Abstract
The aim of the study was to evaluate the ruminal function and microbial community of lamb under different nutrient levels. Sixty-four lambs with similarity body weight were randomly assigned to four groups after weaning off ewe’s milk on the 17th day (6.2 ± 0.2 kg). The lambs of the control group (CON) were fed a basal diet, and the other three groups were subjected to a diet of decreased protein (PR), digestible energy (ER) or both of them at 20% (BR) of basal diet. The decrease of dietary protein or energy level decreased the average daily gain, ruminal weight and mucosal thickness of lambs (P < 0.05). The total volatile fatty acid (TVFA), acetate and propionate concentration of the CON group were significantly higher than that of the other three groups. The dietary protein and energy level affected the microbial diversity, and low energy level increased the relative abundance of phyla of Fibrobacteres, whereas at the genus level, increased the relative abundance of Butyrivibrio and Prevotellaceae. Under different dietary energy and protein levels, 14 genera exhibited significant correlation with ruminal fermentation. These findings supplied new perspective for the understanding of the dietary effect on ruminal microbial community interactions and are of great significance for establishing the optimal nutrient supply strategy for lambs.
Subject terms: Microbial ecology, Animal physiology
Introduction
The rumen is a complex ecosystem that harbors a functional microbial population including bacteria, protozoa, archaea and fungi, which are important for the host’s nutrient uptake and energy metabolism1. The interaction between microorganisms and the host results in a symbiotic relationship that allows ruminants to digest diets rich in fiber2. Within this microbiome, bacteria are the dominant domain and make the greatest contribution to digestion and conversion of feed components to microbial proteins and volatile fatty acids (VFA), such as acetate, propionate, and butyrate during ruminal fermentation.
The microbiome is established as the development of rumen, and it is a widely held view in ruminant nutrition that rumen microbial populations highly adapt to different diets3,4. Studies in deer5,6, cows7,8, and lamb9 showed that interplay patterns between rumen bacterial community composition and metabolic phenotypes were altered by diets, and a large number of work have been performed to investigate how different dietary compositions and feeding programs can improve the ability of ruminal microbiota to degrade forages and feedstuffs for better animal performance and reduce the need for supplements. However, characteristic of ruminal fermentation and microbial community in response to different dietary nutritional levels are poorly understood in lamb.
In this study, we hypothesized that dietary with same ingredients but different nutritional levels could influence the development and function of rumen and microbial diversity of lamb. The aim of this research was to investigate the growth performance, ruminal fermentation and characterize the microbial composition and diversity of weaned lambs in response to dietary energy and protein levels based on high-throughput next generation sequencing. A better understanding of the correlation of nutritional level and ruminal development could provide the basis for the targeted improvement of nutrient levels in ruminants.
Results
Growth performance
No differences were detected in DM intake or nutrient intake of the milk replacer and starter among the four groups (P > 0.05; Table 1). The ADG and FCR of the lambs in BR group were significantly lower than that in the PR and ER group (P < 0.05), and there were no significant differences between PR and ER group (P > 0.05). The ADG and FCR of the three treatment group were significantly lower than the CON group (P < 0.01).
Table 1.
Effects of nutritional levels on growth performance of early-weaned Hu lambs.
| Items | Groups | SEM | P-value | |||
|---|---|---|---|---|---|---|
| CON | PR | ER | BR | |||
| Initial body weight | 6.29 | 6.19 | 6.22 | 6.20 | 0.07 | 0.8241 |
| Final body weight | 15.40 | 13.90 | 13.94 | 13.02 | 0.24 | 0.0062 |
| Average daily gain (g/d) | 228.80a | 186.56b | 197.72b | 168.55c | 6.19 | <0.0000 |
| Milk replacer intake (g/d) | 154.63 | 154.63 | 154.63 | 154.63 | - | - |
| Starter intake (g/d) | 325.96 | 322.26 | 337.22 | 320.26 | 3.36 | 0.3148 |
| Feed conversion ratio | 2.10a | 2.57b | 2.49b | 2.82c | 0.07 | 0.0002 |
a,b,cMeans within same row with the same superscript letter are not significantly different (P > 0.05).
Ruminal morphology and fermentation
The effects of energy and protein level on ruminal development and fermentation are shown in Table 2. The rumen weight of lambs in the CON group was significantly higher than the other three groups (P < 0.05). The decrease of protein or/and energy had no effect on the length and width of papillae of the rumen (P > 0.05). The mucosal thickness of rumen of group PR, ER, and BR was significantly lower than that of the CON group (P < 0.05).
Table 2.
