Abstract
Creation of optogenetic switches for specific activation of cell death pathways can provide insights into apoptosis and could also form a basis for noninvasive, next-generation therapeutic strategies. Previous work has demonstrated that cryptochrome 2 (Cry2)/cryptochrome-interacting β helix–loop–helix (CIB), a blue light–activated protein–protein dimerization module from the plant Arabidopsis thaliana, together with BCL2-associated X apoptosis regulator (BAX), an outer mitochondrial membrane–targeting pro-apoptotic protein, can be used for light-mediated initiation of mitochondrial outer membrane permeabilization (MOMP) and downstream apoptosis. In this work, we further developed the original light-activated Cry2-BAX system (hereafter referred to as OptoBAX) by improving the photophysical properties and light-independent interactions of this optogenetic switch. The resulting optogenetic constructs significantly reduced the frequency of light exposure required for membrane permeabilization activation and also decreased dark-state cytotoxicity. We used OptoBAX in a series of experiments in Neuro-2a and HEK293T cells to measure the timing of the dramatic morphological and biochemical changes occurring in cells after light-induced MOMP. In these experiments, we used OptoBAX in tandem with fluorescent reporters to image key events in early apoptosis, including membrane inversion, caspase cleavage, and actin redistribution. We then used these data to construct a timeline of biochemical and morphological events in early apoptosis, demonstrating a direct link between MOMP-induced redistribution of actin and apoptosis progression. In summary, we created a next-generation Cry2/CIB–BAX system requiring less frequent light stimulation and established a timeline of critical apoptotic events, providing detailed insights into key steps in early apoptosis.
Keywords: optogenetics, actin, cryptochrome, cytoskeleton, Bax, apoptosis, mitochondrial apoptosis, BCL2-associated X apoptosis regulator (BAX), cytoskeletal dynamics, effector molecule, mitochondrial outer membrane permeabilization (MOMP), OptoBAX 2.0 system
Introduction
Among the wide-ranging applications of optogenetic proteins, which include light-mediated control of transcription (1), ion channels (2), and enzymatic activity (3), the use of light for wholesale activation or inhibition of cell signaling pathways is a uniquely powerful approach to determination of cell fate, acting as a rapid noninvasive trigger for cascades of biochemical events that ultimately decide whether a cell proliferates or dies. After many years of effort, numerous cell signaling pathways critical for cell fate decisions have been elucidated, including pathways for the control of cell division (4), cell motility (5), and apoptosis (6). As a result, it is possible to identify individual proteins that serve as highly specific effector biomolecules for the initiation of signaling cascades undergirding these vital processes. Such effector molecules are ideal candidates for incorporation into light-activated protein switches for control of cell signaling pathways (7–9). These switches are useful not only for study of the effector molecules themselves but also of biochemical events that occur in the wake of their activation. In the case of apoptosis, which can be viewed as a singular destination (“cell death”) attainable by various routes (10), numerous effector molecules exist that could serve as candidates for optical control of apoptosis, including the Bcl-2 family of proteins (11). BAX, a 21-kDa protein that is a key effector of mitochondrial membrane permeabilization (MOMP)2 in the intrinsic apoptosis pathway, is one such effector molecule with an in vivo activity profile that makes it ideal for incorporation into an optogenetic switch: phosphorylation-gated activity, predominantly cytosolic localization in the “off” state, and robust initiation of permeabilization of the outer mitochondrial membrane (OMM) (12–15). Prior work has demonstrated that incorporation of BAX into a cryptochrome 2 (Cry2)–cryptochrome-interacting β helix–loop–helix (CIB)–based optogenetic switch enables light-activated MOMP and release of apoptotic effector proteins such as Smac-1 (16). A key step in the engineering of this switch was the implementation of a phosphomimetic mutation in the BAX C terminus at serine 184 (S184E), which maintains BAX in a predominantly cytosolic state by increasing the off rate of localization at the OMM, eliminating one potential source of light-independent background (17). However, another potential source of light-independent (or “dark”) background for this optogenetic switch is the dark-state interaction of the Cry2 photoreceptor with the mitochondrial Tom20–CIB construct. In this work, we created next-generation iterations of the Cry2–CIB BAX system (OptoBAX), hereafter referred to as “OptoBAX 2.0”. The optimized versions of this cell death switch include longer Cry2 variants designed to reduce the dark-state interaction with CIB and incorporate a mutation that extends the photocycle of the Cry2–CIB interaction, promoting longer association times with the OMM (18). This optimized construct requires less frequent light stimulation to maintain OptoBAX and the OMM, an advantage for long-time course experiments where blue light toxicity may be of concern (19). In addition, although the initial studies with OptoBAX were conducted in HeLa cells, this study applied the OptoBAX 2.0 system to the Neuro-2a and HEK293T cell lines, demonstrating light-initiated apoptosis in these cell lines analogous to that observed previously in HeLa cells. Finally, in HEK293T cells, a combination of fluorescently tagged proteins and commercially available dyes are used for multicolor monitoring of critical steps in apoptosis downstream of the initial mitochondrial insult (cytoskeletal rearrangement, chromatin condensation, membrane inversion, and caspase cleavage) and compared with results observed previously with a classic small-molecule initiator of the apoptotic cascade. The resulting data are used to create a timeline of key apoptotic events in HEK293T cells and provide insight into changes in actin localization and degradation of the nuclear/cytoplasmic barrier, both key steps in early apoptosis.
Results and discussion
Creation and validation of new optogenetic constructs
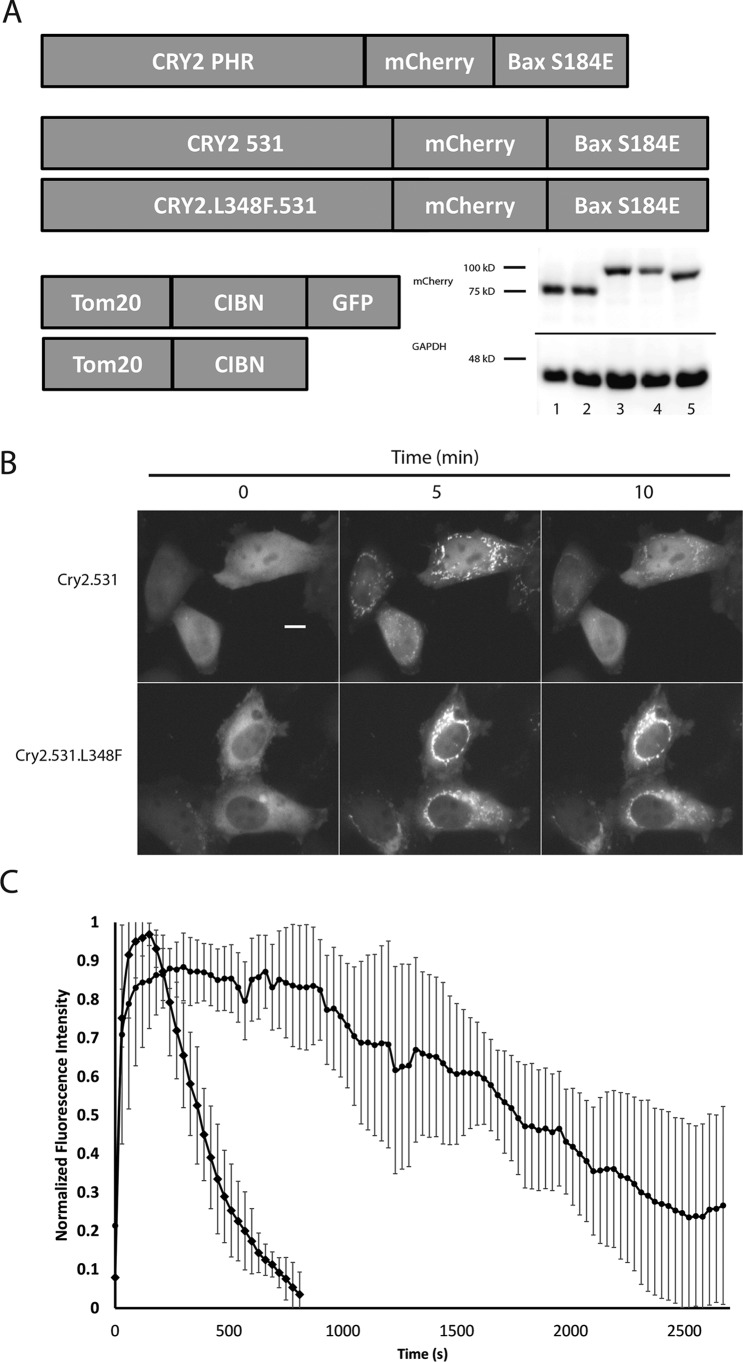
Initial versions of the OptoBAX system were comprised of BAX as a fusion to the N terminus of a Cry2PHR (1–498)–mCherry (BAX.Cry2PHR.mCh) fusion protein or as a fusion to the C terminus of a Cry2PHR (1–498)–mCherry fusion (Cry2PHR.mCh.BAX) in tandem with the Cry2-interacting partner CIB in fusion with a Tom20 OMM localization domain with or without a C-terminal GFP (Tom20.CIB.GFP or Tom20.CIB) (20). Although both BAX fusions have been demonstrated to initiate light-mediated apoptosis in HeLa cells, the C-terminal fusion of a BAX mutant (S184E) has been demonstrated to be a more effective light-activated apoptotic switch due to reduced association of BAX with the OMM because of the phosphomimic S184E and faster induction of the apoptotic cascade versus the N-terminal BAX construct (<1 h (C-terminal) versus 2–3 h (N-terminal)) (16). Furthermore, in the initial study, although the C-terminal BAX construct could be recruited irreversibly to the OMM after 50 min of repeated illumination, the N-terminal construct, after the same duration and pattern of illumination, still reverted to the cytosol, identifying the exposed C terminus of BAX as necessary for efficient induction of apoptosis. However, despite its higher efficacy, light-independent cell death was a significant drawback associated with the C-terminal construct over the N-terminal construct (22% (C-terminal) versus 9% (N-terminal) 48 h post-transfection in HeLa cells, as assessed by trypan blue assay (16)). Taken together, these results implicate dark-state association between Cry2 and CIB in tandem with an exposed C terminus of BAX as the source of undesirable levels of background cell death. To improve these initial constructs, we sought to create an OptoBAX with reduced levels of light-independent cell death and retention of light-dependent pro-apoptotic efficacy.
