rosemont, IL) was used for immunogold staining and immunoblot analysis. Anti-NF-κB p65 (sc-8008, RRID:AB_628017; Santa Cruz Biotechnology) was used for immunocytochemical analysis. The following secondary antibodies were used: anti-rabbit IgG (H+L) horseradish peroxidase (HRP) conjugated (no. 20320) from Alpha Diagnostic International (San Antonio, TX) and anti-mouse IgG (H+L) HRP conjugated (no. NB7539) from Novus Biologicals (Centennial, CO). The RAGE inhibitor TTP448 was from Med Koo Biosciences (Morrisville, NC). The IKKα/β inhibitor TPCA-1 was from Selleck Chemical (Houston, TX). Recombinant mouse S100A11 protein (CR31) was from Novoprotein (Shanghai, China).
Cell cultures.
The SV40-transformed, nonmalignant mouse cholangiocyte cell line 603B was cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 0.2% Primocin (Invivogen, San Diego, CA). The normal human cholangiocyte cell line NHC was cultured as previously described (6). 603B and NHC were authenticated by short tandem repeat analysis. Bone marrow-derived macrophages (BMDM) from 3- to 4 mo-old male and female C57BL/6 (Jackson Laboratories, Bar Harbor, ME) and Rage−/− (a gift from Dr. A. M. Schmidt, New York University School of Medicine, New York, NY) mice were isolated and cultured as previously described (22). BMDM were isolated in accordance with protocols approved by the Mayo Clinic Institutional Animal Care and Use Committee. All cultures underwent Mycoplasma contamination testing periodically using a PlasmoTest-Mycoplasma Detection kit (Invivogen).
Generation of stable clones expressing S100A11 short hairpin RNA.
603B cells were transfected with short hairpin (sh) RNA lentiviral plasmids against S100A11 obtained from Millipore-Sigma (GenBank accession numbers: NM_016740.3-221s21c1; NM_016740.3-282s21c1; NM_016740.3-323s21c1; and NM_016740.3-434s21c1). Transfection and clone selection were performed as previously described by us (7). Knockdown efficiency was evaluated by immunoblot analysis.
EV isolation and EV treatment of BMDM.
603B and NHC were grown to 90% confluence on 150-mm culture dishes, washed twice with PBS to remove FBS-derived EVs, and then incubated with each specific medium with 10% extracellular vesicle-free FBS, which was prepared by overnight centrifugation at 100,000 g at 4°C according to a standard protocol (33). The cell culture medium was collected after 24 h. for EV analysis. Organoids were cultured in complete organoid medium diluted 1:1 with DMEM/F-12, and the medium was collected after 48–72 h for EV analysis. EVs isolated from the culture media were resuspended in PBS or lysed in RIPA buffer for immunoblot analysis or fixed in 4% paraformaldehyde for immunogold staining. EV concentration and size distribution were assessed by nanoparticle tracking analysis (NTA) using the NanoSight NS300 instrument (NanoSight, Amesbury, UK), as previously described (15).
BMDM were preincubated with serum-free RPMI for 1 h and then the medium was replaced with serum-free RPMI containing EVs at a final concentration of 3–4 × 109 particles/mL, with or without the RAGE inhibitor TTP448 (10 μM) or the IKKα/β inhibitor TPCA-1 (5 μM). Total RNA and/or protein were isolated from the BMDM after 6 h treatment.
Isolation of EpCAM-positive mouse cholangiocytes, generation, and culture of cholangiocyte organoids.
Mouse liver cholangiocytes were isolated from 3- to 4 mo-old male and female C57BL/6 and C57BL/6.Mdr2−/− mice (a gift from Dr. Oude Elferink, Tytgat Institute, Amsterdam, The Netherlands) using a magnetic bead/affinity antibody approach. Mice were anesthetized, and the livers were procured and collagenase-digested as previously described for isolating hepatocytes (4). Once the liver was properly digested, hepatocytes were mechanically dissociated from the vasculature and biliary tree. The biliary tree was rinsed once with DMEM and placed in a 50 ml conical tube containing 30 ml DMEM, 20 mg collagenase II (Roche, Basel, Switzerland), 20 mg protease from Streptomyces griseus (Millipore Sigma, St. Louis, MO), and 1 mg DNase I (Roche). The tube was placed in a 37°C water bath, and under moderate agitation, the biliary tree was allowed to digest for 1 h, followed by agitation at room temperature for 30–45 min. Once the biliary tree finished the digestion, it was poured over a 70-μm strainer (Corning, Durham, NC) into a 50-ml conical tube. The strainer was rinsed with 10 ml DMEM, and the suspension was centrifuged at 650 g for 7 min. The pellet was resuspended in 500 μl of DMEM and placed on ice. The affinity anti-EpCAM Ab/magnetic bead complex was assembled by incubation in 1 ml PBS, pH 7.4, containing 0.1% bovine serum albumin (BSA), 5 μl Biotin anti-mouse CD326 antibody (118204; Biolegend, San Diego, CA), and 50 μl of dynabeads (Invitrogen/ThermoFisher Scientific, Waltham, MA). After the bead/antibody complex coalesced, the beads were magnetically separated and rinsed three times with PBS + 0.1% BSA. The bead/antibody complex was next resuspended in 500 μl PBS + 0.1% BSA and added to the resuspended cell pellet. The cell/bead/antibody mixture was allowed to moderately agitate at room temperature for 15–20 min, and then, the cells were magnetically separated and gently washed three times with PBS containing 0.1% BSA. After the final wash, the complex was resuspended in 150 μl of cold hepatic progenitor cell (HPC) media (21), added to 150 μl Matrigel (Corning), and gently mixed. Twenty-five microliters of this mixture was placed as a droplet into the center of each well of a 48-well tissue culture plate and allowed to solidify for 20 min at room temperature. Five hundred microliters of hepatic progenitor cell media was then added to each well, and the plates were placed in a 5% CO2 tissue culture incubator at 37°C to allow formation of 3D organoids. Twenty-four hours later, the media were replaced with the following organoid culture media, which we slightly modified from previously published protocols (2, 11): Advanced DMEM/F-12, 100 μg/mL primocin, and 1% GlutaMAX (ThermoFisher Scientific); 10 mM HEPES, 25 ng/mL Noggin, 25 ng/mL recombinant mouse HGF, 100 ng/mL Wnt3a, 100 ng/mL recombinant mouse FGF10, 500 ng/mL R-spondin, and 50 ng/mL recombinant mouse EGF (all from Peprotech, Rocky Hill, NJ); 10 nM gastrin, 1.25 mM N-acetyl-l-cysteine, 10 μM γ27632, 5 μM A83-01, and 10 mM nicotinamide (all from Millipore Sigma); 1% N2 supplement and 2% B27 supplement (both from Invitrogen/ThermoFisher); and 10 μM forskolin (R&D Systems, Minneapolis, MN). The organoids were passaged by breaking down mechanically into small fragments using a Pasteur glass pipet when they reached ~70% confluence and were used up to passage 4.
Western blot analysis.
