Abstract
Throughout history, natural products have significantly contributed to the discovery of novel chemistry, drug leads, and tool molecules to probe and address complex challenges in biology and medicine. Recent microbial genome sequencing efforts have uncovered many microbial biosynthetic gene clusters without an associated natural product. This means that the natural products isolated to date do not fully reflect the biosynthetic potential of microbial strains. This observation has rejuvenated the natural product community and inspired a return to microbial strain collections. Mining large microbial strain collections with the most current technologies in genome sequencing, bioinformatics, and high-throughput screening techniques presents new opportunities in natural product discovery. In this review, we report on the newly expanded microbial strain collection at The Scripps Research Institute, which represents one of the largest and most diverse strain collections in the world. Two complementary approaches, i.e. structure-centric and function-centric, are presented here to showcase how to leverage a large microbial strain collection for natural product discovery and to address challenges and harness opportunities for future efforts. Highlighted examples include the discovery of alternative producers of known natural products with superior growth characteristics and high titers, novel analogs of privileged scaffolds, novel natural products, and new activities of known and new natural products. We anticipate that this large microbial strain collection will facilitate the discovery of new natural products for many applications.
Keywords: biosynthesis, natural product, high-throughput screening (HTS), bioinformatics, drug screening, function-centric approaches, genome mining, microbial strain collection, strain prioritization, structure-centric
Introduction
Natural products have been exquisitely tailored via evolution to elicit potent and unique biological activities, rendering them unrivaled in structural complexity and diversity. Accordingly, natural products have served as drug leads for the pharmaceutical industry, affording antibiotics and anticancer compounds, greatly improving the quality of life for humanity. Of the FDA2-approved small molecule therapeutics, 67% of anti-infective and 83% of anti-cancer drugs are natural products, natural product derivatives, or inspired by natural products (1, 2).
Bacteria and fungi are prolific resources of natural products. Among the half-million of natural products known to date, ∼70,000 of them are of bacterial and fungal origin, of which approximately half (∼33,500) have had their bioactivities identified (3, 4). Of the 70,000 total, ∼30,000 are from fungi and ∼40,000 have been isolated from bacteria, of which about half (∼20,000) are from actinobacteria (4).
Over the last century, many microbial strains have been collected around the world for natural product discovery. The largest commercial strain collections include the American Type Culture Collection (ATCC) and the DSMZ-German collection. ATCC houses 18,000 bacterial strains from over 750 genera and over 110,000 fungal strains from over 1,500 genera (https://www.atcc.org),3 whereas the DSM houses over 24,000 bacterial and 2,571 fungal strains (https://www.dsmz.de).3
Strategies for prioritization have become increasingly important due to the large number of microbial strains available. Genome sequence data are now generally regarded as a very useful starting point for prioritization. Over the last 2 decades, high-throughput capabilities and lower costs of DNA sequencing have enabled genome sequencing of many microbial strains. There are currently over 1,000 fungal and 20,000 actinobacterial whole-genome sequences publicly available from the Joint Genome Institute (JGI, https://genome.jgi.doe.gov/portal) and the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov).
One of the most surprising findings from microbial genome sequencing is that only a very small fraction of the biosynthetic potential of Nature has been realized. On average, a bacterial genome harbors ∼30 biosynthetic gene clusters (BGCs), potentially encoding the production of 30 natural products (5). These numbers are even higher for fungal strains. According to the Atlas of Biosynthetic Gene Cluster within the Integrated Microbial Genome system (IMG-ABC) database (as of August 15, 2019), there are a total of 1,178,352 BGCs, of which 809,297 BGCs are from bacteria; however, only 2,487 BGCs have been associated with characterized natural products (6). Although the annotated BGCs certainly contain redundancies, the number of natural products that remain undiscovered is certainly immense.
Microbial strain collection at The Scripps Research Institute
During the 1990s, many pharmaceutical companies eliminated their natural product discovery programs in pursuit of high-throughput screening (HTS) and combinatorial libraries. As a result, microbial natural product discovery shifted to small companies and academic institutes. In 2011, the Natural Product Library Initiative (NPLI) at The Scripps Research Institute (TSRI) was launched; the goal was to build a microbial strain collection and to develop technologies to construct a natural product library (NPL) consisting of crude extracts, partially purified fractions, and pure natural products, thus complementing the small molecule library at TSRI for HTS. Central to the NPLI is the microbial strain collection, including both our in-house collection and the historic pharma collection that Pfizer contributed to TSRI in 2018. More information on the TSRI collection can be found at https://www.scripps.edu/shen/NPLI/npliattsri.html.3 This large microbial strain collection has enabled natural product discovery efforts that led to the scaffold- and function-directed discovery of alternative producers of known natural products with superior growth characteristics and high titers, novel analogs of privileged scaffolds, novel natural products, and new activities of known and new natural products.
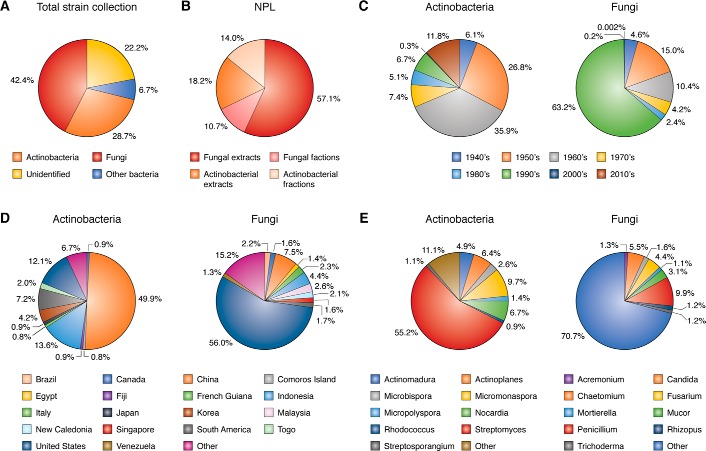
The newly-expanded NPL builds off previous successes and will undoubtedly accelerate natural product research conducted at TSRI. The microbial strain collection at TSRI currently contains a total of 217,352 bacterial and fungal strains, of which 62,328 are actinobacteria, 14,465 other bacteria, 92,225 fungi, and 48,334 unidentified (bacteria or fungi) (Fig. 1A). These strains were isolated over the last eight decades with the majority acquired between 1940 and 2010 (Fig. 1C). The wide time range of collection allows for capture of chemical diversity based on evolution and environmental cues, which change over time and are impossible to reproduce in laboratory settings today. Geographically, these strains were isolated from 109 different countries (Fig. 1D) with factors such as climate and ecology that further increase the chemical and biological diversity. Although these statistics, as well as taxonomic data, are incomplete, the fraction of the strains whose information is available provides a glimpse into the vast spatial, temporal, and taxonomic diversity of the entire collection (Fig. 1).
Figure 1.