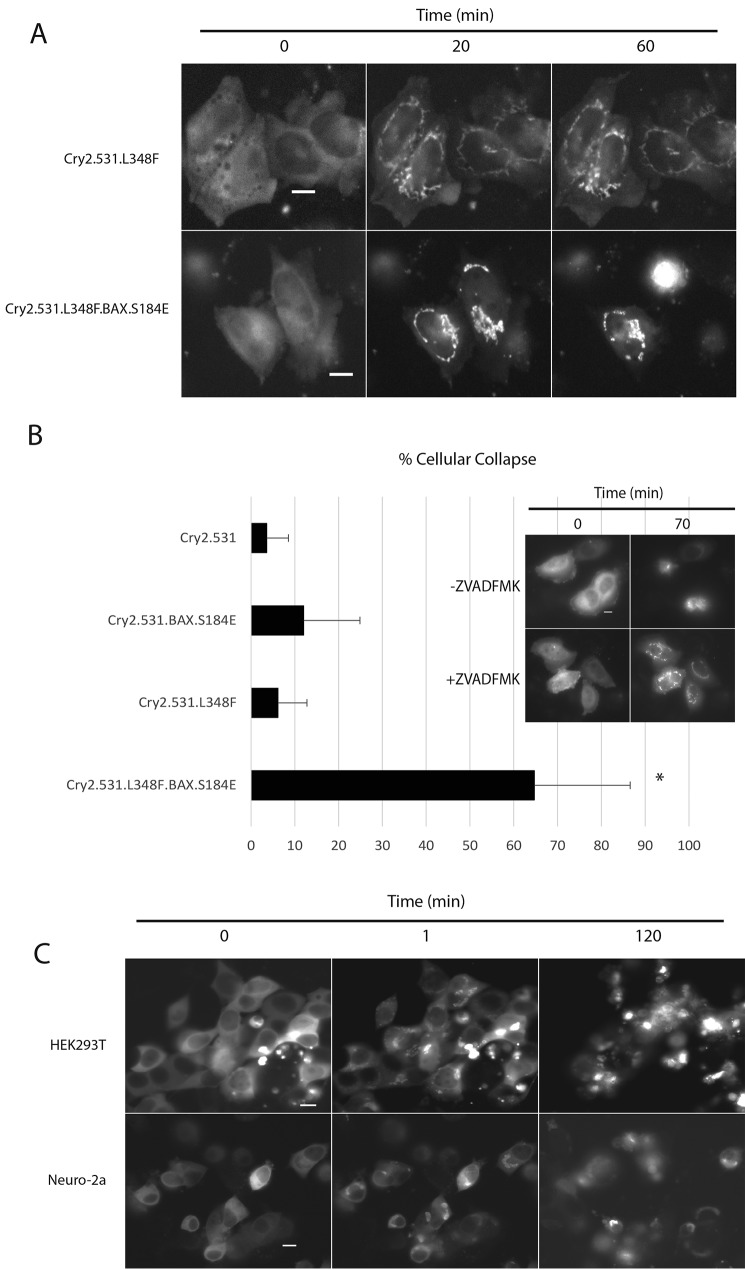
Recently, optimized versions of the Cry2 optogenetic system have been reported, demonstrating, in the context of a yeast two-hybrid assay, that extending the truncated Cry2PHR from 498 to 515 amino acids or longer (up to 535 amino acids) significantly reduced the light-independent background present in the optogenetic system (18). In the same report, a genetic screen also identified a Cry2 mutant (L348F) that extends the photocycle of Cry2 well beyond that of the WT protein, prolonging the half-life of the light-promoted Cry2–CIB interaction from ∼6 min to ∼24 min (18). This mutation prolongs the lifetime of the light-promoted semiquinone form of FAD, the light-responsive Cry2 cofactor. We hypothesized that incorporating similar changes into OptoBAX might result in an improved pro-apoptotic switch via reduced light-independent Cry2–CIB interaction and, with incorporation of the L348F mutant, reduced light input compared with the original OptoBAX, creating a system more easily amenable to incorporation into drug discovery platforms or model organisms. As a result, for the OptoBAX 2.0 system, two additional Cry2–BAX fusions were assembled: Cry2(1–531).mCh.BAX.S184E and Cry2(1–531).L348F.mCh.BAX.S184E, pairing them with the original Tom20.CIB.GFP and Tom20.CIB constructs (Fig. 1A). Assays of light-independent cell death (“dark background”) were subsequently performed in dually transfected HeLa cells using a fluorescent indicator of cell viability (Fig. S1). Compared with the original OptoBAX system, the OptoBAX 2.0 system reduced dark background from ∼36% (Cry2PHR.mCh.BAX.S184E) to ∼20% (Cry2(1–531).mCh.BAX.S184E and Cry2(1–531).L348F.mCh.BAX.S184E), similar to the background cell death observed with the Cry2.mCh fusions without BAX. The effect of the L348F mutant on the Cry2.mCh fusions was further validated in HeLa cells, utilizing a single pulse of 470-nm light to promote association of Cry2 with the OMM-localized CIB domain (Fig. 1, B and C, and Movie S1), confirming that the L348F mutant extended the half-life of association with the OMM from 6.25 (±0.5) min to 32.8 (±1.0) min. The Cry2(1–531).L348F.mCh.BAX.S184E construct was also assayed for its ability to induce apoptosis versus the Cry2(1–531).L348F.mCh fusion (Fig. 2A and Movie S2). We subsequently investigated whether the L348F mutant induces apoptosis with less frequent light stimulation versus the nonmutant Cry2(1–531).mCh.BAX.S184E (Fig. 2B). An experiment using an interval of 10 min between blue light pulses, which is longer than the half-life of the WT construct but significantly shorter than that of the extended photocycle mutant, was designed to demonstrate the result of less frequent light stimulation in the extended photocycle construct. As a result of this illumination sequence, the nonmutant Cry2 construct repeatedly reverted to the cytosolic state during the experiment, whereas the L348F mutant maintained consistent localization at the OMM. The results of this experiment were assessed by manually counting the number of apoptotic cells (based on the loss of adherent cell morphology) after 1 h of simultaneous blue light stimulation and imaging on a Leica Dmi8 wide-field fluorescence microscope. In this study, 65% (±21%) of transfected cells expressing the Cry2(1–531). L348F.mCh.BAX.S184E/Tom20.CIB.GFP fusion became apoptotic versus 12% of transfected cells (±13%) expressing the standard photocycle Cry2(1–531).mCh.BAX.S184E construct, demonstrating that prolonging the photocycle enables more apoptosis with less frequent light input (percent cell collapse and standard deviations determined from 10 replicate measurements). In addition, some background apoptosis was observed with the fusions without BAX (Cry2(1–531).L348F.mCh and Cry2(1–531).mCh; 6% (±7%) and 4% (±5%), respectively, determined from 10 replicate measurements). This background is attributed to the loss of mitochondrial integrity and subsequent apoptotic entry because of repeated recruitment of protein fusions to the OMM. To confirm that the observed background was a result of apoptosis, we conducted identical illumination experiments in the presence of 50 μm Z-VAD(OMe)-fmk. In the presence of this irreversible caspase inhibitor, no background apoptosis was observed for the Cry2 fusions, and OptoBAX-dependent apoptosis was completely inhibited (Fig. 2B, inset; eight replicates per protein fusion). It should be noted that these experiments analyzed apoptotic entry of transfected cells that were fully adherent at the beginning of the illumination time course; a measure of overall cell viability in the absence of light stimulation is shown in Fig. S1.