603B-, NHC-, and organoid-derived EVs and BMDM were lysed in RIPA lysis and extraction buffer (Thermo Fisher Scientific) with protease (cOmplete mini) and phosphatase (PhosSTOP) inhibitors (both from Roche/Millipore Sigma) for 30 min on ice and then centrifuged at 12,000 g for 15 min at 4°C to remove debris. Protein concentration in the supernatants was determined using the Bradford reagent (Millipore Sigma). Equal amounts of cholangiocyte-derived EV or whole cell lysates were loaded onto 4–20% Mini-PROTEANR TGX Precast Gel (Bio-Rad Laboratories, Hercules, CA) and transferred to nitrocellulose membranes. The membranes were blocked with blocking buffer (5% nonfat milk in TBS-Tween) for 1 h at room temperature and then blotted with primary antibodies at 4°C overnight in 5% BSA-TBS Tween. After washing, the membranes were incubated with the appropriate HRP-conjugated secondary antibodies (1:2,000–1:7,000) for 1 h at room temperature. Proteins were visualized with enhanced chemiluminescence reagent (ECL) or ECL prime (GE Healthcare Life Sciences). β-Actin protein levels were used as loading controls.
Reverse transcription/polymerase chain reaction and quantitative polymerase chain reaction.
Total RNA was isolated from cells using RNeasy Mini kit (Qiagen, Hilden, Germany). Quantity and quality of isolated RNA were assessed using a NanoDrop ND1000 (Thermo Fisher Scientific, Waltham, MA). Reverse transcription was performed using Moloney murine leukemia virus reverse transcriptase and random primers (Life Technologies/Thermo Fisher Scientific). The generated cDNA was amplified by conventional (qualitative) PCR consisting of one cycle of 3-min denaturation at 95°C; 32 cycles of 20 s at 95°C, 20 s at 65°C, and 30 s at 72°C, and a final extension step at 72°C for 7 min. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA was also amplified to confirm that an equal amount of total cDNA was used for each sample. Quantitative PCR was performed on Light Cycler 480 (Roche, Indianapolis, IN) using SYBR green (Roche) as the fluorophore. Target gene expression level was calculated using the ΔΔCt method, and expression was normalized to 18s rRNA or hypoxanthine guanine phosphoribosyl transferase (Hprt) RNA. The primers used for either qualitative or quantitative PCR are listed in Table 1.
Table 1.
Human and mouse PCR primers
| Gene Name | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
|---|---|---|
| Mouse | ||
| Tnfα | CCCTCACACTCAGATCATCTTCT | GCTACGACGTGGGCTACAG |
| Il1β | GCAACTGTTCCTGAACTCAACT | ATCTTTTGGGGTCCGTCAACT |
| Il6 | ACCAGAGGAAATTTTCAATAGGC | TGATGCACTTGCAGAAAACA |
| Cd206 | CTCTGTTCAGCTATTGGACGC | CGGAATTTCTGGGATTCAGCTTC |
| Arg1 | AGGAGCTGTCATTAGGGACATC | CTCCCAGCCAAAGTCCTTAGAG |
| Ym1 | GTCTTGCTCATGTGTGTAAGTGA | CAGGTCTGGCAATTCTTCTGAA |
| Pdgfβ | CCCACAGTGGCTTTTCATTT | GTGAACGTAGGGGAAGTGGA |
| Cxcl1 | ACTCAAGAATGGTCGCGAGG | GTGCCATCAGAGCAGTCTGT |
| Cxcl2 | AGGGCGGTCAAAAAGTTTGC | CGAGGCACATCAGGTACGAT |
| Cxcl5 | TGCCCTACGGTGGAAGTCATA | TGCATTCCGCTTAGCTTTCTTT |
| Rage | ACTACCGAGTCCGAGTCTACC | CCCACCTTATTAGGGACACTGG |
| Ck7 | AGGAGATCAACCGACGCAC | GTCTCGTGAAGGGTCTTGAGG |
| Ck19 | GGGGGTTCAGTACGCATTGG | GAGGACGAGGTCACGAAGC |
| Gapdh | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
| 18s | CGCTTCCTTACCTGGTTGAT | GAGCGACCAAAGGAACCATA |
| Hprt | TCCTCCTCAGACCGCTTTT | CCTGGTTCATCATCGCTAATC |
| Human | ||
| IL8 | ACTGAGAGTGATTGAGAGTGGAC | AACCCTCTGCACCCAGTTTTC |
Immunocytochemistry.
Bone marrow-derived macrophages were grown on sterile glass coverslips in six-well plates for 24 h and then treated with serum-free RPMI containing EVs with or without TTP448 (10 μM) or TPCA-1 (5 μM) for 0, 30, 45, and 60 min. Cells were fixed with 4% paraformaldehyde for 20 min, permeabilized with 0.1% Triton X-100 in PBS for 5 min and then blocked with 5% BSA + 0.1% glycine in PBS for 30 min at room temperature. After blocking, the cells were incubated with mouse monoclonal anti-NF-κB p65 primary antibody (dilution 1:100) in blocking buffer overnight at 4°C and then incubated with goat anti-mouse Alexa Fluor 488 (1:1,000; Life Technologies/Thermo Fisher Scientific) diluted in blocking buffer for 1 h at room temperature. The coverslips were mounted on a clean glass slide in Prolong Gold antifade reagent (Thermo Fisher Scientific) with DAPI for nuclear staining. The slides were analyzed by fluorescent confocal microscopy (LSM 780; Zeiss).
High-pressure liquid chromatography/tandem mass spectrometry.
EVs derived from 603B and NHC were lysed in RIPA buffer as described above. The protein lysates were run on a 4–20% gel for SDS-PAGE, sliced, digested, and analyzed by high-pressure liquid chromatography/tandem mass spectrometry (LC-MS/MS) performed by the Mayo Proteomics Core. The following databases were used to match peptides: for 603B cells, Swissprot-Mouse Sept 2017, with a reverse sequence decoy; false discovery rate (FDR): 2.1%; and for NHC, Swissprot-Human/Bovine Nov 2016, with a reverse decoy; FDR: 1.3%. Protein identification criteria: 2 peptides minimum, 95% probability, and 95% peptide threshold.
Immunogold staining and electron microscopy.
The extracellular vesicle pellets were fixed in 4% paraformaldehyde in PBS overnight at 4°C. Five μl of the each sample were placed on formvar-carbon-coated nickel grid (200 mes) and air-dried for 20 min. The grids were transferred to 2% fetal calf serum (FCS)-PBS containing 50 mmol/L glycine for 30 min for saturation of free aldehyde groups and then blocked in 10% FCS-PBS for 30 min, followed by incubation with primary anti-S100A11 antiserum diluted 1:15 in blocking buffer for 2 h at room temperature. After washing with PBS, the grids were incubated with colloidal gold donkey anti-rabbit IgG (Jackson ImmunoResearch, West Grove, PA) diluted 1:20 in blocking buffer for 1 h, fixed in 1% glutaraldehyde/PBS for 5 min, and then contrasted and embedded with a mixture of 4% uranyl acetate and 2% methylcellulose (1:9 ratio) for 5 min. The grids were air-dried and examined with a JEOL 1400 electron microscope (JEOL, Peabody, MA).
Statistical analysis.
All data are expressed as means (or fold change in mean over control) ± SE. Statistical analyses were performed with two-tailed Student’s t-test for comparing groups using GraphPad Prism 7.03 (GraphPad Software) and P < 0.05 was considered statistically significant.
RESULTS
Human and mouse cholangiocytes release S100A11-containing EVs.
