Abstract
Anaerobic ammonium oxidation (anammox) is a microbial process responsible for significant nitrogen loss from the oceans and other ecosystems. The redox reactions at the heart of anammox are catalyzed by large multiheme enzyme complexes that rely on small cytochrome c proteins for electron shuttling. Among the most highly abundant of these cytochromes is a unique heterodimeric complex composed of class I and class II c-type cytochromes called NaxLS, which has distinctive biochemical and spectroscopic properties. Here, we present the 1.7 Å resolution crystal structure of this complex from the anammox organism Kuenenia stuttgartiensis (KsNaxLS). The structure reveals that the heme irons in each subunit exhibit a rare His/Cys ligation, which, as we show by substitution, causes the observed unusual spectral properties. Unlike its individual subunits, the KsNaxLS complex binds nitric oxide (NO) only at the distal heme side, forming 6cNO adducts. This is likely due to steric immobilization of the proximal heme-binding motifs upon complex formation, a finding that may be of functional relevance, because NO is an intermediate in the central anammox metabolism. Pulldown experiments with K. stuttgartiensis cell-free extract showed that the KsNaxLS complex binds specifically to one of the central anammox enzyme complexes, hydrazine synthase, which uses NO as one of its substrates. It is therefore possible that the KsNaxLS complex plays a role in binding the volatile NO to retain it in the cell for transfer to hydrazine synthase. Alternatively, we propose that KsNaxLS may shuttle electrons to this enzyme complex.
Keywords: cytochrome c, bacterial metabolism, oxidation-reduction (redox), nitric oxide, crystal structure, ultraviolet-visible spectroscopy (UV-visible spectroscopy), anaerobic ammonium oxidation, anammox, nitrogen cycle
Introduction
Anaerobic ammonium oxidation (anammox)5 is a major process in the biogeochemical nitrogen cycle, and it is estimated to be responsible for 50–70% of the yearly loss of fixed nitrogen from the ocean to the atmosphere as N2 (1, 2). In this process, chemolithoautotrophic bacteria convert ammonium (NH4+) and nitrite (NO2−) into N2. Based on genetic, physiological, and biochemical information, it was proposed that the anammox process is carried out in three steps (Equations 1–3) involving NO and the unusual intermediate hydrazine (N2H4) (3, 4).
| (Eq. 1) |
| (Eq. 2) |
| (Eq. 3) |
The first step is catalyzed by a nitrite reductase (Nir), the second step by hydrazine synthase (HZS) (5), and the final step by hydrazine dehydrogenase (HDH) (6). These enzymes are localized in a membrane-bound compartment called the anammoxosome (7). In the current model of the anammox process, the four electrons released in the final reaction are transferred to the menaquinone pool inside the anammoxosome membrane to set up a proton-motive quinol cycle (3, 8). The resulting proton gradient is finally utilized by an F0F1-type ATP synthase to generate ATP. Electrons are proposed to be shuttled between soluble and membrane enzyme complexes by small c-type cytochromes (3, 9). Indeed, the genome of the anammox model organism Kuenenia stuttgartiensis encodes 65 c-type cytochromes that are predicted to be located inside the anammoxosome (10). Many of these are members of the low-molecular-weight class I and class II cytochrome c proteins (11), which could perform the proposed functions in electron transfer. However, which of these small c-type cytochromes performs these electron transfer reactions and whether some of them perform other crucial tasks in the central anammox metabolism are unknown. Such other functions might include the binding of highly-reactive and volatile intermediates such as NO and hydrazine to protect the cell and/or to keep them from diffusing out.
Among the most abundant small cytochrome c proteins in anammox organisms are NaxL (a member of class IIb) and NaxS (class I). In the anammox model organism K. stuttgartiensis, the genes encoding NaxL and NaxS (kusta0087 and kust0088, respectively) show the highest transcription levels of all monoheme c-type cytochromes (3) (Table S1) encoded in the genome. Previously, the homologues of these proteins from Jettenia caeni were purified as a heterodimeric complex (JcNaxLS) and were partially spectroscopically characterized (12). The UV-visible spectrum of JcNaxLS shows a blue shift of the Soret and α,β absorption peaks upon reduction, whereas other cytochrome c proteins typically show a red shift. Based on this, as well as on the sequence and electron paramagnetic resonance (EPR) data, it was postulated that the heme in each subunit possesses a rare His/Cys ligation (12). Moreover, the NaxLS complex is the first example of a complex between a class I and a class IIb c-type cytochrome.
Here, we describe the crystal structure of the NaxLS complex from K. stuttgartiensis (KsNaxLS). We provide a full analysis of the UV-visible spectroscopic properties of KsNaxLS, including the molecular origins of these unusual spectral shifts. We also demonstrate that KsNaxLS binds NO and CO. Importantly, we also show that the KsNaxLS complex interacts specifically with the hydrazine synthase complex, suggesting a function in NO and/or electron transport.
Results
KsNaxLS is a heterodimer
KsNaxLS was purified from K. stuttgartiensis as a stable heterodimer (Fig. 1A). Moreover, the recombinantly expressed components KsNaxL and KsNaxS could be reconstituted into a heterodimer in vitro (Fig. 1B). The sedimentation coefficient distribution c(S) of the native KsNaxLS complex obtained by analytical ultracentrifugation sedimentation velocity (AUC-SV) resulted in a single symmetric peak at 2.7 S (Fig. S1). Using the calculated molecular mass of the KsNaxLS heterodimer (24,397 Da), the maximum possible sedimentation coefficient (i.e. the sedimentation coefficient of a perfectly spherical molecule of the same molecular mass) can be calculated as Smax ≈0.00361 × M2/3. The ratio of this value and the measured sedimentation coefficient corrected to water at 20 °C s20, w and Smax indicates a protein complex with a globular shape (Smax/s20, w = 1.11) (13). The sedimentation coefficient distribution c(S) of the recombinantly produced individual components of the KsNaxLS complex also resulted in single symmetric peaks at 1.8 S and 1.7 S for KsNaxL and KsNaxS, respectively (Fig. S1). The comparison of the measured s20, w values and the maximum possible sedimentation coefficients indicate that the components are globular and monomeric in solution (Smax/s20, w of 1.17 and 1.14 for KsNaxL and KsNaxS, respectively). AUC-SV of the reconstituted KsNaxLS complex from its overexpressed subunits (both with C-terminal His6-tags) resulted in a sedimentation coefficient of 2.8 S (s20, w of 2.78 S), which is also consistent with a heterodimeric complex of the NaxL and NaxS components with a Smax/s20, w ratio of 1.16.
Figure 1.
Oligomeric state of KsNaxLS. A, 12% Tris-Tricine SDS-PAGE (51) analysis of the natively purified KsNaxLS complex and its heterologously expressed components (with C-terminal His6-tags). M, molecular mass markers (in kDa), L-His6, recombinant His-tagged KsNaxL; S-His6, recombinant His-tagged KsNaxS; LS (native), as-isolated KsNaxLS complex. B, SEC-MALS analysis of the KsNaxLS complex reconstitution from its individual components. Solid curves show SEC elution profiles measured as 280 nm; dots indicate the molecular mass in Da as determined by MALS. Green, recombinant His-tagged KsNaxL; blue, recombinant His-tagged KsNaxS; black, as-isolated KsNaxLS complex; and red, 2:1 mixture of recombinant His-tagged KsNaxL and NaxS. The red curve shows both the reconstituted KsNaxLS complex at 11.9 ml and the remaining KsNaxL at 13.1 ml.
AUC-SE of the native KsNaxLS complex resulted in a molecular mass of 23.9 kDa, which also corresponds well to the calculated value for the heterodimer (24.4 kDa) (Fig. S2).
An analysis of the native KsNaxLS complex and its reconstitution from heterologously expressed components by size-exclusion chromatography coupled with multiangle light scattering (SEC-MALS) is shown in Fig. 1B. The size-exclusion chromatograms of KsNaxLS and its component KsNaxL all showed single peaks at elution volumes of 11.9 and 13.1 ml, respectively. MALS analysis resulted in 20.4 and 13.4 kDa for the KsNaxLS complex and KsNaxL, respectively. When the KsNaxL and KsNaxS components were mixed in a 2:1 molar ratio, they could be reconstituted into a stable complex, with the excess of KsNaxL remaining in a monomeric state. MALS analysis of the reconstituted complex resulted in a molecular mass of 22.8 kDa.
The results of small-angle X-ray scattering (SAXS) (Fig. S3) are also consistent with a KsNaxLS heterodimer; the radius of gyration as determined by Guinier analysis (Fig. S3B) was 19.9 Å (using data between 0.58 < qRg <1.30), which is close to the value of 17.2 Å calculated from the crystal structure (see below). The Porod volume is 39,880 Å3, which when multiplied by 0.625 (14) results in an estimated molecular mass of 24.9 kDa, which is very close to the theoretical molecular weight of the KsNaxLS heterodimer. Moreover, a dummy-atom reconstruction reproduces the overall dimensions of the complex observed in the crystal (see below) (Fig. S3).
Structures of WT and mutants of the KsNaxLS complex
The 1.7 Å resolution X-ray structure of the WT, natively purified KsNaxLS complex, contains a heterodimer consisting of one KsNaxL and one KsNaxS subunit in the asymmetric unit. The molecular shape of the complex is somewhat elongated and resembles a pointing hand (Fig. 2), where the KsNaxL subunit forms the pointing finger(s) and KsNaxS the rest of the hand. The KsNaxL subunit forms a four-helix bundle, which is a typical characteristic of class II c-type cytochromes. KsNaxS possesses a class I cytochrome c-fold consisting of six α-helices. The contact surface area between the two subunits is 2120 Å2 (out of the total surface area of the complex of 10,950 Å2); it is formed by the regions Asp-82 to Tyr-92 and Arg-115 to Thr-129 of KsNaxL as well as Glu-29 to Gly-61 and Val-119 to Lys-126 of KsNaxS.
