Abstract
tRNA fragments (tRFs) and tRNA halves have been implicated in various cellular processes, including gene silencing, translation, stress granule assembly, cell differentiation, retrotransposon activity, symbiosis, apoptosis, and more. Overexpressed angiogenin (ANG) cleaves tRNA anticodons and produces tRNA halves similar to those produced in response to stress. However, it is not clear whether endogenous ANG is essential for producing the stress-induced tRNA halves. It is also not clear whether smaller tRFs are generated from the tRNA halves. Here, using global short RNA-Seq approach, we found that ANG overexpression selectively cleaves a subset of tRNAs, including tRNAGlu, tRNAGly, tRNALys, tRNAVal, tRNAHis, tRNAAsp, and tRNASeC to produce tRNA halves and tRF-5s that are 26–30 bases long. Surprisingly, ANG knockout revealed that the majority of stress-induced tRNA halves, except for the 5′ half from tRNAHisGTG and the 3′ half from tRNAAspGTC, are ANG independent, suggesting there are other RNases that produce tRNA halves. We also found that the 17–25 bases–long tRF-3s and tRF-5s that could enter into Argonaute complexes are not induced by ANG overexpression, suggesting that they are generated independently from tRNA halves. Consistent with this, ANG knockout did not decrease tRF-3 levels or gene-silencing activity. We conclude that ANG cleaves specific tRNAs and is not the only RNase that creates tRNA halves and that the shorter tRFs are not generated from the tRNA halves or from independent tRNA cleavage by ANG.
Keywords: RNA metabolism, RNA processing, transfer RNA (tRNA), ribonuclease, stress, angiogenin, gene regulation, noncoding RNA, small RNA, tRNA fragment
Introduction
In the last decade, advances in next-generational sequencing have sparked the discovery of groups of previously unnoticed small noncoding RNAs (1, 2). One among these groups includes small RNAs derived from tRNAs, so-called tRNA-derived RNA fragments (tRFs, tDRs)3 (3) or tRNA-derived small RNAs (tsRNAs) (4). tRFs have been discovered in multiple organisms and tissues (5–10). Based on the current knowledge about tRNA fragments, there are six major groups (Fig. 1A): (a) 5′ tRNA halves, (b) 3′ tRNA halves, (c) tRF-3s, (d) tRF-5s, (e) tRF-1s, and (f) misc-tRFs. (a and b) The tRNA halves are also called tRNA-derived stress-induced RNAs (tiRs) that are produced by cleavage at anticodon arm of the mature tRNAs, linked with various stress response in diverse organisms (11–17). The two halves produced by tRNA cleavage are named respectively as 5′ tRNA halves and 3′ tRNA halves. 5′ tRNA halves have been shown to inhibit global translation and promote stress granule assembly (18, 19), whereas 3′ tRNA halves interact with cytochrome c and protect cells from stress-induced apoptosis (20). (c) Similar to 3′ tRNA halves, tRF-3s also end at the 3′ end of mature tRNAs, but start by cleavage within the T-loop. Although these 17- to 25-nt tRF-3s (major peaks at 18 nt and 22 nt) could load into Argonaute complexes to perform microRNA-like gene silencing activities (5, 21–23), they are still generated in Dicer- and Drosha-deficient cells (5, 21). tRF-3s have also been implicated in retrotransposon inhibition (24) and ribosome biogenesis (25). (d) tRF-5s start at the 5′ end of mature tRNAs and end before the anticodon loop. 17- to 25-nt (“short”) tRF-5s have been shown to inhibit protein translation (26–28) and also have been suggested to play potential microRNA-like functions (5). Another distinct group of 26- to 30-nt (“long”) tRF-5s, also called tRNA-derived PIWI-interacting RNAs (td-piRNAs), could enter PIWI complex to regulate downstream pathways (29–31). (e) tRF-1s or 3′U tRFs correspond to the 3′ trailer from the precursor tRNAs during tRNA maturation process. The 5′ end of tRF-1 results from RNaseZ cleavage and the 3′ end correlates with termination by PolIII transcription. tRF-1s have been linked with cancer (3, 32, 33). (f) Misc-tRFs include the other types of tRFs that are not represented by the above groups, such as internal tRFs (i-tRFs, also called tRF-2s) that start and end within the mature tRNAs. Internal tRFs that start at D-loop and end at variable loop or T-loop displace YBX1 from 3′UTR of oncogenic transcripts to suppress breast cancer progression (34). Overall there is a growing interest in the functions of tRNA fragments, and evidence so far suggests they might play divergent roles in different biological pathways via specific mechanisms.
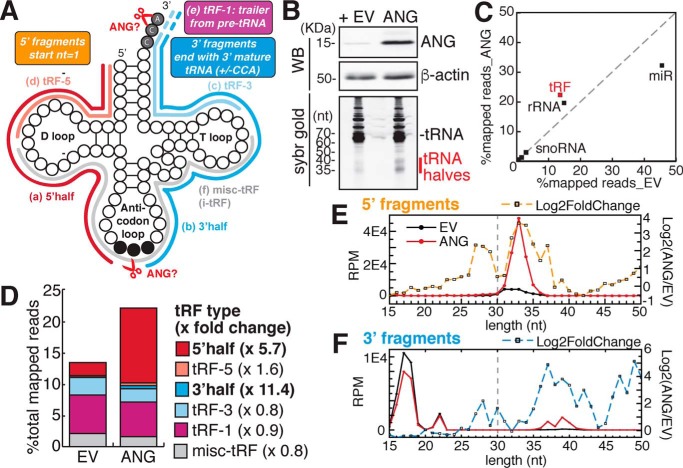
Figure 1.
Angiogenin overexpression increases tRNA halves. A, different types of tRNA fragments. Refer to the Introduction for the description of each type. Potential cleavage sites by ANG are indicated by scissors. B, overexpression of ANG in HEK293T cells (EV, empty vector) induces tRNA half bands. Upper panel, specificity of ANG antibody was shown in Fig. S1B (WB, Western blotting). Lower panel, total RNA was separated on 15% Urea-PAGE and stained by SYBR Gold. C–F, ANG overexpression specifically increases tRNA halves, shown by small RNA-Seq analysis averaged from three biological replicates. C, scatter plot shows log10 of read counts mapped to each category from small RNA-Seq analysis (x-axis: empty vector control, y-axis: ANG overexpression). D, ANG overexpression specifically increases overall reads of 5′ halves and 3′ halves. Here 5′ half refers to 5′ fragments of >30 nucleotides in length, and tRF-5 refers to <30 nucleotides (corresponding to vertical dashed gray line in panel E); similarly, 3′ half and tRF-3 are separated by length (dashed gray line in panel F). E and F, length distribution of all tRF reads that map to 5′ or 3′ end of mature tRNAs by small RNA-Seq (y-axis: reads per mapped million reads, black line: empty vector, red line: ANG overexpression). Dashed line shows log2 -fold change upon ANG overexpression for tRFs of specific length (y-axis on the right).
Despite the emerging interest in tRFs, how specific tRFs are generated still remains largely unknown. Most studies have focused on the biogenesis of the stress-induced tRNA halves. In human cells, stress-induced cleavage in tRNA has been mostly attributed to enzymatic cleavage by angiogenin (ANG or RNase 5), a member of the vertebrate-specific RNase A family (16–20, 35–37). ANG is widely expressed in most tissues (38) and has been linked to angiogenesis, hematopoiesis, oncogenesis, inflammation, and immunity (39); loss-of-function mutations in ANG have been implicated in neurodegenerative diseases such as ALS and Parkinson's disease (39). Under normal conditions, cellular ANG is inactive because of high-affinity binding with RNH1 (RNase inhibitor 1) (40). Under stress conditions, cytoplasmic ANG is activated by dissociation from RNH1, which triggers tRNA cleavage (41). Recombinant ANG added directly to culture media increased tRNA anticodon loop cleavage in multiple cell lines (16–20, 35–37); interestingly, ANG cleavage did not significantly reduce the overall tRNA abundance. One report also suggests stress-activated ANG could cleave the CCA at the 3′ end of mature tRNAs quickly, even before the cleavage at the anticodon loop (42) (Fig. 1A). Most of these previous studies quantified the effect of ANG cleavage only on a subset of tRNAs by Northern blot analysis, so a global view is still lacking. On the other hand, biogenesis of stress-independent tRFs is poorly known, except a recent report of ANG-dependent tRNA halves in nonstressed sex hormone–positive cells (43). We have shown that most 17- to 25-nt tRF-3s are generated by uncharacterized Dicer-independent pathways (5, 21), and it has not been tested whether tRNA halves could serve as precursors for these and other shorter tRFs. This is a possibility, especially considering that ANG could directly generate tRF-3 as suggested by in vitro reactions (44) or detach amino acids from tRNA 3′ CCA end (42), which might be a prerequisite for generating a fraction of tRF-3s.
