Acquired endocrine therapy resistance is a significant clinical problem for breast cancer patients. In recent years, increasing attention has been paid to long noncoding RNA (lncRNA) as a critical modulator for cancer progression. Based on RNA-sequencing data of breast invasive carcinomas in The Cancer Genome Atlas database, we identified thymopoietin antisense transcript 1 (TMPO-AS1) as a functional lncRNA that significantly correlates with proliferative biomarkers.
KEYWORDS: RNA stability, breast cancer, estrogen, long noncoding RNA
ABSTRACT
Acquired endocrine therapy resistance is a significant clinical problem for breast cancer patients. In recent years, increasing attention has been paid to long noncoding RNA (lncRNA) as a critical modulator for cancer progression. Based on RNA-sequencing data of breast invasive carcinomas in The Cancer Genome Atlas database, we identified thymopoietin antisense transcript 1 (TMPO-AS1) as a functional lncRNA that significantly correlates with proliferative biomarkers. TMPO-AS1 positivity analyzed by in situ hybridization significantly correlates with poor prognosis of breast cancer patients. TMPO-AS1 expression was upregulated in endocrine therapy-resistant MCF-7 cells compared with levels in parental cells and was estrogen inducible. Gain and loss of TMPO-AS1 experiments showed that TMPO-AS1 promotes the proliferation and viability of estrogen receptor (ER)-positive breast cancer cells in vitro and in vivo. Global expression analysis using a microarray demonstrated that TMPO-AS1 is closely associated with the estrogen signaling pathway. TMPO-AS1 could positively regulate estrogen receptor 1 (ESR1) mRNA expression by stabilizing ESR1 mRNA through interaction with ESR1 mRNA. Enhanced expression of ESR1 mRNA by TMPO-AS1 could play a critical role in the proliferation of ER-positive breast cancer. Our findings provide a new insight into the understanding of molecular mechanisms underlying hormone-dependent breast cancer progression and endocrine resistance.
INTRODUCTION
Long noncoding RNA (lncRNA) is defined as an RNA molecule with a length of >200 nucleotides that does not encode any protein (1). High-throughput sequencing technologies have uncovered the existence of an enormous number of lncRNAs (2). Recent advances of technology for operation of lncRNA have revealed that lncRNAs were involved in biological and pathological processes (3, 4). In particular, some lncRNAs have been reported to associate with development and progression of cancers, including breast cancer (5–7). For example, HOX transcript antisense RNA (HOTAIR) associates with breast cancer metastasis through reprogramming the chromatin status (8), and long intergenic noncoding RNA for kinase activation promotes tumor growth through activating the hypoxia-inducible factor 1 pathway (9).
In breast cancer, estrogen signaling is primarily a critical pathway to regulate proliferation (10, 11). Estrogen receptor α (ERα) is an essential ligand-dependent transcription factor that orchestrates the gene-regulatory network in breast cancer cells (12). The majority of breast cancers are initially ERα positive and treated with endocrine therapy using antiestrogens or aromatase inhibitors (13). During long-term endocrine therapy, however, breast cancers often can acquire drug resistance, and patients can suffer from recurrence and metastasis (14–16). Various molecular mechanisms could contribute to endocrine resistance, as exemplified by glycogen synthase kinase 3β activation (17) and the downregulation of tumor-suppressive microRNAs 378a-3p (miR-378a-3p) (18) and miR-574-3p (19) in tamoxifen-resistant breast cancer cells. Intriguingly, the overexpression of estrogen-inducible estrogen-responsive finger protein (Efp) promotes hormone-naive breast cancer cells even in an estrogen-deprived environment (20, 21). In ERα-positive metastatic breast cancers treated with endocrine therapy, constitutively active estrogen receptor 1 gene (ESR1) mutations are frequently observed, such as Tyr537Ser and Asp538Gly alterations in the ligand-binding domain, favoring agonist-receptor interaction (22). Of note, erb-b2 receptor tyrosine kinase 2 and the known ERα target cyclin D1 are amplified in >20% of ERα-positive metastatic breast cancers after endocrine therapy (22). Thus, the deregulated activation of ERα and its target genes would play a central role in the acquisition of endocrine resistance and the progression of disease states.
In terms of lncRNAs, HOTAIR expression is transcriptionally regulated by estrogen in breast cancer (23). A recent study also revealed that bidirectional ncRNAs transcribed on enhancers, or eRNAs, function in breast cancer MCF-7 cells even before ligand treatment by stabilizing estrogen/ERα/eRNA-induced enhancer-promoter looping systems (24). Considering that a number of lncRNAs are expressed primarily in cancer cells, the identification of novel tumor growth- and estrogen-related lncRNAs would further facilitate the understanding of breast cancer pathophysiology.
In the present study, we identified that thymopoietin antisense transcript 1 (TMPO-AS1) is a critical lncRNA that substantially associates with the proliferation of breast cancer. Clinicopathological study showed that TMPO-AS1 could be a prognostic factor for the disease. Loss- and gain-of-function studies of TMPO-AS1 demonstrated that TMPO-AS1 promotes cell cycle progression and reduces apoptosis of estrogen-sensitive breast cancer cells. The RNA antisense purification method demonstrates that TMPO-AS1 directly binds to ESR1 mRNA in living cells and stabilizes ESR1 mRNA, activating estrogen signaling and the transcription of proliferation-related genes. In tamoxifen-resistant MCF-7 xenograft models, TMPO-AS1-specific short interfering RNA (siRNA) significantly reduced tumor growth. Taken together, our findings define TMPO-AS1 as a promising diagnostic and therapeutic target for hormone-dependent as well as endocrine therapy-resistant breast cancers.
RESULTS
Cell proliferation-associated lncRNA TMPO-AS1 positivity correlates with poor prognosis of breast cancer patients.