Figure 1.
Redesign of optogenetic actuators and characterization of the extended photocycle mutant L348F. A, optimized versions of OptoBAX created for reduction of light-independent cell death and extension of the photocycle. Western blot (α-mCherry, inset) lanes: 1, Cry2.531.mCherry; 2, Cry2.531.L348F.mCherry; 3, Cry2.531.mCherry.BAX; 4, Cry2.531.L348F.mCherry.BAX; 5, Cry2PHR.mCherry.BAX. B, demonstration of the elongated photocycle in the Cry2 L348F mutant. Images shown are before and 5 and 10 min after activation with a 150-ms pulse of 470-nm light. C, plot of normalized fluorescence data from the experiment shown in B. Error bars represent standard deviations from six replicate measurements of light-initiated recruitment using the method described by Haar et al. (53). Scale bar = 10 μm.
Figure 2.
Minimal light application required for apoptotic cell death and application to other cell lines. A, OMM recruitment of OptoBAX constructs with (Cry2.531.L348F.BAX/Tom20.CIB.GFP) and without (Cry2.531.L348F/Tom20.CIB.GFP) the pro-apoptotic BAX fusion in HeLa cells. Cells were illuminated with a 150-ms pulse of 470-nm light every 10 min over a 2-h time course. B, quantification of cellular collapse over the 1-h time course experiment. Error bars represent standard deviations from a minimum of 10 replicate measurements per condition. *, p < 0.05 for all pairwise comparisons (one-way analysis of variance). Inhibition of Cry2.531.L348F.BAX occurs in the presence of 50 μm Z-VAD(OMe)-fmk. C, demonstration of light-induced cellular collapse with OptoBAX (Cry2.531.L348F.BAX/Tom20.Cib.GFP) in HEK293T and Neuro-2a cell lines over a 2-h time course (470 nm, 150-ms pulse; images were acquired every minute). Scale bars = 10 μm.
Overall, the HeLa cell studies indicate that OptoBAX 2.0 constructs possess two key improvements over the original OptoBAX system: lower dark background and enhanced photocontrol. The lowering of dark background relative to the initially reported system is particularly important for the biological experiments in this paper. In the system reported previously, the higher levels of dark background present predisposed cells to rapid apoptotic entry upon light activation (<15 min until cellular collapse for the Cry2PHR.BAX.S184E construct (16)). In contrast, in our system, lower levels of dark background and, hence, later apoptotic entry upon light activation (>40 min until cellular collapse for the Cry2PHR.BAX.S184E construct in HeLa cells (Movie S2) provide an extended time window suitable for more detailed dissection of the biochemical events occurring prior to, during, and after cellular collapse.
Translation to other cell lines
Given the common mechanism of BAX-mediated MOMP and subsequent apoptosis in mammalian cells (21), it was anticipated that this optogenetic system might readily translate to other mammalian cell lines. To test this assumption, the OptoBAX 2.0 constructs were deployed in two additional cell lines, Neuro-2a and HEK293T (Fig. 2C). Induction of apoptosis upon recruitment of Cry2(1–531).L348F.mCh.BAX.S184E to the OMM with light was observed in both cell lines. In particular, the reliably high transfection efficiency observed with the HEK293T cell line made further exploration of the apoptotic cascade in this cell line particularly appealing. Furthermore, in contrast to HeLa cells, no background cellular collapse was observed in 1-h light activation experiments conducted with Bax-free protein fusions (Cry2(1–531).L348F.mCh and Cry2(1–531).mCh; 0 of 180 and 0 of 167 total cells counted from six replicates) in the HEK293T cell line. As a result, the remainder of the experiments in this work were carried out in the HEK293T cell line.
Tracking the hallmarks of apoptosis with optogenetic techniques
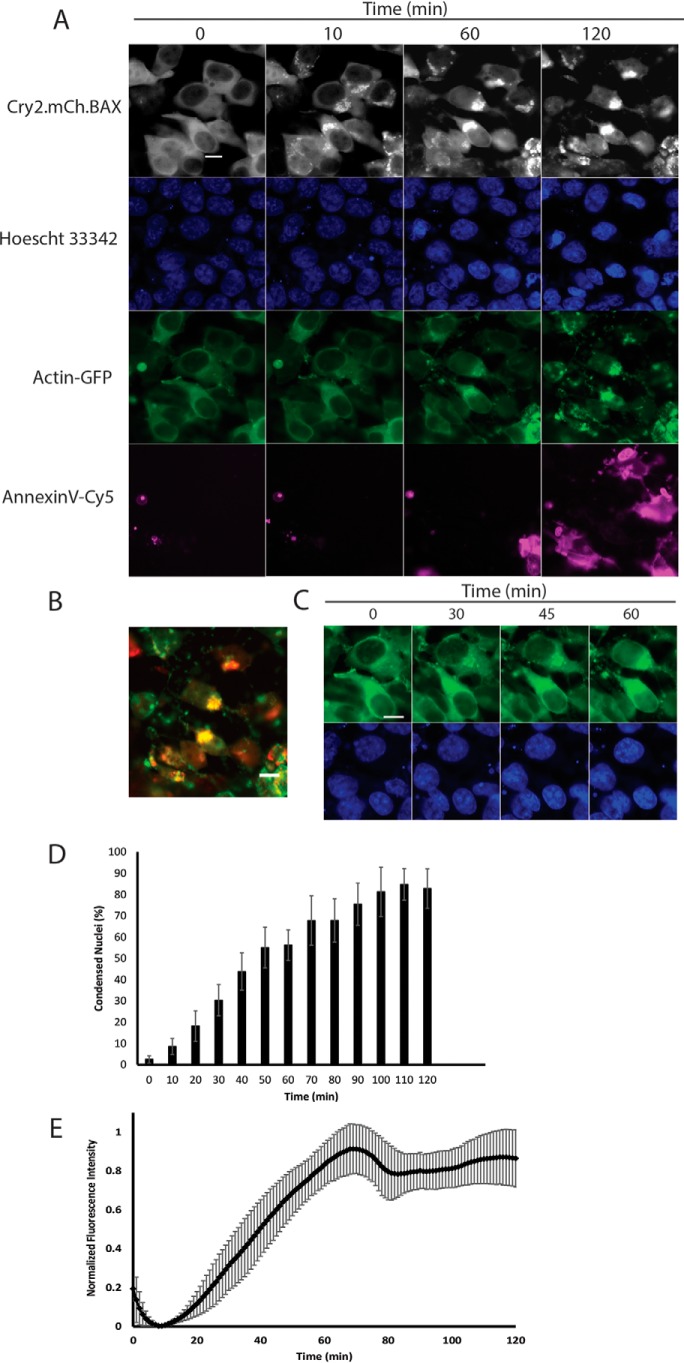
The morphological and biochemical hallmarks of apoptosis include cell blebbing, loss of adhesion and cellular volume, chromatin condensation and nuclear fragmentation, cytoskeletal rearrangement, phosphatidylserine inversion, release of mitochondrial proteins (cytochrome c/Smac/Diablo), and caspase cleavage (22). Fluorescently labeled proteins and small molecules for tracking these key apoptotic events have become widely available in recent years (23–26). This makes practical the prospect of using a light-activated system for induction of apoptosis in conjunction with live-cell–compatible markers on a multichannel fluorescence microscope equipped with a live-cell incubator. To this end, several markers for key apoptotic events were selected for inclusion in multicolor imaging experiments, including a nuclear stain (Hoechst 33342) for monitoring chromatin condensation, a marker of phosphatidylserine (PS) inversion (Annexin V-FITC and Annexin V-Cy5), a cytoskeletal marker (GFP-actin (27)), and a marker for downstream caspase cleavage (CellEvent caspase 3/7). Our first experiment sought to simultaneously monitor chromatin condensation, cytoskeletal rearrangement, and phosphatidylserine inversion in the wake of BAX translocation to the OMM. The HEK293T cell line demonstrated efficient transfection of both components of the OptoBAX 2.0 system (Cry2(1–531).L348F.mCh.BAX.S184E and Tom20.CIB) in addition to a third protein, a cytoskeletal marker for actin (GFP-actin). In the same experiment, chromatin was labeled with Hoechst 33342, and an Annexin V-Cy5 fusion was incorporated as a marker for PS inversion. These additional components provided a four-color system for monitoring light-induced BAX translocation to the mitochondria (mCherry/TxRed), cytoskeletal rearrangement (FITC/GFP), chromatin condensation (4′,6-diamidino-2-phenylindole), and membrane flipping (Cy5). Using this system, the impact of BAX-mediated apoptosis on the cytoskeleton and chromatin was measured over the course of a 2-h illumination experiment (Fig. 3 and Movie S3). In the course of the multicolor imaging experiment, the following changes were observed: redistribution of the GFP-actin construct, indicative of cytoskeletal rearrangement; nuclear collapse and intensification of the fluorescence intensity of the Hoechst 33342 dye, indicative of chromatin condensation; and incorporation of the Annexin V-Cy5 dye, indicative of phosphatidylserine inversion (Fig. 3A).
