Abstract
The 2′-C-methyl ribonucleosides are nucleoside analogs representing an important class of antiviral agents, especially against positive-strand RNA viruses. Their value is highlighted by the highly successful anti-hepatitis C drug sofosbuvir. When appropriately phosphorylated, these nucleotides are successfully incorporated into RNA by the virally encoded RNA-dependent RNA polymerase (RdRp). This activity prevents further RNA extension, but the mechanism is poorly characterized. Previously, we had identified NMR signatures characteristic of formation of RdRp–RNA binary and RdRp–RNA–NTP ternary complexes for the poliovirus RdRp, including an open-to-closed conformational change necessary to prepare the active site for catalysis of phosphoryl transfer. Here we used these observations as a framework for interpreting the effects of 2′-C-methyl adenosine analogs on RNA chain extension in solution-state NMR spectroscopy experiments, enabling us to gain additional mechanistic insights into 2′-C-methyl ribonucleoside-mediated RNA chain termination. Contrary to what has been proposed previously, poliovirus RdRp that was bound to RNA with an incorporated 2′-C-methyl nucleotide could still bind to the next incoming NTP. Our results also indicated that incorporation of the 2′-C-methyl nucleotide does not disrupt RdRp–RNA interactions and does not prevent translocation. Instead, incorporation of the 2′-C-methyl nucleotide blocked closure of the RdRp active site upon binding of the next correct incoming NTP, which prevented further nucleotide addition. We propose that other nucleotide analogs that act as nonobligate chain terminators may operate through a similar mechanism.
Keywords: antiviral agent, hepatitis C virus (HCV), NMR, nucleoside/nucleotide analogue, plus-stranded RNA virus, poliovirus, RNA polymerase, viral polymerase, 2′-C-methyl ribonucleoside, positive-strand RNA virus
Introduction
RNA viruses represent some of the greatest threats to global health systems, including positive-strand RNA viruses like those belonging to the Flaviviridae (1) and Picornaviridae (2, 3) families. Although vaccines have been or are currently being developed against many of these viruses (4, 5), there has also been a recognized need for small-molecule therapeutic agents to complement or even overtake these approaches (6, 7). One important drug target is the viral RNA-dependent RNA polymerase (RdRp)2 (8, 9), responsible for virus RNA replication. Replication inhibitors that target the conserved core of the RdRp may have broad antiviral activity. For example, 2′-C-methyl nucleotide analogs have wide antiviral activity against many flaviviruses, picornaviruses, and other positive-strand RNA viruses (10–12), although they may have limited utility against negative-strand RNA or DNA viruses (13). This class includes the highly successful anti-hepatitis C drug sofosbuvir (14). The RdRp incorporates the triphosphorylated 2′-C-methyl nucleotide analogs into RNA, but further RNA extension is prevented, despite these molecules containing a 3′-hydroxyl necessary for further nucleotide incorporation (10, 15, 16); such molecules are known as nonobligate chain terminators. The antiviral activity of these compounds is likely due to RNA chain termination (10, 13, 16), but the molecular mechanisms behind RNA chain termination are poorly understood. A better appreciation of these molecular mechanisms may provide insights into further development of this important class of antiviral agents.
The RdRp three-dimensional structure has been described as a cupped right hand with fingers, thumb, and palm subdomains, similar to the overall architecture of other single-subunit nucleic acid polymerases (17). Viral RdRps are unique compared with other nucleic acid polymerases in that there are extensive interactions between the fingers and thumb subdomains, providing completely encircled active sites (18) (Fig. 1). The enclosure of the RdRp active site means that mechanisms of nucleotide selection, addition, and translocation may differ from other nucleic acid polymerases. There are available X-ray crystal structures for a number of RdRps, including those for flaviviruses and picornaviruses (18). Unfortunately, the only X-ray crystal structures with RdRp (in this case, from hepatitis C virus (HCV)) in complex with 2′-C-methyl nucleotides represent a step before nucleotide incorporation, with the triphosphorylated 2′-C-methyl nucleotide acting as the next incoming NTP (19). Although these structures have revealed molecular determinants important for nucleotide analog recognition, they provide little insight into the mechanism of RNA termination.
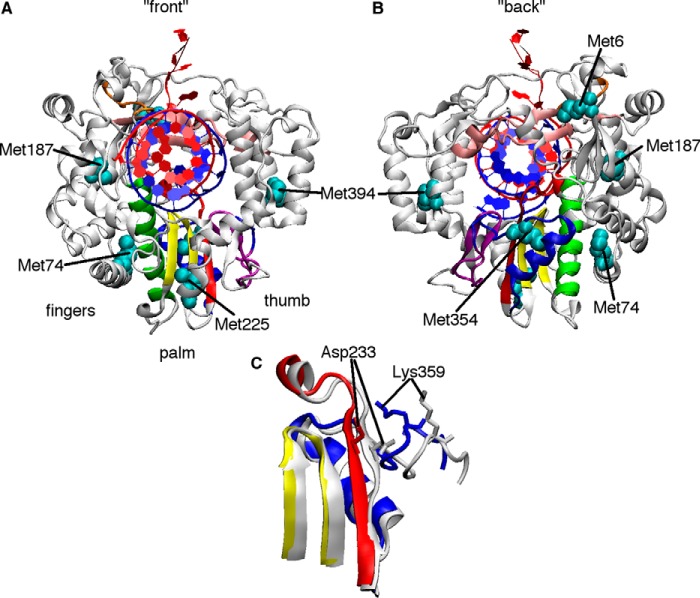
Figure 1.
Structural changes in PV RdRp necessary for nucleotide selection and incorporation. A and B, the X-ray crystal structure of PV RdRp (PDB code 3OL7) (21) with the conserved structural motifs colored (A, red; B, green; C, yellow; D, blue; E, purple; F, pink) and the locations of Met probes indicated (cyan). C, close-up of motifs A, C, and D, showing structural changes before NTP binding (white, PDB code 3OL6) (21) and after NTP binding and incorporation (colored, PDB code 3OL7). The white and colored structures represent the open and closed conformations according to X-ray crystallography. These and other structural changes may be monitored by solution-state NMR analysis of [methyl-13C]Met probes.
A series of X-ray crystal structures of poliovirus (PV) and related RdRps provides insights into the steps of nucleotide incorporation and subsequent translocation (20–22), including insights into the functional roles of the seven conserved structural motifs in RdRps (i.e. motifs A–G). After base-pairing with the template nucleotide, the 2′- and 3′-hydroxyls of the incoming NTP make key hydrogen bonding interactions with Ser-288 and Asn-297 in motif B (PV numbering), triggering structural rearrangements to “close” the active site and prepare for the phosphoryl transfer reaction (21, 22). Critically, structural motif A is realigned to form a complete three-stranded β-sheet with motif C, which causes motif A residue Asp-233 to swing toward the RNA, allowing it to coordinate the magnesium ions necessary for catalysis (21) (Fig. 1). The triphosphate of the incoming NTP is also realigned to promote inline nucleophilic attack from the primer 3′-hydroxyl, aided in part by conserved residues in motifs D and F (21–23). The phosphoryl transfer reaction then takes place, according to the two metal ion mechanism shared by other nucleic acid polymerases (24), with Asp-233 and motif C residue Asp-328 coordinating the metals (21). We have also proposed that there is a repositioning of motif D residue Lys-359 so that it can act as a general acid to protonate the pyrophosphate leaving the group (25–27). Following nucleotide addition, the active site reopens to release the pyrophosphate, but this conformational change is not tightly coupled to translocation as it is in other nucleic acid polymerases (22, 28). It has been proposed that additional structural changes in motif B then mechanically aid translocation (29, 30) to eventually reset the protein to accept the next incoming NTP.
The 2′-C-methyl ribonucleotides may act at any of these steps to terminate RNA synthesis. Initial modeling studies have proposed that the next incoming NTP is prevented from binding because of a steric clash with the 2′-C-methyl group (13). Excision of the incorporated 2′-C-methyl nucleotide through pyrophosphorolysis (i.e. the reverse of the nucleotide addition reaction) was not sensitive to the next NTP, which, the authors suggested, was also evidence that NTP binding was impeded (31). Incorporation of the 2′-C-methyl nucleotide might also disrupt interactions between the RdRp and RNA to promote RNA release, diminish the ability of the RdRp to form the catalytically competent closed conformation, and/or inhibit translocation after nucleotide analog incorporation.