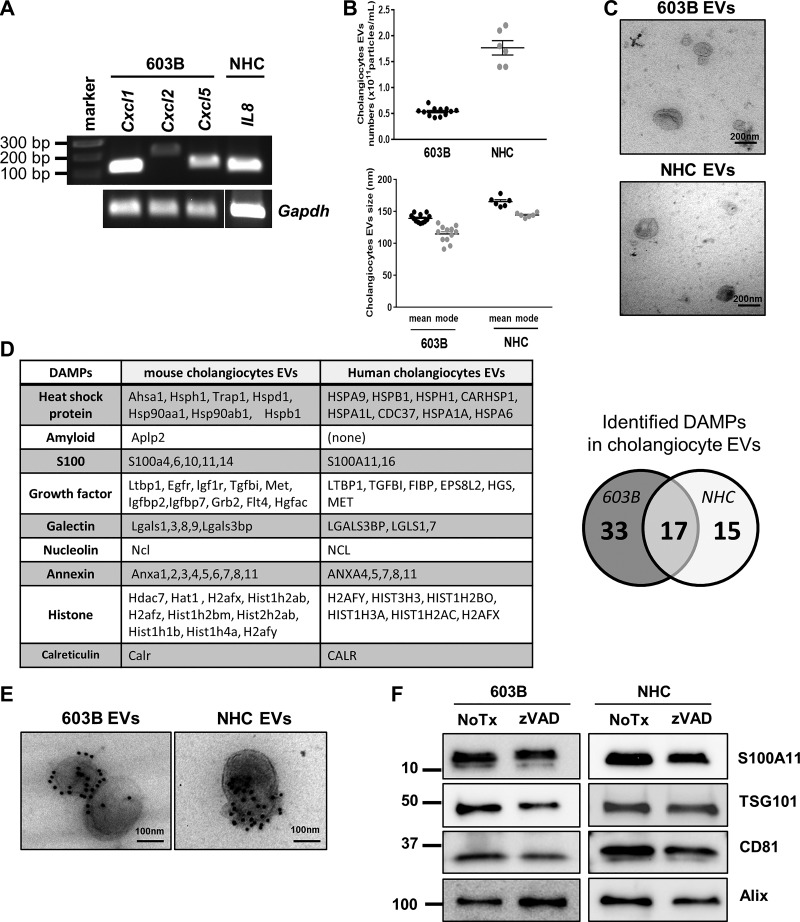
To examine the nature of the EVs released by cholangiocytes, mouse 603B and human NHC cholangiocyte cell lines were employed. First, we determined that these cells lines were in an activated state by measuring expression of the chemokine IL-8, a marker of activated cholangiocytes/ductular reactive cells (24, 25, 38); indeed, expression of the mouse Il8 gene homologs C-X-C motif chemokine ligand 1 (Cxcl1), Cxcl2, and Cxcl5 in 603B cells, and human IL8 expression in NHC cells was readily detected (Fig. 1A). Having established the proliferating cell lines were activated, we next examined EV release into the cell culture media after 24 h by nanoparticle tracking analysis (NTA). The average number of released EVs was 0.5 × 1011 particles/mL and 1.6 × 1011 particles/mL for 603B and NHC, respectively (Fig. 1B). The size distribution of the EVs from both cell lines was 120–150 nm, with a mode of ~150 nm, excluding the possibility that they may be originating from ongoing apoptotic cell death (Fig. 1B). EV morphology was further confirmed by transmission electron microscopy (TEM), showing the characteristic cup shape appearance and lipid bilayer membrane enclosing the EVs (Fig. 1C). To characterize the EV cargo, we next performed proteomic analysis by tandem mass spectrometry (LC-MS/MS) on EVs isolated from 603B and NHC culture media. The analysis identified a total of 1,886 peptides in the mouse cholangiocyte EVs and 1,810 peptides in the human cholangiocyte EVs. Among them, we recognized 50 putative DAMPs in 603B and 32 DAMPs in NHC EVs, with 17 DAMPs being identified in both EVs (Fig. 1D). Of the 17 common DAMPs, we chose to concentrate on S100A11, a member of the S100 family of calcium-binding proteins with a known role in regulating macrophage migration and differentiation (29, 37). The presence of S100A11 as cargo on the mouse and human cholangiocyte-derived EVs was readily confirmed by immunogold electron microscopy (Fig. 1E) and by Western blot analysis (Fig. 1F). Moreover, treatment of 603B cells or NHC with the pan-caspase inhibitor zVAD-fmk did not affect the number of EVs released into the culture media, the abundance of the cargo S100A11, or the EV markers tumor susceptibility gene 101 (TSG101), cluster of differentiation 81 (CD81), and Alix/AIP1; these observations further confirm that apoptotic bodies were not contributing to the EV pool (Fig. 1F). Thus these data demonstrate that human and mouse cholangiocytes release S100A11-containing EVs in the absence of cell death.
Fig. 1.
Human and mouse cholangiocytes release S100A11-containing extracellular vesicles (EVs). A: RNA from 603B mouse cholangiocyte cell line and normal human cholangiocyte (NHC) cell line was extracted and subjected to conventional/qualitative RT-PCR for Cxcl1, Cxcl2, and Cxcl5 gene expression (mouse) or IL8 gene expression (human). Gapdh was used as housekeeping gene. Gapdh in NHC was run on a separate gel. B: EVs were isolated from 603B and NHC cultures by differential ultracentrifugation and analyzed by nanoparticle tracking analysis. Concentration (top) and size distribution (bottom) are shown (603B n = 12; NHC n = 6, from independent EV preparations). C: transmission electron microscopy (TEM) images of 603B and NHC EVs. D: list of damage-associated molecular patterns (DAMPs) identified by liquid chromatography/tandem mass spectrometry on EVs isolated from 603B and NHC culture media. E: representative TEM images of 603B and NHC EVs following immunogold labeling with an anti-S100A11 antibody. F: immunoblot analysis for EV markers and S100A11 in 603B and NHC EVs after incubation with or without (NoTx) the pan-caspase inhibitor zVAD-fmk (10 μM) for 24 h.
Cholangiocyte-derived EVs induce S100A11-mediated proinflammatory macrophage polarization.
