Abstract
The intracellular accumulation of aggregated misfolded proteins is a cytopathological hallmark of neurodegenerative diseases. However, the functional relationship between protein misfolding or aggregation and the cellular proteostasis network that monitors and maintains proteome health is poorly understood. Previous studies have associated translational suppression and transcriptional remodeling with the appearance of protein aggregates, but whether these responses are induced by aggregates or their misfolded monomeric or oligomeric precursors remains unclear. Because aggregation in cells is rapid, nonlinear, and asynchronous, it has not been possible to deconvolve these kinetically linked processes to determine the earliest cellular responses to misfolded proteins. Upon removal of the synthetic, biologically inert ligand shield-1 (S1), AgDD, an engineered variant FK506-binding protein (FKBP1A), rapidly (t½ ∼5 min) unfolds and self-associates, forming detergent-insoluble, microscopic cytoplasmic aggregates. Using global diglycine-capture (K-GG) proteomics, we found here that this solubility transition is associated with immediate increases in ubiquitylation of AgDD itself, along with that of endogenous proteins that are components of the ribosome and the 26S proteasome. We also found that the earliest cellular responses to acute S1 removal include recruitment of ubiquitin protein ligase E3C (UBE3C) to the 26S proteasome and ubiquitylation of two key proteasomal ubiquitin receptors, 26S proteasome regulatory subunit RPN10 (RPN10) and Rpn13 homolog (RPN13 or ADRM1). We conclude that these proteasomal responses are due to AgDD protein misfolding and not to the presence of detergent-insoluble aggregates.
Keywords: protein misfolding, protein aggregation, proteasome, ubiquitin, E3 ubiquitin ligase, ADRM1, RPN13, UBE3C
Introduction
To perform their biological functions, proteins must fold into and maintain a correct three-dimensional structure in a crowded intracellular environment (1). Moreover, the concentration of each protein must be precisely maintained by a carefully orchestrated balance of protein synthesis and degradation. This balance is maintained by the proteostasis network (PN),5 composed of over 2000 proteins that comprise molecular chaperones, proteolytic systems, protein synthesis machinery, and regulators of these processes (2). Failure to maintain this homoeostatic balance leads to the deposition of aggregated proteins into intracellular inclusion bodies (IBs) in the nucleus and cytosol of diseased neurons (3). Although the nearly ubiquitous association of IBs in neurodegenerative diseases has long suggested a functional link between PN disruption and pathogenesis, the precise mechanistic relationship between protein misfolding and aggregation and neuropathology has been difficult to sort out. Indeed, there is no consensus whether IBs are toxic, neutral, or even cytoprotective (4).
Because the PN is a highly interconnected, multilayered network that controls all aspects of protein metabolism, its disruption has pleiotropic consequences that have been extremely challenging to deconvolve. Although conformationally compromised proteins accumulate in cells slowly and asynchronously and conformational diseases typically take decades to manifest, the conversion of a folding-compromised protein from a soluble to an insoluble state occurs rapidly and acutely both in cell culture (5) and in animal models (6) of disease. However, although genetically encoded conformational derangement (misfolding) of a single defined protein, such as huntingtin or tau, is sufficient to initiate a cascade of events leading ultimately to massive PN collapse (5, 7, 8), the asynchronous nature of protein misfolding and aggregation in animal and cell models of disease has hitherto precluded the ability to accurately identify the early (minute time scale) responses to conformationally induced PN challenge that precede the well-studied transcriptional responses to protein aggregation.
In this study, we exploit a model of ligand-inducible protein misfolding and aggregation that permits highly synchronous, population-wide analysis of global PN perturbation at the proteome level with exquisite temporal precision (9). This approach uses cell lines that express variants of the FK506-binding protein (FKBP1A) termed degradation domains, whose stability is absolutely dependent on the binding of a high-affinity synthetic ligand, Shield-1 (S1) (10, 11). In the presence of S1, degradation domains are well-behaved, folded globular proteins. In the absence of ligand, degradation domains rapidly misfold and, when expressed in mammalian cells, are rapidly degraded by the ubiquitin proteasome system (UPS), together with any protein to which they are attached (10). This work exploits “AgDD,” a variant degradation domain that differs from the widely used “DD” by the presence of an N-terminal decapeptide that is enriched in hydrophobic and aliphatic residues (Fig. 1A) (9). Both DD and AgDD are conjugated to superfolder GFP (sfGFP), are monomeric, and are uniformly distributed in the cell when bound to their ligand, S1 (9, 10). Upon acute removal of S1 from the culture medium (“washout” (w/o)), both proteins unfold, sampling different conformations and potentially transitioning dynamically among them (9, 11). These partially unfolded proteins are targeted for ubiquitylation and, in the case of DD, rapid proteasomal degradation (t½ = 90 min) (12, 13). By contrast, S1 w/o causes AgDD to rapidly aggregate into puncta that are refractory to degradation and coalesce into cytoplasmic IBs (9). Our data indicate that one of the earliest cellular responses (<10 min) to acute protein misfolding is ubiquitylation of RPN13, one of the key Ub receptors on the apical cap of the 26S proteasome. We find that partially unfolded rather than aggregated AgDD induces recruitment of the Ub ligase, UBE3C to the proteasome as well as UBE3C-dependent multiubiquitylation of RPN13 and polyubiquitylation of proteasome-associated AgDD.
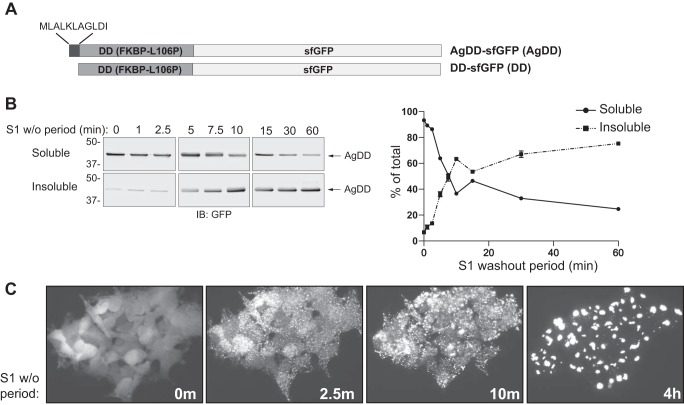
Figure 1.
Synchronous aggregation of AgDD induced by S1 w/o. A, AgDD-sfGFP and DD-sfGFP protein domains (“AgDD” and “DD” throughout for brevity). B, time course of AgDD partitioning into the Triton X-100–insoluble fraction of cell lysates following S1 w/o. C, quantification of the data in C (n = 4). D, time course of AgDD transition from diffuse to punctate subcellular distribution following S1 w/o. Error bars, S.D.
Results
Ubiquitylation of key PN nodes in response to acute misfolding of a single protein
To study the earliest global changes to the PN that occur in response to acute protein folding stress, we exploited the ability of AgDD to unfold and aggregate following acute removal of the stabilizing ligand, S1 (9). To evaluate the suitability of AgDD for such global analysis, the spatial distribution of sfGFP fluorescence was monitored by time-lapse imaging of HEK293 cells stably expressing AgDD-sfGFP (heretofore, AgDD) following acute w/o of S1 (Fig. 1B). In the presence of ligand, AgDD fluorescence was diffuse, indistinguishable from that typically observed for unmodified GFP. Upon S1 w/o, AgDD rapidly shifted to a highly granular, punctate distribution observable at the earliest imaged time point, 2.5 min (Fig. 1C). Importantly, this pattern was observed in all cells at this time point, indicating that acute removal of S1 leads to simultaneous changes in AgDD distribution across an entire population of cells. Over the next 60 min, the puncta continued to increase in size and decrease in number, as reported previously (9), suggesting that individual AgDD IBs grow by coalescence of puncta. Coincident with this, diffuse AgDD fluorescence declined and was largely undetectable at later time points, suggesting that newly synthesized AgDD molecules, in the absence of S1, efficiently add onto pre-existing IBs. This transition of AgDD fluorescence from a diffuse to punctate appearance was accompanied by a rapid decline in the Triton X-100 solubility of AgDD, as assessed by sedimentation analysis (Fig. 1, C and B). The half-time for this solubility transition was ∼5.2 min, with 63% insoluble after 10 min of S1 w/o. These data indicate that AgDD rapidly, reproducibly, and synchronously transitions into detergent-insoluble aggregates upon acute S1 w/o. This approach is, therefore, well-suited to conduct a systems-level interrogation of temporally resolved PN responses to acute unfolding and aggregation of a single, defined cytosolic protein.
