Abstract
Sialic acid-binding immunoglobulin-like lectins (Siglecs) are a family of cell-surface immune receptors that bind to sialic acid at terminal glycan residues. Siglecs also recognize nonsialic acid ligands, many of which remain to be characterized. Here, we found that Siglec5 and Siglec14 recognize lipid compounds produced by Trichophyton, a fungal genus containing several pathogenic species. Biochemical approaches revealed that the Siglec ligands are fungal alkanes and triacylglycerols, an unexpected finding that prompted us to search for endogenous lipid ligands of Siglecs. Siglec5 weakly recognized several endogenous lipids, but the mitochondrial lipid cardiolipin and the anti-inflammatory lipid 5-palmitic acid-hydroxystearic acid exhibited potent ligand activity on Siglec5. Further, the hydrophobic stretch in the Siglec5 N terminus region was found to be required for efficient recognition of these lipids. Notably, this hydrophobic stretch was dispensable for recognition of sialic acid. Siglec5 inhibited cell activation upon ligand binding, and accordingly, the lipophilic ligands suppressed interleukin-8 (IL-8) production in Siglec5-expressing human monocytic cells. Siglec14 and Siglec5 have high sequence identity in the extracellular region, and Siglec14 also recognized the endogenous lipids. However, unlike Siglec5, Siglec14 transduces activating signals upon ligand recognition. Indeed, the endogenous lipids induced IL-8 production in Siglec14-expressing human monocytic cells. These results indicated that Siglec5 and Siglec14 can recognize lipophilic ligands that thereby modulate innate immune responses. To our knowledge, this is the first study reporting the binding of Siglecs to lipid ligands, expanding our understanding of the biological function and importance of Siglecs in the innate immunity.
Keywords: lectin, cell-surface receptor, lipid-protein interaction, cellular immune response, innate immunity, cardiolipin, cytokine response, lipophilic ligands, Siglec, Trichophyton
Introduction
Pattern recognition receptors play crucial roles in the sensing of potential dangers, such as invading pathogens and altered self in innate immune cells (1). Among them, Toll-like receptors, RIG-I–like receptors, Nod-like receptors, and C-type lectin receptors are extensively studied; numerous of their ligands have been identified; and their importance for innate immunity has been demonstrated (2–4). However, other pattern recognition receptors also contribute to innate immune responses through interaction with specific ligands. Sialic acid–binding immunoglobin-like lectins (Siglecs)3 are receptors for sialic acid at terminal glycan residues on glycoproteins that mediate cell–cell interactions and immunoregulation (5). In humans, 14 Siglec members (Siglec1 to -16 except Siglec12 and -13) have been identified and are expressed mainly on hematopoietic cells in a cell type–specific manner. Innate immune cells predominantly express Siglec3, -5, -7, -9, and -14 (6). Siglec3, -5, -7, and -9 function as inhibitory receptors through their cytoplasmic immunoreceptor tyrosine-based inhibitory motif (ITIM), which recruits phosphatases such as SHP-1 and SHP-2 and suppresses kinase-dependent activating signals (7). Siglec14 lacks an ITIM but is associated with immunoreceptor tyrosine-based activation motif (ITAM)-containing molecule DAP12 and transduces activating signals through Syk-family kinases, eventually activating transcription factors such as NFAT and NF-κB (8). Siglec5 and -14 share most of the extracellular region, indicating partial gene conversion, and thus show similar binding preferences (8). Therefore, Siglec5 and -14 are thought to function as paired receptors.
Sialic acids are predominantly and abundantly expressed on vertebrate cells (9), and Siglecs are constitutively associated with endogenous sialic acid on the surface of the same cell (cis-interaction) or of the neighboring cell (trans-interaction) (10, 11). Certain pathogens uniquely express sialic acid on their surface and bind to Siglecs to modulate host immune responses (12). Lipopolysaccharide (LPS) of certain Gram-positive bacteria, such as Campylobacter jejuni, group B Streptococcus, and Neisseria meningitidis, are sialylated and bind to Siglec1, -5, -7, and -9 (13–15). Pseudomonas aeruginosa ingests host sialic acids to decorate its glycoproteins and dampens neutrophil activation through Siglec9 (16). Siglecs also interact with several viruses and parasites in a sialic acid–dependent manner (17–20).
Siglecs also recognize nonsialic acid ligands. Siglec5 and -14 recognize β-protein of group B Streptococcus and modulate host immunity (21, 22). Heat-shock protein 70 released from mammalian cells engages Siglec5 and -14 and controls the immune status of monocytes (23). High-molecular-weight hyaluronan on group A Streptococcus binds to Siglec9 to suppress neutrophil activation (24). However, we have not found any evidence of an interaction of Siglecs with lipophilic ligands in the literature.
In this study, we discovered lipophilic ligands of Siglec5 and -14 in fungi and mammalian cells. These lipophilic ligands interact with Siglec5 through a unique hydrophobic stretch, which differs from the sialic acid–binding site, and modulate cellular activation in innate immune cells.
Results
Human Siglec5 and -14 recognize pathogenic Trichophyton spp.
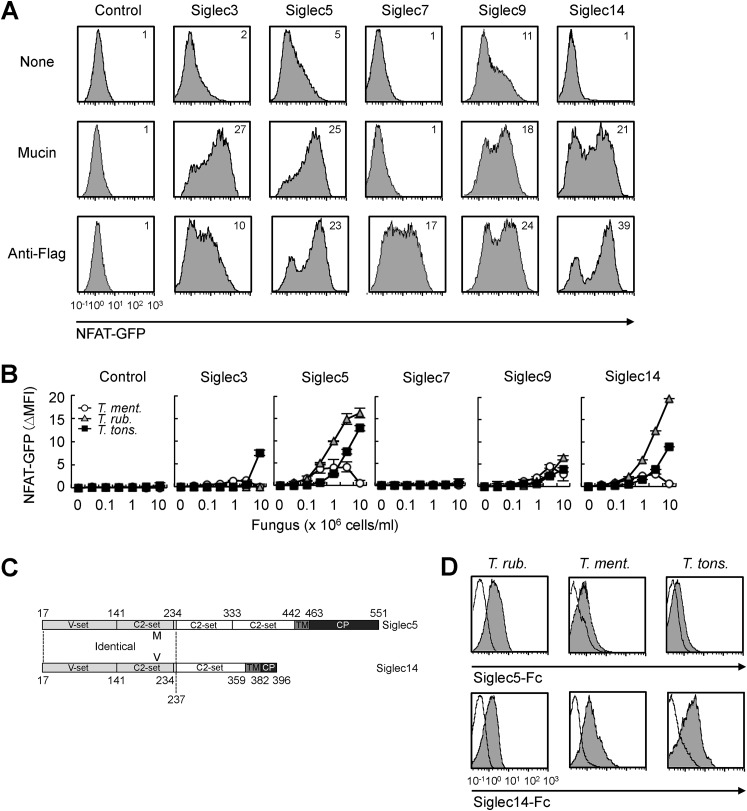
To detect potential ligand recognition activity of Siglecs, we generated Siglec reporter cells, in which GFP is expressed upon ligand binding. The cytoplasmic region of inhibitory Siglec receptor, which includes the ITIM sequence, was replaced with that of CD3ζ, which contains an ITAM sequence (Fig. S1A, left). FLAG-tagged Siglec-CD3ζ chimeric receptor was then transfected into NFAT-GFP reporter cells (2B4 T cell hybridoma) (25), which allow detection of ITAM signaling activation through GFP expression (Fig. S1A, right). We established four reporter cell lines for human Siglec3, -5, -7, and -9, which are expressed on innate immune cells, such as macrophages and monocytes (6). GFP expression by the Siglec reporter lines was confirmed by stimulation with plate-coated anti-FLAG antibody or the sialic acid–rich glycoprotein mucin (Fig. 1A). Mucin failed to activate GFP expression in Siglec7 reporter cells. Siglec7 uniquely recognizes disialylated glycans or gangliosides (26, 27). Mucin may not contain these Siglec7 ligands. Using the Siglec reporter lines, we carried out a screening for Siglec-binding pathogens in a pathogenic fungi library and found that Siglec5 recognized several pathogenic fungi. Among them, Trichophyton spp., including Trichophyton rubrum, Trichophyton mentagrophytes, and Trichophyton tonsrans showed potential ligand activity (Fig. 1B). Siglec3 and -9 weakly recognized Trichophyton spp.
Figure 1.
Human Siglec5 and -14 recognize Trichophyton spp. A, Siglec reporter cells and nontransfected NFAT-GFP reporter cells (control) were stimulated with plate-coated mucin (2 mg/ml) or anti-FLAG (10 μg/ml) for 18 h. Induction of NFAT-GFP was analyzed by flow cytometry. Numbers in histograms indicate the mean fluorescence intensity (MFI) of NFAT-GFP. B, Siglec reporter cells were stimulated with the indicated numbers of T. mentagrophytes, T. rubrum, or T. tonsrans cells for 18 h. Induction of NFAT-GFP was calculated as ΔMFI = MFI of treated − MFI of nontreated. C, schematic domain structures of Siglec5 and Siglec14. Siglec5 contains four Ig-like domains (V-set domain and three C2-set domains), a transmembrane domain (TM), and a cytoplasmic domain (CP). Siglec14 contains three Ig-like domains (V-set domain and two C2-set domains), a TM, and a CP. The N-terminal regions (amino acids 17–237) of Siglec5 and -14 are identical except for one amino acid residue positioned at 215 (Met and Val, respectively). D, Trichophyton spp. were incubated with recombinant Siglec5-Fc, Siglec14-Fc (filled histogram), or Fc (open histogram). Binding Fc proteins were detected by fluorescence-labeled secondary antibody and analyzed by flow cytometry. Data are representative of at least three independent experiments and are presented as the mean ± S.D. (error bars).