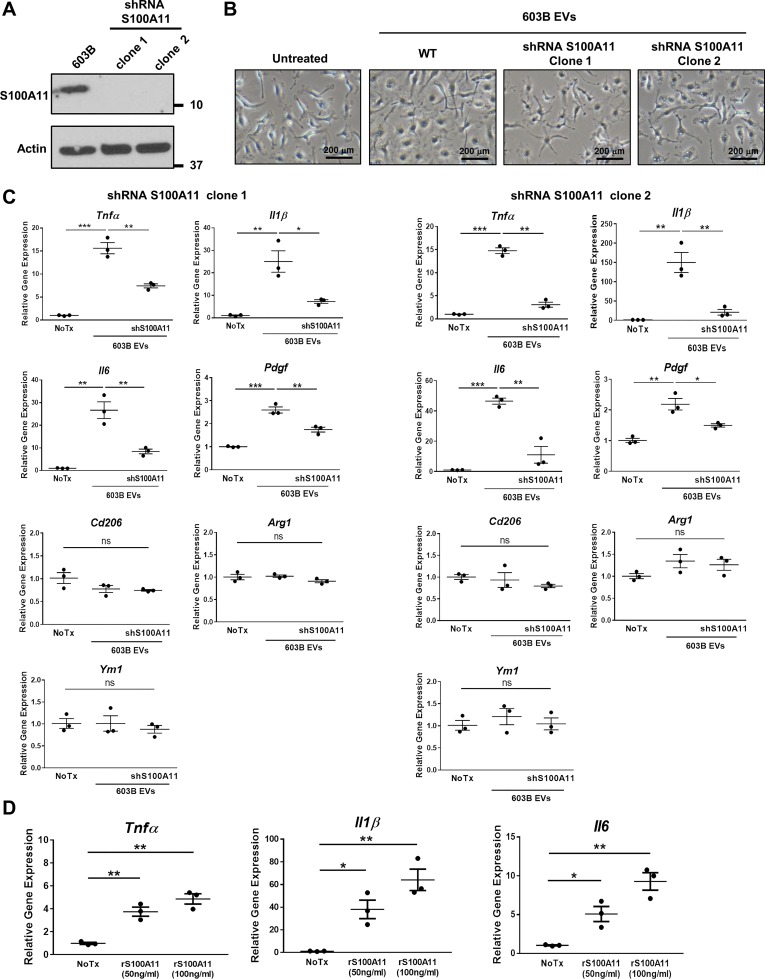
Because S100A11 has been implicated in macrophage activation, we next sought to investigate whether S100A11-laden cholangiocyte-derived EVs have an effect on macrophage gene expression and polarization. EVs were collected from the supernatant of both nontargeted and S100A11 shRNA-targeted 603B cells; successful knockdown of S100A11 in two separate clones (1, 2) was confirmed by immunoblot analysis of cell lysates (Fig. 2A). Bone marrow-derived macrophages (BMDM) isolated from C57BL/6 mice were incubated with purified EVs by S100A11 shRNA-targeted and -nontargeted 603B cells (Fig. 2B), and gene expression was evaluated after 6 h by quantitative PCR. 603B-derived EVs significantly increased BMDM gene expression of the proinflammatory cytokines TNFα, IL-1β and IL-6 associated with the classically activated M1-like phenotype but had no effect on the expression of genes (Cd206, Arg1, and Ym1) associated with the alternatively activated M2-like phenotype (Fig. 2C). Expression of the profibrogenic gene Pdgf in BMDM was also enhanced by the 603B-derived EV treatment (Fig. 2C). However, expression of the proinflammatory cytokines was strongly reduced when BMDM were incubated with S100A11-deficient EVs (Fig. 2C). Similar results were replicated when BMDM were incubated with mouse recombinant S100A11, suggesting S100A11 plays a significant role in EV-mediated regulation of gene expression in BMDM (Fig. 2D). Collectively, these results demonstrate that cholangiocyte-derived EVs can induce proinflammatory macrophage polarization through the activation of a S100A11-mediated signaling pathway.
Fig. 2.
Cholangiocyte-derived extracellular vesicles (EVs) induce S100A11-mediated proinflammatory macrophage polarization. A: immunoblot analysis for S100A11 of cell lysates from 603B and S100A11 shRNA 603B (clone 1 and 2) cells. Actin was used as loading control. B: representative phase contrast microphotographs of wild-type mouse bone marrow-derived macrophages (BMDM) incubated without (untreated) or with EVs purified from the culture media of 603B and shRNA S100A11 603B cells (clone 1 and 2) at a concentration of 109/mL for 6 h. WT, wild type. C: expression of proinflammatory (Tnfα, Il1β, Il6), profibrogenic (Pdgf), and anti-inflammatory genes (Cd206, Arg1, Ym1) analyzed by quantitative (q)RT-PCR in BMDM treated as in B (n = 3, from independent BMDM and EV isolations). Target gene expression normalized to 18s. D: proinflammatory gene expression in wild-type BMDM treated with recombinant mouse S100A11 at the indicated concentrations for 6 h by qRT-PCR (n = 3, from independent BMDM isolations). Target gene expression normalized to 18s. Data represent mean of fold increase over control (untreated BMDM) ± SE. Two-tailed Student’s t-test: *P < 0.05, **P < 0.005, ***P < 0.001.
S100A11-enriched cholangiocyte EVs stimulate macrophage via engagement of its receptor RAGE.
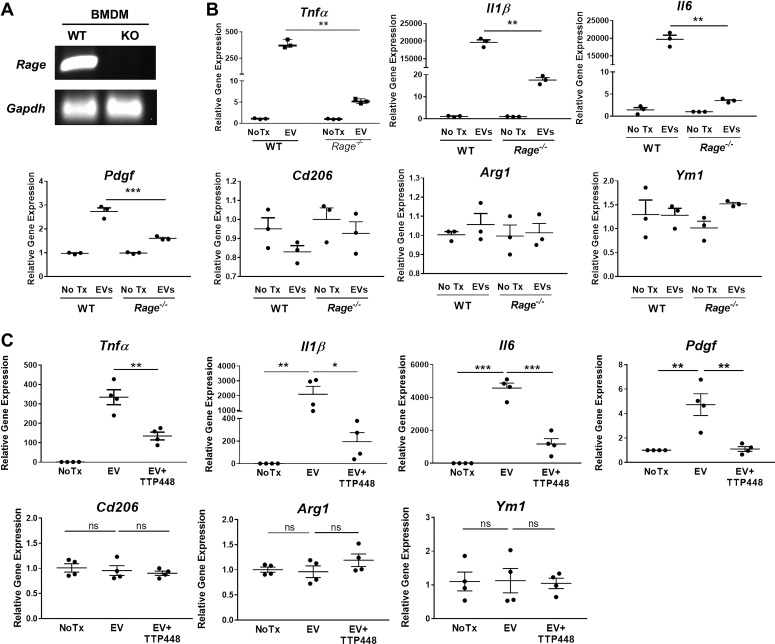
We next investigated how S100A11 in cholangiocyte EVs influence BMDM gene expression and polarization. S100 family members, including S100A11, have been shown to interact with receptor for advanced glycation end products (RAGE) (20, 30): therefore, we postulated that binding of EV S100A11 to RAGE on the BMDM plasma membrane would trigger a signaling cascade resulting in the upregulation of proinflammatory genes. BMDM were isolated from wild-type and Rage−/− mice, and expression of RAGE was verified by conventional RT-PCR analysis (Fig. 3A). Next, wild-type and Rage−/− BMDM were incubated with 603B-derived EVs and gene expression was analyzed after 6 h. Tnfα, Il1β, Il6, and Pdgf gene expression was significantly reduced in Rage−/− compared with wild-type BMDM (Fig. 3B). Conversely, the expression of the genes that were not induced by the EV treatment (Cd206, Arg1, and Ym1) was unchanged in Rage−/− BMDM (Fig. 3B). Consistently, pharmacological RAGE inhibition by TTP448 (10 μM) also reduced proinflammatory gene expression, but had no effect on anti-inflammatory genes associated with the “M2-like” phenotype (Fig. 3C), underlining the importance of the S100A11-RAGE signaling pathway in EV-induced macrophage activation and polarization. These results demonstrate that cholangiocyte EV-mediated proinflammatory polarization of BMDM is dependent on RAGE.
Fig. 3.
S100A11-enriched cholangiocyte-derived extracellular vesicles (EVs) stimulate macrophages via engagement of its receptor for advanced glycation end products (RAGE). A: mRNA obtained from bone marrow-derived macrophages (BMDM) from wild-type (WT) and Rage−/− mice were analyzed by conventional RT-PCR for RAGE gene expression. Gapdh was used as housekeeping gene. KO, knockout. B: gene expression analysis by quantitative (q)RT-PCR in BMDM from WT and Rage−/− mice incubated with 603B-derived EVs for 6 h (n = 3, from independent BMDM and EV isolations). Target gene expression normalized to 18s. C: gene expression analysis by qRT-PCR in wild-type BMDM treated with 603B-derived EVs in the presence or absence of the RAGE inhibitor TTP448 (10 μM) for 6 h (n = 4, from independent BMDM and EV isolations). Target gene expression normalized to 18s. Data represent mean of fold increase over control (untreated BMDM) ± SE. Two-tailed Student’s t-test: *P < 0.05, **P < 0.005, ***P < 0.001.