Figure 3.
Four-color experiment. A, multicolor imaging (Leica DmI8 wide-field microscope equipped with an OKOLab stage-top incubator) of mitochondrial recruitment, chromatin condensation, actin rearrangement, and phosphatidylserine exposure in HEK293T cells. B, overlay of actin/mitochondria after 2 h of imaging. C, time lapse of actin rearrangement and accumulation at mitochondria. D and E, percent chromatin condensation versus time for the chromatin marker (Hoechst 33342) (D) and fluorescence intensity changes for the PS marker (Annexin V-Cy5) (E). Images were acquired every minute over a 2-h time course. Error bars represent standard deviations from four replicate measurements (Fig. 3D) and six replicate measurements (Fig. 3E) per condition. Scale bars = 10 μm.
Actin rearrangement
Significant redistribution of the GFP-Actin marker from the cytosol to both the nucleus and the mitochondria occurred in the wake of OptoBAX activation, with 44% of cells (111 of 252) clearly demonstrating mitochondrial GFP-actin 70 min post-activation (Fig. 3, B and C). The accumulation of actin and numerous actin-binding proteins (cofilin 1, CAP1) at the OMM in the wake of small-molecule-induced apoptosis is often characterized as an early event that occurs well prior to BAX translocation (28–30). In our system, however, membrane permeabilization with OptoBAX precipitates downstream actin redistribution to the OMM well after BAX recruitment (∼30 min post-OptoBAX activation). Although the accumulation of actin at mitochondria remains a poorly understood event in the BAX-mediated apoptosis pathway (28, 30–32), this result points to a closer relationship between cytoskeletal dynamics, Bcl-2 family translocation events, and apoptotic progression than described previously (31).
Inhibition of actin dynamics
The multicolor OptoBAX experiments were repeated in the presence of compounds known to have opposing effects on actin dynamics: jasplakinolide, which inhibits actin depolymerization, maintaining the F-actin state, and latrunculin A, which inhibits actin polymerization by sequestering G-actin (33). As expected, treatment of cells with these compounds resulted in strikingly different distributions of our actin-GFP reporter (Fig. S2). However, despite the differences in actin distribution, treatment with either compound delayed the entry of cells into light-activated apoptosis compared with the no-treatment control, as assessed by both the reduction of chromatin condensation and PS externalization during a 2-h time course (percent condensed nuclei 70 min post-OptoBAX activation: jasplakinolide, 12% (±2%) condensed nuclei (14 of 115 total cells); Latrunculin A, 5% (±1%) condensed nuclei (8 of 148 total cells); untreated cells, 55% (±2%) condensed nuclei (54 of 98 total cells); Fig. S2; assessed from three replicate measurements). Although the redistribution of actin to mitochondria occurred as described previously in untreated cells, the same event was infrequently observed with the jasplakinolide-treated (3% (±1%), 3 of 115 total cells) and Latrunculin A–treated (1% (±0.02%), 2 of 148 total cells) populations post-OptoBAX activation. It should be noted that, although these compounds delayed apoptotic onset in our system, both compounds have been demonstrated to be pro-apoptotic in cell culture over long incubation periods, precluding lengthier experiments (34–36). Nonetheless, it is clear that perturbations of actin dynamics delay key components of our light-activated apoptotic cascade. Furthermore, the observed correlation between BAX-induced MOMP and actin rearrangement may not be entirely causal. It is possible that the rapid mitochondrial depolarization induced by small-molecule-initiated apoptosis (i.e. staurosporine (STS)) triggers events that lead to actin redistribution in advance of endogenous BAX translocation. For example, in previous studies of STS-induced apoptosis, mitochondrial depolarization occurred rapidly after treatment, whereas BAX maximally translocated to the OMM several hours post-STS treatment (37, 38). As a result, in our system, it cannot be ruled out that BAX-dependent mitochondrial depolarization drives actin redistribution via a partially MOMP-independent mechanism.
Nuclear accumulation of actin
In addition to actin redistribution, nuclear accumulation of GFP-actin was also observed in the wake of OptoBAX activation, occurring just after the appearance of mitochondrial GFP-actin (Fig. 3C). The transition of cytosolic actin to nuclear actin has been described previously and may be part of a general cellular stress response (39, 40). The question of whether this freshly nuclear localized actin binds to chromatin may also have interesting implications for actin's role in the repression or activation of transcription of genes vital for completing the apoptotic cascade in mammalian cells, reinforcing the notion of actin as an important part of the apoptotic transcriptome, as previously observed in yeast (41–44). Furthermore, redistribution of cytoskeletal actin into the nucleus may also provide insight into the timing of disruption of the nuclear–cytoplasmic barrier, a key apoptotic event that precipitates many of the later events in apoptosis by providing access of cytoplasmic proteins to the nuclear compartment (45–47). Interestingly, both increasing chromatin condensation (Fig. 3D) and increasing phosphatidylserine inversion (Fig. 3E) occur in tandem with nuclear accumulation of actin. Both chromatin condensation and phosphatidylserine inversion peaked 70–90 min after OptoBAX activation.