In this report, we systematically characterize angiogenin's role in tRNA cleavage in nonstressed and stressed human cell lines. We test whether ANG is sufficient and/or required for tRNA cleavage. By small RNA-Seq, we profiled the global tRNA cleavage pattern by ANG overexpression, which leads to specific increase of both tRNA 5′ halves and 3′ halves. Although increase of tRNA 5′ halves correlates with an increase in the long (26–30 nt) tRF-5s, tRF-1s, short (17–25 nt) tRF-5s and tRF-3s were unaffected by ANG cleavage. Surprisingly, the majority of tRFs were unaffected by ANG knockout and only a few specific tRNA halves (5′ tRNAHisGTG and 3′ tRNAAspGTC) were decreased by loss of ANG. CRISPR knockout of ANG cells still displayed arsenite stress-induced tRNA anticodon cleavage and translational arrest, suggesting other RNase(s) contribute extensively to the overall stress-induced tRNA cleavage. We conclude that human angiogenin is sufficient but not essential for tRNA anticodon cleavage and that the short tRF-3s and tRF-5s are generated by ANG-independent mechanisms.
Results
ANG generates tRNA halves
To identify which tRNAs are cleaved and how by ANG, we overexpressed full-length human ANG in HEK293T cells (Fig. 1B and Fig. S1A). Western blotting in these cells confirms that a 14-kDa mature ANG protein is overexpressed in the cell lysate (Fig. 1B and Fig. S1B). We noted ANG mRNA was overexpressed over 500-fold (Fig. S1A), whereas ANG protein appears to be overexpressed in cell lysates to ∼10- to 20-fold (Fig. 1B), which could be because of the secretion of ANG protein from the cells. SYBR Gold staining of total RNA detects RNA bands around 35 and 40 nucleotides upon ANG overexpression (Fig. 1B), consistent with the tRNA halves in previous reports (17, 36, 45).
To characterize the tRNA fragmentation pattern induced by ANG overexpression, we performed small RNA-Seq of 15- to 50-nt–long RNAs from total RNA (workflow shown in Fig. S1C). This size range will capture tRNA fragments but not the full-length tRNAs (Fig. S1D). Sequencing of tRNA fragments is known to be sensitive to both terminal and internal modifications on the RNA molecules. To make quantitative comparison of tRNA fragments between two conditions, we assumed the cloning efficiency for each tRF is not changed by different conditions of cell treatment. This is why we compare relative levels of the same tRF across conditions, rather than the levels of different tRFs in a given condition. ANG overexpression specifically increased counts of fragments derived from tRNAs, and fragments from ribosomal RNAs (rRNAs) to a lesser extent (Fig. 1C). The slight increase in rRNA fragment reads is consistent with previous reports on ANG's role in rRNA transcription and processing (46–48). Despite the relative global decrease in microRNA reads upon ANG overexpression (Fig. 1C), no microRNA showed significant decrease upon ANG over-expression by differential analysis using DESeq2 (49) on the union of microRNAs and tRFs (Table S2), which suggests ANG is unlikely to decrease specific microRNA levels.
Among the different types of tRFs (Fig. 1A), 5′ halves (≥30 nt) increase by 5.7-fold upon ANG overexpression and tRF-5s (<30 nt) are only mildly increased (Fig. 1D). Similarly, 3′ halves (≥30 nt) increase by 11-fold, whereas tRF-3s (<30 nt) are not increased (Fig. 1D). Here we define 3′ fragments irrespective of CCA presence to account for potential cleavage of the 3′ CCA at CA dinucleotide by ANG (42). Despite this precaution, it is worth noting that most of the 3′ fragments contain intact 3′ CCA sequence (examples shown in Fig. S2A). tRF-1s, produced from the trailer sequence through cleavage by RNase Z, was also unchanged by ANG overexpression (Fig. 1D). When we looked at tRFs by size, ANG overexpression most specifically increases 5′ tRNA halves in the size range of 31–36 nucleotides (Fig. 1E), and 3′ halves of 36–41 nucleotides (Fig. 1F). In contrast, tRF-5s and tRF-3s of 17–25 nt were not much affected by ANG (Fig. 1, E and F, dashed line represents log2-fold change). Overall, results from ANG overexpression suggest that ANG does not have a role in producing tRF-1s and the short (17–25 nt) tRF-3s and tRF-5s. Interestingly, long tRF-5s of 26–30 nt (also called td-piRNAs) were also increased by ANG overexpression (Fig. 1D), although it is not clear whether they are directly created by cleavage in the anticodon stem or are further processed from the 3′ end of the 5′ tRNA halves.
ANG specifically increases tRNA halves from a subset of parental tRNAs
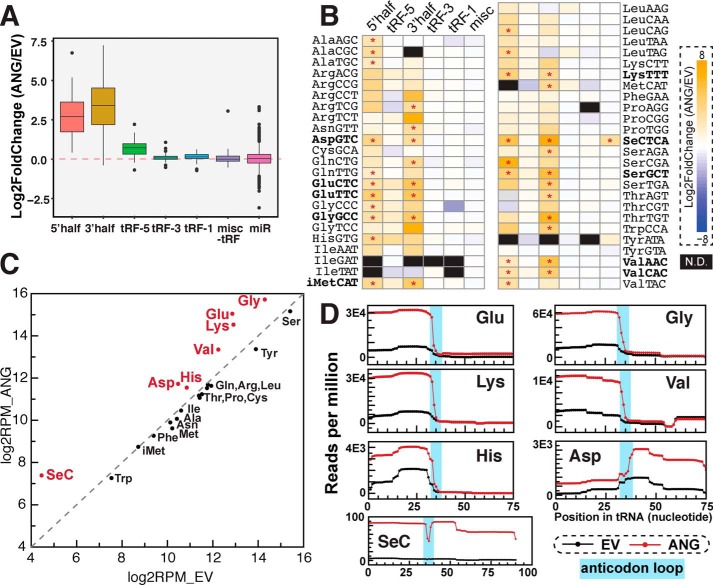
To analyze whether ANG cleaves particular groups of tRNAs, we grouped tRNA fragments by the parental tRNAs. ANG overexpression up-regulates most of the tRNA halves detected by small RNA-Seq (Fig. 2, A and B). Interestingly, both 5′ and 3′ halves from certain tRNAs (including tRNAGlu, tRNAGly, tRNAAsp, tRNAVal, tRNALys, tRNASer, tRNAiMet, and tRNASeC), and 5′ half from tRNAHis, are up-regulated significantly (adjusted p value less than 0.05) upon ANG overexpression (Fig. 2B). When we sum up the different types of tRFs by their parental tRNAs, a group of tRNAs (tRNAGly, tRNAGlu, tRNALys, tRNAVal, tRNAHis, tRNAAsp, and tRNASeC) are clearly more ANG-responsive than other tRNAs (Fig. 2C). Using Northern blotting, we validated that 5′ halves from tRNAGluCTC and tRNAValmAC are indeed increased by ANG overexpression (Fig. S3A and B). The 5′ tRNA halves have much higher cloning frequency than 3′ halves for most of these tRNAs (e.g. tRNAGlu, tRNAGly, tRNAHis, tRNALys, tRNAVal) in both control cells and ANG overexpression cells (Fig. 2D and Fig. S2B). This could be a result of higher stability or higher clonability of the 5′ halves (further discussed in “Discussion”). Furthermore, these ANG-responsive tRNAs (tRNAGly, tRNAGlu, tRNALys, tRNAVal, tRNAHis, tRNAAsp, and tRNASeC) also have higher relative levels of tRNA halves even without any ANG overexpression (Fig. S2B), corroborating the existence of tRNA halves in nonstressed cells. Why these tRNAs are more responsive to ANG is later discussed and awaits future investigation.
Figure 2.
tRNA fragments up-regulated by angiogenin overexpression. A–D, tRF changes upon ANG overexpression by small RNA-Seq. A and B, tRNA halves are increased by ANG overexpression. A, Box-and-whisker plot showing relative -fold change of reads per million mapped reads on log2 scale of ANG overexpression compared with the empty vector control (averaged from three biological replicates). tRFs are grouped by types and parental tRNAs. Box plot shows median value as middle line, first and third quartile as box outline, and data range (excluding outliers represented by individual dots) as whiskers. B, heat map shows log2 -fold change of tRFs by ANG overexpression. tRFs are grouped by tRF types and parental tRNAs. Significant changes determined by DESeq2 (adjusted p value less than 0.05) are labeled by asterisks. N.D., not detected. C and D, ANG overexpression increases tRF reads from certain parental tRNAs, including tRNAGlu, tRNAGly, tRNALys, tRNAVal, tRNAHis, tRNAAsp, and tRNASeC. For C, tRF reads (all types) were summed by parental tRNAs (x-axis: empty vector, y-axis: ANG overexpression). For D, coverage plot shows where accumulated tRF reads (excluding tRF-1s) map to the parental mature tRNAs (black line: empty vector control, red line: ANG overexpression). Anticodon loop region for each tRNA (predicted by tRNA covariance model -fold in GtRNAdb) is highlighted in cyan.