Effects of nutritional levels on ruminal fermentation of early-weaned Hu lambs.
| Items | Groups | SEM | P-value | |||
|---|---|---|---|---|---|---|
| CON | PR | ER | BR | |||
| Rumen morphometrics | ||||||
| Rumen weight (g) | 327.9a | 285.73b | 241.48c | 237.43c | 11.49 | 0.0003 |
| Papillae Length (μm) | 3247.34 | 3107.44 | 2741.46 | 2945.79 | 129.70 | 0.595 |
| Papillae Width (μm) | 394.28 | 383.85 | 341.63 | 322.21 | 14.00 | 0.2195 |
| Mucosal thickness (μm) | 211.31a | 175.18b | 140.89b | 156.33b | 8.53 | 0.0059 |
| Rumen fermentation | ||||||
| PH | 6.78 | 6.89 | 6.96 | 6.95 | 0.16 | 0.8540 |
| NH3-N (mmol/L) | 17.47 | 15.60 | 13.42 | 12.39 | 3.60 | 0.8718 |
| TVFA (mmol/L) | 36.25a | 10.14b | 13.26b | 14.16b | 4.31 | 0.0055 |
| Acetate (mmol/L) | 18.23a | 5.84b | 5.27b | 7.78b | 1.70 | 0.0013 |
| Propionate (mmol/L) | 17.53a | 3.06b | 3.42b | 4.23b | 1.34 | 0.0001 |
| Butyrate (mmol/L) | 1.68a | 0.58b | 0.86b | 1.01ab | 0.18 | 0.0512 |
a,b,cMeans within same row with the same superscript letter are not significantly different (P > 0.05).
Ruminal pH and NH3-N concentrations showed no significant difference among the four groups (P > 0.05). The TVFA, acetate and propionate concentration of the CON group was significantly higher than that of the other three groups (P < 0.05). Compared to the CON group, butyrate concentration of PR, ER and BR group showed a decreased tendency (P = 0.0512). No significant difference was observed among the PR, ER, and BR group (P > 0.05).
Index of microbial community
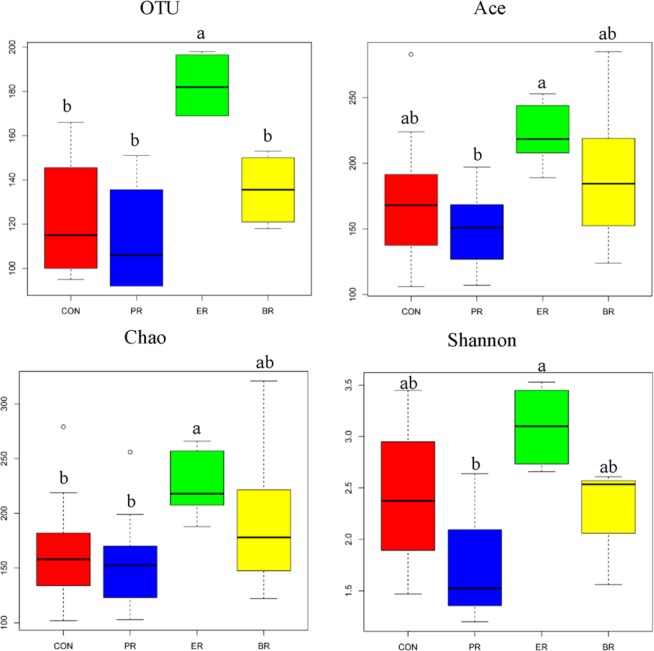
The saturation plateau of rarefaction curves indicated that the sampling effort had sufficient sequence coverage to accurately describe the bacterial composition of each group (Fig. S1). Based on OTU, indices of bacterial richness were estimated by the index of Ace and Chao, and indices of bacterial diversity were determined using the index of Simpson and Shannon. Dietary energy levels significantly altered the rumen bacterial community (Fig. 1). At the 0.03 dissimilarity level, the OTU numbers of ER group was significantly higher than the other three groups (P < 0.05). The indices of Chao and Shannon were significantly affected by the protein and energy levels (P < 0.05).
Figure 1.
Community richness estimates and diversity indices for different treatments.
The relative abundance and diversity of bacterial communities
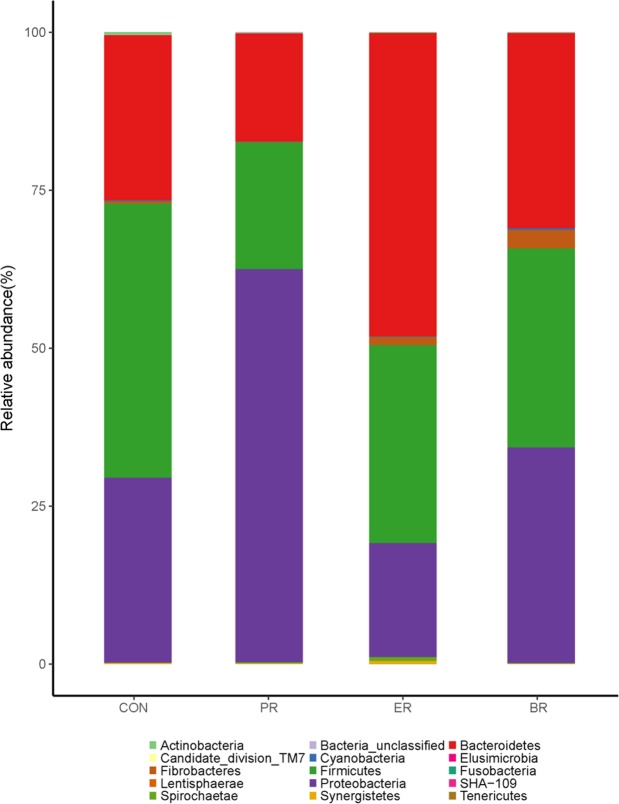
All sequences were classified from phylum to species based on the SILVA taxonomic database. Fifteen different phyla were detected in these samples. The four groups showed dissimilar taxonomic compositions at the phylum-level distributions and the major sequences obtained from the samples belonged to Firmicutes, Bacteroidetes, and Proteobacteria (Fig. 2, Table 3). The relative abundance of these predominant phyla varied considerably among the four groups. Compared with the CON and PR group, the phylum Bacteroidetes and Fibrobacteres were abundant in the samples taken from the ER and BR groups while the phylum Fibrobacteres of BR group was significantly higher than that of the other three groups (P < 0.05). The abundance of phylum Firmicutes and Proteobacteria showed no significant difference among the four groups while the abundance of phylum Proteobacteria showed an increased tendency (P = 0.0973). The other 9 phyla were relatively minor (<1% of total sequences) in abundance in comparison.
