Abstract
Mucin 5B (MUC5B) has an essential role in mucociliary clearance that protects the pulmonary airways. Accordingly, knowledge of MUC5B structure and its interactions with itself and other proteins is critical to better understand airway mucus biology and improve the management of lung diseases such as asthma, cystic fibrosis, and chronic obstructive pulmonary disease (COPD). The role of an N-terminal multimerization domain in the supramolecular organization of MUC5B has been previously described, but less is known about its C-terminal dimerization domain. Here, using cryogenic electron microscopy (cryo-EM) and small-angle X-ray scattering (SAXS) analyses of recombinant disulfide-linked dimeric MUC5B dimerization domain we identified an asymmetric, elongated twisted structure, with a double globular base. We found that the dimerization domain is more resistant to disruption than the multimerization domain suggesting the twisted structure of the dimerization domain confers additional stability to MUC5B polymers. Size-exclusion chromatography-multiangle light scattering (SEC-MALS), SPR-based biophysical analyses and microscale thermophoresis of the dimerization domain disclosed no further assembly, but did reveal reversible, calcium-dependent interactions between the dimerization and multimerization domains that were most active at acidic pH, suggesting that these domains have a role in MUC5B intragranular organization. In summary, our results suggest a role for the C-terminal dimerization domain of MUC5B in compaction of mucin chains during granular packaging via interactions with the N-terminal multimerization domain. Our findings further suggest that the less stable multimerization domain provides a potential target for mucin depolymerization to remove mucus plugs in COPD and other lung pathologies.
Keywords: mucin, mucus, von Willebrand factor, lung, cryo-electron microscopy, small-angle X-ray scattering (SAXS), Mucus obstruction
Introduction
Polymeric mucins are the major structural component of mucus; a hydrogel that plays a pivotal role in protecting mucosal surfaces. In healthy airways, mucus in conjunction with cilia, protect the airway epithelium from inhaled pathogens, particles, and toxins (1, 2). The polymeric mucins confer the correct rheological properties for the gel to facilitate removal of these inhaled components by mucociliary clearance (MCC).4 However, during airway diseases such as asthma, cystic fibrosis (CF), and chronic obstructive pulmonary fibrosis (COPD), mucus concentration is increased which results in an increase in viscoelasticity. The abnormal mucus impairs effective MCC, generating an environment that harbors pathogenic microbes (3–5).
MUC5B and MUC5AC are the major polymeric mucins in respiratory tract mucus (6–9). The focus of this study is MUC5B, which is essential for mucociliary clearance of the lung (10, 11). MUC5B is a large (2–50 MDa), multidomain gel-forming glycoprotein that shares high sequence homology with the cysteine-rich domains of von Willebrand factor (vWF) at its N- and C-terminal regions (12, 13). MUC5B consists of an N-terminal region composed of vWF-like D1-D2-D′-D3 domains, a central proline-threonine-serine-rich mucin domain interspersed with cysteine-rich (Cys) domains, and C-terminal region composed of vWF-like D4-B-C-CK domains (14). The N- and C-terminal regions are important for disulfide bond-mediated polymer assembly.
MUC5B biosynthesis is a multistep process involving CK-domain disulfide-mediated C-terminal dimerization in the endoplasmic reticulum, and extensive O-glycosylation of the central mucin domain followed by D3-domain disulfide-mediated N-terminal multimerization in the Golgi apparatus (15–19). The assembled, linear MUC5B polymer is packaged in a compact and cross-linked form inside secretory granules, via noncovalent interactions between N-terminal dimerization domains controlled by calcium ions and acidic pH (19–21). Post-secretion, uncoupling of these mucin-mucin interactions allows transition to an expanded linear chain that is critical for formation of a flowing mucus gel that facilitates lung protection by MCC (1). The supramolecular organization of MUC5B within mucus is not completely described, although recent studies have shown that submucosal gland-derived MUC5B forms assemblies of the linear mucin strands, termed bundles (22).
Compromised MCC is a feature of muco-obstructive disease and there are multiple mechanisms proposed to generate the pathogenic mucus associated with asthma, CF, and COPD; important contributors include concentration of mucin, defective mucin expansion, and covalent cross-linking of the secreted mucin network (20, 23–27). Understanding how airway obstruction can be reversed is crucial to tackling mucus accumulation in the airways. Therefore, elucidating the molecular details of mucin structure, assembly, packaging, and post-secretory expansion is critical in the development of mucolytic therapies that can directly target the mucin polymers that underpin the adherent mucus plugs/plaques. Although the structure and roles of the MUC5B N-terminal multimerization domain in MUC5B assembly and intragranular packaging have been described (19, 21) the roles of other protein-rich regions of MUC5B in airway mucus biology have not been elucidated. Therefore, we have focused on elucidating the structure and functional roles of the C-terminal dimerization domain of MUC5B in polymer stability and intragranular packaging.
Here, using recombinant protein (D4-B-C-CK), we describe the detailed structure of the C-terminal dimerization domain of MUC5B using single particle cryo-electron microscopy (cryo-EM) and small-angle X-ray scattering (SAXS). We demonstrate that the C-terminal dimerization domain of MUC5B has similar structure to that of the related glycoprotein, vWF (28, 29). Moreover, we show that the C-terminal dimerization domain confers extra stability on the MUC5B polymer compared with the N-terminal dimerization domain. Finally, we have gained new insight into the molecular details of MUC5B packaging in secretory granules by using recombinant C-terminal protein to investigate the effect of calcium and pH on (a) homotypic interaction between C-terminal dimerization domain and (b) heterotypic interaction with recombinant N-terminal multimerization domain.
Results
Expression and characterization of MUC5B C-terminal dimerization domain
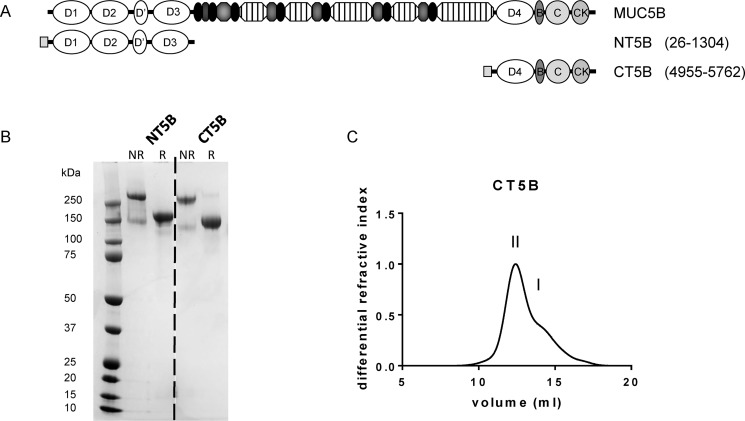
To investigate the structure and further define the role of the C-terminal dimerization domain of MUC5B, we stably expressed a C-terminal construct of MUC5B (CT5B), consisting of D4-B-C-CK domains (Fig. 1A). We also expressed the previously published N-terminal dimerization domain of MUC5B (NT5B protein) consisting of D1-D2-D′-D3 domains (19). The expressed proteins were purified from conditioned 293-EBNA medium using a combination of nickel affinity, size exclusion, and anion exchange chromatography. The reduced (R) and nonreduced (NR) proteins were analyzed by SDS-PAGE (Fig. 1B) and showed that CT5B was expressed mainly as a disulfide-linked dimer with a small proportion of monomer. The molecular mass for the CT5B dimer (∼245 kDa) and monomer (∼147 kDa) was determined by size exclusion chromatography multi-angle static light scattering (SEC-MALS) (Fig. 1C).
Figure 1.
Expression of recombinant MUC5B protein domains. A, schematic showing the domain structure of N- and C-terminal constructs of MUC5B. The constructs were composed of a N-terminal His6 tag (gray square) coupled to the MUC5B domains, NT5B (D1-D2-D′-D3) and CT5B (D4-B-C-CK). B, N- and C-terminal constructs, reduced (R) with 10 mm DTT and nonreduced (NR), were analyzed by SDS-PAGE, and stained with InstantBlue. Gel splicing occurred at the position indicated by the dashed line, the data came from the same gel. C, recombinant CT5B in 0.2 m NaCl, pH 7.4, was analyzed by SEC-MALS and the representative graph shows the differential refractive index. The chromatograph showed two peaks corresponding to dimer (peak II) and monomer (peak I). Experiments were repeated at least 3 times.