A, total number of strains in the current microbial strain collection at TSRI is as follows: actinobacteria (62,328, 28.7%); fungi (92,225, 42.4%); other bacteria (14,465, 6.7%); and unidentified bacteria or fungi (48,334, 22.2%). B, total number of extracts and fractions in the current NPL at TSRI is as follows: fungal extracts (136,123 made from 12,060 strains, 57.1%); fungal fractions (24,980 made from 2,498 strains, 10.7%); actinobacterial extracts (42,612 made from 8,020 strains, 18.2%); and actinobacterial fractions (32,832 made from 3,025 strains, 14.0%). C, collection dates of strains by decade: 1940s (A, 6.1%; F, 4.6%); 1950s (A, 26.8%; F, 15.0%); 1960s (A, 35.9%; F, 10.4%); 1970s (A, 7.4%; F, 4.2%); 1980s (A, 5.1%; F, 2.4%); 1990s (A, 6.7%; F, 63.2%); 2000s (A, 0.3%; F, 0.2%); and 2010s (A, 11.8%; F, 0.002%), where A indicates actinobacteria; F indicates fungi. These statistics are based on 77% of the actinobacteria, 46% of fungi, 80% of other bacteria, and 12% of unidentified bacteria or fungi from the total collection, for which collection dates could be traced. D, location of collection of strains is as follows: Brazil (F, 2.2%); Canada (A, 0.9%; F, 1.6%); China (A, 49.9%; F, 7.5%); Comoros Island (A, 0.8%); Egypt (F, 1.4%); Fiji (A, 0.9%); French Guiana (F, 2.3%); Indonesia (A, 13.6%; F, 4.4%); Italy (A, 0.8%); Japan (A, 0.9%); Korea (A, 4.2%); Malaysia (F, 2.6%); New Caledonia (F, 2.1%); Singapore (F, 1.6%); South America (A, 7.2%; F, 1.7%); Togo (A, 2.0%); United States (A, 12.2%; F, 56.0%); Venezuela (F, 1.3%); and other countries (A from 58 countries, 6.7%; F from 89 countries, 15.2%). These statistics are based on 20% of the actinobacteria and 41% of fungi from the total collection, for which collection locations could be traced. E, taxonomic information for actinobacteria is as follows: Actinomadura, 4.9%; Actinoplanes, 6.4%; Microbispora, 2.6%; Micromonaspora, 9.7%; Micropolyspora, 1.4%; Nocardia, 6.7%; Rhodococcus, 0.9%; Streptomyces, 55.2%; Streptosporangium, 1.1%; and the 79 other genera, 11.1%. Taxonomic information for fungi is as follows: Acremonium, 1.3%; Candida, 5.5%; Chaetomium, 1.6%; Fusarium, 4.4%; Mortierella, 1.1%; Mucor, 3.1%; Penicillium, 9.9%; Rhisopus, 1.2%; Trichoderma, 1.2%; and the 2,665 other genera; 70.7%. These statistics are based on 20% of the actinobacteria and 66% of fungi from the total collection for which some forms of the taxonomic data could be traced.
The actinobacterial strains are very diverse in terms of taxonomy, geographical origin, and collection time. The 62,328 actinobacterial strains span 88 different genera. The most represented are Streptomyces (6,650), Micromonospora (1,170), Nocardia (806), Actinoplanes (766), Actinomadura (592), Microbispora (311), Micropolyspora (172), Streptosporangium (137), and Rhodococcus (111) (Fig. 1E). These strains were isolated from 69 different countries with the majority coming from China (6,399), Indonesia (1,744), United States (1,547), South America (922), Korea (535), Togo (261), Fiji (118), Canada (115), and Japan (111) (Fig. 1D).
The fungal strains offer a great deal of diversity, similarly to the actinobacteria, in terms of taxonomy, geographical origin, and collection time. The 92,225 fungal strains span 2,614 different genera. The most represented are Penicillium (4,198), Candida (2,337), Fusarium (1,876), Chaetomium (669), Acremonium (558), Trichoderma (519), Rhizopus (512), Mortierella (475), and Mucor (475) (Fig. 1E). These strains were isolated from 101 different countries with the majority coming from the United States (21,304), China (2,868), Indonesia (1,683), Malaysia (993), French Guiana (890), Brazil (835), and New Caledonia (793) (Fig. 1D).
The potential number of natural products awaiting discovery from the microbial strain collection at TSRI is immense. Estimated on the assumption of ∼30 BGCs per strain, the strains in the collection could encode more than 6 million BGCs, i.e. the potential of producing more than 6 million natural products. In reference to the ∼70,000 microbial natural products known to date, the current number of known natural products is only ∼1% of this value, leaving millions of compounds to be discovered. Of course, there will be redundancies with many strains producing the same natural products or structurally similar congeners. However, these redundancies are unlikely to fundamentally reduce the total number of natural products waiting for discovery. In fact, redundancies in the forms of alternative producers or producers of congeners could allow for systematic study of gene regulation of natural product biosynthesis and production, on both the pathway-specific and host-genome wide levels. Furthermore, these redundancies can be used to exploit the evolution of natural product biosynthetic pathways in Nature for combinatorial biosynthesis.
A fraction of the strain collection has also been cultured in multiple media, under varying fermentation conditions, to capture the natural product diversity as a part of the NPL for HTS. The current NPL contains 42,612 crude extracts from 8,020 actinobacteria and 136,123 crude extracts from 12,060 fungi. Of these strains cultured, 3,025 actinobacteria and 2,498 fungi have been selected for large-scale fermentation, the crude extracts of which have been subjected to C-18 chromatography, affording 32,832 and 24,980 partially-purified fractions, respectively (Fig. 1B). Again, the cultured ∼20,000 strains could produce up to ∼600,000 natural products (based on ∼30 BGCs per strain estimation). Even if only 10% of the natural product diversity is captured within the crude extracts and partially-purified fractions, there could be ∼60,000 natural products waiting to be discovered by HTS against emerging biology.
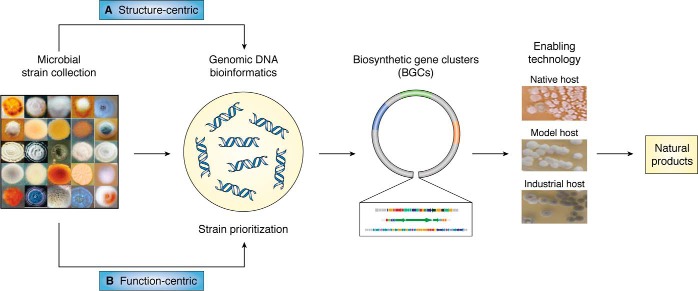
Detailed herein are examples from our current efforts showcasing two complementary strategies to leverage the microbial strain collection at TSRI for natural product discovery. Structure-centric approaches rely on genomics and bioinformatics, whereas function-centric approaches rely on the innate biological activity of the natural products.