Caspase 3/7 cleavage
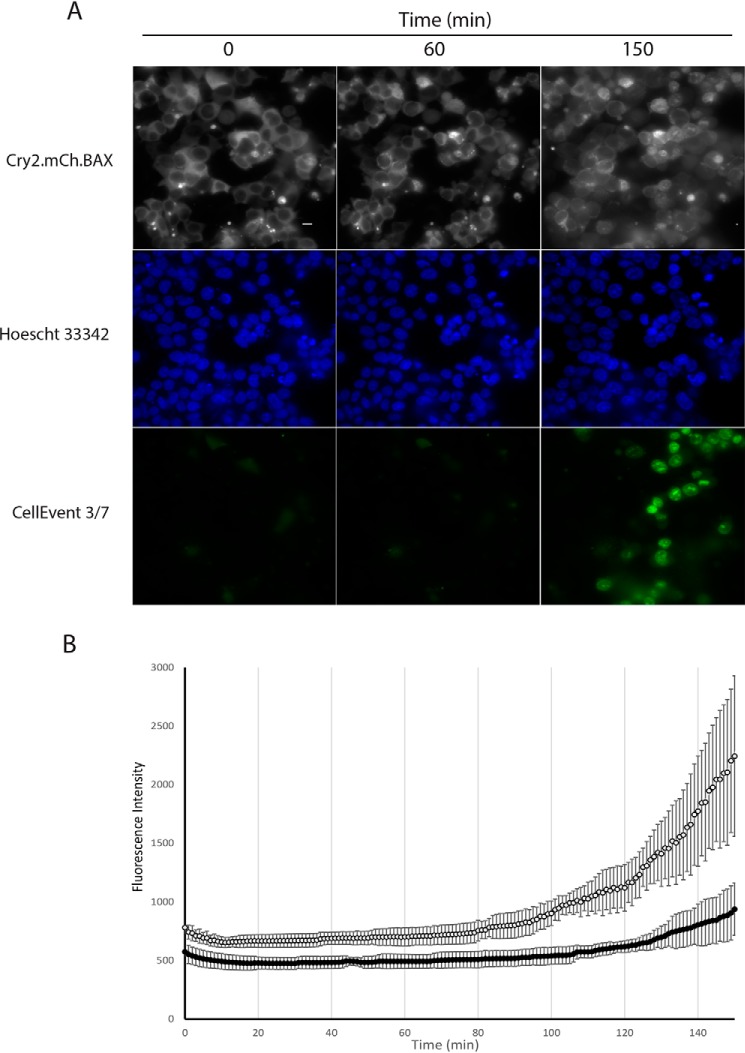
In our final multicolor experiment, a fluorescent indicator of caspase cleavage was used to assess caspase 3/7 cleavage downstream of OptoBAX activation (Fig. 4A). Caspase activity was observed within 1 h of BAX recruitment and increased steadily thereafter (Fig. 4B). Although this event begins relatively early, it peaks later (>2 h post-BAX recruitment), consistent with previous Western blot assessment of caspase cleavage in the OptoBAX system (16). In contrast, control experiments utilizing Cry2(1–531).L348F.mCh and Tom20.CIB did not activate the caspase activity indicator dye or the marker for phosphatidylserine inversion within the same time period of recruitment to the OMM and imaging (Fig. S3). Furthermore, experiments with OptoBAX in the presence of 50 μm Z-VAD(OMe)-fmk showed significant suppression of caspase activity (Fig. 4B). Interestingly, although chromatin condensation was observed during our experiments, significant nuclear fragmentation was not observed during the same timeframe (<1% of cells over a 120-min time course), suggesting that, like peak caspase cleavage, nuclear fragmentation occurs later in BAX-mediated apoptosis. As a corollary to these results, longer time course experiments in HeLa cells utilizing a programmable light emitting diode (LED) device for cellular illumination, followed by nuclear staining and immunostaining, demonstrated nuclear fragmentation ∼7 h post-apoptosis induction (Figs. S4 and S5).
Figure 4.
Three-color experiment. A, multicolor imaging (Leica DmI8 wide-field microscope equipped with an OKOLab stage-top incubator) of mitochondrial recruitment, chromatin condensation, and caspase cleavage in HEK293T cells. B, fluorescence intensity versus time for the caspase activity marker (CellEvent 3/7). Images were acquired every minute over a 2.5-h time course. Cells were either treated with 50 μm Z-VAD-fmk (dark circles) or left untreated (empty circles) for 50 min prior to the experiment. Error bars represent standard deviations from four replicate measurements. Scale bar = 10 μm.
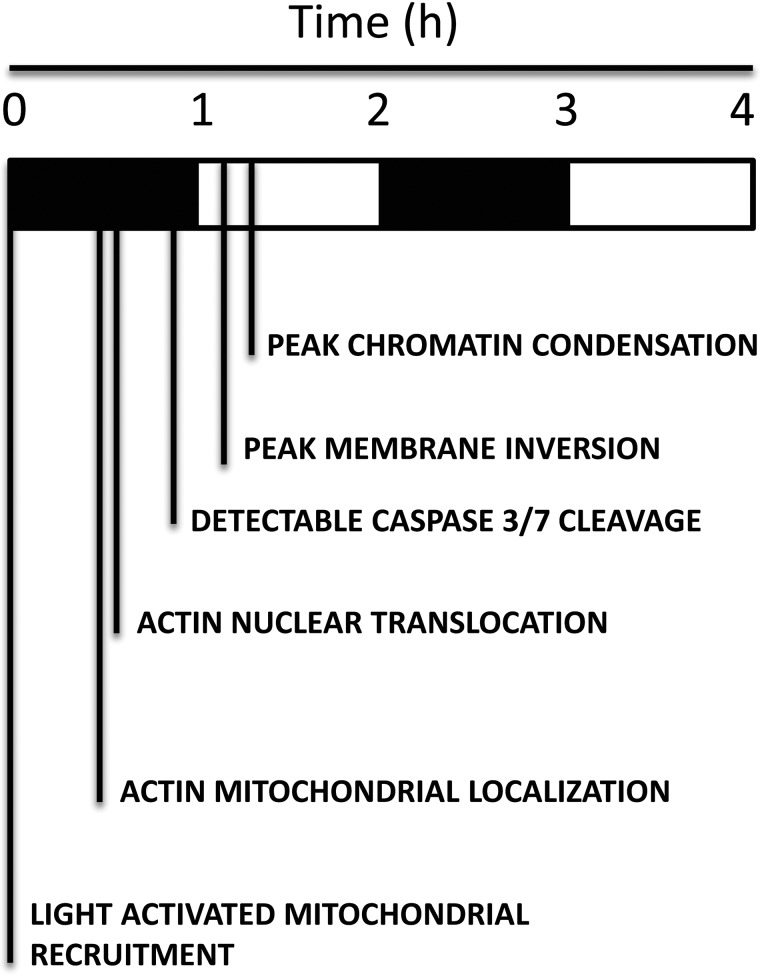
A timeline of apoptotic events
One potentially useful application of the OptoBAX system, in conjunction with the aforementioned markers, is establishment of a timeline of key apoptotic events to define not only the order but also the interplay between key steps in the apoptotic cascade. In turn, establishment of a timeline around a specific event (i.e. BAX translocation to the OMM) may provide novel insights into their interdependence. Utilizing data from our multicolor imaging experiments, a timeline of these early apoptotic events relative to OptoBAX activation was assembled (Scheme 1). This timeline demonstrates that much happens within the first 2 h of the apoptotic cascade, with certain events appearing to be synchronized. For example, the actin cytoskeleton, long implicated as a key player in apoptotic progression, undergoes redistribution from the cytosol to the mitochondria in addition to movement from the cytosol to the nucleus. This redistribution event generally precedes phosphatidylserine inversion. It is possible that wholesale redistribution of actin, a key structural component of the cytoskeleton, contributes to membrane instability and subsequent inversion. Interestingly, the increase in accumulation of Annexin V is also concurrent with an increase in chromatin condensation. These events (chromatin condensation and Annexin V accumulation) occur within the same timeframe as actin redistribution, indicating collapse and rearrangement of the actin cytoskeleton as a common thread linking these events in the progression of apoptosis (31). Finally, previous studies of staurosporine-induced apoptosis have reported that actin redistribution to the OMM occurs well before BAX translocation. In our system, recruitment of OptoBAX to the OMM precipitates the actin OMM localization event. Staurosporine has been shown to affect multiple apoptotic pathways (intrinsic and extrinsic), which may be the basis for the timing of the BAX localization event relative to that of actin. (48) Utilization of an optogenetic system such as OptoBAX for control of a single event in the apoptotic cascade may be a more precise way of examining these translocation events. For example, although previous studies have focused on defining the pro-apoptotic role of cofilin at the OMM (49), the universal importance of the role played by cofilin and other ABPs at the OMM during apoptosis remains a matter of debate (28). Future studies in our laboratory using these optogenetic tools will attempt to resolve the complicated web of translocation events at the OMM occurring in early apoptosis.
SCHEME 1.
Timeline of apoptotic events.
Experimental procedures
Cloning
The Cry2(1–531) gene was amplified by PCR (New England Biolabs Q5 polymerase) from the full-length Cryptochrome-2 gene using a forward primer encoding an XhoI restriction site with the sequence GGCCAACTCGAGATGAAGATGGACAAAAAGAC and a reverse primer encoding an XmaI restriction site with the sequence TGATATCCCCGGGCTACTTGTTGGTCATTAGAAG. The resulting PCR amplicon was then digested, gel-purified, and ligated into a mCherry-N1 vector (Clontech) containing BAX S184E as a fusion to the C terminus of mCherry. Site-directed mutagenesis was used to introduce the L348F mutation using a forward primer with the sequence CCGGAATGAGAGAGTTTTGGGCTACCGGATGG and reverse primer with the sequence CCATCCGGTAGCCCAAAACTCTCTCATTCCGG. Cloning methods for constructs used in prior work (Cry2PHR.mCh.BAX, Tom20.CIB.GFP, and Tom20.CIB.STOP) have been described elsewhere (16). The actin-GFP (pCAG-mGFP-actin; Addgene, 21948) construct was a generous gift from Ryohei Yasuda (27).