Short tRF-3s and tRF-5s are not increased in parallel with the tRNA halves
Short tRFs in the size range compatible with Argonaute loading (17–25 nt) were predicted to play microRNA-like functions because of their direct association with Argonaute proteins by PAR-CLIP (photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation) and CLASH (cross-linking, ligation and sequencing of hybrids) experiments (5). Recently we showed three short tRF-3s from tRNALeu and tRNACys can enter RISC (RNA-induced silencing complex) to repress target genes and are generated in a Dicer-independent mechanism (21). To examine whether a tRNA half could be a precursor to a shorter tRF, we plotted 5′ or 3′ fragments by length from the ANG-responsive tRNAs (tRNAGlu, tRNAGly, tRNALys, tRNAVal, tRNAAsp, and tRNAHis) and another two tRNAs that produce abundant tRF-3s (tRNASer and tRNALeu) (Fig. S2C). For the ANG-responsive tRNAs, the increase of a tRNA half did not lead to an increase of a smaller tRF from the same tRNA. For example the 23-nt tRF-5s from tRNAVal did not change upon ANG overexpression, despite the significant increase in the corresponding 5′ tRNA half; similarly, increase of 3′ half in tRNAAsp did not lead to increase of its 22-nt tRF-3. For the other tRNAs (e.g. tRNASer and tRNALeu) that did not produce abundant tRNA halves, shorter tRF-3s were not increased by ANG (Fig. S2C). Together with the above analysis showing global tRF-3 levels unaltered by ANG overexpression (Figs. 1F and 2A and Fig. S2C), this suggests there is no precursor–product relationship between a tRNA half and a smaller tRF (17–25 nt), which are most likely generated by different biogenesis pathways.
All tRFs, including tRNA halves, are unchanged in unstressed ANG knockout cells
To determine whether endogenous ANG is essential for generating tRNA halves, we knocked out the ANG gene in HEK293T and U2OS cells by Cas9/CRISPR technology. ANG (also known as RNase 5) and RNase 4 are transcribed from the same promoter and each protein is translated from one unique exon; therefore, we designed guide RNA against the ANG-specific exon to deplete ANG specifically (scheme shown in Fig. S4A). Genomic PCR spanning the ANG-specific exon shows that we obtained several homozygous clones that have deleted the majority of ANG coding sequence (Fig. 3A), further confirmed by Sanger sequencing (Fig. S4B) and quantitative RT-PCR (Fig. S4C). Immunoblotting by ANG-specific antibody (Fig. S1B) confirmed the complete loss of ANG protein in these clones (Fig. 3, B and C). Interestingly, RNase 4 transcript level is increased 4- to 6-fold in ANG KO clones (Fig. S4C), without affecting RNase 4 protein (Fig. S4D).
Figure 3.
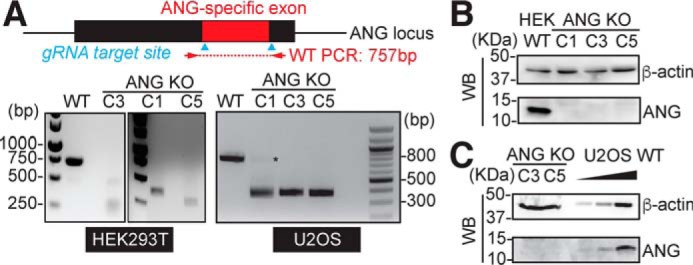
Characterization of angiogenin knockout cells. A, PCR of genomic DNA confirms homozygous deletion of majority of ANG coding sequence in HEK293T and U2OS. Guide RNA target sites are shown by cyan arrows, and primer pair (indicated by red arrow) spans the ANG-specific exon (expected amplicon in WT is 757 bp). U2OS clone 1 was excluded in later experiments because of the residual WT band detected by PCR (indicated by asterisk). B and C, Western blotting to confirm loss of ANG protein in the ANG KO clones (B for HEK293T and C for U2OS). B, 20 μg whole cell lysate for HEK293T WT and KO cells were loaded. C, 30 μg whole cell lysate for ANG KO clones were compared with a gradient amount (1.5, 6, and 30 μg) of WT U2OS lysate.
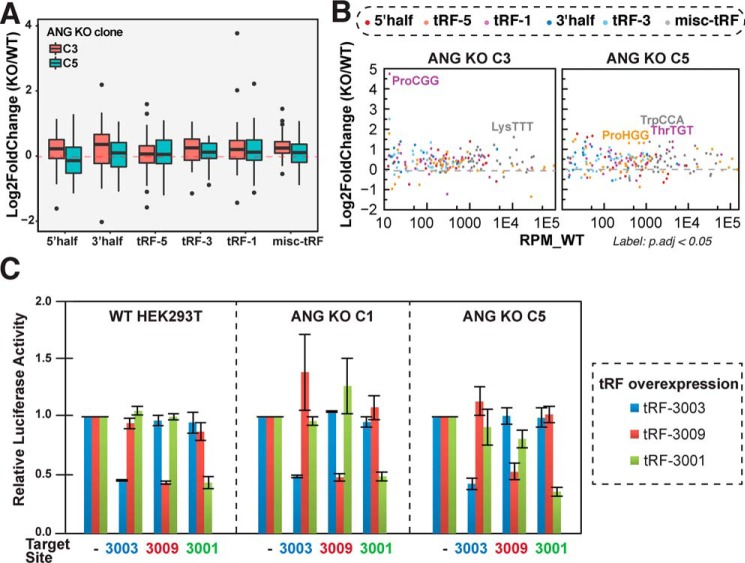
Small RNA profiling of ANG knockout cells suggests there is no global change in any of the six types of tRFs at endogenous levels (without stress) (Fig. 4A). There are also no significant changes in specific tRFs that are consistent between different ANG KO clones (Fig. 4B). In particular, tRF-3 levels are unaltered in ANG KO cells (Fig. 4, A and B). We have earlier shown that tRF-3s induced by overexpressing the parental tRNA could specifically repress a target reporter bearing tRF-complementary sequence in the 3′UTR (21). Using this experimental system, we compared the gene silencing activity of three tRF-3s, tRF-3001, -3003, and -3009 (naming based on tRFdb) (50), in both WT and ANG knockout cells (Fig. 4C). The overproduced tRF-3s robustly and specifically repressed the luciferase reporter that bears a matching target site (Fig. 4C, left panel), as shown previously (21). Such tRF-3–dependent repression continues in the ANG knockout cells (Fig. 4C, middle and right panels), confirming that functional tRF-3 can be produced in the absence of ANG.
Figure 4.
tRF-3s are unchanged in ANG KO cells. A and B, changes of tRFs in ANG KO cells without stress by small RNA-Seq (A, box-and-whisker plot; B, MA plot). Y-axis shows relative -fold change of reads per million mapped tRF reads on log2 scale of ANG knockout cells compared with the U2OS WT control (averaged from two biological replicates). B, significant changes determined by DESeq2 (adjusted p value less than 0.05) are labeled. C, gene silencing activity of three tRF-3s (tRF-3003, -3009, and -3001) were measured by Dual-Luciferase assay in WT HEK293T cells and ANG KO cells (two clones C1 and C5). tRFs were overexpressed by corresponding parental tRNAs and relative repression was observed when 3′UTR of Renilla luciferase gene has a paired complementary tRF target site (relative to no target site luciferase gene). Error bars represent S.D. from three biological replicates.
ANG is dispensable for translation and proliferation arrest induced by high-concentration arsenite stress
ANG has been proposed to be the enzyme responsible for the generation of tRNA halves in stressed human cells (16, 17). Similar to previous studies, we stressed U2OS cells with high concentration (1 mm) of sodium arsenite (SA) for 1 h, which has been shown to induce abundant tRNA halves and inhibit translation (17, 19, 42, 45). Indeed the sodium arsenite stress-induced tRNA cleavage and produced tRNA halves that can be detected by SYBR Gold (Fig. 5A and Fig. S5A). In addition, high concentration (0.2 mm or 1 mm) of sodium arsenite treatment leads to complete protein translational arrest in both WT and ANG KO cells as measured by puromycin pulse labeling (Fig. 5B and Fig. S5B), suggesting ANG is dispensable for the translational arrest induced by high concentration of arsenite. Consistent with previous observations, WT cells did not show much translational repression at a lower concentration (50 μm) of arsenite for 1 h (Fig. S5C) (42). Although ANG KO cells displayed mild translational repression at 50 μm of arsenite (Fig. S5C), suggesting ANG KO cells might be more sensitive to low concentration of arsenite, the cell growth arrest at 24 h was similar in WT and ANG KO cells (Fig. 5C) regardless of arsenite concentrations (Fig. S5C). Thus, ANG is not required for stress-induced growth arrest.