Structural analysis of MUC5B dimerization domain
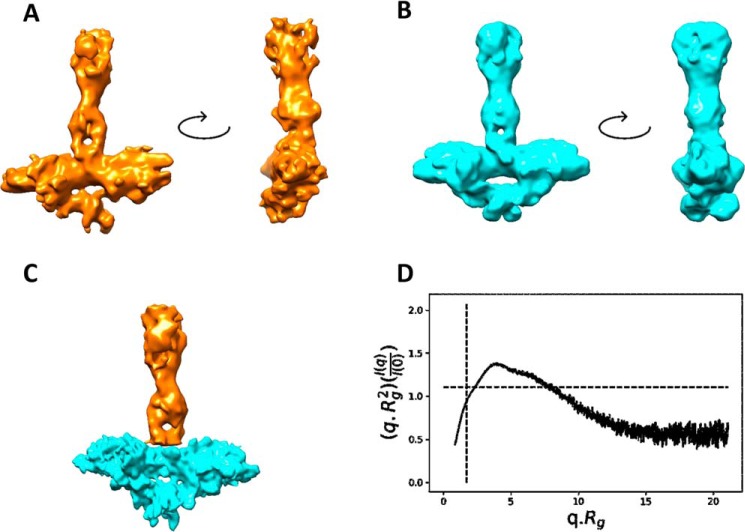
We investigated the structure of dimeric CT5B using cryo-EM and SAXS (Fig. 2 and Figs. S1 and S2). 2D classification of CT5B showed an elongated shape consisting of a globular base connected to an extended stalk region (Fig. 3C and Fig. S1B). There appeared to be flexibility between the globular base and the stalk as visualized in the 2D classes (Fig. S1B). As CT5B is a dimer, the data were processed using either C1 or C2 symmetry. The C1 structure refined to a resolution of 9.3 Å, where the twist in the stalk was clearly visible (Fig. 2A), and the C2 structure refined to a resolution of 8.9 Å where the base benefitted from the application of 2-fold symmetry, but the flexible stalk structure deteriorated when symmetry was applied (Fig. 2B). Therefore, the stalk was subtracted from the C2 refined structure and locally refined resulting in a gain in resolution to 7.5 Å (Fig. S1C and Fig. 3). A composite map of the refined C2 base and the C1 stalk is shown in Fig. 2C, where CT5B is a “T” shaped dimeric molecule with dimensions 21 × 18 × 5 nm (height × width × depth).
Figure 2.
Structural analysis of MUC5B multimerization domain. Cryo-EM maps of CT5B refined using either C1 (A) or C2 (B) symmetry shown with a rotation through 90 degrees. C, a composite map of CT5B showing the stalk from the C1 model (yellow) and the C2 base (blue) from an independent subtracted refinement. D, normalized Kratky plot calculated from the SAXS data of CT5B showing a biphasic profile indicating that CT5B has rigid and flexible regions.
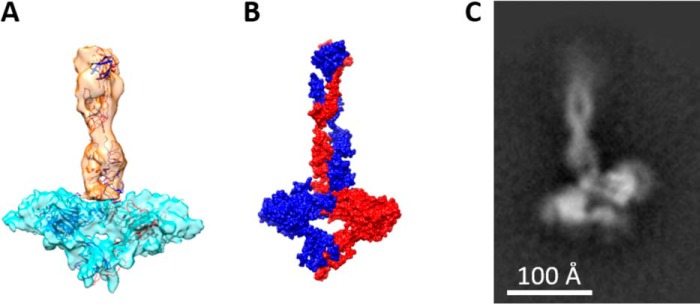
Figure 3.
Structural overview of C-terminal MUC5B. A, the composite map (from Fig. 2C) is shown with docked models of each domain. A dimer of the D4 domain fits within the base, with the monomers colored red and blue. In the stalk, the B-C-CK domains are fitted twisting along the stalk axis, with each monomer colored orange and blue. B, a surface rendered representation of the docked models shown in A with each monomer chain color coded either red or blue. C, a representative 2D class of CT5B showing this twist along the length of the stalk.
Analysis of the SAXS data confirmed the flexible nature of CT5B, with the normalized Kratky plot indicating a biphasic system with both globular and elongated regions connected by limited flexibility (Fig. 2D). Based on the similarity to the vWF structure and crystal structure of the vWFD domain (28, 29), it was clear that the D4 region was located in the globular base structure. A homology model of the D4 region was therefore generated, which readily docked into the symmetric base (Fig. 3). The D4 domains form a butterfly shape with the termini connecting through the central axis. The CT5B region has 13 predicted N-glycans per monomer so some areas where the EM density is not fully occupied by protein structure may reflect the locations of N-glycans, which could also add flexibility to the structure (Fig. S3B). A resolution of 7.5 Å was achieved despite the heterogeneity contributed by complex N-glycosylation and conformational flexibility through the stalk region.
The B-C-CK domains form the stalk structure. It was apparent from the 2D class averages (Fig. 3C and Fig. S1B) and 3D reconstruction (Fig. 2, A and C) that the two molecules within the stalk region twist around each other. This may increase the interface and the number of domain-domain interactions between the monomers in the stalk region, potentially increasing the stability of this C-terminal region (Fig. 3). To determine whether the structure of the C-terminal dimerization domain is pH regulated, we analyzed the CT5B dimer by size exclusion chromatography at different pH values (Fig. S4). Results showed no evidence of a change in CT5B structure at different pH values. Taken together, our data suggest that the twist in the stalk of the molecule, which represents the B-C domains, could be a feature that provides enhanced stability to the CT5B dimer.
Stability of the dimerization and multimerization domains
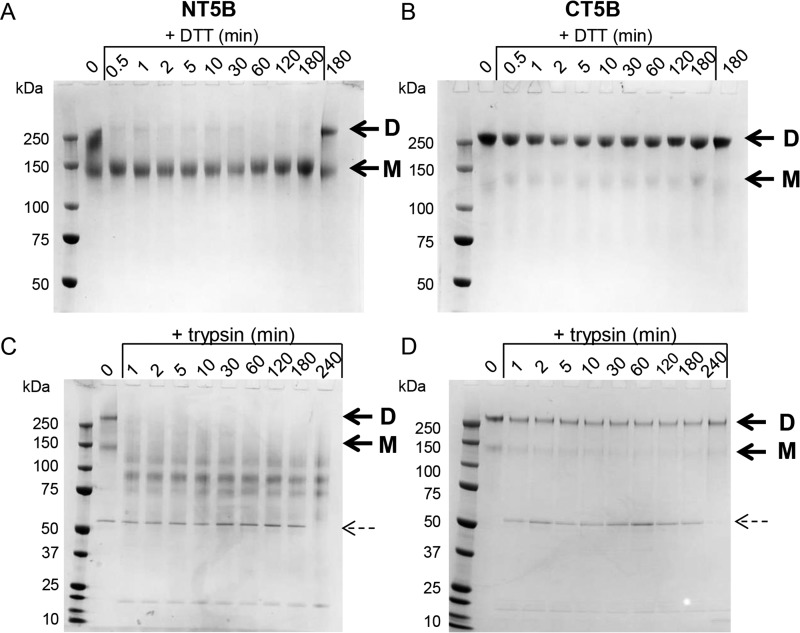
MUC5B forms linear polymers via disulfide-linked N-N and C-C–terminal interactions. We compared the relative stability of the expressed protein representing the N-terminal multimerization domain of MUC5B (NT5B) (Fig. 1B) (19) with CT5B by using limited proteolysis and partial reduction approaches. The NT5B and CT5B proteins were treated with 1 mm DTT on ice (partial reduction; Fig. 4, A and B) or with 1 μg/ml of trypsin at room temperature (partial proteolysis; Fig. 4, C and D) and samples collected at different time points were analyzed by SDS-PAGE. Dimer and monomer bands are indicated by arrows and trypsin inhibitor (∼50 kDa) is indicated by a dashed arrow. Results showed that CT5B dimer was resistant to limited proteolysis and partial reduction, maintaining a dimer throughout the time course of the experiment (Fig. 4, B and D). In contrast, NT5B dimer was sensitive to both treatments, being reduced to monomer in the presence of DTT after ∼0.5 min (Fig. 4A) and lower molecular weight species by the action of trypsin within 1 min (Fig. 4C).
Figure 4.