To assess the immediate (i.e. ≤10 min) effects of acute AgDD unfolding on the PN that occur prior to transcriptional responses, we used an unbiased quantitative K-GG proteomic strategy to comprehensively assess the impact of S1 w/o on the cellular ubiquitylome (Fig. 2A). Because the UPS tags conformationally altered proteins with Ub chains that direct them for degradation by the 26S proteasome (14), changes to the stability or ubiquitylation status of cellular proteins constitute a sensitive measure of proteome stress (15). K-GG proteomics exploits an antibody against the diglycine signature remnant on peptides generated from ubiquitylated proteins after complete digestion with trypsin (due to cleavage of the C-terminal Arg-Gly-Gly sequence of ubiquitin) (16, 17). Thus, it allows global specific profiling of ubiquitin post-translational modification on proteins present in a cell population at a given time or under a given stress. By combining K-GG immunocapture with tandem mass tagging (TMT) technology (a combination of highly multiplexed isobaric tags), it is possible to obtain precise time-resolved assessments of changes to the ubiquitylome in a single experiment. Lysates from AgDD cells subjected to S1 w/o for 0, 2.5, 5, 7.5, and 10 min (Fig. 2A) were digested with trypsin, and K-GG immunoprecipitates from each time point were isolated, labeled with 11-plex isobaric tags, pooled, fractionated, and analyzed by nLC-MS/MS as described previously (18). In total, we identified and quantified 4,549 common unique sites of ubiquitylation across all 11 experimental conditions in two independent experiments (Table S1). Parallel analyses of control samples from WT HEK293 cells not expressing AgDD cultured in the presence or absence of S1 or subjected to S1 w/o were performed to ensure that in the absence of AgDD, S1 and S1 w/o have no significant biological impact on protein ubiquitylation (Fig. S1 (B and C) and Table S2).
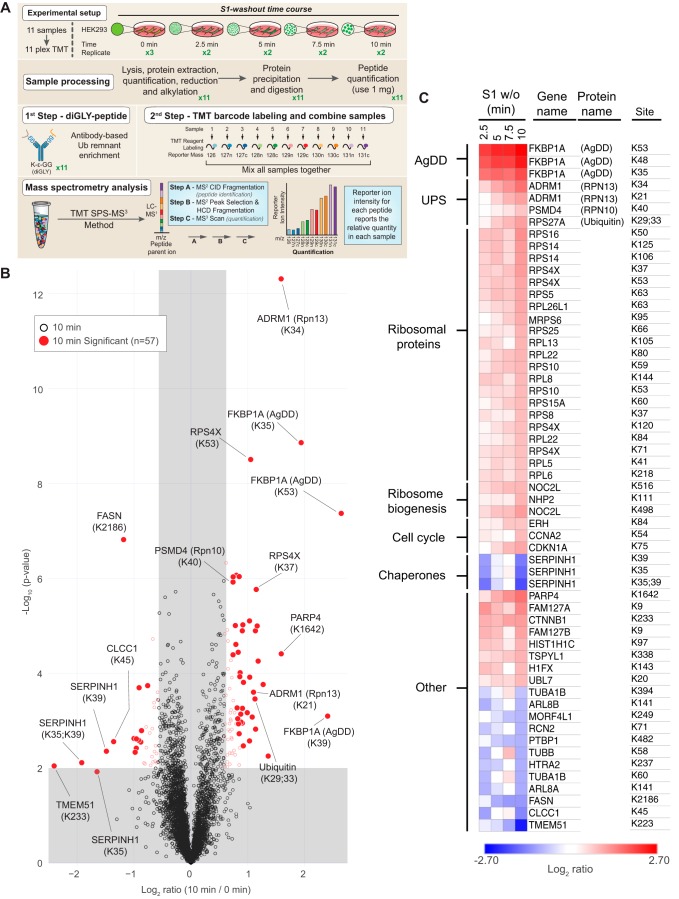
Figure 2.
S1 w/o in AgDD cells rapidly alters the ubiquitylation of PN components. A, experimental design and workflow: TMT labeling of S1 w/o time course sample and replicate. B, volcano plot (−log10 p value versus log2 ratio to untreated cells) of K-GG sites at 10 min following S1 w/o. Proteins with log2 ratios <−0.585 or >0.585 (p < 0.01) are indicated as colored empty circles, and filled colored circles indicate statistically significant hits (Welch's t test (S0 = 0.5), corrected for multiple comparison by permutation-based FDR (1%)). C, heat map of log2 scaled changes in K-GG sites at all time points for sites determined to be statistically altered at 10 min. Gene names are listed with select protein names in parentheses. Ubiquitylated AgDD lysines are numbered according to their position in native FKBP1A. The numbering of these sites in AgDD is shifted relative to FKBP1A by the N-terminal decapeptide, becoming Lys-45, Lys-58, and Lys-63.
Unsurprisingly, in AgDD cells, the earliest and most substantial changes in ubiquitylation (3.8–5.2-fold increase after 10-min S1 w/o) were detected on AgDD itself, with three sites (Lys-35, Lys-48, and Lys-53) significantly modified (Fig. 2B; note that ubiquitylated AgDD lysines are numbered according to their position in native FKBP1A). This result is consistent with previous publications showing that the DD domain undergoes partial unfolding and becomes rapidly ubiquitylated following S1 w/o (11).
In addition to ubiquitylation of AgDD itself, significant changes to the endogenous ubiquitylome were also observed following the acute 10-min S1 w/o, with some sites exhibiting increased or decreased levels of ubiquitylation and many sites trending monotonically over the time course (Fig. 2C and Fig. S1A). At 10 min, we identified 57 ubiquitylation sites corresponding to 48 proteins exhibiting statistically significant changes, including 42 sites with increased, and 15 sites with decreased modification (Fig. 2, B and C). The sites exhibiting significantly decreased ubiquitylation correspond to a group of cytosolic and organellar proteins with no obvious functional relationship to one another or to known stress-response pathways. By contrast, the 42 sites exhibiting statistically significant increased ubiquitylation in response to S1 w/o are strikingly enriched in functionally related PN proteins, including chaperones, proteasome subunits, ribosomal proteins, and ribosome biogenesis factors, suggesting that acute unfolding and aggregation of a single abundant protein leads to rapid (≤10-min) Ub modification of both protein degradation and protein synthesis machinery. In addition to proteins with documented roles in the PN, which will be discussed below, our analysis identified a significant increase in PARP4 ubiquitylation that increased monotonically across all time points following S1 w/o. PARP4 is a cytosolic poly-ADP ribosylase and one of three protein components of mammalian vault complexes. Purified vaults demonstrate ADP-ribosylase activity, confirming that PARP4 is enzymatically active in vaults (19). Whereas many PARP proteins are active in stress-response pathways (20), the relationship of PARP4 and potentially vault-associated PARP4 to the PN remains unknown.
The most striking early changes to the ubiquitylome in response to S1 w/o were increased Ub modification of components of the UPS itself. We observed a significant monotonic increase in the modification of Ub at Lys-29 and Lys-33, suggesting that increased production or decreased turnover of these noncanonical Ub chains is an early and progressive response to acute unfolding and aggregation of AgDD. K-GG proteomic profiling also revealed that S1 w/o induces immediate 2–3-fold increases in Ub modification of the proteasome itself, notably on the Ub receptor, RPN13 (ADRM1), at two sites (Lys-21 and Lys-34) that were second in magnitude only to modification of AgDD itself. A single site in RPN10 (PSMD4, Lys-40) also exhibited a modest (1.5-fold) increase in ubiquitylation, reaching statistical significance only at 10 min following S1 w/o. These data reveal that acute unfolding and/or aggregation of a single highly expressed cytosolic protein leads to immediate and progressive increases in ubiquitylation of key nodes in the PN network, particularly the ribosome and the 26S proteasome.
AgDD unfolding/aggregation do not impair global 26S proteasome activity
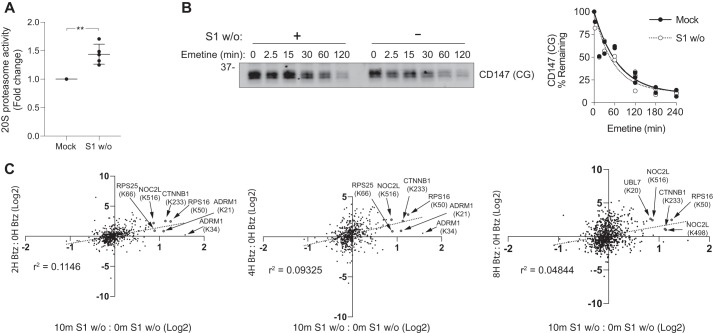
Acute unfolding and aggregation of AgDD could impair proteasome function by overwhelming substrate binding capacity or by “choking” the proteasome with hard-to-unfold aggregates (5, 21, 22). Moreover, ubiquitylation of 19S Ub receptors RPN13 and RPN10 could interfere with binding and degradation of UPS substrates, leading to their accumulation. Indeed, nearly stoichiometric modification of RPN13 through in vitro ubiquitylation of isolated proteasomes has been reported to impair substrate binding and degradation (23, 24), although it is unclear whether those effects are entirely due to modification of RPN13 as opposed to other proteasome subunits. To determine whether the observed increase in endogenous protein ubiquitylation induced by AgDD aggregation is the consequence of inhibited proteasome activity, we evaluated cleavage of a bioluminescent peptide reporter of chymotrypsin-like proteasome activity in cleared lysates from AgDD cells subjected to S1 w/o. Intriguingly, instead of a decrease, we observed a 40% increase in chymotrypsin-like activity relative to mock-treated controls after a 10-min S1 w/o (Fig. 3A), suggesting that unfolding and aggregation of AgDD stimulates, rather than inhibits, peptidase activity of the 20S proteasome. Despite this curious effect on 20S peptidase activity, S1 w/o impacted neither the steady-state level nor the degradation kinetics of core-glycosylated CD147 (CD147 (CG); Fig. 3B), an endogenous proteasome substrate whose degradation is strictly dependent on Ub conjugation and on 26S proteasome function (25). To assess whether acute AgDD unfolding causes global proteasome impairment, we compared the impact of acute S1 w/o on the K-GG ubiquitylome of AgDD cells with the published (17) effects of proteasome inhibition by bortezimib (Btz) in HCT116 cells. Overall, there was very poor (r2 < 0.11) correlation between the ubiquitylation sites altered at 10 min following S1 w/o and published Btz-responsive sites (Fig. 3C). Among the 57 significantly altered ubiquitylation sites identified in our K-GG proteomic analysis, only five—nuclear complex protein 2 homolog (NOCL2, Lys-516), 40s ribosomal protein S25 (RPS25, Lys-66), 40s ribosomal protein S16 (RPS16, Lys-50), catenin β-1 (CTNNB1, Lys-233), and RPN13 (ADRM1, Lys-21)—were also increased following direct proteasome inhibition. The poor correlation between the S1 w/o and Btz-induced changes to the K-GG ubiquitylome suggests that acute S1 w/o does not lead to globally impaired proteasome function. Together, these data indicate that the altered ubiquitylation status of proteins following S1 w/o might reflect activation of a cellular stress response and not a secondary consequence of proteasome impairment.