Human Siglec14 and -5 have high sequence identity in the N-terminal extracellular region (Fig. 1C) and thus have similar ligand preference (8). Siglec14 lacks the ITIM sequence but instead associates with ITAM adaptor molecule DAP12 and transduces activating signals upon ligand binding. To determine whether Siglec14 also recognizes the above pathogenic fungi, we generated Siglec14 reporter cells by introducing Siglec14 and DAP12 into NFAT-GFP reporter cells (Fig. S1B). Anti-FLAG antibody and mucin effectively induced GFP expression in Siglec14 reporter cells (Fig. 1A). As expected, Siglec14 recognized Trichophyton spp. as potently as Siglec5 (Fig. 1B). We confirmed the direct interaction between Siglecs and Trichophyton spp. by using recombinant Siglec-Fc fusion proteins in which the extracellular region of Siglec was fused with the fragment crystallizable (Fc) region of human IgG1 (Fig. 1D). Collectively, these results suggested that Siglec5 and -14 recognize T. rubrum, T. mentagrophytes, and T. tonsrans.
Identification of Siglec5 and -14 ligands in Trichophyton spp.
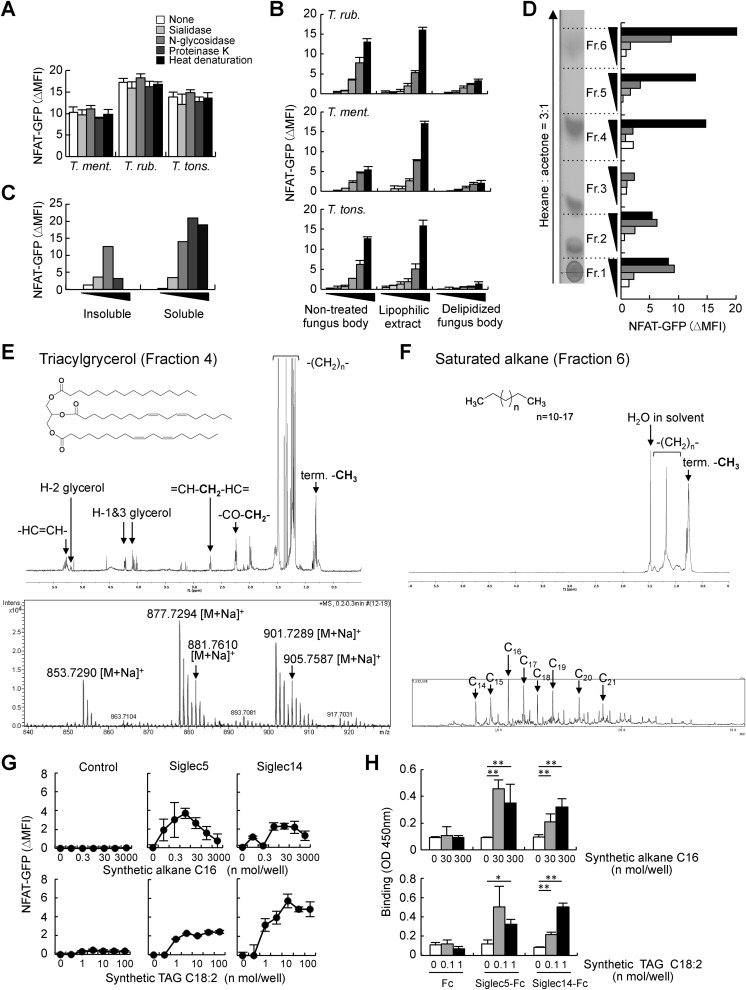
We next aimed to identify the ligands of Siglec5 in Trichophyton spp. A previous study demonstrated that T. rubrum and T. mentagrophytes lack sialic acids (28). Consistently, sialidase or N-glycosidase treatment had no impact on the ligand activity of Trichophyton spp. (Fig. 2A). Siglec5 and -14 reportedly recognize proteinaceous ligands in a sialic acid–independent manner (21–23). However, ligand activity was still observed upon proteinase K treatment or heat denaturation (Fig. 2A). These results strongly suggested that Trichophyton spp. have previously unknown ligands. We therefore tried to purify these ligands using biochemical approaches. Trichophyton spp. were treated with organic solvent (CHCl3/MeOH, 2:1 (v/v)) and separated the lipophilic extract and delipidized fungal biomass. Ligand activity was dominantly observed in the lipophilic extract (Fig. 2B). As we did not find any reports on lipophilic ligands of Siglec5 in the previous literature, we further purified the ligands in the lipophilic extract of T. mentagrophytes. The lipophilic extract was separated in hexane-soluble and -insoluble fractions; the hexane-soluble fraction potently activated Siglec5 reporter cells (Fig. 2C). The hexane-soluble fraction was further separated into six subfractions by high-performance TLC (HPTLC). Fractions 4 and 6 showed strong ligand activity and clear spots upon iodide staining (Fig. 2D).
Figure 2.
Identification of lipophilic ligands of Siglec5 and -14 in T. mentagrophytes. A, Trichophyton spp. were treated with sialidase, N-glycosidase, or Proteinase K for 18 h or incubated at 95 °C for 10 min (heat denaturation) and then cocultured with Siglec5 reporter cells for 18 h. Induction of NFAT-GFP was analyzed by flow cytometry. B, Trichophyton spp. were treated with CHCl3/MeOH (2:1, v/v) and separated into lipophilic extract and delipidized fungal biomass. Siglec5 reporter cells were stimulated with nontreated fungus, delipidized fungal biomass, or plate-coated lipophilic extract. C, Siglec5 reporter cells were stimulated with hexane-soluble and -insoluble fractions of lipophilic extract of T. mentagrophytes. D, hexane-soluble fraction was subjected to HPTLC, stained with I2, and separated into six fractions. Each fraction was coated on a plate for Siglec5 reporter cell stimulation. E, 1H NMR spectrum (top) and ESI-TOF-MS of Fraction 4 (bottom). F, 1H NMR spectrum (top) and GC-MS chromatogram (bottom) of Fraction 6. G, Siglec5 and -14 reporter cells and nontransfected NFAT-GFP reporter cells (control) were stimulated with plate-coated synthetic alkane or TAG. H, recombinant Siglec5-Fc, Siglec14-Fc, and Fc proteins were incubated with plate-coated synthetic alkane or TAG. Bound proteins were detected with anti-hIgG-HRP. Data are representative of at least three independent experiments and are presented as the mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01.
To determine the chemical structures of the compounds present in Fractions 4 and 6, we used NMR and high-resolution electrospray ionization TOF MS (ESI-TOF-MS) analyses. The 1H NMR spectrum of Fraction 4 showed proton signals characteristic of triacylglycerol (TAG), and ESI-TOF-MS spectrum showed molecular-related ion peaks at m/z 853.7290, 877.7294, 881.7610, 901.7289, and 905.7587 (Fig. 2E). Next, we determined the fatty acid composition by GC-MS following methanolysis. The GC-MS chromatogram of fatty acid methyl esters (FAMEs) revealed the presence of C16:0, C18:1, C18:2, and C18:0 FAMEs (Fig. S2A). The structures of the TAGs identified in Fraction 4 were shown in Fig. S2B.
The compounds present in Fraction 6 were elucidated by 1H NMR and GC-MS analysis. The 1H NMR spectrum of Fraction 6 showed proton signals characteristic of saturated alkane, and GC-MS revealed molecular ion peaks assigned to a mixture of saturated alkanes consisting of C14H30 to C21H44 (Fig. 2F and Fig. S2 (C and D)).
Chemically synthesized saturated alkane (C16) and TAG (C18:2) coated in 96-well plates activated both Siglec5 and Siglec14 reporter cells (Fig. 2G). To confirm the direct interaction between Siglecs and lipophilic ligands, we carried out an ELISA-based binding assay using recombinant Siglec-Fc fusion proteins. Siglec5-Fc and Siglec14-Fc substantially bound to plate-coated synthetic alkane and TAG (Fig. 2H). Taken together, these data indicated that alkane and TAG are novel lipophilic ligands of Siglec5 and -14.