Strategies to leverage strain collections
Structure-centric approaches
Advances in DNA sequencing and bioinformatics, along with an exponential increase in available microbial genome sequence data, have highlighted the biosynthetic potential of Nature and allow for targeted natural product discovery via genome mining. Correlation of a natural product structure to its BGC has enabled the identification of alternative producing strains for the natural product of interest, as well as the targeted discovery of natural products based on their structural features, such as scaffold or key functional groups.
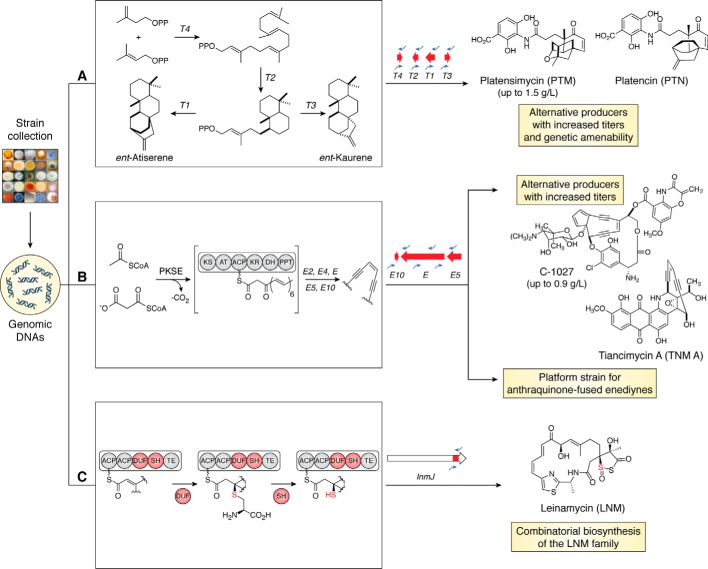
Because the strain collection at TSRI has not been sequenced, we developed a real-time PCR (RT-PCR) high-throughput screening method to identify strains harboring BGCs encoding the biosynthesis of a given class of natural products (7). Representative hits are then subjected to whole-genome sequencing. This method has been successfully utilized to screen the strain collection for three families of natural products, platensimycin/platencin (PTM/PTN), enediynes, and leinamycin (LNM) (Figs. 2 and 3).
Figure 2.
Schematic representation of how to leverage a microbial strain collection for natural product discovery by two complementary approaches. A, structure-centric approach utilizes genomic information from the strain collection, along with bioinformatics, to prioritize privileged strains based on a unique pharmacophore or scaffold of the target natural products. B, function-centric approach utilizes biological activity, via HTS against targeted biology, to prioritize privileged strains based on unique targets or mechanism of action. Upon identification of these privileged strains, correlation of the targeted natural products or biology to specific BGCs and exploitation of these BGCs via enabling technology, such as cluster activation or heterologous expression, allow for the characterization of novel natural products, alternative producers of known natural products, and novel enzymes for combinatorial biosynthesis and biocatalysis.
Figure 3.
Examples of structure-centric approaches to leverage the microbial strain collection at TSRI for natural product discovery. A, PTM and PTN contain unique diterpenoid scaffolds, the biosynthesis of which is encoded by terpene cyclases, T1, T2, T3, and T4. An RT-PCR screen of a fraction of the actinobacteria strains in the collection prioritized strains containing all four cyclase genes, resulting in the identification of alternative PTM and PTN producers with high titers and superior growth characteristics. B, enediyne natural products contain a unique pharmacophore, consisting of two triple bonds in conjugation with a double bond or incipient double bond, the biosynthesis of which is encoded by the enediyne PKS gene cassette, E, E10, E3, E4, and E5. An RT-PCR screen of a fraction of the actinobacterial strains in the collection prioritized strains containing the enediyne PKS gene cassette, resulting in the identification of alternative C-1027 producers with high titers and new anthraquinone-fused enediynes TNMs. The TNM producer, with its high TNM titers and superior growth characteristics, has enabled the development of a platform strain for engineered biosynthesis and production of the anthraquinone-fused family of enediyne natural products. C, LNM contains a unique sulfur-containing heterocycle, where the sulfur incorporation chemistry is encoded by a DUF-SH didomain. An RT-PCR screen of a fraction of the actinobacterial strains in the collection prioritized strains containing the DUF-SH didomain, resulting in the identification of a family of modular biosynthetic pathways that exemplify how Nature does combinatorial biosynthesis for the LNM family of natural products.
PTM and PTN are highly sought-after natural products with unique, but related, structures, as well as unprecedented activity inhibiting bacterial fatty acid biosynthesis (12, 13). PTM and PTN both contain the same 3-amino-4-hydroxybenzoic acid moiety connected via an amide bond to distinct diterpenoid-derived ketolides. PTM was discovered from Streptomyces platensis MA7327, originally isolated from South Africa (8), whereas PTN was discovered from S. platensis MA7339, which was isolated from Spain (9). We subsequently identified both the PTM and PTN BGCs from the S. platensis MA7327 and MA7339, respectively, and discovered that the BGC encoding PTM biosynthesis in the S. platensis MA7327 strain included all necessary genes for PTN production and an additional five genes encoding a “PTM” cassette (14). The S. platensis MA7327 genome therefore contains a PTM–PTN dual BGC, encoding the proteins necessary for the biosynthesis of both PTM and PTN (10). The wild type S. platensis MA7327 and MA7339 strains produced PTM and PTN in trace quantities (∼1–4 mg/liter). Although we were successful in generating the first generation of PTM and PTN overproducers by inactivating the pathway-specific negative regulator PtmR1 or PtnR1 (11, 12), the resultant mutant strains failed to sporulate and suffered from inferior growth characteristics. This resulted in our inability to further manipulate the PTM and PTN biosynthetic machinery for titer improvement and structural diversity.
We next turned to the strain collection at TSRI and applied our RT-PCR strain prioritization strategy to search for alternative producers (10). Using primers specific for the terpene cyclase genes in PTM and PTN BGCs (i.e. ptmT4, ptmT2, ptmT1, and ptmT3), 1,911 actinobacteria were screened, affording six strains as potential PTM and PTN producers, all differing in the geographical distribution, morphology, and growth characteristics (Fig. 3A) (8). Representative hits were subjected to whole-genome sequencing, which confirmed that they were all PTM–PTN dual producers. The genetic amenability was confirmed by inactivation of the PtmR1 negative regulator to afford mutant strains that overproduced PTM and PTN. Although the PTM–PTN dual BGCs are highly homologous, with DNA sequence identity varying between 90 and 99%, these strains produce PTM with varying titers, with the highest titer reaching 1.6 g/liter upon fermentation optimization, which is more than a 1,000-fold improvement from the original MA7327 and MA7339 strains (11–14). Identification of alternative producers with high titers and genetic amenability allowed for detailed biosynthetic studies, as well as isolation of sufficient quantities of PTM to enable medicinal chemistry for structural diversity and further biological evaluation. This example highlights how a single genotype, i.e. a BGC, can exhibit different phenotypes depending on the genomic context.
