Abstract
Lipid droplets (LDs) are evolutionarily conserved organelles that play critical roles in mammalian lipid storage and metabolism. However, the molecular mechanisms governing the biogenesis and growth of LDs remain poorly understood. Phosphatidic acid (PA) is a precursor of phospholipids and triacylglycerols and substrate of CDP-diacylglycerol (CDP-DAG) synthase 1 (CDS1) and CDS2, which catalyze the formation of CDP-DAG. Here, using siRNA-based gene knockdowns and CRISPR/Cas9-mediated gene knockouts, along with immunological, molecular, and fluorescence microscopy approaches, we examined the role of CDS1 and CDS2 in LD biogenesis and growth. Knockdown of either CDS1 or CDS2 expression resulted in the formation of giant or supersized LDs in cultured mammalian cells. Interestingly, down-regulation of cell death-inducing DFF45-like effector C (CIDEC), encoding a prominent regulator of LD growth in adipocytes, restored LD size in CDS1- but not in CDS2-deficient cells. On the other hand, reducing expression of two enzymes responsible for triacylglycerol synthesis, diacylglycerol O-acyltransferase 2 (DGAT2) and glycerol-3-phosphate acyltransferase 4 (GPAT4), rescued the LD phenotype in CDS2-deficient, but not CDS1-deficient, cells. Moreover, CDS2 deficiency, but not CDS1 deficiency, promoted the LD association of DGAT2 and GPAT4 and impaired initial LD maturation. Finally, although both CDS1 and CDS2 appeared to regulate PA levels on the LD surface, CDS2 had a stronger effect. We conclude that CDS1 and CDS2 regulate LD dynamics through distinct mechanisms.
Keywords: lipid droplet (LD), phospholipid, enzyme, phosphatidic acid, metabolic disease, CDP-diacylglycerol synthase (CDS), cell death-inducing DFF45-like effector C (CIDEC), Fsp27, diacylglycerol O-acyltransferase 2 (DGAT2), glycerol-3-phosphate acyltransferase 4 (GPAT4), triacylglycerol (TAG), phosphatidic acid (PA), SEIPIN
Introduction
Lipid droplets (LDs)2 are important metabolic organelles, which are used as cellular storage sites for neutral lipids in virtually all organisms, from bacteria to humans (1–5). Nearly all LDs comprise a hydrophobic core of neutral lipids, predominantly triacylglycerols (TAGs) and/or sterol esters, which are encapsulated by a monolayer of phospholipids. The phospholipid monolayer is embedded with LD-associated proteins, which regulate the dynamics of LDs, including biogenesis, growth, and turnover (1, 2). The neutral lipids provide a buffer for energy fluctuations and a reservoir for membrane lipid precursors. Both deficient and excessive storage of neutral lipids in LDs is associated with human diseases, including lipodystrophy, nonalcoholic fatty liver disease, atherosclerosis, and obesity/type 2 diabetes (6). Aberrant LD dynamics can also be a contributing factor to cancer progression (7) or Alzheimer's disease (8).
LDs are highly dynamic organelles. Although the molecular mechanisms of LD biogenesis and expansion remain to be elucidated, the prevailing model of LD growth is that LDs originate from the endoplasmic reticulum (ER) before they expand and mature (5). This process is regulated by metabolic enzymes, nonbilayer lipids, and LD-associated proteins (2, 5, 9–17). LDs can be classified into initial LDs (iLDs) and expanding LDs (eLDs) according to their size and stage in life cycle. iLDs are those LDs of 400–800-nm diameter that are formed from the ER. eLDs are converted from iLDs through a maturation/growth process, mediated by specific proteins such as triglyceride synthesis enzymes (12), Arf1/COPI (13), and CIDEC/Fsp27 (18, 19). Recent studies also showed that loss of SEIPIN, an ER protein implicated in human genetic lipodystrophy, can delay the maturation of iLDs and accumulate supersized LDs (sLDs) (16, 17, 20–22). Finally, pre-LDs that give rise to iLDs must exist, but these LDs cannot be stained by common neutral lipid dyes, such as BODIPY and LipidTox Deep Red, due to their tiny size and lack of neutral lipids. Thus, markers, such as Livedrop and Hpos, have been artificially made to label these pre-LDs (11, 17).
The size of LDs varies in different tissues and even in the same cell types (1). How LDs grow is not fully understood, although lipid transfer from smaller LDs to bigger ones, a process mainly mediated by CIDE family proteins, and increased lipid synthesis are two driving factors. Under specific conditions, CIDE proteins can generate “fusion pores” between LDs and facilitate the transfer of neutral lipids from small to large LDs (18). LDs can also grow by locally incorporating lipids. For example, some TAG synthesis enzymes, such as glycerol-3-phosphate acyltransferase 3/4 (GPAT3/4) and diacylglycerol O-acyltransferase 2 (DGAT2), may translocate from the ER to LDs to synthesize lipids locally for the expansion of LDs (12, 23). In addition, phospholipids, such as phosphatidic acid (PA) and phosphatidylcholine (PC), are also important regulators of LD growth. Giant LDs can be formed when reducing the amount of PC and/or increasing the level of PA (24–27). Also, PA was suggested to be involved in the formation of supersized lipid droplets (sLDs) caused by the abnormal expression of some key proteins, such as SEIPIN and GPAT3/4 (14, 15, 23, 25).
CDP-diacylglycerol (CDP-DAG) synthase (CDS) enzymes convert PA, the precursor of all phospholipids and TAG synthesis, into CDP-DAG for the synthesis of phosphatidylinositol (PI), phosphatidylglycerol, and cardiolipin (28). Although the biochemical function of CDS1 and CDS2 has been characterized, little is known about their involvement in cellular lipid storage (LD formation). In a genome-wide screen, we previously reported that the depletion of CDS1 in the budding yeast Saccharomyces cerevisiae and mammalian cells causes the formation of sLDs (15, 25). Here, we show that CDS1 and CDS2 control LD growth through distinct mechanisms.
Results
CDS deficiency results in sLDs
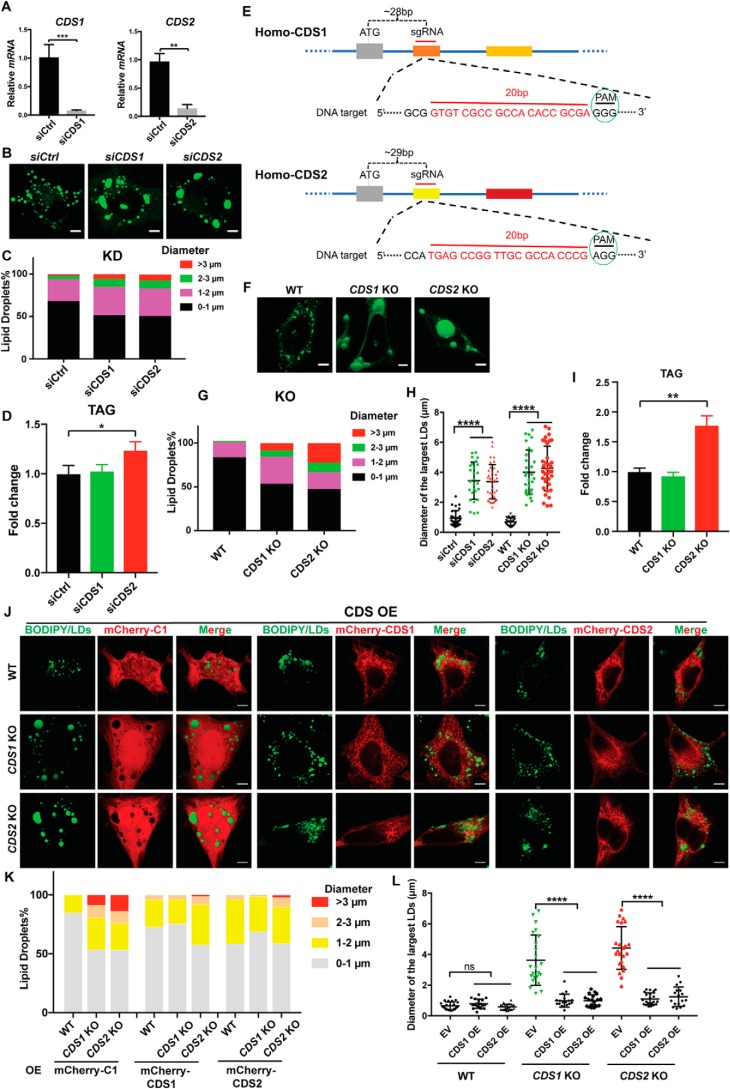
We confirmed our previous findings in CDS1 and CDS2 knockdown (KD) cells (Fig. 1, A–D and H), and here, we further extended those observations in CDS1 and CDS2 knockout (KO) cells generated by the CRISPR/Cas9 system (Fig. 1, E–G), that CDS depletion resulted in sLD formation (15). The percentage of cells with LD diameters larger than 3 μm increased dramatically in the KO cells compared with the KD cells (Fig. 1F). Moreover, the average diameter of three largest LDs per cell in the KO and KD cells increased significantly over the control cells (Fig. 1H). The increase in TAG also became more obvious in CDS2 KO cells (Fig. 1I). With the KO cells, we were able to more accurately examine the rescue effect of CDS1 or CDS2. CDS1 overexpression could restore normal LD morphology in both CDS1- and CDS2-deficient cells. Likewise, CDS2 overexpression could also restore LD morphology in both cells (Fig. 1, J–L). Thus, CDS1 and CDS2 appear to share overlapping functions in relation to regulating LD size.
Figure 1.