Stability of the dimerization and multimerization domains of recombinant MUC5B. A, recombinant NT5B and B, CT5B were partially reduced by incubation with 1 mm DTT on ice for 0.5 to 180 min. Samples without DTT were incubated for 0 and 180 min. C, recombinant NT5B and D, CT5B were subjected to limited proteolysis with 1 μg/ml of trypsin for 1 to 240 min at room temperature. Samples without trypsin were incubated for 0 min. The bands visible at approximately 50 and 20 kDa represent the trypsin inhibitor and trypsin, respectively. Dimer (D) and monomer (M) of NT5B and CT5B are indicated by arrows, and trypsin inhibitor is indicated by a dashed arrow. The experiments were repeated 3 times.
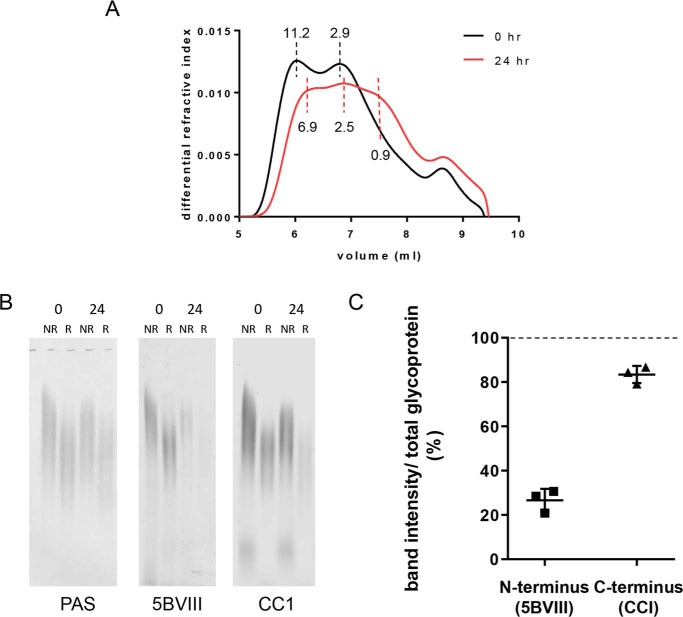
To assess whether this differential stability demonstrated with the N- and C-terminal recombinant proteins was observed with native polymers, we analyzed purified MUC5B following proteolysis. SEC-MALS analysis showed that proteolysis caused a reduction in the average molecular mass of MUC5B from 6.8 to 3.5 MDa (Fig. 5A). Western blot analysis before and after proteolysis was performed and blots were probed for D3 domain (5BVIII) in the N terminus, and B domain (CC1) in the C terminus, and total glycoprotein was determined by PAS stain (Fig. 5B). Results for the unreduced samples showed ∼70% loss of N-terminal antibody signal following proteolysis, whereas the C-terminal antibody signal was only decreased by ∼15% (Fig. 5C). A similar trend was observed for the samples that were reduced prior to electrophoresis. These data are consistent with results following trypsin digestion of the recombinant MUC5B proteins (Fig. 4, C and D) and demonstrate that in native MUC5B polymers the C-terminal dimerization domain is more stable than the N-terminal multimerization domain.
Figure 5.
Stability of the dimerization and multimerization domains of polymeric MUC5B. MUC5B was incubated with 1 μg/ml of trypsin for 0 and 24 h at 37 °C. A, samples were analyzed by SEC-MALS on a Shodex OHpak SB-806 M HQ column. The molecular mass of peaks is indicated (MDa). This experiment was repeated twice. B, samples (reduced (R) and nonreduced (NR)) were analyzed on an agarose gel and transferred to nitrocellulose, then probed with MAN-5BVIII (D3 domain) or CC1 (B domain) antisera or PAS stained. C, nonreduced bands were quantified, then MAN-5BVIII and CC1 signals were divided by PAS stain signal (total glycoprotein) and expressed as a percentage compared with the 0 h sample (dashed line). Error bars represent the S.D. This experiment was repeated 3 times.
Effect of calcium and pH on the dimerization domain of MUC5B
We next investigated the potential role of CT5B in MUC5B intragranular packaging. Prior to secretion, linear disulfide-stabilized MUC5B polymers are condensed and packaged within secretory granules that have an acidic pH and high calcium content (1, 19). We previously demonstrated that the disulfide-linked dimeric N-terminal multimerization domains of MUC5B were noncovalently assembled into tetramers through calcium-mediated cross-links, active at acidic pH (pH 5–6) (21). This interaction is proposed to change the organization of the linear polymeric chains and aid their ordered packaging inside secretory granules (19). To determine whether calcium and acidic pH could cause noncovalent interactions between the recombinant C-terminal multimerization domain; dimeric CT5B was incubated in the presence or absence of calcium (5 mm) at pH 7.4 or 6 and analyzed by SEC-MALS (Fig. S5A). Results in the presence of EGTA at pH 7.4 and 6 (Fig. 5A, i and ii) showed a peak of dimer (peak II) and monomer (peak I). In the presence of calcium at pH 7.4, 6, or 5, the profiles of CT5B were similar to the data for EGTA and showed that calcium and pH did not facilitate noncovalent interactions between CT5B (Fig. S5A, iii–vi). These results were also confirmed using analytical ultracentrifugation, which showed monomer (I) and dimer (II) peaks with similar sedimentation profiles for CT5B under all conditions studied (Fig. S5B, i–iii). These data show that unlike the N-terminal multimerization domain, the C-terminal dimerization domain of MUC5B does not form homotypic calcium or pH-dependent multimers and therefore, does not have the same role as the N-terminal multimerization region in the organized packaging of MUC5B.
Effect of calcium and pH on the interaction between the dimerization and multimerization domains of MUC5B
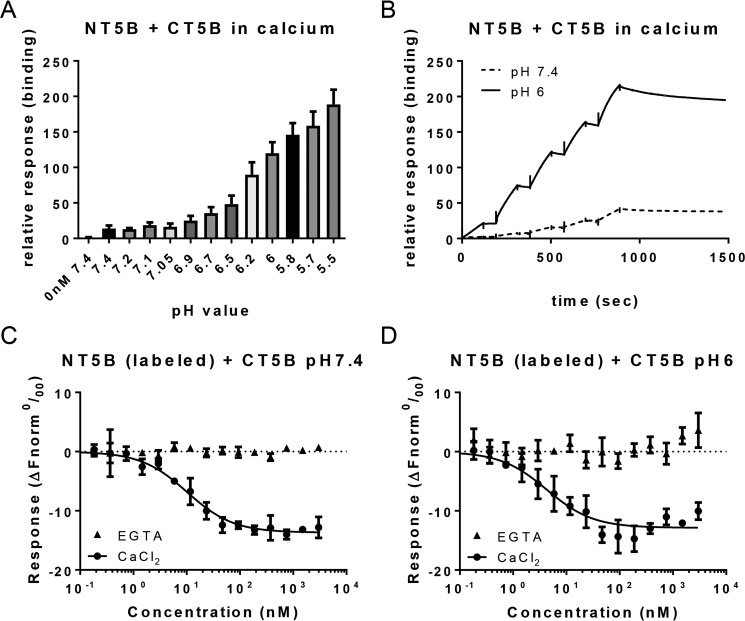
Although the presence of calcium and acidic pH did not engender homotypic interaction between CT5B, we hypothesized that the C-terminal multimerization domains of MUC5B may interact with N-terminal dimerization domains during packaging to further facilitate the organization of the mucin polymer inside the secretory granule. Using surface plasmon resonance (SPR), the interaction of immobilized dimeric CT5B with 50 nm dimeric NT5B was analyzed in the presence of 5 mm CaCl2 over a range of pH (Fig. 6A). This pH scouting assay showed that there was some low background binding between NT5B and CT5B in the presence of calcium at pH 7.4. Binding was shown to increase as a function of decreasing pH, the most binding was at pH ∼ 6.2, and binding was abolished following EDTA treatment. Single cycle kinetic assays were performed with increasing concentrations of NT5B (5-40 nm) flowed over immobilized CT5B in the presence of 5 mm CaCl2 at pH 6 and 7.4 (Fig. 6B, solid black line and dashed line, respectively). Analysis showed a strong binding interaction between NT5B and CT5B at pH 6, with a KD value of ∼1 ± 0.04 nm. At pH 7.4, NT5B flowed over immobilized CT5B with 5 mm CaCl2 and produced some binding with a KD value in a similar range (∼3 nm), however, the binding response was 5-fold lower; suggesting the same binding affinity, but a much lower amount of NT5B bound.
Figure 6.