Figure 2.
Phylum-level composition of the rumen microbiome. A color-coded bar plot showing the average bacterial phylum distribution across the different age groups that were sampled.
Table 3.
Comparison of the dominant phylum (average relative abundance ≥1% for at least one group) within the rumen.
| Items | Groups | SEM | P-value | |||
|---|---|---|---|---|---|---|
| CON | PR | ER | BR | |||
| Bacteroidetes | 26.10 | 17.09 | 38.54 | 30.92 | 6.27 | 0.2038 |
| Fibrobacteres | 0.43b | 0.12b | 1.25b | 3.90a | 0.72 | 0.0325 |
| Firmicutes | 43.37 | 20.05 | 24.48 | 20.44 | 7.11 | 0.1374 |
| Proteobacteria | 29.26 | 62.26 | 18.04 | 34.15 | 11.45 | 0.0973 |
a,bValues in the same row with different superscripts differ significantly (P < 0.05).
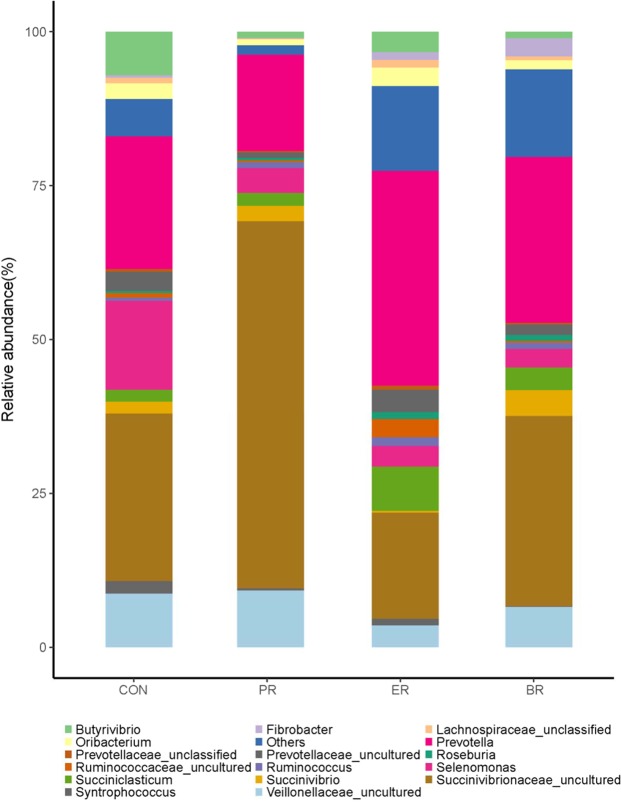
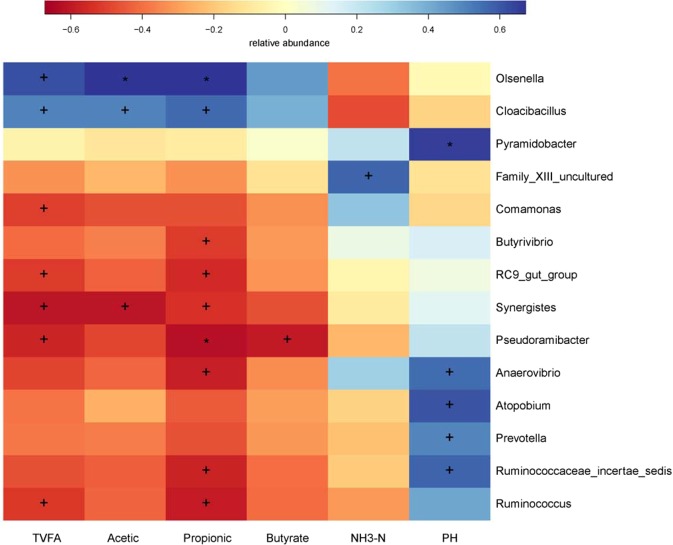
At the genus level, 103 genera were detected in the samples. The most abundant genera (with a relative abundance ≥2% of the four libraries) were used to determine which bacteria might be the most important (Fig. 3, Table 4). The most abundant taxa of the CON group included Prevotella, Selenomonas, Succiniclasticum, as well as the unclassified taxa derived from Succinivibrionaceae (family) and Veillonellaceae (family). In the PR group, the Succinivibrionaceae_uncultured was predominant with an abundance of 59.62%, followed by Prevotella, Veillonellaceae_uncultured, Selenomonas, Succiniclasticum and Succinivibrio. In the ER group, the most dominant genera were Prevotella, Succinivibrionaceae_uncultured, Succiniclasticum, Veillonellaceae_uncultured, Prevotellaceae_uncultured and Butyrivibrio, which together accounted for 69.89% of the total sequences. The abundance of genus Butyrivibrio was abundant in the samples taken from the ER group when compared with the CON and PR groups. The samples of both protein and digestible energy reduced group showed that most of the dominant taxa were Succinivibrionaceae_uncultured, Prevotella, Veillonellaceae_uncultured, Selenomonas, Succinivibrio, and Fibrobacter. We evaluated the correlation between the ruminal fermentation parameters and bacteria at genus level by performing Spearman correlation analysis. Almost 14 genera were exhibited significant correlation with TVFA, acetic, butyrate, propionic, NH3-N and pH, respectively (Fig. 4).