The enediyne class of natural products are known for their unique pharmacophore consisting of two alkynes in conjugation with a double bond or incipient double bond within either a 9- or 10-membered macrocycle (15, 16). The 9-membered enediynes are found as a chromoprotein complex, whereas the 10-membered enediynes are discrete natural products. This privileged scaffold gives rise to the extremely potent antitumor activity, via DNA damage (Fig. 3B). Currently there are 13 members of the enediyne family of natural products. Comparative characterization of the known 9- and 10-membered enediyne BGCs led to the identification of a conserved set of genes, implicated in the biosynthesis of the enediyne core. This set of genes, known as the enediyne polyketide synthase (PKS) gene cassette, consists of E3, E4, E5, E, and E10.
C-1027, first isolated from Streptomyces globisporus in 1993 (17), has been studied as a model for 9-membered enediyne biosynthesis and mechanism of action (18). Additionally, C-1027 is undergoing phase II clinical trials, requiring a reliable and sufficient supply of the material to support further development. Detailed study into the regulation of the C-1027 BGC in S. globisporus has enabled the construction of C-1027 overproducing recombinant strains by the combination of inactivating the negative regulator SgcR and overproducing the positive regulator SgcR1. The resultant recombinant strains produced C-1027 with a titer of 466 mg/liter, representing a 6-fold increase over the WT (74 mg/liter) (19, 20).
Adopting the same RT-PCR strain prioritization strategy, we screened 3,400 actinobacteria strains from the TSRI strain collection, using degenerate primers designed to target the enediyne PKS cassette. This effort led to the identification of 81 strains harboring the enediyne PKS cassettes. Further bioinformatic analysis grouped these strains into 28 distinct clades. From these, 31 representatives were subjected to whole-genome sequencing, confirming that each of them contained a distinct enediyne BGC (21, 22).
One of the 28 clades contained four strains, Streptomyces sp. CB02366, CB00657, CB02329, and CB03608, together with the original C-1027 producer S. globisporus, indicating that they were alternative C-1027 producers (21, 22). Close examination of the five strains within this clade showed that they were from distinct geographic locations, were morphologically different, and harbored the C-1027 BGC with high DNA sequence identity (83–99%) (23). Remarkably, under the optimized conditions for C-1027 production, developed for the engineered C-1027 overproducer from the WT S. globisporus, the four alternative producers produced C-1027 with titers up to 900 mg/liter, which is ∼12-fold higher than the original S. globisporus WT (19, 20). This example exemplifies once again that identical or similar BGCs can be found from distinct geographical locations. Within the varying genomic context, strains harboring similar BGCs can exhibit different phenotypes and produce the desired natural products in evolutionary-optimized high titers.
In addition to alternative producers of C-1027, this study also enabled the engineered production of the anthraquinone-fused enediyne family of natural products. As of 2015, there were two known anthraquinone-fused enediyne family members, dynemicin (DYN) from Micromonospora chersina (24) and uncialamycin (UCM) from Streptomyces uncialis (25). Studies of this class of enediyne natural products were hampered by the low titers from both native strains, along with genetic intractability for DYN in M. chersina and the inability to produce UCM in submerged fermentation of S. uncialis. To overcome these challenges, we set out to search for strains encoding BGCs of novel anthraquinone-fused enediynes, with the ultimate goal of developing them into platform strains. This platform can be utilized for the engineered production of all anthraquinone-fused enediyne family of natural products.
Genome mining of the sequenced 31 representative strains identified Streptomyces sp. CB03234 as a potential anthraquinone-fused enediyne producer (21). We also mined all of the sequenced bacterial genomes publicly available at JGI and NCBI to complement our own strain collection, identifying Micromonospora yangpuensis as another potential anthraquinone-fused enediyne producer (26). Fermentation of Streptomyces sp. CB03234 afforded tiancimycin A (TNM), whereas M. yangpuensis afforded yangpumicin A (YPM) (26), two new anthraquinone-fused enediynes. Although YPM was from genetically intractable Micromonaspora, similar to DYN, the TNM-producing CB03234 strain proved to be an ideal platform strain with titers 10–300-fold higher than DYN, UCM, and YPM (27). Moreover, Streptomyces sp. CB03234 was shown to be genetically amenable. Extensive manipulation of the TNM biosynthetic machinery in Streptomyces sp. CB03234 resulted in characterization of six additional TNM family members, thus allowing for the proposal of a unified anthraquinone-fused biosynthetic pathway common to DYN, UCM, YPM, and TNM (28). Identification of Streptomyces sp. CB03234, as a novel anthraquinone-fused enediyne producer, has overcome the challenges associated with other strains, such as the low titer and genetic amenability, thus providing a platform strain enabling further biosynthetic and biological investigations and engineered biosynthesis.
LNM was originally isolated from Streptomyces atroolivaceus S-140 in 1989 (29, 30) and is a hybrid peptide–polyketide natural product (31). LNM contains a 1,3-dioxo-1,2-dithiolane heterocycle that causes the molecule to react with DNA in a reductive environment containing thiols (32). Since its debut ∼30 years ago, no LNM congeners have been discovered. We have also not been very successful in generating LNM analogs by engineering the LNM biosynthetic machinery, despite exhaustive efforts. The biosynthetic origin of the sulfur atom at C-3 within the heterocycle has been linked to a unique PKS didomain (domain of unknown function-sulfhydralase, DUF-SH) (33, 34) within the LNM BGC (35). Again, adopting the same RT-PCR strain prioritization strategy, we designed PCR primers according to the DUF-SH sequence and screened ∼5,000 actinobacteria from the TSRI strain collection, as well as virtually screened the ∼48,780 bacterial genomes publicly available at JGI and NCBI (as of March, 2017) (36). This screen led to the discovery of 49 new LNM-type BGCs, which fell into 17 distinct clades upon phylogenetic analysis, and representatives from each clade were subjected to whole-genome sequencing. Importantly, this allowed access to an alternative producer of LNM, as well as new scaffolds within the LNM family of natural products for the first time since LNM's discovery three decades ago. Upon fermentation optimization of the prioritized strains, two new members of the LNM family of natural products were isolated, guangnanmycin and weishanamycin.
Strikingly, upon close examination of the hybrid NRPS-PKS and PKS genes in each of the 17 LNM-like BGC clades, it was apparent that Nature is the ultimate combinatorial biosynthetic chemist in maximizing natural product structural diversity (36). The predicted scaffolds of the LNM family of natural products are similar but differ in structural features, such as starter and extender unit selection by the hybrid NPRS-PKS and PKS, and β-alkylation and α/β-modifications by the rest of the biosynthetic machinery. The fact that LNM-type biosynthetic machinery divergently evolved to yield distinct yet structurally-related hybrid peptide–polyketides is a great inspiration to the field of combinatorial biosynthesis and synthetic biology. As more natural product scaffolds are studied within the context of large microbial strain collections, more combinatorial biosynthesis examples are likely to be discovered.