CDS1/2 deficiency resulted in the formation of sLDs. A, efficiency of CDS1 and CDS2 KD in HeLa cells. B, LD morphology upon CDS KD in HeLa cells. LDs were stained by BODIPY. Bars, 5 μm. C, distribution of LDs according to their sizes in CDS KD cells. LDs from ∼50 cells/cell type were used. D, total TAG level in HeLa cells upon CDS KD. E, CDS KO strategy diagram by CRISPR/Cas9. F, LD morphology upon CDS KO in HeLa cells. LDs were stained by BODIPY. Bars, 5 μm. G, distribution of LDs according to their sizes in CDS KO cells. LDs from ∼50 cells/cell type were used. H, quantification of diameters of the three largest LDs in each cell as shown in B and F. Two-tailed Student's t test was used: mean ± S.D. (error bars); n = 45 LDs from 15 cells for each cell type; ****, p < 0.0001. I, total TAG level in HeLa cells upon CDS KO. J, overexpression of CDS restored normal LD morphology in CDS KO cells. HeLa cells with CDS KO were transfected with mCherry-C1 empty vector or mCherry-tagged CDS1 or CDS2 for 24 h when cell confluence reached ∼60%. Bars, 5 μm. OE, overexpression. K, distribution of LDs according to their sizes in J. LDs from ∼50 cells/cell type were used. L, quantification of the diameters of three largest LDs in each cell as shown in J. Two-tailed Student's t test was used: mean ± S.D.; n = 45 LDs from 15 cells for each cell type; ****, p < 0.0001; ns, no significance. For CDS KD and KO cells, two different siRNAs and KO colonies were examined for each experiment. For B, D, F, I, and J, 200 μm oleate was added to cells to induce LD formation for 16 h. For A, D, and I, two-tailed Student's t test was used: mean ± S.D.; n = 3; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Although CDS1 and CDS2 share certain core functions, it remains a key question as to why mammalian cells have two isoforms of the same enzyme. One possibility is different substrate preferences (29), and another possibility could be distinct cellular localization. For instance, CDS enzymes were proposed to function at ER–plasma membrane (PM) contact sites during the synthesis and transfer of PI (30). We therefore carefully examined the localization of CDS1 and CDS2 in relation to the ER, LDs, ER–PM contact sites, and mitochondria using markers of Sec61β (31), LipidTox, MAPPER (32), Nir2 (33), and MitoTracker (34), respectively. We found that CDS1 and CDS2 mainly localized to the ER (Fig. S1A, top array), with CDS1 also localizing to mitochondria (Fig. S1A, bottom array). In addition, we also examined the distribution of LDs in CDS KD cells by labeling the ER (DsRed-ER) (Fig. S1B), Golgi (DsRed-Golgi) (Fig. S1C), and mitochondria (MitoTracker Deep Red) (Fig. S1D). Interestingly, the giant LDs in CDS-deficient cells colocalized with mitochondria, especially in CDS1 KD cells (Fig. S1, D and E). These data indicate that the ablation of CDS, especially CDS1, may affect mitochondrial function. Overall, the increase in LD size in CDS1- or CDS2-deficient cells was rather striking, and it is important to understand the underlying molecular mechanisms.
Knockdown of CIDE family proteins impairs sLD formation in CDS1-deficient cells
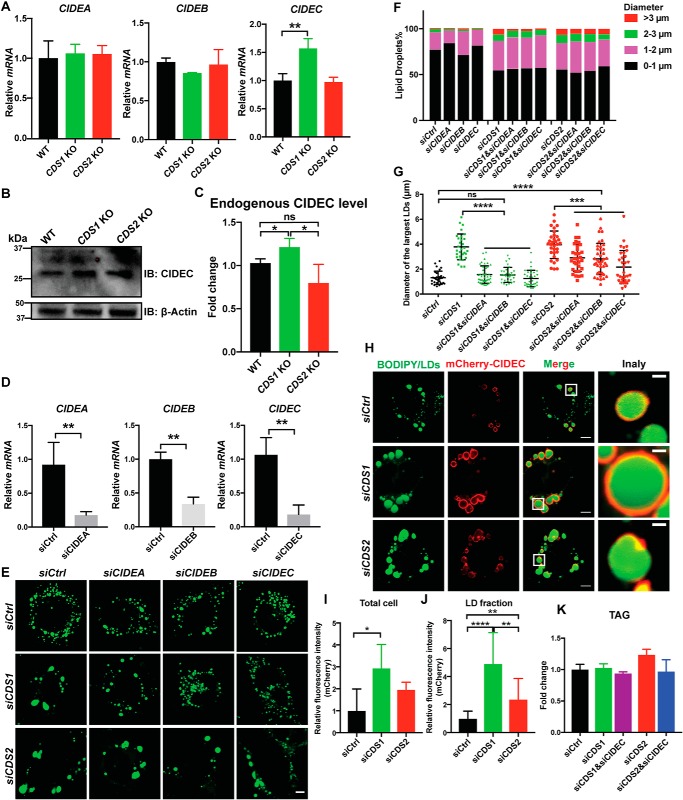
LD growth can be mediated by CIDE family proteins, including CIDEA, CIDEB, and CIDEC, which mediate lipid transfer from small to large LDs. It is possible that CIDE proteins promote sLD formation in CDS1/2-deficient cells. We first examined the expression of CIDE genes in CDS1/2-deficient cells. Whereas CDS1 KD had minor effects on the mRNA levels of CIDEA, -B, and -C (Fig. S2), CDS1 KO in HeLa cells significantly increased CIDEC mRNA and protein levels (Fig. 2, A–C). Interestingly, neither CDS2 KD nor KO had any effects on the expression of CIDE genes (Fig. 2A and Fig. S2). We next sought to investigate whether the depletion of CIDE genes (Fig. 2D) has any impacts on LD size in CDS1/2-deficient cells. Down-regulation of CIDE genes almost completely eliminated sLDs in CDS1-deficient cells, where LD size became similar to that of control cells, with silencing CIDEC showing the strongest rescue effect (Fig. 2, E–G). LD size in CDS2-deficient cells was also decreased by down-regulation of CIDE proteins, but remained significantly larger than that of control cells (Fig. 2G). Furthermore, there was a significantly stronger expression of CIDEC protein localizing on LDs in CDS1-deficient cells, but not in CDS2-deficient cells, based on fluorescence intensity (Fig. 2, H–J). Finally, no significant changes were observed in the total TAG level when CIDEC was knocked down in CDS1- or CDS2-deficient cells (Fig. 2K). This indicates that CIDEC regulates the growth of LDs through the fusion-transfer mechanism without affecting TAG metabolism. Together, these data suggest that sLD formation in CDS1-deficient cells mainly results from an enhanced LD fusion event mediated by CIDE proteins, especially CIDEC, whereas sLD formation in CDS2-deficient cells may be driven by different factors.
Figure 2.
Knocking down CIDEC reduced the occurrence of sLDs in CDS1-deficient cells. A, CDS1 KO increased the mRNA level of CIDEC in HeLa cells. Two-tailed Student's t test was used: mean ± S.D. (error bars); n = 3; **, p < 0.01. B, immunoblot analysis of endogenous CIDEC in HeLa cells upon CDS depletion. C, quantification of CIDEC level in B. Two-tailed Student's t test was used: mean ± S.D.; n = 3; *, p < 0.05; ns, no significance. D, the KD efficiency of CIDEA, -B, and -C in HeLa cells. Two-tailed Student's t test was used: mean ± S.D.; n = 3; **, p < 0.01. E, morphology of LDs upon CIDEA/B/C KD in CDS-deficient cells. 200 μm oleate was added to cells to induce LD formation for 16 h. LDs were stained by BODIPY. Two different siRNA sequences were used for each gene. Bars, 5 μm. F, distribution of LDs according to their sizes in E. LDs from ∼50 cells/cell type were used. G, quantification of the diameters of three largest LDs in each cell as shown in F. One-way analysis of variance was used: mean ± S.D.; n = 45 LDs from 15 cells for each cell type; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, no significance. H, localization of CIDEC in CDS KD cells. Bars, 5 μm. Inset, bars, 1 μm. I, protein level of CIDEC upon CDS depletion in whole cell examined by fluorescence. Two-tailed Student's t test was used: mean ± S.D.; n = 20; *, p < 0.05. J, protein level of CIDEC upon CDS depletion in LD fraction examined by fluorescence. Two-tailed Student's t test was used: mean ± S.D.; n = 20; **, p < 0.01; ****, p < 0.0001. K, TAG levels when CIDEC was knocked down in CDS-deficient HeLa cells. Values are mean ± S.D.; n = 3. 200 μm oleate was added to cells to induce LD formation for 16 h.