Figure 3.
Phylum-level composition of the rumen microbiome. A color-coded bar plot showing the average bacterial phylum distribution across the different age groups that were sampled.
Table 4.
Comparison of the dominant genus (average relative abundance ≥2% for at least one group) within the rumen.
| Taxa | Groups | SEM | P-value | |||
|---|---|---|---|---|---|---|
| CON | PR | ER | BR | |||
| Succinivibrionaceae_uncultured | 35.44 | 59.62 | 22.96 | 41.16 | 11.15 | 0.2473 |
| Prevotella | 25.18 | 15.66 | 34.84 | 26.95 | 6.44 | 0.2703 |
| Veillonellaceae_uncultured | 11.56 | 9.25 | 4.75 | 6.58 | 3.86 | 0.7089 |
| Selenomonas | 14.43 | 4.03 | 3.34 | 3.02 | 0.61 | 0.2723 |
| Succiniclasticum | 1.96 | 2.07 | 7.19 | 3.67 | 1.61 | 0.1294 |
| Butyrivibrio | 0.29b | 0.34b | 1.43a | 1.06ab | 0.21 | 0.0187 |
| Prevotellaceae_uncultured | 3.12a | 0.92b | 3.68a | 1.72ab | 0.65 | 0.0404 |
| Succinivibrio | 1.91 | 2.53 | 0.27 | 4.21 | 1.59 | 0.4658 |
| Oribacterium | 2.59 | 0.97 | 3.01 | 1.44 | 1.13 | 0.6016 |
| Fibrobacter | 0.43 | 0.12 | 1.25 | 2.93 | 0.86 | 0.1798 |
| Ruminococcaceae_uncultured | 0.72 | 0.27 | 2.95 | 0.28 | 1.11 | 0.3083 |
| Syntrophococcus | 2.03 | 0.36 | 1.05 | 0.14 | 0.88 | 0.4530 |
a,bValues in the same row with different superscripts differ significantly (P < 0.05).
Figure 4.
Spearman correlation analysis of VFA and microbiome at genus level. The depth of the color indicates the correlation between species and environmental factors. The “+” and “*” indicates the different level at 0.05 and 0.01, respectively.
Discussion
In this study, we evaluated the growth performance, ruminal development and fermentation, and microbial diversity of weaned lambs under different dietary energy and protein levels. Hossain et al. (2003) and Negesse et al. (2001) reported that energy and protein level of diets influenced the growth performance and N retention of goats, respectively9–11. Results from this study indicated that a lower level of protein, energy, or both protein and energy significantly decreased the average daily gain and feed efficiency. This result was consisted with the fact that high protein and energy intake was necessary to meet the demand of rapid growth of lamb. Atti et al. (2004) reported that lambs supplemented with high protein level diet exhibited higher growth rates than those fed diets of low protein level during the first 6 weeks12. Previous study indicated that high concentrate level (600 g) of diet improved the ADG and FCR of lambs13.
The establishment of rumen function is the symbol of the transition from functioning as a monogastric to a ruminant14. Rumen is the dominant place where bacteria contributed to digest and convert plant materials to volatile fatty acids and microbial proteins. The development of rumen is critical for the utilization of solid feed. In this study, low protein and energy diet significantly decreased the rumen weight and mucosal thickness. This finding is in consistent with the fact that young animals may require high protein and energy intakes to ensure the organ development. These observations are consistent with analogous trials on digestive tract development. In ruminants, gastrointestinal tissues are affected by changes in ME intake14, protein intake, as well as dietary energy density15,16. Most studies report height and width of ruminal papillae as an estimate of ruminal epithelium growth17. Length, uniformity, and general appearance of papillae are greatly influenced by composition and particle size of the diet. Previous studies showed that encouraging solid feed intake by restricting milk access increases the size of the rumen and the morphological structure of rumen papillae18. Shen et al. (2004) reported that an energy-rich diet caused ruminal papillae proliferation in young goats19. Wan et al. (2012) proved that the high concentrate level significantly increased both ruminal papillae length and width, compared with the hay and low concentrate feeding group20. In our research, decrease of protein or energy decreased the length and width of papillae, but no obvious difference was observed among the groups. The insufficiently limitation of protein or energy at 20% of basal diet might account for this fact. Previous studies have confirmed the VFA concentration tended to increase with increasing dietary energy level by adding more non-structure carbohydrates (NSC)21. Compared to the control group, low energy or protein level decreased the concentration of volatile fatty acid (VFA). These results are consistent with other researches22.
