Abstract
Cystic fibrosis (CF) is the most common life-threatening genetic disease among Caucasians. CF patients suffer from chronic lung infections due to the presence of thick mucus, caused by cftr gene dysfunction. The two most commonly found bacteria in the mucus of CF patients are Staphylococcus aureus and Pseudomonas aeruginosa. It is well known that early-infecting P. aeruginosa strains produce anti-staphylococcal compounds and inhibit S. aureus growth. More recently, it has been shown that late-infecting P. aeruginosa strains develop commensal-like/coexistence interaction with S. aureus. The aim of this study was to decipher the impact of P. aeruginosa strains on S. aureus. RNA sequencing analysis showed 77 genes were specifically dysregulated in the context of competition and 140 genes in the context of coexistence in the presence of P. aeruginosa. In coexistence, genes encoding virulence factors and proteins involved in carbohydrates, lipids, nucleotides and amino acids metabolism were downregulated. On the contrary, several transporter family encoding genes were upregulated. In particular, several antibiotic pumps belonging to the Nor family were upregulated: tet38, norA and norC, leading to an increase in antibiotic resistance of S. aureus when exposed to tetracycline and ciprofloxacin and an enhanced internalization rate within epithelial pulmonary cells. This study shows that coexistence with P. aeruginosa affects the S. aureus transcriptome and virulence.
Subject terms: Gene expression, Bacteriology
Introduction
Most microorganisms are frequently embedded within communities of mixed species where different microbial interactions can occur between individual species. In the case of infection, these interactions between species can influence pathogenic behavior such as virulence, biofilm formation and antibiotic tolerance1–4.
One of the most well-known examples of pathologies in which many bacterial interactions are described are lung diseases occurring during Cystic Fibrosis (CF). The airways of CF patients are colonized by multiple microorganisms whose prevalence varies with the age of the patients. Among them, Staphylococcus aureus and Pseudomonas aeruginosa are the most prevalent pathogens and are acquired in subsequent order. The typical pattern of chronic infection establishment begins with the early acquisition of S. aureus, (60% prevalence among children aged <2 years and the highest prevalence in children of 11–17 years (80%)), while prevalence slowly declines in adults (50%)5. In contrast, infections by P. aeruginosa occur later with the highest prevalence in adults (70% among 35–44 year-old patients). Although these bacteria seem to succeed one another, they are not mutually exclusive since patients are frequently diagnosed as being co-infected by S. aureus and P. aeruginosa (from 35% to 50%)6,7.
While P. aeruginosa is recognized as the leading cause of lung function decline, the significance of S. aureus in the course of CF disease is still being debated. It has been shown that one of the risk factors for initial P. aeruginosa airway infection includes S. aureus pre-colonization8–10. However, the impact of coinfection by the two pathogens on the evolution of the disease remains unclear11–13.
S. aureus and P. aeruginosa have been identified in the same lobe of CF lungs14,15, suggesting that both pathogens are present in the same niche and can in fact interact in vivo. Interactions have been widely studied and it is commonly admitted that P. aeruginosa outcompetes S. aureus. Different mechanisms have been described16: for example, P. aeruginosa secreted products can inhibit the growth or lyse S. aureus as well as induce epithelial cells to kill S. aureus and other Gram-positive bacteria8,17,18.
However, these interactions can evolve during chronic colonization. Indeed, P. aeruginosa strains isolated from early infection outcompete S. aureus, as previously described, while strains isolated from chronic infection are less aggressive and can be co-cultivated with S. aureus19,20. Furthermore, P. aeruginosa isolates from mono-infected patients are more competitive towards S. aureus than isolates from coinfected patients21.
In contrast to antagonistic interactions, nothing is known about the effects of P. aeruginosa and S. aureus interactions in this context of coexisting bacteria within the same infectious niche. Using a transcriptomic approach, we analyzed how co-cultivation with non-competitive P. aeruginosa altered S. aureus gene expression, especially genes encoding Nor family efflux pumps. In the presence of P. aeruginosa, over-expression of these genes increased S. aureus antibiotic tolerance and the rate of internalization into epithelial cells, two key determinants of chronic infection.
Results
Coexistence interaction involves more than half of the S. aureus and P. aeruginosa isolates from co-infected CF patients
Two types of interactions between S. aureus and P. aeruginosa could be observed with CF patient isolates: the well-described competitive phenotype, where P. aeruginosa inhibits S. aureus growth16, and the newly described phenotype of coexistence, where P. aeruginosa is unable to outcompete S. aureus19–21. In order to quantify the importance of this last phenotype, we collected 50 pairs of S. aureus and P. aeruginosa from 36 co-infected CF patients. The interaction between the two pathogens was analyzed by a competitive test on trypticase soy agar (TSA) plates (Table 1 - Fig. 1A). We observed that 61% of strain pairs presented a coexistence phenotype whereas 39% were in competitive interaction. To determine whether the pairs of coexisting strains and competitive strains were phenotypically different, we measured colony size, analyzed the hemolytic properties of each strain, and searched for pigmentation and mucoid phenotype for all P. aeruginosa strains. No significant differences were observed between coexisting and competitive strains with respect to pigment production, mucoid phenotype and hemolysis (Fig. S1). We observed a significant difference in the size of S. aureus colonies in which those of coexisting strains were larger than those of competition strains after 24 h of TSA plate culture. The significance of such a difference and its impact on interaction with P. aeruginosa remain to be explored. As others have already described that early infectious strains of P. aeruginosa are more aggressive for S. aureus than the late infectious strains19,20, we wondered if the type of interaction could be related to the duration of colonization. To answer this question, we determined the duration of co-colonization of S. aureus and P. aeruginosa for each patient. The average duration of colonization for coexisting strains was 744.8 ± 97.64 days and for competing strains 941.2 ± 137 days. The difference was statistically non-significant (Fig. S1).
Table 1.
S. aureus and P. aeruginosa clinical strains used in this study.
| Patients | S. aureus | P. aeruginosa | co-colonisation length (days) | type of interaction | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| strain number | hemolyse type | Colony size (mm) | pigmentation | MLST | strain number | hemolyse type | Colony size (mm) | pigmentation | mucoidy | |||