Knockdown of DGAT2/GPAT4 impairs sLD formation in CDS2-deficient cells
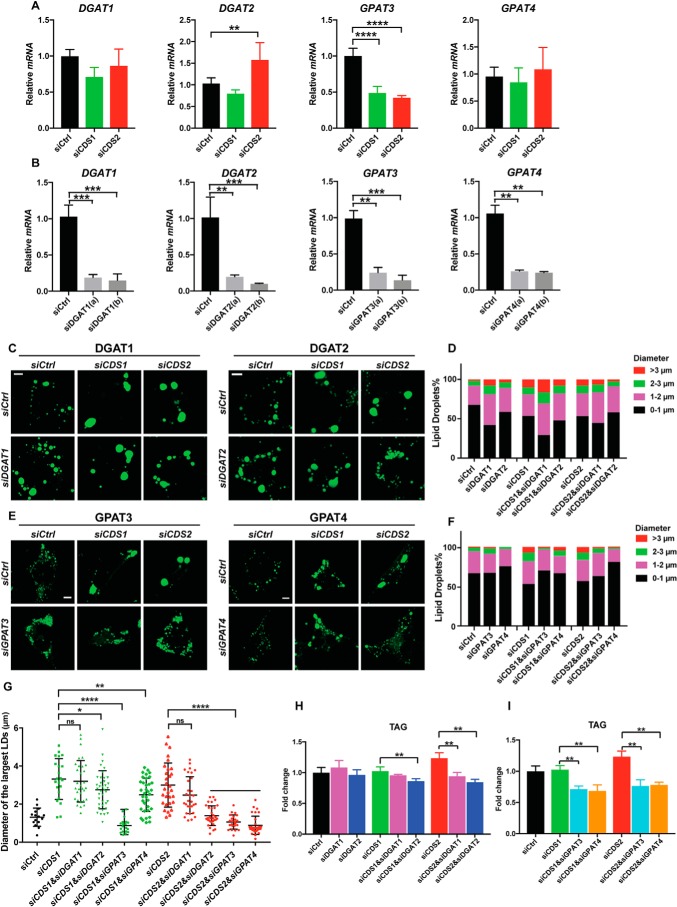
We next sought to understand how sLDs are formed in CDS2-deficient cells. Apart from LD fusion, increased TAG synthesis is another factor that drives LD expansion (1, 12). Enzymes such as DGAT2 and GPAT3/4 can relocate from the ER to LD surface, where they locally synthesize TAG to meet the requirement of LD growth (12, 23). Hence, we first examined the mRNA levels of DGAT1/2 and GPAT3/4 in CDS1/2-deficient cells (Fig. 3, A) and found that CDS1 or CDS2 KD had no effect on the mRNA expression of DGAT1, whereas CDS2 KD significantly increased the mRNA expression of DGAT2. The expression of GPAT3 was dramatically decreased in both CDS1- and CDS2-deficient cells, whereas that of GPAT4 was not affected. PA is the substrate of CDS enzymes and plays important roles in the regulation of LD dynamics (25, 35). The discrepancy in the changes of mRNA expression between DGAT1/2 and GPAT3/4 when depleting CDS enzymes may be linked to their respective roles in the TAG synthesis pathway, in which GPAT3/4 produces PA, whereas DGAT1/2 consumes PA (36). Increased mRNA expression of DGAT2 in CDS2-deficient, but not CDS1-deficient, cells implies the involvement of TAG synthesis in the formation of sLDs when ablating CDS2. Thus, it is necessary to examine the dynamics of LDs in CDS-deficient cells when reducing TAG synthesis. We silenced DGAT1/2 and GPAT3/4 (Fig. 3B) and examined LD size in both CDS1- and CDS2-deficient cells. Whereas knocking down DGAT1 had little effect on sLD formation in either CDS1- or CDS2-deficient cells, DGAT2 knockdown specifically impaired sLD formation in CDS2-deficient cells, but not in CDS1-deficient cells (Fig. 3, C, D, and G). In the case of GPAT3/4, the formation of sLDs in CDS-deficient cells was clearly blocked by down-regulation of GPAT3. However, only sLD numbers in CDS2- but not CDS1-deficient cells were drastically reduced by GPAT4 depletion (Fig. 3, E–G). As expected, down-regulation of DGAT1/2 and GPAT3/4 in CDS2 KD cells significantly decreased the total TAG level compared with CDS2 KD alone (Fig. 3, H and I). Importantly, similar effects of CIDE family proteins, DGAT2, and GPAT4 on sLD formation caused by CDS1 or CDS2 deficiency were also found in HEK293T cells (Fig. S3, A–C), 3T3-L1 cells (Fig. S3, D–F), and CDS KO HeLa cells (Fig. S3, G–I). CDS KO HeLa cells displayed much larger LDs (>5 μm) than that in KD cells. Down-regulation of CIDEC or DGAT2/GPAT4 effectively reduced the proportion of the largest LDs (>5 μm) in CDS1 and CDS2 KO cells, respectively (Fig. S3H). These results strongly suggest that increased TAG synthesis mediated by DGAT2 and/or GPAT4 may be mainly responsible for sLD formation upon CDS2 depletion.
Figure 3.
Knocking down DGAT2 or GPAT4 restored normal LD morphology in CDS2-deficient cells. A, mRNA levels of DGAT1/2 and GPAT3/4 upon CDS KD in HeLa cells. Two-tailed Student's t test was used: mean ± S.D. (error bars); n = 3; **, p < 0.01; ****, p < 0.0001. B, KD efficiency of DGAT1/2 and GPAT3/4 in HeLa cells. Two-tailed Student's t test was used: mean ± S.D.; n = 3; **, p < 0.01; ***, p < 0.001. Two different sequences (a and b) of siRNA against each gene were used. C, LD morphology when DGAT1/2 were knocked down in CDS-deficient HeLa cells. 200 μm oleate was added to cells to induce LD formation for 16 h. LDs were stained by BODIPY. Bars, 5 μm. D, distribution of LDs according to their sizes in C. LDs from ∼50 cells/cell type were used. E, LD morphology when DGAT3/4 were knocked down in CDS-deficient HeLa cells. 200 μm oleate was added to cells to induce LD formation for 16 h. LDs were stained by BODIPY. Bars, 5 μm. F, distribution of LDs according to their sizes in E. LDs from ∼50 cells/cell type were used. G, quantification of the diameters of the three largest LDs in each cell as shown in C and E. Two-tailed Student's t test was used: mean ± S.D.; n = 45 LDs from 15 cells for each cell type; *, p < 0.05; **, p < 0.01; ****, p < 0.0001; ns, no significance. H, total TAG level in CDS1/2 and DGAT1/2 KD cells. Two-tailed Student's t test was used: mean ± S.D.; n = 3; **, p < 0.01. I, total TAG level in CDS1/2 and GPAT3/4 KD cells. Two-tailed Student's t test was used: mean ± S.D.; n = 3; **, p < 0.01.
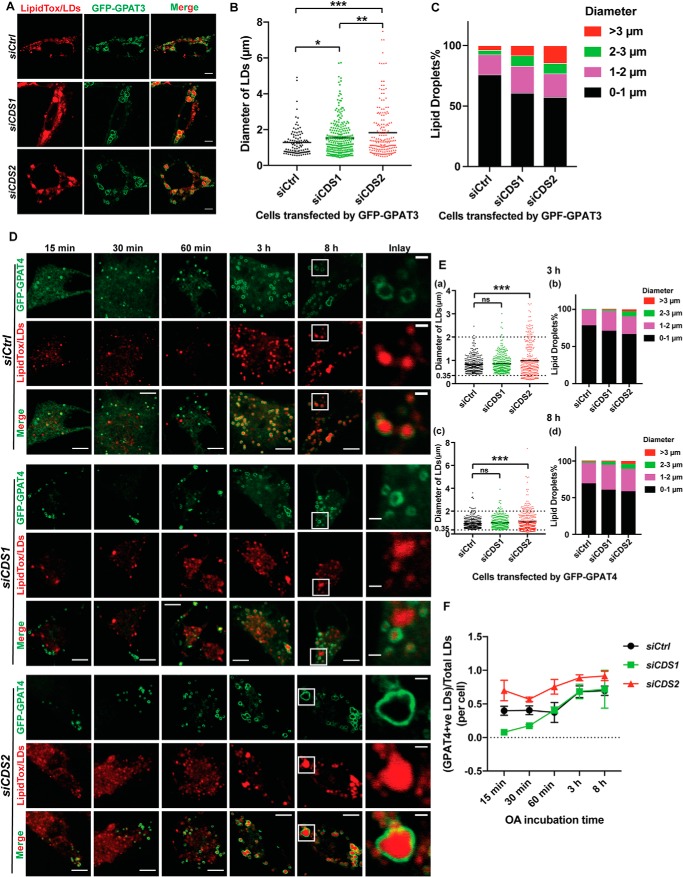
CDS2 deficiency promotes LD association of DGAT2 and GPAT4
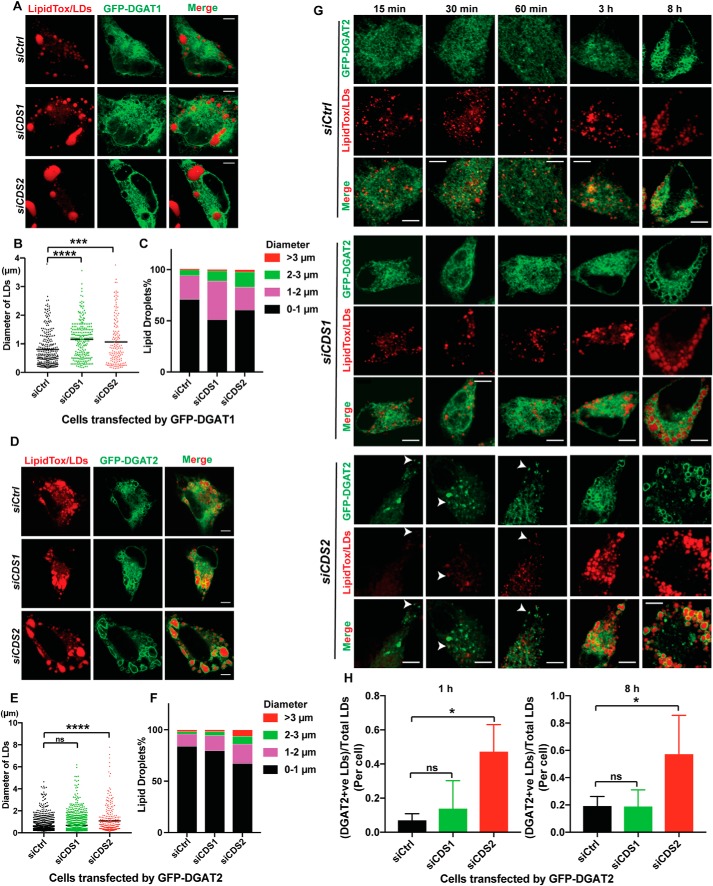
sLDs caused by CDS2 deficiency are most likely generated through the incorporation of lipids synthesized in situ on the LD surface (12). The involvement of DGAT2 and GPAT4 in sLD formation prompted us to examine the localization of these enzymes in CDS2-deficient cells. Whereas DGAT1 had no colocalization with LDs (Fig. 4A), DGAT2 was clearly associated with LDs in control, CDS1, and CDS2 KD cells (Fig. 4D). Interestingly, DGAT2 almost completely localized to the LD surface in CDS2 KD cells, whereas only a fraction of DGAT2 was found to associate LDs in control and CDS1 KD cells after incubation with oleate for 16 h, with the majority of DGAT2 showing a pattern resembling the ER network (Fig. 4D). Quantification of LD sizes in cells overexpressing DGAT1 or DGAT2 revealed that DGAT1 mainly contributed to the growth of middle-sized LDs (about 2–3 μm in diameter) in CDS1 or CDS2 KD cells (Fig. 4, B and C), whereas DGAT2 increased the sLD percentage (>3 μm in diameter) in CDS2 KD cells but only slightly enhanced the growth of middle-sized LDs in CDS1 KD cells (Fig. 4, E and F). All of these results implied that CDS2 depletion promotes more DGAT2 association with the LD surface, where it produces TAG to support LD expansion. We next investigated the localization of DGAT2 at early stages of LD formation. In control or CDS1 KD cells, DGAT2 did not translocate to LDs until 3 h after incubation with oleate (Fig. 4G). Strikingly, in CDS2 KD cells, DGAT2 started to translocate from the ER to LDs at the very beginning of LD formation (i.e. as early as 30 min after oleate treatment) (Fig. 4G). The dramatic differences of DGAT2 localization between control or CDS1 KD and CDS2 KD cells could be observed either at the early stage of 1 h or at the late stage of 8 h after oleate treatment (Fig. 4H). The phenotype of enhanced DGAT2 localization on the LD surface under CDS2 deficiency was consistently observed in CDS KO cells (Fig. S4).