Ruminal microorganisms and host have evolved together for millions of years. The primary function of ruminal microbiome is the conversion of fibre into digestible compounds for the utilization of the ruminants. In the present study, 16S rRNA sequencing method was selected to evaluate the diversity of the ruminal bacterial community of lambs under different dietary energy and protein levels. The results of the present study revealed that Bacteroidetes, Firmicutes and Proteobacteria were the dominant phyla among the groups. Our findings were in agreement with previous studies that Bacteroidetes and Firmicutes are numerically the most dominant phyla in the microbiome of terrestrial mammals23–26.
The current research found that the genera Succinivibrionaceae_uncultured, Prevotella, Veillonellaceae_uncultured, Selenomonas, Succiniclasticum, and Butyrivibrio dominated in the four groups. Prevotella has been reported as the most abundant genus in the rumen of adult cows, which is related to ruminal carbohydrate and protein fermentation27. This research showed that the proportion of Prevotellaceae_uncultured in the PR group was significantly lower than that in the CON and ER group. Mao et al. (2012) indicated the abundance of Prevotella was highly correlated with the content of CP7. Since the content of CP in the alfalfa hay is greater than in the rice straw, this provided a reasonable explanation for the higher abundance of Prevotella observed in the alfalfa samples28. In this study, the Butyrivibrio abundance of ER and BR group was higher than the CON and PR group. The Butyrivibrio species which involved in a number of ruminal functions are common in the ruminants, such as deer, cows and sheep29. Of particular importance to ruminant digestion, the degradation of structural carbohydrates of plant materials is the most important role of Butyrivibrio. The dietary crude fiber content of ER and BR group was more than twice when compared to the CON and PR group, and the increased fiber level might be the reason for the high abundance of Butyrivibrio.
In conclusion, the results presented here provide new information regarding the effects of different dietary energy and protein levels on growth performance and ruminal development and microbiota communities. Low level of protein or energy retarded the growth performance and ruminal development of lambs. Low dietary energy levels significantly decreased the concentration of volatile fatty acids. Based on 16S rRNA gene sequencing method, this study indicated the changes of the overall composition of the bacterial communities in the rumen ecosystem. The dietary protein and energy level affected the microbial diversity and the low level of energy increased the relative abundance of Fibrobacteres at the phylum level and increased the relative abundance of Butyrivibrio and Prevotellaceae at the genus level, and 14 genera exhibited significant correlation with ruminal fermentation. These findings are of great importance for the targeted improvement of nutrient levels in ruminants.
Materials and Methods
Statement
This research was conducted at the Hai Lun sheep industry Co., Ltd., Jiangsu, China (latitude 32.30′N, longitude 119.54′E). All experiments were performed in accordance with relevant guidelines and regulations. The experimental protocol and all methods were approved by the Animal Ethics Committee of Chinese Academy of Agricultural Sciences, and humane animal care and handling procedures were performed throughout the experiment (AEC-CAAS-FRI-CAAS20180602).
Animals, diets and management
Sixty-four lambs with an average birth weight of 2.5 ± 0.2 kg were randomly divided into four groups (n = 16/group) after weaning off ewe’s milk on the 17th day (6.2 ± 0.2 kg), with 4 replicates of each group and 4 lambs per replicate. The control group (CON) was fed a basal diet and the other three groups were subjected to a diet of decreased protein (PR), digestible energy (ER) or both protein and digestible energy at 20% (BR) of basal diet. Each group was fed the assigned milk replacer and starter from 21 to 60 days after 4 days transitional period of milk replacer and pelleted starter (Diameter, 4 mm; Length, 10 mm). These lambs were fed 3 times daily (08:00, 12:00, and 18:00 h) from day 21 to 30 and then twice daily (09:00, 18:00 h) from day 31 to 60, and the feed amount of milk replacer was adjusted in direct accordance with 2% of the lamb’s body weight. Milk replacer consumption was equal in all four groups. The pelleted starter supplement of the CON group was assigned to be fed ad libitum, and the feed amounts of the other three groups were according to the intake of the CON group. The feed intake of pelleted starter was kept consistent among the four groups. The components and chemical composition of the milk replacer and pelleted starter are presented in Table 5.
Table 5.
Composition and nutrient levels of milk replacer and starters (DM basis).
| Items | Milk replacer | Starter | ||||||
|---|---|---|---|---|---|---|---|---|
| CON | PR | ER | BR | CON | PR | ER | BR | |
| Ingredients | ||||||||
| Corn | — | — | — | — | 53 | 62 | 25 | 38 |
| Soybean meal | — | — | — | — | 27 | 14 | 27 | 16 |
| Powdered rice hulls | — | — | — | — | 0 | 0 | 16 | 17 |
| Wheat bran | — | — | — | — | 6 | 10 | 18 | 15 |
| Premixa | — | — | — | — | 4 | 4 | 4 | 4 |
| Alfalfa meal | — | — | — | — | 10 | 10 | 10 | 10 |
| Total | — | — | — | — | 100 | 100 | 100 | 100 |
| Nutrient levels b | ||||||||
| DM | 94.35 | 94.51 | 93.33 | 93.48 | 86.59 | 86.50 | 87.35 | 87.25 |
| CP | 24.21 | 19.13 | 24.45 | 19.26 | 20.80 | 16.35 | 20.68 | 16.10 |
| ME,MJ/Kg | 14.83 | 14.77 | 12.55 | 12.55 | 10.59 | 10.61 | 8.52 | 8.52 |
| EE | 20.58 | 20.78 | 11.06 | 10.92 | 2.89 | 3.12 | 2.67 | 2.83 |
| Ash | 4.99 | 4.84 | 4.81 | 4.88 | 9.71 | 9.82 | 9.85 | 9.82 |
| NDF | — | — | — | — | 18.49 | 18.18 | 28.89 | 28.28 |
| Calcium | 0.95 | 0.95 | 0.95 | 0.95 | 0.41 | 0.40 | 0.51 | 0.46 |
| Phosphorus | 0.68 | 0.68 | 0.68 | 0.68 | 0.24 | 0.21 | 0.26 | 0.22 |
aThe premix provided the following nutrients per kilogram of the diet: VA 12000IU, VD 20 00IU, VE 30IU, Cu 12 mg, Fe 64 mg, Mn 56 mg, Zn 60 mg, I 1.2 mg, Se 0.4 mg, Co 0.4 mg, Ca 3.2 g, P 1.2 g, NaCl 6.4 g.