Figure 3.
AgDD unfolding and aggregation do not cause global inhibition of 26S proteasome activity. A, 20S chymotrypsin-like activity increases in cleared Nonidet P-40 lysates following 10-min S1 w/o. Error bars, S.D. **, p = 0.0051 (paired t test). B, S1 w/o has no detectable impact on degradation of core-glycosylated CD147 (CD147 (CG)). Lysates of AgDD cells treated with emetine for the indicated times were assessed by immunoblot analysis with an antibody to endogenous CD147 (left) and quantified (right). C, site-specific ubiquitylation patterns induced by 2-, 4-, and 8-h Btz treatment in HCT116 cells (17) as compared with 10-min S1 w/o in AgDD HEK293 cells. Sites significantly (0.2% FDR (17) and 1% FDR (this work)) altered by both S1 w/o and Btz treatment are colored red and labeled.
AgDD and RPN13 are ubiquitylated in response to S1 w/o
To directly assess the effects of S1 w/o on RPN13 ubiquitylation, we used a mixture of Halo-tagged Ub-association domain (UBA) reagents that bind Ub conjugates without preference for chain linkage topology (26, 27) to enrich ubiquitylated proteins from detergent lysates of AgDD cells (Fig. 4). GFP immunoblot analysis of Halo-UBA–captured proteins from mock-treated AgDD cells revealed a faint “ladder” of bands migrating more slowly than unmodified AgDD (Fig. 4A). S1 w/o caused a substantial increase in the intensity of this ladder and led to the appearance of a prominent high-molecular weight (HMW) GFP-immunoreactive smear. All of these species, both in mock-treated and S1 w/o cells, were eradicated by pretreatment of lysates with the deubiquitylase USP2-cc (28) (Fig. 4B), demonstrating that they correspond to multiubiquitylated and polyubiquitylated forms of AgDD. As the proteomic analysis revealed substantially increased abundance of the same three K-GG peptides following S1 w/o, rather than the appearance of additional K-GG peptides, it is likely that the HMW ubiquitylated species produced in response to S1 w/o are produced by extension of existing oligoubiquitin species on AgDD. These data demonstrate that AgDD, like its nonaggregating counterpart, DD (11, 13), is rapidly and extensively polyubiquitylated in response to removal of the synthetic stabilizing ligand S1.
Figure 4.
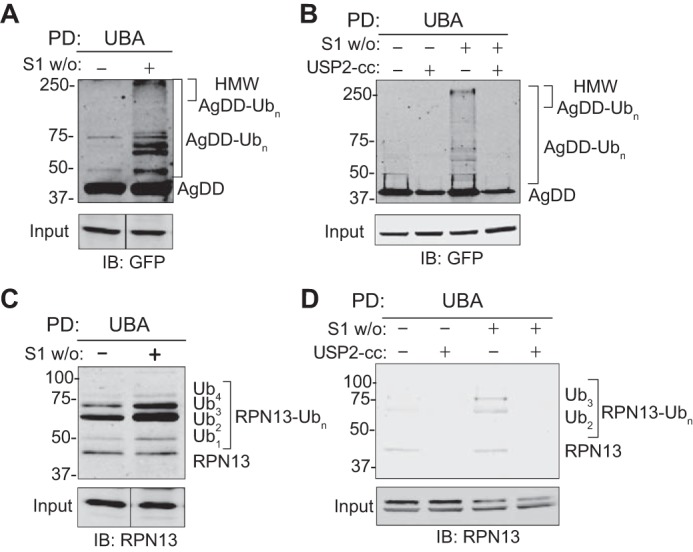
Multiubiquitylation of RPN13 in response to 10-min S1 w/o. A, AgDD-Ubn conjugates captured by UBA pulldown from mock– and S1 w/o–treated AgDD cells. B, pretreatment with the deubiquitinase domain USP2-cc pretreatment eliminates UBA capture of HMW AgDD bands. C, RPN13-Ubn conjugates captured by UBA pulldown from mock– and S1 w/o–treated AgDD cells. D, USP2-cc pretreatment eliminates UBA capture of HMW RPN13 bands. Immunoblots were probed with the indicated antibodies.
Immunoblot analysis of Halo-UBA–captured proteins with RPN13 antibody also identified a ladder of slower migrating species in AgDD cells that increased in intensity following S1 w/o (Fig. 4C) and were lost following pretreatment with USP2-cc (Fig. 4D). Unlike AgDD, we did not observe evidence of HMW polyubiquitin chains on RPN13 in response to S1 w/o.
Together with the K-GG proteomic analysis, we conclude that basal ubiquitylation of RPN13 is augmented by acute unfolding and/or aggregation of AgDD. In contrast to AgDD and RPN13 ubiquitylation, we were unable to detect a slower-migrating form of RPN10 by immunoblotting among affinity-captured ubiquitylated conjugates or associated with affinity-captured proteasomes, suggesting that the stoichiometry of this modification is below the threshold for immunodetection (Fig. S2A).
UBE3C mediates RPN13 multiubiquitylation and AgDD polyubiquitylation
RPN13 is a substoichiometric proteasome subunit that dynamically exchanges between 26S proteasomes and cytosol (29). To determine whether ubiquitylated forms of RPN13 are proteasome-associated, we used streptavidin affinity capture to isolate intact proteasomes under native conditions from AgDD cells stably expressing the 19S proteasome subunit RPN11 tagged with a C-terminal HTBH sequence (29, 30) (RPN11-HTBH). RPN13 immunoblots of streptavidin-captured proteasomes detected a ladder of RPN13 species, confirming that multiply ubiquitylated (Ub < 5) RPN13 is associated with 26S proteasomes (Fig. 5A). The increase in total RPN13-Ubn signal intensity captured by RPN11-HTBH pulldown also increased following S1 w/o (Fig. 5A, right-hand panels), consistent with our observations in cells not expressing RPN11-HTBH (Fig. 4, C and D).
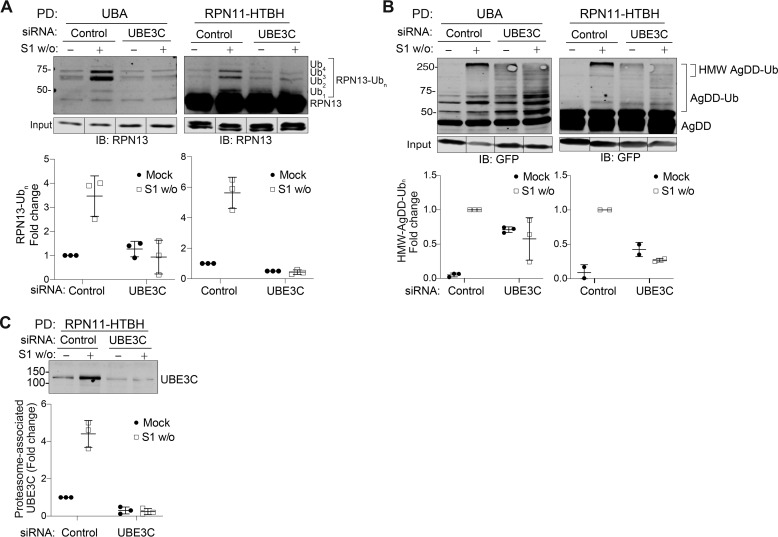
Figure 5.
UBE3C-catalyzed ubiquitylation of RPN13 and AgDD at the proteasome. A, RPN13-Ubn captured by UBA PD (left) or co-precipitated with affinity-captured proteasomes (right) increases ∼4-fold following 10-min S1 w/o. UBE3C knockdown blocks S1 w/o–stimulated increase in RPN13-Ubn. B, AgDD-Ubn captured by UBA PD (left) or co-precipitated with affinity-captured proteasomes (right). UBE3C knockdown prevents the formation of only HMW-AgDD-Ubn that associates with the proteasome. C, proteasome-associated UBE3C increases ∼4-fold following S1 w/o. Error bars, S.D.