To dissect functional lncRNAs that closely associate with proliferation signature in clinical breast cancers, we screened an RNA-sequencing data set retrieved from The Cancer Genome Atlas (TCGA) invasive breast carcinoma database (25). In terms of RNA expression levels analyzed by RNA sequencing for 816 breast cancer tissues, including both invasive ductal and lobular carcinomas, we found that TMPO-AS1 (26) is the only lncRNA that commonly associates with the proliferative biomarkers marker of proliferation Ki-67 (MKI67) and proliferating cell nuclear antigen (PCNA) at a threshold >0.5 by Spearman’s correlation (see Data Sets S1 and 2 in the supplemental material).
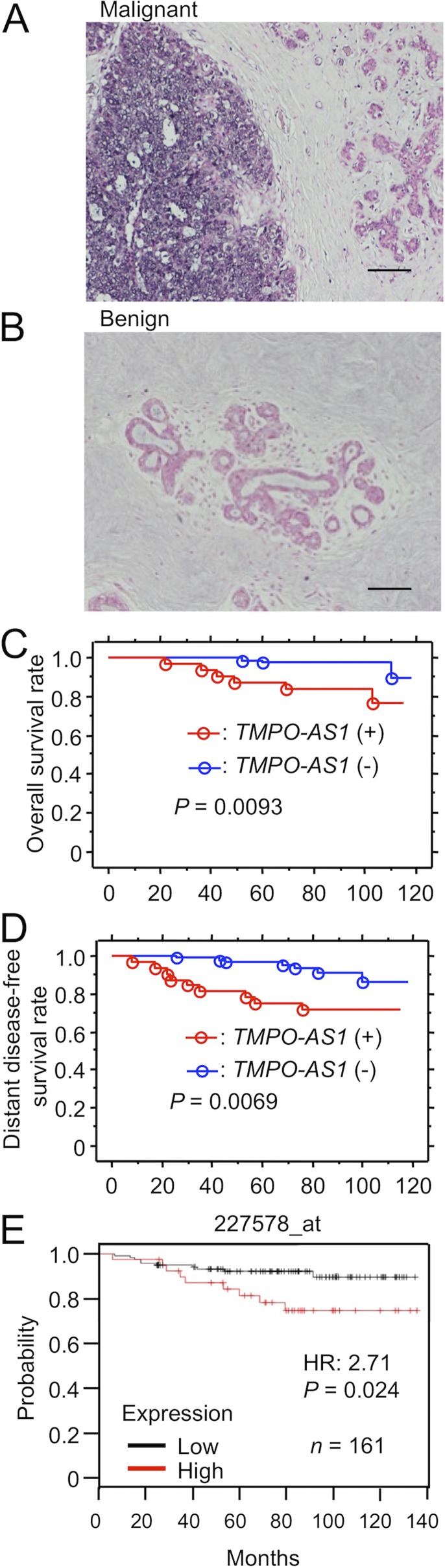
We evaluated the pathophysiological relevance of TMPO-AS1 in clinical ER-positive breast cancer specimens obtained from 115 Japanese patients who underwent surgical treatment for primary breast tumors. In in situ hybridization (ISH) analysis, intense signals of TMPO-AS1 in the nucleus and cytoplasm were often observed in some solid breast cancer lesions and were defined as ISH positive (Fig. 1A). For benign mammary ductal tissues, ISH signals of TMPO-AS1 were not detected and were defined as ISH negative (Fig. 1B). Based on the positivity criteria, 32 of 115 patients (28%) had tumors with positive ISH signal of TMPO-AS1, whereas the rest of the 83 patients had tumors with negative ISH signal (Table 1). We next analyzed the relationship between TMPO-AS1 positivity and clinicopathological parameters (Table 1). TMPO-AS1 positivity was significantly associated with stage (P = 0.0074), pathological T factor (pT; P = 0.022), histological grade (P = 0.018), and HER2 status (P = 0.026).
FIG 1.
Cell proliferation-associated lncRNA TMPO-AS1 positivity correlates with poor prognosis of breast cancer patients. (A and B) Representative results of in situ hybridization (ISH) analysis for TMPO-AS1 in malignant (A) and benign (B) mammary tissues. Scale bars, 100 μm. (C and D) Kaplan-Meier plot analysis showing the relationship between TMPO-AS1 ISH signals in cancer tissues and overall (C) and distant disease-free (D) survival of breast cancer patients (blue, ISH negative, n = 91; red, ISH positive, n = 37). (E) Relapse-free survival curve, analyzed by the Kaplan-Meier Plotter platform (http://kmplot.com/analysis/). TMPO-AS1 expression data were retrieved from 161 breast cancer patients treated with tamoxifen. P values and hazard ratios (HR) are shown.
TABLE 1.
Association between TMPO-AS1 status and clinicopathological factors in 115 breast carcinomas
| Parameter | Value by TMPO-AS1 status |
P valuea | |
|---|---|---|---|
| + (n = 32) | − (n = 83) | ||
| Age (yr) | |||
| <50 | 11 | 37 | |
| ≧50 | 21 | 46 | 0.32 |
| Stage | |||
| I | 9 | 50 | |
| II | 20 | 30 | |
| III | 3 | 3 | 0.0074 |
| Pathological T factor (pT) | |||
| pT1 | 15 | 58 | |
| pT2–4 | 17 | 25 | 0.022 |
| Pathological N factor (pN) | |||
| pN0–1 | 23 | 65 | |
| pN2–3 | 9 | 18 | 0.47 |
| Histological grade | |||
| 1–2 | 24 | 76 | |
| 3 | 8 | 7 | 0.018 |
| HER2 status | |||
| Positive | 7 | 6 | |
| Negative | 25 | 77 | 0.026 |
P value of <0.05 was considered significant.