bNutrient levels are all measured values except ME. ME of milk replacer and starter was calculated according to Tables of Feed Composition and Nutritive Values in China 2012 and Feeding Standard of sheep(NY/T 816-2004); DM, dry matter; CP, crude protein; ME, metabolic energy; EE, ether extract; NDF, neutral detergent fiber.
Chemical analyses
The lambs were weighed at 20, 40, and 60 days, and the intake of milk replacer and starter feed was measured daily to calculate the average daily gain (ADG), average daily feed intake (ADFI), and feed conversion ratio (FCR).
The composition and nutrient levels of milk replacer and starters were analyzed according to the official methods of analysis (Association of Official Analytical Chemists: Washington, DC)30. The dry matter was determined by drying the samples in an oven at 105 °C for 24 h (method 930.15; AOAC1990). The nitrogen (N) content was determined by the Kjeldahl method and crude protein (CP) was calculated as 6.25 × N (method 984.13; AOAC1990). The ether extract was measured by the weight loss of the dry matter upon extraction with diethyl ether in a Soxhlet extraction apparatus for 8 h (method 920.85; AOAC 1990). The contents of ash (550 °C in a muffle furnace for 6 h, method 942.05; AOAC1990), neutral detergent fiber (NDF; method 962.09; AOAC 1990) and gross energy (GE; Bomb calorimeter, C200; IKA Works Inc., Staufen, Germany) were determined using appropriate protocols. The calcium (Ca) was analysed using an atomic absorption spectrophotometer (M9 W-700; Perkin-Elmer Corp., Norwalk, CT, USA; method 968.08; AOAC1990). The phosphorus (P) was analysed by the molybdovanadatecolourimetric method (method 965.17; AOAC1990) using a spectrophotometer (UV-6100; Mapada Instruments Co., Ltd, Shanghai, China).
Ruminal morphometrics
Four lambs (healthy and bodyweight close to the average bodyweight of the group) from each group were selected at the age of 60 days and slaughtered to collect the ruminal tissues and digesta. A 2-cm2 fragment of each rumen was collected from ventral sac and fixed in 4% paraformaldehyde (Sigma-Aldrich) for histological assessment. The samples were dehydrated with an ethanol and toluene (Beijing Chemical Works) series and embedded in paraffin (Leica, Wetzlar, Germany). Serial sections (6 μm thickness) were mounted on gelatin-coated glass slides and stained with hematoxylin and eosin (H&E)31. For histomorphometry, five ruminal papillae from each section were examined and measured (in µm) considering their papillae height, papillae width, and mucosal thickness. Both measurements were taken at 200x utilizing an Olympus BX51 photomicroscope, equipped with a DP software for the image analysis (Olympus, Italy).
Ruminal fermentation parameters
Samples of ruminal digesta were collected at the age of 60 days after slaughtered. The pH of the ruminal fluid was determined using a pH metre (Model 144 PB-10, Sartorius Co., Germany). Approximately 10-ml sample of the strained fluid was collected and stored frozen at −20 °C for analysis of VFA and NH3-N after acidified with 2 ml 25% (w/v) metaphosphoric acid. The concentration of VFA in ruminal fluid was measured by gas chromatography (GC) using methyl valerate as an internal standard in an Agilent 6890 series GC equipped with a capillary column (30 m x0.53 mm internal diameter, film thickness 1/-Lm)32. Ammonia-N was measured with the method of the colorimetric method described by Ma et al. (2015)33.
DNA extraction, amplification of 16S rRNA and Hiseq sequencing
Microbial DNA was extracted after ruminal samples were thoroughly homogenized with a commercial DNA extraction Kit (Omega Bio-tek, Norcross, GA, U.S.) according to manufacturer’s protocols. Amplification and sequencing were performed as described by Mao et al. (2012)7. The V3-V4 region of the bacteria 16S ribosomal RNA genes were amplified using primers 338F (5’-barcode-ACTCCTRCGGGAGGCAGCAG)-3’ and 806R (5’-GGACTACCVGGGTATCTAAT-3’), where the barcode is an eight-base sequence unique to each sample.
Amplicons were purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, U.S.) according to the manufacturer’s instructions and quantified using QuantiFluor™ -ST (Promega, U.S.). Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 250) on an Illumina HiSeq platform (Illumina Inc., San Diego, CA) according to the standard protocols.