Previous studies have suggested a role for the HECT-domain Ub ligase UBE3C in ubiquitylating proteasome-bound RPN13 in response to a variety of acute stresses (24). Like RPN13, UBE3C is a nonstoichiometric 26S proteasome subunit that partitions between cytosolic and proteasome-bound pools (31). The yeast UBE3C ortholog, Hul5, was previously shown to play a central role in ubiquitin-dependent degradation of low-solubility cytosolic proteins (32) and to increase the processivity of stalled proteasomes (33). To assess the role of UBE3C in ubiquitylating RPN13 in response to S1 w/o, we used siRNA to deplete UBE3C from AgDD cells expressing RPN11-HTBH and assessed both total (Halo-UBA) and proteasome-associated (RPN11-HTBH PD) RPN13 by immunoblot analysis of the affinity-captured eluates. UBE3C-targeted siRNAs reduced UBE3C expression by ∼95% in Triton X-100 lysates and reduced the amount of UBE3C captured by RPN11-HTBH PDs by ∼75% (Fig. 5C). UBE3C knockdown blocked S1 w/o-induced increases of total and proteasome-associated RPN13-Ubn conjugates but did not reduce basal RPN13 ubiquitylation (Fig. 5A). These data suggest that S1 w/o activates UBE3C-dependent RPN13 multiubiquitylation as reported previously for pleiotropic stresses, such as heat shock and pharmacological proteasome inhibition (23, 24).
The effect of UBE3C knockdown on AgDD ubiquitylation differed markedly from that observed for RPN13 (Fig. 5B). As with RPN13, AgDD-Ubn conjugates were co-enriched with RPN11-HTBH-captured proteasomes. Whereas nonubiquitylated AgDD binds nonspecifically to streptavidin beads, Ubn-AgDD capture by RPN11-HTBH depends upon association with 26S proteasomes (Fig. S4A). However, unlike with RPN13, UBE3C knockdown resulted in increased levels of AgDD multiubiquitylation in the presence of S1, suggesting that UBE3C is dispensable for basal ubiquitylation of folded AgDD (Fig. 5B). Importantly, UBE3C deletion prevented the enhanced multiubiquitylation and HMW polyubiquitylation in response to S1 w/o (Fig. 5B). These data suggest that UBE3C can extend oligoubiquitin chains on AgDD, perhaps by virtue of the E4 activity reported for Hul5 (34).
UBE3C is recruited to the 26S proteasome in response to AgDD unfolding
Increased RPN13 ubiquitylation in response to S1 w/o could result from either globally increased UBE3C activity or from recruitment of UBE3C to the proteasome from a cytosolic pool. To distinguish between these possibilities, we used RPN11-HTBH capture to assess the impact of S1 w/o on UBE3C proteasome association (Fig. 5C). The amount of UBE3C that coprecipitated with RPN11-HTBH increased ∼4-fold following S1 w/o, closely matching the magnitude of increase in total RPN13 ubiquitylation (Ub1–4) under the same conditions (Fig. 5C). At the same time, S1 w/o increased the fraction of proteasome-associated UBE3C from ∼25 to 60% (Fig. S3, A and B). Thus, S1 w/o in AgDD cells stimulates UBE3C recruitment to the proteasome from the cytosol, leading to increased RPN13 ubiquitylation and AgDD polyubiquitylation.
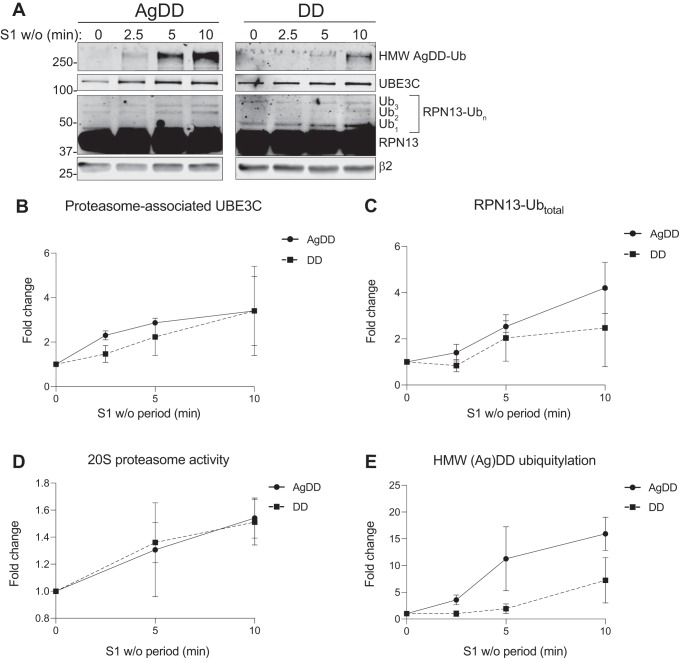
In principle, UBE3C recruitment and consequent ubiquitylation events in response to S1 w/o could be due to either unfolding or aggregation of AgDD. Because AgDD aggregation occurs rapidly upon S1 w/o, it is not possible to kinetically determine whether these events are the consequence of the sudden appearance of a large bolus of a partially unfolded, aggregation-prone protein or its subsequent aggregation. To distinguish between these possibilities, we assessed the effects on the proteasome of S1 w/o in HEK293 cells stably expressing DD, a variant of FKBP1A that differs from AgDD only by the absence of an N-terminal decapeptide (Fig. 1A). In the absence of S1, DD adopts a partially folded “molten globule”–like state (11) that is efficiently degraded by the UPS (10) without generating fluorescent puncta, detergent-insoluble aggregates, or IBs (10, 11, 13). Remarkably, we found that S1 w/o from DD cells recapitulated all of the proteasome-associated effects that we observed in AgDD cells, including UBE3C recruitment (Fig. 6, A and B), RPN13 ubiquitylation (Fig. 6, A and C), enhanced 20S peptidase activity (Fig. 6D), and formation of HMW Ub conjugates (Fig. 6, A and E), although these events proceed with slower kinetics in DD compared with AgDD cells. Together, these data suggest that the sudden generation of partially unfolded AgDD and DD proteins is sufficient to drive UBE3C recruitment and activity increases at the proteasome.
Figure 6.
AgDD unfolding but not aggregation drives PN changes. A and B, S1 w/o were performed in AgDD-RPN11-HTBH (A) and DD-RPN11-HTBH (B) cells for the indicated periods. Cells were harvested, and proteasome PDs were performed to assess the change in UBE3C recruitment, RPN13 ubiquitylation, and the increase of AgDD ubiquitylation at the proteasome (normalized to β2 intensity). C–E, quantification of S1 w/o–stimulated UBE3C recruitment to the proteasome (C), RPN13 ubiquitylation (D), and AgDD and DD HMW ubiquitylation (E) relative to β2 density over time. F, 20-s proteasome activity increases in response to S1 w/o in DD and AgDD cells stably expressing RPN11-HTBH. All time points reflect an average of three independent replicates. Error bars, S.D.
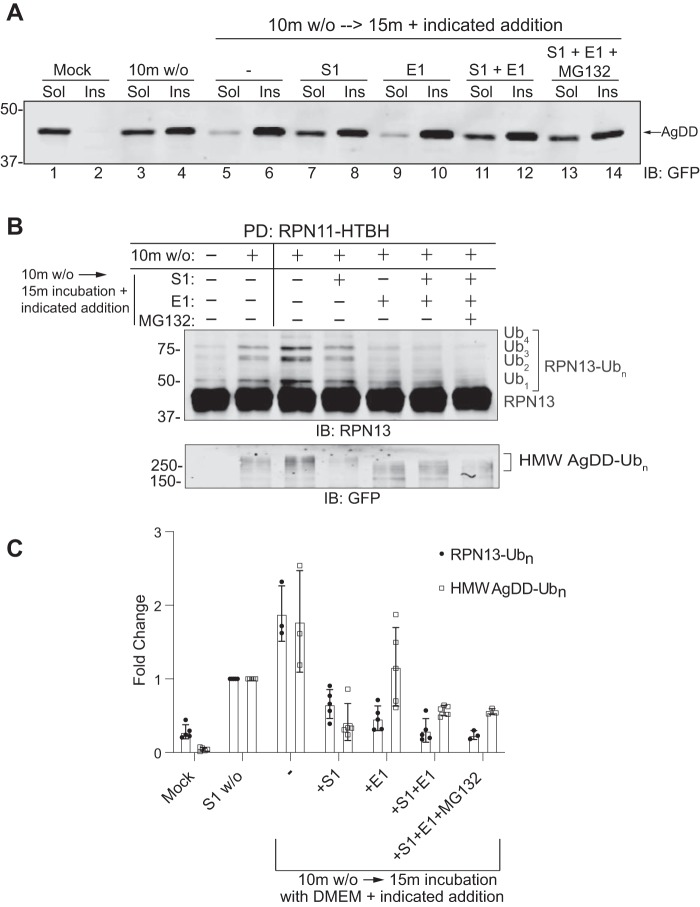
Because these effects appear to occur more rapidly in AgDD compared with DD cells, it is possible that detergent-insoluble AgDD microaggregates could contribute to these responses. Alternatively, it is possible that the enhanced kinetics in AgDD cells simply reflect AgDD's aggregation propensity or the more efficient degradation of ubiquitylated DD. To distinguish among these possibilities, we used an S1 addback approach (9) to arrest AgDD unfolding by exchanging cells that had been subjected to 10-min S1 w/o into a fresh medium “chase” containing S1 and incubating for an additional 15 min (Fig. 7). Without S1 re-addition, this additional 15-min chase substantially increased the fraction of detergent-insoluble AgDD (Fig. 7A; compare lanes 5 and 6 with lanes 3 and 4). In contrast, the fraction of insoluble AgDD in cells that had been chased in the presence of S1 was comparable with the fraction in cells subjected to 10-min S1 w/o alone (Fig. 7A; compare lanes 7 and 8 with lanes 3 and 4). Blocking ubiquitylation with an inhibitor of the Ub-activating enzyme E1 did not influence either the kinetics of aggregation or the effect of S1 re-addition (Fig. 7A, lanes 9–12); nor did inclusion of a proteasome inhibitor (Fig. 7A, lanes 13 and 14). These data confirm the previous report (9) that S1 re-addition prevents new aggregate formation but does not cause dissolution of already-formed aggregates. S1 re-addition also prevented the formation of additional proteasome-associated RPN13-Ubn and HMW AgDD-Ubn (Fig. 7, B and C), suggesting that these events are responses to the presence of partially unfolded AgDD and not to the presence of AgDD aggregates (7, B and C). By contrast, inhibiting E1 during the chase led to a decrease in multiubiquitylated RPN13 and to an increase in the mobility, but not the abundance, of polyubiquitylated HMW AgDD. These observations suggest that proteasome-associated RPN13 and AgDD are subject to rapid de-ubiquitylation, a conclusion that is supported by the lack of effect of MG132. Taken together, these data suggest that ubiquitylation of RPN13 and UBE3C recruitment to the proteasome are the consequence of acute and rapid unfolding of a single abundant cytosolic protein and not a response to aggregation.