We further examined the relationship between TMPO-AS1 positivity and the clinical prognosis of breast cancer patients. Based on Kaplan-Meier plot analysis, the positive ISH signal of TMPO-AS1 was significantly correlated with poorer overall survival (Fig. 1C) and distant disease-free survival (Fig. 1D) of breast cancer patients. Univariate analysis of overall and distant disease-free survival using the Cox proportional hazard model demonstrated that TMPO-AS1 positivity could be a significant prognostic factor for overall and distant disease-free survival, in addition to the known prognostic factors, such as pT and pathological N factor (pN) (Table 2 and 3). Multivariate analysis for 3 factors, including TMPO-AS1 positivity, pT, and pN, showed that all these factors are independent prognostic factors for overall and distant disease-free survival. Kaplan-Meier plot analysis with a publicly available breast cancer data set in the Kaplan-Meier Plotter platform (http://kmplot.com/analysis/) showed that TMPO-AS1 overexpression was associated with lower levels of relapse-free survival in tamoxifen-treated breast cancer patients (Fig. 1E).
TABLE 2.
Univariate and multivariate analyses of overall survival in 115 breast cancer patientsa
| Variable | Univariate P valueb | Multivariate |
|
|---|---|---|---|
| P valueb | Relative risk (95% CId ) | ||
| pN (pN0–1/pN2–3) | 0.0089c | 0.015 | 5.65 (1.39–22.97) |
| pT (pT1/pT2–4) | 0.0025c | 0.071 | 4.43 (0.88–22.31) |
| TMPO-AS1 status (negative/positive) | 0.020c | 0.030 | 34.69 (1.16–18.94) |
| HER2 status (negative/positive) | 0.91 | ||
| Histological grade (1–2/3) | 0.0.42 | ||
| Age (<50/≧50 yr) | 0.0.87 | ||
Statistical analysis was evaluated by a proportional hazard model (Cox).
P value of <0.05 was considered significant.
Significant (P < 0.05) univariate values were examined in the multivariate analyses in this study.
95% CI, 95% confidence interval.
TABLE 3.
Univariate and multivariate analyses of distant disease-free survival in 115 breast cancer patientsa
| Variable | Univariate P valueb | Multivariate |
|
|---|---|---|---|
| P value | Relative risk (95% CId ) | ||
| pN (pN0–1/pN2–3) | 0.0019c | 0.0040 | 4.40 (1.60–12.11) |
| pT (pT1/pT2–4) | 0.0071c | 0.048 | 2.97 (1.01–8.74) |
| TMPO-AS1 status (negative/positive) | 0.011c | 0.013 | 3.54 (1.30–9.65) |
| HER2 status (negative/positive) | 0.45 | ||
| Histological grade (1–2/3) | 0.50 | ||
| Age (<50/≧50 yr) | 0.70 | ||
Statistical analysis was evaluated by a proportional hazard model (Cox).
P value of <0.05 was considered significant.
Significant (P < 0.05) univariate values were examined in the multivariate analyses in this study.
95% CI, 95% confidence interval.
TMPO-AS1 is upregulated in tamoxifen-resistant breast cancer cells and is estrogen inducible.
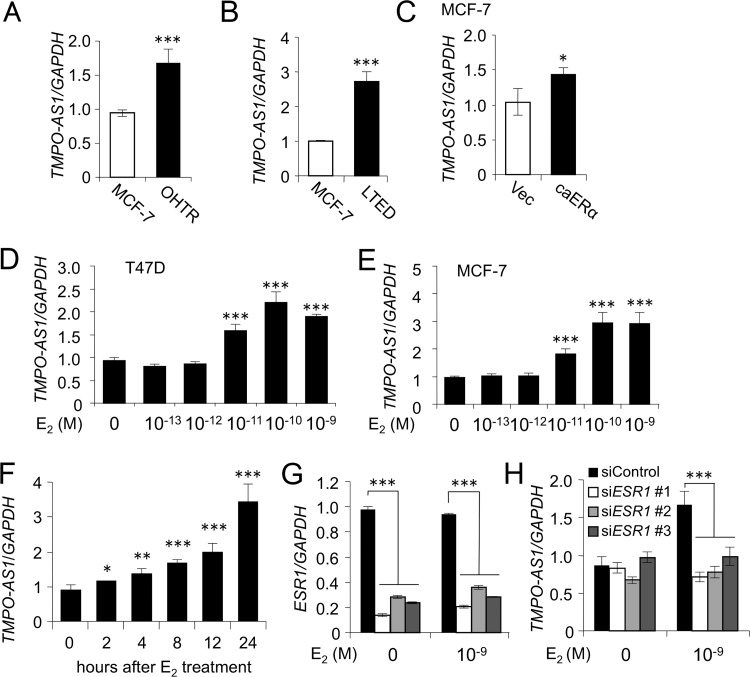
As all clinical specimens from the Japanese cohort were ERα positive and subjected to adjuvant endocrine therapy, we next questioned whether TMPO-AS1 expression associates with acquired tamoxifen resistance. As endocrine therapy-resistant ERα-positive breast cancer models, we previously generated 4-hydroxytamoxifen (OHT)-resistant MCF-7 cells, denoted OHTR cells (19), and long-term estrogen-deprived (LTED) MCF-7 cells (18). In addition to these models, we established MCF-7 cells overexpressing constitutively active ERα (caERα) with Y537S substitution, and their biological activity was previously characterized (27). Compared within parental MCF-7 cells, we found that the TMPO-AS1 expression level was significantly elevated in OHTR, LTED, and caERα-overexpressing MCF-7 cells (Fig. 2A to C). Because TMPO-AS1 could be functionally involved in the tumor proliferation and acquisition of tamoxifen resistance, we next questioned whether the expression of lncRNA associates with estrogen signaling. In ERα-positive MCF-7 and T47D cells, TMPO-AS1 expression was induced by 24 h of treatment with 17β-estradiol (E2) at a concentration of >10 pM (Fig. 2D and E). The elevation of TMPO-AS1 expression was significant >2 h after 10 nM E2 treatment in MCF-7 cells (Fig. 2F). Using specific siRNAs against ESR1 (Fig. 2G), we showed that E2-dependent TMPO-AS1 upregulation was repressed to its basal expression level (Fig. 2G).