Analysis of heterotypic interaction of MUC5B dimerization and multimerization domains. A, a pH scan of 50 nm NT5B in different pH buffers in the presence of 5 mm CaCl2, flowed over CT5B immobilized to a chip and analyzed using SPR. There was some background binding at pH 7.4, but strong binding occurred between NT5B and CT5B at pH ∼ 6.2. This experiment was performed once with triplicate samples. B, single cycle kinetic analysis with increasing concentrations of NT5B in 5 mm CaCl2 (0, 5, 10, 15, 20, and 40 nm) at pH 6 (black line) or pH 7.4 (dashed line). C and D, serially diluted recombinant CT5B protein was mixed with an equal volume of fluorescently labeled NT5B recombinant protein, and interactions were measured by fluorescence using MST. Samples were prepared in HBS buffer containing 0.05% Tween 20 and either 5 mm CaCl2 (circles) or 5 mm EGTA (triangles) at pH 7.4 (C) or 6 (D). Error bars represent the mean ± S.E. Experiments in B–D were repeated at least twice.
We investigated the heterotypic binding further by performing in solution analysis of the interaction between NT5B (fluorescently labeled) and CT5B (serially diluted) using microscale thermophoresis (MST). The data were analyzed using a KD model of fit using the NT Affinity Analysis software (Fig. 6, C and D). The results confirmed that NT5B and CT5B interact in calcium at pH 6, with a KD of 3 ± 1.4 nm (Fig. 6D, circles), which was comparable with the result obtained from SPR. The results also showed an interaction at pH 7.4 with a similar binding affinity (KD 7.4 ± 1.5 nm; Fig. 6C). Importantly, there was no interaction detected between NT5B and CT5B in the presence of EGTA at either pH (Fig. 6, C and D, triangles). These results establish that MUC5B dimerization and multimerization domains form reversible pH-sensitive and calcium-dependent intermolecular associations between polymeric mucins.
Discussion
Determining the nature of polymeric mucin chain interactions during packaging into secretory granules and their subsequent uncoupling post-secretion is critical to understanding the mechanisms that control mucus gel formation in health and disease. Abnormal mucus gel properties may arise due to mucin chains not fully expanding upon secretion, resulting in altered molecular structure. Indeed, airway mucins with aberrant macromolecular structure are a feature of hyper-concentrated mucus that obstructs the CF airways (23). We have previously described a role for the N-terminal multimerization domain of MUC5B in forming noncovalent, reversible calcium-dependent tetramers that are involved in the organization of the mucin chains within the secretory granule (19, 21). Here we have extended understanding of MUC5B intragranular packaging by describing a role for the MUC5B C-terminal dimerization domain.
During packaging of MUC5B within secretory granules, the mucin chains are organized around protein nodes formed from pH and calcium-dependent, noncovalent, reversible, homotypic interactions between N-terminal multimerization domains (19, 21). In marked contrast, we demonstrate that the C-terminal dimerization domain of MUC5B does not form homotypic multimers in the presence of calcium at acidic pH, suggesting it has a different function in intragranular packaging. Indeed, using SPR and MST we demonstrated the potential for reversible, calcium-dependent heterotypic interactions between the dimerization and multimerization domains of MUC5B most active at acidic pH. We propose that these heterotypic interactions aid further compaction of MUC5B during packaging, adding an additional level of organization of the mucin polymers inside the secretory granule (Fig. 7). Further work is required to investigate whether there are any other protein-protein interactions, perhaps with the internal Cys domains of MUC5B (7 in total), that contribute to the full mechanism of packaging and subsequent unfurling of mucins into mucus.
Figure 7.
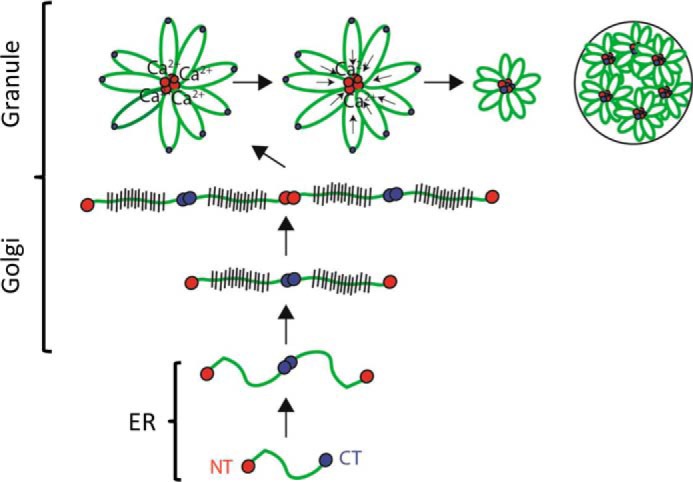
Model for MUC5B intracellular assembly and packaging. In the endoplasmic reticulum, MUC5B forms homotypic disulfide-linked dimers via its C-terminal dimerization domains (blue circles). The dimer is transported to the Golgi where it undergoes O-glycosylation prior to linear polymer formation via disulfide linkage between N-terminal multimerization domains (red circles). A reduction in pH and an increase in free calcium concentration occur across the secretory pathway. This results in noncovalent, calcium-mediated interactions between N termini, which appear as proteinaceous nodes, and aid organization of MUC5B for intragranular packaging. The new findings in this paper extend this model and establish that the C-terminal dimerization domains have the potential to interact with the multimerization domains at acidic pH in the presence of calcium, which may aid further compaction of the MUC5B molecule for intragranular packaging. The order of assembly of these N- and C-terminal containing nodes has yet to be elucidated. Uncoupling of these noncovalent interactions in mucus is critical for the transition to an expanded linear chains underpinning normal flowing mucus that can be transported by MCC to keep the lung free from infection. Addition of agents to mucus that displace calcium from mucins has been shown to normalize mucin conformation and decrease mucus viscoelasticity and improve MCC (64, 65).
Although the structure of the N-terminal multimerization domain of MUC5B has been described (19, 21), there is little information on the C-terminal dimerization domain. To address the gap in our knowledge, the structure of the C-terminal dimerization domain of MUC5B was determined using SAXS and cryo-EM analysis, and was identified as an anisotropic, extended structure with globular domains. The stalk section of the structure contained a twist, which we interpret as a key structural feature that is likely to contribute to stabilization of the dimerization domain. The related C-terminal region of vWF has a similar shape, described as an extended dimeric stem with bouquet structure (28). However, there are certain differences between MUC5B and vWF C-terminal structures. In vWF, there is no evidence of a twist in the stem-like structure, although interactions have been hypothesized between each pair of tandem C domains (30). Also, at neutral pH encountered during secretion, the vWF stem-like structure unzips, remaining disulfide-bonded at the CK domain (28). Our results showed that the C-terminal structure of MUC5B was not subject to pH-dependent changes, and maintained a stable conformation, potentially aided by the noncovalent bonded, twisted structure of the C domain. These differences between the two molecules may be attributed to the presence of a larger number of B and C domains in the C terminus of vWF (D4-B-B-B-C-C-CK) compared with MUC5B (30–33) and the distinct roles of the two molecules. The resolution of our cryo-EM data are most likely limited by the flexibility of the protein, as shown in the 2D classes (Fig. S1B) and by the normalized Kratky plot (Fig. 2D), and the large proportion of heterogeneity as a result of glycosylation across the molecule (18).
This study is the first to describe the structure of the MUC5B mucin dimerization domain and it may form a model for other members of the polymeric mucin family. The C-terminal of MUC5B shares some sequence homology with the other polymeric mucins MUC2 and MUC5AC (34–37). However, MUC2 and MUC5AC contain a GDPH autocatalytic cleavage sequence in their D4 domain that is absent in MUC5B (the equivalent sequence is GGSH). The GDPH site is cleaved at acidic pH for MUC2, and neutral to acidic pH for MUC5AC, producing reactive ends that may be involved in forming self-interactions, interactions with other molecules and/or the epithelial cell surface (38, 39). There is no evidence in the literature for C-terminal cleavage in MUC5B. Therefore, there are potential differences in the stability of the C-terminal dimerization domains of MUC2 and MUC5AC compared with MUC5B, which may affect their function.
On comparing the stability of the N- and C-terminal regions of MUC5B, results showed that the C-terminal dimerization domain was more resistant to degradation than the N-terminal multimerization domain, this was seen with both recombinant proteins and the native MUC5B polymer. We have shown here that the central stalk of the dimerization domain has a twisted structure, whereas the multimerization domain has a more open conformation in a boomerang-like shape (19). Thus, the differences in structure correlate well with the differences noted in their stability. These findings provide new insight into the structure and stability of the MUC5B polymer, which may benefit future therapeutic development to alleviate pathology associated with airway mucus obstruction. Indeed, there are already reports of mucolytic agents that directly target the integrity of airway mucin polymers to improve airway clearance by reducing the viscosity of abnormal mucus (40–42). The determination of the differential structure and stability of the critical regions involved in MUC5B polymer assembly offers potential to design more targeted mucoactive agents. In this case, the multimerization domains of MUC5B appear to provide a better target, than the more stable dimerization domains, for production of a mucolytic agent to breakdown mucin polymers and thereby reduce mucus accumulation and improve lung function.