| 1 | SA2597 | γ | 1 | none | PA2596 | β | 4 | Yellow | yes | 1544 | competition | |
| 2 | SA2599 | β | 3 | none | CC188 | PA2600 | γ | 5 | Yellow | yes | 1902 | coexistence |
| 3 | SA27 | β | 2 | none | CC398 | PA27 | γ | 3 | none | no | 295 | coexistence |
| 4 | SA30 | β | 2 | none | CC188 | PA30 | γ | 1,2 | none | no | 133 | coexistence |
| 5 | SA31 | β | 3 | none | CC15 | PA31 | γ | 1,1 | none | yes | 921 | coexistence |
| 6 | SA42 | γ | 2 | none | CC30 | PA42 | γ | 20 | Yellow | yes | ? | coexistence |
| 7 | SA69 | β | 3 | none | CC25 | PA69 | β | 2,1 | none | yes | 92 | coexistence |
| 8 | SA80 | β | 2 | none | CC15 | PA80A | α | 2,2 | Green | yes | 623 | coexistence |
| none | PA80B | α | 6 | none | yes | 623 | coexistence | |||||
| 9 | SA82 | β | 4 | none | CC45 | PA82 | γ | 4 | none | yes | ? | coexistence |
| 10 | SA146 | γ | 2 | none | CC88 | PA146 | γ | 4,5 | Green | yes | 928 | coexistence |
| 11 | SA147 | β | 2 | none | PA147A | γ | 2 | Green | no | ? | competition | |
| none | PA147B | γ | 1,4 | none | no | competition | ||||||
| 12 | SA148 | γ | 1 à 2 | none | PA148A | β | 7 | none | yes | 839 | competition | |
| none | PA148B | β | 14 | Green | yes | coexistence | ||||||
| 13 | SA150 | β | 2 | none | PA150 | γ | 1 | none | no | 380 | coexistence | |
| 14 | SA151 | β | 2 | none | PA151A | α | 4,5 | Brown | yes | 538 | coexistence | |
| none | PA151B | β | 4 | Yellow | yes | coexistence | ||||||
| 15 | SA152 | β | 3 | none | CC8 | PA152 | γ | 13 | none | yes | 991 | coexistence |
| 16 | SA153 | β | 3 | none | CC398 | PA153A | β | 3 | none | no | 967 | coexistence |
| PA153B | β | 2,5 | Green | yes | coexistence | |||||||
| 17 | SA154 | γ | 2 | none | PA154A | γ | 35 | none | yes | 595 | coexistence | |
| none | PA154B | γ | 5 | Green | yes | coexistence | ||||||
| 18 | SA155 | β | 2 | none | PA155 | α | 11 | Green | yes | 1256 | competition | |
| 19 | SA156 | β | 2 | none | CC398 | PA156 | α | 2,1 | none | yes | 49 | coexistence |
| 20 | SA157 | β | 2 | none | PA157 | β | 5 | Green | yes | ? | competition | |
| 21 | SA158 | γ | 1 à 2 | none | PA158 | β | 6 | Green | yes | ? | competition | |
| 22 | SA159 | β | <1 SCV (1,5)* | none | PA159A | β | 5 | none | yes | 1353 | competition | |
| none | PA159B | β | 18 | Green | yes | competition | ||||||
| 23 | SA160 | γ | <1 SCV (1,1)* | none | PA160A | β | 5 | Green | yes | 1332 | competition | |
| none | PA160B | β | 10 | Yellow | yes | competition | ||||||
| 24 | SA161 | β | 2 | none | PA161 | α | 2 | none | no | 1043 | coexistence | |
| 25 | SA165 | β | 3 | none | PA165A | β | 3,9 | none | yes | 343 | coexistence | |
| none | PA165B | β | 4,1 | none | yes | coexistence | ||||||
| 26 | SA166 | γ | 2 | none | PA166A | α | 4 | Green | yes | 1355 | coexistence | |
| none | PA166B | α | 4,5 | Green | yes | coexistence | ||||||
| 27 | SA167 | β | 3 | none | PA167 | α | 13 | Green | yes | 938 | competition | |
| 28 | SA168A | β | 2 | none | PA168 | γ | 4 | none | yes | 294 | competition | |
| SA168B | β | 2 | none | competition | ||||||||
| 29 | SA169 | β | 4 | none | PA169 | γ | 3,5 | none | no | 904 | coexistence | |
| 30 | SA171 | β | 1 à 2 | none | PA171 | β | 1,1 | none | yes | 601 | coexistence | |
| 31 | SA177 | β | 3 | none | PA177 | γ | 2 | none | yes | 1380 | coexistence | |
| 32 | SA178 | β | 3 | none | PA178 | β | 2,5 | none | yes | 519 | coexistence | |
| 33 | SA179 | β | 1 à 2 | none | PA179 | β | 3 | none | yes | 91 | competition | |
| 34 | SA181 | β | 4 | none | PA181 | β | 6 | none | yes | 1096 | competition | |
| 35 | SA182 | γ | 2 | none | PA182A | β | 5 | Yellow | yes | 987 | competition | |
| none | PA182B | α | 7 | none | yes | competition | ||||||
| none | PA182C | β | 4,2 | none | yes | coexistence | ||||||
| 36 | SA186 | β | 2 | none | PA186 | β | 3 | none | yes | 623 | competition | |
50 couples of S. aureus and P. aeruginosa were collected from 36 patient sputum samples. Some patients presented several P. aeruginosa isolates, and one patient presented two S. aureus isolates. Colony size, pigmentation and mucoid phenotype were determined on TSA. Hemolysis type was determined on COS. Interaction type was determined by agar plate competition assay as described in the materials and methods section. The isolates in bold correspond to those used for the RNAseq and RT-qPCR analyses. MLST type were determined only for these isolates.
*Colony size after 48 h incubation - ? colonization time before sampling could not be determined for these patients.
Figure 1.
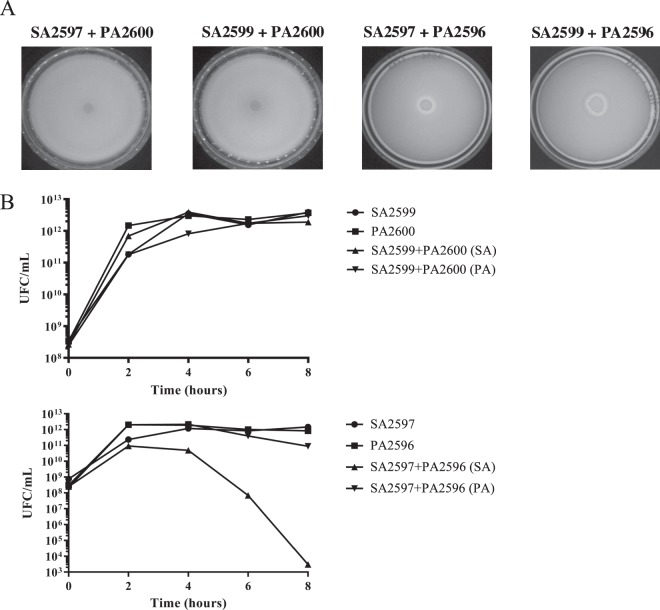
Competition assay between S. aureus and P. aeruginosa. (A) Competition test on agar plate. S. aureus and P. aeruginosa were grown on BHI for 8 hours at 37 °C. A layer of S. aureus was added on a TSA. After drying, a drop of P. aeruginosa was spotted. The inhibition halo indicates a competition state (SA2597 + PA2596 and SA2599 + PA2596). (B) Competition assay in planktonic culture. S. aureus and P. aeruginosa were mono-cultivated and co-cultivated for 8 hours. Every two hours, bacteria were plated on mannitol salt agar (MSA) and cetrimide to count S. aureus and P. aeruginosa, respectively. The results show one representative experiment from a triplicate. Upper panels, pairs in coexistence. Lower panels, pairs in competition.
Planktonic cultures were conducted on two pairs of strains: one competitive pair (SA2597/PA2596) and one coexisting pair (SA2599/PA2600). In both cases, we observed that P. aeruginosa growth was not altered by S. aureus. On the other hand, in the case of the competitive pair, P. aeruginosa had a negative effect on S. aureus growth after 4 hours of coculture (Fig. 1B).
Agar plate competition assays mixing respectively PA2600 (from coexisting pair) and PA2596 (from competitive pair) with both SA2597 and SA2599 were performed (Fig. 1A). PA2596 outcompeted both SA strains whereas PA2600 was unable to inhibit any of the S. aureus strains, suggesting that the interaction phenotype is dependent on the P. aeruginosa strains. These results were confirmed with other combinations of strains (Fig. S2).
P. aeruginosa differentially dysregulates S. aureus transcriptome according to coexistence/competition
To obtain an overview of the impact of P. aeruginosa on the expression of S. aureus genes, a comparative transcriptomic study was conducted between SA2597 and SA2599 in monocultures, and the same strains in coculture with a competition PA strain (PA2596) and a coexisting PA strain (PA2600). Thus, for each interaction state, we tested two pairs of strains, namely SA2597/PA2596 and SA2599/PA2596 for the competition and the SA2597/PA2600 and SA2599/PA2600 pairs for coexistence. Gene expression was considered dysregulated when dysregulation was common to both pairs of strains. Therefore, seventy-seven S. aureus genes were specifically dysregulated in the context of competition and 140 genes in the context of coexistence while only 16 genes were dysregulated both in competition and in co-existence (Table S4).
