Abstract
Nutrient sensing is a critical cellular process controlling metabolism and signaling. mTOR complex 1 (mTORC1) is the primary signaling hub for nutrient sensing and, when activated, stimulates anabolic processes while decreasing autophagic flux. mTORC1 receives nutrient status signals from intracellular amino acid sensors. One of these sensors, Sestrin-2, functions as an intracellular sensor of cytosolic leucine and inhibitor of mTORC1 activity. Genetic studies of Sestrin-2 have confirmed its critical role in regulating mTORC1 activity, especially in the case of leucine starvation. Sestrin-2 is known to be transcriptionally controlled by several mechanisms; however, the post-translational proteolytic regulation of Sestrin-2 remains unclear. Here, we explored how Sestrin-2 is regulated through the ubiquitin proteasome system. Using an unbiased screening approach of an siRNA library targeting ubiquitin E3 ligases, we identified a RING-type E3 ligase, ring finger protein 186 (RNF186), that critically mediates the Sestrin-2 ubiquitination and degradation. We observed that RNF186 and Sestrin-2 bind each other through distinct C-terminal motifs and that Lys-13 in Sestrin-2 is a putative ubiquitin acceptor site. RNF186 knockdown increased Sestrin-2 protein levels and decreased mTORC1 activation. These results reveal a new mechanism of E3 ligase control of mTORC1 activity through the RNF186-Sestrin-2 axis, suggesting that RNF186 inhibition may be a potential strategy to increase levels of the mTORC1 inhibitor Sestrin-2.
Keywords: E3 ubiquitin ligase, autophagy, high-throughput screening (HTS), ubiquitylation (ubiquitination), mechanistic target of rapamycin (mTOR), nutrient sensing, ring finger protein 186 (RNF186), SESN2
Introduction
Nutrient sensing is a critical process controlling metabolism and growth function for cells and tissues (1). The primary signaling pathway controlling nutrient sensing is the mTOR3 complex 1 (mTORC1) pathway (2). During nutrient availability, mTORC1 phosphorylates several key signaling proteins, such as P70S6K, 4E-BP1, and ULK1, working to activate anabolic processes and inhibit processes such as autophagy (3). Specific amino acid sensors, such as Sestrin-2, sense intracellular nutrient levels and integrate this signal to the mTORC1 complex (4). Sestrin-2 (also known as Hi95) exerts inhibitory control over mTORC1 by associating with an mTORC1-activating protein complex called GATOR2 (5). This function is in a leucine-dependent manner, such that when leucine is abundant, the Sestrin-2–GATOR2 interaction is impeded, allowing activation and anabolic mTORC1 activity (5, 6). Sestrin-2 loss-of-function and mutational studies demonstrate the sensor's criticality for mTORC1 activity, as Sestrin-2–depleted cells result in constitutively active mTORC1, even in the absence of nutrients such as leucine (5). Further, Sestrin-2 has been shown to regulate autophagic flux in numerous cell types (7–9). Transcriptional control of Sestrin-2 has been well-characterized (10–12); however, the post-translational mechanisms of Sestrin-2 regulation remain unclear.
Ubiquitination is a major cellular mechanism to degrade proteins, often through the proteasome (13). This process entails the stepwise shuttling of the small protein ubiquitin to target substrates. This process is accomplished in an enzymatic cascade ending with a family of proteins called ubiquitin E3 ligases. E3 ligases play the critical role in identifying and binding the substrate protein fated for ubiquitination. Research has shown that ubiquitination controls various aspects of development and disease (14). Our group and others have noted distinct mechanisms of E3 ligase control of disease, such as lung innate immunity and fibrosis, and of mTORC1 activity (15–17). E3 ligases are also a growing field for therapeutic targeting and inhibition (18). We sought to determine whether Sestrin-2 was subject to ubiquitin E3 ligase–mediated degradation and the effect of this control on mTORC1 activity.
Here, we report the mechanistic study of Sestrin-2 ubiquitination and degradation by the RING E3 ligase RNF186. We observed that Sestrin-2 has a short half-life in Beas-2b cells that is prolonged by inhibiting the proteasome or the ubiquitination cascade. We used unbiased high-throughput screening of Sestrin-2-GFP–expressing cells with a library of siRNA-targeting ubiquitin proteins and E3 ligases to uncover the E3 ligase RNF186 as a regulator of Sestrin-2 stability. RNF186 silencing prolongs Sestrin-2 stability, whereas RNF186 expression accelerated Sestrin-2 degradation. RNF186 ubiquitinates and binds Sestrin-2 through specific domains. Silencing of RNF186 leads to decreased mTORC1 activity, potentially through increased Sestrin-2 protein abundance. This study adds a new mechanism for control of nutrient sensing through protein ubiquitination and suggests that RNF186 may be a potential target for inhibition to increase the level of the mTORC1 inhibitor Sestrin-2.