In the absence of X-ray crystal structure data, we used solution-state NMR studies of PV RdRp, along with steady-state kinetics, to provide insight into the mechanism of RNA chain termination by 2′-C-methyl ribonucleotides. These studies suggest that incorporation of 2′-C-methyl nucleotide prevents subsequent active-site closure, a prerequisite step for incorporation of the next incoming nucleotide.
Results
Incorporation of 2′-C-Me-AMP by PV RdRp terminates RNA synthesis and inhibits virus replication
We first tested the ability of PV RdRp to incorporate 2′-C-methyl nucleotide. Here we used 2′-C-methyladenosine (2′-C-Me-A) as a representative member of the 2′-C-methyl nucleotide analogs (Fig. 2), and the ssAU RNA, which contains a 6-bp duplex flanked by four nucleotide 5′ overhangs (i.e. 5′-GCAUGGGCCC-3′) (32) (Fig. 2). For the ssAU RNA, the next correct incoming nucleotides would be ATP followed by UTP. PV RdRp was able to incorporate 2′-C-Me-AMP, but this resulted in RNA chain termination, preventing the second nucleotide from being incorporated (Fig. 2). Previous kinetics studies had indicated that incorporation of 2′-C-Me-AMP occurs with reduced catalytic efficiency (33), as evidenced by a decreased second-order rate constant kpol/Kd,app, where kpol is the maximum polymerase rate constant and Kd,app is the apparent dissociation constant for the incoming NTP (Fig. 2). Interestingly, the second-order rate constants for 2′-C-Me-AMP and 2′-dAMP incorporation were very similar, but 2′-C-Me-AMP was more efficiently incorporated than GMP (33) (Fig. 2). As shown previously (13), our studies also indicated that 2′-C-Me-A has anti-PV activity (Fig. 2).
Figure 2.

Incorporation of 2′-C-Me-AMP by PV RdRp terminates RNA synthesis and inhibits virus replication. A, chemical structure of 2′-C-Me-adenosine. B, sequence and base-pairing within the ssAU RNA used in many of the described studies. C, incorporation of 2′-C-Me-AMP terminates RNA synthesis. In brief, PV RdRp (1 μm) was incubated with 20 μm ssAU RNA (i.e. 10 μm duplex RNA) and appropriate NTPs (500 μm). RNA reaction products were resolved by denaturing PAGE and visualized by phosphorimaging. When 2′-C-Me-ATP and UTP were incubated with ssAU RNA, there was no band corresponding to two nucleotide addition events (i.e. +2 position) for over 5 h (i.e. under the time necessary for the NMR experiments). D, the second-order rate constants for single-nucleotide incorporation based on the ssAU RNA as determined previously (33, 51). The kinetic parameters kpol and Kd are the maximum polymerase rate constant and the apparent dissociation constant for the incoming NTP, respectively. For the kinetics studies with mispaired termini, studies were conducted using the RNA sequence 5′-GCAUGGGCCCG-3′ so that the RNA terminus contained a G:U mismatch and the next correct incoming NTP was UTP (27). E, 2′-C-Me-adenosine has antiviral activity. HeLa S3 cells were incubated with 2′-C-methyladenosine for 1 h at the concentrations shown and subsequently infected with 106 pfu of PV. Fifteen minutes after the infection, fresh medium containing 2′-C-methyladenosine at the appropriate concentration was added, and the infection progressed for 6 h. Cell-associated virus was titered with plaque assays.
13C-Met probes report on ligand binding and conformational changes throughout the RdRp structure
PV RdRp is 52 kDa in size, which has made analysis of backbone NMR spectra challenging. Nonetheless, it is now well-established that larger proteins (>50 kDa) can be studied by solution-state NMR using 13C isotopically labeled methyl groups, in part because of their favorable relaxation properties, which can result in higher signal to noise (34). Indeed, we have previously used [methyl-13C]Met groups in PV RdRp to gain insight into structural and dynamic changes important for nucleotide selection and catalysis (27, 35). The [methyl-13C]Met probes are conveniently located in or near regions that have been shown by X-ray crystallography structures and/or molecular dynamics simulations to undergo structural and/or dynamic changes upon binding RNA and/or NTP (20–22, 36, 37) (Fig. 1). In this study, we were especially interested in the Met-6, Met-74, Met-187, Met-225, Met-354, and Met-394 probes, as their associated resonances have been shown to undergo chemical shift and/or peak intensity changes upon binding RNA and/or NTP (27, 35) (Fig. 3). Other Met probes are more solvent-exposed and so do not respond to ligand binding and/or associated structural changes (35).
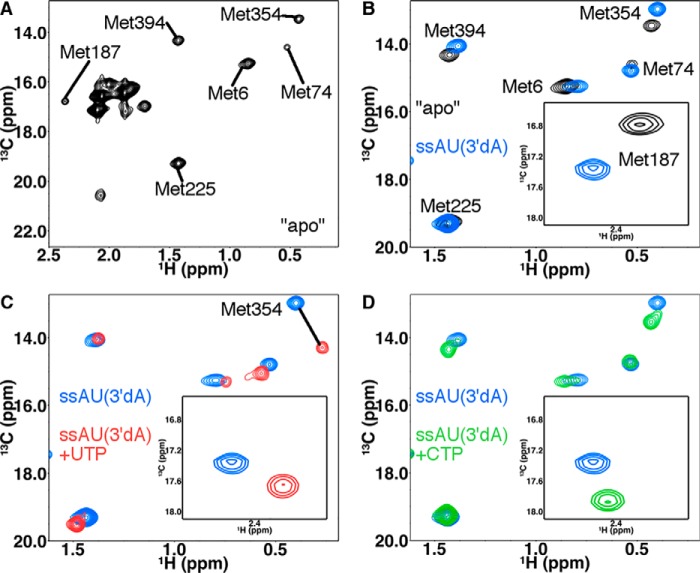
Figure 3.
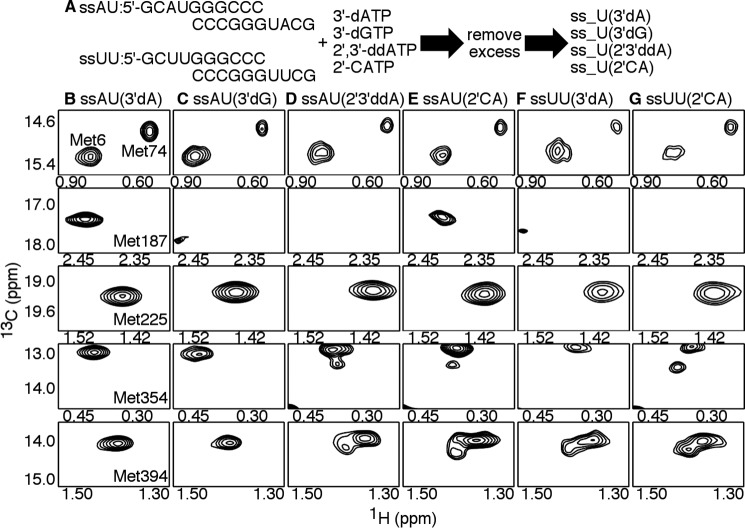
[Methyl-13C]Met chemical shift perturbations provide insight into structure and dynamic changes in PV RdRp upon binding RNA and nucleotide. A, 1H-13C HSQC of PV RdRp in the absence of RNA and nucleotide. Important Met resonances are labeled. Other Met resonances do not show chemical shift changes upon RNA and/or nucleotide addition and so were not considered further. B–D, 1H-13C HSQC comparisons between PV RdRp in the absence of RNA and nucleotide (black), PV RdRp bound to ssAU(3′dA) RNA (blue), PV RdRp bound to ssAU(3′dA) RNA and UTP (red), and PV RdRp bound to ssAU(3′dA) RNA and CTP (green). The resonances for Met-187 are indicated in the insets. For these experiments, PV RdRp (250 μm) is first incubated with 500 μm duplex RNA (ssAU (Fig. 2)) and 4 mm 3′-dATP so that 3′-dAMP is incorporated but lack of the 3′-hydroxyl prevents further nucleotide addition. Excess 3′-dATP is removed through a desalting column before addition of the second NTP (4–12 mm) to generate the ternary RdRp–RNA–NTP complexes. The D2O-based buffer consisted of 10 mm HEPES (pH 8.0), 200 mm NaCl, 0.02% NaN3, 5 mm MgCl2, and 10 μm ZnCl2. NMR spectra were collected at 293 K using a Bruker Avance III 600 MHz spectrometer. Aspects of these NMR experiments have been reported previously (27, 35, 36).