Figure 2.

Crystal structure of KsNaxLS. A, cartoon representation of the KsNaxLS complex. KsNaxL and KsNaxS are colored in green and blue, respectively. The hemes are shown a stick models. The interactions between the two subunits are predominantly polar, with 23 hydrogen bonds and five salt bridges. B, stereo figure showing a representative portion of the simulated annealing 2mFo− DFc composite omit map (beige surface, contoured at 1.0 σ) overlaid onto the heme-binding site of KsNaxL.
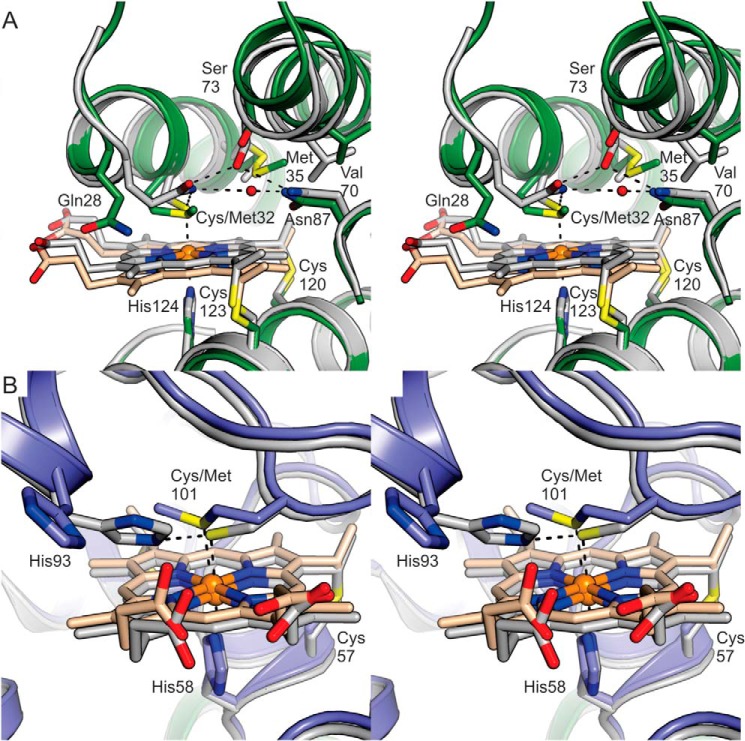
The c-type heme moieties of KsNaxL and KsNaxS are 19 Å apart in the complex (using the edge–to–edge distance (15)), and both are hexa-coordinated, with histidines serving as the proximal ligands (i.e. located on the side of the CXXC heme-binding motif), although cysteine residues, rarely acting as heme ligands in other c-type cytochromes, serve as the distal ligands (i.e. on the opposite side, Fig. 3, A and B). The distances between the heme iron and histidine Nϵ atoms are 2.0 Å in both KsNaxL and KsNaxS, and the heme-iron–to–cysteine–sulfur distances are 2.4 Å in both subunits. As expected for a class II c-type cytochrome, the heme-binding motif (120CRNCH124) in KsNaxL is located near the C terminus of the protein sequence, whereas the distal cysteine (Cys-32) ligand originates from the N-terminal part. The opposite is the case in KsNaxS where the heme-binding motif (54CYYCH58) is located close to the N terminus and the distal cysteine ligand (Cys-101) is located close to the C terminus, as is the case in other class I c-type cytochromes. The distal environment of the heme in KsNaxL is amphipathic, with several hydrophilic residues such as Gln-28, Ser-73, and Asn-7 at the side of the B and C pyrrole rings. Together with a solvent molecule, these residues form a hydrogen-bonding network. Interestingly, the Gln-28 Nϵ atom is within hydrogen-bonding distance (3.1 Å) of the iron-coordinating Sγ atom of Cys-32. The opposite side (above the A and B pyrrole rings) contains hydrophobic residues such as Met-35, Trp-36, and Val-70. The environment at the proximal face of the NaxL heme is mainly hydrophobic, containing residues such as Leu-133, Val-134, and the terminal Pro-135 residue (Fig. 3A). The heme environment in KsNaxS is distinctly hydrophobic, being made up of several aromatic amino acids (such as Tyr-43, Phe-71, Phe-76, Phe-81, and Tyr-104) as well as other hydrophobic residues, e.g. Met-35, Val-73, and Ile-94 at both distal and proximal sides. Interestingly, the iron-coordinating Sγ of the distal Cys-101 is within hydrogen-bonding distance (3.1 Å) of the Nϵ atom of His-93 (Fig. 3B).
Figure 3.
Details of the heme-binding sites in KsNaxL (A) and KsNaxS (B). Heme groups and several amino acid side chains are shown as sticks, and the rest of the molecule is shown in cartoon representation. In both heme-binding sites, the distal cysteine Sγ is within hydrogen-bonding distance from a nitrogen atom: the Gln-28 Nϵ in KsNaxL and the His-93 Nϵ in KsNaxS. In KsNaxL, this atom is further involved in a hydrogen bonding network with a solvent molecule, Ser-73 and Asn-87.
Because cysteine as a distal heme ligand is relatively rare, we made several mutants for structural and spectroscopic investigations by replacing one or both distal cysteines with either methionine or glycine. Of these, mutants KsNaxL C32G/NaxS WT and KsNaxL C32M/NaxS C101M formed crystals, and we determined their X-ray crystal structures to 2.0 and 1.9 Å resolution, respectively, to assess the mutations' impact on the overall structure and heme environment. For the most part, there are no major structural differences between WT and mutant KsNaxLS complexes, and the cysteine–to–methionine mutations resulted in the methionine coordinating the heme iron as expected. Only the mutation of Cys-32 to methionine in KsNaxL caused some larger-scale structural effects: Gln-28 moves away from the heme, destroying the hydrogen bond network with Ser-73 and Asn-7 (Fig. 4A), which results in helix α1 moving outward compared with the situation in the WT. This in turn allows residues 33–38 on the opposite helix to move slightly inward, with the side chain of Met-35 adopting a more stretched-out conformation than in the WT protein. Other than that, the mutations cause local changes only. In the KsNaxL C32G/NaxS WT crystal structure, a water molecule acts as a sixth heme ligand, which is within hydrogen-bonding distance from Gln-28 (Fig. S4). Moreover, upon mutation of Cys-101 to methionine in KsNaxS, His-93 rotates away causing the formation of a hydrogen bond with one of the propionate groups of the heme (Fig. 4B).
Figure 4.
KsNaxLS mutants. Stereo figures showing the structural effects of the distal cysteine to methionine mutations. The WT protein subunits are shown in gray, and the mutant proteins are in shades of green and blue. A, details of the KsNaxL heme-binding site in the KsNaxL C32M/NaxS C101M mutant. The introduction of the bulkier methionine residue has caused the Gln-28 side chain to rotate away from the heme, disrupting the hydrogen-bonding network with Asn-87 and Ser-73, which causes minor rearrangements of the helices containing these residues. B, structure of the KsNaxS heme-binding site in the KsNaxL C32M/NaxS C101M mutant. Mutation of Cys-101 results in the loss of the hydrogen bond between Cys-101 and His-93, causing the latter residue to rotate away from the heme. Otherwise, no large differences with the WT protein are observed.
UV-visible spectroscopy of WT and mutant proteins
The heterologously expressed KsNaxLS complex and its components were reddish brown. In all proteins (from which His-tags were removed), the Soret band in the as-isolated state was found to be located at 419–420 nm (Fig. 5, A–C). Upon reduction with 0.5 mm Ti(III)citrate, the Soret band showed a clear blue shift and the typical α- and β-bands appeared at 553 nm (551 nm for KsNaxL and KsNaxS) and 523 nm (521 nm for KsNaxL and 522 nm for KsNaxS), respectively (Fig. 5, A–C).
Figure 5.
Spectroscopy of KsNaxLS. A–C, UV-visible spectra of recombinantly produced proteins after removal of the His-tag. A, natively purified, WT KsNaxLS, as well as recombinant, His-tagged; B, KsNaxL; C, KsNaxS; D–F, UV-visible spectra of His-tagged mutants. D, KsNaxL C32M/NaxS C101M; E, KsNaxL C32M/NaxS; and F, KsNaxL/NaxS C101M. Spectra of as-isolated and Ti(III)citrate-reduced samples are shown in black and red, respectively.
Interestingly, when Ti(III)-reduced KsNaxLS and it constituents were re-oxidized by sparging the sample with 1–2 ml of air, the Soret band further blue-shifted to 412 nm with simultaneous disappearance of α- and β-bands. Re-oxidation with 0.5 mm potassium ferricyanide, however, resulted in a red shift of the Soret bands back to 420 nm for KsNaxLS and its constituent KsNaxL and to 419 nm for KsNaxS (Fig. 6, A–C).
Figure 6.
Reoxidation after reduction. A–C, spectra of KsNaxLS and its constituents, reoxidized with air (red lines) or potassium ferricyanide (black lines) after reduction with Ti(III)citrate. Reoxidation by air results in a blue shift, whereas with ferricyanide the Soret band returns to the same position as in the as-isolated state. D–F, spectra of KsNaxLS distal Cys–to–Met mutants reoxidized with air after reduction with Ti(III)citrate.