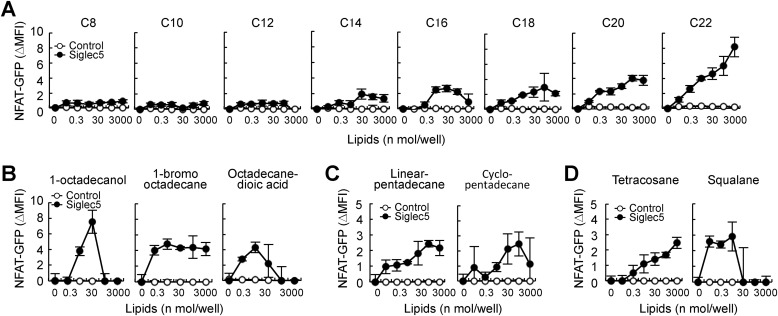
Ligand activity of alkanes
To investigate the structure–activity relationship of alkanes and Siglec5, we first compared the ligand activity of saturated alkanes of different lengths. Long-chain alkanes (C14–C22) activated Siglec5 reporter cells, whereas short-chain alkanes (C8–C12) did not, suggesting that a certain length is required for ligand activity (Fig. 3A). Next, we examined the effect of alkane end-group modification on ligand activity (Fig. S3A). Siglec5 recognized alkane derivatives with one end modification, including 1-octadecanol and 1-bromooctadecane (Fig. 3B). Octadecanedioic acid, which contains hydroxyl groups at both ends, still activated Siglec5 reporter cells, suggesting that end-group modification does not interfere with the ligand activity of alkanes. GFP expression was reduced upon high-dose treatment with 1-octadecanol and octadecanedioic acid because of cytotoxicity. Cyclo-pentadecane and linear pentadecane showed similar ligand activity (Fig. 3C). Squalane, which is multiple methyl-branched tetracosane, also activated Siglec reporter cells (Fig. 3D and Fig. S3B). These results suggested that Siglec5 can broadly recognize alkanes of a certain length.
Figure 3.
Ligand activity of alkanes and derivatives. Siglec5 reporter cells were stimulated with plate-coated lipids for 18 h. Induction of NFAT was analyzed by flow cytometry. A, saturated alkanes of different lengths. B, octadecane derivatives. Chemical structures are shown in Fig. S3A. C, linear or cyclic pentadecane. D, tetracosane and multimethylated tetracosane, squalane. Chemical structures are shown in Fig. S3B. Data are representative of at least three independent experiments and are presented as the mean ± S.D. (error bars).
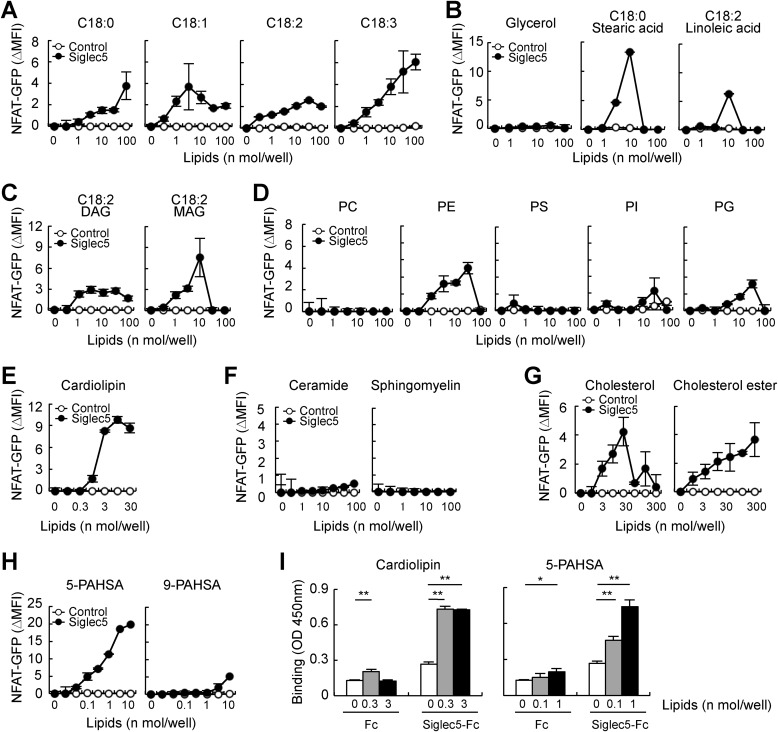
Ligand activity of TAGs and related glycerolipids
Next, we analyzed the ligand activity of TAGs and related glycerolipids. Comparison of TAGs with unsaturated or saturated C18 fatty acid chains (Fig. S3C) revealed that C18:3 TAG activated Siglec5 reporter cells the most potently, although all TAGs showed ligand activity (Fig. 4A). TAGs consist of glycerol and three fatty acid chains. Consistent with the above finding that Siglec5 recognized alkane derivatives with end modification, fatty acids, but not glycerol, activated Siglec5 reporter cells (Fig. 4B). Both diacylglycerol (DAG) and monoacylglycerol (MAG) (Fig. S3D) also showed ligand activity, suggesting that the presence of multiple alkyl chains is not required for ligand activity (Fig. 4C). Stearic acid and MAG potently activated Siglec5 reporter cells; however, they also showed strong cytotoxicity.
Figure 4.
Ligand activity of TAG-related molecules and other endogenous lipids. Siglec5 reporter cells were stimulated with plate-coated lipids for 18 h. Induction of NFAT was analyzed by flow cytometry. A, C18 TAGs with saturated or unsaturated fatty acid chains. Chemical structures are shown in Fig. S3C. B, glycerol and C18 saturated or unsaturated fatty acids. C, C18:2 DAG and MAG. Chemical structures are shown in Fig. S3D. D, glycerophospholipids, such as phosphatidylcholine (PC), PE, phosphatidylserine (PS), phosphatidylinositol (PI), and PG. E, cardiolipin. F, C16 ceramide and sphingomyelin. G, cholesterol and cholesterol ester (cholesteryl linoleate). H, 5-PAHSA and 9-PAHSA. Chemical structures are shown in Fig. S3E. I, recombinant Siglec5-Fc and Fc proteins were incubated with plate-coated cardiolipin or 5-PAHSA. Bound proteins were detected with anti-hIgG-HRP. Data are representative of at least three independent experiments and are presented as the mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01.
Ligand activity of other endogenous lipids
Our unexpected finding that Siglec5 recognized lipophilic ligands prompted us to further investigate the ligand activity of other endogenous lipids. Glycerophospholipids, which are major components of the mammalian cell membrane, are synthesized from cytidine diphosphate-DAG and thus share a common structure with DAG. Siglec5 reporter cells were activated by phosphatidylethanolamine (PE) and phosphatidylglycerol (PG), but not by other glycerophospholipids, suggesting that polar headgroups may affect ligand activity (Fig. 4D). Cardiolipin, which is abundantly found in the mitochondrial inner membrane and is synthesized from PG and cytidine diphosphate-DAG, potently activated Siglec5 reporter cells (Fig. 4E). Like glycerophospholipids, sphingolipids contain long alkyl chains and are widely distributed in the cell membrane. Sphingomyelin and C16 ceramide failed to activate Siglec5 reporter cells (Fig. 4F). Cholesterol is another major lipid component in the cell membrane, with a sterol structure and a short alkyl chain. Most ingested cholesterol is esterified with long-chain fatty acids and stored in lipid droplets along with TAG. Siglec5 recognized cholesterol and cholesterol ester (Fig. 4G). Palmitic acid hydroxystearic acids (PAHSAs) are branched fatty acid esters with anti-inflammatory and anti-diabetic activities (29) (Fig. S3E). 5-PAHSA showed the strongest ligand activity toward Siglec5 among all lipids tested, whereas 9-PAHSA only weakly activated Siglec5 reporter cells (Fig. 4H). Taken together, these data indicated that Siglec5 recognizes several endogenous lipids. Because cardiolipin and 5-PAHSA were particularly potent ligands of Siglec5, they were studied in detail in further experiments. Direct interaction between Siglec5 and cardiolipin/5-PAHSA was confirmed by an ELISA-based binding assay (Fig. 4I).