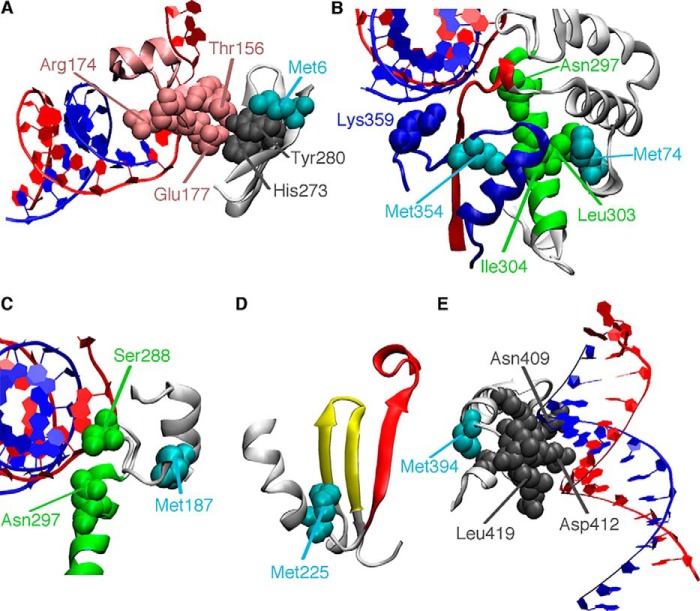
Met-6, Met-74, and Met-187 are located in the fingers subdomain and likely respond to structural rearrangements in motifs A, B, and F. Met-6 is part of the N-terminal β-strand that makes hydrogen bond interactions with palm structural motifs A and B. Met-6 is also part of the three-strand β-sheet that makes contact with residues in structural motif F (Fig. 4). Motif F is important for RNA template and incoming NTP binding (20, 21). Arg-174 in motif F is likely responsible for properly aligning the α-phosphate with the primer 3′-hydroxyl (38); the R174K-substituted variant provides the highest nucleotide selection incorporation fidelity for any PV variant characterized so far (32), and Coxsackievirus encoding the R174K substitution is not viable (39). His-273 is also part of this interaction network, and it has been shown that the H273R substitution results in a lower nucleotide selection fidelity; PV encoding the H273R substitution has reduced virulence in a mouse model (36, 40). Met-74 makes van der Waals contact with the motif B α-helix (Fig. 4) and with residues on the α1-α2 loop that also interact with motifs A and B. Met-187 makes contact with the motif B loop important for recognition of the ribose of the incoming NTP (Fig. 4), and residues in this loop may be important for inducing translocation along the RNA (29).
Figure 4.
Methionine probes report on ligand binding and conformational changes throughout the RdRp structure. The template and primer RNA strands are colored red and blue, respectively. A, Met-6 is part of the three-stranded β-sheet that makes interactions with residues in motif F (pink). B, Met-74 is in the fingers subdomain and makes van der Waals contact with residues on the motif B helix (green). Met-354 is in motif D (blue). Other important residues include the proposed general acid Lys-359 and Asn-297, which makes hydrogen-bonding interactions with the 2′-hydroxyl of the incoming NTP. C, Met-187 is near motif B, including Ser-288 and Asn-297, and so likely reports on RNA and NTP binding. D, Met-225 is near motifs A and C (red and yellow, respectively) and likely reports on the realignment of motif A to form the three-stranded β-sheet important in active-site closure. E, Met-394 is near residues important for RNA binding.
Met-225 and Met-354 are both located in the palm subdomain. Met-225 makes contact with residues in motifs A and C (Fig. 4) and likely responds to structural changes in these regions, including realignment of the β-strand in motif A as part of active-site closure (21) (Fig. 1). Met-354 is in motif D (Fig. 4), and we have proposed that this probe is responsive to structural changes that reposition the general acid Lys-359 for catalysis (27). This probe might also be responsive to accompanying structural changes in motifs A and B. Met-394 is on the thumb subdomain, near the nascent RNA channel (Fig. 4), and likely reports on structural changes important for binding and positioning the RNA primer/template.
Our previous NMR studies of RdRp–RNA binary and RdRp–RNA–NTP ternary complexes provide a framework for comparing this study with the 2′-C-Me-A analogs. The 1H and 13C chemical shifts for all complexes analyzed in this manuscript can be found in Table S1. Upon addition of ssAU RNA to ligand-free PV RdRp, there were chemical shift changes to the Met-6, Met-187, Met-225, Met-354, and Met-394 resonances and a peak intensity change in the Met-74 resonance (35), suggesting structural and/or dynamics changes in all of the associated regions (Fig. 3 and Fig. S1). To gain insight into ternary RdRp–RNA–NTP complexes, we first added 3′-dATP, which the RdRp incorporates into the ssAU RNA to produce the ssAU(3′dA) RNA, and then the RdRp–RNA complex was passed through a desalting column to remove excess 3′-dATP. Previous studies indicated minor chemical shift differences for Met-187 and Met-354 between the RdRp–ssAU and RdRp–ssAU(3′dA) binary complexes (35) (Fig. S1). The RdRp–ssAU(3′dA) RNA complex can bind but not incorporate the next incoming NTP. Addition of the next correct NTP to form the RdRp–ssAU(3′dA)–UTP ternary complex resulted in additional chemical shift changes in the Met-6, Met-74, Met-187, Met-225, and Met-354 resonances (Fig. 3 and Fig. S1). Addition of incorrect NTP (2′-dUTP or CTP) resulted in chemical shift changes to the Met-6, Met-187, Met-354, and Met-394 resonances compared with what was observed for the RdRp–RNA(3′dA) binary complex, indicating that the incorrect NTP still binds but does not induce the same conformational changes as the correct NTP (Fig. 3). Notably, the chemical shift patterns were different between the RdRp–ssAU(3′dA)–UTP ternary complex and the RdRp–ssAU(3′dA)–2′-dUTP and RdRp–ssAU(3′dA)–CTP ternary complexes (Figs. 3 and 5). These studies established NMR “fingerprints” for when the enzyme is in an “open” (e.g. RdRp–ssAU(3′dA)–CTP) or a closed (e.g. RdRp–ssAU(3′dA)–UTP) conformation. In the open conformation, it is likely that Asp-233, the triphosphate of the incoming NTP, and/or the general acid Lys-359 are not properly positioned for the nucleotide addition reaction.
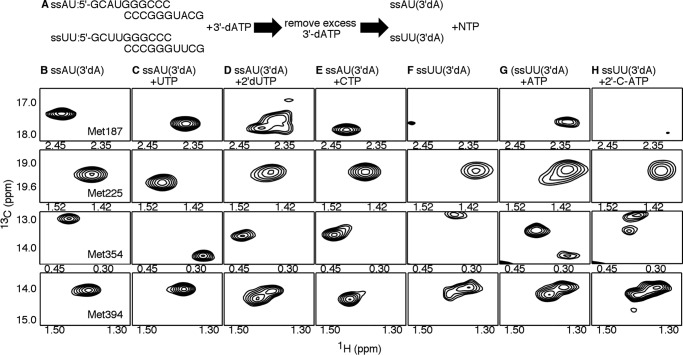
Figure 5.