Results
The ubiquitin-proteasome system potently controls Sestrin-2 stability in airway cells
We first investigated the stability of Sestrin-2 in human primary airway epithelial Beas-2B cells. Cycloheximide (CHX) time course revealed that endogenous Sestrin-2 has a half-life of around 4 h, which is prolonged with proteasomal inhibition by MG132 (Fig. 1A). Treatment with the lysosomal inhibitor leupeptin did not increase Sestrin-2 protein stability. MG132 treatment also resulted in a time-dependent increase in endogenous Sestrin-2 level (Fig. 1B). Whereas the proteasome is the end point for most protein degradation, the ubiquitination pathway is a major upstream mechanism controlling proteins fated for proteasomal degradation. Co-treatment of Beas-2B cells with CHX and the ubiquitin E1 inhibitor MLN7243 resulted in enhanced Sestrin-2 stability, suggesting a role for ubiquitin control of Sestrin-2 stability (19). Expression of HA-tagged ubiquitin with CHX treatment led to Sestrin-2 degradation, which was also prevented with proteasomal inhibition (Fig. 1D). Ubiquitin is often assembled in polymeric chains upon proteins to be degraded; this polyubiquitination is a canonical signal for degradation (13). Sestrin-2 pulldown in the presence of MG132 displayed an extensive high-molecular weight ubiquitin signal, characteristic of polyubiquitination (Fig. 1E). These polyubiquitin chains have the capacity to assemble different linkage types, forming a cellular code (20). We used the UbiCREST assay to unravel the specific polyubiquitin chain type assembled on Sestrin-2. UbiCREST utilizes the differential reactivity of deubiquitinase enzymes for specific polyubiquitin linkage types (21). We observed that Sestrin-2 assembles predominately Lys-48 and -63 polyubiquitin chains, as evidenced by the degradation of Sestrin-2 polyubiquitin signal upon treatment with deubiquitinases specific for these linkage types (Fig. 1F).
Figure 1.
Sestrin-2 is ubiquitinated and degraded in the proteasome. A, immunoblot analysis of Sestrin-2 protein from Beas-2b cells following CHX time course with MG132 or leupeptin co-treatment. Shown are data and means ± S.D. (error bars) of three independent experiments. B, Sestrin-2 blotting from Beas-2b cells treated with vehicle, MG132, or leupeptin for the indicated times. Shown are data and means ± S.D. of three independent experiments. C, immunoblot analysis of Sestrin-2 protein from Beas-2b cells following CHX time course with the ubiquitin E1 inhibitor MLN7243 or leupeptin co-treatment. Shown are data and means ± S.D. of three independent experiments. D, HA-tagged ubiquitin was expressed prior to CHX time course with MG132 co-treatment and Sestrin-2 immunoblotting. Shown are data and means ± S.D. of three independent experiments. E, Sestrin-2-V5-HIS was expressed in Beas-2b cells prior to MG132 treatment, HIS pulldown, and ubiquitin immunoblotting. F, UbiCREST analysis of Sestrin-2 polyubiquitination pattern. Sestrin-2-V5-HIS was expressed in Beas-2b cells, treated with MG132, and pulled down prior to digestion with different deubiquitinases and immunoblot analysis. *, p < 0.05, by F-test comparisons of nonlinear regression relative to controls (A–D). WCL, whole-cell lysate.
The ubiquitin E3 ligase RNF186 ubiquitinates and degrades Sestrin-2
Ubiquitin E3 ligases are the critical substrate-recognition mechanism of the ubiquitination pathway. To understand the mechanism of Sestrin-2 degradation, we prepared a stable Sestrin-2-GFP–expressing cell line and utilized an RNAi library targeting proteins involved in ubiquitination (E1, E2, E3, etc.) to screen for Sestrin-2 regulators. Sestrin-2-GFP cells were transfected with the siRNA library, and GFP fluorescence was measured after 72 h and ranked by median absolute deviation Z-score. We observed several key regulators of Sestrin-2 stability (Fig. 2A). Specifically, the ubiquitin E3 ligases RNF186 and PRFP19 resulted in potent Sestrin-2-GFP signal relative to control siRNA (Fig. 2B). Top hits from the screen were evaluated by immunoblotting, and RNF186 knockdown resulted in increased SESN2 protein signal (Fig. 2C). Further, RNF186 expression resulted in dose-dependent decrease in Sestrin-2 protein (Fig. 2D). In contrast, mutation of a key residue within the RNF186 active site, His-60 to Trp (22), led to an inability to decrease Sestrin-2 protein (Fig. 2E). Beas-2B cells were treated with siRNA against RNF186 prior to CHX chase, and we observed that RNF186 knockdown resulted in prolonged Sestrin-2 stability relative to control siRNA (Fig. 2F). Conversely, RNF186 overexpression led to accelerated Sestrin-2 degradation in CHX chase (Fig. 2G). Finally, co-expression of RNF186 with ubiquitin and Sestrin-2 in an in vivo ubiquitination assay resulted in increased polyubiquitination signal detected from Sestrin-2 pulldown, suggesting that RNF186 facilitates the ubiquitination of Sestrin-2 (Fig. 2H).