UV-visible spectra of different distal-cysteine–to–methionine mutants (with His-tags) in their as-isolated state as well as after reduction with 0.5 mm Ti(III)citrate are shown in Fig. 5, D–F. The Soret bands of the as-isolated states of all these mutants were located around 413–415 nm. Upon reduction with Ti(III)citrate, a red shift of the Soret band to 417–418 nm was observed in these mutants, with concomitant appearance of α- and β-bands at around 553 and 523 nm (Fig. 5, D–F). The double methionine mutant KsNaxL C32M/NaxS C101M was apparently isolated in a partially reduced state given the presence of weak α- and β-bands (Fig. 5D). The Soret peaks shifted back to around 408–414 nm upon re-oxidation by sparging with air (except for the mutant KsNaxL WT/NaxS C101M, which has a Soret maximum at 416 nm in the re-oxidized state, Fig. 6, D–F). Moreover, after full reduction the re-oxidation by air of this mutant took more than 1 h, and it resulted in a noticeable decrease in intensity of the α- and β-bands.
Although the mutant KsNaxL WT/NaxS C101G (Fig. S5) appeared to be in a completely oxidized state after purification, re-oxidation after full reduction yielded still partially reduced species as evident from minor α- and β-bands (in this case the spectra were measured immediately after sparging with air). Conversely, the single mutant KsNaxL C32G/NaxS WT (Fig. S5), which was isolated in its oxidized state, was immediately fully re-oxidized by air starting from the completely reduced state.
Redox potential of KsNaxLS
Spectropotentiometric measurements with KsNaxLS in an optically transparent thin-layer electrochemical cell resulted in irreversible reactions at the working electrode. Ti(III)-reduced KsNaxLS was therefore titrated with ferricyanide under anoxic conditions while monitoring the redox potential and UV-visible spectrum. The absorption at 417 nm showed a single, sharp transition during titration between −300 and +50 mV versus SHE, from which a redox potential of −175 mV versus SHE could be extracted by fitting with the Nernst equation assuming a 2-electron transition (Fig. 7). Nearly identical results were obtained at absorption wavelengths of 552 and 454 nm. Redox potentials much lower than −300 mV or much higher than +50 mV versus SHE could not be reached without using amounts of reducing/oxidizing agents that resulted in protein aggregation.
Figure 7.
Redox potential determination. Redox titration of KsNaxLS with Ti(III)citrate while following the absorption change at 417 nm (open circles) and fitted (solid line) with the 2-electron Nernst equation resulting in a redox potential of −175 mV versus SHE.
KsNaxLS binds NO and CO
The KsNaxLS complex and its components were tested for their ability to bind several small molecule ligands such as nitric oxide (NO), carbon monoxide (CO), hydroxylamine (NH2OH), hydrazine (N2H4), nitrite (NO2−), azide (N3−), and cyanide (CN−). Of these ligands, NO and CO were found to coordinate to the c-type hemes in the KsNaxLS complex as judged by UV-visible spectroscopy. When non-His–tagged, reduced KsNaxLS was incubated with NO, the Soret band decreased in intensity and shifted from 417 to 415 nm, and the intensity of the α- and β-bands decreased, too (Fig. 8A). These latter bands did not, however, disappear completely. With His-tagged protein, the Soret intensity only decreased slightly, pointing to an effect of the presence of the His-tag on the NO-binding properties of the protein (Fig. 8B). The individual recombinantly-produced components of the complex displayed much larger spectral changes upon NO exposure. Upon incubation with NO, Ti(III)-reduced KsNaxL showed a prominent shift of the Soret band to ∼395 nm and formation of a shoulder at ∼416 nm indicating coordination of the nitrosyl ligand to the heme iron (Fig. 8C). In reduced KsNaxS, the Soret band shifted to 415 nm with formation of a shoulder at 396 nm (Fig. 8D). Titration of reduced KsNaxLS complex with NO, monitored by UV-visible spectroscopy, indicated a dissociation constant (Kd) for NO of ∼12 μm (Fig. 8E).
Figure 8.
NO binding to reduced KsNaxLS and its components. A, effect of NO on the UV-visible spectra of recombinant, reduced KsNaxLS after removal of the His-tag; B, His-tagged KsNaxLS; C, KsNaxL (His-tag removed); and D, KsNaxS (His-tag removed). Reduced and NO-incubated reduced states are shown in blue and black, respectively. E, determination of binding constant of reduced KsNaxLS for NO, by titration with NO, and observation of the spectral change at 553 nm. Two series of measurements are shown, using protein concentrations of 11 and 4.5 μm protein (black and gray data points, respectively). The solid lines show the fits to each series using the average Kd of 12 μm.
When WT KsNaxLS as isolated from Kuenenia biomass was exposed to NO, the Soret band increased in intensity and showed a blue shift from 420 to 417 nm (Fig. S6A). With the recombinantly-produced His-tagged KsNaxLS protein, exposure to NO resulted in a much smaller increase in intensity of the Soret peak, although its position showed a clear blue shift from 419 to 417 nm (Fig. S6B). Thus, in both the reduced and as-isolated states, a difference in behavior toward NO was observed between His-tagged and non-His–tagged protein. At present, it is not clear what causes this difference in behavior.
Titration of as-isolated KsNaxLS complex with NO followed by UV-visible spectroscopy did not allow the Kd value to be determined, as no saturation of any spectral feature could be reached using final NO concentrations up to 1 mm. This points to a low affinity with a Kd in the millimolar range or higher in the as-isolated state.
Upon exposure to CO, the Soret bands of the reduced states of the KsNaxLS complex and its components sharpened and increased in their intensities (Fig. S7). The Soret band shifted from 417 to 415–416 nm for the recombinant KsNaxLS complex, as well as its subunits. Moreover, in all cases two broad bands around 540 and 570 nm appeared, replacing the α- and β-bands.
KsNaxLS binds to hydrazine synthase
The KsNaxLS complex (with a C-terminal His-tag on KsNaxS), individual KsNaxL and KsNaxS, as well as Kustc0563, another highly-expressed small cytochrome c from Kuenenia, were used as “bait” proteins in a pulldown assay. To this end, they were bound to Ni-IDA beads, which were then incubated with a cell-free extract of K. stuttgartiensis. After elution, SDS-PAGE (Fig. 9) followed by mass spectrometric analyses of the co-eluted proteins bands showed that only the KsNaxLS complex was able to pull down all three subunits of the HZS complex. In addition, a protein band at around 200 kDa appeared in all lanes when lysate was used. Peptide mass fingerprinting showed that this band belongs to KsHDH (6, 16). However, KsHDH was most likely bound nonspecifically, as it was also bound to the Ni-IDA beads without any bait protein. No co-eluting bands were observed when the single components KsNaxL or KsNaxS or the reference cytochrome c Kustc0563 was used as bait. Similar results were obtained when immobilized bait proteins were incubated with purified HZS protein solution.
Figure 9.

SDS-PAGE analysis of pulldown experiments. Upper panel shows a Coomassie-stained gel, and the lower panel shows a heme-stained gel. The arrowheads indicate protein bands belonging to the three subunits of the potential interaction partner K. stuttgartiensis hydrazine synthase (HzsA, HzsB, and HzsC). The HzsB subunit does not contain a heme group and therefore does not show up in the heme-stained gel. Lane 63 indicates C-terminally 6×His-tagged Kustc0563 used as a control.
Discussion and conclusions
KsNaxLS provides the first crystal structure of a complex between class I and II c-type cytochromes
The crystal structure of KsNaxLS at 1.7 Å resolution provides the first molecular model of a complex between class I and class II c-type cytochromes. Moreover, based on its hexa-coordinated heme iron along with its four-helix bundle fold, NaxL can be assigned to subclass IIb of monoheme c-type cytochromes (11). The structure of KsNaxL presented here is the first high-resolution structure of a member of this subclass. A molecular model, partially validated by NMR spectroscopy (17), exists of the His/Met-ligated cytochrome c-556 from Rhodopseudomonas palustris, another class IIb cytochrome. Its four-helix bundle fold shows an RMSD of 3.0 Å for 97 Cα-atoms with that of KsNaxL.
The purification of KsNaxLS from K. stuttgartiensis biomass as a stable heterodimeric complex, which was also the case for its orthologue from J. caeni (12), indicates that this oligomeric state is indeed physiologically relevant. Moreover, we show here that the recombinantly produced individual components KsNaxL and KsNaxS readily assemble into the heterodimer in vitro. Biophysical characterization in solution by AUC, SAXS, and SEC-MALS further corroborates that KsNaxLS exists as a heterodimer in solution. The crystal structure of the KsNaxLS complex shows that the interaction surface between the subunits constitutes a large fraction (∼24%) of the total surface area. This strongly suggests that the heterodimeric assembly found in the crystal structure indeed represents the oligomeric state in solution. Our high-resolution structure shows that the c-type hemes in both KsNaxL and KsNaxS are coordinated by proximal histidine and distal cysteine ligands. This His/Cys heme ligation is relatively rare in nature with only a few examples known so far (18–24).