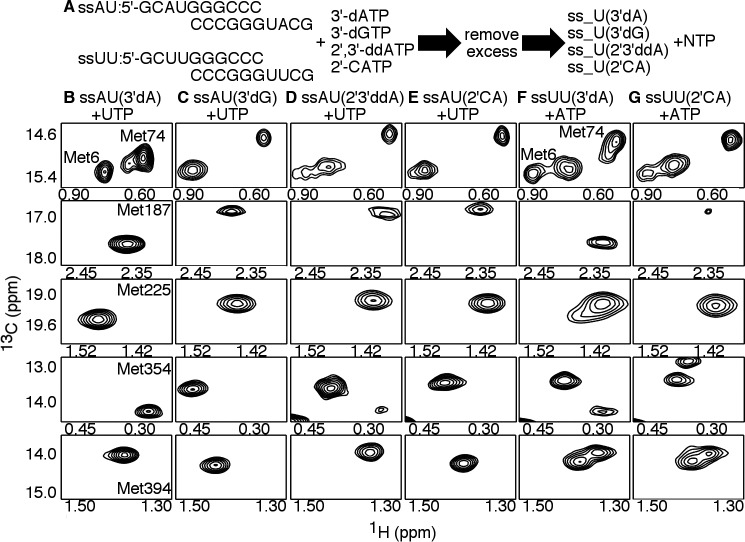
The RdRp–RNA ternary complex with 2′-C-Me-ATP does not achieve the closed conformation. A, experimental design. PV RdRp (250 μm) is first incubated with 500 μm duplex RNA (ssAU or ssUU) and 4 mm 3′-dATP so that 3′-dAMP is incorporated, but lack of the 3′-hydroxyl prevents further nucleotide addition. Excess 3′-dATP is removed through a desalting column before addition of the second NTP (4–12 mm) to generate the ternary RdRp–RNA–NTP complexes. The D2O-based buffer consisted of 10 mm HEPES (pH 8.0), 200 mm NaCl, 0.02% NaN3, 5 mm MgCl2, and 10 μm ZnCl2. B–H, [13C-methyl]Met 1H-13C HSQC NMR spectra of different RdRp–RNA binary and RdRp–RNA–NTP ternary complexes, including the RdRp–ssAU(3′dA) (i.e. ssAU RNA with incorporated 3′-dAMP) (B), RdRp–ssAU(3′dA)–UTP (C), RdRp–ssAU(3′dA)–2′-dUTP (D), RdRp:ssAU(3′dA)–CTP (E), RdRp–ssUU(3′dA) (i.e. ssUU RNA with incorporated 3′-dAMP) (F), RdRp–ssUU(3′dA)–ATP (G), and RdRp–ssUU(3′dA)–2′-C-Me-ATP (H) complexes. Resonances belonging to the ϵ-13CH3 groups of Met-187, Met-225, Met-354, and Met-394 are highlighted. NMR spectra were collected at 293 K using a Bruker Avance III 600 MHz spectrometer.
The RdRp–RNA ternary complex with 2′-C-methyl-ATP does not achieve the closed conformation
To better understand how 2′-C-methyl-ATP interacts with PV RdRp, binary and ternary complexes were evaluated using the ssUU RNA (5′-GCUUGGGCCC-3′), which was 3′-H–terminated according to previous procedures. There were some differences in the NMR spectra between the RdRp–ssAU(3′dA) and RdRp–ssUU(3′dA) binary complexes (Fig. 5). In particular, there were minor chemical shift differences for the Met-225 and Met-354 resonances. Perhaps more interestingly, there was evidence of conformational exchange in regions associated with Met-187 and Met-394. Met-394 was associated with two resonances, and the Met-187 resonance was at very low intensity, suggestive of conformational exchange on the intermediate NMR timescale. These findings suggested that the RdRp complexes with ssUU(3′dA) were more structurally dynamic, which complicated spectral analysis.
The ternary complex with correct incoming NTP (i.e. RdRp–ssUU(3′dA)–ATP) also exhibited signs of conformational exchange (Fig. 5). In this case, there were (at least) two resonances associated with Met-225, Met-354, and Met-394. The two resonances for Met-354 in the RdRp–ssUU(3′dA)–ATP complex had similar chemical shift positions as those observed for Met-354 in the RdRp–ssAU(3′dA)–UTP and RdRp–ssAU(3′dA)–CTP complexes. This result suggested that the RdRp–ssUU(3′dA)–ATP complex fluctuated between the closed and open states, although, in this case, it appeared that the open state was more favored. Although this result was not expected, it should be noted that previous single-nucleotide incorporation studies indicated that AMP incorporation templated against U had the lowest kpol/Kd,app value for all “correct” nucleotide incorporations (41). This result may be due to a more limited ability of PV RdRp to fluctuate into the more active closed conformation, consistent with the results observed here. NMR studies with other RNA templates may provide more insight. Nonetheless, the Met-187 resonance for the RdRp–ssUU(3′dA)–ATP complex was at a similar chemical shift as that observed for the RdRp–ssAU(3′dA)–UTP complex, suggesting that motif B and the surrounding region still recognized ATP as the correct incoming nucleotide.
The RdRp–ssUU(3′dA)–2′-C-Me-ATP ternary complex had substantial spectral differences compared with the RdRp–ssUU(3′dA)–ATP complex (Fig. 5). The Met-187 resonance for the RdRp–ssUU(3′dA)–2′-C-Me-ATP complex had a very low intensity, again suggestive of conformational exchange, although the chemical shift position was different from that for the RdRp–ssUU(3′dA) complex (Fig. 5). The Met-225 probe did not show evidence of conformational exchange. In contrast, both Met-354 and Met-394 were associated with two (or more) resonances, but neither of the Met-354 resonances corresponded to chemical shift positions characteristic of the closed conformation, as observed for the RdRp–ssAU(3′dA)–UTP and RdRp–ssUU(3′dA)–ATP complexes.
A mispaired RNA terminus prevents active-site closure
Because we have established that PV RdRp can successfully incorporate 2′-C-Me-AMP but then cannot extend RNA synthesis (Fig. 2), we wanted to better understand the downstream consequences of 2-C-Me-AMP incorporation. We had previously analyzed the consequences of GMP misincorporation (templated against U) (27). For the NMR studies, we incubated ssAU RNA with 3′-dGTP and then removed excess 3′-dGTP by passage across a desalting column to generate the RdRp–ssAU(3′dG) binary complex. We did not note substantial differences between the RdRp–ssAU(3′dA) and RdRp–ssAU(3′dG) complexes (Fig. 6), except for the Met-187 resonance. This resonance had a very low peak intensity for the RdRp–ssAU(3′dG) complex, suggesting conformational exchange on the intermediate NMR timescale for the associated region. The small differences between the RdRp–ssAU(3′dA) and RdRp–ssAU(3′dG) complexes suggested that these complexes reached similar conformations, except perhaps in or near motif B.
Figure 6.
Incorporation of 2′-C-Me-AMP does not substantially affect the structure and/or dynamics of the RdRp–RNA binary complex. A, experimental design. PV RdRp (250 μm) is first incubated with 500 μm duplex RNA (ssAU or ssUU) and 4 mm nucleotide analogs, which, when incorporated, terminate RNA synthesis. The nucleotide analogs include the obligate chain terminators 3′-dATP, 3′-dGTP, and 2′,3′-ddATP and the nonobligate chain terminator 2′-C-Me-ATP (2′-CATP). Excess nucleotide analog was later removed through a desalting column to generate the RdRp–RNA binary complexes. The D2O-based buffer consisted of 10 mm HEPES (pH 8.0), 200 mm NaCl, 0.02% NaN3, 5 mm MgCl2, and 10 μm ZnCl2. B–G, [13C-methyl]Met 1H-13C HSQC NMR spectra of different RdRp–RNA binary complexes, including RdRp–ssAU(3′dA) (i.e. ssAU RNA with incorporated 3′-dAMP) (B), RdRp–ssAU(3′dG) (i.e. ssAU RNA with incorporated 3′-dGMP) (C), RdRp–ssAU(2′3′ddA) (i.e. ssAU RNA with incorporated 2′,3′-ddAMP) (D), RdRp–ssAU(2′CA) (i.e. ssAU RNA with incorporated 2′-C-Me-AMP) (E), RdRp–ssUU(3′dA) (i.e. ssUU RNA with incorporated 3′-dAMP) (F), and RdRp–ssUU(2′CA) (i.e. ssUU RNA with incorporated 2′-C-Me-AMP) (G). Resonances belonging to the ϵ-13CH3 groups of Met-6, Met-74, Met-187, Met-225, Met-354, and Met-394 are highlighted. NMR spectra were collected at 293 K using a Bruker Avance III 600 MHz spectrometer.
There were additional chemical shift changes for the Met-187, Met-354, and Met-394 resonances upon addition of the next correct nucleotide to form the RdRp–ssAU(3′dG)–UTP ternary complex (Fig. 7 and Fig. S1). The chemical shift positions for the Met-6, Met-74, Met-225, Met-354, and Met-394 resonances were very similar between RdRp–ssAU(3′dA)–CTP (i.e. matched RNA terminus, incorrect incoming NTP) and RdRp–ssAU(3′dG)–UTP (i.e. mismatched RNA terminus, correct incoming NTP), suggesting that the RdRp–ssAU(3′dG)–UTP complex also did not form the closed conformation (Fig. S1). Consistent with this result was the finding that nucleotide incorporation onto RNA with a terminal mispair was slowed to a similar extent as incorrect nucleotide incorporation onto RNA with a properly paired terminus (Fig. 2).