S100A11-RAGE signaling pathway regulates gene expression through activation of NF-κB.
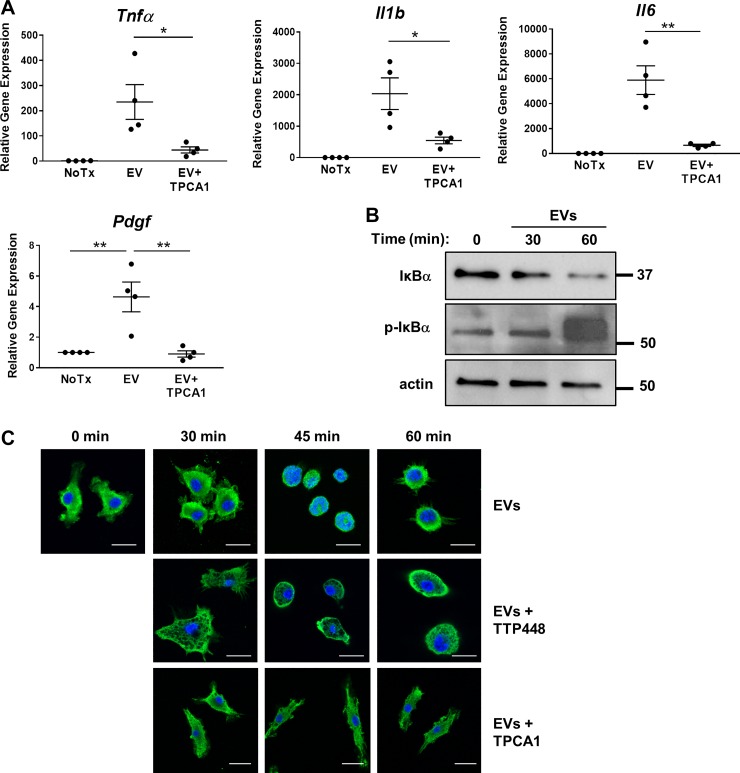
RAGE is known to activate multiple downstream intracellular signaling pathways, including mitogen-activated protein kinases (ERK 1/2, p38, SAPK/JNK), Akt, and JAK. These, in turn, activate downstream transcription factors, including NF-κB (by Akt, ERK1/2, and p38), which regulate the expression of several proinflammatory genes (12). To elucidate whether the NF-κB signaling pathway was engaged by binding of the S100A11-laden EVs to RAGE, we analyzed the effect of the selective IKKα/β inhibitor TPCA-1 on EV-induced gene expression. Inhibition of NF-κB by TPCA-1 (5 μM) significantly reduced Tnfα, Il1β, Il6, and Pdgf gene expression in BMDM (Fig. 4A). Activation of NF-κB was confirmed by Western blot evidence of phosphorylation and degradation of IκBα, the NF-κB regulatory subunit (Fig. 4B) and by detecting the transient translocation of the NF-κB p65 subunit into the nucleus 45 min after EVs treatment by immunocytochemistry (Fig. 4C). Similarly to TPCA-1, inhibition of RAGE by TTP448 prevented p65 translocation to the nucleus, demonstrating that NF-κB activation is indeed downstream of RAGE signaling (Fig. 4C). Therefore, these data are consistent with the activation of a signaling cascade triggered by RAGE binding to S100A11, resulting in the activation of the NF-κB pathway and subsequent upregulation of proinflammatory genes.
Fig. 4.
S100A11-receptor for advanced glycation end products (RAGE) signaling pathway regulates gene expression through activation of NF-κB. A: gene expression analysis by quantitative RT-PCR in wild-type bone marrow-derived macrophages (BMDM) treated with 603B-derived extracellular vesicles (EVs) in the presence or absence (NoTx) of the IKKα/β inhibitor TPCA-1 (5 μM) for 6 h (n = 4, from independent BMDM and EV isolations). Target gene expression normalized to 18s. Data represent mean of fold increase over control (untreated BMDM) ± SE. Two-tailed Student’s t-test: *P < 0.05, **P < 0.005. B: representative immunoblot analysis of total and phosphorylated IκBα in total cell lysates from wild-type BMDM incubated with 603B-derived EVs for the indicated time. Actin was used as a loading control. C: representative immunocytochemistry for the NF-κB subunit p65 on wild-type BMDM incubated with 603B-derived EVs for the indicated time, in the presence or absence of TTP448 or TPCA-1 (p65: green; DAPI: blue). Scale bar = 10 μm.
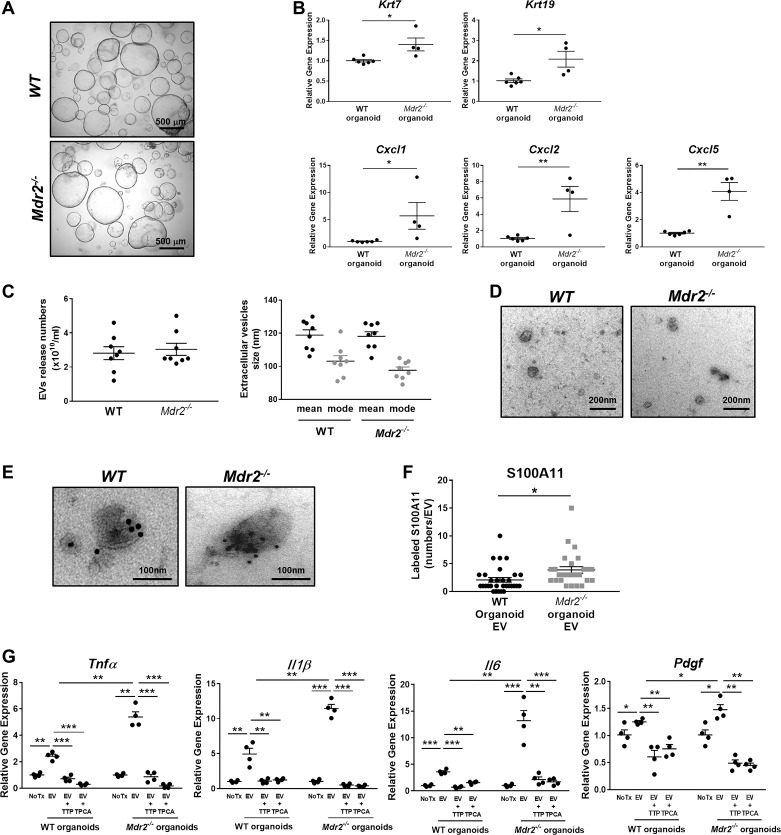
EVs from Mdr2−/− mouse cholangiocytes promote proinflammatory polarization expression in BMDM.