Figure 7.
Partially unfolded but not aggregated AgDD induces UBE3C-dependent ubiquitylation at the proteasome. S1 w/o was performed on AgDD-RPN11-HTBH cells, after which medium was aspirated and replaced with new medium (DMEM) in combination with S1, an E1 inhibitor (TAK-243) or MG132 for 15 min. A, AgDD content in both soluble (Sol) and insoluble (Ins) fractions under the indicated conditions. B, RPN13 ubiquitylation (top) and HMW AgDD ubiquitylation (bottom) co-precipitated with affinity-captured proteasomes from the indicated treatment conditions. C, quantification of signal intensities in B (n = 3). Error bars, S.D.
Discussion
Much of our understanding of cellular protein homeostasis networks comes from studies in which cells are subjected to insults like heat shock (35), Hsp90 inhibitors (36), or chemical denaturants or oxidants (37, 38) that cause immediate disruption to global protein conformation. Although such approaches have been instrumental in revealing the existence of homeostatic networks that promote stereotypical, temporally orchestrated restoration of homeostasis, animal cells are rarely, if ever, subjected to such acute, massive, proteome-wide insults. Instead, most conformational diseases arise from the production of a single, conformationally impaired, aggregation-prone protein, such as the mutant proteins linked to genetic diseases (39). Although cell culture models expressing such mutant proteins have been valuable tools in examining the impact on the PN of producing a single conformationally defective protein, the slow and asynchronous nature of protein expression and aggregation has limited the utility of these models to investigate the temporal orchestration of the PN responses. Consequently, there is a substantial gap in our understanding of the relationship between acute global insults like heat shock and the more physiologically relevant but slow PN responses to the presence of a single conformationally impaired protein. In this study, we have combined a systems-level analysis of PN perturbation with a chemical biological approach to define the earliest cellular responses to acute unfolding and aggregation of a single, defined protein. Our data reveal that cells respond to acute misfolding of a single protein with broad changes to protein synthesis and protein degradation machinery on a time scale of minutes, well in advance of known transcriptional responses.
The most striking response of the PN to the induced unfolding and aggregation of AgDD was the ubiquitylation of two proteasomal Ub receptors, RPN10 and RPN13. Both proteins are located near the apex of the 19S regulatory particle (RP) and are proposed to facilitate cooperative binding of polyubiquitylated conjugates in a manner that optimally orients them for downstream processing and degradation (40–43). Although the identification of RPN10 is intriguing, because of the borderline statistical significance of this identification in our K-GG analysis and our inability to biochemically confirm this modification, we focused our attention on ubiquitylation of RPN13, a protein that binds to the 19S regulatory particle via an interaction between its N-terminal domain and RPN2 (44). RPN13 also binds and activates the deubiquitylase UCH37 via its C-terminal domain (45–47). The two K-GG–modified lysines on RPN13 map to its Ub-binding surface (48). Whereas ubiquitylation of both RPN10 and RPN13 has been previously reported to occur in response to a number of stressors, including heat shock, acute oxidation, and proteasome inhibition (23, 24), our data indicate that the 19S proteasome is a major target of early ubiquitylation events stimulated by protein unfolding.
Our study establishes a key role for UBE3C in modifying both RPN13 and AgDD. It is noteworthy that yeast RPN10 is monoubiquitylated by Hul5 (49). UBE3C and Hul5 possess E4 activities that can extend Ub chains on substrates bound to 26S proteasomes, perhaps antagonistically with proteasome-bound deubiquitylases (34); this antagonism is proposed to facilitate degradation of difficult-to-degrade proteins (13). UBE3C was previously identified in an RNAi screen for DD degradation (13). However, the modest stabilization of DD observed upon UBE3C depletion suggests that it does not act alone and, together with the findings presented here, suggests a model wherein partially unfolded clients like DD and AgDD are first multiubiquitylated in the cytosol by a yet unidentified E3 and subsequently polyubiquitylated by the E4 activity of 26S-associated UBE3C.
UBE3C can extend Lys-48–linked Ub chains on substrates with Lys-29 linkages (50, 51). In this regard, it is noteworthy that the K-GG analysis identified a significant increase in doubly modified Lys-29, Lys-33 Ub–Ub linkages in response to S1 w/o. UBE3C-dependent RPN13 ubiquitylation is one of the most robust and immediate changes to global ubiquitylation following S1 w/o in both AgDD and DD cells (Fig. 2, B and C), occurring simultaneously with proteasomal recruitment of UBE3C (Fig. 6B). These rapid kinetics pose intriguing questions about the mechanism by which UBE3C “senses” protein unfolding and the means by which it rapidly redistributes to the proteasome to aid in degradation.
Our finding that the changes to the proteasome in response to S1 w/o in AgDD cells are fully recapitulated in cells expressing aggregation-resistant DD strongly suggests that protein unfolding is sufficient to induce a proteasome stress response. This conclusion is supported by the observation that S1 re-addition following 10-min S1 w/o results in rapid deubiquitylation of both RPN13 and proteasome-associated AgDD, despite the persistence of detergent-insoluble AgDD aggregates. Moreover, these data suggest that the ubiquitylation states of both RPN13 and AgDD are dynamic, maintained by antagonism between 26S-associated Ub conjugation and deubiquitylation activities. Whereas these observations are consistent with previous data suggesting antagonism between Hul5 and the proteasome-associated deubiquitylase, Ubp6 (34), additional studies will be needed to identify the deubiquitylases that maintain this equilibrium for proteins like AgDD, which are refractory to degradation.
Experimental procedures
Cell culture and S1 w/o
HEK293 cells stably expressing AgDD-sfGFP or DD-sfGFP were cultured in DMEM supplemented with 10% heat-inactivated FBS and 1 μm S1. S1 w/o was performed by adding purified 500 μm recombinant FKBP1A F36V protein (rDD, diluted in PBS + 4% glycerol) (9) to the media of cultured DD or AgDD cells to create a final concentration of 5 μm. Mock washouts were performed by adding an equal volume of PBS + 4% glycerol to cell culture media.
UBA affinity capture
Purified Halo-TUBEDsk2 and Halo-TUBEubiquilin-1 (52) were separately conjugated to Magne® HaloTag® beads (Promega) by rotating at room temperature. Following mock or S1 w/o, cells were washed in ice-cold PBS, harvested by scraping, and stored at −80 °C. For UBA pulldowns, cells were lysed in UBA lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, 50 mm chloroacetamide, 2 mm NEM, 2.5 mm EDTA, 1 mm PMSF, cOmpleteTM EDTA-free protease inhibitor mixture (Roche Applied Science)). 45 μl of both 10% bead-TUBE slurries was added to 500 μg of cleared AgDD cell lysates and rotated overnight at 4 °C. Beads were then washed four times with 1 ml of wash buffer (50 mm Tris, pH 7.5, 500 mm NaCl, 1% Nonidet P-40, 50 mm chloroacetamide, 2 mm NEM, 2.5 mm EDTA, 1 mm PMSF, cOmpleteTM EDTA-free protease inhibitor mixture (Roche Applied Science)). Protein was eluted from these beads with two sequential 10-min incubations at 95 °C with 25 μl of 2× protein sample buffer, resolved on SDS-PAGE gels, and analyzed by Western blotting using the indicated antibodies.