Figure 7.
Incorporation of 2′-C-Me-AMP prevents active-site closure upon binding the next incoming nucleotide. A, experimental design. PV RdRp (250 μm) is first incubated with 500 μm duplex RNA (ssAU or ssUU) and 4 mm nucleotide analogs, which, when incorporated, terminate RNA synthesis. The nucleotide analogs include the obligate chain terminators 3′-dATP, 3′-dGTP, and 2′,3′-ddATP and the nonobligate chain terminator 2′-C-Me-ATP (2′-CATP). Excess nucleotide analog was later removed through a desalting column, followed by addition of the second NTP (4–12 mm) to generate the RdRp–RNA–NTP ternary complexes. The D2O-based buffer consisted of 10 mm HEPES (pH 8.0), 200 mm NaCl, 0.02% NaN3, 5 mm MgCl2, and 10 μm ZnCl2. B–G, [13C-methyl]Met 1H-13C HSQC NMR spectra of different RdRp–RNA–NTP ternary complexes, including the RdRp–ssAU(3′dA)–UTP (B), RdRp–ssAU(3′dG)–UTP (C), RdRp–ssAU(2′3′ddA)–UTP (D), RdRp–ssAU(2′CA)–UTP (E), RdRp–ssUU(3′dA)–ATP (F), and RdRp–ssUU(2′CA)–ATP (G) complexes. Designations for the chain-terminated RNA can be found in the legend for Fig. 6, including the ssAU(2′CA) and ssUU(2′CA) RNA, which are ssAU and ssUU RNA with incorporated 2′-C-Me-AMP. Resonances belonging to the ϵ-13CH3 groups of Met-6, Met-74, Met-187, Met-225, Met-354, and Met-394 are highlighted. NMR spectra were collected at 293 K using a Bruker Avance III 600 MHz spectrometer.
2′-C-Me-AMP incorporation has minor effects on the RdRp–RNA binary complex
We collected NMR spectra for additional RdRp–RNA binary and RdRp–RNA–NTP ternary complexes with mismatched RNA termini by incorporating 2′,3′-ddATP (i.e. ssAU(2′3′ddA)) and 2′-C-methyl-ATP (i.e. ssAU(2′CA) and ssUU(2′CA)) (Figs. 6 and 7 and Fig. S2). The RdRp–ssAU(2′3′ddA), RdRp–ssAU(2′CA), and RdRp–ssUU(2′CA) binary complexes had similar NMR spectra and only minor differences compared with the NMR spectra of the RdRp–ssAU(3′dA) and RdRp–ssUU(3′dA) complexes (Fig. 6). Along with the Met-354 resonance observed for the RdRp–ssAU(3′dA) and RdRp–ssUU(3′dA) complexes, there was another lower-intensity resonance observed for the RdRp–ssAU(2′3′ddA), RdRp–ssAU(2′CA), and RdRp–ssUU(2′CA) complexes, suggesting some conformational exchange in motif D and associated regions. There were also two resonances associated with Met-394, although similar behavior was observed for the RdRp–ssUU(3′dA) complex. Perhaps most interesting, the Met-187 resonance for the RdRp–ssAU(2′CA) complex was more intense than that observed in either the RdRp–ssAU(3′dG) and RdRp–ssAU(2′3′ddA) complexes, although at a slightly different chemical shift position than that observed for the RdRp–ssAU(3′dA) complex. This finding suggests that motif B and surrounding regions are less conformationally dynamic when the RdRp is bound to ssAU(2′CA) compared with when bound to either ssAU(3′dG) or ssAU(2′3′ddA). The incorporated 2′-C-Me-AMP may still make appropriate interactions with the RdRp, including a hydrogen bond between its 2′-hydroxyl and the backbone carbonyl Tyr-326, to limit conformational exchange in this region.
Incorporation of 2′-C-Me-AMP prevents active-site closure
The NMR spectra for the RdRp–RNA–NTP ternary complexes with mismatched RNA termini provided insight into 2′-C-Me-AMP–induced chain termination (Fig. 7). The NMR spectrum for the RdRp–ssAU(2′CA)–UTP complex was very similar to that of the RdRp–ssAU(3′dG)–UTP and RdRp–ssAU(3′dA)–CTP complexes. The RdRp–ssUU(2′CA)–UTP complex also had similar resonances, although analysis of this spectrum was complicated by conformational exchange events observed in the ssUU-bound complexes that appeared to be absent in the ssAU-bound complexes. These results imply that UTP was capable of binding to the RdRp–ssAU(2′CA) and RdRp–ssUU(2′CA) complexes but that the ternary complex was not able to fluctuate into the closed conformation necessary for nucleotide incorporation.
The next incoming NTP can still bind after 2′-C-Me-AMP incorporation
Our NMR studies suggested that the next incoming NTP could still bind after 2′-C-Me-AMP incorporation, although the ternary complex is not able to fluctuate into the closed conformation necessary for the phosphoryl transfer reaction. This proposal is in contrast to the previous suggestion that 2′-C-Me-AMP incorporation sterically prevents the next NTP from binding (13). To verify our proposal, we conducted steady-state nucleotide incorporation assays (32) to monitor the effects of the next incoming NTP on RdRp–RNA complex dissociation (Fig. 8). Under steady-state conditions, the rate-determining step for nucleotide incorporation is dissociation of the RdRp–RNAn+1 complex so that the RdRp can bind to another RNAn for another nucleotide incorporation reaction (32, 42) (Fig. 8A). Addition of the next incoming NTP may have an effect on the rate of RdRp–RNA complex dissociation (Fig. 8). For example, the steady-state incorporation of 3′-dAMP was reduced in the presence of UTP, likely because the RdRp becomes trapped in a tight closed conformation (i.e. RdRp–ssAU(3′dA)–UTP) that prevents complex dissociation. Formation of the closed conformation would also block the entrance of pyrophosphate and protect the 3′-dAMP from pyrophosphorolysis, as observed with the HCV RdRp (31). With 2′-C-Me-AMP misincorporation, the presence of UTP increased the steady-state rate, implying that UTP reduced the stability of the RdRp–RNA complex (Fig. 8). However, this result also indicates that UTP is still able to bind and form the RdRp–ssAU(2′CA)–UTP complex to exert such an effect. In contrast, addition of the noncognate nucleotide CTP had no effect on the steady-state rate (Fig. 8), likely because CTP would not bind sufficiently to form a ternary complex. RNA incorporated with 2′-C-Me nucleotide prevents the RdRp from forming the closed conformation and likely compromises the ability of the next incoming NTP to protect the 2′-C-Me nucleotide from pyrophosphorolytic excision, as observed with HCV RdRp (31).
Figure 8.
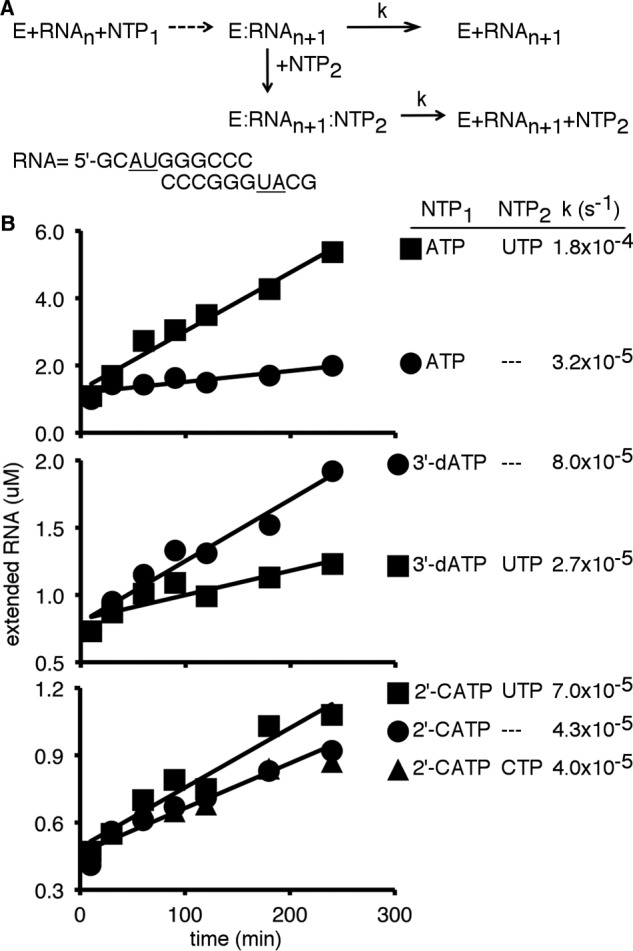
Incorporation of 2′-C-Me-AMP does not prevent the next nucleotide from binding. A, experimental design. The rate-determining step in steady-state nucleotide incorporation is dissociation of the RdRp–RNAn+1 complex (42), in which RNAn is the ssAU RNA, and RNAn+1 is the ssAU RNA following a single-nucleotide incorporation event. Addition of a second NTP may affect RdRp–RNA complex dissociation to affect the steady-state rate constant. B, steady-state incorporation of nucleotides. Reactions contained 20 μm ssAU RNA (i.e. 10 μm duplex), 5 mm MgCl2, 1 μm PV RdRp, and 500 μm NTPs. Reactions were initiated by addition of NTP(s), incubated at 30 °C, and quenched by addition of EDTA to a final concentration of 50 mm at the indicated times. The steady-state rate constants (k) were determined by dividing the slope by the y-intercept of the line indicated. Addition of UTP (i.e. the next correct incoming NTP) increased the rate of dissociation between the RdRp and RNA with incorporated 2′-C-Me-AMP (2′-CATP, 2′-C-Me-ATP). This result suggests that UTP is still able to bind to form the RdRp–ssAU(2′CA)–UTP ternary complex.