FIG 2.
TMPO-AS1 is upregulated in tamoxifen-resistant breast cancer cells and is estrogen inducible. (A to C) TMPO-AS1 levels in 4-hydroxytamoxifen (OHT)-resistant (OHTR) (A), long-term estrogen-deprived (LTED) (B), or caERα-overexpressing (C) MCF-7 cells and parental or control vector-transfected (Vec) MCF-7 cells analyzed by qRT-PCR. Relative RNA levels were determined by normalization to GAPDH levels based on the ΔΔCT method, and values for OHTR cells are presented as mean fold changes ± standard deviations (SD) versus values for MCF-7 cells. (D and E) Concentration-dependent effect of estrogen on TMPO-AS1 expression in MCF-7 (D) and T47D (E) cells. Cells were treated with 17β-estradiol (E2) at the indicated concentrations for 24 h. Data are presented as mean fold changes ± SD versus the basal level at 0 M (n = 3). (F) Time-dependent effect of E2 on TMPO-AS1 expression in MCF-7 cells. Cells were treated with 10 nM E2 for the indicated durations. (G and H) Effects of ESR1 siRNAs on ESR1 (G) and TMPO-AS1 (H) expression in MCF-7 cells. Relative RNA levels are shown as mean fold changes ± SD versus levels for siControl (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
TMPO-AS1 knockdown attenuates the proliferation and viability of primary and hormone-refractory breast cancer cells.
To understand the function of TMPO-AS1 in ERα-positive breast cancer cell growth, we next used siRNAs targeting TMPO-AS1 to knock down TMPO-AS1 in ERα-positive MCF-7, T47D, and endocrine-resistant model OHTR cells (Fig. 3A). TMPO-AS1 knockdown significantly suppressed the proliferation of these cells (Fig. 3B) and decreased the percentages of S-phase cells (Fig. 3C to E). The knockdown of TMPO-AS1 also increased the percentages of apoptosis-related annexin V-positive fractions (Fig. 3F to H).
FIG 3.
TMPO-AS1 knockdown attenuates the proliferation and viability of primary and hormone-refractory breast cancer cells. TMPO-AS1 associates with the proliferation and viability of ERα-positive breast cancer cells. (A) Knockdown efficiency of TMPO-AS1 siRNAs in MCF-7, OHTR, and T47D cells analyzed by qRT-PCR. Relative RNA levels are presented as mean fold changes ± SD versus levels for siControl in each cell type (n = 3). (B) Viability of MCF-7, OHTR, and T47D cells on day 5 after siRNA treatment, analyzed by DNA assay. Values are presented as means ± SD versus levels for siControl in each cell type (n = 5). (C to E) Cell cycle profiles with propidium iodide (PI) of MCF-7 (C), OHTR (D), and T47D (E) cells treated with the indicated siRNAs, analyzed by flow cytometry. Percentages of cell populations in G1, S, and G2/M phases are shown (n = 3). (F to H) Percentages of annexin V-positive populations in MCF-7 (F), OHTR (G), and T47D (H) cells treated with the indicated siRNAs, analyzed by flow cytometry (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
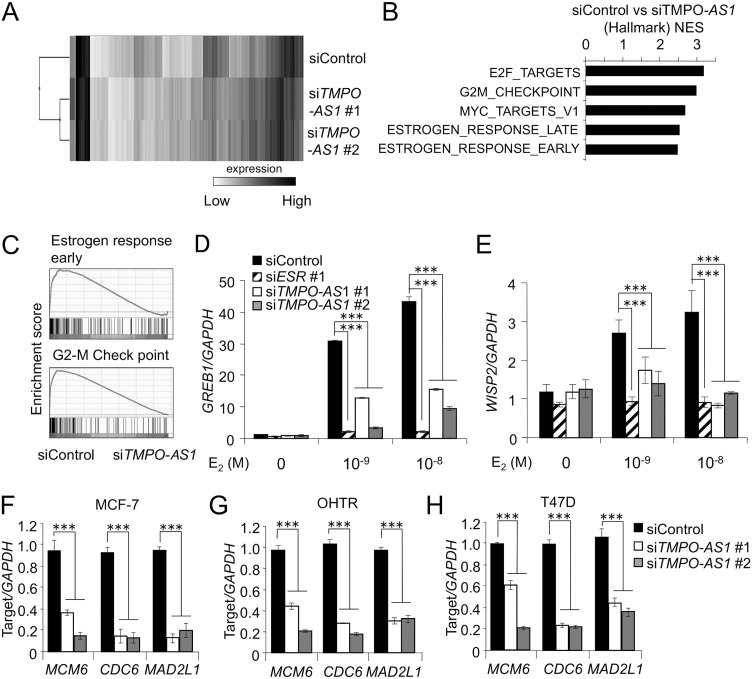
TMPO-AS1 knockdown represses estrogen signaling and proliferation-related gene expression.
To clarify the effects of TMPO-AS1 on the transcriptional profiles, we performed microarray analysis in MCF-7 cells with or without knockdown of TMPO-AS1 (Fig. 4A). Pathway analysis based on RNA expression showed that decrease of TMPO-AS1 was associated with the repression of cell proliferation and estrogen signaling (Fig. 4B and C). In terms of the effects of TMPO-AS1 on estrogen signaling, lncRNA knockdown substantially impaired the estrogen-dependent upregulation of growth-regulating estrogen receptor binding 1 (GREB1) and WNT1-inducible signaling pathway 2 (WISP2) (Fig. 4D and E). TMPO-AS1 knockdown also suppressed the expression of proliferation-related genes, including minichromosome maintenance 6 (MCM6), cell division cycle 6 (CDC6), and mitotic arrest deficient 2-like 1 (MAD2L1) in MCF-7 (Fig. 4F), T47D (Fig. 4H), and even in OHTR (Fig. 4G) cells.