The KsNaxLS complex possesses unusual UV-visible spectral properties
In this study, we show that the mutation of the distal cysteines to either methionine or glycine in either one or both subunits of the KsNaxLS complex leads to blue-shifted Soret bands (413–415 nm in the as-isolated state) with respect to the WT complex (at 420 nm). Moreover, in all the mutants, a red shift to 417 nm takes place upon reduction, as opposed to the blue shift in WT KsNaxLS. These observations demonstrate that the unusual position of the Soret band and its blue shift upon reduction indeed originate from the His/Cys ligation in the KsNaxLS complex. The spectral features of the KsNaxLS complex and its components are reminiscent of those of cytochrome c in which the axial Met had been replaced by Cys, suggesting a thiolate-Fe(III) ligation in both subunits of the KsNaxLS complex that is lost upon reduction (25). This was also pointed out for JcNaxLS (12). The crystal structure shows that in both subunits, a hydrogen bond is formed between the coordinating cysteine and another side chain (Gln-28 in KsNaxL and His-93 in KsNaxS). These would likely weaken the thiolate-Fe bond by reducing the σ-donor capability of the cysteine (26) making it easier for the thiolate to dissociate from the heme iron in the reduced state. It must be pointed out, however, that other authors have suggested an iron-bound, neutral thiol ligation in the reduced state of His/Cys coordinated heme groups (27, 28).
Re-oxidation of the WT KsNaxLS complex and its individual components with air yields spectra with even further blue-shifted Soret maxima. This could be explained by a dissociated cysteine ligand that does not rebind, leaving a His/H2O heme ligation, or one that has been replaced by another ligand. The position of the Soret band in the air-oxidized form of the distal glycine and methionine mutants of KsNaxLS around 410 nm is consistent with both possibilities. However, neither subunit contains a residue that is sufficiently close to the heme iron to take the place of the thiolate ligand upon its dissociation without a considerable structural rearrangement.
Reoxidizing the complex with ferricyanide, however, brings the Soret band position back to 420 nm, the same position as in the as-isolated state. The same holds for KsNaxS, where the Soret band goes back to 419 nm on re-oxidation with ferricyanide. In KsNaxL, ferricyanide incubation results in a Soret band at 420-nm, very close to the 421-nm maximum observed for the as-isolated component. Thus, it appears that when ferricyanide is used as the oxidizing agent, re-oxidation does not result in an irreversible modification of the protein, whereas re-oxidation with air does.
Small gaseous ligand binding by KsNaxLS
Crystallization experiments with NO- or CO-bound KsNaxLS or subunits thereof were unsuccessful, and incubation of crystals with either of these gases did not result in complex formation. However, our spectroscopic data provide detailed information on CO and NO binding. The UV-visible spectroscopic features of CO-bound KsNaxLS, its components, and its mutants (Soret band around 415–417 nm with increased intensity and flattening of α- and β-bands) are similar to the ferrous six-coordinated CO adducts of other cytochromes (29). This implies that the distal cysteine ligand is flexible enough to move out, enabling distal CO binding.
Upon NO incubation, the as-isolated state of KsNaxL shows a broadening of the Soret band around 395–400 nm. This has been associated in several studies with the formation of a five-coordinated NO (5cNO) adduct in which NO coordinates at the proximal side as the only axial ligand. The additional Soret peak at ∼416 nm is suggestive of a six-coordinated NO (6cNO) adduct, where NO binds at the distal side as the sixth axial ligand (30–33). Thus, in the as-isolated state, the spectra of the NO-bound individual components combine features typical for 5cNO and 6cNO complexes and may represent a mixture of the two. Strikingly, however, the spectrum of the as-isolated KsNaxLS complex incubated with NO only shows features typical for a 6cNO adduct, i.e. an increase in its intensity and a shift of the Soret peak from 420 to 417 nm, whereas none of the features indicative of a 5cNO adduct appear. Thus, in the as-isolated state, complex formation appears to result in the exclusive production of 6cNO adducts.
In the reduced state, where the affinity for NO is much higher, the situation is somewhat different. Although the KsNaxL subunit in the reduced state also shows both 5cNO- and 6cNO spectral features upon incubation with NO, the isolated KsNaxS subunit displays a slight decrease in intensity as well as a blue-shift of the Soret band, features typical for 6cNO adduct formation by reduced cytochromes c. When the reduced KsNaxLS complex is treated with NO, the Soret band decreases in intensity and shifts to 415 nm, again indicating the formation of a 6cNO adduct, but a small shoulder appears around 395 nm. This could be explained by the formation of a small amount of 5cNO complex, whereas most of the NO-bound protein forms a 6cNO adduct. The data thus suggest that in the reduced state, too, the formation of the complex favors the formation of 6cNO adducts. This may be explained by comparing the structure of the KsNaxLS complex with the structures of the individual components: the secondary structure elements that contain the proximal histidines are on the surface of the molecules in both individual monomers, but face each other in the complex (Fig. 10). The end of helix α4 containing the heme-binding motif of KsNaxL is in contact with helix α2, which contains the heme-binding motif of KsNaxS. Moreover, although the loop between α2 and α3 in KsNaxS is very rich in conserved glycine residues (Fig. S8), and thus likely confers considerable flexibility to the loop containing the proximal histidine in the monomeric state, in the complex it is in close contact with helix α4 of KsNaxL. The KsNaxLS complex thus appears specifically geared toward the exclusive formation of 6cNO complexes.
Figure 10.
Proximity of heme-binding motifs in the complex. The heme-binding motif on helix α4 of KsNaxL (120CRNCH124) is in contact with the heme-binding motif on helix α2 of KsNaxS 54CYYCH58, as well as with the loop between α2 and α3.
Binding to hydrazine synthase; possible functions of the KsNaxLS complex
Two possible roles come to mind for a protein such as KsNaxLS that binds specifically to HZS: nitric oxide and electron transfer. As one of the substrates of HZS, a central anammox enzyme, nitric oxide is a precious yet volatile and highly-reactive commodity for the cell. Thus, particularly under routine growth conditions where nitrite is limiting (3) and therefore NO is scarce, a means of retaining NO in the cell and delivering it to HZS would be beneficial. This would require a protein that is highly abundant, capable of NO binding, and interacts with the HZS complex. Indeed, based on transcriptomic data (Table S1) (3) and its ready purification from anammox biomass as reported in this study, it is evident that KsNaxLS is highly expressed in Kuenenia. Moreover, as reported by Ukita et al. (12), the JcNaxLS complex represents ∼10% of the total molar protein content in Jettenia cells. Interestingly, however, a recent study (34) reports that when K. stuttgartiensis is grown on NO rather than on nitrite the transcription levels of the genes encoding KsNaxL and KsNaxS are reduced 20- and 10-fold, respectively. Additionally, the current study suggests that KsNaxLS predominantly forms a 6cNO adduct upon NO binding, similar to other NO-shuttling proteins. Our binding studies showed that in the reduced state KsNaxLS has a dissociation constant for NO of ∼12 μm. The concentration of NO in anammox cells is not known. Rathnayake et al. (35) have reported NO concentrations of up to 2.7 μm in active anammox granules. However, the intracellular NO concentration is unlikely to be much higher than the low micromolar range, given the Ki of 2.5 μm for hydrazine dehydrogenase reported by Maalcke et al. (6). However, given the large amount of the KsNaxLS complex in the cell, it remains possible that NO binding by KsNaxLS assists in retaining the volatile NO inside the cell.
Alternatively or additionally, the NaxLS complex could function as an electron shuttle between HZS and an electron donor. Earlier, Ukita et al. (12) had noted that excess sodium dithionite was not able to effect complete reduction of JcNaxLS (which we also observed for KsNaxLS) and suggested that the JcNaxLS complex likely has an unusually low redox potential, given the low redox potential of −660 mV versus SHE of dithionite (36). Because the reaction catalyzed by the HZS complex has a much higher midpoint potential (E′0 = +60 mV) (9), for HZS to accept electrons from a binding partner at such an extremely low redox potential would appear to be “wasteful” from a bioenergetic point of view. However, our redox titrations show that, unexpectedly, KsNaxLS has a redox transition at a much higher potential of only −175 mV versus SHE (although additional redox transitions at a much lower redox potential cannot be excluded). This makes electron transfer from KsNaxLS to HZS much more economical in terms of energy conservation. Thus, given the properties of the KsNaxLS complex presented here, at least two possible functions for KsNaxLS present themselves, which are not mutually exclusive: NO retention/transfer to HZS and electron transfer to HZS. In vivo research on anammox organisms or in vitro reconstitution of the central anammox pathway could help settle this issue.