Figure 2.
Ubiquitin RING E3 ligase RNF186 controls Sestrin-2 ubiquitination and degradation. A, screening of an siRNA library targeting ubiquitin E3 ligases for effect on Sestrin-2-GFP signal. Fluorescence was measured by Cytation 5 automated microcopy. Sestrin-2-GFP signal was reported as median absolute deviation Z-score. Top hits are ranked and displayed. B, representative Sestrin-2-GFP images from scramble siRNA and candidate E3 ligase siRNA from screen in A. Scale bar, 100 μm. C, immunoblot analysis of candidate E3 ligases identified from the screen in A. D, RNF186 was expressed dose-wise in Beas-2b prior to Sestrin-2 immunoblotting. E, immunoblot analysis of Sestrin-2 following expression of RNF186 H60W mutant. F, CHX chase of Sestrin-2 protein following siRNA knockdown of RNF186 in Beas-2b cells. Shown are data and means ± S.D. (error bars) of three independent experiments. G, CHX chase of Sestrin-2 protein following expression of RNF186 in Beas-2b cells. Shown are data and means ± S.D. of three independent experiments. H, immunoblot analysis of Sestrin-2 ubiquitination following in vivo ubiquitination assay and pulldown of Sestrin-2 protein. *, p < 0.05, by F-test comparisons of nonlinear regression compared with controls (F and G). WCL, whole-cell lysate.
RNF186 and Sestrin-2 bind each other through C-terminal motifs
To further examine the mechanism of the RNF186/Sestrin-2 interaction, we conducted reductionist mapping experiments to find the critical protein motif for binding. We prepared RNF186 deletion mutants (Fig. 3A), synthesized the protein fragments in vitro, and exposed them to immunoprecipitated Sestrin-2 in binding assays. We observed that the RNF186 ΔC27 mutant lost binding with Sestrin-2, and upon further deletion mapping, we observe that the C10–20 region of RNF186 is critical for Sestrin-2 binding (Fig. 3, B and C). We also prepare deletion mutants of Sestrin-2 encompassing critical protein domains (Fig. 3D). Sestrin-2 mutants lost binding with RNF186 with the ΔC173 mutant (Fig. 3E). Further mapping studies show that the key region for RNF186 engagement exists between residues 308 and 380 of Sestrin-2 (Fig. 3F).
Figure 3.
RNF186 and Sestrin-2 bind through discrete C-terminal motifs. A, schematic of RNF186 deletion mutant strategy and individual binding status with Sestrin-2. B and C, binding assay of in vitro synthesized RNF186 deletion mutants and Sestrin-2. D, Sestrin-2 deletion mutant strategy and binding status with RNF186. E and F, immunoblot analysis of Sestrin-2 mutant binding with RNF186 protein. G, WT or K13R mutant Sestrin-2 was expressed in Beas-2b cells without or with expression of RNF186 prior to blotting. Shown are data and means ± S.D. of three independent experiments. H, immunoblot analysis of Sestrin-2 WT or K13R expression prior to CHX time course. Shown are data and means ± S.D. (error bars) of three independent experiments. I, immunoblot analysis of Sestrin-2 K452R mutant in CHX assay. NS, p > 0.05; *, p < 0.05; ***, p < 0.001 by ANOVA, with Tukey's test for multiple comparisons (G), or by F-test comparisons of nonlinear regression relative to control (H). PD, pulldown.
Lysine 13 is a critical ubiquitin acceptor site for Sestrin-2
Substrates are ubiquitinated through a covalent isopeptide bond between ubiquitin's C terminus and an acceptor amino acid on the substrate. The key substrate amino acid is often lysine (23). We sought to identify the critical ubiquitin acceptor lysine site within Sestrin-2. Previous ubiquitin proteomics studies detected a ubiquitinated peptide corresponding to Sestrin-2 Lys-13 (24). To confirm this, we prepared Sestrin-2 point mutants and co-expressed them with RNF186 in Beas-2B cells. Sestrin-2 K13R mutant demonstrated resistance to RNF186 expression relative to WT (Fig. 3G). Further, Sestrin-2 K13R mutant displayed a prolonged half-life in CHX time course (Fig. 3H). As a negative control, mutation of nonubiquitin-associated Sestrin-2 lysine (K452R) did not confer protection from degradation (Fig. 3I).