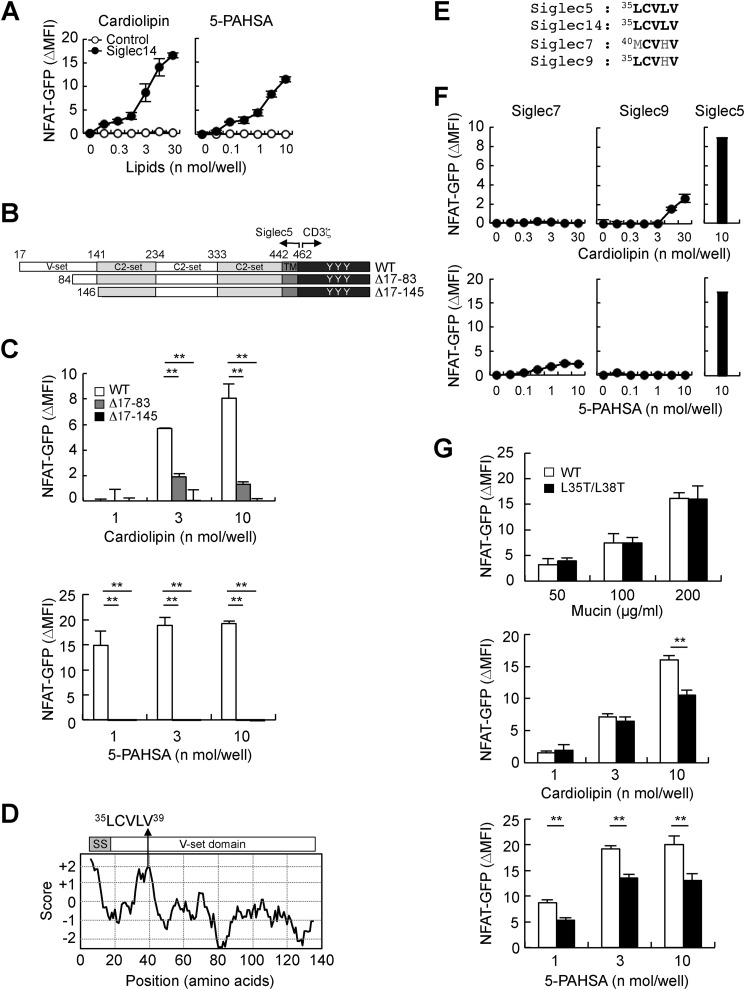
Siglec5 binds to lipid ligands through a hydrophobic stretch in the V-set domain
We next investigated the binding site in Siglec5 for lipid ligand recognition. Siglec5 contains four putative Ig-like domains, referred to as the V-set domain, and three C2-set domains (7) (Fig. 1D). Siglec14 shares the V-set domain and the first C2-set domain with Siglec5, except one amino acid residue positioned at 215 (Met in Siglec5 and Val in Siglec14) (8) (Fig. 1D). Both cardiolipin and 5-PAHSA also activated Siglec14 reporter cells (Fig. 5A), indicating that the V-set and/or first C2-set domains mediate lipid ligand recognition. To clarify which domain is required for the recognition of lipid ligands, we generated Siglec5 reporter cells lacking the V-set domain in the N terminus (Fig. 5B, Δ17–145). This deletion mutant completely lost recognition activity toward cardiolipin and 5-PAHSA, indicating that the V-set domain is necessary to recognize these lipid ligands (Fig. 5C, black columns). To further narrow down the ligand-binding site in the V-set domain, we generated an additional deletion mutant lacking approximately half of the N-terminal region of the V-set domain (Fig. 5B, Δ17–83). This mutant showed compromised responses against lipid ligands (Fig. 5C, gray columns). Thus, the N-terminal region of the V-set domain (amino acids 17–83) is required for efficient recognition of lipid ligands.
Figure 5.
The hydrophobic stretch in the N terminus of Siglec5 is required for efficient recognition of lipophilic ligands. A, Siglec14 reporter cells were stimulated with plate-coated cardiolipin or 5-PAHSA for 18 h. Induction of NFAT-GFP was analyzed by flow cytometry. B, schematic representations of N-terminal deletion mutants of Siglec5-CD3ζ chimeric receptors. C, WT and N-terminus–deleted Siglec5 reporter cells were co-cultured with plate-coated cardiolipin or 5-PAHSA. D, hydrophobicity of N-terminal region of Siglec5 was analyzed using the ExPASy-ProtScale online tool. SS, signal sequence. E, sequence alignment of hydrophobic stretches of Siglecs. F, Siglec7, -9, and -5 reporter cells were exposed to plate-coated cardiolipin or 5-PAHSA. G, Siglec5 reporter cells expressing WT and L35T/L38T mutant were stimulated with plate-coated mucin, cardiolipin, or 5-PAHSA. Data are representative of at least three independent experiments and are presented as the mean ± S.D. (error bars). **, p < 0.01.
Several receptors bind to lipophilic ligands through a hydrophobic binding pocket (30–32). Therefore, we analyzed hydrophobic features in the N-terminal region of Siglec5 using the ExPASy-ProtScale online tool (http://web.expasy.org/protscale/), and we thus identified an 35LCVLV sequence (Fig. 5D). The crystal structure of Siglec5 revealed that this hydrophobic stretch was exposed on the surface and formed a pocket-like groove (Fig. S4, blue arrows) opposite of the sialic acid–binding site, Arg-119 (Fig. S4, red arrow) (33). This hydrophobic stretch is well-conserved in Siglec5 and -14 of primates and several mammalian species (Fig. S5, A and B). In contrast, this hydrophobic stretch is compromised in human Siglec7 and -9 (Fig. 5E). Human Siglec7 and -9 reporter cells showed only weak responses to cardiolipin and 5-PAHSA, suggesting that this hydrophobic stretch mediates lipid ligand recognition (Fig. 5F). To address this possibility, we replaced two hydrophobic Leu residues with hydrophilic Thr residues (L35T/L38T). Siglec5 L35T/L38T mutant normally recognized the sialic acid ligand, mucin (Fig. 5G, top). In contrast, recognition of cardiolipin and 5-PAHSA were significantly reduced in the Siglec5 L35T/L38T mutant (Fig. 5G, middle and bottom). These results suggested that the 35LCVLV hydrophobic stretch of Siglec5 is critically involved in the recognition of lipid ligands such as cardiolipin and 5-PAHSA.
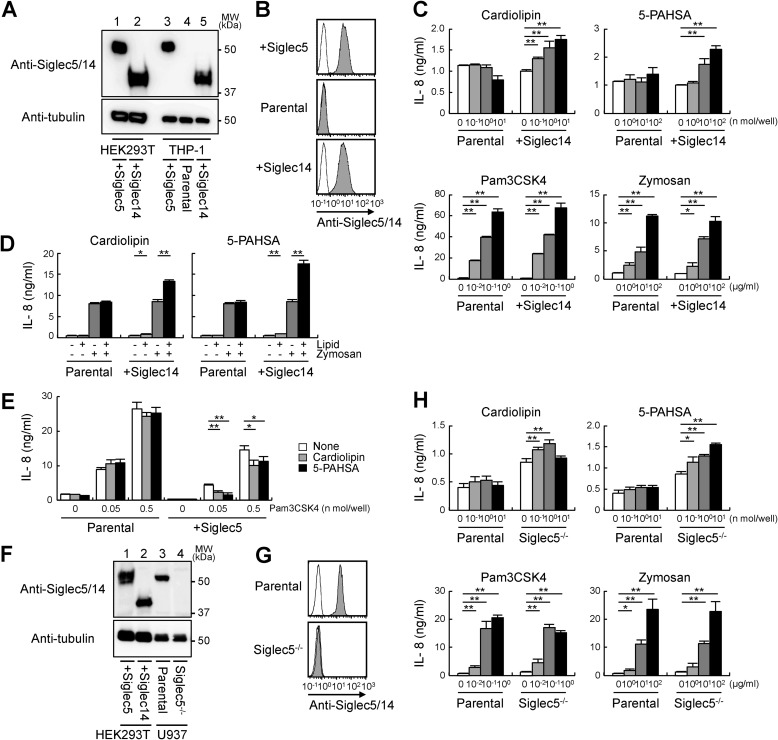
Lipid ligands modulate immune cell activation through Siglec5 and -14
We examined whether lipid ligands modulate immune cell activation. Siglec5 and -14 are mainly expressed on innate immune cells, including macrophages, monocytes, and neutrophils. Currently available antibodies recognize both Siglec5 and -14 because of their extensive sequence identity in the extracellular region (34). Although Western blot analysis can discriminate Siglec5 and -14 based on their different molecular weights (Fig. 6A, lanes 1 and 2), human monocytic THP-1 cells expressed undetectable levels of Siglec5 and -14 (Fig. 6A, lane 4). Therefore, we transfected THP-1 cells with Siglec5 or -14 expression plasmid (Fig. 6A, lanes 3 and 5). Surface expression of Siglec5 or -14 was confirmed by flow cytometry (Fig. 6B). Cardiolipin and 5-PAHSA induced IL-8 production in Siglec14-transfected cells, but not in parental cells, suggesting that they activated immune cells via Siglec14 (Fig. 6C, top). Notably, IL-8 production induced by Siglec-independent ligands Pam3CSK4 and zymosan was not enhanced in Siglec14-transfected cells (Fig. 6C, bottom). The induction of IL-8 in response to lipid ligands was not impressive compared with that in response to Psm3CSK4 and zymosan. However, lipid ligands synergistically enhanced IL-8 production induced by zymosan (Fig. 6D), suggesting that lipid ligands may serve as priming signal in the innate immune cells. Cardiolipin and 5-PAHSA suppressed IL-8 production upon Pam3CSK4 stimulation only in Siglec5-transfected cells (Fig. 6E). Collectively, these findings indicated that lipid ligands such as cardiolipin and 5-PAHSA modulate IL-8 production through Siglec5 and -14.
Figure 6.
Lipid ligands modulate innate immune responses through Siglec5 and -14. A, Siglec5 or -14 was transfected into HEK293T cells or THP-1 cells. Lysates were subjected to Western blotting with anti-Siglec5/14 or anti-tubulin as a loading control. B, THP-1 cells expressing Siglec5 or -14 were labeled (filled) or not (open) with anti-Siglec5/14 in combination with PE-labeled anti-mouse IgG and analyzed by flow cytometry. C, parental and Siglec14-transfected THP-1 cells were stimulated with plate-coated cardiolipin and 5-PAHSA or soluble LPS and Pam3CSK4 for 18 h. Concentrations of IL-8 in the supernatants were determined by ELISA. D, parental and Siglec14-transfected THP-1 cells were stimulated with plate-coated cardiolipin (1 nmol/well) and 5-PAHSA (30 nmol/well) in combination with zymosan (10 μg/ml). E, Pam3CSK4 was coated on a plate alone or in combination with cardiolipin or 5-PAHSA to stimulate parental and Siglec5-transfected THP-1 cells. F, lysates of parental or Siglec5-deficient U937 cells were subjected to Western blotting with anti-Siglec5/14 or anti-tubulin. G, parental or Siglec5-deficient U937 cells were labeled (filled) or not (open) with anti-Siglec5/14 and analyzed by flow cytometry. H, parental and Siglec5-deficient U937 cells were stimulated with plate-coated cardiolipin and 5-PAHSA or with soluble LPS and Pam3CSK4 for 18 h. Data are representative of at least three independent experiments and are presented as the mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01.