Figure 4.
CDS2 deficiency promoted LD-association of DGAT2. A, localization of DGAT1 in CDS KD cells after incubation with 200 μm OA for 16 h. Bars, 5 μm. B, quantification of LD sizes in cells transfected by GFP-DGAT1 upon CDS deficiency. Two-tailed Student's t test was used: mean; n = 150–300 LDs from ∼20 cells; ***, p < 0.001; ****, p < 0.0001. C, distribution of LDs in cells transfected by GFP-DGAT1 upon CDS deficiency. LDs from ∼20 cells/cell type were used. D, localization of DGAT2 in CDS KD cells after incubation with 200 μm OA for 16 h. Bars, 5 μm. E, quantification of LD sizes in cells transfected by GFP-DGAT2 upon CDS deficiency. Two-tailed Student's t test was used: mean; n = ∼1000 LDs for siCtrl and siCDS1 and 300 LDs for siCDS2; ****, p < 0.0001; ns, no significance. F, distribution of LDs in cells transfected by GFP-DGAT2 upon CDS deficiency. LDs from ∼20 cells/cell type were used. G, localization of DGAT2 at the early stage of LD formation in CDS KD cells. 200 μm oleate was added to cells for the indicated time. Bars, 5 μm. White arrows, DGAT2 puncta colocalizing with LDs. H, quantification of DGAT2-LD associations during LD growth upon CDS deficiency. Two-tailed Student's t test was used: mean ± S.D. (error bars); n = 20 cells; *, p < 0.05; ns, no significance.
In the case of GPAT3/4, GPAT3 was able to relocate from the ER to LDs in all cell types (Fig. 5A and Fig. S5, A (a and d) and B (a and d)). GPAT3 contributed to sLD formation in either CDS1- or CDS2-deficient cells, especially in CDS2-deficient cells (Fig. 5, B and C). However, this contribution was not apparent until the late stage of LD formation (8-h oleate treatment) (Fig. S5, A (b and c) and B (b and c)). In contrast, GPAT4 specifically contributed to sLD formation in CDS2 KD cells (Fig. 5, D and E). GPAT4 showed more contribution to sLD formation (>3 μm) in CDS2 KD cells from the early stage of LD growth (3-h oleate treatment) (Fig. 5E (a and b)), whereas in CDS1 KD cells, GPAT4 seemed to mainly support the middle-size (1–3 μm) LD growth at the late stage of LD growth (Fig. 5E (c and d)). Specifically, LDs associated with GPAT4 in CDS1 KD cells were smaller than those in CDS2 KD cells (Fig. 5, D and E (a and c)). Similar observations were also made in CDS1 or CDS2 KO cells (Fig. S5, C and D). There was more GPAT4-LD colocalization in CDS2 KD cells, although GPAT4 translocation to LDs started at the very early stage of LD formation under all conditions (Fig. 5, D and F). In summary, these results demonstrated that CDS2 deficiency specifically promoted the translocation of DGAT2 and GPAT4 to the LD surface, indicating that continuous TAG production on the LD surface might be the main driving force for sLD formation in CDS2-deficient cells.
Figure 5.
CDS2 deficiency promoted LD-association of GPAT4. A, localization of GPAT3 in CDS KD cells after incubation with 200 μm oleate for 16 h. Bars, 5 μm. B, quantification of LD sizes in cells transfected by GFP-GPAT3 upon CDS deficiency. Two-tailed Student's t test was used: mean, n = ∼200–300 LDs from ∼20 cells. *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, distribution of LDs in cells transfected by GFP-GPAT3 upon CDS deficiency. LDs from ∼20 cells/cell type were used. D, localization of GPAT4 in CDS KD cells at the early stage of LD formation in CDS KD cells. 200 μm oleate was added to cells for the indicated time. Bars, 5 μm. Inset, bars, 1 μm. E, quantification of size (a and c) and distribution (b and d) of GPAT4-positive LDs at 3 and 8 h after adding oleate. Two-tailed Student's t test was used: mean; n = ∼200–350 LDs from ∼20 cells; ***, p < 0.001; ns, no significance. F, quantification of GPAT4-LD associations during LD growth upon CDS deficiency: mean ± S.E. (error bars); n = 20 cells. OA, oleate.
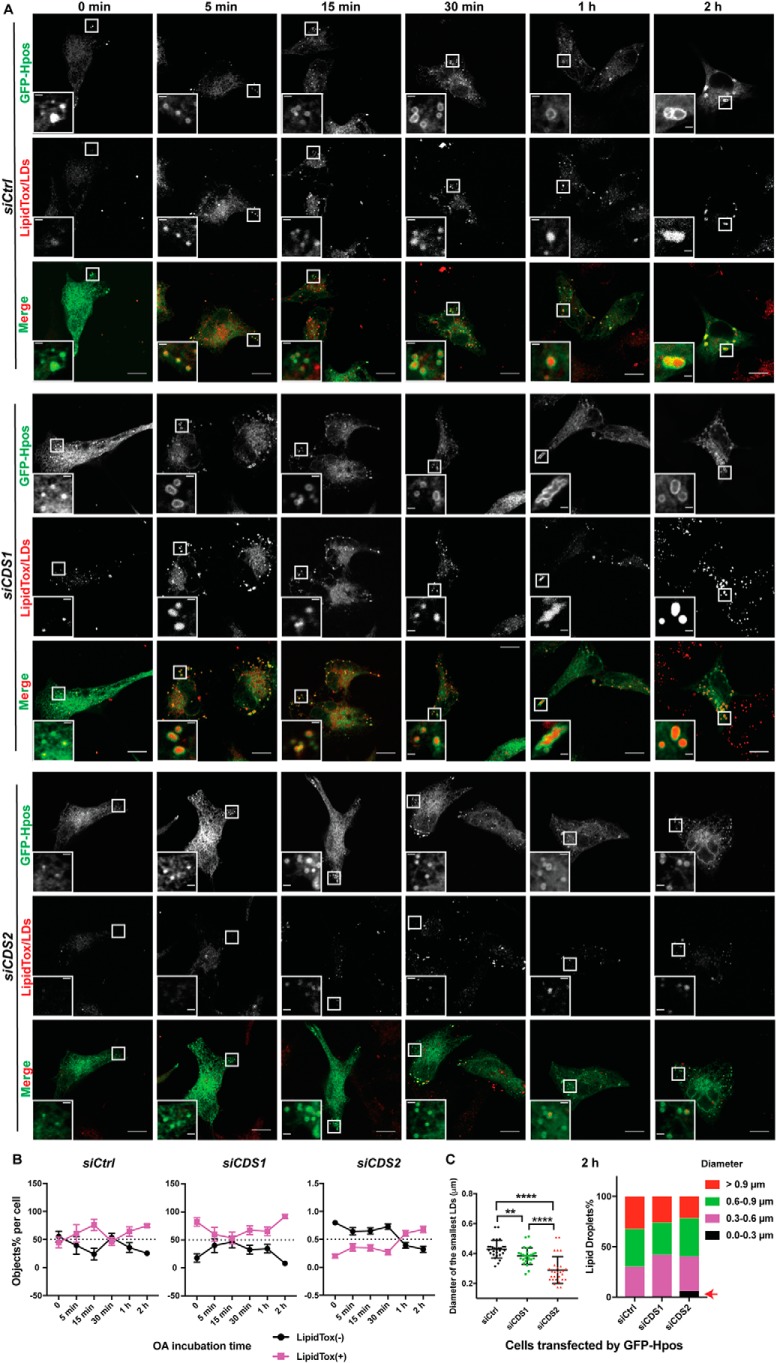
CDS2 deficiency results in a delay in iLD maturation
We next investigated the role of CDS1 and CDS2 in the biogenesis of LDs at the very initial stage. An artificial LD marker, Hpos, was used to label the nascent LDs that were not big enough to be labeled by the lipophilic dye LipidTox Deep Red, which can only stain LDs of a certain size (11). In control or CDS1 KD cells, iLDs were quickly generated and could be stained by LipidTox Deep Red as early as at 5 min after oleate treatment (Fig. 6, A and B). However, in CDS2 KD cells, most iLDs accumulated could not be labeled by LipidTox until 1–2 h after oleate treatment (Fig. 6, A and B). Based on the distribution of LD sizes at 2-h oleate incubation, there was an obvious population (∼10%) of smaller LDs with diameter below 0.3 μm in CDS2 KD cells (Fig. 6C, indicated by a red arrow). This delay in iLD maturation in CDS2-deficient cells was more striking in KO cell lines (please note that large LipidTox-positive particles are pre-existing LDs that remained after starvation) (Fig. S6). The LipidTox-negative Hpos puncta accumulation in CDS2-deficient cells indicates that CDS2 may play an important role in promoting the maturation of iLDs. This difference in LD formation at the early stage between CDS1- and CDS2-deficient cells also suggests that CDS1 and CDS2 may function differently in the process of LD biogenesis.