Experimental procedures
Protein expression and purification
The native KsNaxLS complex was purified from K. stuttgartiensis cultured as suspended cells in a membrane bioreactor (37, 38). Cells were collected by centrifugation at 8000 rpm for 15 min. The pellet was resuspended in 20 ml of 20 mm potassium phosphate buffer (KPi), pH 7.0. Cells were disrupted by passing them through a French press three times at 1200 p.s.i., and the lysate was incubated with 1% (w/v) sodium deoxycholate for 1 h under mild stirring. Soluble proteins were separated from debris by ultracentrifugation for 1 h at 150,000 × g (Discovery 100 centrifuge with a T-1270 rotor; Sorvall, Newtown, CT). FPLC was performed with an ÄKTA purifier (GE Healthcare, Uppsala, Sweden). A 30-ml Q-Sepharose XL column (GE Healthcare, Uppsala, Sweden) was equilibrated with 20 mm Tris-HCl buffer, pH 8.0. Cell extract was loaded at 2 ml/min. Nonbinding proteins were washed off with Tris-HCl buffer until the A280 signal was stable. KsNaxLS was eluted isocratically at 200 mm NaCl in 20 mm Tris-HCl buffer, pH 8.0 (flow rate 2 ml/min, collected at 2 ml/fraction). Fractions were pooled, desalted, and concentrated. The resulting sample was loaded onto a 10-ml ceramic hydroxyapatite packed column (Bio-Rad), which had been previously equilibrated with 20 mm KPi buffer, pH 7.0. KsNaxLS was eluted isocratically with 500 mm KPi buffer, pH 7.0 (flow rate 2 ml/min, collected at 2 ml/fraction). Pooled fractions were desalted and concentrated using Vivaspin 20 spin filters. For heterologous expression of the KsNaxLS complex and its individual components, the kusta0087 (KsNaxL) and kusta0088 (KsNaxS) genes were cloned either as bicistronic operon or individually into a pUC19-derived vector (39); the primers and vectors used are given in Tables S2 and S3. The native N-terminal signal peptides (Fig. S8) were replaced by the strong signal peptide from the small tetraheme cytochrome c (STC, SO2727) obtained from the expression host Shewanella oneidensis MR-1 (40) (in case of the bicistronic operon, this was done only for the upstream gene kusta0087). The pUC19-derived vectors containing the inserts were transformed into S. oneidensis MR-1 ΔendA (41) by electroporation at 0.55 kV, 25 microfarads, 200 ohms using a Gene Pulser II (Bio-Rad). For large-scale purification, 2-liter batches (6–10 liters total) of LB medium were supplemented with 50 μg/ml kanamycin and inoculated with 1% (v/v) of S. oneidensis MR-1 overnight culture. Initially, large-scale cultures were grown in 5-liter Erlenmeyer flasks (without baffles) at 30 °C and 100 rpm for 5–6 h until an OD600 of 0.6–0.8, upon which the temperature was lowered to 20 °C and the cultures were shaken at 60 rpm for a further 60–70 h. The cells were harvested by centrifugation at 6000 rpm, 4 °C for 10 min in a FiberliteTM F9–6 × 1000 LEX rotor (Thermo Fisher Scientific, Darmstadt, Germany). The cell pellets were either directly used for purification or frozen in liquid nitrogen and stored at −80 °C. The expressed proteins were purified using immobilized metal ion-affinity chromatography using nickel-nitrilotriacetic acid–agarose (Qiagen, Hilden, Germany) followed by size-exclusion chromatography using a Superdex 75 (10/300 GL) column connected to an Äkta Purifier FPLC system (GE Healthcare, Uppsala, Sweden). For spectroscopic, electrochemical, and NO-binding studies of WT proteins, the hexahistidine tags were removed by incubation with tobacco etch virus protease overnight prior to the size-exclusion step.
For reference purposes, another typical monoheme c-type cytochrome from K. stuttgartiensis, Kustc0563, was cloned, expressed, and purified with a C-terminal hexahistidine tag in the same way as described above for the individual components of KsNaxLS. Protein purity was assessed by 15% SDS-PAGE analysis. The identity of the proteins was confirmed by MALDI-MS peptide mass fingerprinting, and total molecular masses were determined by ESI-MS.
Protein crystallization
Initial precipitant screenings with the native KsNaxLS complex and its mutants (KsNaxL C32G/NaxS WT and KsNaxL C32M/NaxS C101M) were performed using the sitting drop vapor-diffusion method. 100 nl of concentrated proteins were screened against 100-nl precipitant solutions from commercial crystallization screens pre-pipetted into 96-well sitting drop trays (Greiner XTL low-profile, Greiner Bio One, Frickenhausen, Germany) using a Mosquito crystallization robot (TTP Labtech Ltd., Melbourne, UK) at 20 °C. For larger-scale hanging drop vapor-diffusion crystallization setups, 1 μl of KsNaxLS (WT complex or mutants) protein stock (A2801 cm = 6–7 in 25 mm HEPES/KOH, pH 7.5, 25 mm KCl) was mixed with 1 μl of precipitant solution containing 20–25% (w/v) PEG 3000 and 0.1 m sodium citrate, pH 5.5. The resulting drops were equilibrated against 800-μl reservoir solution in 24-well Linbro plates. All the crystals from WT and mutant complexes were cryo-protected by short soaking in reservoir solution supplemented with 20% (v/v) ethylene glycol, followed by flash-cooling in liquid nitrogen.
Diffraction data collection and crystal structure determination
High-resolution native data and high-redundancy single-wavelength anomalous diffraction data were collected at beamline X10SA at the Swiss Light Source (SLS) of the Paul-Scherrer-Institute (Villigen, Switzerland). Diffraction data sets were collected using the rotation method with oscillation ranges of 0.1° (fine-slicing) while the crystals were kept at 100 K. Diffraction images were recorded using a PILATUS 6M detector (Dectris, Baden, Switzerland). Diffraction data were processed using XDS (42), and data statistics are given in Table 1. Experimental phases using data from single-wavelength anomalous diffraction experiments were obtained using AutoSHARP 2.0 (Global Phasing Ltd, Cambridge, UK). Density modification was performed using DM (43). Structural models were iteratively built using COOT (44) and refined using REFMAC (45). Model statistics are given in Table 2. Structure validation was performed using the validation tools in COOT and with MolProbity (http://molprobity.biochem.duke.edu/)6 (52). Figures were prepared using PyMOL (Schrödinger LLC).
Table 1.
Data collection and processing statistics
Values for the outer shell are given in parentheses.
| KsNaxLS Fe-SAD | KsNaxLS native, PDB code 6R6M | KsNaxLS C32M/C101 M, PDB code 6R6N | KsNaxLS C32G/WT, PDB code 6R6O | |
|---|---|---|---|---|
| Diffraction source | SLS PXII | SLS PXII | SLS PXII | SLS PXII |
| Wavelength (Å) | 1.7433 | 1.000 | 1.000 | 1.000 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Detector | Pilatus 6 M | Pilatus 6 M | Pilatus 6 M | Pilatus 6 M |
| Crystal-detector distance (mm) | 166 | 300 | 400 | 300 |
| Rotation range per image (°) | 0.1 | 0.15 | 0.2 | 0.1 |
| Total rotation range (°) | 360 | 180 | 180 | 120 |
| Exposure time per image (s) | 0.1 | 0.15 | 0.2 | 0.2 |
| Space group | P21212 | P21212 | P21212 | P21212 |
| a, b, c (Å) | 44.1, 131.1, 46.1 | 44.3, 130.4, 45.4 | 44.7, 130.4, 45.7 | 44.2, 131. 5,45.3 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution range (Å) | 40.0–2.3 (2.4–2.3) | 40.0–1.7 (1.8–1.7) | 50.0–2.0 (2.1–2.0) | 50.0–1.9 (2.0–1.9) |
| Total no. of reflections | 454,148 (47,909) | 184,876 (21,515) | 116,845 (15,895) | 182,263 (25,654) |
| No. of unique reflections | 22,941 (2,727) | 29,424 (4,287) | 18,722 (2,465) | 21,517 (2,996) |
| Completeness (%) | 100 (99.9) | 98.8 (93.4) | 99.6 (99.4) | 99.6 (99.5) |
| Redundancy | 19.8 (17.6) | 6.3 (5.0) | 6.2 (6.4) | 8.5 (8.6) |
| 〈I/σ(I)〉 | 28.1 (5.6) | 17.6 (2.2) | 13.7 (2.0) | 19.2 (3.0) |
| Rmeas. (%) | 8.6 (56.5) | 6.0 (61.0) | 12.3 (73.6) | 7.3 (96.0) |
Table 2.
Refinement and model statistics
| KsNaxLS native, PDB code 6R6M | KsNaxLS C32M/C101M, PDB code 6R6N | KsNaxLS C32G/WT, PDB code 6R6O | |
|---|---|---|---|
| Resolution range (Å) | 65.2–1.75 | 65.7–2.0 | 65.7–1.9 |
| Completeness (%) | 98.8 | 99.5 | 99.5 |
| σ-Cutoff | None | None | None |
| No. of reflections, working set | 27949 | 17705 | 20427 |
| No. of reflections, test set | 1450 | 960 | 1081 |
| Final Rcryst | 0.188 | 0.200 | 0.182 |
| Final Rfree | 0.210 | 0.236 | 0.206 |
| ML-based coordinate error (Å) | 0.13 | 0.24 | 0.20 |
| No. of non-H atoms | |||
| Protein | 1632 | 1626 | 1620 |
| Heme groups | 86 | 86 | 86 |
| Water | 192 | 129 | 173 |
| Total | 1910 | 1841 | 1879 |
| RMSD | |||
| Bonds (Å) | 0.010 | 0.008 | 0.007 |
| Angles (°) | 0.972 | 1.063 | 0.979 |
| Average B factors (Å2) | |||
| Protein | 34.4 | 41.2 | 38.6 |
| Heme groups | 26.4 | 34.7 | 30.0 |
| Water | 40.0 | 44.0 | 45.5 |
| Ramachandran plot | |||
| Most favored (%) | 98.5 | 97.6 | 98.5 |
| Allowed (%) | 1.5 | 2.4 | 1.5 |
| Outliers (%) | 0.0 | 0.0 | 0.0 |
Analytical ultracentrifugation
AUC-SV was used to determine the oligomeric state of KsNaxLS in solution. For each sample, the buffer was exchanged to 25 mm KCl, 25 mm HEPES/KOH, pH 7.5, by repeated washings in 10-kDa MWCO Amicon tubes (Millipore, Schwalbach, Germany). The samples were diluted to A4201 cm ∼0.42. The measurement cells (Beckman Coulter Inc., Palo Alto, CA) were assembled according to the manufacturer's instructions. The two sectors of the cells were filled with 400 μl of protein sample and 420 μl of buffer, respectively. Samples were sedimented at 30,000 rpm for 24 h at 20 °C in an An-60 Ti rotor in an XL-1 analytical ultracentrifuge (Beckman Coulter Inc.). Absorbance scans were recorded at 280 and 420 nm wavelength. The first 150 scans were processed using SEDFIT (46) to obtain the c(S) distributions, s20, w values (sedimentation coefficients corrected to pure water), and molecular masses assuming a fixed frictional ratio (f/fmin) of 1.2. Sednterp was used to calculate the density and viscosity of the buffer. A partial specific volume of 0.73 ml/g for the proteins was used to fit the data (13). AUC-SE measurements were performed to determine the molecular mass of the KsNaxLS complex and its components in solution. AUC-SE experiments were carried out in the same cells but with 200 μl of sample and 220 μl of buffer. Two angular velocities (20,000 and 30,000 rpm) were used to equilibrate the samples. Five photometric scans at 280 nm were recorded at each speed after 22 h of equilibration at 20 °C. AUC-SE data were processed using SEDPHAT (46).