Cell culture and transfection
Midi prep quantities of DNA of each construct were then created from Escherichia coli and collected for cell transfection. Transfections were then performed with Lipofectamine 3000 reagent (Invitrogen) following the manufacturer's suggested protocols. Briefly, for dual transfections in 35-mm glass-bottom dishes, plasmid DNA was combined at a 1:1 ratio (1,250 ng/plasmid) in 125 μl of Opti-MEM, followed by addition of 5 μl of P3000 reagent. In a separate vial, 3.75 μl of Lipofectamine 3000 was added to 125 μl of Opti-MEM. The two 125-μl solutions were combined and allowed to incubate at room temperature for 10 min, followed by dropwise addition to cell culture. For triple transfections, plasmid DNA was combined at a 1:1:1 ratio (1,000 ng/plasmid), followed by an identical transfection procedure. Transfection solutions were allowed to remain on cells overnight. Cells were maintained at 37 °C and 5% CO2 in a humidified tissue culture incubator. HeLa and HEK293T cells were maintained in DMEM supplemented with 10% FBS and 1% Pen-Strep (Penicillin-Streptomycin). Neuro-2a cells were maintained in minimum Eagle's medium supplemented with 10% FBS and 1% Pen-Strep.
Cell viability measurements
Background cell death experiments were performed to determine the cell toxicity of OptoBAX constructs in the absence of light exposure. Transfections were performed on HeLa cells in 6-well plates and then maintained in complete darkness for 2 days prior to incubation with a cell viability stain (Live-or-Dye 488/515, Biotium). Briefly, cells were washed once with DPBS containing calcium and magnesium, followed by incubation at room temperature with the viability stain (diluted 1:1,000 in DPBS) for 30 min. Afterward, cells were fixed with 4% paraformaldehyde in PBS for 15 min at room temperature, washed three times with 1 ml of DPBS, and maintained in DPBS for subsequent imaging. Images of fixed cells were acquired on a Leica Dmi8 fluorescence microscope.
Application of fluorescent dyes
For the four-color experiment, triple-transfected cells (Cry2.mCh.BAX/Tom20.CIB/GFP-Actin) in a 35-mm glass-bottom dish (Mattek) were removed from the incubator, cell culture medium was removed by pipette, and cells were gently washed once with DPBS supplemented with calcium and magnesium. 1 ml of PBS containing Hoechst 33342 was then applied to the cells, which were returned to the incubator for 10 min. After the elapsed time, the medium was removed, and cells were washed with DPBS (2 × 1 ml). A prewarmed buffer containing DPBS supplemented with 10% FBS and Annexin V-Cy5 (10 μl/1 ml of buffer; Thermo Fisher, A23204) was added to the cells, which were promptly placed on the microscope for initiation of the light-activated experiment. For the three-color experiment, dually transfected cells (Cry2.mCh.BAX/Tom20.CIB.STOP) were removed from the incubator, cell culture medium was removed by pipette, and cells were gently washed once with DPBS supplemented with calcium and magnesium. 1 ml of PBS containing Hoechst 33342 was then applied to the cells, which were returned to the incubator for 10 min. After the elapsed time, the medium was removed, and cells were washed with DPBS (2 × 1 ml). A prewarmed buffer containing DPBS supplemented with 10% FBS and CellEvent Caspase 3/7 Green (1 μl/1 ml of buffer; Thermo Fisher, C10427) was added to the cells, which were promptly placed on the microscope for initiation of the light-activated experiment.
Cell treatments with actin-binding compounds
Cells were pretreated with actin-binding compounds for 1 h prior to imaging. Jasplakinolide (Santa Cruz Biotechnology) was diluted from a 1 mm stock in DMSO to a working concentration of 200 nm in DMEM supplemented with 10% FBS. Prior to imaging, the cells were incubated with Hoechst 33342 for 10 min, followed by a 1-ml PBS wash. Cells were then resupplied with medium for live-cell imaging (DPBS/10% FBS/200 nm jasplakinolide/Annexin V-Cy5). Latrunculin A (Santa Cruz Biotechnology) was diluted from a 1 mm stock in DMSO to a working concentration of 2 μm in DMEM supplemented with 10% FBS. Prior to imaging, the cells were incubated with Hoechst 33342 for 10 min, followed by a 1-ml PBS wash. Cells were then resupplied with medium for live-cell imaging (DPBS/10% FBS/2 μm Latrunculin A/Annexin V-Cy5).
Cell treatments with caspase inhibitor
Cells were pretreated with the pan-caspase inhibitor Z-VAD(OMe)-fmk (Santa Cruz Biotechnology) for 50 min prior to imaging. The compound was diluted from a 10 mm stock in DMSO to a working concentration of 50 μm in DMEM supplemented with 10% FBS. Prior to imaging, the cells were incubated with Hoechst 33342 for 10 min, followed by a 1-ml PBS wash. Cells were then resupplied with medium for live-cell imaging (L-15/10% FBS/50 μm Z-VAD(OMe)-fmk).
Microscopy
A Leica DMi8 live-cell imaging system equipped with an OKOLab stage-top live-cell incubation system, LASX software, Leica HCX PL APO ×63/1.40–0.60 numerical aperture oil objective, Lumencor LED light engine, CTRadvanced+ power supply, and Leica DFC900 GT camera was used to acquire images. Exposure times were set at 150 ms (GFP, 470 nm), 150 ms (4′,6-diamidino-2-phenylindole, 395 nm), 300 ms (mCherry, 550 nm), and 300 ms (Cy5, 640 nm) with LED light sources at 50% power, and images were acquired every minute or every 10 min.
Microscopy data analysis
Analysis of microscopy date was performed in Fiji equipped with the BioFormats package. Fluorescence intensity measurements were normalized on a scale from 0–1 prior to being averaged. Average normalized intensities were computed from four to six fields of view on a ×63 objective, with each field of view encompassing 100 (±10) transfected cells. Mitochondrial residence time plots were generated by quantifying the change in fluorescence intensity at the mitochondria over time after light recruitment. Fluorescence intensity measurements were normalized and averaged from six mitochondrial clusters per transfection condition. After normalizing the change in fluorescence over time, the mitochondrial association half-life was defined as the time for fluorescence intensity of the light-recruited reagent to reach 50% maximum intensity at the mitochondria. Image overlays were created using the Merge Channels function in Fiji.
Western blotting
HeLa cells were lysed post-transfection with 200 μl of mammalian protein extraction reagent lysis buffer (Thermo Scientific) plus protease inhibitors. After 10 min on a rotary shaker at room temperature, lysates were collected and centrifuged for 15 min (1,000 rpm at 4 °C), and supernatants were reserved. The resulting lysates were subjected to electrophoresis on a 10% SDS-PAGE gel and then transferred to PVDF membranes (20 V overnight at 4 °C). Membranes were then blocked for 1 h with 5% BSA in TBS with 1% Tween (TBST), followed by incubation with the primary antibody (Rockland anti-mCherry antibody, 600-401-P16) at 1:1,000 dilution in 5% milk and TBST overnight at 4 °C on a platform rocker. The membranes were then washed three times for 5 min each with TBST and incubated with anti-rabbit IgG HRP secondary antibody (Abcam, 6721) at 1:3,000 in 5% BSA and TBST for 2 h at room temperature. After washing three times for 5 min each with TBST, the membranes were exposed to a chemiluminescent substrate for 5 min and imaged using an Azure cSeries imaging station.
Programmable LED device
The device (designed and constructed by Hoffman and Hughes) combines four wavelength ranges (440–460 nm, 520–540 nm, 650–670 nm, and 720–750 nm) into a single package using surface mount technology and is powered with an Arduino Uno system. Luxeon Rebel and Luxeon Color series LEDs were soldered on 20-mm mounts with a hot air gun designed for surface-mounted devices. The LEDs/metal core printed circuit board aluminum LED bases were fixed to a 130 × 70 mm, rectangular, 15-mm-high heat sink to prevent overheating and allow longer exposure of samples. The intensity of the light was controlled with a combination of variable resistors, positive-negative-positive transistors, and Zener diodes. The LED arrays are controlled using an Arduino controller to control illumination, time, intensity, and wavelength. Light intensity was measured with a Fisherbrand digital light meter; effects of illumination protocols on medium temperature were determined with a Fisherbrand digital thermometer using a type K beaded probe.