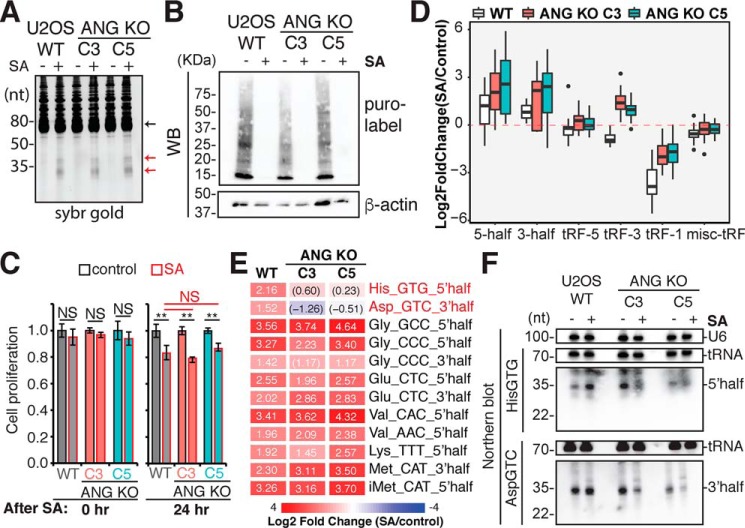
Figure 5.
ANG-dependent and ANG-independent stress response. A–F, WT and ANG KO U2OS cells were treated with 1 mm sodium arsenite for 1 h to identify ANG-dependent stress response. A, SYBR Gold staining of 5 μg total RNA detected arsenite-induced tRNA half bands (indicated by red arrows) in both WT and ANG KO U2OS cells (black arrow points to tRNA band). B, puromycin pulse labeling was used to detect translation arrest. C, cell proliferation immediately (0 h) and 24 h after SA treatment was measured by MTT assay and normalized to WT control cells. Error bars represent S.D. from three biological replicates. Student's t test was performed between control and SA for each condition, or between WT and ANG KO (**, p < 0.05). D and E, small RNA-Seq was used to quantify stress-responsive tRFs (D, box-and-whisker plot; E, heat map). D, y-axis shows relative -fold change of stress versus control on log2 scale in WT and ANG knockout cells (two clones C3 and C5). E, arsenite-induced tRNA half changes in WT and ANG KO cells. Only significant changes in WT are shown here (determined by DESeq2, adjusted p value less than 0.05). Numbers inside each box show Log2 -fold change (parentheses indicate p.adj > 0.05, otherwise p.adj ≤ 0.05). For a more complete list of tRNA halves, refer to Table S3. F, Northern blotting of ANG-dependent stress-induced tRNA halves, 5′ tRNAHisGTG half and 3′ tRNAAspGTC half, in WT and ANG KO U2OS cells.
ANG is required for generating specific tRNA halves induced by high-concentration arsenite
Upon ANG overexpression we observed significant and specific increase of both 5′ and 3′ tRNA halves from multiple isoacceptors (Figs. 1 and 2). However, to our surprise, the stress-induced tRNA half bands did not diminish in ANG KO cells (Fig. 5A). Thus, although ANG can cleave tRNAs in the anticodon loop, most of the tRNA cleavage seen after high-concentration arsenite-induced stress is ANG independent.
To identify whether any stress-induced tRNA halves were dependent on ANG, small RNA-Seq was carried out in WT and ANG KO cells with and without sodium arsenite stress. To avoid normalization artifacts from arsenite-induced changes in other types of small RNAs (such as rRNA fragments and miRs), total reads mapped to tRNAs (all tRFs) were used for normalization. Small RNA-Seq revealed a global up-regulation of 5′ halves and 3′ halves by SA stress in WT cells (Fig. 5D and Fig. S6, A and B), consistent with our SYBR Gold staining (Fig. 5A) and previous publications (17, 19, 42, 45). Such induction remained the same in ANG KO cells (Fig. 5D). Consistent with the SYBR Gold staining (Fig. 5A) and global trend by small RNA-Seq (Fig. 5D), differential analysis reveals that most stress-induced tRNA halves (such as from tRNAGlu, tRNAGly, tRNALys, and tRNAVal) are still up-regulated to similar degree in ANG KO cells (Fig. 5E and Table S3). However, there are specific tRNA halves (mainly 5′ half from tRNAHisGTG and 3′ half from tRNAAspGTC) that are significantly induced by SA stress in WT but not in ANG KO cells (Fig. 5E and Table S3). The role of ANG in producing the 5′ half of tRNAHisGTG and 3′ half of tRNAAspGTC were confirmed by Northern blotting, which also shows that the levels of the parental tRNAs are not changed by arsenite or by ANG KO (Fig. 5F and Fig. S7). Interestingly, these fragments were decreased by arsenite treatment in ANG KO cells, suggesting unknown tRNA half destabilizing/degradation pathway(s) might be more active in the ANG KO cells upon stress. Meanwhile, Northern blotting for 5′ tRNA halves from tRNAGluCTC and tRNAGlyGCC still shows similar levels of induction by high concentration arsenite in ANG KO cells (Fig. S6C). Altogether our analysis reveals that specific stress-induced tRNA halves are altered in ANG KO cells, and the majority of stress-induced tRNA halves can be generated by ANG-independent pathways.
Interestingly, short tRF-3s were slightly decreased by arsenite stress in WT cells but were increased globally by arsenite stress in ANG KO cells (Fig. 5D and Fig. S6A). This suggests that ANG may have a role in regulating the levels of tRF-3s following stress. As a side-note, tRF-1s are globally down-regulated 8- to 16-fold by arsenite stress, which is not ANG dependent (Fig. 5D). The mechanism for this decrease after stress is unclear.
Discussion
In this study, we profiled tRNA cleavage pattern after overexpression or knockout of human ANG. The results suggest ANG can induce but is not required for tRNA cleavage to produce tRNA halves. It has been known that ANG is specific toward tRNA whereas it has rather low RNase activity toward other RNAs in vitro (51), and such specificity was explained by ANG's unique structural features (52, 53). However, it was not entirely clear whether ANG cleaves all tRNAs in cells equally well. Here we show by overexpressing human ANG that its cleavage has specificity toward certain tRNAs, including tRNAGlu, tRNAGly, tRNALys, tRNAVal, tRNAHis, tRNAAsp and tRNASeC (Fig. 1 and Fig. 2). This is consistent with a previous report showing some specificity of recombinant ANG toward certain tRNAs by a customized tRF microarray approach (45) and another report showing that ANG does not produce halves from tRNATyr (16). Our results largely agree with the previous report (45) showing ANG cleaves most but not all tRNAs, but there are slight differences. For example, we found 5′ half from tRNAValmAC is increased by ANG overexpression by small RNA-Seq (Fig. 2B) and Northern blotting (Fig. S3B), which was negative by the microarray approach (45). Such differences could be because of differences in experimental cell lines, ANG overexpression approaches, and detection methods. Base resolution information from small RNA-Seq has its own advantages for tRF detection because tRFs often have very similar sequences that might not be distinguishable by hybridization-based methods. Small RNA-Seq might also offer better sensitivity, since 5′ half from tRNASeC detected by small RNA-Seq falls below detection limit by Northern blotting (Fig. S3C). Overall, the apparent preference for ANG cleavage could come from a combination of (a) ANG's substrate specificity, which could be affected by base sequence (54) and tRNA modifications (55–58) and (b) higher stability of these tRNA halves. 5′ halves from tRNAGly and tRNAGlu have recently been shown to have higher stabilities because of dimerization (59). It is also interesting that cloning frequency of the 5′ half is higher than the corresponding 3′ half for most tRNAs except tRNAAsp, suggesting that either the stability or the cloneability of the two halves are different. This might be because of the 3′ attachment of amino acids on 3′ halves that prevent their cloning (43). The effect of all these factors on tRNA half-induction by ANG (and other RNases) needs further mechanistic investigation.
tRNA regulation is important in stress biology (60, 61) and stress-induced tRNA cleavage seems to be a conserved feature. Oxidative stress (17, 62), hyperosmotic stress (45), nutrient deprivation, ER stress (63), virus infection (64, 65), and other types of stress increase the levels of tRNA halves. Stress-induced tRNA halves have been suggested to be signaling molecules, rather than by-products to reduce tRNA levels (66). Indeed, tRNA halves have been shown to play various roles in stress pathways. For example, 5′ halves from tRNAAla and tRNACys could interact with YBX-1 to inhibit protein translation and promote stress granule formation independent of eIF2 phosphorylation (17–19). Interestingly, these two 5′ tRNA halves can form stable intermolecular G-quadruplex structure (67), and yet neither of them was induced by sodium arsenite in our experiment. In addition, tRNA halves (especially 3′ halves) from various tRNAs interact with cytochrome c and protect cells from apoptosis during stress (20). Recently stress-induced 3′ half from tRNAThr in Trypanosoma brucei has been found to stimulate translation (68). It is highly likely that tRNA halves have divergent functions and many tRNA halves (including the ones we identify from selenocystein) have not been investigated in details.
Our results suggest ANG is not the only RNase that mediates stress-induced tRNA cleavage. This is not surprising because RNase A family is vertebrate-specific, yet stress-induced tRNA cleavage is a well-conserved phenomenon in organisms that do not have RNase A. For example, In Tetrahymena, Saccharomyces cerevisiae, and Arabidopsis, stress-induced generation of tRNA halves has been attributed to the RNase T2 family (12, 69, 70). In mammalian cells, other RNases have also been described to cleave tRNAs, such as Dicer (2, 22, 71), RNase 1 (37), RNase L (74), and Schlafen13/SFLN13 (75). RNase L was suggested to act independently from ANG by cleaving anticodon loop of tRNAPro, tRNAHis, and tRNAGln (74). Whether different RNases are regulated by different stresses and whether one RNase could have different specificity in different cell types still needs to be determined.