In summary, this study has elucidated a potentially new role for the C-terminal dimerization domain of MUC5B in aiding compaction of mucin chains during granular packaging, via heterotypic interactions with the N-terminal multimerization domains. Results have determined a detailed structure for the dimerization domain of MUC5B, which has shown some similarity with the related vWF C-terminal structure. The structural data have highlighted a central twist in the structure of the dimerization domain, and we propose that this feature contributes to enhanced stability of this region of the MUC5B polymer.
Experimental procedures
Expression and purification of recombinant human MUC5B protein domains
MUC5B (UniProtKB accession number Q9HC84) C-terminal construct consisting of D4-B-C-CK (CT5B; residues 4955–5762) (12) was created with an N-terminal His6 tag using the mammalian episomal expression vector pCEP-His in 293-EBNA cells (19, 43, 44) (Fig. 1A). The N-terminal construct (NT5B consisting of D1-D2-D′-D3) has been described previously (19) (Fig. 1A). A Superose 6 10/300 GL column was used for size fractionation. Protein identity and purity were confirmed by tandem MS. Samples, with and without treatment with 10 mm for 4 min at 95 °C, were analyzed by SDS-PAGE using 4–12% BisTris gels (Life Technologies) and stained with InstantBlue (Expedeon).
SEC-MALS
For determination of molecular mass by SEC-MALS analysis, recombinant proteins were applied to a Superose 6 10/300 GL column and MUC5B was applied to a Shodex OHpak SB-806 M HQ column, in 0.2 m NaCl, 0.05 m EDTA, 0.05% sodium azide. For experiments in the presence of calcium and EGTA, recombinant CT5B protein was incubated overnight at 4 °C in 5 mm CaCl2 or 5 mm EGTA at pH 7.4 or 6 in 25 mm HEPES, 150 mm NaCl, before applying to a Superose 6 10/300 GL column at room temperature. For samples at pH 6 and 5, the HEPES was replaced with 25 mm MES. Column eluents passed through an in-line Helios 18-angle laser photometer and a T-rEX refractometer with QELS dynamic light scattering attachment (Wyatt Technology). Analysis was performed using ASTRA version 6 software.
Analytical ultracentrifugation
Recombinant CT5B protein was incubated in the presence of 5 mm CaCl2 overnight at 4 °C in either HEPES or MES buffer (25 mm HEPES, 150 mm NaCl, pH 7.4, or 25 mm MES, 150 mm NaCl, pH 6 or pH 5). Samples were analyzed using velocity experiments on an Optima XL-A ultracentrifuge (Beckman Instruments), as previously described (19). The sedimentation coefficients were determined using SedFit version 13.0b (45).
Cryo-EM
Grid prep, regular grids
Quantifoil R1.2/1.3 holey carbon grids were cleaned in 2× chloroform soaks on filter paper in a glass Petri dish and then glow discharged for 2 min at 25 mA. 3 μl of purified CT5B at 0.25 mg/ml was applied to the grids and blotted for 8–10 s at 22 °C in a Vitrobot Mark IV (Thermo Fisher Scientific), before plunge freezing in liquid ethane.
Data acquisition
Cryo-EM data were collected on a Titan Krios electron microscope operating at 300 kV equipped with a K2 Summit direct detector (Gatan) and a FEI Volta phase plate (Thermo Fisher Scientific) at the Electron Bio-Imaging Centre (eBIC) (Didcot, UK). EPU (Thermo Fisher Scientific) was used to automate the collection of 1441 movies, comprising of 40 frames, 10 s exposures, and a total dose of 40 e− Å2 were recorded on the detector in counting mode at a calibrated magnification of 93,000 corresponding to a magnified pixel size of 1.043 Å. Defocus was maintained at −0.5 μm as suggested previously (46).
Data processing
The recorded movies were processed in cryoSPARC v2.8 (47, 48). Movies were aligned using local patch-based motion correction. CTF estimation of the aligned movie stacks was carried out using the local patch-based CTF estimation. Images were then excluded based on motion distance, resolution fit, defocus, and phase shift resulting in 1093 images. Several hand-picked particles were submitted to 2D classification and used for template-based picking over the entire data set. 231,589 particles were extracted after per particle motion correction in a 500 pixel box. Particles were submitted to two rounds of 2D classification and resulted in 16,590 particles. These particles were used to generate an ab initio model for further refinement. Refinement was carried out using C1 and C2 symmetries resulting in structures with resolutions of 10.0 and 9.4 Å, respectively. Heterogeneous refinement was then applied to each result with modest differences between the classes. The particles from the best classes were then used to re-refine the structure resulting in lower resolution structures. Non-uniform refinement was then used to refine the C1 and C2 refined models resulting in a resolution of 9.3 and 8.9 Å, respectively. Particle subtraction was then employed to enhance the resolution of each of the base or stalk regions using masks around the base and stalk. Further refinement of both subtracted symmetry models were completed yielding a base with a resolution of 7.5 Å. The stalk, in the absence of the base, was refined but resulted in a much lower resolution map.
Model generation
The sequence of CT5B (residues 4955–5762) was submitted to the HHpred server (49–51) through the new MPI Bioinformatics Toolkit online portal (52). HHpred was able to identify structures in the PBD (53) with homology equating to 97% of the CT5B sequence (784 residues of 807). The homologous structures showed high probability with values greater that 95%. The corresponding PDBs (Table 1) and sequence were then submitted to Modeller (54) though the online portal. The resulting models were downloaded and manually placed into the cryo-EM density map using Chimera (55). As the C-CK domains contain an intramolecular disulfide bond in the cysteine knot, a homology model of the C-CK domain was generated in SWISS-MODEL (56), which generated a homology model based on chain B of the second biological assembly of PDB 5BPU (57); the C-CK homology model was produced with an intramolecular disulfide bond. Glycans were identified on the full sequence using NetNGlyc server (www.cbs.dtu.dk/services/NetNGlyc/).5
Table 1.
Modeling the domains of CT5B based on structural homology
The table shows the percentage identity between domains in CT5B and the corresponding structures from the PDB.
SAXS
SAXS intensity data, I(q) versus q (q = 4π. sin 2θλ), of CT5B were collected using SEC-SAXS on beamline B21 (Diamond Light Source, UK). 50 μl of BMPER was loaded onto a Superdex 200 Increase 3.2/300 column (GE Healthcare) at 0.075 ml/min. SAXS data were collected at 3-s intervals on an Eiger X 4M detector (Dectris) at a distance of 2.7 m and wavelength of 0.95 Å.
Data were reduced using in-house software. Subtractions of the SEC-SAXS data were completed for each frame across the elution peak and the radius of gyration (Rg) and the integral of intensity ratio to background were plotted. The data were scaled, merged, and averaged for each frame with a consistently similar Rg. All further processing and analysis of data were carried out using ScÅtter (www.bioisis.net/scatter)5 (66).
Partial reduction and limited proteolysis of recombinant MUC5B proteins
For partial reduction experiments, NT5B and CT5B proteins were incubated with 1 mm DTT on ice between 0.5 and 180 min. At each time point, 2.5 mm iodoacetamide was added and incubated for 15 min in the dark at room temperature. For limited proteolysis, NT5B and CT5B proteins were incubated with 1 μg of sequencing-grade trypsin (Promega) at room temperature for 1 to 240 min. At each time point, 1 μl of trypsin neutralizing solution (Lonza) was added to stop the reaction. Samples were analyzed by SDS-PAGE using 4–12% BisTris gels (Life Technologies), staining with InstantBlue (Expedeon).