Next, we investigated whether lipid ligands modulate cell activation through endogenous Siglec receptors. Human monocytic U937 cells abundantly express Siglec5, whereas expression of Siglec14 was undetectable (Fig. 6, F (lane 3) and G (top)). To examine cellular responses through interaction between lipid ligands and endogenous Siglec5, we generated Siglec5-deficient cells by using the CRISPR-Cas9 system (Fig. 6, F (lane 4) and G (bottom)). Both cardiolipin and 5-PAHSA had no impact on IL-8 production in U937 parental cells, whereas these lipid ligands did induce IL-8 production in Siglec5-deficient cells (Fig. 6H, top). This may be because U937 cells express both the inhibitory Siglec5 receptor and activating receptors for lipid ligands such as Toll-like receptors (35, 36). Siglec5 deficiency compromised inhibitory signals and potentiated IL-8 production. Siglec-independent ligands such as LPS, zymosan, and Pam3CSK4 stimulated IL-8 production equally in U937 parental and Siglec5-deficient cells (Fig. 6H, bottom). Taken together, these findings indicated that endogenous Siglec5 negatively regulates cellular responses against cardiolipin and 5-PAHSA.
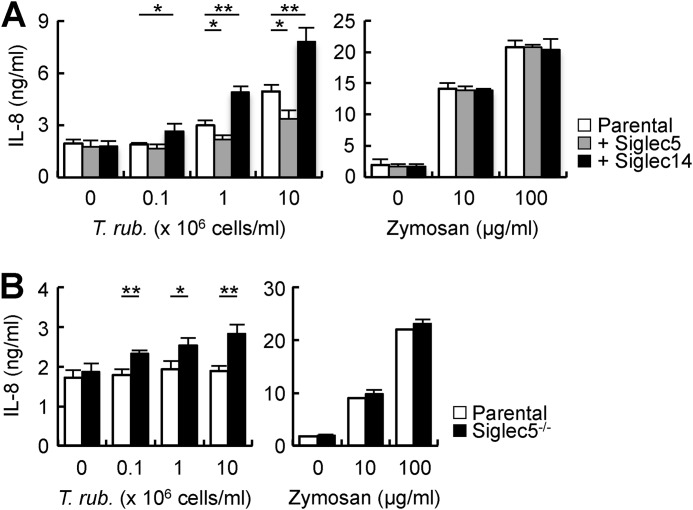
Siglec5 and -14 modulated innate immune responses against T. rubrum
Finally, we examined the effect of Siglec on the innate immune responses against Trichophyton spp. T. rubrum–induced IL-8 production was reduced in Siglec5-expressing THP-1 cells and enhanced in Siglec14-expressing THP-1 cells (Fig. 7A). T. rubrum failed to induce IL-8 production in U937 parental cells. However, Siglec5-deficient U937 cells showed a slight but significant response to T. rubrum (Fig. 7B). These data suggested that Siglec5 and -14 modulated innate immune responses against T. rubrum.
Figure 7.
Siglec5 and -14 modulated innate immune responses against T. rubrum. A, THP-1 cells expressing Siglec5 or -14 were stimulated with T. rubrum and zymosan for 18 h. Concentrations of IL-8 in the supernatants were determined by ELISA. B, parental and Siglec5-deficient U937 cells were stimulated with T. rubrum and zymosan for 18 h.
Discussion
In this study, we found that Siglec5 and -14 recognize Trichophyton spp., and we identified alkanes and TAGs as novel lipophilic ligands. Although TAGs and alkanes clearly show ligand activity toward Siglec5 and -14, it is still questionable whether TAGs and alkanes are actually involved in the recognition of Trichophyton spp. by Siglec5 and -14 because TAGs are generally stored in cytoplasmic lipid droplets as an energy source in eukaryotic and prokaryotic cells (37). Some fungi and bacteria incorporate or synthesize alkanes in cytosolic compartments to utilize them as a carbon source (38, 39). Thus, TAGs and alkanes are not likely to be exposed on the cell surface and to be recognized by Siglec5 and -14. In addition, TAGs and alkanes are broadly distributed in eukaryotic and prokaryotic cells. Actually, Siglec5 and -14 also recognized hydrophobic extracts from other fungi that failed to activate Siglec5 and -14 reporter cells (data not shown). We cannot exclude the possibility that TAGs and alkanes are exposed on the cell surface of Trichophyton spp. in the steady state or in certain conditions, such as during hypha formation or upon plasma membrane rupture. Further investigation is required to conclude whether TAGs and alkanes mediate ligand activity in Trichophyton spp. Structure–activity relationship analysis of alkanes and derivatives revealed that Siglec5 can broadly recognize alkyl chains of a certain length and their derivatives, and TAGs do contain multiple alkyl chains.
Siglec5 also interacted with DAGs and MAGs. DAGs are anchored in the inner layer of the plasma membrane and function as a second messenger to activate protein kinase C. A certain portion of DAGs is also found in the outer layer; however, the functions of exposed DAGs remain unknown (40). Siglecs may bind to these DAGs in cis- or trans-mode. MAGs and DAGs are added to diverse food products as emulsifiers (41). It is possible that these free MAGs and DAGs might interact with Siglec receptors and modulate immune responses.
Most of the lipid ligands showed marginal ligand activity. Although MAGs and some fatty acids had moderate ligand activity, they also compromised cell viability. Some Siglec receptors mediate cell death upon ligand ligation (42–44). However, the cytotoxic effect of MAGs and fatty acids was independent of Siglecs because they also killed NFAT-GFP reporter cells that did not express Siglecs (data not shown). Thus, a detergent effect or modulation of a metabolic pathway may have reduced cell viability.
Through extensive screening, we finally identified two potent lipid ligands, cardiolipin and 5-PAHSA. Both modulated innate immune responses through Siglec5 and -14 in human monocytic cell lines. Cardiolipin is preferentially located in the inner membranes of mitochondria (45). During apoptosis, cardiolipin relocates to the outer mitochondrial membranes and interacts with Bid to promote mitochondrial dysfunction (46, 47). A portion of cardiolipin is exposed on the cell surface during apoptosis and triggers the production of anti-phospholipid antibodies (48). Siglec5 and -14 may recognize cardiolipin on apoptotic cells. In agreement with this assumption, phagocytosis of apoptotic bodies by macrophages was inhibited with anti-Siglec5 antibody and sialo-oligosaccharide ligands of Siglec5 (49). Oxidized but not native cardiolipin had a pro-inflammatory effect in monocytes and neutrophils (50). It is of interest to examine the binding selectivity of Siglec5 and -14 toward oxidized and native cardiolipins.
PAHSAs are endogenous lipids with anti-inflammatory and anti-diabetic properties detected in a various organs, such as adipose tissue, the kidneys, and serum. They act on multiple cell linages, including adipocytes, pancreatic beta cells, enteroendocrine cells, and macrophages. GP120 and GP40 have been reported to mediate their functions in adipocytes and beta cells, respectively (29, 51); however, functional receptors in other cells remain unclear. Our data suggest that Siglec5 may contribute to the anti-inflammatory effects of PAHSA, especially in macrophages and other myeloid cells.
Siglec is constitutively associated with endogenous sialic acid in cis-interaction (52, 53). Given that lipid ligands bind to Siglec5 and -14 via a binding site distinct from that for sialic acid, they may access Siglecs in a noncompetitive manner with sialic acid. Indeed, C14 alkane and mucin additively activated Sigelc5 reporter cells (data not shown). Thus, lipid ligands may interact with Siglec5 and -14 even if they are bound with sialic acid.
Mycobacteria contain a wide variety of hydrophobic components on the cell surface. Actually, the mycobacteria H37Ra and BCG weakly activated Siglec5 and -14 reporter cells (data not shown). It is tempting to speculate that Siglec5 and -14 may modulate anti-mycobacterial immunity. Although Siglec5 and -14 failed to recognize trehalose dimycolate and mycolic acids, which are major hydrophobic lipids in the mycobacterial cell wall (data not shown), it would be interesting to search for novel ligands for Siglec5 and -14 in mycobacteria.
Our finding that Siglecs have a potential to recognize lipophilic ligands may expand our understanding of the biological function and importance of Siglecs both in steady state and in pathological conditions, including metabolic disorders.