Proteasome affinity capture
Cells were lysed in proteasome PD lysis buffer (50 mm sodium phosphate, pH 7.5, 100 mm NaCl, 1% Nonidet P-40, 10% glycerol, 5 mm ATP, 5 mm MgCl2, cOmpleteTM EDTA-free protease inhibitor mixture (Roche Applied Science)). Lysates were cleared by centrifugation at 13,000 × g for 15 min at 4 °C. Cleared lysates were then combined with magnetic streptavidin resin (10 μg of total protein/1 μl of resin) (Pierce) and rotated for 30 min at 4 °C. Resin was washed twice in lysis buffer (vortexing to suspend resin in each wash). Protein was eluted from resin with two sequential incubations with 25 μl of 2× protein sample buffer at 95 °C for 10 min, resolved on SDS-polyacrylamide gels, and analyzed by Western blotting using the indicated antibodies.
20S proteasome activity assays
Cleared lysates from AgDD cells were normalized by total protein. Samples of cleared lysates were combined in a 1:1 ratio with reconstituted Proteasome GloTM (Promega) solution, and bioluminescence was detected in a BioTekTM SynergyTM NEO HTS multimode microplate reader.
Solubility shift assays
S1 w/o was performed in AgDD cells for the indicated time periods. Cells were harvested by aspirating media and immediately washing cells with ice-cold PBS. Cells were held on ice while PBS was aspirated and cells were collected by scraping. Cells were lysed in ice-cold buffer containing 50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, 2 μg/ml aprotinin, 1 μg/ml pepstatin, 10 mm NEM. The lysate was rotated at 4 °C for 15 min and cleared by centrifugation at 20,000 × g for 30 min at 4 °C. The resulting pellet was washed two times with lysis buffer and repelleted after each wash at 20,000 × g. After all supernatant was removed, the pellet was solubilized in buffer containing 50 mm Tris, 1% SDS, 150 mm NaCl, 2 μg/ml aprotinin, 1 μg/ml pepstatin, 10 mm NEM.
S1 w/o pulse and S1 addback chase
S1 w/o was performed on AgDD cells stably expressing Rpn11-HTBH. At 10 min following S1 w/o, medium was aspirated and replaced with DMEM alone or DMEM containing either 2 μm S1, 10 μm E1 inhibitor (TAK-243, MedChem Express), both together, or both together in combination with 10 μm MG132. All replacement medium was warmed to 37 °C prior to the exchange. Following medium replacement, cells were incubated at 37 °C for an additional 15 min and then harvested and processed according to the proteasome affinity capture protocol (see above).
Live-cell imaging
Live-cell imaging was performed on AgDD cells, plated in DMEM with 10% heat-inactivated FBS, and incubated at 37 °C and 5% CO2 in a glass-bottom 8-well chambered coverglass (Nunc). For time-lapse imaging experiments, S1 w/o was performed immediately prior to the start of imaging. Cells were imaged by time-lapse fluorescence video microscopy using an inverted microscope (Zeiss Axiovert 200M) encased in a Perspex chamber heated to 37 °C. The chambered coverglass was placed in a secondary chamber perfused with humidified 5% CO2. A motorized stage allowed imaging of multiple fields. Images were acquired with a cooled charge-coupled device (Cool-SNAP HQ) and UV light source (X-Cite; Lumen Dynamics), filters for visualization of GFP, and a ×20 air objective. MetaMorph software was used to control image acquisition, and ImageJ (version 1.6, National Institutes of Health) was used for image analysis.
Emetine chase
AgDD cells were left untreated (0-h time point) or treated with final concentrations of either 5 μm rDD (S1 w/o) or vehicle and 1 μm emetine for the indicated times before collection in cold PBS. Cells were pelleted with a tap spin and lysed in 1% Triton X-100, 50 mm Tris, pH 7.5, 150 mm NaCl, 1 μm pepstatin A, and 1 mm PMSF, and lysates were cleared by centrifugation at 20,000 × g. Protein concentration was measured using a BCA Protein Assay Kit (Thermo Fisher Scientific), and concentrations were normalized prior to SDS-PAGE and immunoblotting. Band intensities were quantified using Image Studio Lite software (LI-COR Biosciences). The protein remaining was calculated as a percentage of untreated, and one-phase exponential decay curves were fit using Prism version 7 (GraphPad Software).
Immunoblotting
Protein samples were resolved on 12% acrylamide SDS-polyacrylamide gels and transferred to nitrocellulose membranes using a Bio-Rad Trans-Blot Turbo transfer system. Membranes were blocked in PBS + 5% BSA and then incubated with primary antibodies for varying time periods. Proteins were stained with the following primary antibodies: mouse anti-RPN10 (Santa Cruz Biotechnology, Inc., sc-514990 (1:400)), rabbit anti-UBE3C (Bethyl Laboratories, A304-122A-A (1:1000)), mouse anti-β2 (Santa Cruz Biotechnology, sc-58410 (1:400)), rabbit anti-RPN13 (Cell Signaling Technology, 12019S (1:1000)), and mouse anti-GFP (Clontech, 632381 (1:1000)). All blots were developed using proprietary LI-COR secondary antibodies and imaged on a LI-COR Odyssey® CLx scanner. Band intensities were quantified using Image Studio Lite software (LI-COR Biosciences).
K-GG proteomics
Protein isolation
S1 w/o was performed in AgDD cells for the indicated time period. At the indicated times, cells were washed twice with ice-cold PBS, collected, and snap-frozen. Cell pellets were lysed in lysis buffer (50 mm EPPS, pH 8.0, 8 m urea, 0.25% (v/v) Triton X-100, 1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 10 μm PR-619, 0.5 mm tris(2-carboxyethyl)phosphine, 50 mm chloroacetamide), to produce whole-cell extracts. Whole-cell extracts were sonicated and clarified by centrifugation (16,000 × g for 10 min at 4 °C) followed by filtration through a 0.45-μm filter, and protein concentrations were determined by the Bradford assay.
General sample preparation
Protein extracts (1.5 mg) were subjected to disulfide bond reduction with 5 mm tris(2-carboxyethyl)phosphine (room temperature, 10 min) and alkylation with 25 mm chloroacetamide (room temperature, 20 min). Methanol-chloroform precipitation was performed prior to protease digestion. In brief, four parts undiluted methanol were added to each sample and vortexed, one part chloroform was then added to the sample and vortexed, and finally three parts water were added to the sample and vortexed. The sample was centrifuged at 6,000 rpm for 2 min at room temperature and subsequently washed twice with 100% methanol. Samples were resuspended in 100 mm EPPS, pH 8.5, containing 6 m urea and digested at 37 °C for 2 h with LysC protease at a 200:1 protein/protease ratio. Samples were then diluted with 100 mm EPPS, pH 8.5, to a final urea concentration of 0.75 m. Trypsin was then added at a 100:1 protein-to-protease ratio, and the reaction was incubated for a further 6 h at 37 °C. Samples were acidified with 1% formic acid for 15 min and subjected to C18 solid-phase extraction (Sep-Pak, Waters). The Pierce quantitative colorimetric peptide assay (catalog no. 23275) was used to quantify the digest and to accurately aliquot 1 mg of peptides for diGLY enrichment.
Immunoprecipitation of diGLY-containing peptides
diGLY capture was performed largely as described (18). The diGly mAb (Cell Signaling Technology; D4A7 clone) (32 μg of antibody/1 mg of peptide) was coupled to Protein A Plus Ultralink resin (1 μl slurry to 1 μg of antibody) (Thermo Fisher Scientific) overnight at 4 °C prior to its chemical cross-linking reaction. Dried peptides (1 mg) were resuspended in 1.5 ml of ice-cold IAP buffer (50 mm MOPS (pH 7.2), 10 mm sodium phosphate, and 50 mm NaCl) and centrifuged at maximum speed for 5 min at 4 °C to remove any insoluble material. Supernatants (pH ∼7.2) were incubated with the antibody beads for 2 h at 4 °C with gentle end-over-end rotation. After centrifugation at 215 × g for 2 min, beads were washed three more times with ice-cold IAP buffer and twice with ice-cold PBS. The diGLY peptides were eluted twice with 0.15% TFA, desalted using homemade StageTips, and dried via vacuum centrifugation, prior to TMT labeling.
diGLY proteomics analysis using TMT
TMT-labeled diGLY peptides were fractionated according to the manufacturer's instructions using a High pH reversed-phase peptide fractionation kit (Thermo Fisher Scientific) for a final six fractions and subjected to C18 StageTip desalting prior to MS analysis. Mass spectrometry data were collected using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled to a Proxeon EASY-nLC1200 liquid chromatography (LC) pump (Thermo Fisher Scientific). Peptides were separated on a 100-μm inner diameter microcapillary column packed in-house with ∼35 cm of Accucore150 resin (2.6 μm, 150 Å, Thermo Fisher Scientific) with a gradient consisting of 5–26% (0–155 min), 26–32% (155–170 min) (acetonitrile, 0.1% formic acid) over a total 180-min run at ∼600 nl/min. For analysis, we loaded one-half of each fraction onto the column. Each analysis used the Multi-Notch MS3-based TMT method (53). The scan sequence began with an MS1 spectrum (Orbitrap analysis; resolution 120,000 at 200 Th; mass range 400–1250 m/z; automatic gain control (AGC) target 1 × 106; maximum injection time 100 ms). Precursors for MS2 analysis were selected using a top 4 second method. MS2 analysis consisted of collision-induced dissociation (quadrupole Orbitrap analysis; AGC 1 × 105; isolation window 0.8 Th; normalized collision energy 35; maximum injection time 300 ms; resolution was 15,000 at 200 Th). Monoisotopic peak assignment was used, and previously interrogated precursors were excluded using a dynamic window (120 s ± 10 ppm). As described previously, only precursors with a charge state between 3 and 6 were selected for downstream analysis (18). Following acquisition of each MS2 spectrum, a synchronous precursor selection MS3 scan was collected on the top 10 most intense ions in the MS2 spectrum (53). MS3 precursors were fragmented by high-energy collision-induced dissociation and analyzed using the Orbitrap (normalized collision energy 65; AGC 2 × 105; maximum injection time 500 ms; resolution was 50,000 at 200 Th).