RNF186 affects Sestrin-2 regulation of mTORC1
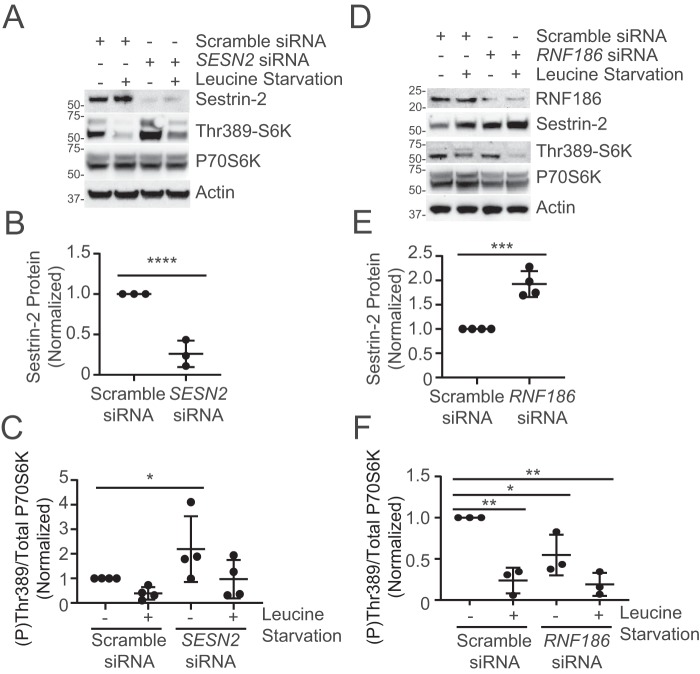
We next sought to explore the effect of the RNF186-Sestrin-2 axis on downstream mTORC1 signaling. As a leucine sensor, Sestrin-2 acts to inhibit mTORC1 activity by interacting with GATOR2 and preventing its downstream regulation of the Rag GTPases (5). The inhibitory effect of Sestrin-2 is most potent during leucine starvation. SESN2 silencing in Beas-2B cells led to increased mTORC1 activity as measured by phosphorylation of P70S6K (Thr-389) (Fig. 4, A–C). This effect persists even during cellular leucine starvation. Silencing of RNF186 led to increased SESN2 protein relative to control (Fig. 4, D and E). Of note, RNF186 knockdown led to decreased mTORC1 activity relative to control (Fig. 4F).
Figure 4.
RNF186 knockdown impairs leucine-dependent nutrient sensing. A, immunoblot analysis of mTORC1 activity and autophagic flux from Beas-2b cells treated with control or SESN2 siRNA prior to leucine starvation. Protein densitometry of Sestrin-2 (B) and ratio of phosphorylated (Thr-389) to total P70S6K (C) were quantified. Shown are data and means ± S.D. (error bars) of 3–4 independent experiments. D, immunoblot analysis of mTORC1 activity and autophagic flux from Beas-2b cells treated with control or RNF186 siRNA prior to leucine starvation. Protein densitometry of Sestrin-2 (E) and ratio of phosphorylated (Thr-389) to total P70S6K (F) was quantified. Shown are data and means ± S.D. of 3–4 independent experiments. *, p < 0.05; **, p < 0.01; ****, p < 0.0001 by two-sided unpaired t test (B and E) or by one-way ANOVA, with Tukey's test for multiple comparisons (C and F).
As a complementary assay, we utilized LC3-GFP-RFP reporter cells to measure the effect of RNF186 and SESN2 interaction. This autophagic tool exploited the instability of GFP at low pH to create a reporter sensitive to lysosomal maturation. The LC3 construct also has constitutively cleaved RFP fusion protein, serving as a loading control. The ratio of GFP/RFP is a surrogate for autophagic flux in a cell (25). As autophagy is an end point of mTORC1 regulation, we utilized this tool as a readout for mTORC1 activity. We knocked down SESN2 and RNF186 in Beas-2B cells that stably express LC3-GFP-RFP reporter prior to leucine starvation, fixation, and automated microscopy. We observed control siRNA during leucine starvation, resulting in a 70% decrease in GFP/RFP ratio relative to baseline, consistent with previous studies on the effect of amino acid starvation on autophagic flux (Fig. 5, A and B) (25). SESN2 knockdown resulted in a significantly higher GFP/RFP ratio, suggesting less autophagic flux (Fig. 5, A and C). Interestingly, RNF186 silencing decreased the GFP/RFP further, suggestive of increased autophagic flux (Fig. 5, A and D). These assays suggest that the RNF186/Sestrin-2 axis plays a role in regulating cellular nutrient sensing and metabolic flux.
Figure 5.

Sestrin-2 and RNF186 depletion affect starvation-mediated autophagic flux. A, quantification of LC3B GFP to RFP ratio in cells starved of leucine. Data are normalized to control siRNA baseline ratio. Shown are data and means ± S.D. (error bars) (n = 4–8). B, representative image of LC3B-GFP-RFP reporter cells transfected with scramble siRNA at baseline conditions. C–E, representative images of LC3B-GFP-RFP reporter cells starved of leucine and transfected with siRNA targeting scramble (C), SESN2 (D), or RNF186 (E). ***, p < 0.001; ****, p < 0.0001 by ANOVA, with Tukey's test for multiple comparisons (A). Scale bar, 50 μm.