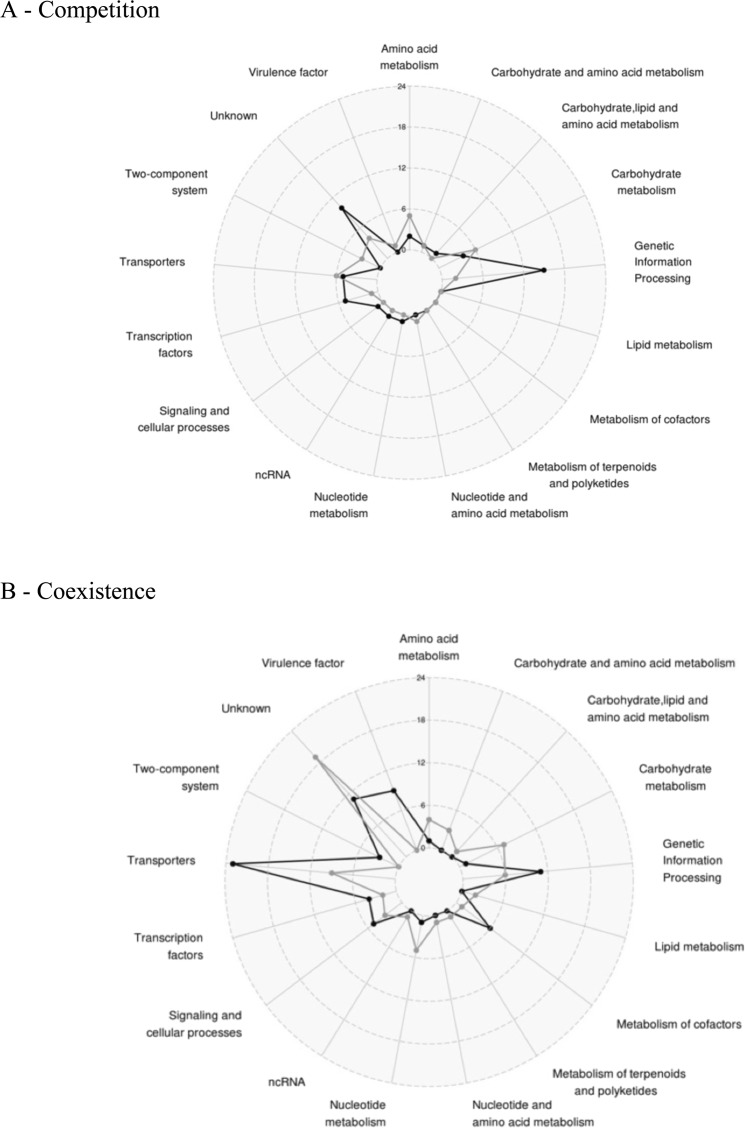
KEGG analyses were performed on dysregulated genes to associate each gene with a functional class (Fig. 2). In competition state, the main dysregulated class of genes belongs to genetic information and processing, with an increase of tRNA and ribosomal RNA (Fig. 2A). We also observed the dysregulation of genes involved in major metabolism pathways of carbohydrates and amino acids. The down-regulation of the Acetyl-coenzyme A synthetase encoding gene (acsA) was noted. Other genes involved in energetic metabolism were up-regulated in the presence of P. aeruginosa, especially dehydrogenase enzymes such as the lactate dehydrogenase (ldhA), the alanine dehydrogenase (ald1), the glutamate dehydrogenase (gluD), the 1-pyrroline-5-carboxylate dehydrogenase (rocA), the 2-oxoglutarate dehydrogenase (odhA) and the aldehyde-alcohol dehydrogenase (adhE). The upregulation of the ldh gene is consistent with the up regulation of the L-lactate permease (lctP) encoding gene. All these factors, as well as acetyl-coA, are involved in energetic metabolism and redox reactions conducted to feed the Krebs cycle and ensure the production of ATP.
Figure 2.
Number and functions of differentially expressed staphylococcal genes in the presence of P. aeruginosa when both species are: (A) in competition (SA2596 and SA2599 were co-cultivated with PA2597) or (B) in coexistence (SA2596 and SA2599 were co-cultivated with PA2600). RNAs were extracted at 4 hours and a RNAseq was performed in triplicates. KEGG mapper analysis was conducted on common significantly over-expressed (black) and under-expressed genes (grey) to address a functional classification. A gene was considered as differentially expressed when the fold change was strictly higher than 4 with P_adj < 0.05.
In the context of coexistence, although P. aeruginosa does not appear to impact major metabolic pathways of S. aureus as it does not alter growth, the expression of 140 S. aureus genes was affected by the presence of P. aeruginosa (Fig. 2B). Nine known and predicted virulence factor encoding genes were upregulated, including alpha-hemolysin (hla), staphylokinase (sak), aureolysin (aur), the immunoglobulin-binding protein (sbi) and staphylococcal complement inhibitor (scn) genes. We also observed the overexpression of saeRS genes, coding a two component system that has been described as playing a major role in controlling the production of virulence factors such as those mentioned above22.
Other genes whose expression was affected by the presence of P. aeruginosa in a coexistence situation are involved in carbohydrates, lipids, nucleotides and amino acids metabolism. Most of them were down-regulated as several genes (pgi, fbp, fda) involved in glycolysis and the pentose phosphate pathway. Moreover, two operons (nrdE, nrdI, nrdF and nrdG, nrdD) belonging to ribonucleotide reductase (RNR) systems and converting nucleoside phosphate into deoxynucleotide phosphate, were both down-regulated (Table S4). RNRs are involved in the de novo production of deoxyribonucleotide di- or triphosphates, an essential process for the biosynthesis of DNA and its repair. They catalyze the limiting step of the synthesis of deoxyribonucleotide phosphates and thus control cell concentration23.
Finally, several genes belonging to a transporter family were also over-expressed (polyamines, methionine, iron uptake and antibiotic resistance) in the coexistence state. Notably, all genes from the polyamine operon were over-expressed (potABCD) including potR, the regulator of polyamine genes. Polyamines control the physiology of S. aureus by acting as regulators of several genes involved in metabolism, transport and virulence24,25. In addition, the same pattern was observed for the metQPN operon involved in methionine transport and sirA/B and sstA/BC genes for iron uptake, which may also reflect nutrient competition in coculture. Finally, transporter norb_3 predicted as belonging to the nor family was over-expressed. Pumps from this family export a wide range of antibiotics such as erythromycin, tetracycline and quinolones. Indeed, norb_3 corresponds to the well-described tet38 gene involved in tetracycline resistance and internalization in pulmonary epithelial cells26,27.
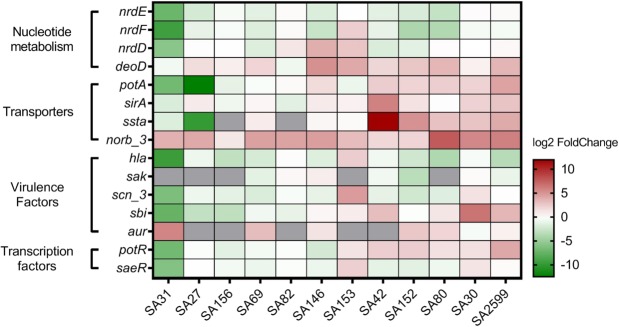
To confirm these results, we performed co-cultivations with 12 different co-existence P. aeruginosa-S. aureus strain pairs from CF patients. The 12 strain pairs came from 12 different patients and presented phenotypic diversity. S. aureus isolates belong to 8 different multilocus sequence typing (MLST) types (Table 1). Ten isolates of P. aeruginosa were mucoid and four secreted pigmentation, which was representative of the collection of all the isolates. Gene expression was assessed by RT-qPCR for the two categories most impacted: virulence factors and transporters (Fig. 3). Regarding virulence factors, we confirmed the over-expression of the aureolysin encoding gene in 6 of the 7 strains that expressed the gene. For sbi, 5 out of 12 strains presented over-expression and 5 out 12 presented decreased expression, meaning that there was no clear profile of P. aeruginosa’s impact on this gene expression. For the other virulence genes tested, we observed reduced expression in the majority of the strains (10/12 for hla, 5/7 for sak, 7/12 for saeRS and 8/12 for scn) (Fig. 3).
Figure 3.
Confirmation of S. aureus gene expression dysregulation by P. aeruginosa. Twelve clinical SA-PA strain pairs were co-cultivated for 4 hours. RNAs were extracted and RT-qPCR were performed on 15 genes. The results are represented as fold change of expression (gene relative expression in coculture/gene relative expression in monoculture) on a heatmap. Under-expressed genes are indicated in green whereas over-expressed genes are indicated in red. No RNA detection is shown in grey. Pairs were hierarchically clustered by the Euclidean method.
For transporter encoding genes, we confirmed the over-expression of pot genes and the sstA gene in 6/12 and 7/10 strains, respectively. Noticeably, we confirmed the up-regulation of tet38 genes in 11/12 strains with a fold change ranging from 3 to 200. In addition, deoD gene upregulation was also confirmed in 9/12 strains, consistent with its operon structure with tet38 gene26.
The over-expression of the tet38 gene is the most predominant transcriptomic alteration in our study and may be of great importance as it can affect the antibiotic susceptibility of S. aureus, an important element in the context of CF disease. Therefore, we aimed to better characterize this transcriptomic alteration.
Over-expression of the tet38 gene is due to the dysregulation of the MgrA regulatory pathway that impacts other nor family genes
To decipher the molecular pathway involved in the over-expression of the tet38 gene, we analyzed the expression of known regulators in the presence or absence of P. aeruginosa. Three transcriptional negative regulators of tet38 have already been described: TetR2126, SarZ28 and MgrA27. The expression of these regulators was quantified by RT-qPCR in coculture and compared to expression in monoculture. None of the tetR21, sarZ and mgrA RNA levels was affected by the presence of P. aeruginosa (Fig. S3).
However, it has been described that regulation by MgrA is dependent on its phosphorylation state29 and that the deletion of mgrA induces increased expression of tet3827. Therefore, we analyzed the impact of P. aeruginosa on tet38 expression using a Newman ΔmgrA mutant (Fig. 4). The wild type Newman strain presented a 20-fold change over-expression of the tet38 gene in the presence of P. aeruginosa, as we previously observed in clinical strains. This fold change was reduced to 6 for the ΔmgrA mutant. Therefore, it appears that the over-expression of the tet38 gene is induced by an alteration of the MgrA regulatory pathway. These results were confirmed in S. aureus Lac isogenic strains (Fig. S4).