SEC-MALS
Protein samples were separated on Superdex 75 gel-filtration columns (GE Healthcare, Uppsala, Sweden). Analyses were performed using a 1260 Infinity II HPLC system (Agilent Inc., Santa Clara, CA) equipped with a photodiode array detector SPD-M20A (Shimadzu, Duisburg, Germany). Static light-scattering analyses were performed in line using a DAWN 8+ Heleos II® multiangle scattered light photometer (Wyatt Technology, Santa Barbara, CA) combined with a refractive index detector Optilab T-rEX (Wyatt Technology, Santa Barbara, CA). The system was equilibrated with a buffer containing 50 mm HEPES, pH 7.5, and 150 mm KCl. A flow rate of 0.5 ml min−1 was used. To calibrate the gel-filtration column, a standard (Bio-Rad) containing several proteins was used. 40 μl of each sample with concentrations up to 150–250 μm were injected using an autosampler. Data analysis was performed using the ASTRA 7.1.2 software (Wyatt Technology, Santa Barbara, CA), providing weight-averaged molar masses of the eluting species.
Small-angle X-ray scattering
Solution SAXS measurements were performed at the cSAXS beamline (X12SA) of the Swiss Light Source at the Paul Scherrer Institute, Villigen (Switzerland). KsNaxLS was concentrated to an A280 of ∼0.9 in 25 mm HEPES/KOH, pH 7.5, 25 mm KCl. Scattering data were measured in a 1-mm diameter quartz capillary kept at 283 K. The X-ray photon energy was 11.2 keV, and 200 measurements of 0.5 s each were recorded over 10 positions along the length of the capillary, which was mounted at a distance of 2.133 m from a Pilatus 2M detector. Background measurements with buffer were performed using the identical capillaries, positions, and measurement protocol. Scattering data were used to a maximum momentum transfer of 0.25 Å−1. Data analysis and three-dimensional reconstruction were performed using PRIMUS (47), GNOM (48), and GASBOR (49) from the ATSAS suite.
UV-visible spectroscopy
All spectroscopic measurements of WT proteins were performed on heterologously-expressed proteins after removal of the hexahistidine tags unless otherwise stated, and for mutant proteins the His-tags were not removed. Spectra in the UV and visible range (200–700 nm, scan speed 400 nm/min) were recorded in 200-μl quartz micro-cuvettes with 1.0-cm path length (Hellma GmbH, Müllheim, Germany) at 0.5 nm bandwidth using JASCO V-650 or V-760 spectrophotometers (Jasco GmbH, Gross-Umstadt, Germany) at room temperature. The data were processed using the Jasco32 software. For qualitative spectra, protein samples were diluted in 25 mm KCl, 25 mm HEPES/KOH, pH 7.5. Ferrous protein samples were prepared in a nitrogen-filled glove box (Belle Technology Ltd., Weymouth, UK, <10 ppm O2) using thoroughly degassed and argon-saturated buffers. Samples of the KsNaxLS complex and its mutants (A2801 cm = 0.25–0.5) were reduced by adding freshly prepared 10 mm Ti(III)citrate (about 1 mm final concentration) in a buffer containing 25 mm KCl, 25 mm HEPES/KOH, pH 7.5. The reduced samples were transferred to stoppered quartz microcuvettes to measure the spectra outside of the glove box. Afterward, the cuvette was opened, and 10 ml of air supplied from a syringe was gently bubbled through the samples to obtain the spectra of the re-oxidized proteins.
Ligand binding
The binding of ligands to the KsNaxLS complex and its mutants was followed using UV-visible spectroscopy as described above. Spectra of the ferric proteins were also recorded in the presence of 10 mm hydroxylamine (NH2OH, as hydrochloride), hydrazine (N2H4, as hydrochloride), phenylhydrazine (C6H5-N2H3), methylhydrazine (CH3-N2H3), and hydroxyethylhydrazine (HO-C2H4-N2H3). These reagents were supplied by dilution from 200 mm stock solutions (pH adjusted to 7.0 with 1 m NaOH or 1 m HCl as required). Nitric oxide (NO) was freshly prepared by mixing 5 ml of anoxic 1.2 m ferrous sulfate in 1.8 m H2SO4 with 3 ml of anoxic 1.2 m sodium nitrite in a syringe inside of the glove box, followed by washing of the resulting colorless gas through 1 m anoxic NaOH to remove NO2 impurities. CO was transferred directly from a lecture bottle (Air Liquide, Ludwigshafen, Germany) into a balloon, which was then transferred into the glove box for further use. To obtain qualitative spectra of CO- or NO-bound cytochromes, 1 ml of buffer (25 mm KCl, 25 mm HEPES/KOH, pH 7.5) was sparged with 20–30 ml of either CO or NO. In the case of CO, this buffer was used for buffer exchanging protein samples immediately. For NO, the buffer was mixed with the protein sample in a 1:1 ratio by volume.
To estimate the Kd value of KsNaxLS for nitric oxide, protein was titrated with NO inside a nitrogen-filled glove box (Belle Technology Ltd., Weymouth, UK, <10 ppm O2) using thoroughly degassed and argon-saturated buffers. For each data point, a 1.0-ml reaction mixture was prepared in a polystyrene cuvette with a 10-mm path length. First, recombinantly produced KsNaxLS (from which the His tags had been removed) dissolved in 25 mm HEPES/KOH, pH 7.5, 25 mm KCl was added to the cuvette and reduced with 0.5 mm Ti(III)citrate as described above. Then, an appropriate amount of NO-containing buffer prepared as described above was added, and the cuvette was closed with a plastic lid and sealed with parafilm. The cuvette was then transferred out of the glove box, and an absorption spectrum was measured immediately using a JASCO V-650 spectrophotometer (Jasco GmbH, Gross-Umstadt, Germany) between 400 and 700 nm, with a 0.5-nm data interval and at a data acquisition rate of 400 nm/min. For each measurement, the NO concentration in the stock solution was determined using the Griess assay (50). Two data sets were measured, one at a KsNaxLS concentration of 4.5 μm and one at 11 μm, and the absorption change at 553 nm was used to determine binding. Each binding curve was fitted independently using the quadratic equation, and the weighted arithmetic mean of the fit results was calculated to arrive at an estimated Kd of 12 μm.
Redox titration
To determine redox potentials, the protein was titrated with Ti(III)citrate or ferricyanide inside a nitrogen-filled glove box (Belle Technology Ltd., Weymouth, UK, <10 ppm O2) using thoroughly degassed and argon-saturated buffers. 1.5 ml of protein solution (15 μm in 25 mm HEPES/KOH, pH 7.0, 100 mm KCl) was mixed with 5 μm each of the following redox mediators: potassium ferricyanide; p-benzoquinone; 2,5-dimethyl-p-benzoquinone; 1,2-naphthoquinone; phenazine methosulfate; 1,4-naphthoquinone; phenazine ethosulfate; 5-hydroxy-1,4-naphthoquinone; 2-methyl-1,4-naphthoquinone; 2,5-dihydroxy-p-benzoquinone; 2-hydroxy-1,4-naphthoquinone; anthraquinone; sodium anthraquinone-2-sulfonate; benzyl viologen; and methyl viologen. The mixture was placed in a quartz cuvette (10 mm path length), and a small magnetic stirring bar was added. The cuvette was placed in a CV100 cuvette holder (Thorlabs, Dachau, Germany) equipped with collimating lenses (LA4647, Thorlabs, Dachau, Germany) and fiberoptic light guides that were fed out of the glove box through silicone-filled plugs. The cuvette holder was placed on a miniature magnetic stirrer, and light was supplied from an CLX500 xenon lamp (Zeiss, Oberkochen, Germany) through one of the fiberoptic cables, whereas the exiting light was fed through the other light guide into an Avaspec ULS2048CL spectrometer (Avantes, Apeldoorn, The Netherlands). The redox potential was monitored using a 0.5-mm diameter platinum wire and a 3.5-mm diameter Ag/AgCl, 4 m KCl reference electrode (Pine Research, Durham, NC) using a custom-built, high-input impedance recording millivoltmeter. The protein was titrated with 10-μl portions of ferricyanide or Ti(III) stock solution (10 mm on redox potential plateaus, 0.5 mm for Ti(III)citrate, and 1 mm for ferricyanide close to redox transitions) using a gas-tight Hamilton syringe. Absorption spectra were recorded when the potential difference had reached a stable value. The Nernst equation was used to fit the data, and the obtained values were corrected to reflect the potentials with respect to the SHE assuming an offset of 200 mV.