Cellular illumination and immunostaining
HeLa cells were plated in 35-mm glass-bottom dishes (Mattek) at a density of 2.0 × 105 cells/dish and maintained at 37 °C in a humidity-controlled incubator with a 5% CO2 atmosphere in DMEM (10% FBS and 1% Pen-Strep). The following day, cells were transfected with Cry2.L348F.531.mCh.BAX and Tom20.CIB.GFP as reported previously. 24 h after transfection, cells were illuminated with 440- to 460-nm light using a programmable LED device prior to being returned to the incubator. At the conclusion of the dark incubation period, cells were washed three times with 1 ml of PBS and then fixed with 1 ml of 4% paraformaldehyde in PBS at room temperature for 10 min. Cells were washed twice with 1 ml of PBS and blocked and permeabilized for 20 min in antibody dilution buffer (1% BSA, 0.3% Triton X-100, and PBS) at 4 °C. Blocking was followed by overnight incubation at 4 °C with rabbit anti-cleaved caspase 3 antibody (Cell Signaling, 9661) at 1:400 dilution in antibody dilution buffer (1% BSA, 0.3% Triton X-100, and PBS). Cells were then washed with PBS (three times for 5 min) before incubation with anti-rabbit Alexa Fluor 647 secondary antibody (Life Technologies, A21245) at 1:500 dilution in antibody dilution buffer. After washing cells with PBS (three times for 5 min), nuclei were stained briefly (5 min) with Hoechst 33342 (1 μg/ml), followed by image acquisition on a confocal microscope (Olympus IX2-DSU tandem spinning disk confocal microscope, ×60 objective).
Corrected total cell fluorescence
Whole-cell fluorescence intensities were measured in Fiji/ImageJ (50) and quantified using the corrected total cell fluorescence (CTCF) method (51, 52). Briefly, individual cells were outlined in Fiji, followed by calculation of the area, integrated density, and mean gray value for each cell. For each image, background measurements of fluorescence intensity were taken from regions without cells. The following formula was then used to calculate the CTCF for each cell: CTCF = integrated density − (cell area × mean background fluorescence).
Plasmids
The plasmids generated for this publication are available through Addgene, Inc., a not-for-profit plasmid DNA repository.
Author contributions
W. C. G., G. F. H., T. J. G., and R. M. H. investigation; W. C. G., G. F. H., T. J. G., and R. M. H. methodology; W. C. G. and R. M. H. writing-original draft; W. C. G. and R. M. H. writing-review and editing; G. F. H. resources; T. J. G. and R. M. H. formal analysis; R. M. H. conceptualization; R. M. H. data curation; R. M. H. supervision; R. M. H. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Karen Litwa (East Carolina University Brody School of Medicine) for the gift of HEK293T cells, Dr. Alex Murashov (East Carolina University Brody School of Medicine) for the gift of Neuro-2a cells, and Dr. Nathan Hudson (East Carolina University Department of Physics) for use of the Leica Dmi8 wide-field microscope.
Funding was provided by the East Carolina University Division of Research, Economic Development and Engagement, Thomas Harriot College of Arts and Sciences, and the Department of Chemistry. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5 and Movies S1–S3.
- MOMP
- mitochondrial outer membrane permeabilization
- OMM
- outer mitochondrial membrane
- BAX
- BCL2-associated X apoptosis regulator
- CIB
- cryptochrome-interacting β helix–loop–helix
- Z-VAD(OMe)-fmk
- benzyloxycarbonyl-VAD(OMe)-fluoromethyl ketone
- PS
- phosphatidylserine
- STS
- staurosporine
- TBS
- Tris-buffered saline
- CTCF
- corrected total cell fluorescence
- LED
- light emitting diode
- Pen-Strep
- Penicillin-Streptomycin
- DPBS
- Dulbecco's phosphate buffered saline.
References
- 1. Hughes R. M. (2018) A compendium of chemical and genetic approaches to light-regulated gene transcription. Crit. Rev. Biochem. Mol. Biol. 53, 453–474 10.1080/10409238.2018.1487382 [DOI] [PubMed] [Google Scholar]
- 2. Boyden E. S., Zhang F., Bamberg E., Nagel G., and Deisseroth K. (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 10.1038/nn1525 [DOI] [PubMed] [Google Scholar]
- 3. Wu Y. I., Frey D., Lungu O. I., Jaehrig A., Schlichting I., Kuhlman B., and Hahn K. M. (2009) A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108 10.1038/nature08241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rhind N., and Russell P. (2012) Signaling pathways that regulate cell division. Cold Spring Harb. Perspect. Biol. 10.1101/cshperspect.a005942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mitra S. K., Hanson D. A., and Schlaepfer D. D. (2005) Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6, 56–68 10.1038/nrm1549 [DOI] [PubMed] [Google Scholar]
- 6. Elmore S. (2007) Apoptosis: a review of programmed cell death. Toxicol. Pathol. 35, 495–516 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bléger D., and Hecht S. (2015) Visible-light-activated molecular switches. Angew. Chem. Int. Ed. Engl. 54, 11338–11349 10.1002/anie.201500628 [DOI] [PubMed] [Google Scholar]
- 8. Stein V., and Alexandrov K. (2015) Synthetic protein switches: design principles and applications. Trends Biotechnol. 33, 101–110 10.1016/j.tibtech.2014.11.010 [DOI] [PubMed] [Google Scholar]
- 9. Mühlhäuser W. W. D., Hörner M., Weber W., and Radziwill G. (2017) in Synthetic Protein Switches: Methods and Protocols (Stein V., ed.), pp. 257–270, Springer, New York, NY [Google Scholar]
- 10. Green D. R. (1998) Apoptotic pathways: the roads to ruin. Cell 94, 695–698 10.1016/S0092-8674(00)81728-6 [DOI] [PubMed] [Google Scholar]
- 11. Adams J. M., and Cory S. (1998) The Bcl-2 protein family: arbiters of cell survival. Science 281, 1322–1326 10.1126/science.281.5381.1322 [DOI] [PubMed] [Google Scholar]
- 12. Westphal D., Kluck R. M., and Dewson G. (2014) Building blocks of the apoptotic pore: how Bax and Bak are activated and oligomerize during apoptosis. Cell Death Differ. 21, 196–205 10.1038/cdd.2013.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Westphal D., Dewson G., Czabotar P. E., and Kluck R. M. (2011) Molecular biology of Bax and Bak activation and action. Biochim. Biophys. Acta 1813, 521–531 10.1016/j.bbamcr.2010.12.019 [DOI] [PubMed] [Google Scholar]
- 14. Lalier L., Cartron P.-F., Juin P., Nedelkina S., Manon S., Bechinger B., and Vallette F. M. (2007) Bax activation and mitochondrial insertion during apoptosis. Apoptosis 12, 887–896 10.1007/s10495-007-0749-1 [DOI] [PubMed] [Google Scholar]
- 15. Gross A., Jockel J., Wei M. C., and Korsmeyer S. J. (1998) Enforced dimerization of BAX results in its translocation, mitochondrial dysfunction and apoptosis. EMBO J. 17, 3878–3885 10.1093/emboj/17.14.3878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hughes R. M., Freeman D. J., Lamb K. N., Pollet R. M., Smith W. J., and Lawrence D. S. (2015) Optogenetic apoptosis: light-triggered cell death. Angew. Chem. Int. Ed. Engl. 54, 12064–12068 10.1002/anie.201506346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nechushtan A., Smith C. L., Hsu Y. T., and Youle R. J. (1999) Conformation of the Bax C-terminus regulates subcellular location and cell death. EMBO J. 18, 2330–2341 10.1093/emboj/18.9.2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taslimi A., Zoltowski B., Miranda J. G., Pathak G. P., Hughes R. M., and Tucker C. L. (2016) Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat. Chem. Biol. 12, 425–430 10.1038/nchembio.2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Godley B. F., Shamsi F. A., Liang F.-Q., Jarrett S. G., Davies S., and Boulton M. (2005) Blue light induces mitochondrial DNA damage and free radical production in epithelial cells. J. Biol. Chem. 280, 21061–21066 10.1074/jbc.M502194200 [DOI] [PubMed] [Google Scholar]