Figure 4.
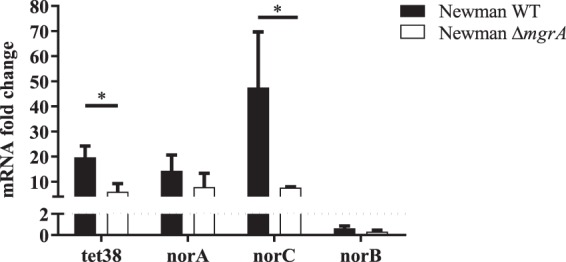
The S. aureus MgrA regulator is important for nor gene over-expression. Cocultures of S. aureus Newman wild type (WT) and ΔmgrA mutant with PA30 were performed. RNAs were extracted at 8 hours and nor gene expression was monitored by RT-qPCR. The results are shown as the mean + standard deviation of three independent experiments. Dotted lines indicate fold change = 2. Statistical analysis was performed by unpaired t-test (*P < 0.05).
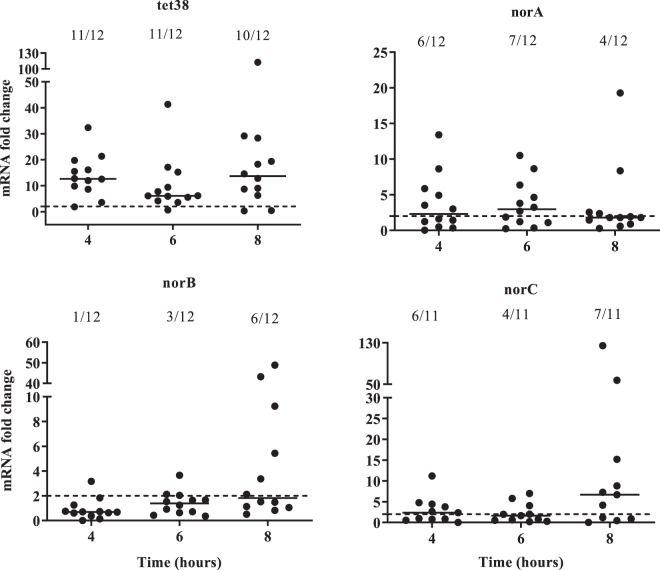
MgrA is also a transcriptional regulator of other nor family protein genes such as norA, norB and norC27,29,30. Hence, the expression of nor genes in S. aureus was monitored throughout cocultures of the 12 S. aureus strains with P. aeruginosa (Fig. 5). The tet38 gene was significantly overexpressed in the presence of P. aeruginosa throughout the 8 hours of the culture. The norA gene was over-expressed in at least 50% of cocultures (6/12 and 7/12) after 4 and 6 hours of culture and norC expression was increased at 4 and 8 hours in 6/11 and 7/11 strains, respectively (Fig. 5). norB was overexpressed only at 8 hours of culture in 50% of the strains tested.
Figure 5.
The over-expression of S. aureus nor family genes induced by P. aeruginosa. Mono- and cocultures with twelve clinical strain pairs were performed. RNAs were extracted and gene expression was monitored by RT-qPCR at 4, 6 and 8 hours. The results are represented as fold change expression. Dotted lines indicate fold change = 2. Numbers above each hour indicate the number of pairs with a fold change strictly higher than 2.
From the analysis of expression in the ΔmgrA mutant in the S. aureus Newman strain, we concluded that over-expression of norC was dependent on MgrA integrity (Fig. 4). A milder effect was observed on norA over-expression. No overexpression of norB gene was observed with the S. aureus Newman strain. In the S. aureus Lac strain, we observed a diminution of norA and norC overexpression in the ΔmgrA mutant but the effect was less significant than on tet38 gene expression (Fig. S4). Therefore, we concluded that the overexpression of norA and norC genes in the presence of P. aeruginosa was partially due to mgrA dysregulation.
The presence of P. aeruginosa induces over-expression of nor genes by specific and direct interaction
To determine if a secreted product of P. aeruginosa induced the over-expression of nor genes, a transwell experiment was conducted in which cultures of P. aeruginosa and S. aureus were separated by a 0.4μm filter. In these conditions, nor genes were not overexpressed (Fig. 6). The same results were obtained when S. aureus culture was exposed to supernatant of P. aeruginosa (Fig. S5), suggesting that at least a close interaction between the two species was necessary. Finally, to determine if the over-expression was specific to the interaction with P. aeruginosa, cocultures were conducted with other bacteria frequently associated with S. aureus in CF patients, such as Burkholderia cenocepacia and Stenotrophomonas maltophilia (Fig. 7). No over-expression was observed, suggesting that the effect was specific to the presence of P. aeruginosa. These results were confirmed with other clinical S. aureus strains (Fig. S6).
Figure 6.
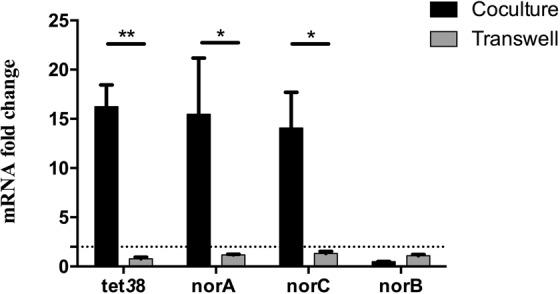
S. aureus nor gene over-expression requires close contact with P. aeruginosa. S. aureus was deposited onto the bottom of wells. P. aeruginosa was added either with S. aureus (black) or into the insert of transwells (gray). RNAs were extracted and nor gene expression was monitored by RT-qPCR. Dotted lines represent fold change = 2. The results are shown as the mean + standard deviation of three independent experiments on SA30-PA30 pair. Statistical analysis was performed by unpaired t-test (*P < 0.05, **P < 0.01).
Figure 7.
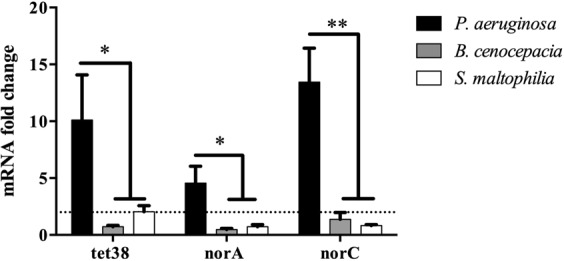
The overexpression of S. aureus nor genes is specifically induced by P. aeruginosa. S. aureus was mono- and cocultivated with P. aeruginosa, B. cenocepacia or S. maltophilia. RNAs were extracted and gene expression was monitored by RT-qPCR. Dotted lines indicate fold change = 2. The results represent the mean + standard deviation of three independent experiments on SA30-PA30 pair. Statistical analysis was performed by One-way Anova with Dunnett’s multiple test correction (*P_adj < 0.05, **P_adj < 0.01).
Over-expression of nor genes induces an increase of antibiotic resistance and internalization of S. aureus into epithelial cells
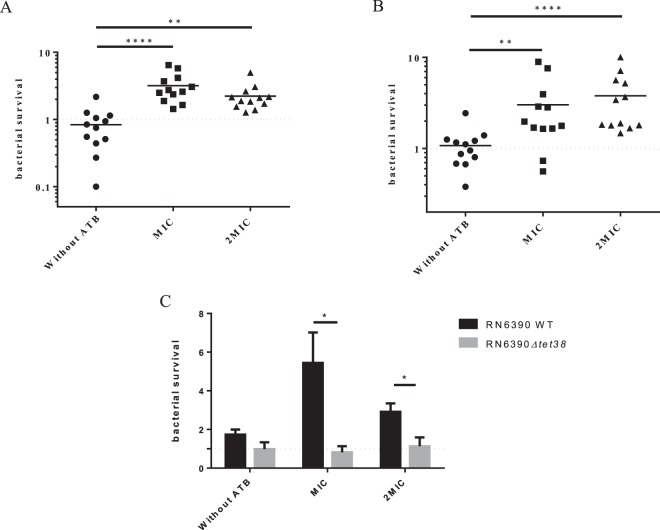
As Tet38 is involved in tetracycline resistance27 and NorA and NorC are also implicated in quinolones (such as ciprofloxacin) uptake30,31, the impact of coculture with P. aeruginosa on S. aureus antibiotic susceptibility was tested. Firstly, the MIC was determined for each of the 12 S. aureus strains used (Table S3). Monocultures and cocultures were then exposed to tetracycline and ciprofloxacin at MIC or 2xMIC. After plating on selective agar and numeration, the survival rate was determined by dividing the number of S. aureus after antibiotic treatment by the number of S. aureus without antibiotic treatment. A 3-fold increase in survival rate was observed at MIC and 2xMIC in the presence of P. aeruginosa (Fig. 8A,B).