Finally, we sought to verify whether our observations could be reproduced using EVs isolated from cultures of EpCAM-positive cholangiocyte organoids (cholangioids), a three-dimensional model that better recapitulates the cell-to-cell interactions occurring in a physiologic environment (Fig. 5A). First, we confirmed that cholangioids derived from either wild-type or Mdr2−/− mouse cholangiocytes, a well-recognized model of sclerosing cholangitis, retain expression of cholangiocyte differentiation genes [cytokeratin 7 (Krt7) and Krt19; Fig. 5B]. Next, we assessed cholangiocyte activation in the cultured cholangioids by measuring gene expression of the IL8 mouse homologs Cxcl1, Cxcl2, and Cxcl5 and determined that Mdr2−/− mouse cholangioids are significantly activated compared with wild-type cholangioids (Fig. 5B). These observations are consistent with a recently published report that cholangiocyte organoids are activated (27). We then isolated EVs from cultured wild-type and Mdr2−/− mouse cholangioids and analyzed them by NTA. The average number of released EVs was similar in wild-type and Mdr2−/− cholangioids (~2.5 × 1010 particles/mL), with a size distribution of 110 nm and a mode of 95 nm (Fig. 5C), and displayed the characteristic EV morphology by electron microscopic analysis (Fig. 5D). Interestingly, immunogold electron microscopy and subsequent quantification of the labeled particles determined that S100A11 was significantly more abundant in EVs from Mdr2−/− cholangioids than in those from wild-type cholangioids (Fig. 5, E and F). More importantly, when BMDM were exposed to Mdr2−/−cholangioid-derived EVs, Tnfα, Il1β, Il6, and Pdgf gene expression was significantly increased compared with wild-type cholangioid EVs (Fig. 5G). Consistent with our previous observations with 603B-derived EVs, upregulation of gene expression was prevented by cotreatment with TTP448 or TPCA-1 (Fig. 5G). Our results demonstrated that cholangioid EVs from cholestatic livers induce macrophage proinflammatory polarization via RAGE-mediated activation of NF-κB signaling pathway.
Fig. 5.
Extracellular vesicles (EVs) from Mdr2−/− mouse cholangiocyte-derived organoids promote proinflammatory polarization in bone marrow-derived macrophages (BMDM). A: representative phase-contrast photomicrographs of wild-type (WT) and Mdr2−/− mouse cholangiocyte organoids (×4). B: Krt7, Krt19, Cxcl1, Cxcl2, and Cxcl5 gene expression by quantitative RT-PCR in WT and Mdr2−/− mouse cholangiocyte-derived organoids at passage 1 (WT n = 6; Mdr2−/− n = 4, from independent isolations). Target gene expression normalized to Hprt and expressed as fold increase over WT cholangiocyte-derived organoids. Data represent mean of fold increase over control (WT) ± SE. Two-tailed Student’s t-test: *P < 0.05, **P < 0.005. C: concentration (left) and size distribution (right) of EVs isolated from WT and Mdr2−/− mouse cholangiocyte-derived organoids analyzed by nanoparticle tracking analysis (n = 8, from independent isolations). D: transmission electron microscopy (TEM) images of EVs from WT and Mdr2−/− mouse cholangiocyte-derived organoids. E: representative TEM images of EVs from WT and Mdr2−/− mouse cholangiocyte-derived organoids following immunogold labeling with an anti-S100A11 antibody. F: quantification of number of S100A11-positive particles on EVs from WT and Mdr2−/− mouse cholangiocyte-derived organoids (10 EV/genotype from 3 independent isolations). *P < 0.05. G: gene expression analysis by quantitative RT-PCR in WT BMDM treated with EVs from WT and Mdr2−/− mouse cholangiocyte-derived organoids in the presence or absence of TPCA-1 or TTP448 for 6 h. Target gene expression was normalized to Hprt. Data represent mean of fold increase over control (untreated BMDM) ± SE. Two-tailed Student’s t-test: *P < 0.05, **P < 0.005, ***P < 0.001 (n = 4, from independent EV isolations).
DISCUSSION
The results of this study provide mechanistic insights regarding macrophage activation by cholangiocyte-derived EVs. The principal findings indicate that 1) activated cholangiocyte cell lines release EVs containing the cargo S100A11, an established DAMP; 2) cholangiocyte-derived EVs activate BMDM by engaging the RAGE signaling pathway; and 3) the observations were verified in cholangiocyte-derived organoids, termed cholangioids. These finding are discussed in greater detail below.
In health, the rate of cholangiocyte turnover is minimal and requires occasional self-replication. However, during liver injury, reparative processes initiate cholangiocyte replication, often resulting in expansion of a proliferative cholangiocyte-derived compartment (31). Given that inflammation and regeneration are interlinked mechanistically (16), it is not surprising that cholangiocyte proliferation is associated with their activation and secretion of chemokines and cytokines (34). This activation state maybe a characteristic of proliferating cholangiocytes, as we observed cytokine generation by immortalized, but nontransformed, human and mouse cholangiocyte cell lines and primary mouse cholangioids. Having verified this activated state of proliferating cholangiocytes in vitro, we employed these models for examining the relationship between activated cholangiocytes and macrophage activation.
Both human and mouse cholangiocyte cell lines released extracellular vesicles, a characteristic of virtually all cells. The EVs were between 100 and 150 nm in size and, therefore, were not apoptotic bodies from dying cells that are >500 nm. The biogenesis of the EVs was not elucidated in the current study, and they are likely heterogeneous, consisting of exosomes due to release of multivesicular bodies into the extracellular milieu or budding and scission of microparticles from the plasma membrane. Further studies will be necessary to clarify the heterogeneity and biogenesis of cholangiocyte EVs, questions that have been challenging for the EVs research community in general (23).
We observed that both human and mouse cholangiocyte cell lines released EVs containing S100A11 as cargo in the absence of cell death. This calcium-binding, normally cytosolic protein has been recognized as a DAMP. For example, it has been shown to initiate chemokine responses in monocytes during pathogen-induced inflammatory responses (29) and to activate macrophages (37). We have previously reported that proteomic analysis revealed DAMPs associated with EVs originating from sublethally injured hepatocytes (14), and others have reported that endoplasmic reticulum stress in choriocarcinoma cells results in EVs carrying DAMPs (3). Recent studies have also identified a subfraction of exosome-like vesicles released by apoptotic cells, but much smaller than apoptotic bodies, called apoptotic exosome-like vesicles, which contain macrophage-activating proinflammatory DAMPs (26); however, our data show no significant difference in the magnitude of released EVs in the presence or absence of a pan-caspase inhibitor, supporting the interpretation that these EVs do not originate from apoptotic cells. Our current observations, together with the limited, but emerging literature, suggest that DAMP-mediated sterile inflammation (e.g., innate immune cell recruitment and activation) may occur in the absence of cell necrosis or apoptosis but rather may be instigated by EV cargo.
We observed that EVs stimulated proinflammatory cytokine gene expression in BMDM. Macrophages may either be resident (Kupffer cells in the liver) or recruited from bone marrow-derived monocytes (35). In both acute and chronic models of mouse bile duct injury, we observed that the periductal macrophages were largely recruited, and hence, we employed BMDM, as opposed to liver resident macrophages for our studies (8). Several studies have reported proinflammatory macrophage activation by EVs, but the mechanisms have remained unclear. In the current study, we implicate RAGE-mediated macrophages activation as a mechanism for our observations. First, S100A11 has been reported by others to be a potent agonist of RAGE (37). Second, the genetic deletion or selective RAGE inhibition by TTP448 blocked expression of proinflammatory cytokines by BMDM incubated with cholangiocyte-derived EVs. Finally, RAGE is known to signal through an NF-κB signaling pathway, and we observed activation of this pathway in BMDM exposed to EVs. Accordingly, the NF-κB pathway inhibitor TPCA-1 reduced proinflammatory cytokine expression in these studies. Although other processes for EV activation of BMDM likely exist, stimulation of RAGE appears to be a potent mechanism.