Partial proteolysis of native MUC5B
MUC5B was purified from A549MUC5Bm (A549 cell line in which MUC5AC was knocked down by gene editing) cell-conditioned medium using cesium chloride isopycnic centrifugation in 0.1 m NaCl, as previously described (58). The MUC5B was further purified on a Sepharose CL2B size exclusion column. Partial proteolysis of purified MUC5B was achieved by incubation with 1 μg of trypsin at 37 °C for 0 and 24 h. Trypsin neutralizing solution (1 μl; Lonza) was added to samples to stop the reaction. Samples (reduced (R) and nonreduced (NR)) were equally loaded in triplicate and analyzed on 0.7% (w/v) agarose gels in 40 mm Tris acetate, 1 mm EDTA, pH 8.0, containing 0.1% SDS, for 16 h. Following electrophoresis, gels with unreduced samples were incubated with 10 mm DTT in 4× SSC, pH 8.0, for 15 min. MUC5B was transferred to nitrocellulose membrane by vacuum blotting at 45 millibars for 1.5 h. The membrane was cut into strips and either probed with polyclonal MAN-5BVIII antiserum (19), which detects the N-terminal D3 domain and CC1 antiserum, which detects C-terminal B domain, at 1:2000 dilution. A secondary IRDye® 800CW antibody (LI-COR Biosciences) was used for detection and blots were analyzed on the LI-COR Odyssey® CLx and quantified using Image Studio Lite v5.2 software (LI-COR Biosciences). The final strip was stained using PAS as previously described (59), and bands were imaged on a Bio-Rad ChemiDocTM MP and quantified using Image Lab v5.2 software (Bio-Rad Laboratories). The nonreduced bands were quantified, and a ratio of the signal was determined for 5BVIII and CC1 relative to PAS signal.
SPR
To investigate interactions between NT5B and CT5B proteins, a Biacore T200 biosensor with Biacore Control software was used (GE Healthcare). Initially 25 μg/ml of CT5B diluted in 50 mm sodium acetate, pH 5.5, was immobilized onto a CM5 sensor chip by amine coupling using 1-ethyl-3(3-dimethylaminopropyl)-carbodiimide hydrochloride, N-hydroxysuccinimide, and ethanolamine-HCl (GE Healthcare). The immobilized CT5B gave ∼3400 response units. To perform a pH scan experiment, purified NT5B protein (50 nm) was injected over the immobilized CT5B in varying pH buffers containing 5 mm CaCl2 and 0.05% Tween 20 for 120 s at a flow rate of 30 μl/min and dissociation time of 60 s. The CM5 sensor chip was regenerated after each sample application with 10 mm EDTA, pH 7.4, at a flow rate of 30 μl/min for 30 s and stabilization of 1 min after regeneration. For single cycle kinetic analysis, 0, 5, 10, 15, 20, and 40 nm NT5B analyte was flowed over the immobilized CT5B in HBS containing 5 mm CaCl2 and 0.05% Tween 20, at pH 6 or 7.4. Curves were fitted using 1:1 Langmuir association/dissociation model (Biacore Evaluation 4.1 software; GE Healthcare) to obtain kinetic values of binding affinity.
MST
Purified NT5B protein in 25 mm HEPES, 150 mm NaCl, pH 7.4, was fluorescently labeled using the MonolithTM NT protein labeling kit RED-NHS (Amine Reactive) dye, following the manufacturers' instructions (NanoTemper Technologies). All stock protein samples were centrifuged for 10 min prior to setting up the experiment. The labeled NT5B protein was diluted to 2.5 nm final concentration, in the appropriate buffer for each experiment (25 mm HEPES, 150 mm NaCl, 0.05% Tween 20, with either 5 mm CaCl2 or 5 mm EGTA, at pH 7.4 or 6). The nonlabeled recombinant CT5B protein was serially diluted 1:1 in the same buffer (starting concentration of 6 μm) and an equal volume of the labeled protein was mixed with each dilution. The samples were loaded onto MonolithTM NT.115 Standard Treated Capillaries (NanoTemper Technologies) and thermophoresis was measured using a MonolithTM NT.115PICO instrument (NanoTemper Technologies) with NT Control software version 1.0.1 at room temperature with 5 s/30 s/5 s laser off/on/off times, respectively, and 15% LED power and 40% IR-laser (MST) power. Experiments were carried out in triplicate and data from three independently pipetted measurements were analyzed using NT Affinity Analysis software version 2.0.2, using the KD model of fit.
Author contributions
C. R., M. P. L.-C., R. F. C., and T. A. J. data curation; C. R., M. P. L.-C., R. F. C., T. A. J., and C. B. formal analysis; C. R., M. P. L.-C., R. F. C., and T. A. J. validation; C. R., M. P. L.-C., R. F. C., and T. A. J. investigation; C. R., M. P. L.-C., and R. F. C. visualization; C. R. and D. J. T. writing-original draft; C. R. and D. J. T. project administration; C. R., M. P. L.-C., R. F. C., T. A. J., C. B., and D. J. T. writing-review and editing; D. B. S. and M. K. resources; C. B. and D. J. T. supervision; D. J. T. conceptualization; D. J. T. funding acquisition.
Supplementary Material
Acknowledgments
We thank Professor Tim Hardingham (University of Manchester) for critical reading of the manuscript. We acknowledge support from the Electron Microscopy, Biomolecular Analysis and Biological Mass Spectrometry core facilities at the University of Manchester. We acknowledge Diamond for access and support of the Cryo-EM facilities at the UK national electron bio-imaging center (eBIC), proposal EM16619 (3 sessions), funded by the Wellcome Trust, MRC, and BBSRC. We also thank Diamond for access to SAXS beamline B21, proposal mx17773-50. The Wellcome Trust Centre for Cell-Matrix Research, University of Manchester, is supported by core funding from the Wellcome Trust Grant 203128/Z/16/Z.
This work was supported by Medical Research Council Grant MR/R002800/1 and Cystic Fibrosis Foundation Therapeutics Grants THORNT07XXX0 and SUBRAM17I0. The authors declare that they have no conflicts of interest with the contents of this article.
The EM Map Validation Reports are available from the EMData Bank EMBD ID EMD-10264, EMD-10336, and EMD-10339.
This article contains Figs. S1–S5.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- MCC
- mucociliary clearance
- CF
- cystic fibrosis
- COPD
- chronic obstructive pulmonary disease
- vWF
- von Willebrand factor
- SAXS
- small angle X-ray scattering
- SEC-MALS
- size exclusion chromatography multi-angle static light scattering
- PAS
- periodic acid-Schiff
- SPR
- surface plasmon resonance
- MST
- microscale thermophoresis
- PDB
- Protein Data Bank
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Ridley C., and Thornton D. J. (2018) Mucins: the frontline defence of the lung. Biochem. Soc. Trans. 46, 1099–1106 10.1042/BST20170402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Symmes B. A., Stefanski A. L., Magin C. M., and Evans C. M. (2018) Role of mucins in lung homeostasis: regulated expression and biosynthesis in health and disease. Biochem. Soc. Trans. 46, 707–719 10.1042/BST20170455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonser L. R., and Erle D. J. (2017) Airway mucus and asthma: the role of MUC5AC and MUC5B. J. Clin. Med. 6, 112 10.3390/jcm6120112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Button B., Anderson W. H., and Boucher R. C. (2016) Mucus hyperconcentration as a unifying aspect of the chronic bronchitic phenotype. Ann. Am. Thorac Soc. 13, S156–S162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hill D. B., Long R. F., Kissner W. J., Atieh E., Garbarine I. C., Markovetz M. R., Fontana N. C., Christy M., Habibpour M., Tarran R., Forest M. G., Boucher R. C., and Button B. (2018) Pathological mucus and impaired mucus clearance in cystic fibrosis patients result from increased concentration, not altered pH. Eur. Respir. J. 52, 1801297 10.1183/13993003.01297-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davies J. R., Svitacheva N., Lannefors L., Kornfält R., and Carlstedt I. (1999) Identification of MUC5B, MUC5AC and small amounts of MUC2 mucins in cystic fibrosis airway secretions. Biochem. J. 344, 321–330 [PMC free article] [PubMed] [Google Scholar]
- 7. Hovenberg H. W., Davies J. R., Herrmann A., Lindén C. J., and Carlstedt I. (1996) MUC5AC, but not MUC2, is a prominent mucin in respiratory secretions. Glycoconj. J. 13, 839–847 10.1007/BF00702348 [DOI] [PubMed] [Google Scholar]