Figure 8.
S. aureus antibiotic resistance increases when co-cultivated with P. aeruginosa. Twelve clinical S. aureus strains were mono- and cocultivated with coexisting P. aeruginosa strains and exposed to tetracycline (A) and ciprofloxacin (B) at MIC and 2xMIC. After 5 hours, cultures were plated on MSA to count remaining S. aureus. Bars represent the median and dotted lines bacterial survival equal to 1. Statistical significance was determined by One-way Anova with Dunnett’s multiple test correction (**P_adj < 0.01, and ****P_adj < 0.0001). (C) Tet38 is responsible for the increase of tetracycline resistance induced by P. aeruginosa. RN6390 and isogenic Δtet38 derivative were cultivated with and without P. aeruginosa and susceptibility to tetracycline was monitored. Statistical significance was determined by unpaired t-test (*P < 0.05) from three independent experiments. All the results are expressed as the number of surviving bacteria in coculture divided by the number of surviving bacteria in monoculture.
In order to demonstrate that the over-expression of the tet38 gene was responsible for tetracycline resistance, the impact of P. aeruginosa was tested on the RN6390 wild type strain and its isogenic Δtet38 mutant upon exposure to tetracycline. As expected, P. aeruginosa induced a higher survival rate of the RN6390 wild type strain after tetracycline exposure. On the contrary, it had no impact on the bacterial survival of the Δtet38 mutant after tetracycline exposure (Fig. 8C), confirming the role of the tet38 gene in the enhancement of tetracycline resistance in the presence of P. aeruginosa.
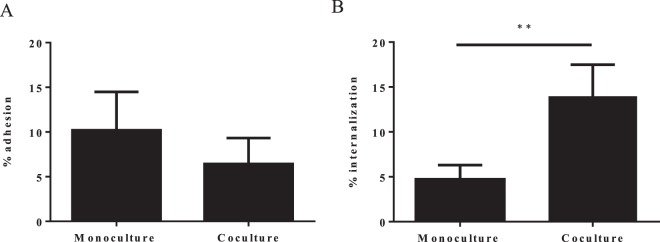
Tet38 has also been described as being involved in pulmonary epithelial cell internalization26, so the impact of the presence of P. aeruginosa on S. aureus cell internalization was tested using the Gentamicin protection assay. When A549 epithelial cells were infected with a monoculture of S. aureus, five percent of adherent bacteria were internalized into cells. When S. aureus was co-cultivated with P. aeruginosa before cell infection, 15% of S. aureus were internalized, meaning a 3-fold increase of the S. aureus internalization rate in the presence of P. aeruginosa (Fig. 9). No difference was observed in terms of bacterial adhesion onto A549 cells. To ensure that the effect we observed was not due to an alteration of the A549 cell layer by P. aeruginosa that could have facilitated S. aureus internalization, we performed an LDH measurement on the cell supernatant as an indicator of A549 cell viability (Fig. S7B). Although the LDH level was slightly higher for cells infected only with P. aeruginosa, we found no significant difference between the S. aureus infected and S. aureus plus P. aeruginosa co-infected cells. Indeed, the A549 co-infected cells had the lowest level of LDH. Moreover, microscopic observation of the cells revealed no difference between the mono- and co-infected cells (Fig. S7A). Therefore, it appeared that the presence of P. aeruginosa did not alter the A549 cells and the highest rate of S. aureus internalization was due to its direct impact on S. aureus. However, we could not be sure that the increase in the internalization rate was directly related to tet38 overexpression. It could be the result of a modification of different factors involved in the internalization process, although we did not identify such factors in our transcriptomic analysis apart from the tet38 gene.
Figure 9.
S. aureus internalization is increased within A549 epithelial pulmonary cells in the presence of P. aeruginosa. (A) Adhesion of S. aureus onto epithelial cells. A549 cells were infected at MOI 10:1 for S. aureus monoculture and 20:1 for S. aureus/P. aeruginosa coculture. After 2 hours of contact, cells were washed with phosphate buffer saline (PBS) to remove unattached bacteria and lysed with sterile water. Supernatants were plated on MSA to count S. aureus. The results are represented as the percentage of inoculum that adhered. (B) Internalization of S. aureus within epithelial cells. After 2 hours of contact, cells were treated with antibiotics and lysostaphine for one hour, lysed with sterile water and bacteria plated on MSA. The results are represented as the percentage of adhered cells that have internalized. All values represent the mean + standard deviation from three independent experiments with three strain pairs (SA27-PA27, SA31-PA31 and SA69-PA69). Statistical significance was determined by unpaired t-test (**P < 0.01).
Discussion
The goal of this study was to investigate the impact of the interaction of coexisting S. aureus and P. aeruginosa on S. aureus at the transcriptional and phenotypical levels.
Firstly, we collected isolates from co-colonized CF patients and demonstrated that in 61% of cases, S. aureus was able to coexist with P. aeruginosa with no alteration of its growth. So far, it appears that coexistence of the two pathogens may be a frequent situation in the context of CF patients’ lung infection. Previous studies described that early infectious strains of P. aeruginosa are more aggressive for S. aureus than the late infectious strains19,20. In the present study, we did not find any correlation between the interaction type and the duration of S. aureus and P. aeruginosa co-colonization. In the first studies, limited numbers of patients were studied (1 in Michelsen et al., 2014 and 8 in Baldan et al., 2014). Even in our present study, only 11 patients had competitive strain pairs, which might be not be enough to reach a conclusion. Up to now, it has been difficult to conclude whether the interaction type between the two species is linked to the evolution of the P. aeruginosa strain over the time of co-colonization. A larger cohort of patients would be needed to answer this question. Also, longitudinal clinical studies would be appropriate to analyze the kinetics of interaction evolution over time and determine how it could affect patients’ health. Furthermore, the conditions and environmental factors leading to co-existence instead of competition require clarification, particularly through studies such one conducted recently that demonstrated the positive impact of hypoxia found in static mucus within CF airways on a coexisting interaction32.
The type of interaction may have an impact on the physiology of the two pathogens involved. In order to answer this question, we conducted a transcriptional study of the impact of P. aeruginosa on S. aureus.
In the context where P. aeruginosa inhibits S. aureus growth, transcriptomic modifications affect major metabolism pathways such as translation, Krebs cycle and genes involved in oxidative stress. The increase in the amount of tRNAs and ribosomal RNAs observed could be attributed to a decrease in translation efficiency. Regarding energetic metabolic pathways, we observed a down-expression of Acetyl-coA synthetase, a key factor metabolized into pyruvate to feed the Krebs cycle and produce energy. The down-regulation of expression observed may certainly lead to a defect in ATP production. Conversely, we observed the increased expression of several dehydrogenase enzymes, suggesting a shift from aerobic respiration to lactic acid fermentation to feed the Krebs cycle, as shown previously in laboratory strains33,34. Some dehydrogenases, such as adhE and gluD genes, are also implicated in oxidative stress responses35. All these major dysregulations observed are consistent with the lethal effect of P. aeruginosa on S. aureus in competitive interaction.
More genes were dysregulated when S. aureus and P. aeruginosa were coexisting. We observed a drastic modification in the nucleotide synthesis pathway with a down-regulation of genes involved in the de novo pathway (nrd operon) and upregulation of the deoD gene encoding a purine nucleoside phosphorylase involved in an alternative metabolic pathway for nucleotides when the de novo pathway is altered. We also observed a down-expression of genes involved in the classical energetic metabolism pathways: glycolysis and pentose phosphate pathways (pgi, fbp, fda). These results suggest nutritional competition between the two pathogens and indicate that in our conditions, S. aureus preferentially produced energy and nucleotides from sources other than glucose.
Finally, we observed the increased expression of several transporters, especially tet38, norA and norC genes. Curiously, the tet38 gene belongs to the same transcription unit as the deoD gene. It is tempting to speculate that the overexpression of tet38-deoD operon may be linked to the down regulation of the nrd genes to compensate for the alteration of the de novo nucleotide synthesis pathway.