Function-centric approaches
Function-centric approaches, unlike structure-centric, rely on the biological function of a compound in a structure-agnostic manner. This allows for the discovery of completely novel scaffolds, whose hallmark features for their BGCs have yet to be defined, or natural products, whose bioactivity has not been linked to a single pharmacophore and/or scaffold. Enabling technologies in assay development, such as multiplexing and HTS, have allowed for the rapid advances in these approaches.
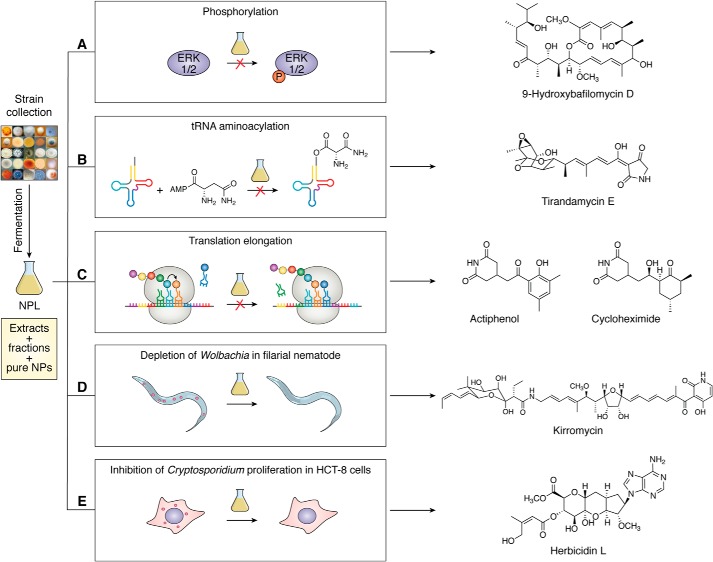
HTS platforms that screen for cytotoxicity have been highly successful throughout history, and these types of approaches still hold value for large microbial strain collections, such as the NPL at TSRI. Screening the NPL against cancer cell lines, an extract from an endophytic Streptomyces sp. YIM56209 exhibited considerable cytotoxicity (37). Bioassay-guided dereplication led to the discovery of two novel bafilomycins (Fig. 4A) and nine known congeners of this natural product family. Bafilomycins exhibit a wide range of bioactivities. They hold promise as antitumor agents (38) and are important chemical probes to study V-ATPase enzymes (39).
Figure 4.
Examples of function-centric approaches to leverage the microbial strain collection at TSRI for natural product discovery. Selected strains from the collection are fermented in diverse media to make the crude extracts, and upon HPLC analysis, the most chemically diverse conditions are selected, scaled up, and subjected to chromatography to afford the partially purified fractions. The current NPL consists of crude extracts, partially purified fractions, and pure natural products (also see Fig. 1B) and has been subjected to HTS, against emerging biology, for natural product discovery. A, NPL was screened for cytotoxicity and prolactin-initiated phosphorylation of ERK1/2. Bioassay-guided dereplication of the active hits led to the isolation of two new bafilomycin congeners, along with nine known ones. B, NPL was screened for parasitic AsnRS inhibition. Bioassay-guided dereplication of the hits identified TAMs as potent and selective AsnRS inhibitors, three of which were new congeners, and two were previously characterized. C, NPL was screened for inhibition of protein translation initiation via a high-throughput bicistronic mRNA translation assay. Bioassay-guided dereplication of the hits led to the identification of actiphenol and cycloheximide as potent inhibitors, importantly, from the same Streptomyces strain, shedding new insights into their biosynthesis. D, pure natural product collection of the NPL was subjected to a high-content image-based screening to discover inhibitors of Wolbachia via a Drosophila infection model, resulting in the identification of kirromycin as a potent and specific inhibitor of Wolbachia. E, partially-purified fraction collection of the NPL was subjected to a high-content image-based screen to search for inhibitors of Cryptosporidium via a JW18 infection model, leading to the discovery of the herbicidins as a promising scaffold for anti-Cryptosporidium drug development.
Lymphatic filariasis, caused by the parasitic nematodes Brugia malayi and Wuchereria bancrofti, represents a worldwide health crisis. Discovery of effective macrofilaricides, drugs that can kill adult female worms, is a top priority (40). This is especially relevant due to the realization that the parasites have started to become resistant to the well-distributed standard-of-care medications ivermectin and albendazole. Structural characterization of the first parasitic asparaginyl-tRNA synthetase (AsnRS), compared with the well-characterized human AsnRS, revealed new opportunities to target parasitic AsnRS, offering an orthogonal mechanism of action to ivermectin and albendazole (41). Accordingly, extracts from the NPL were screened for AsnRS-inhibitory activity, and activity-guided dereplication led to the eventual discovery of several natural product classes that showed promising in vitro activities (42, 43), including the tirandamycins (TAMs) (44). Two TAMs identified in this study were previously known (TAMs A and B), and three were new (TAMs E, F, and G, see Fig. 3B). Despite the highly-similar structures of the TAMs, only TAM B exhibited potent inhibitory activity against B. malayi AsnRS. In addition, TAM B was shown to have 10-fold selectivity for B. malayi AsnRS over human AsnRS and killed the adult B. malayi parasite very efficiently. Therefore, TAM B represents a new lead scaffold to discover and develop antifilarial drugs (44).
Breakthroughs in assay development have enabled efforts for our ability to screen for many desirable mechanisms of action. Eukaryotic protein translation has emerged as a promising target for cancer chemotherapy. Development of HTS for this phenotype (45, 46) enabled the screening of the NPL leading to the discovery of Streptomyces spp. YIM56132 and YIM56141, alternative producers of actiphenol and cycloheximide (Fig. 4B) (47). Follow-up studies in the newly-discovered producing strains allowed biosynthetic connections between these important molecules to be made, specifically the realization that actiphenol is a biosynthetic intermediate en route to cycloheximide (48).
High-content image-based HTS has recently become a valuable method for phenotypic screening of small molecule libraries. This type of HTS involves a high-content image readout to determine the biological activity of a compound allowing for complex phenotypes to be rapidly screened. We first subjected the pure natural product collection in our NPL to a high-content image-based screen for anti-Wolbachia activity (49, 50), leading to the identification of the kirromycins as a lead scaffold (Fig. 4C) (51). Impressively, the three kirromycins identified all depleted Wolbachia in Drosophila cells in vitro with IC50 in the nanomolar range, whereas doxycycline, a registered drug with anti-Wolbachia activity, showed lower activity with an IC50 of ∼150 nm. Furthermore, the kirromycins eliminated the Wolbachia endosymbiont in Brugia pahangi ovaries ex vivo with higher efficiency (65–90%) at 1 μm than that of doxycycline (50%) (51). This suggests that kirromycin is an effective lead scaffold, further exploration of which could potentially lead to the development of novel antifilarial drugs.