- 8. Kirkham S., Sheehan J. K., Knight D., Richardson P. S., and Thornton D. J. (2002) Heterogeneity of airways mucus: variations in the amounts and glycoforms of the major oligomeric mucins MUC5AC and MUC5B. Biochem. J. 361, 537–546 10.1042/0264-6021:3610537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thornton D. J., Carlstedt I., Howard M., Devine P. L., Price M. R., and Sheehan J. K. (1996) Respiratory mucins: identification of core proteins and glycoforms. Biochem. J. 316, 967–975 10.1042/bj3160967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grubb B. R., Livraghi-Butrico A., Rogers T. D., Yin W., Button B., and Ostrowski L. E. (2016) Reduced mucociliary clearance in old mice is associated with a decrease in Muc5b mucin. Am. J. Physiol. Lung Cell Mol. Physiol 310, L860–L867 10.1152/ajplung.00015.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roy M. G., Livraghi-Butrico A., Fletcher A. A., McElwee M. M., Evans S. E., Boerner R. M., Alexander S. N., Bellinghausen L. K., Song A. S., Petrova Y. M., Tuvim M. J., Adachi R., Romo I., Bordt A. S., Bowden M. G., et al. (2014) Muc5b is required for airway defence. Nature 505, 412–416 10.1038/nature12807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Desseyn J. L., Buisine M. P., Porchet N., Aubert J. P., and Laine A. (1998) Genomic organization of the human mucin gene MUC5B: cDNA and genomic sequences upstream of the large central exon. J. Biol. Chem. 273, 30157–30164 10.1074/jbc.273.46.30157 [DOI] [PubMed] [Google Scholar]
- 13. Gum J. R. Jr., Hicks J. W., Toribara N. W., Siddiki B., and Kim Y. S. (1994) Molecular cloning of human intestinal mucin (MUC2) cDNA: identification of the amino terminus and overall sequence similarity to prepro-von Willebrand factor. J. Biol. Chem. 269, 2440–2446 [PubMed] [Google Scholar]
- 14. Thornton D. J., Rousseau K., and McGuckin M. A. (2008) Structure and function of the polymeric mucins in airways mucus. Annu. Rev. Physiol 70, 459–486 10.1146/annurev.physiol.70.113006.100702 [DOI] [PubMed] [Google Scholar]
- 15. Bell S. L., Xu G., and Forstner J. F. (2001) Role of the cystine-knot motif at the C-terminus of rat mucin protein Muc2 in dimer formation and secretion. Biochem. J. 357, 203–209 10.1042/0264-6021:3570203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perez-Vilar J., Eckhardt A. E., and Hill R. L. (1996) Porcine submaxillary mucin forms disulfide-bonded dimers between its carboxyl-terminal domains. J. Biol. Chem. 271, 9845–9850 10.1074/jbc.271.16.9845 [DOI] [PubMed] [Google Scholar]
- 17. Perez-Vilar J., and Hill R. L. (1998) The carboxyl-terminal 90 residues of porcine submaxillary mucin are sufficient for forming disulfide-bonded dimers. J. Biol. Chem. 273, 6982–6988 10.1074/jbc.273.12.6982 [DOI] [PubMed] [Google Scholar]
- 18. Ridley C., Kirkham S., Williamson S. J., Davis C. W., Woodman P., and Thornton D. J. (2016) Biosynthesis of the polymeric gel-forming mucin MUC5B. Am. J. Physiol. Lung Cell Mol Physiol 310, L993–L1002 10.1152/ajplung.00046.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ridley C., Kouvatsos N., Raynal B. D., Howard M., Collins R. F., Desseyn J. L., Jowitt T. A., Baldock C., Davis C. W., Hardingham T. E., and Thornton D. J. (2014) Assembly of the respiratory mucin MUC5B: a new model for a gel-forming mucin. J. Biol. Chem. 289, 16409–16420 10.1074/jbc.M114.566679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kesimer M., Makhov A. M., Griffith J. D., Verdugo P., and Sheehan J. K. (2010) Unpacking a gel-forming mucin: a view of MUC5B organization after granular release. Am. J. Physiol. Lung Cell Mol. Physiol 298, L15–L22 10.1152/ajplung.00194.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trillo-Muyo S., Nilsson H. E., Recktenwald C. V., Ermund A., Ridley C., Meiss L. N., Bähr A., Klymiuk N., Wine J. J., Koeck P. J. B., Thornton D. J., Hebert H., and Hansson G. C. (2018) Granule-stored MUC5B mucins are packed by the non-covalent formation of N-terminal head-to-head tetramers. J. Biol. Chem. 293, 5746–5754 10.1074/jbc.RA117.001014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ermund A., Meiss L. N., Rodriguez-Pineiro A. M., Bähr A., Nilsson H. E., Trillo-Muyo S., Ridley C., Thornton D. J., Wine J. J., Hebert H., Klymiuk N., and Hansson G. C. (2017) The normal trachea is cleaned by MUC5B mucin bundles from the submucosal glands coated with the MUC5AC mucin. Biochem. Biophys. Res. Commun. 492, 331–337 10.1016/j.bbrc.2017.08.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Abdullah L. H., Evans J. R., Wang T. T., Ford A. A., Makhov A. M., Nguyen K., Coakley R. D., Griffith J. D., Davis C. W., Ballard S. T., and Kesimer M. (2017) Defective postsecretory maturation of MUC5B mucin in cystic fibrosis airways. JCI Insight 2, e89752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gustafsson J. K., Ermund A., Ambort D., Johansson M. E., Nilsson H. E., Thorell K., Hebert H., Sjovall H., and Hansson G. C. (2012) Bicarbonate and functional CFTR channel are required for proper mucin secretion and link cystic fibrosis with its mucus phenotype. J. Exp. Med. 209, 1263–1272 10.1084/jem.20120562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kesimer M., Ehre C., Burns K. A., Davis C. W., Sheehan J. K., and Pickles R. J. (2013) Molecular organization of the mucins and glycocalyx underlying mucus transport over mucosal surfaces of the airways. Mucosal Immunol. 6, 379–392 10.1038/mi.2012.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shah V. S., Meyerholz D. K., Tang X. X., Reznikov L., Abou Alaiwa M., Ernst S. E., Karp P. H., Wohlford-Lenane C. L., Heilmann K. P., Leidinger M. R., Allen P. D., Zabner J., McCray P. B. Jr, Ostedgaard L. S., Stoltz D. A., Randak C. O., and Welsh M. J. (2016) Airway acidification initiates host defense abnormalities in cystic fibrosis mice. Science 351, 503–507 10.1126/science.aad5589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang X. X., Ostedgaard L. S., Hoegger M. J., Moninger T. O., Karp P. H., McMenimen J. D., Choudhury B., Varki A., Stoltz D. A., and Welsh M. J. (2016) Acidic pH increases airway surface liquid viscosity in cystic fibrosis. J. Clin. Invest. 126, 879–891 10.1172/JCI83922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Y. F., Eng E. T., Nishida N., Lu C., Walz T., and Springer T. A. (2011) A pH-regulated dimeric bouquet in the structure of von Willebrand factor. EMBO J. 30, 4098–4111 10.1038/emboj.2011.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou Y. F., and Springer T. A. (2014) Highly reinforced structure of a C-terminal dimerization domain in von Willebrand factor. Blood 123, 1785–1793 10.1182/blood-2013-11-523639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhou Y. F., Eng E. T., Zhu J., Lu C., Walz T., and Springer T. A. (2012) Sequence and structure relationships within von Willebrand factor. Blood 120, 449–458 10.1182/blood-2012-01-405134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shelton-Inloes B. B., Titani K., and Sadler J. E. (1986) cDNA sequences for human von Willebrand factor reveal five types of repeated domains and five possible protein sequence polymorphisms. Biochemistry 25, 3164–3171 10.1021/bi00359a014 [DOI] [PubMed] [Google Scholar]
- 32. Verweij C. L., Diergaarde P. J., Hart M., and Pannekoek H. (1986) Full-length von Willebrand factor (vWF) cDNA encodes a highly repetitive protein considerably larger than the mature vWF subunit. EMBO J. 5, 1839–1847 10.1002/j.1460-2075.1986.tb04435.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wagner D. D., and Marder V. J. (1983) Biosynthesis of von Willebrand protein by human endothelial cells: identification of a large precursor polypeptide chain. J. Biol. Chem. 258, 2065–2067 [PubMed] [Google Scholar]
- 34. Desseyn J. L., Aubert J. P., Van Seuningen I., Porchet N., and Laine A. (1997) Genomic organization of the 3′ region of the human mucin gene MUC5B. J. Biol. Chem. 272, 16873–16883 10.1074/jbc.272.27.16873 [DOI] [PubMed] [Google Scholar]