These genes are members of the Nor family and encode efflux pumps involved in antibiotic resistance. Tet38 was the most impacted gene with 11 pairs of 12 for which we observed an increased expression throughout the coculture kinetic, whereas the over-expression of other norA and norC genes appeared on 7 pairs of 12 and 11 of 12 at 6 and 8 hours, respectively. Given that the pair 2599/2600 used for the RNA sequencing (RNAseq) was unable to upregulate norA and norC at 4 hours of coculture, it was expected that norA and norC genes would not appear in the RNAseq results. The over-expression of tet38, norA and norC genes appeared to be at least due to a dysregulation of the MgrA pathway. Indeed, the ΔmgrA mutant provoked a strong effect on tet38 over-expression but only a slight effect on norA and norC. Thus, mgrA seems to be essential for tet38 over-expression and other regulators must be implicated for the norA and norC genes. In addition, we were unable to observe clear norB up-regulation in the presence of P. aeruginosa. Indeed, MgrA acts as a repressor of tet38, norA and norC and an activator of norB in an rsbU positive background strain36,37. This discrepancy may explain our results. Despite its role in tet38 induction during coculture, mgrA expression was not affected by the presence of P. aeruginosa. However, it has been shown that the phosphorylation state of MgrA, regulated by RsbU and PknB factors, was a key mechanism for regulation of nor family gene expression29. Thus, P. aeruginosa may induce a variation of MgrA phosphorylation leading to a modification of nor gene expression.
Nor proteins are responsible for antibiotic efflux (tetracycline and fluoroquinolone), and we demonstrated that P. aeruginosa increased the survival rate of S. aureus after exposure to tetracycline and ciprofloxacin. For tetracycline, the effect appears to be mainly due to tet38, as antibiotic resistance in presence of P. aeruginosa was eliminated in a tet38 mutant. The same analysis could not be performed for nor genes due to the functional redundancy of the norA, norB and norC genes and the difficulty in obtaining triple mutants. Tet38 is also able to interact with the CD36 receptor on pulmonary epithelial cells to favor S. aureus internalization38. Indeed, in the presence of P. aeruginosa, we observed a higher rate of S. aureus internalization into epithelial cells. Internalized bacteria are more resistant to antibiotics and less detectable by the immune system39. Our results suggest that by coexisting with P. aeruginosa, S. aureus could hide from the host immune system and be more resistant to antibiotics.
We did not identify the P. aeruginosa specific signal responsible for S. aureus gene expression dysregulation. However, we demonstrated that it seems to be specific to P. aeruginosa (no other species tested had the same effect) and requires very close proximity between S. aureus and P. aeruginosa to be effective. Transcriptomic analysis revealed that the S. aureus potRABCD operon for polyamine uptake and regulation exhibited significant fold change expression upon exposure to P. aeruginosa. The same effects were observed by Yoa and Lu25 when exposing S. aureus to polyamines. Moreover, the exposure of S. aureus to spermine induces transcriptional modifications including over-expression of antibiotic efflux pumps such as norA and tetM genes and the decreased expression of many genes involved in carbohydrate metabolism and transport24. These results are consistent with the reduced expression of genes involved in glycolysis and the pentose phosphate cycle described previously24,25. Indeed, we observed the same profile after exposure to P. aeruginosa as other authors observed after exposure to spermine. Finally, P. aeruginosa presents polyamines at the outer surface of its membrane, more precisely putrescine and spermidine40. Therefore, we suggest that the P. aeruginosa polyamines present at the outer surface may be a signal for S. aureus transcriptional modifications. Further investigation will be necessary to confirm this hypothesis.
To the best of our knowledge, this study is the first to characterize the transcriptomic profile of coexisting S. aureus and P. aeruginosa pairs in a clinical context. We demonstrate that this commensal-like interaction induces phenotypical changes in S. aureus such as increased antibiotic resistance and host cell internalization. These phenotypes may favor the persistence of S. aureus in the context of chronic infection. Since this state of coexistence is apparently solely attributable to P. aeruginosa, the selective advantage for P. aeruginosa leads to questions. Indeed, previous studies showed that cocultivation with S. aureus induces LPS mutation in P. aeruginosa associated with fitness gain and antibiotic resistance41, and that S. aureus exoproducts restore and enhance P. aeruginosa motility32. The state of coexistence could thus represent a trade-off allowing both pathogens to benefit mutually and maintain equilibrium. However, the impact of S. aureus on P. aeruginosa in this state of coexistence warrants further investigation.
Materials and Methods
Bacterial strains and culture growth
The bacterial strains and plasmids used in this study are listed in Tables 1 and S1. The clinical strains were originally isolated by the Institute for Infectious Agents from sputum samples of patients followed-up in the two CF Centers of Lyon (Hospices civils de Lyon, France). The strains were collected between May 2016 and June 2017 from 36 different patients. The size of the colonies, pigmentation and mucoid phenotype were determined after 24 h of culture on TSA. Hemolysis type was determined after 24 h of culture on Columbia agar (COS). MLST clonal complex assignment was inferred from microarray analysis42.
The Δtet38 mutant of S. aureus RN6390 strain was obtained using the pMAD vector43. The two DNA fragments corresponding to the chromosomal regions upstream and downstream of the tet38 coding sequence were amplified by PCR using primers listed in Table S2. They were subsequently cloned into the pMAD vector using the In-Fusion® HD Cloning Kit (Clonetech). The resulting plasmid was electroporated into the RN4220 recipient strain and then transferred to RN6390. Growth at non-permissive temperature (44 °C) was followed by several subcultures at 30 °C and 37 °C to promote double crossing over as previously described44.
All the strains were grown in Brain Heart Infusion (BHI, BBLTM Difco) with shaking at 200 rpm at 37 °C overnight. Cultures were diluted to 0.1 OD600nm and incubated for 2.5 hours (37 °C, 200 rpm). Bacteria were spun down at 4000 rpm for 10 min and re-suspended in fresh BHI medium to 2 OD600nm. Ten ml of S. aureus, P. aeruginosa, B. cenocepacia and S. maltophilia suspension were added to 10 ml of BHI for monocultures. Ten ml of S. aureus suspension were mixed with respectively 10 ml of P. aeruginosa, B. cenocepacia or S. maltophilia for cocultures. Cultures were grown for 8 h. Every two hours, cultures were plated on mannitol salt agar (MSA, BBLTM Difco) and cetrimide (DifcoTM) for S. aureus or P. aeruginosa counts, respectively.
For supernatant exposure, 10 mL of S. aureus culture were added to 10 ml of supernatant from 8 hours culture of P. aeruginosa.
Transwell® (Corning) preliminary experiment demonstrated that bacteria were not able to cross the 0.4 μm filter of the insert. S. aureus and P. aeruginosa suspensions from 2.5 h culture were pelleted and re-suspended to OD600nm = 1 for P. aeruginosa and OD600nm = 0.33–0.5 for S. aureus. The Transwell® experiment was carried out as previously described45,46 with a few modifications. For wells without insert, 400 µL of S. aureus OD600nm = 0.5 suspension and 200 µL of either BHI or P. aeruginosa were added. For wells with insert, 600 µL of S. aureus 0.33 OD600nm were deposed into the wells while 200 µL of either BHI or P. aeruginosa were placed onto the insert. The Transwell® system was incubated at 37 °C for 8 hours.
Staphylococcus aureus growth inhibition on TSA
From overnight cultures, S. aureus and P. aeruginosa suspensions were diluted to OD600 = 0.5 and 100 µl of S. aureus suspension were spread uniformly onto TSA plates. Then, 5 µl of P. aeruginosa were added at the center of the plates. The plates were incubated at 37 °C. The competitive phenotype was characterized by an inhibition halo of S. aureus growth, which was measured. The strains were considered as coexisting in the absence of inhibition halo.
Genome sequencing and annotation
Sequencing libraries were prepared from 1 ng of SA2597 and SA2599 DNA extracted using the DNA Isolation Kit (MO BIO). Library preparation was performed with the Nextera XT DNA sample preparation kit (Illumina) and index kit (Illumina). Library validation was performed on a 2100 Bioanalyzer (Agilent Technologies) to control the distribution of fragmented DNA. WGS was performed with an Illumina HiSeq (Illumina) to generate 150-bp paired end reads. Genomes were sequenced with an average coverage of 130x. Adapters and other illumina-specific sequences were cut from the reads for each set of raw data. Furthermore, Trimmomatic v0.3647 was used to perform an additional trimming step using a sliding window with an average quality threshold of 20. Data were checked for quality by FastQC v0.11.6 (S. Andrews, 2010. FastQC: a quality control tool for high throughput sequence data; available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Assemblies were performed using SPAdes v3.11.148. Contigs smaller than 200 bp or with a coverage threshold smaller than 2 were removed manually. Assembly quality control was performed using Quast v4.6.149. Genome annotation was processed through Prokka v1.13 including ncRNA prediction50.