Discussion
2′-C-Me ribonucleotides have become a very successful class of antiviral agents in the clinic, exemplified by the anti-HCV drug sofosbuvir (14). However, the molecular mechanism by which they terminate RNA synthesis has remained poorly understood. We chose PV RdRp (Fig. 1) as a model system, considering the vast amount of kinetic and structural data available, including our previous solution-state NMR studies (27, 35). PV RdRp can incorporate 2′-C-Me-AMP into RNA, which then leads to RNA chain termination (Fig. 2). We had previously established [methyl-13C]Met probes that report on RNA and NTP binding, including structural/dynamic changes representative of the less active open and more active closed conformations (Figs. 3 and 4). The NMR studies indicated that binding of 2′-C-Me-ATP as the next incoming NTP did not result in achievement of the closed conformation (Fig. 5), which was consistent with the reduced second-order rate constant (kpol/Kd,app) for 2′-C-Me-AMP incorporation compared with AMP incorporation. After 2′-C-Me-AMP was incorporated, the resulting RdRp–RNA binary complex resembled that of other RdRp–RNA complexes (Fig. 6). However, addition of the next correct incoming NTP did not result in formation of the closed conformation (Fig. 7). Steady-state kinetics studies also indicated that the next correct NTP can still bind following 2′-C-Me-AMP incorporation (Fig. 8). It has been proposed previously that incorporation of 2′-C-Me ribonucleotides terminates RNA synthesis because the 2′-C-methyl group sterically prevents the next incoming NTP from binding (13). Although the 2′-C-methyl group may still interfere with the binding of the next NTP and may increase its apparent dissociation constant, our studies indicate that binding of the next NTP is not completely abolished. These results also imply that translocation to the next register is not impeded, considering that, in PV RdRp, translocation occurs before binding of the next incoming NTP (28). Altogether, our results indicate RNA synthesis is terminated because incorporation of 2′-C-Me-AMP prevents the RdRp from fluctuating into the closed conformation necessary for incorporation of the next NTP.
It has been shown previously that the S282T mutation in HCV confers decreased susceptibility to most 2′-C-Me ribonucleotides, including sofosbuvir (43). This Ser is important in binding and recognition of the 2′-hydroxyl group of the incoming ribonucleotide. It has been suggested that this amino acid substitution may interfere with initial 2′-C-Me-NTP binding because of the steric clash from the additional Thr side-chain methyl group (44). However, it is also worth noting that the resistance variant has some ability to extend RNA synthesis past the incorporated nucleotide analog (13). Moreover, the S282T polymerase has lower catalytic efficiency and is associated with a fitness deficit (44, 45). These findings suggest that the S282T substitution may also have an effect on the open-to-closed conformational change necessary to incorporate the next incoming NTP, which would affect both polymerase efficiency and the ability to extend past the incorporated nucleotide analog. Additional NMR studies of the corresponding PV RdRp variant may provide further insight.
Other nonobligate chain terminators may likewise prevent the RdRp from fluctuating into the closed conformation. Nucleoside analogs may also interfere with other events of nucleic acid synthesis. For example, it has been shown that incorporation of the antiviral entecavir by the HIV reverse transcriptase results in “delayed” termination when the polymerase reaches the third nucleotide position downstream of the incorporated analog (46). Incorporation of the Thr-1106 analog results in backtracking of PV RdRp, which likely interferes with processivity and RNA yield (47). NMR studies of these analogs may provide new insights into molecular mechanisms behind translocation and processivity and provide new NMR signatures diagnostic of these mechanistic classes of antiviral nucleotides.
Experimental procedures
Materials
[γ-32P]ATP (>7000 Ci/mmol) was from VWR-MP Biomedical. Nucleoside 5′-triphosphates and 2′-deoxynucleoside 5′-triphosphates (all nucleotides were ultrapure solutions) were from GE Healthcare. 2′3′-Dideoxyadenosine 5′-triphosphate, 3′-deoxyadenosine 5′-triphosphate (cordycepin), and 3′-deoxyguanosine 5′-triphosphate were from Trilink Biotechnologies. 2′-C-methyl nucleotides were purchased from Carbosynth US LLC (San Diego, CA). RNA oligonucleotides were from Dharmacon Research, Inc. (Boulder, CO) or Integrated DNA Technologies (San Diego, CA). [Methyl-13C]methionine was from Cambridge Isotope Laboratories. HisPur nickel-nitrilotriacetic acid resin was from Thermo Scientific. Q-Sepharose Fast Flow was from GE Healthcare. All other reagents were of the highest grade, available from Sigma or Fisher.
Production and purification of PV RdRp from heterologous expression in Escherichia coli
PV RdRp was expressed in E. coli B834(DE3) pRARE cells using autoinduction and then purified as described previously (35, 48, 49). Protein samples used in NMR experiments were 13C-labeled by addition of [methyl-13C]Met to the bacterial growth medium.
NMR sample preparation and spectroscopy
NMR samples were prepared as described previously (27, 35) with NMR buffer consisting of 10 mm HEPES (pH 8.0), 200 mm NaCl, 0.02% NaN3, 5 mm MgCl2, and 10 μm ZnCl2. For generation of the RdRp–RNA binary complexes, PV RdRp (250 μm) was incubated with ssAU or ssUU RNA (500 μm duplex RNA) and 4 mm chain-terminating nucleoside triphosphate for 3–4 h before passage across a Zeba desalting column. The second NTP (4–12 mm) was then added to generate the RdRp–RNA ternary complexes. NMR experiments were performed on a Bruker Avance III 600 MHz spectrometer equipped with a 5-mm “inverse detection” triple-resonance (1H/13C/15N) single-axis gradient TCI cryoprobe. 1H-13C heteronuclear single quantum coherence (HSQC) spectra were generally acquired as 64 (t1) × 512 (t2) complex matrix, with 64–128 scans/increment and 1.0-s recovery delay at a temperature of 293 K (35).
PV RdRp assays
Steady-state kinetics assays were performed and analyzed as described previously (32). Reactions contained 50 mm HEPES (pH 7.5), 10 mm β-mercaptoethanol, 5 mm MgCl2, 60 μm ZnCl2, 1 μm PV RdRp, 20 μm ssAU RNA (i.e. 10 μm duplex RNA), and 500 μm appropriate NTPs. Reactions were initiated with addition of NTP(s), incubated at 30 °C, and then quenched at appropriate times by addition of EDTA to a final concentration of 50 mm. Products were analyzed by denaturing PAGE, and gels were visualized through use of a Phosphor Imager and quantified by using ImageQuant TL software (GE Healthcare).