Pulldown assay
The KsNaxLS complex (with a C-terminal His-tag on KsNaxS), its individual subunits, and Kustc0563, another small soluble cytochrome c from K. stuttgartiensis, were used as bait by immobilizing them on Ni-IDA Profinity beads (Bio-Rad, Munich, Germany), which were pre-equilibrated with wash buffer (150 mm NaCl, 50 mm HEPES/NaOH, pH 7.5). For the preparation of a cell lysate from K. stuttgartiensis biomass, 180–200 mg of cell pellet was resuspended in 1 ml of lysis buffer (25 mm HEPES, pH 7.5, 150 mm NaCl and 0.4% (v/v) Triton X-100). The cells were disrupted by sonication using a Branson 250 sonifier (G. Heinemann, Schwäbisch Gmünd, Germany) equipped with a sonication microtip at 50% amplitude, 0.5 s bursts, and a total of 20 pulses that was repeated three times. The lysate was incubated for 15 min while rotating in a cold room at 8 °C. Then, the lysate was cleared by centrifugation at 16,000 × g, 15–20 min at 4 °C. 50 μl of Ni-IDA beads loaded with the individual immobilized bait proteins were incubated with 100 μl of K. stuttgartiensis cell lysate in 2-ml Eppendorf tubes while rotating at 8 °C for 60 min. In parallel, another set of bait-loaded Ni-IDA beads (50 μl) was incubated with 100 μl of purified K. stuttgartiensis hydrazine synthase (KsHZS) protein solution (A2801 cm = 1.0). Moreover, both cleared cell lysate and KsHZS protein solutions were incubated with Ni-IDA beads without any bait as a control. Excess lysate and protein solution were removed by centrifugation afterward, and the beads were washed three times with 1 ml of wash buffer. Proteins were eluted using 50 μl of elution buffer (50 mm Tris-Cl, pH 8.0, 300 mm NaCl and 250 mm imidazole). Eluted samples were analyzed by 15% SDS-PAGE. Protein bands of interest were subjected to peptide mass fingerprinting by MALDI-MS for their identification. For heme-stained SDS-PAGE, 50 mm tris(2-carboxyethyl)phosphine was used as the reducing agent, as dithioerythritol and β-mercaptoethanol inhibit the chemiluminescence on which the staining is based, and a prestained molecular weight marker was loaded into one of the pockets of the gel. After electrophoresis, proteins were fixed by immersion of the gel in 40% (v/v) ethanol for 10 min. The gel was then soaked in 20 ml of 0.25 mg/ml luminol, 0.1 mg/ml p-coumaric acid, 100 mm CAPS/NaOH, pH 11.0, for 15 min after which 6 μl of 30% (w/w) hydrogen peroxide was added. After 5 min of further incubation, the gel was briefly rinsed in deionized water and placed inside a transparent plastic envelope. The positions of the prestained marker bands were indicated on the envelope using green phosphorescent acrylic paint (Lukas Acryl-Paint effect, Dr. Fr. Schoenfeld GmbH, Düsseldorf, Germany). The chemiluminescence arising from heme-containing protein bands was then recorded using a ChemiDoc MP CCD detection system (Bio-Rad).
Author contributions
M. A. and T. R. B. data curation; M. A. and A. D. formal analysis; M. A., J. R., A. D., W. V., and T. R. B. investigation; M. A., A. M., and T. R. B. methodology; M. A. and T. R. B. writing-original draft; M. A., J. R., A. D., B. K., M. S. J., and T. R. B. writing-review and editing; J. R., B. K., M. S. J., and T. R. B. conceptualization; T. R. B. supervision; T. R. B. validation; T. R. B. visualization; T. R. B. project administration.
Supplementary Material
Acknowledgments
We thank Prof. Dr. Kai Thormann (University of Giessen) for the kind gift of S. oneidensis MR-1, and the staff at the Paul Scherrer Institute for their excellent support and facilities. We are indebted to Dr. Jan Keltjens and Ilme Schlichting for stimulating discussions and critical reading of the manuscript. We also thank Melanie Mueller for expert MALDI-MS and peptide map fingerprinting, Chris Roome for excellent computing support, and the Heidelberg/Dortmund data collection team for synchrotron measurements. We are grateful to Clara Brieke and Marie Luise Grünbein for help building the spectrometer for the anaerobic redox titrations. T. R. M. B. is very grateful to Ilme Schlichting for continuous support.
This work was supported by the Max Planck Society. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Tables S1–S3.
The atomic coordinates and structure factors (codes 6R6M, 6R6N, and 6R6O) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- anammox
- anaerobic ammonium oxidation
- NO
- nitric oxide
- Nir
- nitrite reductase
- HZS
- hydrazine synthase
- HDH
- hydrazine dehydrogenase
- CO
- carbon monoxide
- AUC-SV
- analytical ultracentrifugation sedimentation velocity
- AUC-SE
- analytical ultracentrifugation sedimentation equilibrium
- SEC-MALS
- size-exclusion chromatography with multiangle static light scattering
- SAXS
- small-angle X-ray scattering
- Ti(III)citrate
- titanium (III) citrate
- Ni-IDA
- nickel(II) iminodiacetic acid
- RMSD
- root-mean-square deviation
- 5cNO
- 5-coordinate NO complex
- 6cNO
- 6-coordinate NO complex
- KPi
- potassium phosphate
- PDB
- Protein Data Bank
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- SHE
- standard hydrogen electrode
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid.
References
- 1. Arrigo K. R. (2005) Marine microorganisms and global nutrient cycles. Nature 437, 349–355 10.1038/nature04159 [DOI] [PubMed] [Google Scholar]
- 2. Lam P., and Kuypers M. M. M. (2011) Microbial nitrogen cycling processes in oxygen minimum zones. Annu. Rev. Mar. Sci. 3, 317–345 10.1146/annurev-marine-120709-142814 [DOI] [PubMed] [Google Scholar]
- 3. Kartal B., Maalcke W. J., de Almeida N. M., Cirpus I., Gloerich J., Geerts W., Op den Camp H. J., Harhangi H. R., Janssen-Megens E. M., Francoijs K.-J., Stunnenberg H. G., Keltjens J. T., Jetten M. S., and Strous M. (2011) Molecular mechanism of anaerobic ammonium oxidation. Nature 479, 127–130 10.1038/nature10453 [DOI] [PubMed] [Google Scholar]
- 4. Strous M., Pelletier E., Mangenot S., Rattei T., Lehner A., Taylor M. W., Horn M., Daims H., Bartol-Mavel D., Wincker P., Barbe V., Fonknechten N., Vallenet D., Segurens B., Schenowitz-Truong C., et al. (2006) Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440, 790–794 10.1038/nature04647 [DOI] [PubMed] [Google Scholar]
- 5. Dietl A., Ferousi C., Maalcke W. J., Menzel A., de Vries S., Keltjens J. T., Jetten M. S., Kartal B., and Barends T. R. (2015) The inner workings of the hydrazine synthase multiprotein complex. Nature 527, 394–397 10.1038/nature15517 [DOI] [PubMed] [Google Scholar]
- 6. Maalcke W. J., Reimann J., de Vries S., Butt J. N., Dietl A., Kip N., Mersdorf U., Barends T. R., Jetten M. S., Keltjens J. T., and Kartal B. (2016) Characterization of anammox hydrazine dehydrogenase, a key N2-producing enzyme in the global nitrogen cycle. J. Biol. Chem. 291, 17077–17092 10.1074/jbc.M116.735530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Almeida N. M., Neumann S., Mesman R. J., Ferousi C., Keltjens J. T., Jetten M. S., Kartal B., and van Niftrik L. (2015) Immunogold localization of key metabolic enzymes in the anammoxosome and on the tubule-like structures of K. stuttgartiensis. J. Bacteriol. 197, 2432–2441 10.1128/JB.00186-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Almeida N. M., Wessels H. J., de Graaf R. M., Ferousi C., Jetten M. S., Keltjens J. T., and Kartal B. (2016) Membrane-bound electron transport systems of an anammox bacterium: a complexome analysis. Biochim. Biophys. Acta 1857, 1694–1704 10.1016/j.bbabio.2016.07.006 [DOI] [PubMed] [Google Scholar]
- 9. Kartal B., de Almeida N. M., Maalcke W. J., Op den Camp H. J., Jetten M. S., and Keltjens J. T. (2013) How to make a living from anaerobic ammonium oxidation. FEMS Microbiol. Rev. 37, 428–461 10.1111/1574-6976.12014 [DOI] [PubMed] [Google Scholar]
- 10. van Niftrik L., Geerts W. J., van Donselaar E. G., Humbel B. M., Webb R. I., Fuerst J. A., Verkleij A. J., Jetten M. S., and Strous M. (2008) Linking ultrastructure and function in four genera of anaerobic ammonium-oxidizing bacteria: cell plan, glycogen storage, and localization of cytochrome c proteins. J. Bacteriol. 190, 708–717 10.1128/JB.01449-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ambler R. (1991) Sequence variability in bacterial cytochromes c. Biochim. Biophys. Acta 1058, 42–47 10.1016/S0005-2728(05)80266-X [DOI] [PubMed] [Google Scholar]
- 12. Ukita S., Fujii T., Hira D., Nishiyama T., Kawase T., Migita C. T., and Furukawa K. (2010) A heterodimeric cytochrome c complex with a very low redox potential from an anaerobic ammonium-oxidizing enrichment culture. FEMS Microbiol. Lett. 313, 61–67 10.1111/j.1574-6968.2010.02122.x [DOI] [PubMed] [Google Scholar]