FIG 4.
TMPO-AS1 knockdown represses estrogen signaling and proliferation-related gene expression. (A) Clustering microarray results of MCF-7 cells treated with control or TMPO-AS1-specific siRNAs. (B) Top 5 pathways enriched in genes downregulated by siTMPO-AS1 versus siControl determined by gene set enrichment analysis (GSEA). (C) GSEA enrichment plots for dominant pathways in MCF-7 cells treated with siTMPO-AS1, including estrogen response early and the G2-M checkpoint. (D and E) Effects of TMPO-AS1 knockdown on GREB1 (D) and WISP2 (E) levels in MCF-7 cells. Data are normalized to GAPDH levels and presented as mean fold changes ± SD versus levels for siControl in the absence of E2 (n = 3). (F to H) Effects of TMPO-AS1 knockdown on MCM6, CDC6, and MAD2L1 levels in MCF-7 (F), OHTR (G), and T47D (H) cells. Relative RNA levels are presented as mean fold changes ± SD versus levels for siControl (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
TMPO-AS1 stabilizes ESR1 mRNA.
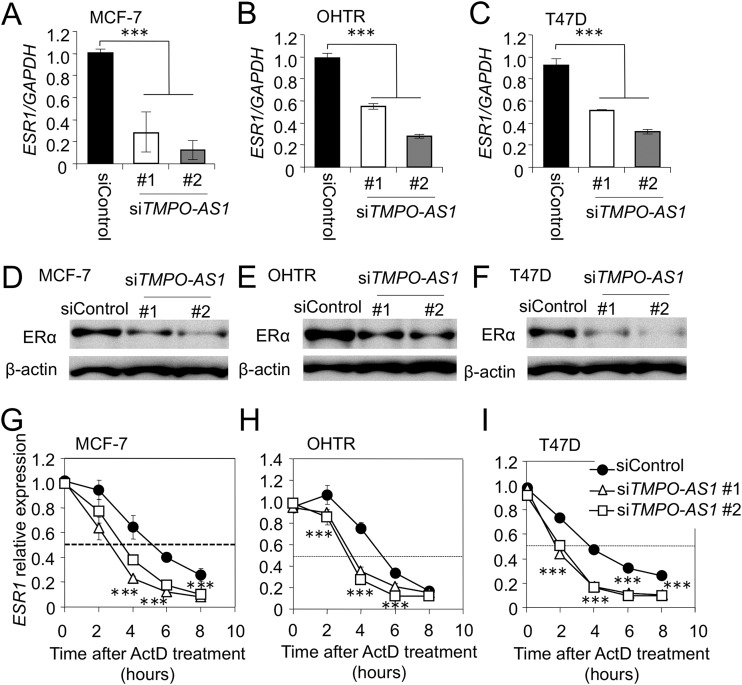
Because we found that TMPO-AS1 closely associates with estrogen signaling (Fig. 4), we next focused on how this lncRNA regulates the estrogen signaling pathway. We studied whether TMPO-AS1 regulates estrogen receptor function or expression, because several lncRNAs were reported to regulate functions of hormone receptors (28–30). Knockdown experiments of TMPO-AS1 in MCF-7, T47D, and OHTR cells showed that the expression of ESR1 (Fig. 5A to C) and its encoded protein, ERα (Fig. 5D to F), was suppressed by siRNAs specific to TMPO-AS1. We next examined the effects of TMPO-AS1 on ESR1 mRNA expression in the presence of the transcription inhibitor actinomycin D (ActD). In TMPO-AS1-repressed cells, ESR1 mRNA was quickly degraded compared with levels for control cells (Fig. 5G to I).
FIG 5.
TMPO-AS1 stabilizes ESR1 mRNA. (A to C) Effects of TMPO-AS1 knockdown on ESR1 mRNA expression in MCF-7 (A), OHTR (B), and T47D (C) cells. Data are normalized to GAPDH levels and presented as mean fold changes ± SD versus levels for siControl (n = 3). (D to F) TMPO-AS1 knockdown represses ERα protein levels in MCF-7 (D), OHTR (E), and T47D (F) cells. ERα and β-actin protein levels were evaluated by immunoblot analysis. β-Actin was used as a loading control. (G to I) TMPO-AS1 siRNAs more rapidly and severely decrease the stability of ESR1 mRNA in MCF-7 (G), OHTR (H), and T47D (I) cells. Actinomycin D (ActD; 10 nM) was added to culture medium 24 h after siRNA transfection. Cells were collected at the indicated times (0, 2, 4, 6, and 8 h after ActD treatment). ESR1 levels were normalized to GAPDH levels and are presented as mean fold changes ± SD versus basal values at 0 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
TMPO-AS1 overexpression promotes breast cancer cell proliferation.