Inhibition of PV replication by 2′-C-methyladenosine
The antiviral activity of 2′-C-methyladenosine was measured using procedures outlined previously (50). Infection with PV employed HeLa S3 host cells (1 × 105) plated 1 day prior to treatment in 24-well plates. Cells were pretreated by addition of 2′-C-methyladenosine nucleoside at the specified concentration in fresh medium adjusted to a final concentration of 1% DMSO. After a 1-h incubation at 37 °C, the medium was removed, and cells were infected with PV (1 × 106 pfu). Plates were incubated for 15 min at 23 °C, PBS was removed by aspiration, and fresh prewarmed (37 °C) medium containing the specified amount of nucleoside was added. The infection was allowed to proceed at 37 °C for 6 h. Cells were washed with PBS and collected after treatment with trypsin. Cells were pelleted by centrifugation, resuspended in PBS, and subjected to three freeze–thaw cycles. Cell debris was removed by centrifugation, and the supernatant containing the cell-associated virus was saved. Titer was determined by applying serial dilutions of supernatant to HeLa S3 monolayers (plated in 6-well plates 1 day before at 5 × 105 cells/well) and overlaying with growth medium containing low-melting-point agarose (1%). Plates were incubated for 2 days at 37 °C. Then the agar was removed, and plaques were visualized by staining with crystal violet (1%) in aqueous ethanol (20%).
Author contributions
A. K. B., J. J. A., H. S. O., and D. D. B. investigation; A. K. B., J. J. A., H. S. O., and D. D. B. methodology; A. K. B., J. J. A., C. E. C., and D. D. B. writing-review and editing; J. J. A., H. S. O., C. E. C., and D. D. B. conceptualization; J. J. A., C. E. C., and D. D. B. formal analysis; C. E. C. and D. D. B. supervision; C. E. C. and D. D. B. funding acquisition; C. E. C. and D. D. B. project administration; D. D. B. data curation; D. D. B. visualization; D. D. B. writing-original draft.
Supplementary Material
Acknowledgments
We thank the Boehr and Cameron laboratories for discussions regarding PV RdRp.
This work was supported by NIAID, National Institutes of Health Grants AI104878 (to D. D. B.) and AI45818 (to C. E. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2 and Table S1.
- RdRp
- RNA-dependent RNA polymerase
- HCV
- hepatitis C virus
- PV
- poliovirus
- 2′-C-A
- 2′-C-methyladenosine
- HSQC
- heteronuclear single quantum coherence.
References
- 1. Wang L. S., D'Souza L. S., and Jacobson I. M. (2016) Hepatitis C: a clinical review. J. Med. Virol. 88, 1844–1855 10.1002/jmv.24554 [DOI] [PubMed] [Google Scholar]
- 2. Sun J., Hu X. Y., and Yu X. F. (2019) Current understanding of human enterovirus D68. Viruses 11, E490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Racaniello V. R. (2006) One hundred years of poliovirus pathogenesis. Virology 344, 9–16 10.1016/j.virol.2005.09.015 [DOI] [PubMed] [Google Scholar]
- 4. Bandyopadhyay A. S., Garon J., Seib K., and Orenstein W. A. (2015) Polio vaccination: past, present and future. Future Microbiol. 10, 791–808 10.2217/fmb.15.19 [DOI] [PubMed] [Google Scholar]
- 5. Collins M. H., and Metz S. W. (2017) Progress and works in progress: update on flavivirus vaccine development. Clin. Ther. 39, 1519–1536 10.1016/j.clinthera.2017.07.001 [DOI] [PubMed] [Google Scholar]
- 6. McKinlay M. A., Collett M. S., Hincks J. R., Oberste M. S., Pallansch M. A., Okayasu H., Sutter R. W., Modlin J. F., and Dowdle W. R. (2014) Progress in the development of poliovirus antiviral agents and their essential role in reducing risks that threaten eradication. J. Infect. Dis. 210, S447–S453 10.1093/infdis/jiu043 [DOI] [PubMed] [Google Scholar]
- 7. Zając M., Muszalska I., Sobczak A., Dadej A., Tomczak S., and Jelińska A. (2019) Hepatitis C: new drugs and treatment prospects. Eur. J. Med. Chem. 165, 225–249 10.1016/j.ejmech.2019.01.025 [DOI] [PubMed] [Google Scholar]
- 8. D'Ambrosio R., Degasperi E., Colombo M., and Aghemo A. (2017) Direct-acting antivirals: the endgame for hepatitis C? Curr. Opin. Virol. 24, 31–37 10.1016/j.coviro.2017.03.017 [DOI] [PubMed] [Google Scholar]
- 9. Deval J., Symons J. A., and Beigelman L. (2014) Inhibition of viral RNA polymerases by nucleoside and nucleotide analogs: therapeutic applications against positive-strand RNA viruses beyond hepatitis C virus. Curr. Opin. Virol. 9, 1–7 10.1016/j.coviro.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carroll S. S., Tomassini J. E., Bosserman M., Getty K., Stahlhut M. W., Eldrup A. B., Bhat B., Hall D., Simcoe A. L., LaFemina R., Rutkowski C. A., Wolanski B., Yang Z., Migliaccio G., De Francesco R., et al. (2003) Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278, 11979–11984 10.1074/jbc.M210914200 [DOI] [PubMed] [Google Scholar]
- 11. Van Dycke J., Arnoldi F., Papa G., Vandepoele J., Burrone O. R., Mastrangelo E., Tarantino D., Heylen E., Neyts J., and Rocha-Pereira J. (2018) A single nucleoside viral polymerase inhibitor against norovirus, rotavirus, and sapovirus-induced diarrhea. J. Infect. Dis. 218, 1753–1758 10.1093/infdis/jiy398 [DOI] [PubMed] [Google Scholar]
- 12. Eyer L., Fojtíková M., Nencka R., Rudolf I., Hubálek Z., and Ruzek D. (2019) Viral RNA-dependent RNA polymerase inhibitor 7-deaza-2′-C-methyladenosine prevents death in a mouse model of West Nile virus infection. Antimicrob. Agents Chemother. 63, e02093–e02118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Migliaccio G., Tomassini J. E., Carroll S. S., Tomei L., Altamura S., Bhat B., Bartholomew L., Bosserman M. R., Ceccacci A., Colwell L. F., Cortese R., De Francesco R., Eldrup A. B., Getty K. L., Hou X. S., et al. (2003) Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278, 49164–49170 10.1074/jbc.M305041200 [DOI] [PubMed] [Google Scholar]
- 14. Keating G. M., and Vaidya A. (2014) Sofosbuvir: first global approval. Drugs 74, 273–282 10.1007/s40265-014-0179-7 [DOI] [PubMed] [Google Scholar]
- 15. Fung A., Jin Z., Dyatkina N., Wang G., Beigelman L., and Deval J. (2014) Efficiency of incorporation and chain termination determines the inhibition potency of 2′-modified nucleotide analogs against hepatitis C virus polymerase. Antimicrob. Agents Chemother. 58, 3636–3645 10.1128/AAC.02666-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jin Z., Tucker K., Lin X., Kao C. C., Shaw K., Tan H., Symons J., Behera I., Rajwanshi V. K., Dyatkina N., Wang G., Beigelman L., and Deval J. (2015) Biochemical evaluation of the inhibition properties of favipiravir and 2′-C-methyl-cytidine triphosphates against human and mouse norovirus RNA polymerases. Antimicrob. Agents Chemother. 59, 7504–7516 10.1128/AAC.01391-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hansen J. L., Long A. M., and Schultz S. C. (1997) Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5, 1109–1122 10.1016/S0969-2126(97)00261-X [DOI] [PubMed] [Google Scholar]
- 18. Ferrero D., Ferrer-Orta C., and Verdaguer N. (2018) Viral RNA-dependent RNA polymerases: a structural overview. Subcell. Biochem. 88, 39–71 10.1007/978-981-10-8456-0_3 [DOI] [PubMed] [Google Scholar]
- 19. Appleby T. C., Perry J. K., Murakami E., Barauskas O., Feng J., Cho A., Fox D. 3rd, Wetmore D. R., McGrath M. E., Ray A. S., Sofia M. J., Swaminathan S., and Edwards T. E. (2015) Viral replication: structural basis for RNA replication by the hepatitis C virus polymerase. Science 347, 771–775 10.1126/science.1259210 [DOI] [PubMed] [Google Scholar]
- 20. Gong P., Kortus M. G., Nix J. C., Davis R. E., and Peersen O. B. (2013) Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts. PLoS ONE 8, e60272 10.1371/journal.pone.0060272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gong P., and Peersen O. B. (2010) Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 22505–22510 10.1073/pnas.1007626107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shu B., and Gong P. (2016) Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl. Acad. Sci. U.S.A. 113, e4005–e4014 10.1073/pnas.1602591113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gohara D. W., Crotty S., Arnold J. J., Yoder J. D., Andino R., and Cameron C. E. (2000) Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B. J. Biol. Chem. 275, 25523–25532 10.1074/jbc.M002671200 [DOI] [PubMed] [Google Scholar]