Experimental procedures
Lipids
Tetradecane (172456), pentadecane (P3406), octadecane (O0652), eicosane (219274), docosane (134457), tetracosane (T8752), squalane (234311), cardiolipin (C1649), C16 ceramide (43799), sphingomyelin (S0756), cholesterol (C8667), cholesteryl linoleate (C0289), and linoleic acid (L1376) were purchased from Sigma-Aldrich. Triacylglycerol 18:3 (10009758), diacylglycerol 18:2 (16481), monoacylglycerol 18:2 (10008869), phosphatidylcholine (10009473), phosphatidylethanolamine (15092), phosphatidylserine (15088), phosphatidylglycerol (15085), 5-PAHSA (17043), and 9-PAHSA (17037) were obtained from Cayman. Cyclopentadecane (C1224), 1-octadecanol (O0006), 1-bromooctadecane (B0625), octadecanedioic acid (O0222), hexadecane (H0066), and stearic acid (S0163) were from Tokyo Chemical Industry Co. Triacylglycerol 18:0 (B-41), 18:1 (B-43), and 18:2 (B-44) were purchased from SRL. Octane (05-0800) and decane (05-1000) were obtained from Larodan Fine Chemicals. Dodecane was from Nacalai Tesque. Phosphatidylinositol (ab146179) was purchased from Abcam. Glycerol (075000616) was obtained from Wako.
Fungi
Trichophyton spp. were provided by Dr. T. Yaguchi (Chiba University, Japan). The fungus was cultured on potato dextrose agar plates (Eiken Chemical) at 30 °C for 4 days. For lipid extraction, cells were collected and treated with CHCl3/MeOH (2:1, v/v) at room temperature for 1 h, and the CHCl3:MeOH layer was collected.
Cells
NFAT-GFP reporter cells were described previously (54). Siglec3, -5, -7, and -9 genes were cloned from a cDNA library of human peripheral blood cells. The extracellular domain of Siglecs was fused with transmembrane and cytoplasmic domains of CD3ζ. Siglec reporter cell lines were prepared by introducing Siglec-CD3ζ chimeric receptors into NFAT-GFP reporter cells by retrovirus-mediated gene transfer (Fig. 1A). Siglec14 reporter cells were prepared by introducing Siglec14 and DAP12 into NFAT-GFP reporter cells (Fig. 1E). Cells were stimulated in 96-well plates coated with mucin (porcine stomach, WAKO), anti-FLAG mAb (WAKO), or lipids. Mucin and anti-FLAG mAb were diluted in borate buffer and added to the plate. Then the plate was incubated for at least 2 h at 37 °C. The wells were then washed with PBS two times. Lipid ligands were diluted in isopropyl alcohol and added to the plate, followed by evaporation of the solvent in a laminar flow cabinet. Siglec reporter cell lines were cultured for 18 h on the coated plate at 37 °C. Induction of NFAT-GFP was analyzed by flow cytometry (MACS Quant, Miltenyi Biotec).
Siglec5- or Siglec14-transfected THP-1 cells were established by lentivirus-mediated gene transfer. Siglec5-deficient U937 cells were generated by using the CRISPR-Cas9 system. gRNA sequence was gcagatgctcctttcgagcc. THP-1 and U937 cells were stimulated with plate-coated lipids or soluble Pam3CSK4 and zymosan for 24 h. For the experiment shown in Fig. 6D, Pam3CSK4 and lipids were diluted in isopropyl alcohol and coated on the plate. Concentrations of IL-8 in the supernatants were measured by ELISA (R&D Systems).
Siglec-Fc binding assay
For cell-surface staining, fungal cells were treated with 10 μg/ml Fc, Siglec5-Fc, or Siglec14-Fc (R&D Systems) on ice for 1 h. Fungal cells were then washed with Hanks' buffer, and bound proteins were detected by staining with anti-human IgG-Cy3 pAb (Jackson ImmunoResearch). The fluorescence intensity was measured by flow cytometry (FACSCalibur, BD Biosciences). For the ELISA-based binding assay, 10 μg/ml Fc, Siglec5-Fc, or Siglec14-Fc was incubated with plate-coated lipid ligands at room temperature for 1 h. Plates were then washed with PBS-Tween 20 (0.05%) three times, and bound proteins were detected by labeling with anti-human IgG-HRP pAb (Jackson ImmunoResearch) followed by the addition of TMBZ colorimetric substrate (Sumilon). Peroxidase activity was measured with an iMark microplate absorbance reader (Bio-Rad).
Detection of Siglecs
Surface expression of Siglec5/14 was analyzed by flow cytometry using anti-Siglec5/14 mAb (clone 194128, R&D Systems) in combination with anti-mouse IgG-PE pAb (Jackson ImmunoResearch). Total expression of Siglec5/14 was analyzed by Western blotting using anti-Siglec5/14 pAb (R&D Systems) and anti-goat IgG-HRP pAb (Jackson ImmunoResearch).
Isolation of Siglec5 ligands in Trichophyton spp.
For enzymatic removal of glycan ligands, fungal cells were treated with 100 milliunits/ml sialidase (Nacalai Tesque) at 50 °C or 5,000 units/ml N-glycosidase F (New England BioLabs) at 37 °C for 3 h. To inactivate protein ligands, fungal cells were treated with 50 μg/ml Proteinase K (Roche Applied Science) at 37 °C for 30 min or heated at 95 °C for 10 min. Fungal cells were then extensively washed with PBS to avoid contamination with enzymatic activity in further experiments.
To isolate Siglec5 ligands, fungal cells were separated into lipophilic extract and delipidized biomass using CHCl3/MeOH (2:1, v/v). The lipophilic extract was concentrated in a SpeedVac and was dissolved in hexane, resulting in a soluble extract and an insoluble precipitate. The hexane-insoluble precipitate was dissolved in CHCl3/MeOH (2:1, v/v). The hexane-soluble extract was further separated into six fractions by HPTLC (Merck) with CHCl3/MeOH/H2O (65:25:4, v/v/v). Lipophilic components were visualized by staining with iodide. Each fraction was extracted from the HPTLC plate and dissolved in CHCl3/MeOH (2:1, v/v). Ligand activity was examined by using Siglec5 reporter cells.
Chemical analysis of Siglec5 ligands
1H NMR spectra were recorded on an Agilent INOVA 600 spectrometer. The operating conditions were as follows: 1H: 600 MHz, 298 K, CDCl3. The 1H chemical shift was referenced to that of tetramethylsilane (δH 0). All spectra were processed using MestReNova version 11.0.4 (Mestrelab Research S.L.). ESI-TOF-MS was conducted with a Bruker micro-TOF mass spectrometer in the positive ESI mode (Bruker Daltonics).
ESI-TOF-MS of Fraction 4: m/z = 853.7290 (calcd. for C53H98NaO6 Δppm = +3.4), m/z = 877.7294 (calcd. for C55H98NaO6 Δppm = +3.8), m/z = 881.7610 (calcd. for C55H102NaO6 Δppm = +4.1), m/z = 901.7289 (calcd. for C57H98NaO6 Δppm = +3.3), m/z = 905.7587 (calcd. for C57H102NaO6 Δppm = +1.8).
For methanolysis, Fraction 4 (∼100 μg) was heated with 10% HCl/MeOH (100 μl) in a sealed tube at 80 °C for 3 h. The reaction mixture was diluted with MeOH (1.0 ml) and extracted twice with n-hexane (100 μl). The n-hexane extract was concentrated in vacuo to yield a mixture of FAMEs. The FAMEs were dissolved in acetone and subjected to GC-MS (Shimadzu QP-2010SE with INERTCAP 5MS/SIL (0.25-mm inner diameter, 30-m length), GL Science Inc. (Tokyo, Japan), column temperature 100–280 °C, rate of temperature increase: 10 °C/min). The retention time (tR), proportion (%), and m/z value (M+) of each FAME were as follows: methyl hexadecanoate (16:0), tR (min) = 14.5, 33.6%, m/z = 270 (M+); methyl octadecanediate (C18:2), tR (min) = 16.2, 30.0%, m/z = 294 (M+); methyl octadecanoate (C18:1), tR (min) = 16.3, 12.5%, m/z = 296 (M+); methyl octadecanoate (18:0), tR (min) = 16.5, 24.6%, m/z = 298 (M+).
Fraction 5 was diluted with n-hexane and subjected to GC-MS. The retention time (tR), proportion (%) and m/z value (M+) of each saturated alkane were as follows: tetradecane (C14H30), tR (min) = 8.2, 9.2%, m/z = 198 (M+); pentadecane (C15H32), tR (min) = 9.4, 7.8%, m/z = 212 (M+); hexadecane (C16H34), tR (min) = 10.8, 15.1%, m/z = 226 (M+); heptadecane (C17H36), tR (min) = 12.0, 12.2%, m/z = 240 (M+); octadecane (C18H38), tR (min) = 13.1, 7.2%, m/z = 254 (M+); nonadecane (C19H40), tR (min) = 14.4, 10.4%, m/z = 268 (M+); icosane (C20H42), tR (min) = 15.3, 4.3%, m/z = 282 (M+); henicosane (C21H44), tR (min) = 16.5, 6.2%, m/z = 296 (M+).
Statistical analysis
All experiments were repeated at least three times independently. Data are expressed as the mean ± S.D. All data were analyzed using an unpaired two-tailed Student's t test. A p value of <0.05 was considered significant.
Author contributions
Y. M. designed experiments; R. S., T. M., and Y. M. performed experiments; S. S. and S. Y. provided materials; S. Y., Y. T., and H. Y. supervised the study; Y. M. wrote the manuscript with T. M. and R. S. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Fumika Mi-ichi for discussion of this work; Ritsuko Yoshida for technical assistance; and Sizuko Furukawa and Keiko Imokawa for secretarial assistance. The flow cytometric analysis was performed using a MACSQuant and FACSCalibur at the Analytical Research Center for Experimental Sciences, Saga University.