Statistical and bioinformatics analysis
As described by Caporaso et al. (2010), the raw Illumina sequences data were demultiplexed, quality filtered, and analyzed using the Quantitative Insights into Microbial Ecology (QIIME, v.1.8.0)34. The assembled sequences were assigned to operational taxonomic units (OTU) at a 97% identity level using UPARSE after quality control35. The phylogenetic affiliation of 16S rRNA gene sequence was analyzed against the SILVA (SSU115) 16S rRNA database using Ribosomal Database Project (RDP) Classifier (http://rdp.cme.msu.edu/) with a confidence threshold of 70%36,37. Rarefaction curves, α diversity, and β diversity calculations were also performed using QIIME38. Spearman correlation analysis between bacterial and ruminal fermentation parameters was performed using R corrplot39. The sequencing data of this research was submitted to the Sequence Read Archive (SRA) with an accession number of SRP145573.
Differences in growth performance and ruminal morphology, fermentation parameters, and microbiota diversity among the four groups were analyzed using the SAS (version 9.1, SAS Institute, Inc., Cary, NC, USA; 2004) general linear model (GLM). The following model was fitted to the data: Yi = μ + αi + ei, where Yi is the dependent variable; μ represents the overall mean; αi represents the fixed effect of treatment, and ei is the random residual error due to the replicate. Duncan’s Multiple Range Test was used to the statistical differences analysis among the means of the treatments. Treatment differences with P < 0.05 were considered statistically significant while 0.05 ≤ P < 0.10 was defined as a tendency.
Acknowledgements
The National Key R&D Program of China (2018YFD0502104) supported this research. We are thankful to Realbio Genomics Institute (Shanghai, China) for 16S rRNA gene sequencing services. The authors declare that no conflict of interest.
Author contributions
Conceived and designed the experiments: N.F.Z. and Q.Y.D. Performed the experiments: K.C. and M.L.Q. Analyzed the data: K.C. Contributed reagents/materials/analysis tools: K.C. Wrote and revised the paper: K.C. and S.Q.W. All authors approved the manuscript before submission.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kai Cui and Minli Qi.
References
- 1.Han X, et al. Rumen Bacterial Diversity of 80 to 110-Day-Old Goats Using 16S rRNA Sequencing. Plos One. 2015;10:e117811. doi: 10.1371/journal.pone.0117811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mackie RI. Mutualistic fermentative digestion in the gastrointestinal tract: diversity and evolution. Integr Comp Biol. 2002;42:319. doi: 10.1093/icb/42.2.319. [DOI] [PubMed] [Google Scholar]
- 3.Mullins CR, et al. Analysis of rumen microbial populations in lactating dairy cattle fed diets varying in carbohydrate profiles and Saccharomyces cerevisiae fermentation product. J Dairy Sci. 2013;96:5872. doi: 10.3168/jds.2013-6775. [DOI] [PubMed] [Google Scholar]
- 4.Pitta DW, et al. Bacterial diversity dynamics associated with different diets and different primer pairs in the rumen of Kankrej cattle. Plos One. 2014;9:e111710. doi: 10.1371/journal.pone.0111710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z, et al. Bacterial community composition and fermentation patterns in the rumen of sika deer (Cervus nippon) fed three different diets. Microb Ecol. 2015;69:307. doi: 10.1007/s00248-014-0497-z. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez-Sanabria E, et al. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl Environ Microbiol. 2012;78:1203. doi: 10.1128/AEM.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao S, Zhang R, Wang D, Zhu W. The diversity of the fecal bacterial community and its relationship with the concentration of volatile fatty acids in the feces during subacute rumen acidosis in dairy cows. Bmc Vet Res. 2012;8:237. doi: 10.1186/1746-6148-8-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu HW, et al. Effects of Suaeda glauca crushed seed on rumen microbial populations, ruminal fermentation, methane emission, and growth performance in Ujumqin lambs. Anim Feed Sci Tech. 2015;210:104. doi: 10.1016/j.anifeedsci.2015.10.007. [DOI] [Google Scholar]
- 9.Hossain, M.E., Shahjalal, M., Khan, M.J. & Hasanat, M.S. Effect of Dietary Energy Supplementation on Feed Intake, Growth and Reproductive Performance of Goats under Grazing Condition. Pakistan Journal of Nutrition (2003).