Increasing evidence suggests tRFs and tRNA halves exist in conditions beyond stress. In particular, 3′ tRNA fragments that are 17–25 bases long, tRF-3s, have been detected at high abundance, enter into Argonaute complexes, and implicated in important biological functions including gene silencing, retrotransposon silencing, ribosome biogenesis, and recently symbiosis (5, 21, 23–25). These tRF-3s are clearly not regulated by ANG in unstressed cells from our analysis; further investigation is required to identify factors involved in the biogenesis of these tRF-3s. The results also argue against the tRNA halves serving as precursors for the biogenesis of the shorter tRFs that enter into Argonaute complexes.
ANG overexpression specifically induces tRNA halves from tRNAGlu, tRNAGly, tRNALys, tRNAVal, tRNAHis, tRNAAsp, and tRNASeC (Fig. 2), whereas ANG knockout in stressed cells decreases tRNA halves from tRNAHis and tRNAAsp (Fig. 5). Recently, tRNA halves including both halves of tRNAHisGTG and tRNAAspGTC (named as SHOT-RNAs, sex hormone–dependent tRNA-derived RNAs) were found in estrogen or androgen receptor–positive cells and the 5′ halves enhance cell proliferation (43). Notably, levels of two SHOT-RNAs (5′ half of tRNAHisGTG and 5′ half of tRNAAspGTC) were decreased by ANG knockdown in BT-474 breast cancer cells (43), suggesting ANG's role in nonstressed sex hormone receptor–positive cells. More recently, 5′ tRNA halves were found to be up-regulated during stem cell differentiation and appeared to be ANG independent (76). Abundant tRNA halves have also been detected in body fluids (e.g. blood/serum, sperm, cerebrospinal fluid, urine) and exosomes (77–83). Intriguingly, 5′ halves from tRNAGly, tRNAGlu, and tRNAVal in mouse sperm are responsive to paternal diet change and mediate transgenerational change to the offspring (84, 85). Whether ANG or another RNase is involved in the generation of these tRNA halves in body fluids and exosomes is unclear.
Another distinct group of tRFs is tRF-5s of 26–30 nt, also called td-piRNAs, that interact with PIWI proteins (29–31). td-piRNA from tRNAGlu is highly expressed in human monocytes and could be down-regulated by IL-4. This td-piRNA complexed with PIWIL4 plays a role in epigenetic regulation to inhibit CD1A transcription (29). This td-piRNA and the corresponding tRNA half are both increased by ANG overexpression (Fig. S2C). Because of the concurrent increase in specific tRNA halves and the tRF-5s of 26–30 nt from the same tRNAs, our results suggest td-piRNAs might be derived from tRNA halves, as recently shown for two td-piRNAs in Bombyx mori germ cells (30). Future studies about tRF and tRNA half biogenesis and function would bring new insights into this new category of noncoding RNAs.
Experimental procedures
Cell culture, treatment, and proliferation assay
HEK293T and U2OS cells were obtained from ATCC and maintained in HyClone Dulbecco's High Glucose Modified Eagles Medium (GE Healthcare Life Sciences, no. SH30081.01) with 10% FBS. For sodium arsenite treatment, U2OS cells were grown to ∼80% confluency and replaced with fresh media, supplemented with 0.05–1 mm (final concentration in water) sodium arsenite (Sigma, no. S7400) in CO2 incubator at 37 °C for 1 h. Cell proliferation was measured with CellTiter 96 Non-Radioactive Cell Proliferation Assay kit (Promega, no. G4100) at 590-nanometer absorbance on plate reader (BioTek Synergy HT) according to manufacturer's instructions.
Plasmid construction and transfection
Human ANGIOGENIN cDNA sequence (corresponding to 147 amino acids, UniProt P03950) was PCR amplified from CCSB human ORFeome collection v5.1 (forward primer, ATGGTGATGGGCCTGGGCGTT; reverse primer, TTACGGACGACGGAAAATTGACTG) and cloned into pcDNA3.1 (digested by BamHI and XhoI) by In-Fusion HD Cloning Plus (Takara Bio, no. 638910). Transient overexpression of pcDNA3.1-ANG and control empty plasmid was performed with Lipofectamine 2000 (Thermo Fisher, no. 11668027) for 72 h accordingly to manufacturer's instructions. tRNA overexpression and luciferase reporter assay in HEK293T cells was performed as previously (21).
CRISPR knockout cell generation and confirmation
CRISPR knockout was generated following previous protocol with slight modifications (86). WT U2OS and HEK293T cells were plated in 6-well plates at ∼80% confluency and co-transfected with human codon-optimized Cas9 plasmid (Addgene, no. 41815), two guide RNA (gRNA)–containing plasmids, and puromycin selection plasmid pPUR. Two gRNAs were cloned separately into pCR-BluntII-TOPO vector (Addgene, no.41820). We initially designed six gRNA target sequence and screened in HEK293T for the best knockout efficiency on population of cells, based on the screening we used the combination of gRNA target sequence CCCACCGACCCTGGCTCAGGATA (gRNA2, located early in the coding exon) and CCGTCGTCCGTAACCAGCGGGCC (gRNA4, located immediately after stop codon). Transfection was performed with Lipofectamine 2000 (Thermo Fisher, no. 11668027) for HEK293T and Lipofectamine LTX plus (Thermo Fisher, no. 15338100) for U2OS according to manufacturer's instructions. 24 h post transfection, cells were expanded into 10-cm dish and selected with 1 μg/ml puromycin up to when control (nontransfected) cells die. Single cell cloning was subsequently performed on puromycin-selected cells and checked by genomic DNA PCR (QuickExtract DNA Extraction Solution, Epicenter, no. QE09050; OneTaq polymerase, New England Biolabs, no. M0482) for successfully deleted genomic region (primers for checking ANG genomic deletion: forward, ATTTGGTGATGCTGTTCTTGGGTCT; reverse, AGGGAGCAGCCAAGTAGAGAAAATG). Clones with successfully genomic deletion as determined by a lower band after PCR and Sanger sequencing were checked by Western blotting to confirm loss of protein.
RNA extraction and Northern blotting
Cells were grown to ∼80% confluency and washed briefly twice with ice-cold PBS. Total RNA was extracted by TRIzol reagent (Invitrogen, no. 15596018) and purified with column-based Direct-zol RNA Miniprep kit (Zymo Research, no. R2051). For SYBR Gold detection, 5 μg total RNA was separated on 15% Novex Urea-TBE gel (Invitrogen, no. EC6885BOX) and stained with SYBR Gold (Invitrogen, no. S11494). For nonradiolabeled Northern blot analysis, 5–10 μg total RNA was separated on 10–15% Novex Urea-TBE gel and transferred to Amersham Hybond-N+ membrane (GE Healthcare Life Sciences, no. RPN203B) by Trans-Blot S.D. semi-dry transfer apparatus (Bio-Rad). After transfer, the membrane was cut to separate the upper part for tRNA plus U6 detection from the lower part for tRF detection. The upper and lower parts were probed and developed separately to avoid saturation of tRNA signals. After the membrane was crosslinked with Stratalinker (Stratagene) at 254-nanometer wavelength for 12,000 μJ and further baked at 70 °C for at least 30 min, the membrane was blocked with ExpressHyb hybridization solution (Clontech, no. 636831) at 42 °C for 30 min. After incubation with 10–50 pmol/ml biotinylated DNA probe at 42 °C overnight, the membrane was washed three times at room temperature with 1× SSC, 0.1% SDS and then detected with Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher, no. 89880) on G:BOX imaging system (Syngene). Northern probe sequences are U6 (BIO-TGCGTGTCATCCTTGCGCAG), 5′ tRNAHisGTG (BIO-CAGAGTACTAACCACTATACGATCACGGC), 3′ tRNAAspGTC (BIO-GTCGGGGAATCGAACCCCGGTC), 5′ tRNAGluCTC (BIO-CCTAACCACTAGACCACCAGGGA), 5′ tRNAGlyGCC (BIO-TCTACCACTGAACCACCCATGC), 5′ tRNAValmAC (BIO-GATAACCACTACACTACGGAAAC), and 5′ tRNASeCTCA (BIO-GACCACTGAGGATCATCCGGGC).