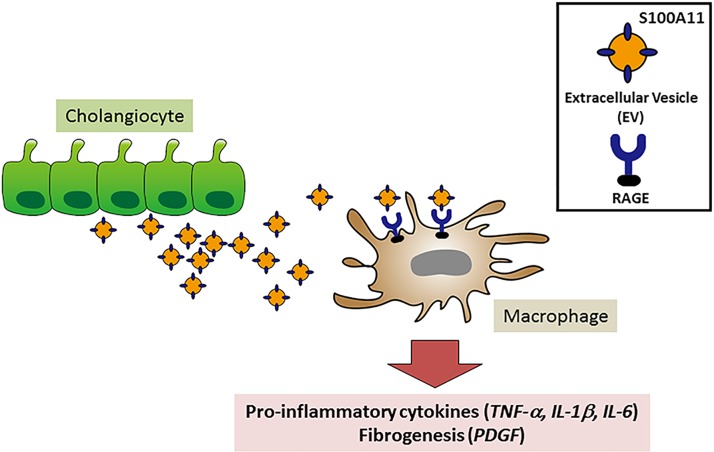
We verified our results obtained with cell lines in primary mouse cholangioids. These cholangioids of primary, nontransformed tissues grow as 3D structures, accurately recapitulate tissue architecture by providing cell-to-cell contact, and preserve cellular function. We do note, however, that the organoids are selected for replicating cells and, hence, may not model nonreplicating cells that may be found in peribiliary areas in vivo. In this context of cholangiocyte proliferation, we also identified chemokine expression by the organoids indicating they may be considered activated. Importantly, the primary mouse cholangiocyte-derived organoids release EVs containing S100A11 and stimulate proinflammatory cytokine expression in BMDM by a RAGE inhibitable pathway. Hence, our observations do not appear to be an artifact of studying cell lines. In vivo confirmation of EV biology has proven challenging, but recently zebra fish embryos have been demonstrated to be a suitable in vivo model for studying the fate of EV (13, 36). These studies confirm that EVs are targeted to monocytes and macrophages in vivo and promote a proinflammatory phenotype (Fig. 6).
Fig. 6.
Schematic depiction of proposed cross talk between activated cholangiocytes and macrophages. Activated cholangiocytes release extracellular vesicles (EVs) containing the damage-associated molecular pattern S100A11 as cargo, which activate bone marrow-derived macrophages following the binding of S100A11 to the receptor for advanced glycation end products (RAGE) receptor, triggering a signaling cascade that culminates in activation of NF-κB and NF-κB-mediated upregulation of proinflammatory cytokines and profibrogenic factors.
The current studies have potential therapeutic implications for human cholestatic liver injury. In particular, RAGE activation by DAMPs has been implicated in hepatic fibrosis, ductular reaction, and hepatocarcinogenesis (5, 9, 17). Given that RAGE inhibitors have been developed for human use and appear to be well tolerated, such inhibitors may be useful for the treatment of human cholangiopathies.
GRANTS
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant P30 DK084567 (to the optical microscopy core of the Mayo Clinic Center for Cell Signaling in Gastroenterology), National Cancer Institute Cancer Center Support Grant 5P30 CA15083-43C1 (to the Mayo Clinic Medical Genome Facility–Proteomics Core), and the Chris M. Carlos and Catharine Nicole Jockisch Carlos Endowment Fund in Primary Sclerosing Cholangitis (PSC).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
T.K., M.E.G., and G.J.G. conceived and designed research; T.K., A.A., and S.F.B. performed experiments; T.K., M.E.G., A.K., and G.J.G. interpreted results of experiments; M.E.G., A.K., and G.J.G. analyzed data; T.K. prepared figures; M.E.G. and G.J.G. drafted manuscript; M.E.G. and G.J.G. edited and revised manuscript; T.K., M.E.G., A.A., S.F.B., A.K., and G.J.G. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Ann Marie Schmidt (New York University School of Medicine) for providing the Rage−/− mice and Courtney Hoover for secretarial assistance.
REFERENCES
- 1.Banales JM, Huebert RC, Karlsen T, Strazzabosco M, LaRusso NF, Gores GJ. Cholangiocyte pathobiology. Nat Rev Gastroenterol Hepatol 16: 269–281, 2019. doi: 10.1038/s41575-019-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarró LM, Bradshaw CR, Allen GE, Arnes-Benito R, Sidorova O, Gaspersz MP, Georgakopoulos N, Koo BK, Dietmann S, Davies SE, Praseedom RK, Lieshout R, IJzermans JN, Wigmore SJ, Saeb-Parsy K, Garnett MJ, van der Laan LJ, Huch M. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med 23: 1424–1435, 2017. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collett GP, Redman CW, Sargent IL, Vatish M. Endoplasmic reticulum stress stimulates the release of extracellular vesicles carrying danger-associated molecular pattern (DAMP) molecules. Oncotarget 9: 6707–6717, 2018. doi: 10.18632/oncotarget.24158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faubion WA, Guicciardi ME, Miyoshi H, Bronk SF, Roberts PJ, Svingen PA, Kaufmann SH, Gores GJ. Toxic bile salts induce rodent hepatocyte apoptosis via direct activation of Fas. J Clin Invest 103: 137–145, 1999. doi: 10.1172/JCI4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge X, Arriazu E, Magdaleno F, Antoine DJ, Dela Cruz R, Theise N, Nieto N. High mobility group box-1 drives fibrosis progression signaling via the receptor for advanced glycation end products in mice. Hepatology 68: 2380–2404, 2018. doi: 10.1002/hep.30093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grubman SA, Perrone RD, Lee DW, Murray SL, Rogers LC, Wolkoff LI, Mulberg AE, Cherington V, Jefferson DM. Regulation of intracellular pH by immortalized human intrahepatic biliary epithelial cell lines. Am J Physiol 266: G1060–G1070, 1994. doi: 10.1152/ajpgi.1994.266.6.G1060. [DOI] [PubMed] [Google Scholar]
- 7.Guicciardi ME, Mott JL, Bronk SF, Kurita S, Fingas CD, Gores GJ. Cellular inhibitor of apoptosis 1 (cIAP-1) degradation by caspase 8 during TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. Exp Cell Res 317: 107–116, 2011. doi: 10.1016/j.yexcr.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guicciardi ME, Trussoni CE, Krishnan A, Bronk SF, Lorenzo Pisarello MJ, O’Hara SP, Splinter PL, Gao Y, Vig P, Revzin A, LaRusso NF, Gores GJ. Macrophages contribute to the pathogenesis of sclerosing cholangitis in mice. J Hepatol 69: 676–686, 2018. doi: 10.1016/j.jhep.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hernandez C, Huebener P, Pradere JP, Antoine DJ, Friedman RA, Schwabe RF. HMGB1 links chronic liver injury to progenitor responses and hepatocarcinogenesis. J Clin Invest 128: 2436–2451, 2018. doi: 10.1172/JCI91786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsova P, Ibrahim SH, Verma VK, Morton LA, Shah VH, LaRusso NF, Gores GJ, Malhi H. Extracellular vesicles in liver pathobiology: small particles with big impact. Hepatology 64: 2219–2233, 2016. doi: 10.1002/hep.28814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, Ellis E, van Wenum M, Fuchs SA, de Ligt J, van de Wetering M, Sasaki N, Boers SJ, Kemperman H, de Jonge J, Ijzermans JN, Nieuwenhuis EE, Hoekstra R, Strom S, Vries RR, van der Laan LJ, Cuppen E, Clevers H. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160: 299–312, 2015. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hudson BI, Lippman ME. Targeting RAGE signaling in inflammatory disease. Annu Rev Med 69: 349–364, 2018. doi: 10.1146/annurev-med-041316-085215. [DOI] [PubMed] [Google Scholar]