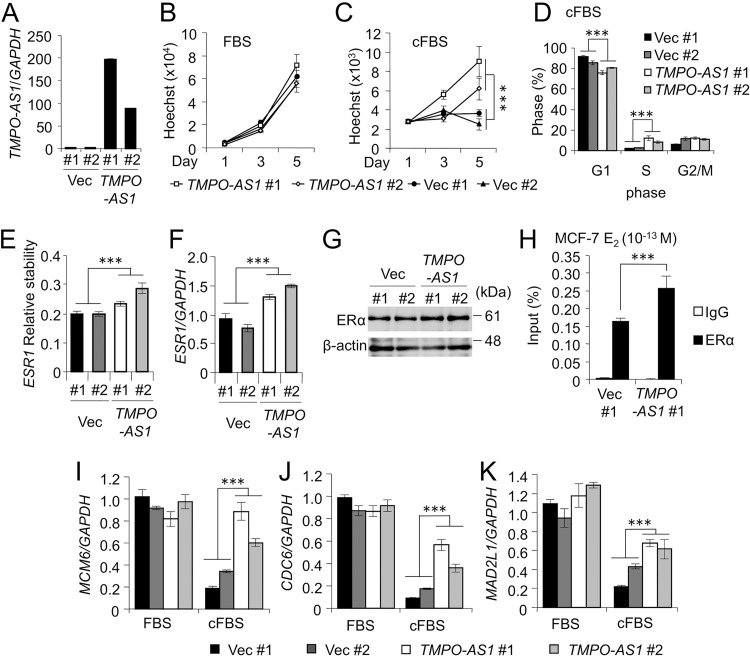
We generated MCF-7 transfectants stably overexpressing TMPO-AS1 and the control vector (Fig. 6A). TMPO-AS1 overexpression did not affect the proliferation of cells under the condition of normal fetal bovine serum (FBS) (Fig. 6B), whereas proliferation (Fig. 6C) and percentages of S-phase populations (Fig. 6D) were significantly increased under the condition of charcoal-stripped FBS (cFBS). TMPO-AS1 overexpression promotes ESR1 mRNA stability and expression (Fig. 6E to G). TMPO-AS1 overexpression promotes stabilization of ERα binding to ERE of the GREB1 gene in the presence of a low concentration of E2 (10−13 M) (Fig. 6H). In addition, we also found that TMPO-AS1 overexpression upregulated MCM6, CDC6, and MAD2L1 mRNA in cells cultured with cFBS-containing medium (Fig. 6I to K).
FIG 6.
TMPO-AS1 overexpression promotes breast cancer cell proliferation. (A) The RNA expression of TMPO-AS1 in cells stably overexpressing TMPO-AS1 and control cells. Data were normalized to GAPDH and are presented as mean fold changes ± SD versus levels for Vec #1 (n = 3). (B and C) Cell growth of MCF-7 cells stably overexpressing TMPO-AS1 and cultured with normal FBS (B) or charcoal-stripped FBS (cFBS) (C) was measured by DNA assay. Data are presented as means ± SD (n = 5). (D) Cell cycle analyses with propidium iodide (PI) in MCF-7 cells stably overexpressing TMPO-AS1 and cultured with cFBS were performed by flow cytometry (n = 3). (E) mRNA stability of ESR1 in the MCF-7 cells stably overexpressing TMPO-AS1 were measured by qRT-PCR. Cells were treated with ActD and collected after 0 and 6 h. Data were normalized to GAPDH and presented as mean fold changes ± SD versus the values at 0 h. (F) The expression levels of ESR1 mRNA in MCF-7 cells stably overexpressing TMPO-AS1 were measured by qRT-PCR. Data were normalized to GAPDH and are presented as mean fold changes ± SD versus levels for Vec #1 (n = 3). (G) ERα protein expression in MCF-7 cells stably overexpressing TMPO-AS1 were measured by Western blotting. β-Actin protein was used as a loading control. (H) ERα occupancy on the GREB1 ERE enhancer region in control and TMPO-AS1-overexpressing MCF7 cells after E2 (10−13 M) treatment for 45 min, analyzed by ChIP assay. Data were normalized by input DNA and are presented as means ± SD (n = 3). (I to K) MCM6 (I), CDC6 (J), and MAD2L1 (K) mRNA levels in MCF-7 cells stably overexpressing TMPO-AS1 cultured for 48 h in medium containing normal FBS or cFBS. Data are presented as mean fold changes ± SD versus values of vector-transfected MCF-7 cells (Vec #1) cultured in FBS (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
TMPO-AS1 overexpression confers tamoxifen resistance.
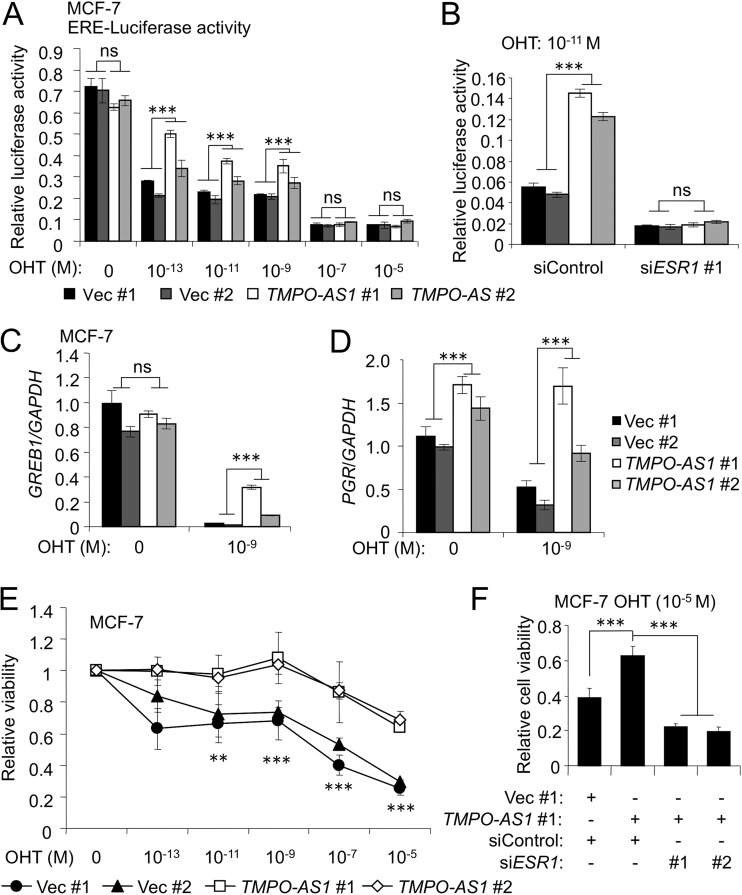
We next clarified whether TMPO-AS1 overexpression is associated with endocrine therapy resistance. Since we showed that stable overexpression of TMPO-AS1 enhanced estrogen signaling in MCF-7 cells (Fig. 6), we next examined whether TMPO-AS1 could increase estrogen signaling activity in the presence of OHT. Estrogen-responsive element (ERE)-based luciferase activities were decreased by OHT treatment in a dose-dependent manner in control vector-transfected MCF-7 cells, whereas TMPO-AS1 overexpression attenuated the suppression activity of OHT compared with that in control cells (Fig. 7A), even though no difference was shown in ERE-luciferase activities between control and TMPO-AS1-overexpressing MCF-7 cells in higher concentrations of OHT (≥10−7 M), suggesting that tamoxifen resistance mediated by TMPO-AS1 is dependent on ligand concentrations. Notably, ESR1 knockdown in cells treated with 10−11 M OHT abolished the enhanced ERE-based luciferase activity by TMPO-AS1 (Fig. 7B). We also showed that the expression of typical estrogen target genes, GREB1 and progesterone receptor (PGR), are upregulated in TMPO-AS1-overexpressing cells treated with OHT (Fig. 7C and D). We further showed that OHT treatment at a concentration of 10−5 M decreased cell viability by >60% in control MCF-7 cells, whereas >60% of TMPO-AS1-overexpressing cells could survive with 10−5 M OHT treatment (Fig. 7E). Knockdown of ESR1 canceled OHT resistance mediated by TMPO-AS1 overexpression (Fig. 7F), suggesting that TMPO-AS1-mediated activation of estrogen signaling would critically contribute to tamoxifen resistance by TMPO-AS1.