This work was supported by Grants-in-Aid for Scientific research 17K08888 (to Y. M.) and 16K08842 (to H. Y.) and the Takeda Science Foundation (to Y. M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
- Siglec
- sialic acid–binding Ig-like lectin
- DAP12
- DNAX-activating protein of 12 kDa
- Fc
- fragment crystallizable
- HPTLC
- high-performance TLC
- ITAM
- immunoreceptor tyrosine-based activation motif
- ITIM
- immunoreceptor tyrosine-based inhibitory motif
- MFI
- fluorescence intensity
- NFAT
- nuclear factor of activated T-cells
- pAb
- polyclonal antibody
- PAHSA
- palmitic acid hydroxystearic acids
- TAG
- triacylglycerol
- LPS
- lipopolysaccharide
- ESI
- electrospray ionization
- FAME
- fatty acid methyl ester
- DAG
- diacylglycerol
- MAG
- monoacylglycerol
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- IL-8
- interleukin-8
- HRP
- horseradish peroxidase
- tR
- retention time.
References
- 1. Takeuchi O., and Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- 2. Toyonaga K., Torigoe S., Motomura Y., Kamichi T., Hayashi J. M., Morita Y. S., Noguchi N., Chuma Y., Kiyohara H., Matsuo K., Tanaka H., Nakagawa Y., Sakuma T., Ohmuraya M., Yamamoto T., et al. (2016) C-type lectin receptor DCAR recognizes mycobacterial phosphatidyl-inositol mannosides to promote a Th1 response during infection. Immunity 45, 1245–1257 10.1016/j.immuni.2016.10.012 [DOI] [PubMed] [Google Scholar]
- 3. Yonekawa A., Saijo S., Hoshino Y., Miyake Y., Ishikawa E., Suzukawa M., Inoue H., Tanaka M., Yoneyama M., Oh-Hora M., Akashi K., and Yamasaki S. (2014) Dectin-2 is a direct receptor for mannose-capped lipoarabinomannan of mycobacteria. Immunity 41, 402–413 10.1016/j.immuni.2014.08.005 [DOI] [PubMed] [Google Scholar]
- 4. Miyake Y., Toyonaga K., Mori D., Kakuta S., Hoshino Y., Oyamada A., Yamada H., Ono K., Suyama M., Iwakura Y., Yoshikai Y., and Yamasaki S. (2013) C-type lectin MCL is an FcRγ-coupled receptor that mediates the adjuvanticity of mycobacterial cord factor. Immunity 38, 1050–1062 10.1016/j.immuni.2013.03.010 [DOI] [PubMed] [Google Scholar]
- 5. Crocker P. R., Paulson J. C., and Varki A. (2007) Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 10.1038/nri2056 [DOI] [PubMed] [Google Scholar]
- 6. Varki A., and Angata T. (2006) Siglecs—the major subfamily of I-type lectins. Glycobiology 16, 1R–27R 10.1093/glycob/cwj008 [DOI] [PubMed] [Google Scholar]
- 7. Crocker P. R. (2005) Siglecs in innate immunity. Curr. Opin. Pharmacol. 5, 431–437 10.1016/j.coph.2005.03.003 [DOI] [PubMed] [Google Scholar]
- 8. Angata T., Hayakawa T., Yamanaka M., Varki A., and Nakamura M. (2006) Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. FASEB J. 20, 1964–1973 10.1096/fj.06-5800com [DOI] [PubMed] [Google Scholar]
- 9. Angata T., and Varki A. (2002) Chemical diversity in the sialic acids and related α-keto acids: an evolutionary perspective. Chem. Rev. 102, 439–469 10.1021/cr000407m [DOI] [PubMed] [Google Scholar]
- 10. O'Reilly M. K., and Paulson J. C. (2010) Multivalent ligands for siglecs. Methods Enzymol. 478, 343–363 10.1016/S0076-6879(10)78017-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lübbers J., Rodríguez E., and van Kooyk Y. (2018) Modulation of immune tolerance via siglec-sialic acid interactions. Front. Immunol. 9, 2807 10.3389/fimmu.2018.02807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khatua B., Roy S., and Mandal C. (2013) Sialic acids siglec interaction: a unique strategy to circumvent innate immune response by pathogens. Indian J. Med. Res. 138, 648–662 [PMC free article] [PubMed] [Google Scholar]
- 13. Avril T., Wagner E. R., Willison H. J., and Crocker P. R. (2006) Sialic acid-binding immunoglobulin-like lectin 7 mediates selective recognition of sialylated glycans expressed on Campylobacter jejuni lipooligosaccharides. Infect. Immun. 74, 4133–4141 10.1128/IAI.02094-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heikema A. P., Bergman M. P., Richards H., Crocker P. R., Gilbert M., Samsom J. N., van Wamel W. J., Endtz H. P., and van Belkum A. (2010) Characterization of the specific interaction between sialoadhesin and sialylated Campylobacter jejuni lipooligosaccharides. Infect. Immun. 78, 3237–3246 10.1128/IAI.01273-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones C., Virji M., and Crocker P. R. (2003) Recognition of sialylated meningococcal lipopolysaccharide by siglecs expressed on myeloid cells leads to enhanced bacterial uptake. Mol. Microbiol. 49, 1213–1225 10.1046/j.1365-2958.2003.03634.x [DOI] [PubMed] [Google Scholar]
- 16. Khatua B., Bhattacharya K., and Mandal C. (2012) Sialoglycoproteins adsorbed by Pseudomonas aeruginosa facilitate their survival by impeding neutrophil extracellular trap through siglec-9. J. Leukoc. Biol. 91, 641–655 10.1189/jlb.0511260 [DOI] [PubMed] [Google Scholar]
- 17. Van Breedam W., Van Gorp H., Zhang J. Q., Crocker P. R., Delputte P. L., and Nauwynck H. J. (2010) The M/GP(5) glycoprotein complex of porcine reproductive and respiratory syndrome virus binds the sialoadhesin receptor in a sialic acid-dependent manner. PLoS Pathog. 6, e1000730 10.1371/journal.ppat.1000730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zou Z., Chastain A., Moir S., Ford J., Trandem K., Martinelli E., Cicala C., Crocker P., Arthos J., and Sun P. D. (2011) Siglecs facilitate HIV-1 infection of macrophages through adhesion with viral sialic acids. PLoS One 6, e24559 10.1371/journal.pone.0024559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Monteiro V. G., Lobato C. S., Silva A. R., Medina D. V., de Oliveira M. A., Seabra S. H., de Souza W., and DaMatta R. A. (2005) Increased association of Trypanosoma cruzi with sialoadhesin positive mice macrophages. Parasitol. Res. 97, 380–385 10.1007/s00436-005-1460-1 [DOI] [PubMed] [Google Scholar]
- 20. Chatterjee M., Chava A. K., Kohla G., Pal S., Merling A., Hinderlich S., Unger U., Strasser P., Gerwig G. J., Kamerling J. P., Vlasak R., Crocker P. R., Schauer R., Schwartz-Albiez R., and Mandal C. (2003) Identification and characterization of adsorbed serum sialoglycans on Leishmania donovani promastigotes. Glycobiology 13, 351–361 10.1093/glycob/cwg027 [DOI] [PubMed] [Google Scholar]
- 21. Carlin A. F., Chang Y. C., Areschoug T., Lindahl G., Hurtado-Ziola N., King C. C., Varki A., and Nizet V. (2009) Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J. Exp. Med. 206, 1691–1699 10.1084/jem.20090691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ali S. R., Fong J. J., Carlin A. F., Busch T. D., Linden R., Angata T., Areschoug T., Parast M., Varki N., Murray J., Nizet V., and Varki A. (2014) Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J. Exp. Med. 211, 1231–1242 10.1084/jem.20131853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fong J. J., Sreedhara K., Deng L., Varki N. M., Angata T., Liu Q., Nizet V., and Varki A. (2015) Immunomodulatory activity of extracellular Hsp70 mediated via paired receptors Siglec-5 and Siglec-14. EMBO J. 34, 2775–2788 10.15252/embj.201591407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Secundino I., Lizcano A., Roupé K. M., Wang X., Cole J. N., Olson J., Ali S. R., Dahesh S., Amayreh L. K., Henningham A., Varki A., and Nizet V. (2016) Host and pathogen hyaluronan signal through human siglec-9 to suppress neutrophil activation. J. Mol. Med. 94, 219–233 10.1007/s00109-015-1341-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohtsuka M., Arase H., Takeuchi A., Yamasaki S., Shiina R., Suenaga T., Sakurai D., Yokosuka T., Arase N., Iwashima M., Kitamura T., Moriya H., and Saito T. (2004) NFAM1, an immunoreceptor tyrosine-based activation motif-bearing molecule that regulates B cell development and signaling. Proc. Natl. Acad. Sci. U.S.A. 101, 8126–8131 10.1073/pnas.0401119101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yamaji T., Teranishi T., Alphey M. S., Crocker P. R., and Hashimoto Y. (2002) A small region of the natural killer cell receptor, Siglec-7, is responsible for its preferred binding to α2,8-disialyl and branched α2,6-sialyl residues: a comparison with Siglec-9. J. Biol. Chem. 277, 6324–6332 10.1074/jbc.M110146200 [DOI] [PubMed] [Google Scholar]