Figure 6.
Accumulation of LipidTox-negative and Hpos-positive puncta in CDS2-deficient HeLa cells. A, LD formation upon CDS deficiency. Control and CDS1/2 KD cells expressing GFP-Hpos were first delipidated by culturing in serum-free medium supplemented with 5% lipoprotein-deficient serum for 48 h. The cells were then treated with 200 μm oleate for the indicated times before fixing. LDs were stained by LipidTox Deep Red. Bars, 5 μm. Inset, bars, 1 μm. B, quantification of Hpos-positive (+)/LipidTox-negative (−) and Hpos-positive (+)/LipidTox-negative (+) objects, respectively, in CDS1/2-deficient cells upon oleate/oleic acid treatment: mean ± S.E. (error bars); n = ∼30 cells. OA, oleic acid/oleate. C, quantification of LD sizes and distribution of LDs in CDS-deficient cells after adding oleate for 2 h. For the LD diameter assay, the three smallest LDs in each cell were analyzed in each cell type. Two-tailed Student's t test was used: mean ± S.D.; n = 45 LDs from 15 cells for each cell type. Red arrow, size and percentage of LDs accumulated in CDS2-deficient cells after incubation with oleate for 2 h.
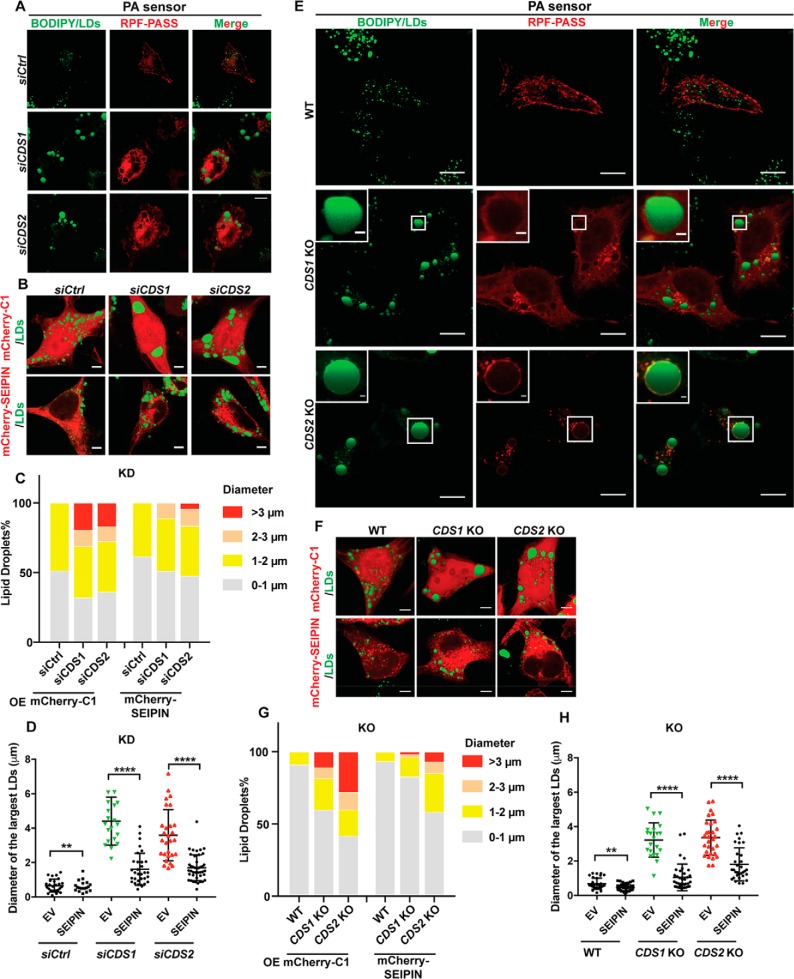
The role of PA in the formation of sLDs in CDS-deficient cells
PA is a key intermediate in the synthesis of all glycerolipids. It is the substrate of CDS1/2, which convert PA into CDP-DAG for the synthesis of other phospholipids, such as PI (37). It has been shown that the accumulation of PA contributes to the giant LD formation and that PA facilitates LD coalescence (25). To investigate whether PA is involved in sLD formation caused by CDS depletion, we used the PA sensor (RFP-PASS) to examine PA localization in CDS-deficient cells. Interestingly, there was a clear localization of PA on the sLD surface in CDS KD cells (Fig. 7A), and this localization became more striking in CDS2 KO cells (Fig. 7E). This was consistent with the lipidomic result that CDS2 KD increased PA level (15). Moreover, our laboratory previously showed that PA accumulation caused by SEIPIN deficiency can regulate LD dynamics (14). Also, we provided biochemical evidence that SEIPIN is capable of binding anionic phospholipids such as PA (38). Fld1/SEIPIN deficiency was also known to increase GPAT activity and cause the formation of giant LDs in yeast and mammalian cells (14, 20, 39). We wondered whether SEIPIN overexpression could inhibit GPAT activity, thereby reducing the level of PA and restoring LD size in CDS-deficient cells. Indeed, our data demonstrated that overexpression of SEIPIN decreased LD size in both CDS1/2 KD (Fig. 7, B–D) and KO cells (Fig. 7, F–H). These data suggest that increased PA level might be an important factor for sLD formation in CDS-deficient cells, especially in CDS2-deficient cells.
Figure 7.
SEIPIN and PA regulated sLD formation in CDS-deficient cells. A, PA accumulation on the surface of sLDs in CDS KD cells. Bars, 5 μm. B, overexpression of SEIPIN reduced sLD size in CDS KD cells. Bars, 5 μm. C, distribution of LDs upon SEIPIN overexpression in CDS KD cells. LDs from ∼20 cells/cell type were used. OE, overexpression. D, quantification of the diameters of the three largest LDs in each cell as shown in B. Two-tailed Student's t test was used: mean ± S.D. (error bars); n = 45 LDs from 15 cells for each cell type; **, p < 0.01; ****, p < 0.0001. E, PA accumulation on the sLD surface in CDS KO cells. Bars, 5 μm. Inset, bars, 1 μm. F, overexpression of SEIPIN reduced sLD size in CDS KO cells. Bars, 5 μm. G, distribution of LDs according to their sizes upon SEIPIN overexpression in CDS KO cells. LDs from ∼20 cells/cell type were used. OE, overexpression. H, quantification of the diameters of three largest LDs in each cell as shown in F. Two-tailed Student's t test was used: mean ± S.D.; n = 45 LDs from 15 cells for each cell type; **, p < 0.01; ****, p < 0.0001. RFP-PASS, empty vector of mCherry-C1, and mCherry-tagged SEIPIN were transfected into CDS KD/KO cells for 24 h when cells reached ∼60% confluence.
Discussion
In a genome-wide screen, we have previously found that yeast cells deficient in CDS1 harbor giant or supersized LDs (25). This was further confirmed in cultured mammalian cells (15). Notably, the giant LDs seen in CDS-deficient cells are among the largest observed in yeast or cultured cells. Work described in this study aims to understand the mechanisms by which sLDs are formed in CDS1- or CDS2-deficient mammalian cells. Although CDS1 and CDS2 catalyze the same biochemical reaction, our data here suggest that CDS1 and CDS2 function differently during LD formation in both early stage (pre-LD formation) and the late stage (LD growth and sLD formation). The CIDE proteins, CIDEC in particular, play a major role in sLD formation under CDS1-deficient conditions, whereas enhanced TAG synthesis mediated by GPAT4/DGAT2 contributes to the formation of sLDs under CDS2 deficiency.