- 24. Steitz T. A. (1993) DNA-dependent and RNA-dependent DNA-polymerases. Curr. Opin. Struct. Biol. 3, 31–38 10.1016/0959-440X(93)90198-T [DOI] [PubMed] [Google Scholar]
- 25. Castro C., Smidansky E., Maksimchuk K. R., Arnold J. J., Korneeva V. S., Götte M., Konigsberg W., and Cameron C. E. (2007) Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 104, 4267–4272 10.1073/pnas.0608952104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Castro C., Smidansky E. D., Arnold J. J., Maksimchuk K. R., Moustafa I., Uchida A., Götte M., Konigsberg W., and Cameron C. E. (2009) Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 16, 212–218 10.1038/nsmb.1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang X., Smidansky E. D., Maksimchuk K. R., Lum D., Welch J. L., Arnold J. J., Cameron C. E., and Boehr D. D. (2012) Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure 20, 1519–1527 10.1016/j.str.2012.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shu B., and Gong P. (2017) The uncoupling of catalysis and translocation in the viral RNA-dependent RNA polymerase. RNA Biol. 14, 1314–1319 10.1080/15476286.2017.1300221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sholders A. J., and Peersen O. B. (2014) Distinct conformations of a putative translocation element in poliovirus polymerase. J. Mol. Biol. 426, 1407–1419 10.1016/j.jmb.2013.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garriga D., Ferrer-Orta C., Querol-Audí J., Oliva B., and Verdaguer N. (2013) Role of motif B loop in allosteric regulation of RNA-dependent RNA polymerization activity. J. Mol. Biol. 425, 2279–2287 10.1016/j.jmb.2013.03.034 [DOI] [PubMed] [Google Scholar]
- 31. Deval J., Powdrill M. H., D'Abramo C. M., Cellai L., and Götte M. (2007) Pyrophosphorolytic excision of nonobligate chain terminators by hepatitis C virus NS5B polymerase. Antimicrob. Agents Chemother. 51, 2920–2928 10.1128/AAC.00186-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arnold J. J., and Cameron C. E. (2000) Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 275, 5329–5336 10.1074/jbc.275.8.5329 [DOI] [PubMed] [Google Scholar]
- 33. Liu X., Musser D. M., Lee C. A., Yang X., Arnold J. J., Cameron C. E., and Boehr D. D. (2015) Nucleobase but not sugar fidelity is maintained in the Sabin I RNA-dependent RNA polymerase. Viruses 7, 5571–5586 10.3390/v7102894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rosenzweig R., and Kay L. E. (2014) Bringing dynamic molecular machines into focus by methyl-TROSY NMR. Annu. Rev. Biochem. 83, 291–315 10.1146/annurev-biochem-060713-035829 [DOI] [PubMed] [Google Scholar]
- 35. Yang X., Welch J. L., Arnold J. J., and Boehr D. D. (2010) Long-range interaction networks in the function and fidelity of poliovirus RNA-dependent RNA polymerase studied by nuclear magnetic resonance. Biochemistry 49, 9361–9371 10.1021/bi100833r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moustafa I. M., Korboukh V. K., Arnold J. J., Smidansky E. D., Marcotte L. L., Gohara D. W., Yang X., Sánchez-Farrán M. A., Filman D., Maranas J. K., Boehr D. D., Hogle J. M., Colina C. M., and Cameron C. E. (2014) Structural dynamics as a contributor to error-prone replication by an RNA-dependent RNA polymerase. J. Biol. Chem. 289, 36229–36248 10.1074/jbc.M114.616193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moustafa I. M., Shen H., Morton B., Colina C. M., and Cameron C. E. (2011) Molecular dynamics simulations of viral RNA-dependent RNA polymerases link conserved and correlated motions of functional elements to fidelity. J. Mol. Biol. 410, 159–181 10.1016/j.jmb.2011.04.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang X., Liu X., Musser D. M., Moustafa I. M., Arnold J. J., Cameron C. E., and Boehr D. D. (2017) Triphosphate reorientation of the incoming nucleotide as a fidelity checkpoint in viral RNA-dependent RNA polymerases. J. Biol. Chem. 292, 3810–3826 10.1074/jbc.M116.750638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gnädig N. F., Beaucourt S., Campagnola G., Bordería A. V., Sanz-Ramos M., Gong P., Blanc H., Peersen O. B., and Vignuzzi M. (2012) Coxsackievirus B3 mutator strains are attenuated in vivo. Proc. Natl. Acad. Sci. U.S.A. 109, E2294–2303 10.1073/pnas.1204022109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Korboukh V. K., Lee C. A., Acevedo A., Vignuzzi M., Xiao Y., Arnold J. J., Hemperly S., Graci J. D., August A., Andino R., and Cameron C. E. (2014) RNA virus population diversity, an optimum for maximal fitness and virulence. J. Biol. Chem. 289, 29531–29544 10.1074/jbc.M114.592303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Freistadt M. S., Vaccaro J. A., and Eberle K. E. (2007) Biochemical characterization of the fidelity of poliovirus RNA-dependent RNA polymerase. Virol. J. 4, 44 10.1186/1743-422X-4-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arnold J. J., and Cameron C. E. (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 43, 5126–5137 10.1021/bi035212y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hedskog C., Chodavarapu K., Ku K. S., Xu S., Martin R., Miller M. D., Mo H., and Svarovskaia E. (2015) Genotype- and subtype-independent full-genome sequencing assay for hepatitis C virus. J. Clin. Microbiol. 53, 2049–2059 10.1128/JCM.02624-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dutartre H., Bussetta C., Boretto J., and Canard B. (2006) General catalytic deficiency of hepatitis C virus RNA polymerase with an S282T mutation and mutually exclusive resistance towards 2′-modified nucleotide analogues. Antimicrob. Agents Chemother. 50, 4161–4169 10.1128/AAC.00433-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Powdrill M. H., Tchesnokov E. P., Kozak R. A., Russell R. S., Martin R., Svarovskaia E. S., Mo H., Kouyos R. D., and Götte M. (2011) Contribution of a mutational bias in hepatitis C virus replication to the genetic barrier in the development of drug resistance. Proc. Natl. Acad. Sci. U.S.A. 108, 20509–20513 10.1073/pnas.1105797108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tchesnokov E. P., Obikhod A., Schinazi R. F., and Götte M. (2008) Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. J. Biol. Chem. 283, 34218–34228 10.1074/jbc.M806797200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dulin D., Arnold J. J., van Laar T., Oh H. S., Lee C., Perkins A. L., Harki D. A., Depken M., Cameron C. E., and Dekker N. H. (2017) Signatures of nucleotide analog incorporation by an RNA-dependent RNA polymerase revealed using high-throughput magnetic tweezers. Cell Rep. 21, 1063–1076 10.1016/j.celrep.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arnold J. J., Bernal A., Uche U., Sterner D. E., Butt T. R., Cameron C. E., and Mattern M. R. (2006) Small ubiquitin-like modifying protein isopeptidase assay based on poliovirus RNA polymerase activity. Anal. Biochem. 350, 214–221 10.1016/j.ab.2005.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gohara D. W., Ha C. S., Kumar S., Ghosh B., Arnold J. J., Wisniewski T. J., and Cameron C. E. (1999) Production of “authentic” poliovirus RNA-dependent RNA polymerase (3D(pol)) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17, 128–138 10.1006/prep.1999.1100 [DOI] [PubMed] [Google Scholar]
- 50. Harki D. A., Graci J. D., Galarraga J. E., Chain W. J., Cameron C. E., and Peterson B. R. (2006) Synthesis and antiviral activity of 5-substituted cytidine analogues: identification of a potent inhibitor of viral RNA-dependent RNA polymerases. J. Med. Chem. 49, 6166–6169 10.1021/jm060872x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu X., Yang X., Lee C. A., Moustafa I. M., Smidansky E. D., Lum D., Arnold J. J., Cameron C. E., and Boehr D. D. (2013) Vaccine-derived mutation in motif D of poliovirus RNA-dependent RNA polymerase lowers nucleotide incorporation fidelity. J. Biol. Chem. 288, 32753–32765 10.1074/jbc.M113.484428 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.