Data analysis
Mass spectra were processed using a Comet-based (2018.01 rev. 2) in-house software pipeline (54, 55). Spectra were converted to mzXML using a modified version of ReAdW.exe. Database searching included all entries from the Human Reference Proteome (2017-05) UniProt database, as well as an in-house curated list of contaminants. This database was concatenated with one composed of all protein sequences in the reverse order. Searches were performed using a 20-ppm precursor ion tolerance for total protein level analysis. The product ion tolerance was set to 0.03 Da. TMT tags on lysine residues and peptide N termini (+229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, whereas oxidation of methionine residues (+15.995 Da) and GlyGly modification (+114.0429 Da) were set as variable modification. Peptide-spectrum matches (PSMs) were adjusted to a 1% false discovery rate (FDR) (56). PSM filtering was performed using a linear discriminant analysis, as described previously (55), while considering the following parameters: Comet Log Expect, Diff Seq. Delta Log Expect, XCorr, missed cleavages, peptide length, charge state, and precursor mass accuracy. The presence of at least one GlyGly modification was required. For TMT-based reporter ion quantitation, we extracted the summed signal/noise ratio for each TMT channel and found the closest matching centroid to the expected mass of the TMT reporter ion (integration tolerance of 0.003 Da). Ubiquitylation site localization was determined using the AScore algorithm (57). AScore is a probability-based approach for high-throughput protein phosphorylation site localization. Specifically, a threshold of 13 corresponded to 95% confidence in site localization. Ubiquitylated peptides were quantified by summing reporter ion counts across all matching PSMs using in-house software, as described previously (55). PSMs with poor quality, MS3 spectra with isolation specificity <0.5 or with TMT reporter summed signal/noise ratios that were less than 110, or PSMs that had no MS3 spectra were excluded from quantification (53).
Peptide quantification values were exported for further analysis in Microsoft Excel and Perseus (58), and the statistical test and parameters used are indicated in the corresponding figure legends. Briefly, Welch's t test analysis was performed to compare two data sets, using the s0 parameter (in essence a minimal -fold change cut-off), and correction for multiple comparison was achieved by the permutation-based FDR method, both functions that are built-in in the Perseus software (58). Tables S1 and S2 list all quantified peptides used as well as the associated TMT reporter ratio to control channels used for quantitative analysis.
Author contributions
C. D. G., A. C. S. T., A. O., J. W. H., and R. R. K. conceptualization; C. D. G., A. C. S. T., and R. R. K. data curation; C. D. G., A. C. S. T., A. O., and R. R. K. formal analysis; C. D. G. and A. C. S. T. validation; C. D. G., A. C. S. T., and A. O. investigation; C. D. G., A. C. S. T., and A. O. methodology; C. D. G., A. C. S. T., and R. R. K. writing-original draft; C. D. G., A. C. S. T., A. O., J. W. H., and R. R. K. writing-review and editing; A. C. T. visualization; J. W. H. resources; J. W. H. software; J. W. H. and R. R. K. supervision; J. W. H. and R. R. K. funding acquisition; J. W. H. and R. R. K. project administration.
Supplementary Material
Acknowledgments
We thank Thomas J. Wandless, Ling-Chun Chen, and Yusuke Miyazaki for generously providing expertise, AgDD cells, plasmids, and S1, and we thank Lan Huang for generously providing RPN11-HTBH plasmids. We are grateful to Joshua Elias, Lichao Zhang, Ryan Leib, Chris Adams, and Eric Bennett for assistance with MS and to Khyati Shah for contributions in the early stages of this project and to members of the Kopito laboratory for discussions of the data and critical reading of the manuscript.
This work was supported by National Institutes of Health Grants 2R01NS042842 (to R. R. K.) and R37NS083524 and RO1AG011085 (to J. W. H.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains Tables S1 and S2 and Figs. S1–S4.
- PN
- proteostasis network
- IB
- inclusion body
- S1
- shield-1
- UPS
- ubiquitin proteasome system
- SfGFP
- superfolder GFP
- w/o
- washout
- Ub
- ubiquitin
- TMT
- tandem mass tagging
- K-GG
- global diglycine-capture
- Btz
- bortezimib
- UBA
- Ub-association domain
- HMW
- high-molecular weight
- DMEM
- Dulbecco's modified Eagle's medium
- PMSF
- phenylmethylsulfonyl fluoride
- NEM
- N-ethylmaleimide
- AGC
- automatic gain control
- PSM
- peptide-spectrum match
- FDR
- false discovery rate.
References
- 1. Balch W. E., Morimoto R. I., Dillin A., and Kelly J. W. (2008) Adapting proteostasis for disease intervention. Science 319, 916–919 10.1126/science.1141448 [DOI] [PubMed] [Google Scholar]
- 2. Klaips C. L., Jayaraj G. G., and Hartl F. U. (2018) Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 217, 51–63 10.1083/jcb.201709072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kopito R. R. (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530 10.1016/S0962-8924(00)01852-3 [DOI] [PubMed] [Google Scholar]
- 4. Ross C. A., and Poirier M. A. (2004) Protein aggregation and neurodegenerative disease. Nat. Med. 10, S10–S17 10.1038/nm1066 [DOI] [PubMed] [Google Scholar]
- 5. Hipp M. S., Patel C. N., Bersuker K., Riley B. E., Kaiser S. E., Shaler T. A., Brandeis M., and Kopito R. R. (2012) Indirect inhibition of 26S proteasome activity in a cellular model of Huntington's disease. J. Cell Biol. 196, 573–587 10.1083/jcb.201110093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meyer-Luehmann M., Spires-Jones T. L., Prada C., Garcia-Alloza M., de Calignon A., Rozkalne A., Koenigsknecht-Talboo J., Holtzman D. M., Bacskai B. J., and Hyman B. T. (2008) Rapid appearance and local toxicity of amyloid-β plaques in a mouse model of Alzheimer's disease. Nature 451, 720–724 10.1038/nature06616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hipp M. S., Park S.-H., and Hartl F. U. (2014) Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24, 506–514 10.1016/j.tcb.2014.05.003 [DOI] [PubMed] [Google Scholar]
- 8. Park S. H., Kukushkin Y., Gupta R., Chen T., Konagai A., Hipp M. S., Hayer-Hartl M., and Hartl F. U. (2013) PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154, 134–145 10.1016/j.cell.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 9. Miyazaki Y., Mizumoto K., Dey G., Kudo T., Perrino J., Chen L.-C. Meyer T., Wandless T. J. (2016) A method to rapidly create protein aggregates in living cells. Nat. Commun. 7, 11689–11689 10.1038/ncomms11689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Banaszynski L. A., Chen L. C., Maynard-Smith L. A., Ooi A. G. L., and Wandless T. J. (2006) A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126, 995–1004 10.1016/j.cell.2006.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Egeler E. L., Urner L. M., Rakhit R., Liu C. W., and Wandless T. J. (2011) Ligand-switchable substrates for a ubiquitin-proteasome system. J. Biol. Chem. 286, 31328–31336 10.1074/jbc.M111.264101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bersuker K., Brandeis M., and Kopito R. R. (2016) Protein misfolding specifies recruitment to cytoplasmic inclusion bodies. J. Cell Biol. 213, 229–241 10.1083/jcb.201511024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chu B. W., Kovary K. M., Guillaume J., Chen L. C., Teruel M. N., and Wandless T. J. (2013) The E3 ubiquitin ligase UBE3C enhances proteasome processivity by ubiquitinating partially proteolyzed substrates. J. Biol. Chem. 288, 34575–34587 10.1074/jbc.M113.499350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hershko A., and Ciechanover A. (1992) The ubiquitin system for protein degradation. Annu. Rev. Biochem. 61, 761–807 10.1146/annurev.bi.61.070192.003553 [DOI] [PubMed] [Google Scholar]
- 15. Goldberg A. L. (2003) Protein degradation and protection against misfolded or damaged proteins. Nature. 426, 895–899 10.1038/nature02263 [DOI] [PubMed] [Google Scholar]
- 16. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., and Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 10.1038/nbt849 [DOI] [PubMed] [Google Scholar]
- 17. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., and Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rose C. M., Isasa M., Ordureau A., Prado M. A., Beausoleil S. A., Jedrychowski M. P., Finley D. J., Harper J. W., and Gygi S. P. (2016) Highly multiplexed quantitative mass spectrometry analysis of ubiquitylomes. Cell Syst. 3, 395–403.e4 10.1016/j.cels.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kickhoefer V. A., Siva A. C., Kedersha N. L., Inman E. M., Ruland C., Streuli M., and Rome L. H. (1999) The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 146, 917–928 10.1083/jcb.146.5.917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gibson B. A., and Kraus W. L. (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 13, 411–424 10.1038/nrm3376 [DOI] [PubMed] [Google Scholar]