- 10.Negesse T, Rodehutscord M, Pfeffer E. The effect of dietary crude protein level on intake, growth, protein retention and utilization of growing male Saanen kids. Small Rumin Res. 2001;39:243. doi: 10.1016/S0921-4488(00)00193-0. [DOI] [PubMed] [Google Scholar]
- 11.Yerradoddi RR, et al. Effect of protein and energy levels in sweet sorghum bagasse leaf residue-based diets on the performance of growing Deccani lambs. Trop Anim Health Prod. 2015;47:743. doi: 10.1007/s11250-015-0788-5. [DOI] [PubMed] [Google Scholar]
- 12.Atti N, Rouissi H, Mahouachi M. The effect of dietary crude protein level on growth, carcass and meat composition of male goat kids in Tunisia. Small Ruminant Res. 2004;54:89. doi: 10.1016/j.smallrumres.2003.09.010. [DOI] [Google Scholar]
- 13.Majdoub-Mathlouthi L, Sa DB, Say A, Kraiem K. Effect of concentrate level and slaughter body weight on growth performances, carcass traits and meat quality of Barbarine lambs fed oat hay based diet. Meat Sci. 2013;93:557. doi: 10.1016/j.meatsci.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Steele MA, Penner GB, Chaucheyras-Durand F, Guan LL. Development and physiology of the rumen and the lower gut: Targets for improving gut health. J Dairy Sci. 2016;99:4955. doi: 10.3168/jds.2015-10351. [DOI] [PubMed] [Google Scholar]
- 15.Wester TJ, et al. Differential effects of plane of protein or energy nutrition on visceral organs and hormones in lambs. J Anim Sci. 1995;73:1674. doi: 10.2527/1995.7361674x. [DOI] [PubMed] [Google Scholar]
- 16.Baldwin RL. The proliferative actions of insulin, insulin-like growth factor-I, epidermal growth factor, butyrate and propionate on ruminal epithelial cells in vitro. Small Ruminant Res. 1999;32:261. doi: 10.1016/S0921-4488(98)00188-6. [DOI] [Google Scholar]
- 17.Steele MA, et al. Technical note: Three-dimensional imaging of rumen tissue for morphometric analysis using micro-computed tomography. J Dairy Sci. 2014;97:7691. doi: 10.3168/jds.2014-8374. [DOI] [PubMed] [Google Scholar]
- 18.Khan MA, Weary DM, von Keyserlingk MA. Invited review: effects of milk ration on solid feed intake, weaning, and performance in dairy heifers. J Dairy Sci. 2011;94:1071. doi: 10.3168/jds.2010-3733. [DOI] [PubMed] [Google Scholar]
- 19.Shen Z, et al. An energy-rich diet causes rumen papillae proliferation associated with more IGF type 1 receptors and increased plasma IGF-1 concentrations in young goats. J NUTR. 2004;134:11. doi: 10.1093/jn/134.1.11. [DOI] [PubMed] [Google Scholar]
- 20.Wan, Y.K. et al. Effects of Concentrate Feeding on Rumen Papillae Development in Hanwoo Calves before Weaning. Journal of Animal Science & Technology54 (2012).
- 21.Keady TW, Mayne CS, Fitzpatrick DA, McCoy MA. Effect of concentrate feed level in late gestation on subsequent milk yield, milk composition, and fertility of dairy cows. J DAIRY SCI. 2001;84:1468. doi: 10.3168/jds.S0022-0302(01)70180-4. [DOI] [PubMed] [Google Scholar]
- 22.Agle M, et al. Effect of dietary concentrate on rumen fermentation, digestibility, and nitrogen losses in dairy cows. J Dairy Sci. 2010;93:4211. doi: 10.3168/jds.2009-2977. [DOI] [PubMed] [Google Scholar]
- 23.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. NAT REV Micro Biol. 2008;6:776. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh KM, et al. Metagenomic analysis of Surti buffalo (Bubalus bubalis) rumen: a preliminary study. Mol Biol Rep. 2012;39:4841. doi: 10.1007/s11033-011-1278-0. [DOI] [PubMed] [Google Scholar]
- 26.de Oliveira MN, et al. Characterizing the microbiota across the gastrointestinal tract of a Brazilian Nelore steer. Vet Microbiol. 2013;164:307. doi: 10.1016/j.vetmic.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Chiquette J, Allison MJ, Rasmussen MA. Prevotella bryantii 25A used as a probiotic in early-lactation dairy cows: effect on ruminal fermentation characteristics, milk production, and milk composition. J Dairy Sci. 2008;91:3536. doi: 10.3168/jds.2007-0849. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Zhang M, Xue C, Zhu W, Mao S. Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants. J DAIRY SCI. 2016;99:9668. doi: 10.3168/jds.2016-11398. [DOI] [PubMed] [Google Scholar]
- 29.Kelly WJ, et al. The glycobiome of the rumen bacterium Butyrivibrio proteoclasticus B316(T) highlights adaptation to a polysaccharide-rich environment. Plos One. 2010;5:e11942. doi: 10.1371/journal.pone.0011942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.AOAC. Official methods of analysis (15th ed). Association of Official Analytical Chemists, Washington, D.C (1990).
- 31.Nie J, et al. Comparative analysis of dynamic proteomic profiles between in vivo and in vitro produced mouse embryos during postimplantation period. J Proteome Res. 2013;12:3843. doi: 10.1021/pr301044b. [DOI] [PubMed] [Google Scholar]
- 32.Cao YC, Yang HJ. Ruminal digestibility and fermentation characteristics in vitro of fenugreek and alfalfa hay combination with or without the inoculation of Neocallimastix sp. YAK11. ANIM FEED SCI TECH. 2011;169:53. doi: 10.1016/j.anifeedsci.2011.05.010. [DOI] [Google Scholar]
- 33.Ma T, Tu Y, Zhang NF, Deng KD, Diao QY. Effect of the Ratio of Non-fibrous Carbohydrates to Neutral Detergent Fiber and Protein Structure on Intake, Digestibility, Rumen Fermentation, and Nitrogen Metabolism in Lambs. Asian-Australas J Anim Sci. 2015;28:1419. doi: 10.5713/ajas.15.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. NAT Methods. 2013;10:996. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 36.Cole JR, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amato KR, et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. Isme J. 2013;7:1344. doi: 10.1038/ismej.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]