FIG 7.
TMPO-AS1 overexpression confers tamoxifen resistance. (A and B) Estrogen-responsive element (ERE)-based luciferase activities with the indicated concentrations of OHT were measured. Firefly luciferase values were normalized to Renilla luciferase values (n = 3). (B) ERE-based luciferase activities after treatment with OHT (10−11 M) and the indicated siRNAs. Firefly luciferase values were normalized to Renilla luciferase values (n = 3). (C and D) GREB1 (C) and PGR (D) mRNA levels in MCF-7 cells stably overexpressing TMPO-AS1 treated with OHT (10−9 M) for 48 h. Data are presented as mean fold changes ± SD versus values of vector-transfected MCF-7 cells (Vec #1) cultured in FBS-containing medium (n = 3). (E) Viability of control and TMPO-AS1-overexpressing MCF-7 cells on day 5 after OHT treatment analyzed by DNA assay. Values are presented as means ± SD versus the value of 0 M OHT for each cell (n = 5). (F) Viability of control and TMPO-AS1-overexpressing MCF-7 cells on day 5 after OHT and siRNA treatment, analyzed by DNA assay. Values are presented as means ± SD versus the value for OHT-free treatment (0 M) under each cell condition (n = 5).
TMPO-AS1 stabilizes ESR1 mRNA through RNA-RNA interaction at the ESR1 3′-UTR.
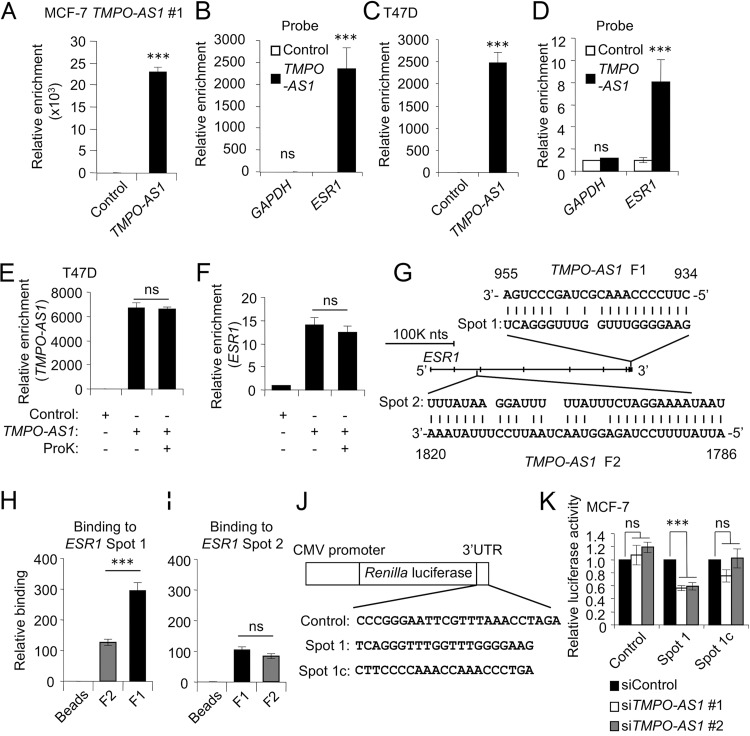
We next questioned how TMPO-AS1 affects ESR1 mRNA stabilization. We hypothesized that TMPO-AS1 directly binds to ESR1 mRNA and forms an RNA-RNA complex. We first performed RNA antisense purification using MCF-7 cells stably overexpressing TMPO-AS1. Probes specific for TMPO-AS1 precipitated TMPO-AS1 RNA at a high level compared with that of a negative-control probe (Fig. 8A). ESR1 mRNA could be coprecipitated by TMPO-AS1 probes, unlike the control probe (Fig. 8B), indicating that TMPO-AS1 could form a complex with ESR1 mRNA. We also showed the interaction between endogenous TMPO-AS1 and ESR1 occurred in living cells (Fig. 8C and D). To examine whether the interaction between TMPO-AS1 and ESR1 is modulated by proteins, we evaluated the TMPO-AS1-ESR1 binding in the presence of proteinase K (ProK). Proteinase K treatment did not change the interaction between TMPO-AS1 and ESR1 (Fig. 8E and F), suggesting that this interaction is not dependent on some intermediate proteins. We next examined whether the interaction between TMPO-AS1 and ESR1 is a direct event. In a screen of the complementary sequences of TMPO-AS1 in ESR1 mRNA by BLAST software, we identified 3 well-matched sequences, including one in the 3′ untranslated region (3′-UTR) of ESR1 and the other two in the intronic regions of ESR1. We denoted a 21-nucleotide sequence in the ESR1 3′-UTR as Spot 1 and another well-matched sequence with TMPO-AS1 in the ESR1 intronic region as Spot 2 (Fig. 8G). We synthesized RNAs including sequences corresponding to the adjacent regions for Spot 1 and Spot 2 from ESR1 as well as for F1 and F2 sequences from TMPO-AS1. An in vitro binding assay showed that the ESR1 Spot 1 RNA was well precipitated with TMPO-AS1 F1 RNA compared to that with TMPO-AS1 F2 (Fig. 8H), whereas no significant differences were observed in the enrichment of ESR1 Spot 2 by TMPO-AS1 F1 and F2 RNAs (Fig. 8I). Because ESR1 Spot 1 was identified from the ESR1 3′-UTR, we questioned whether TMPO-AS1 stabilizes ESR1 mRNA through the interaction with the ESR1 3′-UTR. We constructed luciferase reporter vectors including ESR1 Spot 1 or its complementary sequences (Spot 1c) (Fig. 8J). TMPO-AS1 knockdown significantly suppressed the luciferase activity for ESR1 Spot 1 vector compared with that of Spot 1c or control vectors in MCF-7 (Fig. 8K) cells, suggesting that ESR1 Spot 1 is a critical region for the interaction of ESR1 mRNA with TMPO-AS1.