Puromycin pulse labeling and Western blotting
U2OS cells were grown to ∼ 80% confluency and replaced with fresh media, supplemented with 10 μg/ml (final concentration in water) puromycin (Sigma, no. P8833) in CO2 incubator at 37 °C for 10 min. Cells are washed with ice-cold PBS twice and lysed in RIPA buffer supplemented with Halt Protease Inhibitor Cocktail (Thermo Scientific, no. 78438). For Western blot analysis, 1.5–30 μg (determined by Bradford assay) whole cell lysates were separated on 12% SDS-PAGE and transferred to 0.45 μm Amersham Biosciences Protran NC Nitrocellulose Membrane (GE Healthcare Life Sciences, no. 10600002) by Trans-Blot SD Semi-Dry Transfer apparatus (Bio-Rad). These primary antibodies and corresponding dilutions were used in 3% milk in PBS with 0.05% Tween 20: anti-angiogenin (Santa Cruz Biotechnology, no. sc-74528 C1, mouse monoclonal, used at 1:100), anti-β-actin (Santa Cruz Biotechnology, no. sc-47778 C4, mouse monoclonal, used at 1:2000), anti-RNase4 (Abcam, no. ab200717, rabbit polyclonal, used at 1:500), and anti-puromycin (Millipore, no 12D10, mouse monoclonal, used at 1:10,000). Anti-mouse HRP-linked secondary antibody (Cell Signaling Technology, no. 7076) or anti-rabbit secondary antibody (Cell Signaling Technology, no. 7074) was used at 1:5000 dilution. Chemiluminescence detection was done with Immobilon HRP substrate (Millipore, no. WBKLS0500) on G:BOX imaging system (Syngene).
Small RNA-Seq library preparation
Small RNA-Seq libraries were prepared following NEBNext Small RNA Library Prep kit (New England Biolabs, no. E7300). Briefly, 1 μg total RNA was ligated with 3′ pre-adenylated adaptors and then 5′ adaptors. After reverse transcription and PCR amplification with indexed adaptors, each library was size selected on 8% TBE-PAGE gel (Invitrogen, no. EC6215BOX) to enrich for 15–50 nt insert. A brief workflow and a representative gel before size selection are shown in Fig. S1, C and D. Individual gel-purified libraries were pooled and sequenced on Illumina NextSeq 500 sequencer with mid-output or high-output, 75-cycles single-end mode at Genome Analysis and Technology Core (GATC) of University of Virginia, School of Medicine.
Small RNA-Seq data analysis
For small RNA-Seq analysis, cutadapt v1.15 (Python 2.7.5) was used to trim 3′ adaptor sequence at most 10% errors and remove reads less than 15 nt (cutadapt -nextseq-trim = 20 -a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC -m 15). The reads were then mapped to human genome (gencode GRCh38.p10 Release 27, primary assembly) by STAR aligner v2.5.4 with setting based on a previous paper to allow multi-mapping (87), and the total number of mapped reads were used for normalization. In general, mapped percentage is more than 95% (summarized in Table S1). To quantify microRNA and tRNA fragments' abundance, unitas v.1.7.3 (88) (with SeqMap v1.0.13) (89) was used with setting −species_miR_only−species homo_sapiens to map the reads to human sequence of miRBase Release 22 (90), genomic tRNA database (73), Ensembl Release 95 and SILVA rRNA database Release 132 (with addition of U13369.1 Human ribosomal DNA complete repeating unit). This setting (equivalent to −tail 2 −intmod 1 −mismatch 1 −insdel 0) will allow 2 nontemplated 3′ nucleotides and 1 internal mismatch for microRNA mapping; 1 mismatch and 0 insertion/deletion for tRNA fragments mapping. Overall tRF-3 counts were summed from tRF-3 and tRF-3CCA to count all tRF-3 reads irrespective of 3′CCA addition. tRF reads mapped to specific tRNA are grouped by tRF types (illustrated in Fig. 1A); in the case of multi-mapping, a read is counted as fraction to avoid duplicate counts. For a secondary tRNA fragment mapping method, MINTmap (72) was used to map reads to tRNA without any mismatch. For both unitas and MINTmap, cutoff for tRNA halves (both 5′ and 3′ halves) is 30 nucleotides long. For differential analysis, DESeq2 (49) was used on count matrix of tRFs and miRs.
Author contributions
Z. S. and A. D. conceptualization; Z. S. data curation; Z. S. software; Z. S. formal analysis; Z. S. and A. D. funding acquisition; Z. S. validation; Z. S., C. K., and A. M. investigation; Z. S. visualization; Z. S. and E. S. methodology; Z. S. and A. D. writing-original draft; Z. S. project administration; Z. S., C. K., and A. D. writing-review and editing; E. S. resources; A. D. supervision.
Supplementary Material
Acknowledgments
We thank Dr. Pavel Ivanov and Dr. Paul Anderson (Harvard University) for technical assistance on stress induction of tRNA halves, Dr. David Rosenkranz (Johannes Gutenberg University) for help with unitas, and Dr. Pankaj Kumar and Dr. Manjari Kiran for help with bioinformatics. We thank all Dutta lab members for their helpful discussions, Research Computing at the University of Virginia for computational support, Dr. Yongde Bao at the University of Virginia Genome Analysis and Technology Core (GATC), and the Egelman lab (University of Virginia) for sharing of equipment.
This work was supported by National Institutes of Health Grant R01 AR067712, University of Virginia Brain Institute Seed Funding and University of Virginia Supporting Transformative Autism Research (STAR) Pilot Award (to A. D.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S7 and Tables S1–S3.
The small RNA-Seq data in this paper have been deposited into Gene Expression Omnibus (GEO) with accession number GSE130764.
- tRF
- tRNA fragment
- nt
- nucleotide
- td-piRNA
- tRNA-derived PIWI-interacting RNA
- ANG
- angiogenin
- SeC
- selenocystein
- SA
- sodium arsenite
- gRNA
- guide RNA.
References
- 1. Kawaji H., Nakamura M., Takahashi Y., Sandelin A., Katayama S., Fukuda S., Daub C. O., Kai C., Kawai J., Yasuda J., Carninci P., and Hayashizaki Y. (2008) Hidden layers of human small RNAs. BMC Genomics 9, 157 10.1186/1471-2164-9-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Babiarz J. E., Ruby J. G., Wang Y., Bartel D. P., and Blelloch R. (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 22, 2773–2785 10.1101/gad.1705308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee Y. S., Shibata Y., Malhotra A., and Dutta A. (2009) A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 23, 2639–2649 10.1101/gad.1837609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A. Z., and Kay M. A. (2010) Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 16, 673–695 10.1261/rna.2000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kumar P., Anaya J., Mudunuri S. B., and Dutta A. (2014) Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 12, 78 10.1186/s12915-014-0078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Soares A. R., Fernandes N., Reverendo M., Araújo H. R., Oliveira J. L., Moura G. M., and Santos M. A. (2015) Conserved and highly expressed tRNA derived fragments in zebrafish. BMC Mol. Biol. 16, 22 10.1186/s12867-015-0050-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ba̧kowska-Żywicka K., Mleczko A. M., Kasprzyk M., Machtel P., Żywicki M., and Twardowski T. (2016) The widespread occurrence of tRNA-derived fragments in Saccharomyces cerevisiae. FEBS Open. Bio. 6, 1186–1200 10.1002/2211-5463.12127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cognat V., Morelle G., Megel C., Lalande S., Molinier J., Vincent T., Small I., Duchêne A. M., and Maréchal-Drouard L. (2017) The nuclear and organellar tRNA-derived RNA fragment population in Arabidopsis thaliana is highly dynamic. Nucleic Acids Res. 45, 3460–3472 10.1093/nar/gkw1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gebetsberger J., Zywicki M., Künzi A., and Polacek N. (2012) tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012, 260909 10.1155/2012/260909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luo S., He F., Luo J., Dou S., Wang Y., Guo A., and Lu J. (2018) Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res. 46, 5250–5268 10.1093/nar/gky189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y., Luo J., Zhou H., Liao J. Y., Ma L. M., Chen Y. Q., and Qu L. H. (2008) Stress-induced tRNA-derived RNAs: A novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 36, 6048–6055 10.1093/nar/gkn596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thompson D. M., and Parker R. (2009) The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 185, 43–50 10.1083/jcb.200811119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee S. R., and Collins K. (2005) Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 280, 42744–42749 10.1074/jbc.M510356200 [DOI] [PubMed] [Google Scholar]
- 14. Haiser H. J., Karginov F. V., Hannon G. J., and Elliot M. A. (2008) Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 36, 732–741 10.1093/nar/gkm1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ogawa T., Tomita K., Ueda T., Watanabe K., Uozumi T., and Masaki H. (1999) A cytotoxic ribonuclease targeting specific transfer RNA anticodons. Science 283, 2097–2100 10.1126/science.283.5410.2097 [DOI] [PubMed] [Google Scholar]
- 16. Fu H., Feng J., Liu Q., Sun F., Tie Y., Zhu J., Xing R., Sun Z., and Zheng X. (2009) Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 583, 437–442 10.1016/j.febslet.2008.12.043 [DOI] [PubMed] [Google Scholar]