Discussion
E3 ligases are increasingly appreciated regulators of nutrient sensing and mTORC1 activity (17). A recent study has shown the GATOR1 subunit DEPDC5 to be potently regulated by the Cullin-3 substrate receptor KLHL22 (26). Our data show that RNF186 plays a similar role in targeting Sestrin-2 for ubiquitination and degradation, which in turn affects mTORC1 activity and downstream autophagic flux. Dysfunction of nutrient sensing is recognized as a hallmark of aging; better understanding of the regulation of nutrient sensing will afford new potential targets for inhibition and intervention (27).
RNF186 has been characterized as a RING (really new interesting gene)-type ubiquitin E3 ligase (28). RING E3 ligases function through zinc finger domains that are critical for engaging the E2 ubiquitin-conjugating enzyme (29). We observed that mutation of a critical residue within the RING domain impaired RNF186's ability to degrade Sestrin-2 (Fig. 2E), suggesting that the ubiquitin E3 ligase activity of RNF186 drives its effect on Sestrin-2 (22). We observed that RNF186 and Sestrin-2 binding was lost upon the deletion of a 72-residue region between amino acid 308 and 380 of the Sestrin-2 C terminus (Fig. 3F). Interestingly, this same region has been described as key for regulating mTORC1 activity and for binding leucine (residues 374–377) (6, 30). Future studies are needed to investigate the distinct mechanism of RNF186/Sestrin-2 interaction and whether leucine plays a role.
Lys-48 polyubiquitination is the canonical signal for proteasomal degradation and has been reported as a predominant linkage type among cellular polyubiquitin chains (31). Through the UbiCREST assay, we observed Sestrin-2 polyubiquitination with Lys-48 linkages, consistent with previous studies in neuronal cells (32). We also observed that Sestrin-2 contains polyubiquitin chains with Lys-63 linkages. This linkage type has been implicated in several processes, notably cellular trafficking (33). It remains unclear whether Sestrin-2 ubiquitination plays a nondegradation role in its cellular localization.
mTORC1 plays an important role in lung epithelia and disease. Research has suggested that mTORC1 activity plays a pathogenic role in epithelia during acute lung injury (34). Similarly, mTOR-driven signaling affects the pathophysiology of pulmonary fibrosis in fibroblasts (35). One study suggested Sestrin-2 inhibition to be protective in a mouse model of chronic obstructive pulmonary disease (36). More research is needed to see whether RNF186 and Sestrin-2 play a role in these lung diseases.
In conclusion, we describe a new mechanism of ubiquitin E3 ligase–mediated control of nutrient sensing through the interaction of RNF186 with Sestrin-2.
Experimental procedures
BEAS-2B (catalog no. CRL-9609, RRID:CVCL_0168) was from ATCC. Goat anti-rabbit HRP (catalog no. 170-6515, RRID:AB_11125142), goat anti-mouse HRP (catalog no. 170-6516, RRID:AB_11125547), and rabbit anti-goat HRP (catalog no. 172-1034, RRID:AB_11125144) were from Bio-Rad. UbiCREST assay kit was from BostonBiochem. Mouse monoclonal anti-HA tag (clone 6E2) (catalog no. 2367S RRID:AB_10691311), rabbit anti-phospho-p70 S6 kinase (Thr-389) (108D2) (catalog no. 9234, RRID:AB_2269803), rabbit anti-p70 S6 kinase (49D7) (catalog no. 2708, RRID:AB_390722), rabbit anti-Sestrin-2 (D1B6) (catalog no. 8487, RRID:AB_11178663), mouse anti-ubiquitin (P4D1) (catalog no. 3936, RRID:AB_331292), and rabbit anti-LC3B (D11) XP (catalog no. 3868, RRID:AB_2137707) were from Cell Signaling Technology. Cycloheximide (catalog no. BML-GR310) and the ubiquitinylation kit (catalog no. BML-UW9920-0001) were from Enzo. Sequencing was conducted at Genewiz. DNA primers and DsiRNA were from IDT. pcDNA3.1D-V5-HIS-TOPO (catalog no. K490001) was from Invitrogen. The TNT T7 Quick Coupled Transcription/Translation system (catalog no. L1170) was from Promega. Leupeptin (catalog no. L2884) and mouse monoclonal anti-β-actin (clone AC-15) (catalog no. A5441, RRID:AB_476744) were from Sigma-Aldrich. Mouse monoclonal anti-V5 tag (catalog no. R960-25, RRID:AB_2556564), Protein A/G magnetic beads (catalog no. 88802), and the pcDNA3.1 Directional TOPO Expression Kit (catalog no. K490001) were from Thermo Fisher Scientific. pRK5-HA-ubiquitin-WT (catalog no. 17608, RRID:Addgene17608) was a gift from Ted Dawson (37). Anti-RNF186 antibody (catalog no. ab86547, RRID:AB_2180433) was from Abcam. WesternBright Sirius HRP substrate (catalog no. K-12043) was from Advansta. The QuikChange II XL site-directed mutagenesis kit (catalog no. 200521) was from Agilent Technologies. pLenti CMV GFP Puro (658-5) was a gift from Eric Campeau and Paul Kaufman (Addgene plasmid no. 17448; RRID:Addgene_17448). pMRX-IP-GFP-LC3-RFP was a gift from Noboru Mizushima (Addgene plasmid no. 84573; RRID:Addgene_84573) (25). pRK5-HA-Sestrin2 was a gift from David Sabatini (Addgene plasmid no. 72593, RRID:Addgene_72593) (38).