CDS1 and CDS2 are known to have different substrate preferences and generate different CDP-DAG pools (29). CDS1 has almost no acyl chain preference for PA, whereas CDS2 shows substrate specificity at both the sn-1 and sn-2 positions. For instance, CDS2 prefers 1-stearoyl-2-arachidonoyl-PA. Thus, CDS1 and CDS2 deficiency may accumulate different PA species, which may impact LD biogenesis and growth differently. However, we were not able to detect any specific accumulation of 1-stearoyl-2-arachidonoyl-PA or TAG species enriched in arachidonate in CDS2-deficient cells (data not shown). Alternatively, whereas both CDS1 and CDS2 are believed to localize to the ER (Fig. S1A, top) (30), there were studies showing that CDS1, but not CDS2, is possibly present on mitochondria (Fig. S1A, bottom array), where it regulates mitochondrial lipid composition (40, 41). Consistently, there was much more colocalization between sLDs and mitochondria in CDS1-depleted cells than in CDS2-depleted cells (Fig. S1, D and E). Therefore, the degree of mitochondrial association may explain the phenotypic differences in LD dynamics upon CDS1/2 deficiency.
CIDE proteins are LD-associated proteins and well-known for their role in LD dynamics by generating “fusion pores” between LDs and facilitating the transfer of TAG from small to large LDs (42–44). It has been revealed that CIDEC-deficient white adipocytes lose giant LDs but accumulate many small LDs. Ectopic expression of CIDEC contributes to sLD formation and reduces LD numbers (19, 45–48). Also, CIDEA/C was heavily up-regulated in liver steatosis, which again confirmed their role in promoting lipid storage in the form of giant LDs (49). In this study, we found that CIDE proteins, especially CIDEC, contribute significantly to sLD formation in CDS1-deficient cells, but less so in CDS2-deficient cells (Fig. 2, E–G). It is not clear how CIDEC is up-regulated under CDS1 KO conditions. Moreover, exactly why CIDE proteins are required to form sLDs under CDS1 deficiency is unknown. Given its functional link with mitochondria as mentioned above, CDS1 may regulate CIDE proteins and LD growth through mitochondrial changes. Notably, CIDE proteins were initially found on mitochondria and implicated in apoptosis (50). Thus, CDS1 and CIDE proteins may be functionally connected through mitochondria. Finally, it is somewhat surprising that knocking down each of the three CIDE genes reduced LD size in CDS (especially CDS1)–deficient cells. Perhaps the CIDE proteins need to form functional heterooligomers under CDS deficiency. Future studies will elucidate the exact relationship between CIDE proteins, mitochondria, and CDS1.
Besides the CIDE proteins, enhanced synthesis of TAG is another driving force for giant LD formation (12). DGAT1 had no colocalization with LDs (Fig. 4A), which explained the finding that down-regulating DGAT1 could not restore LD size under CDS1/2-deficient conditions (Fig. 3C). More DGAT2 and GPAT4 were found on the LD surface in CDS2- than CDS1-deficient cells (Figs. 4H and 5F and Fig. S4B), suggesting that these enzymes play a role in sLD formation under CDS2 deficiency. Indeed, knockdown of either DGAT2 or GPAT4 significantly reduced sLD formation in CDS2-deficient cells (Fig. 3, C and E). Interestingly, CDS2, but not CDS1, deficiency also delayed the maturation of iLDs (Fig. 6 and Fig. S6), a phenotype reminiscent of SEIPIN deficiency (17). Moreover, the formation of sLDs in CDS2-deficient cells (Figs. 1 (D and I), 4H, and 5F and Fig. S4B) was also similar to that of SEIPIN deficiency (16, 17, 20, 21, 39, 51, 52). These findings imply that CDS2, but not CDS1, may be functionally related to SEIPIN.
CDS proteins convert PA into CDP-DAG, and either CDS1 or CDS2 deficiency can cause PA accumulation. Indeed, prominent PA accumulation was detected using mass spectrometry (15) and a PA sensor in CDS2-deficient cells (Fig. 7, A and E). PA is a negatively charged, cone-shaped lipid that is known to promote SNARE-dependent and -independent membrane fusion events (35, 53). As a nonbilayer lipid, PA can also impact the local bilayer tension of the ER (54). Thus, accumulation of PA in the ER may increase bilayer tension, thereby delaying early LD growth in CDS2-deficient cells. PA may also promote the formation of sLDs at a later stage, given its fusogenic property. It is possible that a higher level of PA on the monolayer of LDs would promote spontaneous fusion of contacting LDs (Fig. 7, A and E), leading to the formation of sLDs. Interestingly, the loss of SEIPIN function was associated with increased GPAT3/4 activity and the accumulation of PA in the ER (14, 25). As discussed above, CDS2 and SEIPIN deficiency causes similar changes in early and late LD growth. Moreover, SEIPIN overexpression almost completely restored LD morphology in CDS1/2-deficient cells (Fig. 7, B–D and F–H). Thus, CDS1/2 and SEIPIN may be functionally connected by their role in PA metabolism.
In summary, our results here unveil distinct mechanisms by which the two CDS isoforms in mammals, CDS1 and CDS2, may regulate LD growth. Our results also support the notion that phospholipids play a key role in LD biogenesis and growth (5, 35, 54). These results provide mechanistic insights into how cells may generate supersized LDs, which are prominent features of common human metabolic disorders, such as obesity and hepatic steatosis.
Experimental procedures
Mammalian cell culture
HeLa and HEK293T cells were cultured in high-glucose Dulbecco's modified Eagle's medium (Life Technologies, Inc.) supplemented with 10% fetal bovine serum (Life Technologies) and 1% penicillin/streptomycin/glutamine (Life Technologies). 3T3-L1 cells were cultured in high-glucose Dulbecco's modified Eagle's medium supplemented with 10% newborn calf serum (Life Technologies) and 1% penicillin/streptomycin/glutamine. Cells were incubated at 37 °C with 5% CO2. The medium was changed every 2 days. To induce lipid droplet formation, 200 μm oleate (Sigma-Aldrich) was added to cells for the indicated time.
Plasmid construction
cDNA sequences were retrieved from NCBI. Primers were designed in-frame of the multiple cloning site (Table S1). DNA was amplified by PCR using Phusion Hot Start II High-Fidelity DNA polymerase (Thermo Fisher Scientific) according to the manufacturer's instructions. Plasmids constructed in this study and gifted from colleagues are listed in Table S2.
Plasmid transfection
The transient plasmid transfections were performed using Lipofectamine LTX Plus reagent (Life Technologies) based on the manufacturer's instructions. For 6-well plates, 1–2 μg of plasmid DNA was diluted into 250 μl of Opti-MEM medium (Life Technologies), followed by the addition of 2 μl of PLUS reagent. 2 μl of LTX reagent was diluted into a separated tube with the same volume of Opti-MEM medium. After incubation for 5 min, the LTX reagent was transferred into the tube of plasmid, incubating for 10 min. The mixture was then added to the cells. Normally, all of the transfections were performed when cells were at 40–60% confluence. The amount of plasmid DNA and reagent was scaled, depending on the size of the dishes. Cells were harvested 24 or 48 h after transfection.
siRNA transfection
Commonly, 15 × 104 cells were seeded on cover slides in 6-well plates 24 h prior to transfection. Transient transfections of siRNA against CDS1/2 (Sigma-Aldrich), CIDEA/B/C (Shanghai GenePharma Co. Ltd.), GPAT3/4 (Shanghai GenePharma Co. Ltd.), and DGAT1/2 (Sigma-Aldrich) were carried out at 20 nm siRNA using Lipofectamine RNAiMAX reagent (Life Technologies) according to the manufacturer's protocol. The siRNA and RNAiMAX (the volume ratio of RNAiMAX/siRNA was 1.5:1) were dissolved in 50 μl of Opti-MEM medium separately and incubated for 5 min, and then the tubes were combined together gently and incubated for 20 min before transferring to cells. Transfections were commonly carried out at about 40–60% confluence of cells. Cells were harvested 48 h after transfection. The seeding density was increased to around 20 × 104 for the purpose of double knockdown. When both siRNA and plasmid transfections were required, the plasmid transfection would be done at least 6 h later than the siRNA transfection with fresh culture medium.
Generation of knockout cell line using CRISPR/Cas9
Knockout cell lines were generated using CRISPR/Cas9 techniques according to a previous study (55). Single guide RNAs were designed using CRISPR (https://crispor.tefor.net/)3 (56) and constructed into pSpCas9(BB)-2A-GFP vector. HeLa cells were transfected with GFP-tagged CRISPR plasmid for 24 h. The GFP-positive single cell, screened by flow cytometry, was then seeded into 96-well plates, 1 cell/well for 5 plates. After growing cells for 4–6 days, the growing colonies were picked and passaged for screening by Western blotting using antibodies and fluorescence microscopy.
Fluorescence microscopy
Cells grown on coverslips were fixed using 4% paraformaldehyde (ProSciTech) for 15 min at room temperature. LDs were stained by either 1 μg/ml BODIPY 493/503 (Life Technologies) for 15 min or HCS LipidTox Deep Red neutral lipid stain (Life Technologies) for 45 min at room temperature at a dilution of 1:500 in the dark. The mitochondria were labeled with MitoTracker® Deep Red FM (Life Technologies) at a dilution of 1:10,000 by adding the dye into live cells for 16 h at 37 °C with 5% CO2. After washing two times using PBS (Life Technologies), the coverslips were mounted onto slides. Fixed cells were viewed using an Olympus FV1200 confocal microscope. The diameters of the LDs were measured using ImageJ software (National Institutes of Health).
Antibodies
Antibody of rabbit polyclonal to CIDEC is a gift from Prof. Peng Li. It was diluted in 1:1000. For immunoblotting, we obtained horseradish peroxidase–conjugated secondary antibodies from Jackson ImmunoResearch.