- 17. Yamasaki S., Ivanov P., Hu G. F., and Anderson P. (2009) Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185, 35–42 10.1083/jcb.200811106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emara M. M., Ivanov P., Hickman T., Dawra N., Tisdale S., Kedersha N., Hu G. F., and Anderson P. (2010) Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 285, 10959–10968 10.1074/jbc.M109.077560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ivanov P., Emara M. M., Villen J., Gygi S. P., and Anderson P. (2011) Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43, 613–623 10.1016/j.molcel.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Saikia M., Jobava R., Parisien M., Putnam A., Krokowski D., Gao X. H., Guan B. J., Yuan Y., Jankowsky E., Feng Z., Hu G. F., Pusztai-Carey M., Gorla M., Sepuri N. B., Pan T., and Hatzoglou M. (2014) Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol. Cell Biol. 34, 2450–2463 10.1128/MCB.00136-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuscu C., Kumar P., Kiran M., Su Z., Malik A., and Dutta A. (2018) tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24, 1093–1105 10.1261/rna.066126.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maute R. L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., and Dalla-Favera R. (2013) tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 110, 1404–1409 10.1073/pnas.1206761110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ren B., Wang X., Duan J., and Ma J. (2019) Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 365, 919–922 10.1126/science.aav8907 [DOI] [PubMed] [Google Scholar]
- 24. Schorn A. J., Gutbrod M. J., LeBlanc C., and Martienssen R. (2017) LTR-retrotransposon control by tRNA-derived small RNAs. Cell 170, 61–71.e11 10.1016/j.cell.2017.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim H. K., Fuchs G., Wang S., Wei W., Zhang Y., Park H., Roy-Chaudhuri B., Li P., Xu J., Chu K., Zhang F., Chua M. S., So S., Zhang Q. C., Sarnow P., and Kay M. A. (2017) A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552, 57–62 10.1038/nature25005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sobala A., and Hutvagner G. (2013) Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol. 10, 553–563 10.4161/rna.24285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Keam S. P., Sobala A., Ten Have S., and Hutvagner G. (2017) tRNA-derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation. J. Proteome Res. 16, 413–420 10.1021/acs.jproteome.6b00267 [DOI] [PubMed] [Google Scholar]
- 28. Guzzi N., Cieśla M., Ngoc P. C. T., Lang S., Arora S., Dimitriou M., Pimková K., Sommarin M. N. E., Munita R., Lubas M., Lim Y., Okuyama K., Soneji S., Karlsson G., Hansson J., et al. (2018) Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell 173, 1204–1216.e26 10.1016/j.cell.2018.03.008 [DOI] [PubMed] [Google Scholar]
- 29. Zhang X., He X., Liu C., Liu J., Hu Q., Pan T., Duan X., Liu B., Zhang Y., Chen J., Ma X., Zhang X., Luo H., and Zhang H. (2016) IL-4 Inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J. Immunol. 196, 1591–1603 10.4049/jimmunol.1500805 [DOI] [PubMed] [Google Scholar]
- 30. Honda S., Kawamura T., Loher P., Morichika K., Rigoutsos I., and Kirino Y. (2017) The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic Acids Res. 45, 9108–9120 10.1093/nar/gkx537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Keam S. P., Young P. E., McCorkindale A. L., Dang T. H., Clancy J. L., Humphreys D. T., Preiss T., Hutvagner G., Martin D. I., Cropley J. E., and Suter C. M. (2014) The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 42, 8984–8995 10.1093/nar/gku620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Balatti V., Nigita G., Veneziano D., Drusco A., Stein G. S., Messier T. L., Farina N. H., Lian J. B., Tomasello L., Liu C. G., Palamarchuk A., Hart J. R., Bell C., Carosi M., Pescarmona E., et al. (2017) tsRNA signatures in cancer. Proc. Natl. Acad. Sci. U.S.A. 114, 8071–8076 10.1073/pnas.1706908114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pekarsky Y., Balatti V., Palamarchuk A., Rizzotto L., Veneziano D., Nigita G., Rassenti L. Z., Pass H. I., Kipps T. J., Liu C. G., and Croce C. M. (2016) Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc. Natl. Acad. Sci. U.S.A. 113, 5071–5076 10.1073/pnas.1604266113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goodarzi H., Liu X., Nguyen H. C., Zhang S., Fish L., and Tavazoie S. F. (2015) Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161, 790–802 10.1016/j.cell.2015.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Skorupa A., King M. A., Aparicio I. M., Dussmann H., Coughlan K., Breen B., Kieran D., Concannon C. G., Marin P., and Prehn J. H. (2012) Motoneurons secrete angiogenin to induce RNA cleavage in astroglia. J. Neurosci. 32, 5024–5038 10.1523/JNEUROSCI.6366-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goncalves K. A., Silberstein L., Li S., Severe N., Hu M. G., Yang H., Scadden D. T., and Hu G. F. (2016) Angiogenin promotes hematopoietic regeneration by dichotomously regulating quiescence of stem and progenitor cells. Cell 166, 894–906 10.1016/j.cell.2016.06.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thomas S. P., Hoang T. T., Ressler V. T., and Raines R. T. (2018) Human angiogenin is a potent cytotoxin in the absence of ribonuclease inhibitor. RNA 24, 1018–1027 10.1261/rna.065516.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moenner M., Gusse M., Hatzi E., and Badet J. (1994) The widespread expression of angiogenin in different human cells suggests a biological function not only related to angiogenesis. Eur. J. Biochem. 226, 483–490 10.1111/j.1432-1033.1994.tb20073.x [DOI] [PubMed] [Google Scholar]
- 39. Sheng J., and Xu Z. (2016) Three decades of research on angiogenin: A review and perspective. Acta Biochim. Biophys. Sin. 48, 399–410 10.1093/abbs/gmv131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shapiro R., and Vallee B. L. (1987) Human placental ribonuclease inhibitor abolishes both angiogenic and ribonucleolytic activities of angiogenin. Proc. Natl. Acad. Sci. U.S.A. 84, 2238–2241 10.1073/pnas.84.8.2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pizzo E., Sarcinelli C., Sheng J., Fusco S., Formiggini F., Netti P., Yu W., D'Alessio G., and Hu G. F. (2013) Ribonuclease/angiogenin inhibitor 1 regulates stress-induced subcellular localization of angiogenin to control growth and survival. J. Cell Sci. 126, 4308–4319 10.1242/jcs.134551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Czech A., Wende S., Mörl M., Pan T., and Ignatova Z. (2013) Reversible and rapid transfer-RNA deactivation as a mechanism of translational repression in stress. PLoS Genet. 9, e1003767 10.1371/journal.pgen.1003767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Honda S., Loher P., Shigematsu M., Palazzo J. P., Suzuki R., Imoto I., Rigoutsos I., and Kirino Y. (2015) Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. U.S.A. 112, E3816–E3825 10.1073/pnas.1510077112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li Z., Ender C., Meister G., Moore P. S., Chang Y., and John B. (2012) Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 40, 6787–6799 10.1093/nar/gks307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Saikia M., Krokowski D., Guan B. J., Ivanov P., Parisien M., Hu G. F., Anderson P., Pan T., and Hatzoglou M. (2012) Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J. Biol. Chem. 287, 42708–42725 10.1074/jbc.M112.371799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sheng J., Yu W., Gao X., Xu Z., and Hu G. F. (2014) Angiogenin stimulates ribosomal RNA transcription by epigenetic activation of the ribosomal DNA promoter. J. Cell Physiol. 229, 521–529 10.1002/jcp.24477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsuji T., Sun Y. Q., Kishimoto K., Olson K. A., Liu S. M., Hirukawa S., and Hu G. F. (2005) Angiogenin is translocated to the nucleus of HeLa cells and is involved in ribosomal RNA transcription and cell proliferation. Cancer Res. 65, 1352–1360 10.1158/0008-5472.CAN-04-2058 [DOI] [PubMed] [Google Scholar]
- 48. Xu Z. P., Tsuji T., Riordan J. F., and Hu G. F. (2002) The nuclear function of angiogenin in endothelial cells is related to rRNA production. Biochem. Bioph. Res. Co. 294, 287–292 10.1016/S0006-291X(02)00479-5 [DOI] [PubMed] [Google Scholar]
- 49. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kumar P., Mudunuri S. B., Anaya J., and Dutta A. (2015) tRFdb: A database for transfer RNA fragments. Nucleic Acids Res. 43, D141–D145 10.1093/nar/gku1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saxena S. K., Rybak S. M., Davey R. T., Youle R. J., and Ackerman E. J. (1992) Angiogenin is a cytotoxic, transfer RNA-specific ribonuclease in the RNase A superfamily. J. Biol. Chem. 267, 21982–21986 [PubMed] [Google Scholar]
- 52. Acharya K. R., Shapiro R., Allen S. C., Riordan J. F., and Vallee B. L. (1994) Crystal structure of human angiogenin reveals the structural basis for its functional divergence from ribonuclease. Proc. Natl. Acad. Sci. U.S.A. 91, 2915–2919 10.1073/pnas.91.8.2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Russo N., Shapiro R., Acharya K. R., Riordan J. F., and Vallee B. L. (1994) Role of glutamine-117 in the ribonucleolytic activity of human angiogenin. Proc. Natl. Acad. Sci. U.S.A. 91, 2920–2924 10.1073/pnas.91.8.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rybak S. M., and Vallee B. L. (1988) Base cleavage specificity of angiogenin with Saccharomyces cerevisiae and Escherichia coli 5S RNAs. Biochemistry 27, 2288–2294 10.1021/bi00407a007 [DOI] [PubMed] [Google Scholar]