Cell culture
Beas-2b cells were from ATCC and cultured in HITES medium supplemented with 10% fetal bovine serum. Cells were treated with cycloheximide (100 μg/ml), MG132 (20 μm), leupeptin (20 μm), and MLN7243 (10 μm) for the indicated times. Cells were transfected with Nucleofector 2b (Amaxa) or XtremeGene siRNA reagent (Roche Applied Science). Cells were starved by two washes and then incubated with EBSS supplemented with amino acids minus leucine (Gibco).
UbiCREST assay
Ubiquitination linkage-type analysis was measured by UbiCREST assay (21) (BostonBiochem). Briefly, Sestrin-2 was expressed in Beas-2b, treated with MG132 (20 μm, 4 h), and lysed prior to HIS pulldown with HisPur Resin (Thermo Fisher Scientific). Following UbiCREST digestion, resin eluate was assayed by immunoblotting.
Ubiquitination siRNA screen
Stably expressing Beas-2b Sestrin-2-GFP cells in 384-well glass-bottom plates were transfected with MISSION esiRNA targeting ubiquitination proteins (E1, E2, E3 ligases, etc.) (Sigma-Aldrich) using XtremeGene siRNA transfection reagent (Roche Applied Science). Knockdown proceeded for 72 h prior to washing, fixation, and fluorescent imaging using BioTek Cytation 5.
LC3 fluorescent reporter assay
LC3 reporter cells were transfected with DsiRNA against scramble, SESN2, or RNF186 and seeded to 96-well glass-bottle plates for 60 h before starvation in EBSS + amino acids minus leucine for 18 h. Cells were fixed in 4% paraformaldehyde and imaged using ImageXpress Micro XLS. Fluorescence was quantified, and GFP/RFP ratio was calculated with CellProfiler (39).
Statistics
All statistical tests were calculating using GraphPad Prism version 8. p < 0.05 was used to indicate significance. Densitometry was calculated using ImageJ (National Institutes of Health).
In vitro protein-binding assays
Protein binding assays were conducted as described previously (15). Briefly, Sestrin-2 or RNF186 protein was immunoprecipitated from 1 mg of Beas-2b cell lysate using 1:100 antibody dilution. Protein was precipitated in immunoprecipitation buffer (50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.25% (v/v) Triton X-100) for 4 h at +4 ºC and then coupled to protein A/G–agarose resin for an additional 2 h. Binding mutants were in vitro synthesized using TNT expression kits and allowed to bind overnight. Resin was washed, and protein was eluted in 1× Laemmli buffer at 88 °C for 5 min prior to immunoblotting analysis.
Author contributions
T. B. L. and B. B. C. conceptualization; T. B. L., B. L., and Y. L. resources; T. B. L. and M. B. L. data curation; T. B. L., J. W. E., Y. L., and B. B. C. supervision; T. B. L., Y. L., and B. B. C. funding acquisition; T. B. L., K. C. L., Y. O., J. W. E., and M. B. L. investigation; T. B. L. and B. B. C. visualization; T. B. L., J. W. E., M. B. L., B. L., and B. B. C. methodology; T. B. L. writing-original draft; T. B. L. and B. B. C. writing-review and editing; M. B. L. software; Y. L. and B. B. C. project administration.
Acknowledgments
We thank Alison C. McKelvey and Sarah R. Dunn for assistance in cloning the initial RNF186 plasmid construct.
This work was supported by the University of Pittsburgh Aging Institute seed fund (to B. B. C and Y. L.) as well as NIDDK, National Institutes of Health (NIH), Grant 1R01DK119627 (to Y. L.) and NHLBI, NIH, Grants 5F31HL143843–02 (to T. B. L.), 1K08HL144820 (to J. W. E.), 5R01HL14277702 (to Y. L.), and 5R35HL139860–02 and 5R01HL133184 (to B. B. C.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- mTOR
- mechanistic target of rapamycin
- mTORC1
- mTOR complex 1
- RING
- really new interesting gene
- CHX
- cycloheximide
- DsiRNA
- Dicer-substrate siRNA
- EBSS
- Earle's balanced salt solution
- RFP
- red fluorescent protein
- HRP
- horseradish peroxidase
- ANOVA
- analysis of variance.