- 20. Hughes R. M., Freeman D. J., Lamb K. N., Pollet R. M., Smith W. J., and Lawrence D. S. (2015) Optogenetic apoptosis: light-triggered cell death. Angew. Chem. Int. Ed. Engl. 54, 12064–12068 10.1002/anie.201506346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dewson G., and Kluck R. M. (2009) Mechanisms by which Bak and Bax permeabilise mitochondria during apoptosis. J. Cell Sci. 122, 2801–2808 10.1242/jcs.038166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saraste A., and Pulkki K. (2000) Morphologic and biochemical hallmarks of apoptosis. Cardiovasc. Res. 45, 528–537 10.1016/S0008-6363(99)00384-3 [DOI] [PubMed] [Google Scholar]
- 23. Gelles J. D., and Chipuk J. E. (2017) Robust high-throughput kinetic analysis of apoptosis with real-time high-content live-cell imaging. Cell Death Dis. 8, e2758 10.1038/cddis.2017.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gelles J. D., and Chipuk J. E. (2016) Robust high-throughput kinetic analysis of apoptosis with real-time high-content live-cell imaging. Cell Death Dis. 7, e2493 10.1038/cddis.2016.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zeng W., Wang X., Xu P., Liu G., Eden H. S., and Chen X. (2015) Molecular imaging of apoptosis: from micro to macro. Theranostics 5, 559–582 10.7150/thno.11548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Puigvert J. C., de Bont H., van de Water B., and Danen E. H. J. (2010) High-throughput live cell imaging of apoptosis. Curr. Protoc. Cell Biol. Chapter 18, Unit 18.10.1–13 10.1002/0471143030.cb1810s47 [DOI] [PubMed] [Google Scholar]
- 27. Murakoshi H., Lee S.-J., and Yasuda R. (2008) Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol. 36, 31–42 10.1007/s11068-008-9024-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rehklau K., Gurniak C. B., Conrad M., Friauf E., Ott M., and Rust M. B. (2012) ADF/cofilin proteins translocate to mitochondria during apoptosis but are not generally required for cell death signaling. Cell Death Differ. 19, 958–967 10.1038/cdd.2011.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang C., Zhou G.-L., Vedantam S., Li P., and Field J. (2008) Mitochondrial shuttling of CAP1 promotes actin- and cofilin-dependent apoptosis. J. Cell Sci. 121, 2913–2920 10.1242/jcs.023911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang H. L., Le A.-H., and Lung H. L. (2006) The increase in mitochondrial association with actin precedes Bax translocation in apoptosis. Biochem. J. 396, 1–5 10.1042/BJ20060241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Desouza M., Gunning P. W., and Stehn J. R. (2012) The actin cytoskeleton as a sensor and mediator of apoptosis. Bioarchitecture 2, 75–87 10.4161/bioa.20975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Boldogh I. R., and Pon L. A. (2006) Interactions of mitochondria with the actin cytoskeleton. Biochim. Biophys. Acta 1763, 450–462 10.1016/j.bbamcr.2006.02.014 [DOI] [PubMed] [Google Scholar]
- 33. Fenteany G., and Zhu S. (2003) Small-molecule inhibitors of actin dynamics and cell motility. Curr. Top. Med. Chem. 3, 593–616 10.2174/1568026033452348 [DOI] [PubMed] [Google Scholar]
- 34. Martin S. S., and Leder P. (2001) Human MCF10A mammary epithelial cells undergo apoptosis following actin depolymerization that is independent of attachment and rescued by Bcl-2. Mol. Cell Biol. 21, 6529–6536 10.1128/MCB.21.19.6529-6536.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morton W. M., Ayscough K. R., and McLaughlin P. J. (2000) Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat. Cell Biol. 2, 376–378 10.1038/35014075 [DOI] [PubMed] [Google Scholar]
- 36. Posey S. C., and Bierer B. E. (1999) Actin stabilization by jasplakinolide enhances apoptosis induced by cytokine deprivation. J. Biol. Chem. 274, 4259–4265 10.1074/jbc.274.7.4259 [DOI] [PubMed] [Google Scholar]
- 37. Tsuruta F., Masuyama N., and Gotoh Y. (2002) The phosphatidylinositol 3-kinase (PI3K)-Akt pathway suppresses Bax translocation to mitochondria. J. Biol. Chem. 277, 14040–14047 10.1074/jbc.M108975200 [DOI] [PubMed] [Google Scholar]
- 38. Smaili S. S., Hsu Y. T., Sanders K. M., Russell J. T., and Youle R. J. (2001) Bax translocation to mitochondria subsequent to a rapid loss of mitochondrial membrane potential. Cell Death Differ. 8, 909–920 10.1038/sj.cdd.4400889 [DOI] [PubMed] [Google Scholar]
- 39. Pendleton A., Pope B., Weeds A., and Koffer A. (2003) Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J. Biol. Chem. 278, 14394–14400 10.1074/jbc.M206393200 [DOI] [PubMed] [Google Scholar]
- 40. Meijerman I., Blom W. M., de Bont H. J., Mulder G. J., and Nagelkerke J. F. (1999) Induction of apoptosis and changes in nuclear G-actin are mediated by different pathways: the effect of inhibitors of protein and RNA synthesis in isolated rat hepatocytes. Toxicol. Appl. Pharmacol. 156, 46–55 10.1006/taap.1998.8616 [DOI] [PubMed] [Google Scholar]
- 41. Gourlay C. W., and Ayscough K. R. (2005) A role for actin in aging and apoptosis. Biochem. Soc. Trans. 33, 1260–1264 10.1042/BST20051260 [DOI] [PubMed] [Google Scholar]
- 42. Gourlay C. W., and Ayscough K. R. (2005) The actin cytoskeleton: a key regulator of apoptosis and ageing? Nat. Rev. Mol. Cell Biol. 6, 583–589 10.1038/nrm1682 [DOI] [PubMed] [Google Scholar]
- 43. Gourlay C. W., and Ayscough K. R. (2005) Identification of an upstream regulatory pathway controlling actin-mediated apoptosis in yeast. J. Cell Sci. 118, 2119–2132 10.1242/jcs.02337 [DOI] [PubMed] [Google Scholar]
- 44. Zheng B., Han M., Bernier M., and Wen J. (2009) Nuclear actin and actin-binding proteins in the regulation of transcription and gene expression. FEBS J. 276, 2669–2685 10.1111/j.1742-4658.2009.06986.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Strasser C., Grote P., Schäuble K., Ganz M., and Ferrando-May E. (2012) Regulation of nuclear envelope permeability in cell death and survival. Nucleus 3, 540–551 10.4161/nucl.21982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kihlmark M., Rustum C., Eriksson C., Beckman M., Iverfeldt K., and Hallberg E. (2004) Correlation between nucleocytoplasmic transport and caspase-3-dependent dismantling of nuclear pores during apoptosis. Exp. Cell Res. 293, 346–356 10.1016/j.yexcr.2003.10.019 [DOI] [PubMed] [Google Scholar]
- 47. Faleiro L., and Lazebnik Y. (2000) Caspases disrupt the nuclear-cytoplasmic barrier. J. Cell Biol. 151, 951–959 10.1083/jcb.151.5.951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nakamura-López Y., Sarmiento-Silva R. E., Moran-Andrade J., and Gómez-García B. (2009) Staurosporine-induced apoptosis in P388D1 macrophages involves both extrinsic and intrinsic pathways. Cell Biol. Int. 33, 1026–1031 10.1016/j.cellbi.2009.06.010 [DOI] [PubMed] [Google Scholar]
- 49. Chua B. T., Volbracht C., Tan K. O., Li R., Yu V. C., and Li P. (2003) Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat. Cell Biol. 5, 1083–1089 10.1038/ncb1070 [DOI] [PubMed] [Google Scholar]
- 50. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Karpova T. S., McNally J. G., Moltz S. L., and Cooper J. A. (1998) Assembly and function of the actin cytoskeleton of yeast: relationships between cables and patches. J. Cell Biol. 142, 1501–1517 10.1083/jcb.142.6.1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCloy R. A., Rogers S., Caldon C. E., Lorca T., Castro A., and Burgess A. (2014). Partial inhibition of Cdk1 in G2phase overrides the SAC and decouples mitotic events. Cell Cycle 13, 1400–1412 10.4161/cc.28401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Haar L. L., Lawrence D. S., and Hughes R. M. (2019) Optogenetic perturbation of the biochemical pathways that control cell behavior. Methods Enzymol. 622, 309–328 10.1016/bs.mie.2019.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.