- 13. Erickson H. P. (2009) Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 11, 32–51 10.1007/s12575-009-9008-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 10.1107/S0021889812007662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moser C. C., Chobot S. E., Page C. C., and Dutton P. L. (2008) Distance metrics for heme protein electron tunneling. Biochim. Biophys. Acta 1777, 1032–1037 10.1016/j.bbabio.2008.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maalcke W. J., Dietl A., Marritt S. J., Butt J. N., Jetten M. S., Keltjens J. T., Barends T. R., and Kartal B. (2014) Structural basis of biological NO generation by octaheme oxidoreductases. J. Biol. Chem. 289, 1228–1242 10.1074/jbc.M113.525147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bertini I., Faraone-Mennella J., Gray H. B., Luchinat C., Parigi G., and Winkler J. R. (2004) NMR-validated structural model for oxidized Rhodopseudomonas palustris cytochrome c 556. J. Biol. Inorg. Chem. 9, 224–230 10.1007/s00775-003-0511-2 [DOI] [PubMed] [Google Scholar]
- 18. Reijerse E. J., Sommerhalter M., Hellwig P., Quentmeier A., Rother D., Laurich C., Bothe E., Lubitz W., and Friedrich C. G. (2007) The unusual redox centers of SoxXA, a novel c-type heme-enzyme essential for chemotrophic sulfur-oxidation of Paracoccus pantotrophus. Biochemistry 46, 7804–7810 10.1021/bi7003526 [DOI] [PubMed] [Google Scholar]
- 19. Grein F., Venceslau S. S., Schneider L., Hildebrandt P., Todorovic S., Pereira I. A., and Dahl C. (2010) DsrJ, an essential part of the DsrMKJOP transmembrane complex in the purple sulfur bacterium Allochromatium vinosum, is an unusual triheme cytochrome c. Biochemistry 49, 8290–8299 10.1021/bi1007673 [DOI] [PubMed] [Google Scholar]
- 20. Denkmann K., Grein F., Zigann R., Siemen A., Bergmann J., van Helmont S., Nicolai A., Pereira I. A., and Dahl C. (2012) Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome. Environ. Microbiol. 14, 2673–2688 10.1111/j.1462-2920.2012.02820.x [DOI] [PubMed] [Google Scholar]
- 21. Alric J., Tsukatani Y., Yoshida M., Matsuura K., Shimada K., Hienerwadel R., Schoepp-Cothenet B., Nitschke W., Nagashima K. V., and Verméglio A. (2004) Structural and functional characterization of the unusual triheme cytochrome bound to the reaction center of Rhodovulum sulfidophilum. J. Biol. Chem. 279, 26090–26097 10.1074/jbc.M400361200 [DOI] [PubMed] [Google Scholar]
- 22. Van Driessche G., Devreese B., Fitch J. C., Meyer T. E., Cusanovich M. A., and Van Beeumen J. J. (2006) GHP, a new c-type green heme protein from Halochromatium salexigens and other proteobacteria. FEBS J. 273, 2801–2811 10.1111/j.1742-4658.2006.05296.x [DOI] [PubMed] [Google Scholar]
- 23. Lin C.-I., McCarty R. M., and Liu H.-W. (2013) The biosynthesis of nitrogen-, sulfur-, and high-carbon chain-containing sugars. Chem. Soc. Rev. 42, 4377–4407 10.1039/c2cs35438a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Motomura T., Suga M., Hienerwadel R., Nakagawa A., Lai T. L., Nitschke W., Kuma T., Sugiura M., Boussac A., and Shen J. R. (2017) Crystal structure and redox properties of a novel cyanobacterial heme protein with a His/Cys heme axial ligation and a Per-Arnt-Sim (PAS)-like domain. J. Biol. Chem. 292, 9599–9612 10.1074/jbc.M116.746263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raphael A. L., and Gray H. B. (1991) Semisynthesis of axial-ligand (position 80) mutants of cytochrome c. J. Am. Chem. Soc. 113, 1038–1040 10.1021/ja00003a045 [DOI] [Google Scholar]
- 26. Krest C. M., Silakov A., Rittle J., Yosca T. H., Onderko E. L., Calixto J. C., and Green M. T. (2015) Significantly shorter Fe–S bond in cytochrome P450-I is consistent with greater reactivity relative to chloroperoxidase. Nat. Chem. 7, 696–702 10.1038/nchem.2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perera R., Sono M., Sigman J. A., Pfister T. D., Lu Y., and Dawson J. H. (2003) Neutral thiol as a proximal ligand to ferrous heme iron: implications for heme proteins that lose cysteine thiolate ligation on reduction. Proc. Natl. Acad. Sci. U.S.A. 100, 3641–3646 10.1073/pnas.0737142100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Smulevich G., Bjerrum M. J., Gray H. B., and Spiro T. G. (1994) Resonance raman spectra and the active site structure of semisynthetic Met80Cys horse heart cytochrome c. Inorg. Chem. 33, 4629–4634 10.1021/ic00099a012 [DOI] [Google Scholar]
- 29. Hough M. A., and Andrew C. R. (2015) Cytochromes c': structure, reactivity and relevance to haem-based gas sensing. Adv. Microb. Physiol. 67, 1–84 10.1016/bs.ampbs.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 30. Andrew C. R., George S. J., Lawson D. M., and Eady R. R. (2002) Six- to five-coordinate heme-nitrosyl conversion in cytochrome c′ and its relevance to guanylate cyclase. Biochemistry 41, 2353–2360 10.1021/bi011419k [DOI] [PubMed] [Google Scholar]
- 31. Andrew C. R., Kemper L. J., Busche T. L., Tiwari A. M., Kecskes M. C., Stafford J. M., Croft L. C., Lu S., Moënne-Loccoz P., Huston W., Moir J. W., and Eady R. R. (2005) Accessibility of the distal heme face, rather than Fe-His bond strength, determines the heme-nitrosyl coordination number of cytochromes c′: evidence from spectroscopic studies. Biochemistry 44, 8664–8672 10.1021/bi050428g [DOI] [PubMed] [Google Scholar]
- 32. Yoshimura T., Fujii S., Kamada H., Yamaguchi K., Suzuki S., Shidara S., and Takakuwa S. (1996) Spectroscopic characterization of nitrosylheme in nitric oxide complexes of ferric and ferrous cytochrome c′ from photosynthetic bacteria. Biochim. Biophys. Acta 1292, 39–46 10.1016/0167-4838(95)00187-5 [DOI] [PubMed] [Google Scholar]
- 33. Manole A., Kekilli D., Svistunenko D. A., Wilson M. T., Dobbin P. S., and Hough M. A. (2015) Conformational control of the binding of diatomic gases to cytochrome c′. J. Biol. Inorg. Chem. 20, 675–686 10.1007/s00775-015-1253-7 [DOI] [PubMed] [Google Scholar]
- 34. Hu Z., Wessels H. J. C. T., van Alen T., Jetten M. S. M., and Kartal B. (2019) Nitric oxide dependent anaerobic ammonium oxidation. Nat. Comm. 10, 1910 10.1038/s41467-019-09797-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rathnayake R. M. L. D., Oshiki M., Ishii S., Segawa T., Satoh H., and Okabe S. (2018) Experimental evidence for in situ nitric oxide production in anaerobic ammonia-oxidizing bacterial granules. Environ. Sci. Technol. 52, 5744–5752 10.1021/acs.est.8b00876 [DOI] [PubMed] [Google Scholar]
- 36. Mayhew S. G. (1978) The redox potential of dithionite and SO-2 from equilibrium reactions with flavodoxins, methyl viologen and hydrogen plus hydrogenase. Eur. J. Biochem. 85, 535–547 10.1111/j.1432-1033.1978.tb12269.x [DOI] [PubMed] [Google Scholar]
- 37. van der Star W. R. L., Miclea A. I., van Dongen U. G. J. M., Muyzer G., Picioreanu C., and van Loosdrecht M. C. M. (2008) The membrane bioreactor: a novel tool to grow anammox bacteria as free cells. Biotech. Bioeng. 101, 286–294 10.1002/bit.21891 [DOI] [PubMed] [Google Scholar]
- 38. Kartal B., Geerts W., and Jetten M. S. (2011) Cultivation, detection, and ecophysiology of anaerobic ammonium-oxidizing bacteria. Methods Enzymol. 486, 89–108 10.1016/B978-0-12-381294-0.00004-3 [DOI] [PubMed] [Google Scholar]
- 39. Yanisch-Perron C., Vieira J., and Messing J. (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33, 103–119 10.1016/0378-1119(85)90120-9 [DOI] [PubMed] [Google Scholar]
- 40. Shi L., Lin J.-T., Markillie L. M., Squier T. C., and Hooker B. S. (2005) Overexpression of multi-heme c-type cytochromes. BioTechniques 38, 297–299 10.2144/05382PT01 [DOI] [PubMed] [Google Scholar]
- 41. Heun M., Binnenkade L., Kreienbaum M., and Thormann K. M. (2012) Functional specificity of extracellular nucleases of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 78, 4400–4411 10.1128/AEM.07895-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cowtan K. (1994) 'dm': an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34–38 [Google Scholar]
- 44. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 45. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 10.1016/S0006-3495(00)76713-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M., and Svergun D. I. (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 10.1107/S0021889803012779 [DOI] [Google Scholar]
- 48. Svergun D. I. (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 10.1107/S0021889892001663 [DOI] [Google Scholar]
- 49. Svergun D. I., Petoukhov M. V., and Koch M. H. (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 10.1016/S0006-3495(01)76260-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grisham M. B., Johnson G. G., and Lancaster J. R. Jr. (1996) Quantitation of nitrate and nitrite in extracellular fluids. Methods Enzymol. 268, 237–246 10.1016/S0076-6879(96)68026-4 [DOI] [PubMed] [Google Scholar]
- 51. Schägger J. G., and von Jagow G. (1987) Tricine–sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 10.1016/0003-2697(87)90587-2 [DOI] [PubMed] [Google Scholar]
- 52. Chen V. B., Arendall r W. B. Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 10.1107/s0907444903009879 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.