- 13.Hyenne V, Ghoroghi S, Collot M, Bons J, Follain G, Harlepp S, Mary B, Bauer J, Mercier L, Busnelli I, Lefebvre O, Fekonja N, Garcia-Leon MJ, Machado P, Delalande F, López AA, Silva SG, Verweij FJ, van Niel G, Djouad F, Peinado H, Carapito C, Klymchenko AS, Goetz JG. Studying the fate of tumor extracellular vesicles at high spatiotemporal resolution using the zebrafish embryo. Dev Cell 48: 554–572.e7, 2019. doi: 10.1016/j.devcel.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Ibrahim SH, Hirsova P, Gores GJ. Non-alcoholic steatohepatitis pathogenesis: sublethal hepatocyte injury as a driver of liver inflammation. Gut 67: 963–972, 2018. doi: 10.1136/gutjnl-2017-315691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibrahim SH, Hirsova P, Tomita K, Bronk SF, Werneburg NW, Harrison SA, Goodfellow VS, Malhi H, Gores GJ. Mixed lineage kinase 3 mediates release of C-X-C motif ligand 10-bearing chemotactic extracellular vesicles from lipotoxic hepatocytes. Hepatology 63: 731–744, 2016. doi: 10.1002/hep.28252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karin M, Clevers H. Reparative inflammation takes charge of tissue regeneration. Nature 529: 307–315, 2016. doi: 10.1038/nature17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khambu B, Huda N, Chen X, Antoine DJ, Li Y, Dai G, Köhler UA, Zong WX, Waguri S, Werner S, Oury TD, Dong Z, Yin XM. HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers. J Clin Invest 128: 2419–2435, 2018. doi: 10.1172/JCI91814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazaridis KN, LaRusso NF. The cholangiopathies. Mayo Clin Proc 90: 791–800, 2015. doi: 10.1016/j.mayocp.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lazaridis KN, LaRusso NF. Primary sclerosing cholangitis. N Engl J Med 375: 1161–1170, 2016. doi: 10.1056/NEJMra1506330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leclerc E, Fritz G, Vetter SW, Heizmann CW. Binding of S100 proteins to RAGE: an update. Biochim Biophys Acta 1793: 993–1007, 2009. doi: 10.1016/j.bbamcr.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 21.Lu WY, Bird TG, Boulter L, Tsuchiya A, Cole AM, Hay T, Guest RV, Wojtacha D, Man TY, Mackinnon A, Ridgway RA, Kendall T, Williams MJ, Jamieson T, Raven A, Hay DC, Iredale JP, Clarke AR, Sansom OJ, Forbes SJ. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol 17: 971–983, 2015. doi: 10.1038/ncb3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malhi H, Kropp EM, Clavo VF, Kobrossi CR, Han J, Mauer AS, Yong J, Kaufman RJ. C/EBP homologous protein-induced macrophage apoptosis protects mice from steatohepatitis. J Biol Chem 288: 18624–18642, 2013. doi: 10.1074/jbc.M112.442954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 21: 9–17, 2019. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 24.Morland CM, Fear J, Joplin R, Adams DH. Inflammatory cytokines stimulate human biliary epithelial cells to express interleukin-8 and monocyte chemotactic protein-1. Biochem Soc Trans 25: 232S, 1997. doi: 10.1042/bst025232s. [DOI] [PubMed] [Google Scholar]
- 25.Morland CM, Fear J, McNab G, Joplin R, Adams DH. Promotion of leukocyte transendothelial cell migration by chemokines derived from human biliary epithelial cells in vitro. Proc Assoc Am Physicians 109: 372–382, 1997. [PubMed] [Google Scholar]
- 26.Park SJ, Kim JM, Kim J, Hur J, Park S, Kim K, Shin HJ, Chwae YJ. Molecular mechanisms of biogenesis of apoptotic exosome-like vesicles and their roles as damage-associated molecular patterns. Proc Natl Acad Sci USA 115: E11721–E11730, 2018. doi: 10.1073/pnas.1811432115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Planas-Paz L, Sun T, Pikiolek M, Cochran NR, Bergling S, Orsini V, Yang Z, Sigoillot F, Jetzer J, Syed M, Neri M, Schuierer S, Morelli L, Hoppe PS, Schwarzer W, Cobos CM, Alford JL, Zhang L, Cuttat R, Waldt A, Carballido-Perrig N, Nigsch F, Kinzel B, Nicholson TB, Yang Y, Mao X, Terracciano LM, Russ C, Reece-Hoyes JS, Gubser Keller C, Sailer AW, Bouwmeester T, Greenbaum LE, Lugus JJ, Cong F, McAllister G, Hoffman GR, Roma G, Tchorz JS. YAP, but not RSPO-LGR4/5, signaling in biliary epithelial cells promotes a ductular reaction in response to liver injury. Cell Stem Cell 25: 39–53.e10, 2019. doi: 10.1016/j.stem.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200: 373–383, 2013. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Safronova A, Araujo A, Camanzo ET, Moon TJ, Elliott MR, Beiting DP, Yarovinsky F. Alarmin S100A11 initiates a chemokine response to the human pathogen Toxoplasma gondii. Nat Immunol 20: 64–72, 2019. doi: 10.1038/s41590-018-0250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakaguchi M, Sonegawa H, Murata H, Kitazoe M, Futami J, Kataoka K, Yamada H, Huh NH. S100A11, an dual mediator for growth regulation of human keratinocytes. Mol Biol Cell 19: 78–85, 2008. doi: 10.1091/mbc.e07-07-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato K, Marzioni M, Meng F, Francis H, Glaser S, Alpini G. Ductular reaction in liver diseases: pathological mechanisms and translational significances. Hepatology 69: 420–430, 2019. doi: 10.1002/hep.30150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaefer L. Complexity of danger: the diverse nature of damage-associated molecular patterns. J Biol Chem 289: 35237–35245, 2014. doi: 10.1074/jbc.R114.619304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shelke GV, Lässer C, Gho YS, Lötvall J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J Extracell Vesicles 3: 24783, 2014. doi: 10.3402/jev.v3.24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabibian JH, Masyuk AI, Masyuk TV, O’Hara SP, LaRusso NF. Physiology of cholangiocytes. Compr Physiol 3: 541–565, 2013. doi: 10.1002/cphy.c120019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tacke F, Zimmermann HW. Macrophage heterogeneity in liver injury and fibrosis. J Hepatol 60: 1090–1096, 2014. doi: 10.1016/j.jhep.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 36.Verweij FJ, Revenu C, Arras G, Dingli F, Loew D, Pegtel DM, Follain G, Allio G, Goetz JG, Zimmermann P, Herbomel P, Del Bene F, Raposo G, van Niel G. Live tracking of inter-organ communication by endogenous exosomes in vivo. Dev Cell 48: 573–589.e4, 2019. doi: 10.1016/j.devcel.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Xia C, Braunstein Z, Toomey AC, Zhong J, Rao X. S100 proteins as an important regulator of macrophage inflammation. Front Immunol 8: 1908, 2018. doi: 10.3389/fimmu.2017.01908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zweers SJ, Shiryaev A, Komuta M, Vesterhus M, Hov JR, Perugorria MJ, de Waart DR, Chang JC, Tol S, Te Velde AA, de Jonge WJ, Banales JM, Roskams T, Beuers U, Karlsen TH, Jansen PL, Schaap FG. Elevated interleukin-8 in bile of patients with primary sclerosing cholangitis. Liver Int 36: 1370–1377, 2016. doi: 10.1111/liv.13092. [DOI] [PubMed] [Google Scholar]