References
- 1. Yuan H.-X., Xiong Y., and Guan K.-L. (2013) Nutrient sensing, metabolism, and cell growth control. Mol. Cell 49, 379–387 10.1016/j.molcel.2013.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sabatini D. M. (2017) Twenty-five years of mTOR: uncovering the link from nutrients to growth. Proc. Natl. Acad. Sci. U.S.A. 114, 11818–11825 10.1073/pnas.1716173114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saxton R. A., and Sabatini D. M. (2017) mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 10.1016/j.cell.2017.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wolfson R. L., and Sabatini D. M. (2017) The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metab. 26, 301–309 10.1016/j.cmet.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wolfson R. L., Chantranupong L., Saxton R. A., Shen K., Scaria S. M., Cantor J. R., and Sabatini D. M. (2016) Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48 10.1126/science.aab2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saxton R. A., Knockenhauer K. E., Wolfson R. L., Chantranupong L., Pacold M. E., Wang T., Schwartz T. U., and Sabatini D. M. (2016) Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 351, 53–58 10.1126/science.aad2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liang Y., Zhu J., Huang H., Xiang D., Li Y., Zhang D., Li J., Wang Y., Jin H., Jiang G., Liu Z., and Huang C. (2016) SESN2/sestrin 2 induction-mediated autophagy and inhibitory effect of isorhapontigenin (ISO) on human bladder cancers. Autophagy 12, 1229–1239 10.1080/15548627.2016.1179403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kumar A., and Shaha C. (2018) SESN2 facilitates mitophagy by helping Parkin translocation through ULK1 mediated Beclin1 phosphorylation. Sci. Rep. 8, 615 10.1038/s41598-017-19102-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim M. J., Bae S. H., Ryu J. C., Kwon Y., Oh J. H., Kwon J., Moon J. S., Kim K., Miyawaki A., Lee M. G., Shin J., Kim Y. S., Kim C. H., Ryter S. W., Choi A. M., et al. (2016) SESN2/sestrin2 suppresses sepsis by inducing mitophagy and inhibiting NLRP3 activation in macrophages. Autophagy 12, 1272–1291 10.1080/15548627.2016.1183081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Byun J.-K., Choi Y.-K., Kim J.-H., Jeong J. Y., Jeon H.-J., Kim M.-K., Hwang I., Lee S.-Y., Lee Y. M., Lee I.-K., and Park K.-G. (2017) A positive feedback loop between Sestrin2 and mTORC2 is required for the survival of glutamine-depleted lung cancer cells. Cell Rep. 20, 586–599 10.1016/j.celrep.2017.06.066 [DOI] [PubMed] [Google Scholar]
- 11. Ding B., Parmigiani A., Divakaruni A. S., Archer K., Murphy A. N., and Budanov A. V. (2016) Sestrin2 is induced by glucose starvation via the unfolded protein response and protects cells from non-canonical necroptotic cell death. Sci. Rep. 6, 22538 10.1038/srep22538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee J. H., Budanov A. V., Park E. J., Birse R., Kim T. E., Perkins G. A., Ocorr K., Ellisman M. H., Bodmer R., Bier E., and Karin M. (2010) Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 327, 1223–1228 10.1126/science.1182228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hershko A., and Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- 14. Rape M. (2018) Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 19, 59–70 10.1038/nrm.2017.83 [DOI] [PubMed] [Google Scholar]
- 15. Lear T., McKelvey A. C., Rajbhandari S., Dunn S. R., Coon T. A., Connelly W., Zhao J. Y., Kass D. J., Zhang Y., Liu Y., and Chen B. B. (2016) Ubiquitin E3 ligase FIEL1 regulates fibrotic lung injury through SUMO-E3 ligase PIAS4. J. Exp. Med. 213, 1029–1046 10.1084/jem.20151229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen B. B., Coon T. A., Glasser J. R., McVerry B. J., Zhao J., Zhao Y., Zou C., Ellis B., Sciurba F. C., Zhang Y., and Mallampalli R. K. (2013) A combinatorial F box protein directed pathway controls TRAF adaptor stability to regulate inflammation. Nat. Immunol. 14, 470–479 10.1038/ni.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang Y., Su S., Zhang Y., Qian J., and Liu P. (2019) Control of mTOR signaling by ubiquitin. Oncogene 38, 3989–4001 10.1038/s41388-019-0713-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zheng N., and Shabek N. (2017) Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 10.1146/annurev-biochem-060815-014922 [DOI] [PubMed] [Google Scholar]
- 19. Hyer M. L., Milhollen M. A., Ciavarri J., Fleming P., Traore T., Sappal D., Huck J., Shi J., Gavin J., Brownell J., Yang Y., Stringer B., Griffin R., Bruzzese F., Soucy T., et al. (2018) A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med. 24, 186–193 10.1038/nm.4474 [DOI] [PubMed] [Google Scholar]