The herbicidins were discovered to have selective anti-Cryptosporidium parvum activity using a similar image-based HTS approach (52). Discovery of anti-Cryptosporidium compounds has been greatly hampered due to difficulties in working with this intractable parasite. Upon subjecting the partially purified fraction collection of the NPL to our newly-developed image-based assay, six herbicidins were discovered, one of which was new (Fig. 4D) (53). Five of the six herbicidins showed moderate anti-Cryptosporidium activity, with herbicidin K exhibiting comparable activity to the FDA-approved drug nitazoxanide. In addition, herbicidin K showed no toxicity to human cells, highlighting the promise of the herbicidin scaffold for anti-Cryptosporidium drug development (53). This final set of examples demonstrates the value of function-centric approaches, whereby previously characterized natural products can be connected with novel bioactivity.
Conclusions and future perspectives
Natural products have been at the forefront of drug discovery, with about one-third of all FDA-approved drugs being natural products and/or derivatives thereof (2). Advances in microbial genomics and bioinformatics have shed new light on the great biosynthetic potential of Nature, rejuvenating the natural product community and paving the way for a new Golden Age of natural product discovery (54).
Currently, there are numerous challenges for natural product discovery on this path to the new Golden Age. First, consolidation of large microbial strain collections and the construction of a centralized natural product BGC data bank for the community to utilize efficiently needs to be realized. Current data banks such as anti-SMASH and MIBiG have already revolutionized the field; therefore, efforts to expand these by orders of magnitude will be even more revolutionary. Subsequent prioritization of the most promising strains for targeted natural product discovery, based on new enabling technology capable of linking genes to structures to functions, is necessary. Finally, activation of “silent” BGCs in scale and speed, whether in a native or heterologous model host, and bottlenecks in natural isolation, purification, and structural elucidation must be addressed.
These challenges can be overcome by increasing the amount of genetic information available by sequencing large strain collections, such as the microbial strain collection at TSRI. The vast amount of data acquired, ∼6 million BGCs, will allow the establishment of a natural product BGC data bank with >210,000 genomes compared with the ∼20,000 actinobacterial and fungal genomes currently available. This would not only enable the correlation of genes to structures to functions but would allow for systematic study of BGCs and how they interact with their host.
Moreover, the NPL consists of extracts and fractions made from more than 20,000 actinobacterial and fungal strains, harboring great biodiversity (up to 600,000 natural products based on the 30 BGCs per strain estimation). Combining genome mining, prioritization of these extracts based on predicted natural product structures will greatly facilitate efficiency of HTS and increase hit rates, as well as streamline downstream dereplication of the active natural products. The vast potential that a large microbial strain collection may offer can be fully realized for the natural product community by increasing our knowledge of the biosynthetic potential, via sequencing and bioinformatics, and correlation of these structures to their biological functions.
Acknowledgments
Research on natural product discovery, biosynthesis, engineering, and drug discovery in the Shen laboratory is currently supported by National Institutes of Health Grants CA106150, GM114353, and GM115575 and by the Natural Products Library Initiative at The Scripps Research Institute.
This work was supported by National Institutes of Health Postdoctoral Fellowships GM133114 (to A. D. S.) and GM128345 (to C. N. T.). This is manuscript no. 29884 from The Scripps Research Institute. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- FDA
- Food and Drug Administration
- ATCC
- American Type Culture Collection
- BGC
- biosynthetic gene cluster
- DUF-SH
- domain of unknown function-sulfhydralase
- HTS
- high-throughput screening
- JGI
- Joint Genome Institute
- LNM
- leinamycin
- NCBI
- National Center for Biotechnology Information
- NPL
- Natural Product Library
- PTM
- platensimycin
- PTN
- platencin
- RT-PCR
- real-time polymerase chain reaction
- TAM
- tirandamycin
- TNM
- tiancimycin A
- TSRI
- The Scripps Research Institute
- UCM
- uncialamycin
- YPM
- yangpumicin A
- AsnRS
- aparaginyl-tRNA synthetase
- DYN
- dynemicin.
References
- 1. Newman D. J., and Cragg G. M. (2016) Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661 10.1021/acs.jnatprod.5b01055 [DOI] [PubMed] [Google Scholar]
- 2. Newman D. J., and Cragg G. M. (2012) Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335 10.1021/np200906s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bills G. F., and Gloer J. B. (2016) Biologically active secondary metabolites from the fungi. Microbiol. Spectr. 4 10.1128/microbiolspec.FUNK-0009-2016 [DOI] [PubMed] [Google Scholar]
- 4. Bérdy J. (2012) Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiot. 65, 385–395 10.1038/ja.2012.27 [DOI] [PubMed] [Google Scholar]
- 5. Cimermancic P., Medema M. H., Claesen J., Kurita K., and Wieland Brown L. C., Mavrommatis K., Pati A., Godfrey P. A., Koehrsen M., Clardy J., Birren B. W., Takano E., Sali A., Linington R. G., and Fischbach M. A. (2014) Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421 10.1016/j.cell.2014.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hadjithomas M., Chen I. A., Chu K., Huang J., Ratner A., Palaniappan K., Andersen E., Markowitz V., Kyrpides N. C., and Ivanova N. N. (2017) IMG-ABC: new features for bacterial secondary metabolism analysis and targeted biosynthetic gene cluster discovery in thousands of microbial genomes. Nucleic Acids Res. 45, D560–D565 10.1093/nar/gkw1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hindra, Huang T., Yang D., Rudolf J. D., Xie P., Xie G., Teng Q., Lohman J. R., Zhu X., Huang Y., Zhao L. X., Jiang Y., Duan Y., and Shen B. (2014) Strain prioritization for natural product discovery by a high-throughput real-time PCR method. J. Nat. Prod. 77, 2296–2303 10.1021/np5006168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J., Soisson S. M., Young K., Shoop W., Kodali S., Galgoci A., Painter R., Parthasarathy G., Tang Y. S., Cummings R., Ha S., Dorso K., Motyl M., Jayasuriya H., Ondeyka J., et al. (2006) Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441, 358–361 10.1038/nature04784 [DOI] [PubMed] [Google Scholar]