- 21. Kristiansen M., Deriziotis P., Dimcheff D. E., Jackson G. S., Ovaa H., Naumann H., Clarke A. R., van Leeuwen F. W. B., Menéndez-Benito V., Dantuma N. P., Portis J. L., Collinge J., and Tabrizi S. J. (2007) Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol. Cell 26, 175–188 10.1016/j.molcel.2007.04.001 [DOI] [PubMed] [Google Scholar]
- 22. Thibaudeau T. A., Anderson R. T., and Smith D. M. (2018) A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 9, 1097 10.1038/s41467-018-03509-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jacobson A. D., MacFadden A., Wu Z., Peng J., and Liu C. W. (2014) Autoregulation of the 26S proteasome by in situ ubiquitination. Mol. Biol. Cell 25, 1824–1835 10.1091/mbc.e13-10-0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Besche H. C., Sha Z., Kukushkin N. V., Peth A., Hock E. M., Kim W., Gygi S., Gutierrez J. A., Liao H., Dick L., and Goldberg A. L. (2014) Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 33, 1159–1176 10.1002/embj.201386906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tyler R. E., Pearce M. M., Shaler T. A., Olzmann J. A., Greenblatt E. J., and Kopito R. R. (2012) Unassembled CD147 is an endogenous ER-associated degradation (ERAD) substrate. Mol. Biol. Cell 23, 4668–4678 10.1091/mbc.e12-06-0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aillet F., Lopitz-Otsoa F., Hjerpe R., Torres-Ramos M., Lang V., and Rodríguez M. S. (2012) Isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. Methods Mol. Biol. 832, 173–183 10.1007/978-1-61779-474-2_12 [DOI] [PubMed] [Google Scholar]
- 27. Wilson M. D., Saponaro M., Leidl M. A., and Svejstrup J. Q. (2012) MultiDsk: a ubiquitin-specific affinity resin. PLoS One 7, e46398 10.1371/journal.pone.0046398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ryu K. Y., Baker R. T., and Kopito R. R. (2006) Ubiquitin-specific protease 2 as a tool for quantification of total ubiquitin levels in biological specimens. Anal. Biochem. 353, 153–155 10.1016/j.ab.2006.03.038 [DOI] [PubMed] [Google Scholar]
- 29. Wang X., and Huang L. (2008) Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol. Cell Proteomics 7, 46–57 10.1074/mcp.M700261-MCP200 [DOI] [PubMed] [Google Scholar]
- 30. Wang X., Cimermancic P., Yu C., Schweitzer A., Chopra N., Engel J. L., Greenberg C., Huszagh A. S., Beck F., Sakata E., Yang Y., Novitsky E. J., Leitner A., Nanni P., Kahraman A., et al. (2017) Molecular details underlying dynamic structures and regulation of the human 26S proteasome. Mol. Cell Proteomics 16, 840–854 10.1074/mcp.M116.065326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuo C.-L., and Goldberg A. L. (2017) Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c. Proc. Natl. Acad. Sci. U.S.A. 114, E3404–E3413 10.1073/pnas.1701734114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fang N. N., Ng A. H., Measday V., and Mayor T. (2011) Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat. Cell Biol. 13, 1344–1352 10.1038/ncb2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aviram S., and Kornitzer D. (2010) The ubiquitin ligase Hul5 promotes proteasomal processivity. Mol. Cell Biol. 30, 985–994 10.1128/MCB.00909-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Crosas B., Hanna J., Kirkpatrick D. S., Zhang D. P., Tone Y., Hathaway N. A., Buecker C., Leggett D. S., Schmidt M., King R. W., Gygi S. P., and Finley D. (2006) Ubiquitin Chains Are Remodeled at the Proteasome by Opposing Ubiquitin Ligase and Deubiquitinating Activities. Cell 127, 1401–1413 10.1016/j.cell.2006.09.051 [DOI] [PubMed] [Google Scholar]
- 35. Labbadia J., and Morimoto R. I. (2015) The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roe S. M., Prodromou C., O'Brien R., Ladbury J. E., Piper P. W., and Pearl L. H. (1999) Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J. Med. Chem. 42, 260–266 10.1021/jm980403y [DOI] [PubMed] [Google Scholar]
- 37. Grandjean J. M. D., Plate L., Morimoto R. I., Bollong M. J., Powers E. T., and Wiseman R. L. (2019) Deconvoluting stress-responsive proteostasis signaling pathways for pharmacologic activation using targeted RNA sequencing. ACS Chem. Biol. 14, 784–795 10.1021/acschembio.9b00134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Novoa I., Zhang Y., Zeng H., Jungreis R., Harding H. P., and Ron D. (2003) Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 22, 1180–1187 10.1093/emboj/cdg112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bertram L., and Tanzi R. E. (2005) The genetic epidemiology of neurodegenerative disease. J. Clin. Invest. 115, 1449–1457 10.1172/JCI24761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., Hofmann K., Walters K. J., Finley D., and Dikic I. (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488 10.1038/nature06926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peth A., Uchiki T., and Goldberg A. L. (2010) ATP-dependent steps in the binding of ubiquitin conjugates to the 26s proteasome that commit to degradation. Mol. Cell 40, 671–681 10.1016/j.molcel.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deveraux Q., Ustrell V., Pickart C., and Rechsteiner M. (1994) A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 269, 7059–7061 [PubMed] [Google Scholar]
- 43. Huang X., Luan B., Wu J., and Shi Y. (2016) An atomic structure of the human 26S proteasome. Nat. Struct. Mol. Biol. 23, 778–785 10.1038/nsmb.3273 [DOI] [PubMed] [Google Scholar]
- 44. Lu X., Nowicka U., Sridharan V., Liu F., Randles L., Hymel D., Dyba M., Tarasov S. G., Tarasova N. I., Zhao X. Z., Hamazaki J., Murata S., Burke T. R. Jr., and Walters K. J. (2017) Structure of the Rpn13-Rpn2 complex provides insights for Rpn13 and Uch37 as anticancer targets. Nat. Commun. 8, 15540–15540 10.1038/ncomms15540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hamazaki J., Iemura S., Natsume T., Yashiroda H., Tanaka K., and Murata S. (2006) A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 25, 4524–4536 10.1038/sj.emboj.7601338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Qiu X. B., Ouyang S. Y., Li C. J., Miao S., Wang L., and Goldberg A. L. (2006) hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 25, 5742–5753 10.1038/sj.emboj.7601450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yao T., Song L., Xu W., DeMartino G. N., Florens L., Swanson S. K., Washburn M. P., Conaway R. C., Conaway J. W., and Cohen R. E. (2006) Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol. 8, 994–1002 10.1038/ncb1460 [DOI] [PubMed] [Google Scholar]
- 48. Chen X., Lee B. H., Finley D., and Walters K. J. (2010) Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol. Cell 38, 404–415 10.1016/j.molcel.2010.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Isasa M., Katz E. J., Kim W., Yugo V., González S., Kirkpatrick D. S., Thomson T. M., Finley D., Gygi S. P., and Crosas B. (2010) Monoubiquitination of RPN10 regulates substrate recruitment to the proteasome. Mol. Cell 38, 733–745 10.1016/j.molcel.2010.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kristariyanto Y. A., Abdul Rehman S. A., Campbell D. G., Morrice N. A., Johnson C., Toth R., and Kulathu Y. (2015) K29-selective ubiquitin binding domain reveals structural basis of specificity and heterotypic nature of K29 polyubiquitin. Mol. Cell 58, 83–94 10.1016/j.molcel.2015.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Michel M. A., Elliott P. R., Swatek K. N., Simicek M., Pruneda J. N., Wagstaff J. L., Freund S. M. V., and Komander D. (2015) Assembly and specific recognition of K29- and K33-linked polyubiquitin. Mol. Cell 58, 95–109 10.1016/j.molcel.2015.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ordureau A., Paulo J. A., Zhang W., Ahfeldt T., Zhang J., Cohn E. F., Hou Z., Heo J. M., Rubin L. L., Sidhu S. S., Gygi S. P., and Harper J. W. (2018) Dynamics of PARKIN-dependent mitochondrial ubiquitylation in induced neurons and model systems revealed by digital snapshot proteomics. Mol. Cell 70, 211–227.e8 10.1016/j.molcel.2018.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McAlister G. C., Nusinow D. P., Jedrychowski M. P., Wühr M., Huttlin E. L., Erickson B. K., Rad R., Haas W., and Gygi S. P. (2014) MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 86, 7150–7158 10.1021/ac502040v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eng J. K., Jahan T. A., and Hoopmann M. R. (2013) Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 10.1002/pmic.201200439 [DOI] [PubMed] [Google Scholar]
- 55. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., and Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 10.1016/j.cell.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Elias J. E., and Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 10.1038/nmeth1019 [DOI] [PubMed] [Google Scholar]
- 57. Beausoleil S. A., Villén J., Gerber S. A., Rush J., and Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 10.1038/nbt1240 [DOI] [PubMed] [Google Scholar]
- 58. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., and Cox J. (2016) The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.