Immunoblot analysis
Samples were mixed with 2× Laemmli buffer, incubated for 10–15 min at 70 °C, and then subjected to 10% SDS-PAGE. After electrophoresis, the proteins were transferred to Hybond-C nitrocellulose filters (GE Healthcare). Incubations with primary antibodies were performed at 4 °C overnight. Secondary antibodies were peroxidase-conjugated AffiniPure donkey anti-rabbit (H+L; Jackson ImmunoResearch Laboratories) used at a 1:1000 dilution. The bound antibodies were detected by ECL Western blotting detection reagent (GE Healthcare or Merck Millipore) and visualized with Molecular Imager® ChemiDocTM XRS+ (Bio-Rad).
Neutral lipid extraction
HeLa cells were grown in 6-cm dishes with proper density. 200 μm oleate was added to cells for 16 h when cells reached 80–90% confluence. After washing cells once with PBS, the neutral lipids were extracted by a 2-ml mixture of hexane (Ajax FineChem) and isopropyl alcohol (Ajax FineChem) (3:2) for 30 min in the fume hood. The solvent was then transferred into 2-ml glass vials. Another 1 ml of fresh hexane and isopropyl alcohol was used to collect the lipid residues in dishes and then transferred to glass vials together with the previous 2 ml of solvent. The lipids were dried using a SpeedVac centrifuge. The cells were lysed with 0.1 m NaOH (Ajax FineChem) for 15 min at room temperature after the dishes were dried, and the protein concentrations were determined by a bicinchoninic acid (Thermo Fisher Scientific) assay.
TLC assay
The neutral lipids were reconstituted in 60 μl of hexane. The samples were then loaded and separated using a Silica Gel 60 plate (Millipore) and developed in a solvent system consisting of heptane/diethyl ether/glacial acetic acid (90:30:1) (Ajax FineChem). The lipids were stained with iodine for around 15 min. The TLC plate was scanned using the Epson Perfection 4490 Photo, and lipids were quantified using ImageJ software and normalized to protein concentrations.
RNA extraction and quantitative real-time PCR
Total RNA was extracted using TRIzolTM reagent (Sigma-Aldrich). Mammalian cells were grown in 6-well plates. Cells were washed with PBS once and then lysed by the addition of 1 ml of TRIzolTM regent and incubation for 5 min at room temperature. 200 μl of chloroform (Sigma-Aldrich) was added to the lysates, followed by shaking 30 times violently and then incubation for 5 min at room temperature. The mixture was then centrifuged at 12,000 × g for 15 min at 4 °C. The 350-μl upper aqueous phase was carefully removed to a fresh tube, and the same volume of isopropyl alcohol (Sigma-Aldrich) was added and mixed well by the vortex for 5 s. The mixture was centrifuged at 12,000 × g for 10 min at 4 °C. After removing the upper aqueous phase, the pellet was washed twice with 1 ml of 75% ethanol (Sigma-Aldrich). Centrifugation at 7500 × g for 5 min at 4 °C was carried out between each wash. The RNA pellet was dried in a fume hood after removing ethanol and then dissolved in RNase-free water (Life Technologies). RNA concentration and purity were determined using a Nanodrop spectrophotometer (Thermo Fisher Scientific). 1 μg of RNA was adopted for cDNA synthesis using the High Capacity cDNA Reverse Transcription kit (Thermal Fisher Scientific). Quantitative RT-PCR was performed with a Rotor-Gene 6000 real-time PCR machine (Qiagen) using KAPA SYBR® Green mix (KAPA Biosystems). The mRNA levels were normalized against the housekeeping gene and compared with control samples. All quantitative RT-PCR primers used in this study are listed in Table S3.
Statistical analysis
All data are expressed as mean ± S.D. or mean ± S.E. Comparisons between two groups were analyzed using two-tailed Student's t test using GraphPad Prism version 6.0 software. Differences at values of p < 0.05 were considered as significant.
Author contributions
Y. X., X. D., X. H., and H. Y. conceived the project and designed the experiments. Y. X. performed the experiments and analyses with advice from X. D., I. L., K. L. H., and H. Y. X. D., Y. E. L., and H. Y. M. repeated several key experiments. Y. X. and H. Y. wrote the manuscript with input from X. D.
Supplementary Material
Acknowledgment
We thank the Biomedical Imaging Facility at the UNSW Mark Wainwright Analytical Centre, Australia.
This work was supported by National Health and Medical Research Council of Australia (NHMRC) Project Grants 1141939 and 1144726. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3 and Figs. S1–S6.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- LD
- lipid droplet
- iLD
- eLD, and sLD, initial, expanding, and supersized LD, respectively
- TAG
- triacylglycerol
- ER
- endoplasmic reticulum
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- DAG
- diacylglycerol
- CDS
- CDP-DAG synthase
- PI
- phosphatidylinositol
- KD
- knockdown
- KO
- knockout
- PM
- plasma membrane.
References
- 1. Yang H., Galea A., Sytnyk V., and Crossley M. (2012) Controlling the size of lipid droplets: lipid and protein factors. Curr. Opin. Cell Biol. 24, 509–516 10.1016/j.ceb.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 2. Kory N., Farese R. V. Jr., and Walther T. C. (2016) Targeting fat: mechanisms of protein localization to lipid droplets. Trends Cell Biol. 26, 535–546 10.1016/j.tcb.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Henne W. M., Reese M. L., and Goodman J. M. (2018) The assembly of lipid droplets and their roles in challenged cells. EMBO J. 37, e98947 10.15252/embj.201898947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Olzmann J. A., and Carvalho P. (2019) Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gao M., Huang X., Song B. L., and Yang H. (2019) The biogenesis of lipid droplets: lipids take center stage. Prog. Lipid Res. 75, 100989 10.1016/j.plipres.2019.100989 [DOI] [PubMed] [Google Scholar]
- 6. Walther T. C., and Farese R. V. Jr. (2012) Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687–714 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shyu P. Jr., Wong X. F. A., Crasta K., and Thibault G. (2018) Dropping in on lipid droplets: insights into cellular stress and cancer. Biosci. Rep. 38, BSR20180764 10.1042/BSR20180764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Farmer B. C., Kluemper J., and Johnson L. A. (2019) Apolipoprotein E4 alters astrocyte fatty acid metabolism and lipid droplet formation. Cells 8, E182 10.3390/cells8020182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murphy D. J., and Vance J. (1999) Mechanisms of lipid-body formation. Trends Biochem. Sci. 24, 109–115 10.1016/S0968-0004(98)01349-8 [DOI] [PubMed] [Google Scholar]
- 10. Nagle C. A., Vergnes L., Dejong H., Wang S., Lewin T. M., Reue K., and Coleman R. A. (2008) Identification of a novel sn-glycerol-3-phosphate acyltransferase isoform, GPAT4, as the enzyme deficient in Agpat6(−/−) mice. J. Lipid Res. 49, 823–831 10.1194/jlr.M700592-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kassan A., Herms A., Fernández-Vidal A., Bosch M., Schieber N. L., Reddy B. J., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., Gross S. P., Parton R. G., and Pol A. (2013) Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203, 985–1001 10.1083/jcb.201305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wilfling F., Wang H., Haas J. T., Krahmer N., Gould T. J., Uchida A., Cheng J. X., Graham M., Christiano R., Fröhlich F., Liu X., Buhman K. K., Coleman R. A., Bewersdorf J., Farese R. V. Jr., and Walther T. C. (2013) Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell 24, 384–399 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wilfling F., Thiam A. R., Olarte M. J., Wang J., Beck R., Gould T. J., Allgeyer E. S., Pincet F., Bewersdorf J., Farese R. V. Jr., and Walther T. C. (2014) Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. Elife 3, e01607 10.7554/eLife.01607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pagac M., Cooper D. E., Qi Y., Lukmantara I. E., Mak H. Y., Wu Z., Tian Y., Liu Z., Lei M., Du X., Ferguson C., Kotevski D., Sadowski P., Chen W., Boroda S., et al. (2016) SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of glycerol-3-phosphate acyltransferase. Cell Rep. 17, 1546–1559 10.1016/j.celrep.2016.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qi Y., Kapterian T. S., Du X., Ma Q., Fei W., Zhang Y., Huang X., Dawes I. W., and Yang H. (2016) CDP-diacylglycerol synthases regulate the growth of lipid droplets and adipocyte development. J. Lipid Res. 57, 767–780 10.1194/jlr.M060574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salo V. T., Belevich I., Li S., Karhinen L., Vihinen H., Vigouroux C., Magré J., Thiele C., Hölttä-Vuori M., Jokitalo E., and Ikonen E. (2016) Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35, 2699–2716 10.15252/embj.201695170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang H., Becuwe M., Housden B. E., Chitraju C., Porras A. J., Graham M. M., Liu X. N., Thiam A. R., Savage D. B., Agarwal A. K., Garg A., Olarte M. J., Lin Q., Fröhlich F., Hannibal-Bach H. K., et al. (2016) Seipin is required for converting nascent to mature lipid droplets. Elife 5, e16582 10.7554/eLife.16582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gong J., Sun Z., Wu L., Xu W., Schieber N., Xu D., Shui G., Yang H., Parton R. G., and Li P. (2011) Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J. Cell Biol. 195, 953–963 10.1083/jcb.201104142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jambunathan S., Yin J., Khan W., Tamori Y., and Puri V. (2011) FSP27 promotes lipid droplet clustering and then fusion to regulate triglyceride accumulation. PLoS One 6, e28614 10.1371/journal.pone.0028614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A. J., Wenk M. R., Parton R. G., and Yang H. (2008) Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180, 473–482 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Szymanski K. M., Binns D., Bartz R., Grishin N. V., Li W. P., Agarwal A. K., Garg A., Anderson R. G. W., and Goodman J. M. (2007) The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. U.S.A. 104, 20890–20895 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fei W., Du X., and Yang H. (2011) Seipin, adipogenesis and lipid droplets. Trends Endocrinol. Metab. 