- 9. Wang J., Kodali S., Lee S. H., Galgoci A., Painter R., Dorso K., Racine F., Motyl M., Hernandez L., Tinney E., Colletti S. L., Herath K., Cummings R., Salazar O., González I., et al. (2007) Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc. Natl. Acad. Sci. U.S.A. 104, 7612–7616 10.1073/pnas.0700746104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smanski M. J., Yu Z., Casper J., Lin S., Peterson R. M., Chen Y., Wendt-Pienkowski E., Rajski S. R., and Shen B. (2011) Dedicated ent-kaurene and ent-atiserene synthases for platensimycin and platencin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 108, 13498–13503 10.1073/pnas.1106919108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smanski M. J., Peterson R. M., Rajski S. R., and Shen B. (2009) Engineered Streptomyces platensis strains that overproduce antibiotics platensimycin and platencin. Antimicrob. Agents Chemother. 53, 1299–1304 10.1128/AAC.01358-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yu Z., Smanski M. J., Peterson R. M., Marchillo K., Andes D., Rajski S. R., and Shen B. (2010) Engineering of Streptomyces platensis MA7339 for overproduction of platencin and congeners. Org. Lett. 12, 1744–1747 10.1021/ol100342m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi J., Pan J., Liu L., Yang D., Lu S., Zhu X., Shen B., Duan Y., and Huang Y. (2016) Titer improvement and pilot-scale production of platensimycin from Streptomyces platensis SB12026. J. Ind. Microbiol. Biotechnol. 43, 1027–1035 10.1007/s10295-016-1769-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rudolf J. D., Dong L. B., Huang T., and Shen B. (2015) A genetically amenable platensimycin- and platencin-overproducer as a platform for biosynthetic explorations: a showcase of PtmO4, a long-chain acyl-CoA dehydrogenase. Mol. Biosyst. 11, 2717–2726 10.1039/C5MB00303B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van Lanen S. G., and Shen B. (2008) Biosynthesis of enediyne antitumor antibiotics. Curr. Top. Med. Chem. 8, 448–459 10.2174/156802608783955656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adhikari A., Teijaro C. N., Townsend C. A., Shen B. (2019) Biosynthesis of enediyne natural products in Comprehensive Natural Products: Chemistry and Biology, Vol I, Elsevier, in press [Google Scholar]
- 17. Yan X., Hindra, Ge H., Yang D., Huang T., Crnovcic I., Chang C. Y., Fang S. M., Annaval T., Zhu X., Huang Y., Zhao L. X., Jiang Y., Duan Y., and Shen B. (2018) Discovery of alternative producers of the enediyne antitumor antibiotic C-1027 with high titers. J. Nat. Prod. 81, 594–599 10.1021/acs.jnatprod.7b01013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu W., Christenson S. D., Standage S., and Shen B. (2002) Biosynthesis of the enediyne antitumor antibiotic C-1027. Science 297, 1170–1173 10.1126/science.1072110 [DOI] [PubMed] [Google Scholar]
- 19. Chen Y., Yin M., Horsman G. P., Huang S., and Shen B. (2010) Manipulation of pathway regulation in Streptomyces globisporus for overproduction of the enediyne antitumor antibiotic C-1027. J. Antibiot. 63, 482–485 10.1038/ja.2010.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Y., Yin M., Horsman G. P., and Shen B. (2011) Improvement of the enediyne antitumor antibiotic C-1027 production by manipulating its biosynthetic pathway regulation in Streptomyces globisporus. J. Nat. Prod. 74, 420–424 10.1021/np100825y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan X., Ge H., and Huang T., Hindra, Yang D., Teng Q., Crnovčič I., Li X., Rudolf J. D., Lohman J. R., Gansemans Y., Zhu X., Huang Y., Zhao L. X., Jiang Y., et al. (2016) Strain prioritization and genome mining for enediyne natural products. MBio 7, e02104–16 10.1128/mBio.02104-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rudolf J. D., Yan X., and Shen B. (2016) Genome neighborhood network reveals insights into enediyne biosynthesis and facilitates prediction and prioritization for discovery. J. Ind. Microbiol. Biotechnol. 43, 261–276 10.1007/s10295-015-1671-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang L., Wang S., He Q., Yu T., Li Q., and Hong B. (2012), Draft genome sequence of Streptomyces globisporus C-1027, which produces an antitumor antibiotic consisting of a nine-membered enediyne with a chromoprotein. J. Bacteriol. 194, 4144 10.1128/JB.00797-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Konishi M., Ohkuma H., Matsumoto K., Tsuno T., Kamei H., Miyaki T., Oki T., and Kawaguchi H., VanDuyne G. D., and Clardy J. (1989) Dynemicin A, a novel antibiotic with the anthraquinone and 1,5-diyn-3-ene subunit. J. Antibiot. 42, 1449–1452 10.7164/antibiotics.42.1449 [DOI] [PubMed] [Google Scholar]
- 25. Davies J., Wang H., Taylor T., Warabi K., Huang X. H., and Andersen R. J. (2005) Uncialamycin, a new enediyne antibiotic. Org. Lett. 7, 5233–5236 10.1021/ol052081f [DOI] [PubMed] [Google Scholar]
- 26. Yan X., Chen J. J., Adhikari A., Yang D., Crnovcic I., Wang N., Chang C. Y., Rader C., and Shen B. (2017) Genome mining of Micromonospora yangpuensis DSM 45577 as a producer of an anthraquinone-fused enediyne. Org. Lett. 19, 6192–6195 10.1021/acs.orglett.7b03120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yan X., Ge H., Huang T., Hindra, Yang D., Teng Q., Crnovčić I., Li X., Rudolf J. D., Lohman J. R., Gansemans Y., Zhu X., Huang Y., Zhao L. X., Jiang Y., et al. (2016), Strain prioritization and genome mining for enediyne natural products. MBio 7, e02104–16 10.1128/mBio.02104-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan X., Chen J. J., Adhikari A., Teijaro C. N., Ge H., Crnovcic I., Chang C. Y., Annaval T., Yang D., Rader C., and Shen B. (2018) Comparative studies of the biosynthetic gene clusters for anthraquinone-fused enediynes shedding light into the tailoring steps of tiancimycin biosynthesis. Org. Lett. 20, 5918–5921 10.1021/acs.orglett.8b02584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hara M., Asano K., Kawamoto I., Takiguchi T., Katsumata S., Takahashi K., and Nakano H. (1989) Leinamycin, a new antitumor antibiotic from Streptomyces: producing organism, fermentation and isolation. J. Antibiot. 42, 1768–1774 10.7164/antibiotics.42.1768 [DOI] [PubMed] [Google Scholar]
- 30. Hirayama N., and Matsuzawa E. S. (1993) Molecular structure of a novel antitumor antibiotic leinamycin. Chem. Lett. 22, 1957–1958 10.1246/cl.1993.1957 [DOI] [Google Scholar]
- 31. Steele A., Kalkreuter E., Pan G., Meng S., Shen B. (2019) Hybrid peptide-polyketide natural product biosynthesis in Comprehensive Natural Products: Chemistry and Biology, Vol I, Elsevier, in press [Google Scholar]