- 27. Avril T., North S. J., Haslam S. M., Willison H. J., and Crocker P. R. (2006) Probing the cis interactions of the inhibitory receptor Siglec-7 with α2,8-disialylated ligands on natural killer cells and other leukocytes using glycan-specific antibodies and by analysis of α2,8-sialyltransferase gene expression. J. Leukoc. Biol. 80, 787–796 10.1189/jlb.1005559 [DOI] [PubMed] [Google Scholar]
- 28. Esquenazi D., Rozental S., Alviano C. S., Travassos L. R., and Schauer R. (2003) Sialic acids are absent from the dermatophytes Trichophyton mentagrophytes and Trichophyton rubrum. Mycoses 46, 197–202 10.1046/j.1439-0507.2003.00873.x [DOI] [PubMed] [Google Scholar]
- 29. Yore M. M., Syed I., Moraes-Vieira P. M., Zhang T., Herman M. A., Homan E. A., Patel R. T., Lee J., Chen S., Peroni O. D., Dhaneshwar A. S., Hammarstedt A., Smith U., McGraw T. E., Saghatelian A., and Kahn B. B. (2014) Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 159, 318–332 10.1016/j.cell.2014.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zeng Z., Castaño A. R., Segelke B. W., Stura E. A., Peterson P. A., and Wilson I. A. (1997) Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science 277, 339–345 10.1126/science.277.5324.339 [DOI] [PubMed] [Google Scholar]
- 31. Furukawa A., Kamishikiryo J., Mori D., Toyonaga K., Okabe Y., Toji A., Kanda R., Miyake Y., Ose T., Yamasaki S., and Maenaka K. (2013) Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL. Proc. Natl. Acad. Sci. U.S.A. 110, 17438–17443 10.1073/pnas.1312649110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nicholas D. A., Zhang K., Hung C., Glasgow S., Aruni A. W., Unternaehrer J., Payne K. J., Langridge W. H. R., and De Leon M. (2017) Palmitic acid is a toll-like receptor 4 ligand that induces human dendritic cell secretion of IL-1β. PLoS One 12, e0176793 10.1371/journal.pone.0176793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhuravleva M. A., Trandem K., and Sun P. D. (2008) Structural implications of Siglec-5-mediated sialoglycan recognition. J. Mol. Biol. 375, 437–447 10.1016/j.jmb.2007.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamanaka M., Kato Y., Angata T., and Narimatsu H. (2009) Deletion polymorphism of SIGLEC14 and its functional implications. Glycobiology 19, 841–846 10.1093/glycob/cwp052 [DOI] [PubMed] [Google Scholar]
- 35. Rogero M. M., and Calder P. C. (2018) Obesity, inflammation, Toll-like receptor 4 and fatty acids. Nutrients 10, E432 10.3390/nu10040432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Greene C. M., McElvaney N. G., O'Neill S. J., and Taggart C. C. (2004) Secretory leucoprotease inhibitor impairs Toll-like receptor 2- and 4-mediated responses in monocytic cells. Infect. Immun. 72, 3684–3687 10.1128/IAI.72.6.3684-3687.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sturley S. L., and Hussain M. M. (2012) Lipid droplet formation on opposing sides of the endoplasmic reticulum. J. Lipid Res. 53, 1800–1810 10.1194/jlr.R028290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fukuda R. (2013) Metabolism of hydrophobic carbon sources and regulation of it in n-alkane-assimilating yeast Yarrowia lipolytica. Biosci. Biotechnol. Biochem. 77, 1149–1154 10.1271/bbb.130164 [DOI] [PubMed] [Google Scholar]
- 39. Sinha M., Sørensen A., Ahamed A., and Ahring B. K. (2015) Production of hydrocarbons by Aspergillus carbonarius ITEM 5010. Fungal Biol. 119, 274–282 10.1016/j.funbio.2015.01.001 [DOI] [PubMed] [Google Scholar]
- 40. Ueda Y., Makino A., Murase-Tamada K., Sakai S., Inaba T., Hullin-Matsuda F., and Kobayashi T. (2013) Sphingomyelin regulates the transbilayer movement of diacylglycerol in the plasma membrane of Madin-Darby canine kidney cells. FASEB J. 27, 3284–3297 10.1096/fj.12-226548 [DOI] [PubMed] [Google Scholar]
- 41. Suman M., Silva G., Catellani D., Bersellini U., Caffarra V., and Careri M. (2009) Determination of food emulsifiers in commercial additives and food products by liquid chromatography/atmospheric-pressure chemical ionisation mass spectrometry. J. Chromatogr. A 1216, 3758–3766 10.1016/j.chroma.2009.02.055 [DOI] [PubMed] [Google Scholar]
- 42. Nutku E., Aizawa H., Hudson S. A., and Bochner B. S. (2003) Ligation of Siglec-8: a selective mechanism for induction of human eosinophil apoptosis. Blood 101, 5014–5020 10.1182/blood-2002-10-3058 [DOI] [PubMed] [Google Scholar]
- 43. von Gunten S., Yousefi S., Seitz M., Jakob S. M., Schaffner T., Seger R., Takala J., Villiger P. M., and Simon H. U. (2005) Siglec-9 transduces apoptotic and nonapoptotic death signals into neutrophils depending on the proinflammatory cytokine environment. Blood 106, 1423–1431 10.1182/blood-2004-10-4112 [DOI] [PubMed] [Google Scholar]
- 44. Nguyen K. A., Hamzeh-Cognasse H., Palle S., Anselme-Bertrand I., Arthaud C. A., Chavarin P., Pozzetto B., Garraud O., and Cognasse F. (2014) Role of Siglec-7 in apoptosis in human platelets. PLoS One 9, e106239 10.1371/journal.pone.0106239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schlame M., Rua D., and Greenberg M. L. (2000) The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 39, 257–288 10.1016/S0163-7827(00)00005-9 [DOI] [PubMed] [Google Scholar]
- 46. Sorice M., Circella A., Cristea I. M., Garofalo T., Di Renzo L., Alessandri C., Valesini G., and Esposti M. D. (2004) Cardiolipin and its metabolites move from mitochondria to other cellular membranes during death receptor-mediated apoptosis. Cell Death Differ. 11, 1133–1145 10.1038/sj.cdd.4401457 [DOI] [PubMed] [Google Scholar]
- 47. Gonzalvez F., Pariselli F., Dupaigne P., Budihardjo I., Lutter M., Antonsson B., Diolez P., Manon S., Martinou J. C., Goubern M., Wang X., Bernard S., and Petit P. X. (2005) tBid interaction with cardiolipin primarily orchestrates mitochondrial dysfunctions and subsequently activates Bax and Bak. Cell Death Differ. 12, 614–626 10.1038/sj.cdd.4401571 [DOI] [PubMed] [Google Scholar]
- 48. Sorice M., Circella A., Misasi R., Pittoni V., Garofalo T., Cirelli A., Pavan A., Pontieri G. M., and Valesini G. (2000) Cardiolipin on the surface of apoptotic cells as a possible trigger for antiphospholipids antibodies. Clin. Exp. Immunol. 122, 277–284 10.1046/j.1365-2249.2000.01353.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rapoport E. M., Sapot'ko Y. B., Pazynina G. V., Bojenko V. K., and Bovin N. V. (2005) Sialoside-binding macrophage lectins in phagocytosis of apoptotic bodies. Biochemistry 70, 330–338 10.1007/s10541-005-0119-y [DOI] [PubMed] [Google Scholar]
- 50. Wan M., Hua X., Su J., Thiagarajan D., Frostegård A. G., Haeggström J. Z., and Frostegård J. (2014) Oxidized but not native cardiolipin has pro-inflammatory effects, which are inhibited by Annexin A5. Atherosclerosis 235, 592–598 10.1016/j.atherosclerosis.2014.05.913 [DOI] [PubMed] [Google Scholar]
- 51. Syed I., Lee J., Moraes-Vieira P. M., Donaldson C. J., Sontheimer A., Aryal P., Wellenstein K., Kolar M. J., Nelson A. T., Siegel D., Mokrosinski J., Farooqi I. S., Zhao J. J., Yore M. M., Peroni O. D., Saghatelian A., and Kahn B. B. (2018) Palmitic acid hydroxystearic acids activate GPR40, which is involved in their beneficial effects on glucose homeostasis. Cell Metab. 27, 419–427.e4 10.1016/j.cmet.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Razi N., and Varki A. (1998) Masking and unmasking of the sialic acid-binding lectin activity of CD22 (Siglec-2) on B lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 95, 7469–7474 10.1073/pnas.95.13.7469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Han S., Collins B. E., Bengtson P., and Paulson J. C. (2005) Homomultimeric complexes of CD22 in B cells revealed by protein-glycan cross-linking. Nat. Chem. Biol. 1, 93–97 10.1038/nchembio713 [DOI] [PubMed] [Google Scholar]
- 54. Yamasaki S., Ishikawa E., Sakuma M., Hara H., Ogata K., and Saito T. (2008) Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat. Immunol. 9, 1179–1188 10.1038/ni.1651 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.