- 20. Komander D., and Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- 21. Hospenthal M. K., Mevissen T. E. T., and Komander D. (2015) Deubiquitinase-based analysis of ubiquitin chain architecture using ubiquitin chain restriction (UbiCRest). Nat. Protoc. 10, 349–361 10.1038/nprot.2015.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujimoto K., Kinoshita M., Tanaka H., Okuzaki D., Shimada Y., Kayama H., Okumura R., Furuta Y., Narazaki M., Tamura A., Hatakeyama S., Ikawa M., Tsuchiya K., Watanabe M., Kumanogoh A., Tsukita S., and Takeda K. (2017) Regulation of intestinal homeostasis by the ulcerative colitis-associated gene RNF186. Mucosal Immunol. 10, 446–459 10.1038/mi.2016.58 [DOI] [PubMed] [Google Scholar]
- 23. Akutsu M., Dikic I., and Bremm A. (2016) Ubiquitin chain diversity at a glance. J. Cell Sci. 129, 875–880 10.1242/jcs.183954 [DOI] [PubMed] [Google Scholar]
- 24. Udeshi N. D., Svinkina T., Mertins P., Kuhn E., Mani D. R., Qiao J. W., and Carr S. A. (2013) Refined preparation and use of anti-diglycine remnant (K-ϵ-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12, 825–831 10.1074/mcp.O112.027094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaizuka T., Morishita H., Hama Y., Tsukamoto S., Matsui T., Toyota Y., Kodama A., Ishihara T., Mizushima T., and Mizushima N. (2016) An autophagic flux probe that releases an internal control. Mol. Cell 64, 835–849 10.1016/j.molcel.2016.09.037 [DOI] [PubMed] [Google Scholar]
- 26. Chen J., Ou Y., Yang Y., Li W., Xu Y., Xie Y., and Liu Y. (2018) KLHL22 activates amino-acid-dependent mTORC1 signalling to promote tumorigenesis and ageing. Nature 557, 585–589 10.1038/s41586-018-0128-9 [DOI] [PubMed] [Google Scholar]
- 27. López-Otín C., Blasco M. A., Partridge L., Serrano M., and Kroemer G. (2013) The hallmarks of aging. Cell 153, 1194–1217 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang P., Wu Y., Li Y., Zheng J., and Tang J. (2013) A novel RING finger E3 ligase RNF186 regulate ER stress-mediated apoptosis through interaction with BNip1. Cell. Signal. 25, 2320–2333 10.1016/j.cellsig.2013.07.016 [DOI] [PubMed] [Google Scholar]
- 29. Deshaies R. J., and Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- 30. Kim H., An S., Ro S. H., Teixeira F., Park G. J., Kim C., Cho C. S., Kim J. S., Jakob U., Lee J. H., and Cho U. S. (2015) Janus-faced Sestrin2 controls ROS and mTOR signalling through two separate functional domains. Nat. Commun. 6, 10025 10.1038/ncomms10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim W., Bennett E. J., Huttlin Edward L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., and Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kumar A., and Shaha C. (2018) RBX1-mediated ubiquitination of SESN2 promotes cell death upon prolonged mitochondrial damage in SH-SY5Y neuroblastoma cells. Mol. Cell Biochem. 446, 1–9 10.1007/s11010-017-3267-7 [DOI] [PubMed] [Google Scholar]
- 33. Swatek K. N., and Komander D. (2016) Ubiquitin modifications. Cell Res. 26, 399–422 10.1038/cr.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu Y., Lou J., Mao Y. Y., Lai T. W., Liu L. Y., Zhu C., Zhang C., Liu J., Li Y. Y., Zhang F., Li W., Ying S. M., Chen Z. H., and Shen H. H. (2016) Activation of MTOR in pulmonary epithelium promotes LPS-induced acute lung injury. Autophagy 12, 2286–2299 10.1080/15548627.2016.1230584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Romero Y., Bueno M., Ramirez R., Álvarez D., Sembrat J. C., Goncharova E. A., Rojas M., Selman M., Mora A. L., and Pardo A. (2016) mTORC1 activation decreases autophagy in aging and idiopathic pulmonary fibrosis and contributes to apoptosis resistance in IPF fibroblasts. Aging Cell 15, 1103–1112 10.1111/acel.12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wempe F., De-Zolt S., Koli K., Bangsow T., Parajuli N., Dumitrascu R., Sterner-Kock A., Weissmann N., Keski-Oja J., and von Melchner H. (2010) Inactivation of sestrin 2 induces TGF-β signaling and partially rescues pulmonary emphysema in a mouse model of COPD. Dis. Model. Mech. 3, 246–253 10.1242/dmm.004234 [DOI] [PubMed] [Google Scholar]
- 37. Lim K. L., Chew K. C., Tan J. M., Wang C., Chung K. K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C. A., Dawson V. L., and Dawson T. M. (2005) Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J. Neurosci. 25, 2002–2009 10.1523/JNEUROSCI.4474-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chantranupong L., Wolfson R. L., Orozco J. M., Saxton R. A., Scaria S. M., Bar-Peled L., Spooner E., Isasa M., Gygi S. P., and Sabatini D. M. (2014) The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 9, 1–8 10.1016/j.celrep.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J., Golland P., and Sabatini D. M. (2006) CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]