- 32. Hara M., Saitoh Y., and Nakano H. (1990) DNA strand scission by the novel antitumor antibiotic leinamycin. Biochemistry 29, 5676–5681 10.1021/bi00476a005 [DOI] [PubMed] [Google Scholar]
- 33. Ma M., Lohman J. R., Liu T., and Shen B. (2015) C–S bond cleavage by a polyketide synthase domain. Proc. Natl. Acad. Sci. U.S.A. 112, 10359–10364 10.1073/pnas.1508437112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tang G. L., Cheng Y. Q., and Shen B. (2004) Leinamycin biosynthesis revealing unprecedented architectural complexity for a hybrid polyketide synthase and nonribosomal peptide synthetase. Chem. Biol. 11, 33–45 10.1016/j.chembiol.2003.12.014 [DOI] [PubMed] [Google Scholar]
- 35. Cheng Y. Q., Tang G. L., and Shen B. (2002) Identification and localization of the gene cluster encoding biosynthesis of the antitumor macrolactam leinamycin in Streptomyces atroolivaceus S-140. J. Bacteriol. 184, 7013–7024 10.1128/JB.184.24.7013-7024.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pan G., Xu Z., Guo Z., Hindra, Ma M., Yang D., Zhou H., Gansemans Y., Zhu X., Huang Y., Zhao L. X., Jiang Y., Cheng J., Van Nieuwerburgh F., Suh J. W., Duan Y., and Shen B. (2017) Discovery of the leinamycin family of natural products by mining actinobacterial genomes. Proc. Natl. Acad. Sci. U.S.A. 114, E11131–E11140 10.1073/pnas.1716245115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu Z., Zhao L. X., Jiang C. L., Duan Y., Wong L., Carver K. C., Schuler L. A., and Shen B. (2011) Bafilomycins produced by an endophytic actinomycete Streptomyces sp. YIM56209. J. Antibiot. 64, 159–162 10.1038/ja.2010.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yan Y., Jiang K., Liu P., Zhang X., Dong X., Gao J., Liu Q., Barr M. P., Zhang Q., Hou X., Meng S., and Gong P. (2016) Bafilomycin A1 induces caspase-independent cell death in hepatocellular carcinoma cells via targeting of autophagy and MAPK pathways. Sci. Rep. 6, 37052 10.1038/srep37052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mauvezin C., and Neufeld T. P. (2015) Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome–lysosome fusion. Autophagy 11, 1437–1438 10.1080/15548627.2015.1066957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ottesen E. A. (2000) Editorial: the global programme to eliminate lymphatic filariasis. Trop. Med. Int. Health 5, 591–594 10.1046/j.1365-3156.2000.00620.x [DOI] [PubMed] [Google Scholar]
- 41. Kron M., Marquard K., Härtlein M., Price S., and Leberman R. (1995) An immunodominant antigen of Brugia malayi is an asparaginyl-tRNA synthetase. FEBS Lett. 374, 122–124 10.1016/0014-5793(95)01092-S [DOI] [PubMed] [Google Scholar]
- 42. Yu Z., Vodanovic-Jankovic S., Kron M., and Shen B. (2012) New 9326A congeners from Streptomyces sp. 9078 inhibiting Brugia malayi asparaginyl-tRNA synthetase. Org. Lett. 14, 4946–4949 10.1021/ol302298k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rateb M. E., Yang D., Vodanovic-Jankovic S., Yu Z., Kron M. A., and Shen B. (2015) Adipostatins A–D from Streptomyces sp. 4875 inhibiting Brugia malayi asparaginyl-tRNA synthetase and killing adult Brugia malayi parasites. J. Antibiot. 68, 540–542 10.1038/ja.2015.22 [DOI] [PubMed] [Google Scholar]
- 44. Yu Z., Vodanovic-Jankovic S., Ledeboer N., Huang S.-X., Rajski S. R., Kron M., and Shen B. (2011) Tirandamycins from Streptomyces sp. 17944 inhibiting the parasite Brugia malayi asparagine tRNA synthetase. Org. Lett. 13, 2034–2037 10.1021/ol200420u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Novac O., Guenier A. S., and Pelletier J. (2004) Inhibitors of protein synthesis identified by a high throughput multiplexed translation screen. Nucleic Acids Res. 32, 902–915 10.1093/nar/gkh235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cencic R., Robert F., and Pelletier J. (2007) Identifying small molecule inhibitors of eukaryotic translation initiation. Methods Enzymol. 431, 269–302 10.1016/S0076-6879(07)31013-6 [DOI] [PubMed] [Google Scholar]
- 47. Huang S. X., Yu Z., Robert F., Zhao L. X., Jiang Y., Duan Y., Pelletier J., and Shen B. (2011) Cycloheximide and congeners as inhibitors of eukaryotic protein synthesis from endophytic actinomycetes Streptomyces sps. YIM56132 and YIM56141. J. Antibiot. 64, 163–166 10.1038/ja.2010.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yin M., Yan Y., Lohman J. R., Huang S. X., Ma M., Zhao G. R., Xu L. H., Xiang W., and Shen B. (2014) Cycloheximide and actiphenol production in Streptomyces sp. YIM56141 governed by single biosynthetic machinery featuring an acyltransferase-less type I polyketide synthase. Org. Lett. 16, 3072–3075 10.1021/ol501179w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Serbus L. R., Landmann F., Bray W. M., White P. M., Ruybal J., Lokey R. S., Debec A., and Sullivan W. (2012) A cell-based screen reveals that the albendazole metabolite, albendazole sulfone, targets Wolbachia. PLoS Pathog. 8, e1002922 10.1371/journal.ppat.1002922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bakowski M. A., Shiroodi R. K., Liu R., Olejniczak J., Yang B., Gagaring K., Guo H., White P. M., Chappell L., Debec A., Landmann F., Dubben B., Lenz F., Struever D., Ehrens A., et al. (2019) Discovery of short-course antiwolbachial quinazolines for elimination of filarial worm infections. Sci. Transl. Med. 11, eaav3523 10.1126/scitranslmed.aav3523 [DOI] [PubMed] [Google Scholar]
- 51. Xu Z., Fang S. M., Bakowski M. A., Rateb M. E., Yang D., Zhu X., Huang Y., Zhao L. X., Jiang Y., Duan Y., Hull M. V., McNamara C. W., and Shen B. (2019) Discovery of kirromycins with anti-Wolbachia activity from Streptomyces sp. CB00686. ACS Chem. Biol. 14, 1174–1182 10.1021/acschembio.9b00086 [DOI] [PubMed] [Google Scholar]
- 52. Love M. S., Beasley F. C., Jumani R. S., Wright T. M., Chatterjee A. K., Huston C. D., Schultz P. G., and McNamara C. W. (2017) A high-throughput phenotypic screen identifies clofazimine as a potential treatment for cryptosporidiosis. PLoS Negl. Trop. Dis. 11, e0005373 10.1371/journal.pntd.0005373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen J. J., Rateb M. E., Love M. S., Xu Z., Yang D., Zhu X., Huang Y., Zhao L. X., Jiang Y., Duan Y., McNamara C. W., and Shen B. (2018) Herbicidins from Streptomyces sp. CB01388 showing anti-Cryptosporidium activity. J. Nat. Prod. 81, 791–797 10.1021/acs.jnatprod.7b00850 [DOI] [PubMed] [Google Scholar]
- 54. Shen B. (2015) A new golden age of natural products drug discovery. Cell 163, 1297–1300 10.1016/j.cell.2015.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]