- 55. Wang X., Matuszek Z., Huang Y., Parisien M., Dai Q., Clark W., Schwartz M. H., and Pan T. (2018) Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24, 1305–1313 10.1261/rna.067033.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen Z., Qi M., Shen B., Luo G., Wu Y., Li J., Lu Z., Zheng Z., Dai Q., and Wang H. (2019) Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 47, 2533–2545 10.1093/nar/gky1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schaefer M., Pollex T., Hanna K., Tuorto F., Meusburger M., Helm M., and Lyko F. (2010) RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Gene Dev. 24, 1590–1595 10.1101/gad.586710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang Y. F., Zhang X. D., Shi J. C., Tuorto F., Li X., Liu Y. S., Liebers R., Zhang L. W., Qu Y. C., Qian J. J., Pahima M., Liu Y., Yan M. H., Cao Z. H., Lei X. H., et al. (2018) Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol. 20, 535–540 10.1038/s41556-018-0087-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tosar J. P., Gámbaro F., Darré L., Pantano S., Westhof E., and Cayota A. (2018) Dimerization confers increased stability to nucleases in 5′ halves from glycine and glutamic acid tRNAs. Nucleic Acids Res. 46, 9081–9093 10.1093/nar/gky495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huang H. Y., and Hopper A. K. (2016) Multiple layers of stress-induced regulation in tRNA biology. Life (Basel) 6, E16 10.3390/life6020016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Phizicky E. M., and Hopper A. K. (2010) tRNA biology charges to the front. Gene Dev. 24, 1832–1860 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thompson D. M., and Parker R. (2009) Stressing out over tRNA cleavage. Cell 138, 215–219 10.1016/j.cell.2009.07.001 [DOI] [PubMed] [Google Scholar]
- 63. Mesitov M. V., Soldatov R. A., Zaichenko D. M., Malakho S. G., Klementyeva T. S., Sokolovskaya A. A., Kubatiev A. A., Mironov A. A., and Moskovtsev A. A. (2017) Differential processing of small RNAs during endoplasmic reticulum stress. Sci. Rep. 7, 46080 10.1038/srep46080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Selitsky S. R., Baran-Gale J., Honda M., Yamane D., Masaki T., Fannin E. E., Guerra B., Shirasaki T., Shimakami T., Kaneko S., Lanford R. E., Lemon S. M., and Sethupathy P. (2015) Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Sci. Rep. 5, 7675 10.1038/srep07675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang Q., Lee I., Ren J., Ajay S. S., Lee Y. S., and Bao X. (2013) Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther. 21, 368–379 10.1038/mt.2012.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dhahbi J. M. (2015) 5′ tRNA halves: The next generation of immune signaling molecules. Front. Immunol. 6, 74 10.3389/fimmu.2015.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lyons S. M., Gudanis D., Coyne S. M., Gdaniec Z., and Ivanov P. (2017) Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat. Commun. 8, 1127 10.1038/s41467-017-01278-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fricker R., Brogli R., Luidalepp H., Wyss L., Fasnacht M., Joss O., Zywicki M., Helm M., Schneider A., Cristodero M., and Polacek N. (2019) A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat. Commun. 10, 118 10.1038/s41467-018-07949-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Megel C., Hummel G., Lalande S., Ubrig E., Cognat V., Morelle G., Salinas-Giegé T., Duchêne A. M., and Maréchal-Drouard L. (2019) Plant RNases T2, but not Dicer-like proteins, are major players of tRNA-derived fragments biogenesis. Nucleic Acids Res. 47, 941–952 10.1093/nar/gky1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Andersen K. L., and Collins K. (2012) Several RNase T2 enzymes function in induced tRNA and rRNA turnover in the ciliate Tetrahymena. Mol. Biol. Cell 23, 36–44 10.1091/mbc.e11-08-0689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cole C., Sobala A., Lu C., Thatcher S. R., Bowman A., Brown J. W. S., Green P. J., Barton G. J., and Hutvagner G. (2009) Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15, 2147–2160 10.1261/rna.1738409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Loher P., Telonis A. G., and Rigoutsos I. (2017) MINTmap: Fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-Seq data. Sci. Rep. 7, 41184 10.1038/srep41184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chan P. P., and Lowe T. M. (2016) GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44, D184–D189 10.1093/nar/gkv1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Donovan J., Rath S., Kolet-Mandrikov D., and Korennykh A. (2017) Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA 23, 1660–1671 10.1261/rna.062000.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yang J. Y., Deng X. Y., Li Y. S., Ma X. C., Feng J. X., Yu B., Chen Y., Luo Y. L., Wang X., Chen M. L., Fang Z. X., Zheng F. X., Li Y. P., Zhong Q., Kang T. B., et al. (2018) Structure of Schlafen13 reveals a new class of tRNA/rRNA—targeting RNase engaged in translational control. Nat. Commun. 9, 1165 10.1038/s41467-018-03544-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Krishna S., Yim D. G., Lakshmanan V., Tirumalai V., Koh J. L., Park J. E., Cheong J. K., Low J. L., Lim M. J., Sze S. K., Shivaprasad P., Gulyani A., Raghavan S., Palakodeti D., and DasGupta R. (2019) Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 20, e47789 10.15252/embr.201947789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dhahbi J. M., Spindler S. R., Atamna H., Yamakawa A., Boffelli D., Mote P., and Martin D. I. K. (2013) 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics 14, 298 10.1186/1471-2164-14-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wei Z., Batagov A. O., Schinelli S., Wang J., Wang Y., El Fatimy R., Rabinovsky R., Balaj L., Chen C. C., Hochberg F., Carter B., Breakefield X. O., and Krichevsky A. M. (2017) Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat. Commun. 8, 1145 10.1038/s41467-017-01196-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Godoy P. M., Bhakta N. R., Barczak A. J., Cakmak H., Fisher S., MacKenzie T. C., Patel T., Price R. W., Smith J. F., Woodruff P. G., and Erle D. J. (2018) Large differences in small RNA composition between human biofluids. Cell Rep. 25, 1346–1358 10.1016/j.celrep.2018.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ferrero G., Cordero F., Tarallo S., Arigoni M., Riccardo F., Gallo G., Ronco G., Allasia M., Kulkarni N., Matullo G., Vineis P., Calogero R. A., Pardini B., and Naccarati A. (2017) Small non-coding RNA profiling in human biofluids and surrogate tissues from healthy individuals: Description of the diverse and most represented species. Oncotarget 9, 3097–3111 10.18632/oncotarget.23203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shurtleff M. J., Yao J., Qin Y., Nottingham R. M., Temoche-Diaz M. M., Schekman R., and Lambowitz A. M. (2017) Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. U.S.A. 114, E8987–E8995 10.1073/pnas.1712108114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Peng H. Y., Shi J. C., Zhang Y., Zhang H., Liao S. Y., Li W., Lei L., Han C. S., Ning L. N., Cao Y. J., Zhou Q., Chen Q., and Duan E. K. (2012) A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res 22, 1609–1612 10.1038/cr.2012.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Srinivasan S., Yeri A., Cheah P. S., Chung A., Danielson K., De Hoff P., Filant J., Laurent C. D., Laurent L. D., Magee R., Moeller C., Murthy V. L., Nejad P., Paul A., Rigoutsos I., et al. (2019) Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell 177, 446–462.e416 10.1016/j.cell.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G. H., Peng H., Zhang X., Zhang Y., Qian J., Duan E., Zhai Q., and Zhou Q. (2016) Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400 10.1126/science.aad7977 [DOI] [PubMed] [Google Scholar]
- 85. Sharma U., Conine C. C., Shea J. M., Boskovic A., Derr A. G., Bing X. Y., Belleannee C., Kucukural A., Serra R. W., Sun F., Song L., Carone B. R., Ricci E. P., Li X. Z., Fauquier L., et al. (2016) Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396 10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., and Church G. M. (2013) RNA-guided human genome engineering via Cas9. Science 339, 823–826 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Faridani O. R., Abdullayev I., Hagemann-Jensen M., Schell J. P., Lanner F., and Sandberg R. (2016) Single-cell sequencing of the small-RNA transcriptome. Nat. Biotechnol. 34, 1264–1266 10.1038/nbt.3701 [DOI] [PubMed] [Google Scholar]
- 88. Gebert D., Hewel C., and Rosenkranz D. (2017) unitas: The universal tool for annotation of small RNAs. BMC Genomics 18, 644 10.1186/s12864-017-4031-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jiang H., and Wong W. H. (2008) SeqMap: Mapping massive amount of oligonucleotides to the genome. Bioinformatics 24, 2395–2396 10.1093/bioinformatics/btn429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kozomara A., Birgaoanu M., and Griffiths-Jones S. (2019) miRBase: From microRNA sequences to function. Nucleic Acids Res. 47, D155–D162 10.1093/nar/gky1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.