To compare CDS and ncRNA from SA2597 and SA2599, the N315 strain (NC_002745.2) was used as a reference. Refseq numbers were gathered from N315 and used as ID tags for common genes. For non-common genes, CDS and ncRNA from SA2597 and SA2599 were blasted with each other with a coverage and identity of 90%. Finally, refseq numbers were also used to gather KEGG numbers and perform functional classification with the Kyoto database. The complete genome sequences for the SA2597 and SA2599 strains were deposited in GenBank under the accession numbers GCA_005280135.1 and GCA_005280145.1.
Transcriptomic analysis
Cultures and transcriptome sequencing were performed in duplicates or triplicates. The OD600 of each culture was normalized to 1.0 at a time of 4 hours for the mono and cocultures. One mL was centrifuged for 5 minutes at 13,000 rpm. Bacteria were treated with lysostaphin (2.5 mg/mL) and lysozyme (50 mg/mL) prior to RNA extraction using the RNeasy Plus Mini Kit (Qiagen). RNAs were treated with TURBO DNA-free™ (Invitrogen). rRNAs were depleted using the Ribo-Zero rRNA Removal Kit (Illumina). The cDNA libraries were compiled using the TruSeq Stranded Total RNA Library Preparation Kit (Illumina). The quantification and quality of the DNA libraries was evaluated by Bioanalyzer. The libraries were sequenced using Illumina Hi-Seq 2500 with High-Output (HO) mode, using a V4 chemistry sequencing kit (Illumina). Reads were then processed to remove adapter sequences. Poor quality reads were excluded by Trimmomatic47, using a sliding window with an average quality threshold of 20. Each RNAseq read sample was mapped against its own genome through Bowtie2 v2.3.0 with a sensitive local alignment method51. Output files were sorted by read names and converted into BAM format using Samtools v1.3.1. Reads were counted on all feature types (CDS, nc/t/tm/rRNA) using a union mode on Htseq-count v0.6.1 software52. To estimate the enrichment values for the differential expression analysis, statistical analysis was done using R v3.3.3 and DEseq2 v1.14.153. Gene expression was considered as dysregulated when: (i) the fold change between co-culture and monoculture was at least 4-fold, (ii) the dysregulation was observed in the two pairs of strains, (iii) the dysregulation was specific to coexistence or competition state. The RNAseq data that support our findings are available in the SRAdatabase under the BioprojectID PRJNA552713, PRJNA552715, PRJNA552786, PRJNA554237, PRJNA554233, PRJNA554237.
RT-qPCR
RNA extractions were performed using the RNeasy Plus Mini Kit (Qiagen). A DNAse treatment was performed on 10 μg of RNAs using TURBO DNA-free™ (Invitrogen). The absence of contaminating gDNA was controlled by PCR. cDNA was synthetized from 1 µg RNA using the Reverse Transcription system kit (Promega). The qPCR reactions were performed with PowerUpTM SYBRTM Green Master Mix (Thermofisher) following the manufacturer’s instructions. The target genes and primers used are listed in Table S2. The housekeeping genes gyrB and hu were used as endogenous control. Gene expression analyses were performed using the ∆Ct method.
Antibiotic resistance assay
MICs of tetracycline and ciprofloxacin (Sigma) were determined by BHI micro-dilution (Table S3). For the antibiotic resistance assay, 4 hour mono-cultures of S. aureus and cocultures of S. aureus/P. aeruginosa were diluted to OD600nm = 0.002 or 0.004, respectively, and exposed to antibiotics at MIC and 2xMIC for 5 hours at 37 °C at 200 rpm in 1 mL of BHI. Cultures were plated on MSA agar plates for S. aureus counts. The percentage of bacterial survival after antibiotic treatment was determined by dividing the number of S. aureus after antibiotic treatment by the number of S. aureus without antibiotic treatment.
Internalization within A549 cells
S. aureus monocultures and S. aureus/P. aeruginosa cocultures were performed for 4 hours as previously described54. A549 cells were grown in DMEM GlutaMAX™ medium (Gibco) supplemented with 10% of Fetal Bovine Serum (37 °C, 5% CO2). 24-well tissue culture plates were seeded at 80 000 cells per well. After 24 hours, the cells were washed twice with 1 ml of Phosphate Buffered Saline (PBS, Gibco) and infected at a multiplicity of infection (MOI) of 10:1 for mono-culture and 20:1 for coculture. Cells were incubated for 2 hours, then washed once in PBS to remove non-adherent bacteria and incubated for 1 hour in DMEM GlutaMAX™ supplemented with 400 µg/mL gentamicin (Sigma), 100 µg/mL polymyxin B (Sigma) and 10 µg/mL lysostaphin to kill extra-cellular bacteria. Cells were washed again with PBS once and lysed with deionized water. Cell lysates were plated on MSA to quantify the intracellular bacteria.
Ethical statement
All the methods were carried out in accordance with relevant guidelines and regulations.
Supplementary information
Acknowledgements
This work was supported by the Fondation pour la Recherche Médicale, grant number ECO20170637499 to LC; Finovi foundation to KM and FV. We thank Xavier Charpentier of CIRI, Lyon-France, for discussing the work and providing valuable advice.
Author contributions
P.B., L.C., S.B., F.V. and K.M. designed and analyzed the experiments. P.B. conducted the experiments. S.B. conducted and analyzed all bioinformatics works. A.D.J. collected the clinical samples from C.F. patients. K.M. and F.V. coordinated the project. P.B. and K.M. collected the data and wrote the first draft of the manuscript. All the authors contributed to manuscript revision and read and approved the submitted version.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
The complete genome sequences for the SA2597 and SA2599 strains have been deposited in GenBank under the accession number GCA_005280135.1 and GCA_005280145.1.
The RNAseq data that support our findings are available in the SRAdatabase under the BioprojectID PRJNA552713, PRJNA552715, PRJNA552786, PRJNA554237, PRJNA554233, PRJNA554237.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-52975-z.
References
- 1.Armbruster CE, et al. Indirect Pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in Polymicrobial Otitis Media Occurs via Interspecies Quorum Signaling. mBio. 2010;1:e00102-10–e00102-19. doi: 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc. Natl. Acad. Sci. 2013;110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sibley CD, et al. Discerning the Complexity of Community Interactions Using a Drosophila Model of Polymicrobial Infections. PLoS Pathog. 2008;4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vega NM, Allison KR, Samuels AN, Klempner MS, Collins JJ. Salmonella typhimurium intercepts Escherichia coli signaling to enhance antibiotic tolerance. Proc. Natl. Acad. Sci. 2013;110:14420–14425. doi: 10.1073/pnas.1308085110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zolin, Orenti, Naehrlich & van Rens. ECFS Patient Registry Annual Data Report 2017. (2019).
- 6.Hubert D, et al. Association between Staphylococcus aureus alone or combined with Pseudomonas aeruginosa and the clinical condition of patients with cystic fibrosis. J. Cyst. Fibros. 2013;12:497–503. doi: 10.1016/j.jcf.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Wolter DJ, et al. Staphylococcus aureus small-colony variants are independently associated with worse lung disease in children with cystic fibrosis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2013;57:384–391. doi: 10.1093/cid/cit270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman LR, et al. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 2006;103:19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maselli JH, et al. Risk factors for initial acquisition of Pseudomonas aeruginosa in children with cystic fibrosis identified by newborn screening. Pediatr. Pulmonol. 2003;35:257–262. doi: 10.1002/ppul.10230. [DOI] [PubMed] [Google Scholar]
- 10.Junge S, et al. Factors Associated with Worse Lung Function in Cystic Fibrosis Patients with Persistent Staphylococcus aureus. Plos One. 2016;11:e0166220. doi: 10.1371/journal.pone.0166220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raidt L, et al. Increased Prevalence and Resistance of Important Pathogens Recovered from Respiratory Specimens of Cystic Fibrosis Patients During a Decade. Pediatr. Infect. Dis. J. 2015;34:700–705. doi: 10.1097/INF.0000000000000714. [DOI] [PubMed] [Google Scholar]
- 12.Hudson VL, Wielinski CL, Regelmann WE. Prognostic implications of initial oropharyngeal bacterial flora in patients with cystic fibrosis diagnosed before the age of two years. J. Pediatr. 1993;122:854–860. doi: 10.1016/S0022-3476(09)90007-5. [DOI] [PubMed] [Google Scholar]
- 13.Sagel SD, et al. Impact of Pseudomonas and Staphylococcus Infection on Inflammation and Clinical Status in Young Children with Cystic Fibrosis. J. Pediatr. 2009;154:183–188.e3. doi: 10.1016/j.jpeds.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakeman, C. A. et al. The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat. Commun. 7 (2016). [DOI] [PMC free article] [PubMed]
- 15.Hogan DA, et al. Analysis of Lung Microbiota in Bronchoalveolar Lavage, Protected Brush and Sputum Samples from Subjects with Mild-To-Moderate Cystic Fibrosis Lung Disease. PLOS ONE. 2016;11:e0149998. doi: 10.1371/journal.pone.0149998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hotterbeekx, A., Kumar-Singh, S., Goossens, H. & Malhotra-Kumar, S. In vivo and In vitro Interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front. Cell. Infect. Microbiol. 7 (2017). [DOI] [PMC free article] [PubMed]
- 17.Mashburn LM, Jett AM, Akins DR, Whiteley M. Staphylococcus aureus Serves as an Iron Source for Pseudomonas aeruginosa during In Vivo Coculture. J. Bacteriol. 2005;187:554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pernet E, et al. Pseudomonas aeruginosa eradicates Staphylococcus aureus by manipulating the host immunity. Nat. Commun. 2014;5:5105. doi: 10.1038/ncomms6105. [DOI] [PubMed] [Google Scholar]
- 19.Michelsen CF, et al. Staphylococcus aureus Alters Growth Activity, Autolysis, and Antibiotic Tolerance in a Human Host-Adapted Pseudomonas aeruginosa Lineage. J. Bacteriol. 2014;196:3903–3911. doi: 10.1128/JB.02006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldan R, et al. Adaptation of Pseudomonas aeruginosa in Cystic Fibrosis Airways Influences Virulence of Staphylococcus aureus In Vitro and Murine Models of Co-Infection. PLoS ONE. 2014;9:e89614. doi: 10.1371/journal.pone.0089614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Limoli DH, et al. Pseudomonas aeruginosa Alginate Overproduction Promotes Coexistence with Staphylococcus aureus in a Model of Cystic Fibrosis Respiratory Infection. mBio. 2017;8:e00186–17. doi: 10.1128/mBio.00186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, Q., Yeo, W.-S. & Bae, T. The SaeRS Two-Component System of Staphylococcus aureus. Genes7 (2016).