22, 204–210 10.1016/j.tem.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 23. Prinz W. A. (2013) A bridge to understanding lipid droplet growth. Dev. Cell 24, 335–336 10.1016/j.devcel.2013.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J. S., Vale R. D., Walter P., and Farese R. V. (2008) Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453, 657–661 10.1038/nature06928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fei W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T. S., Lin R. C., Dawes I. W., Brown A. J., Li P., Huang X., Parton R. G., Wenk M. R., Walther T. C., and Yang H. (2011) A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7, e1002201 10.1371/journal.pgen.1002201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Krahmer N., Guo Y., Wilfling F., Hilger M., Lingrell S., Heger K., Newman H. W., Schmidt-Supprian M., Vance D. E., Mann M., Farese R. V. Jr., and Walther T. C. (2011) Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 14, 504–515 10.1016/j.cmet.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang M., Gao M., Wu C., He H., Guo X., Zhou Z., Yang H., Xiao X., Liu G., and Sha J. (2014) Lack of testicular seipin causes teratozoospermia syndrome in men. Proc. Natl. Acad. Sci. U.S.A. 111, 7054–7059 10.1073/pnas.1324025111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuchler K., Daum G., and Paltauf F. (1986) Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. J. Bacteriol. 165, 901–910 10.1128/jb.165.3.901-910.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. D'Souza K., Kim Y. J., Balla T., and Epand R. M. (2014) Distinct properties of the two isoforms of CDP-diacylglycerol synthase. Biochemistry 53, 7358–7367 10.1021/bi501250m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim Y. J., Guzman-Hernandez M. L., and Balla T. (2011) A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev. Cell 21, 813–824 10.1016/j.devcel.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cui X. A., Zhang H., Ilan L., Liu A. X., Kharchuk I., and Palazzo A. F. (2015) mRNA encoding Sec61β, a tail-anchored protein, is localized on the endoplasmic reticulum. J. Cell Sci. 128, 3398–3410 10.1242/jcs.168583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang C. L., Hsieh T. S., Yang T. T., Rothberg K. G., Azizoglu D. B., Volk E., Liao J. C., and Liou J. (2013) Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 5, 813–825 10.1016/j.celrep.2013.09.038 [DOI] [PubMed] [Google Scholar]
- 33. Kim Y. J., Guzman-Hernandez M. L., Wisniewski E., and Balla T. (2015) Phosphatidylinositol-phosphatidic acid exchange by Nir2 at ER-PM contact sites maintains phosphoinositide signaling competence. Dev. Cell 33, 549–561 10.1016/j.devcel.2015.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chazotte B. (2011) Labeling mitochondria with MitoTracker dyes. Cold Spring Harb. Protoc. 2011, 990–992 10.1101/pdb.prot5648 [DOI] [PubMed] [Google Scholar]
- 35. Qi Y., Sun L., and Yang H. (2017) Lipid droplet growth and adipocyte development: mechanistically distinct processes connected by phospholipids. Biochim. Biophys. Acta 1862, 1273–1283 10.1016/j.bbalip.2017.06.016 [DOI] [PubMed] [Google Scholar]
- 36. Barneda D., and Christian M. (2017) Lipid droplet growth: regulation of a dynamic organelle. Curr. Opin. Cell Biol. 47, 9–15 10.1016/j.ceb.2017.02.002 [DOI] [PubMed] [Google Scholar]
- 37. Lykidis A., Jackson P. D., Rock C. O., and Jackowski S. (1997) The role of CDP-diacylglycerol synthetase and phosphatidylinositol synthase activity levels in the regulation of cellular phosphatidylinositol content. J. Biol. Chem. 272, 33402–33409 10.1074/jbc.272.52.33402 [DOI] [PubMed] [Google Scholar]
- 38. Yan R., Qian H., Lukmantara I., Gao M., Du X., Yan N., and Yang H. (2018) Human SEIPIN binds anionic phospholipids. Dev. Cell 47, 248–256.e4 10.1016/j.devcel.2018.09.010 [DOI] [PubMed] [Google Scholar]
- 39. Fei W., Li H., Shui G., Kapterian T. S., Bielby C., Du X., Brown A. J., Li P., Wenk M. R., Liu P., and Yang H. (2011) Molecular characterization of seipin and its mutants: implications for seipin in triacylglycerol synthesis. J. Lipid Res. 52, 2136–2147 10.1194/jlr.M017566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mok A. Y., McDougall G. E., and McMurray W. C. (1993) Comparative studies of CDP-diacylglycerol synthase in rat liver mitochondria and microsomes. Biochem. Cell Biol. 71, 183–189 10.1139/o93-029 [DOI] [PubMed] [Google Scholar]
- 41. Lai L., Wang M., Martin O. J., Leone T. C., Vega R. B., Han X., and Kelly D. P. (2014) A role for peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1) in the regulation of cardiac mitochondrial phospholipid biosynthesis. J. Biol. Chem. 289, 2250–2259 10.1074/jbc.M113.523654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu K., Zhou S., Kim J. Y., Tillison K., Majors D., Rearick D., Lee J. H., Fernandez-Boyanapalli R. F., Barricklow K., Houston M. S., and Smas C. M. (2009) Functional analysis of FSP27 protein regions for lipid droplet localization, caspase-dependent apoptosis, and dimerization with CIDEA. Am. J. Physiol. Endocrinol. Metab. 297, E1395–E1413 10.1152/ajpendo.00188.2009 [DOI] [PubMed] [Google Scholar]
- 43. Grahn T. H., Kaur R., Yin J., Schweiger M., Sharma V. M., Lee M. J., Ido Y., Smas C. M., Zechner R., Lass A., and Puri V. (2014) Fat-specific protein 27 (FSP27) interacts with adipose triglyceride lipase (ATGL) to regulate lipolysis and insulin sensitivity in human adipocytes. J. Biol. Chem. 289, 12029–12039 10.1074/jbc.M113.539890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Langhi C., and Baldán Á. (2015) CIDEC/FSP27 is regulated by peroxisome proliferator-activated receptor α and plays a critical role in fasting- and diet-induced hepatosteatosis. Hepatology 61, 1227–1238 10.1002/hep.27607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Puri V., Konda S., Ranjit S., Aouadi M., Chawla A., Chouinard M., Chakladar A., and Czech M. P. (2007) Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J. Biol. Chem. 282, 34213–34218 10.1074/jbc.M707404200 [DOI] [PubMed] [Google Scholar]
- 46. Keller P., Petrie J. T., De Rose P., Gerin I., Wright W. S., Chiang S. H., Nielsen A. R., Fischer C. P., Pedersen B. K., and MacDougald O. A. (2008) Fat-specific protein 27 regulates storage of triacylglycerol. J. Biol. Chem. 283, 14355–14365 10.1074/jbc.M708323200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nishino N., Tamori Y., Tateya S., Kawaguchi T., Shibakusa T., Mizunoya W., Inoue K., Kitazawa R., Kitazawa S., Matsuki Y., Hiramatsu R., Masubuchi S., Omachi A., Kimura K., Saito M., et al. (2008) FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets. J. Clin. Invest. 118, 2808–2821 10.1172/JCI34090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Toh S. Y., Gong J., Du G., Li J. Z., Yang S., Ye J., Yao H., Zhang Y., Xue B., Li Q., Yang H., Wen Z., and Li P. (2008) Up-regulation of mitochondrial activity and acquirement of brown adipose tissue-like property in the white adipose tissue of fsp27 deficient mice. PLoS One 3, e2890 10.1371/journal.pone.0002890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matsusue K., Kusakabe T., Noguchi T., Takiguchi S., Suzuki T., Yamano S., and Gonzalez F. J. (2008) Hepatic steatosis in leptin-deficient mice is promoted by the PPARγ target gene Fsp27. Cell Metab. 7, 302–311 10.1016/j.cmet.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen Z., Guo K., Toh S. Y., Zhou Z., and Li P. (2000) Mitochondria localization and dimerization are required for CIDE-B to induce apoptosis. J. Biol. Chem. 275, 22619–22622 10.1074/jbc.C000207200 [DOI] [PubMed] [Google Scholar]
- 51. Grippa A., Buxó L., Mora G., Funaya C., Idrissi F. Z., Mancuso F., Gomez R., Muntanyà J., Sabidó E., and Carvalho P. (2015) The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211, 829–844 10.1083/jcb.201502070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Craveiro Sarmento A. S., de Azevedo Medeiros L. B., Agnez-Lima L. F., Lima J. G., and de Melo Campos J. T. A. (2018) Exploring Seipin: from biochemistry to bioinformatics predictions. Int. J. Cell Biol. 2018, 5207608 10.1155/2018/5207608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Choi S. Y., Huang P., Jenkins G. M., Chan D. C., Schiller J., and Frohman M. A. (2006) A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 8, 1255–1262 10.1038/ncb1487 [DOI] [PubMed] [Google Scholar]
- 54. Ben M'barek K., Ajjaji D., Chorlay A., Vanni S., Forêt L., and Thiam A. R. (2017) ER membrane phospholipids and surface tension control cellular lipid droplet formation. Dev. Cell 41, 591–604.e7 10.1016/j.devcel.2017.05.012 [DOI] [PubMed] [Google Scholar]
- 55. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.-B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J., Joly J.-S., and Concordet J.-P. (2016) Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 10.1186/s13059-016-1012-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.