- 35. Gum J. R. Jr., Hicks J. W., Toribara N. W., Rothe E. M., Lagace R. E., and Kim Y. S. (1992) The human MUC2 intestinal mucin has cysteine-rich subdomains located both upstream and downstream of its central repetitive region. J. Biol. Chem. 267, 21375–21383 [PubMed] [Google Scholar]
- 36. Lesuffleur T., Roche F., Hill A. S., Lacasa M., Fox M., Swallow D. M., Zweibaum A., and Real F. X. (1995) Characterization of a mucin cDNA clone isolated from HT-29 mucus-secreting cells: the 3′ end of MUC5AC? J. Biol. Chem. 270, 13665–13673 10.1074/jbc.270.23.13665 [DOI] [PubMed] [Google Scholar]
- 37. Meezaman D., Charles P., Daskal E., Polymeropoulos M. H., Martin B. M., and Rose M. C. (1994) Cloning and analysis of cDNA encoding a major airway glycoprotein, human tracheobronchial mucin (MUC5). J. Biol. Chem. 269, 12932–12939 [PubMed] [Google Scholar]
- 38. Lidell M. E., and Hansson G. C. (2006) Cleavage in the GDPH sequence of the C-terminal cysteine-rich part of the human MUC5AC mucin. Biochem. J. 399, 121–129 10.1042/BJ20060443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lidell M. E., Johansson M. E., and Hansson G. C. (2003) An autocatalytic cleavage in the C terminus of the human MUC2 mucin occurs at the low pH of the late secretory pathway. J. Biol. Chem. 278, 13944–13951 10.1074/jbc.M210069200 [DOI] [PubMed] [Google Scholar]
- 40. Ehre C., Rushton Z. L., Wang B., Hothem L. N., Morrison C. B., Fontana N. C., Markovetz M. R., Delion M. F., Kato T., Villalon D., Thelin W. R., Esther C. R. Jr., Hill D. B., Grubb B. R., Livraghi-Butrico A., Donaldson S. H., and Boucher R. C. (2019) An improved inhaled mucolytic to treat airway muco-obstructive diseases. Am. J. Respir. Crit. Care Med. 199, 171–180 10.1164/rccm.201802-0245OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Esther C. R. Jr., Muhlebach M. S., Ehre C., Hill D. B., Wolfgang M. C., Kesimer M., Ramsey K. A., Markovetz M. R., Garbarine I. C., Forest M. G., Seim I., Zorn B., Morrison C. B., Delion M. F., Thelin W. R., et al. (2019) Mucus accumulation in the lungs precedes structural changes and infection in children with cystic fibrosis. Sci. Transl. Med. 11, eaav3488 10.1126/scitranslmed.aav3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tarrant B. J., Le Maitre C., Romero L., Steward R., Button B. M., Thompson B. R., and Holland A. E. (2017) Mucoactive agents for chronic, non-cystic fibrosis lung disease: a systematic review and meta-analysis. Respirology 22, 1084–1092 10.1111/resp.13047 [DOI] [PubMed] [Google Scholar]
- 43. Bax D. V., Bernard S. E., Lomas A., Morgan A., Humphries J., Shuttleworth C. A., Humphries M. J., and Kielty C. M. (2003) Cell adhesion to fibrillin-1 molecules and microfibrils is mediated by α5β1 and αvβ3 integrins. J. Biol. Chem. 278, 34605–34616 10.1074/jbc.M303159200 [DOI] [PubMed] [Google Scholar]
- 44. Rock M. J., Cain S. A., Freeman L. J., Morgan A., Mellody K., Marson A., Shuttleworth C. A., Weiss A. S., and Kielty C. M. (2004) Molecular basis of elastic fiber formation: critical interactions and a tropoelastin-fibrillin-1 cross-link. J. Biol. Chem. 279, 23748–23758 10.1074/jbc.M400212200 [DOI] [PubMed] [Google Scholar]
- 45. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 10.1016/S0006-3495(00)76713-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Danev R., Tegunov D., and Baumeister W. (2017) Using the Volta phase plate with defocus for cryo-EM single particle analysis. Elife 6, e23006 10.7554/eLife.23006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Punjani A., Brubaker M. A., and Fleet D. J. (2017) Building proteins in a day: efficient 3D molecular structure estimation with electron cryomicroscopy. IEEE Trans Pattern Anal. Mach. Intell. 39, 706–718 10.1109/TPAMI.2016.2627573 [DOI] [PubMed] [Google Scholar]
- 48. Punjani A., Rubinstein J. L., Fleet D. J., and Brubaker M. A. (2017) CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 49. Hildebrand A., Remmert M., Biegert A., and Söding J. (2009) Fast and accurate automatic structure prediction with HHpred. Proteins 77, 128–132 10.1002/prot.22499 [DOI] [PubMed] [Google Scholar]
- 50. Meier A., and Söding J. (2015) Automatic prediction of protein 3D structures by probabilistic multi-template homology modeling. PLoS Comput. Biol. 11, e1004343 10.1371/journal.pcbi.1004343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Söding J. (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960 10.1093/bioinformatics/bti125 [DOI] [PubMed] [Google Scholar]
- 52. Zimmermann L., Stephens A., Nam S. Z., Rau D., Kübler J., Lozajic M., Gabler F., Soding J., Lupas A. N., and Alva V. (2018) A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 10.1016/j.jmb.2017.12.007 [DOI] [PubMed] [Google Scholar]
- 53. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., and Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., and Sali A. (2006) Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics Chapter 5, Unit 5.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 56. Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., de Beer T. A. P., Rempfer C., Bordoli L., Lepore R., and Schwede T. (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chang T. H., Hsieh F. L., Zebisch M., Harlos K., Elegheert J., and Jones E. Y. (2015) Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan. Elife 4, e06554 10.7554/eLife.06554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Davies H. S., Singh P., Deckert-Gaudig T., Deckert V., Rousseau K., Ridley C. E., Dowd S. E., Doig A. J., Pudney P. D., Thornton D. J., and Blanch E. W. (2016) The secondary structure and glycosylation of mucus glycoproteins by Raman spectroscopies. Anal. Chem. 88, 11609–11615 10.1021/acs.analchem.6b03095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Thornton D. J., Holmes D. F., Sheehan J. K., and Carlstedt I. (1989) Quantitation of mucus glycoproteins blotted onto nitrocellulose membranes. Anal. Biochem. 182, 160–164 10.1016/0003-2697(89)90735-5 [DOI] [PubMed] [Google Scholar]
- 60. Dong X., Leksa N. C., Chhabra E. S., Arndt J. W., Lu Q., Knockenhauer K. E., Peters R. T., and Springer T. A. (2019) The von Willebrand factor D'D3 assembly and structural principles for factor VIII binding and concatemer biogenesis. Blood 133, 1523–1533 10.1182/blood-2018-10-876300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shiltagh N., Kirkpatrick J., Cabrita L. D., McKinnon T. A., Thalassinos K., Tuddenham E. G., and Hansen D. F. (2014) Solution structure of the major factor VIII binding region on von Willebrand factor. Blood 123, 4143–4151 10.1182/blood-2013-07-517086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xu E. R., von Bülow S., Chen P. C., Lenting P. J., Kolšek K., Aponte-Santamaría C., Simon B., Foot J., Obser T., Schneppenheim R., Gräter F., Denis C. V., Wilmanns M., and Hennig J. (2019) Structure and dynamics of the platelet integrin-binding C4 domain of von Willebrand factor. Blood 133, 366–376 10.1182/blood-2018-04-843615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weidauer S. E., Schmieder P., Beerbaum M., Schmitz W., Oschkinat H., and Mueller T. D. (2009) NMR structure of the Wnt modulator protein Sclerostin. Biochem. Biophys. Res. Commun. 380, 160–165 10.1016/j.bbrc.2009.01.062 [DOI] [PubMed] [Google Scholar]
- 64. Ermund A., Recktenwald C. V., Skjåk-Braek G., Meiss L. N., Onsøyen E., Rye P. D., Dessen A., Myrset A. H., and Hansson G. C. (2017) OligoG CF-5/20 normalizes cystic fibrosis mucus by chelating calcium. Clin. Exp. Pharmacol. Physiol 44, 639–647 10.1111/1440-1681.12744 [DOI] [PubMed] [Google Scholar]
- 65. Fernandez-Petty C. M., Hughes G. W., Bowers H. L., Watson J. D., Rosen B. H., Townsend S. M., Santos C., Ridley C. E., Chu K. K., Birket S. E., Li Y., Leung H. M., Mazur M., Garcia B. A., Evans T. I. A., et al. (2019) A glycopolymer improves vascoelasticity and mucociliary transport of abnormal cystic fibrosis mucus. JCI Insight 4, e125954 10.1172/jci.insight.125954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rambo R. P., and Tainer J. A. (2013) Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 10.1038/nature12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.