- 23.Nordlund, P. & Reichard, P. Ribonucleotide Reductases. 29 (2006). [DOI] [PubMed]
- 24.Yao X, Lu C-D. Characterization of Staphylococcus aureus Responses to Spermine Stress. Curr. Microbiol. 2014;69:394–403. doi: 10.1007/s00284-014-0603-y. [DOI] [PubMed] [Google Scholar]
- 25.Yao X, Lu C-D. Functional Characterization of the potRABCD Operon for Spermine and Spermidine Uptake and Regulation in Staphylococcus aureus. Curr. Microbiol. 2014;69:75–81. doi: 10.1007/s00284-014-0556-1. [DOI] [PubMed] [Google Scholar]
- 26.Truong-Bolduc QC, et al. Role of the Tet38 Efflux Pump in Staphylococcus aureus Internalization and Survival in Epithelial Cells. Infect. Immun. 2015;83:4362–4372. doi: 10.1128/IAI.00723-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong-Bolduc QC, Dunman PM, Strahilevitz J, Projan SJ, Hooper DC. MgrA Is a Multiple Regulator of Two New Efflux Pumps in Staphylococcus aureus. J. Bacteriol. 2005;187:2395–2405. doi: 10.1128/JB.187.7.2395-2405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen PR, et al. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol. Microbiol. 2009;71:198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truong-Bolduc QC, Hooper DC. Phosphorylation of MgrA and its effect on expression of the NorA and NorB efflux pumps of Staphylococcus aureus. J. Bacteriol. 2010;192:2525–2534. doi: 10.1128/JB.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truong-Bolduc QC, Strahilevitz J, Hooper DC. NorC, a New Efflux Pump Regulated by MgrA of Staphylococcus aureus. Antimicrob. Agents Chemother. 2006;50:1104–1107. doi: 10.1128/AAC.50.3.1104-1107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fournier B, Aras R, Hooper DC. Expression of the Multidrug Resistance Transporter NorA from Staphylococcus aureus Is Modified by a Two-Component Regulatory System. J. Bacteriol. 2000;182:664–671. doi: 10.1128/JB.182.3.664-671.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pallett R, et al. Anaerobiosis influences virulence properties of Pseudomonas aeruginosa cystic fibrosis isolates and the interaction with Staphylococcus aureus. Sci. Rep. 2019;9:6748. doi: 10.1038/s41598-019-42952-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filkins LM, et al. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa Drives S. aureus towards Fermentative Metabolism and Reduced Viability in a Cystic Fibrosis Model. J. Bacteriol. 2015;197:2252–2264. doi: 10.1128/JB.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tognon, M., Köhler, T., Luscher, A. & van Delden, C. Transcriptional profiling of Pseudomonas aeruginosa and Staphylococcus aureus during in vitro co-culture. BMC Genomics20 (2019). [DOI] [PMC free article] [PubMed]
- 35.Korem M, Gov Y, Rosenberg M. Global gene expression in Staphylococcus aureus following exposure to alcohol. Microb. Pathog. 2010;48:74–84. doi: 10.1016/j.micpath.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 36.Truong-Bolduc QC, Ding Y, Hooper DC. Posttranslational Modification Influences the Effects of MgrA on norA Expression in Staphylococcus aureus. J. Bacteriol. 2008;190:7375–7381. doi: 10.1128/JB.01068-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luong TT, Dunman PM, Murphy E, Projan SJ, Lee CY. Transcription Profiling of the mgrA Regulon in Staphylococcus aureus. J. Bacteriol. 2006;188:1899–1910. doi: 10.1128/JB.188.5.1899-1910.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Truong-Bolduc QC, Khan NS, Vyas JM, Hooper DC. Tet38 Efflux Pump Affects Staphylococcus aureus Internalization by Epithelial Cells through Interaction with CD36 and Contributes to Bacterial Escape from Acidic and Nonacidic Phagolysosomes. Infect. Immun. 2017;85:e00862–16. doi: 10.1128/IAI.00862-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Löffler B, Tuchscherr L, Niemann S, Peters G. Staphylococcus aureus persistence in non-professional phagocytes. Int. J. Med. Microbiol. 2014;304:170–176. doi: 10.1016/j.ijmm.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 40.Johnson L, Mulcahy H, Kanevets U, Shi Y, Lewenza S. Surface-Localized Spermidine Protects the Pseudomonas aeruginosa Outer Membrane from Antibiotic Treatment and Oxidative Stress. J. Bacteriol. 2012;194:813–826. doi: 10.1128/JB.05230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tognon, M. et al. Co-evolution with Staphylococcus aureus leads to lipopolysaccharide alterations in Pseudomonas aeruginosa. ISME J., 10.1038/ismej.2017.83 (2017). [DOI] [PMC free article] [PubMed]
- 42.Tristan A, et al. Rise of CC398 lineage of Staphylococcus aureus among Infective endocarditis isolates revealed by two consecutive population-based studies in France. PloS One. 2012;7:e51172. doi: 10.1371/journal.pone.0051172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnaud M, Chastanet A, Debarbouille M. New Vector for Efficient Allelic Replacement in Naturally Nontransformable, Low-GC-Content, Gram-Positive Bacteria. Appl. Environ. Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boisset S, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen, A. T. et al. Cystic Fibrosis Isolates of Pseudomonas aeruginosa Retain Iron-Regulated Antimicrobial Activity against Staphylococcus aureus through the Action of Multiple Alkylquinolones. Front. Microbiol. 7 (2016). [DOI] [PMC free article] [PubMed]
- 46.Nguyen AT, Jones JW, Ruge MA, Kane MA, Oglesby-Sherrouse AG. Iron Depletion Enhances Production of Antimicrobials by Pseudomonas aeruginosa. J. Bacteriol. 2015;197:2265–2275. doi: 10.1128/JB.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinforma. Oxf. Engl. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinforma. Oxf. Engl. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinforma. Oxf. Engl. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [Google Scholar]
- 51.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinforma. Oxf. Engl. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varet H, Brillet-Guéguen L, Coppée J-Y, Dillies M-A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PloS One. 2016;11:e0157022. doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trouillet S, et al. A novel flow cytometry-based assay for the quantification of Staphylococcus aureus adhesion to and invasion of eukaryotic cells. J. Microbiol. Methods. 2011;86:145–149. doi: 10.1016/j.mimet.2011.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
The complete genome sequences for the SA2597 and SA2599 strains have been deposited in GenBank under the accession number GCA_005280135.1 and GCA_005280145.1.
The RNAseq data that support our findings are available in the SRAdatabase under the BioprojectID PRJNA552713, PRJNA552715, PRJNA552786, PRJNA554237, PRJNA554233, PRJNA554237.