FIG 8.
TMPO-AS1 stabilizes ESR1 mRNA through direct RNA-RNA interaction at the ESR1 3′-UTR. (A and C) In vivo binding of TMPO-AS1 and ESR1 in living cells, analyzed by the RNA antisense purification method. TMPO-AS1 levels were determined by qRT-PCR in RNA samples precipitated with the indicated probes from lysates of MCF-7 cells stably overexpressing TMPO-AS1 #1 (A) and T47D (C) cells. (B and D) ESR1 and GAPDH levels in RNA samples precipitated with the indicated probes from MCF-7 cells stably overexpressing TMPO-AS1 #1 (B) and T47D cells (D). Data are presented as mean fold changes ± SD versus levels for the control probe (n = 3). (E and F) In vivo binding of TMPO-AS1 and ESR1 in T47D cells treated with proteinase K (ProK) or left untreated, analyzed by the RNA antisense purification method. TMPO-AS1 (E) and ESR1 (F) levels in RNA samples were precipitated with the indicated probes and analyzed by qRT-PCR. Data are presented as mean fold changes ± SD versus levels for the control probe (n = 3). (G) Schematic representation of the predicted RNA-RNA interaction spots between TMPO-AS1 and ESR1. ESR1 Spot 1 and ESR1 Spot 2 were predicted to interact with nucleotides 934 to 955 (TMPO-AS1 F1) and 1786 to 1820 (TMPO-AS1 F2), respectively, of TMPO-AS1 RNA. (H and I) Direct interaction between ESR1 and TMPO-AS1 RNA was analyzed by an in vitro binding assay. Levels of synthesized RNAs for ESR1 Spot 1 (H) and ESR1 Spot 2 (I) coprecipitated with TMPO-AS1 F1 or F2 RNAs were evaluated by qRT-PCR. Data are presented as mean fold changes ± SD versus levels for control beads (n = 3). (J) Schematic representation of luciferase reporter vectors. Indicated sequences were inserted into psiCHECK2 plasmid. ESR1 Spot 1 complementary (Spot 1c) sequence is complementary to ESR1 Spot 1 sequence. (K) Luciferase activity of ESR1 Spot1 reporter in MCF-7 cells treated with the indicated siRNAs. Luciferase assay was performed using cells harvested 48 h after siRNA transfection. Renilla luciferase values were normalized to Firefly luciferase values. Data are presented as mean fold changes ± SD versus values in siControl-treated cells for each reporter gene. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
TMPO-AS1-dependent ESR1 mRNA stabilization is important for cell cycle progression.
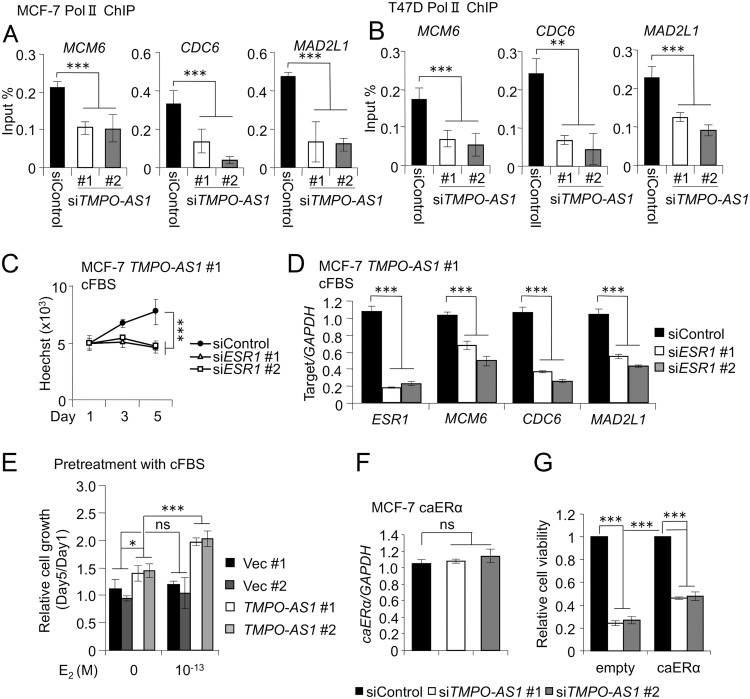
Since we found that TMPO-AS1 is associated with the cell proliferation-associated pathway as well as with the estrogen signaling pathway, we next investigated how TMPO-AS1 regulates the transcription of proliferation-associated genes. We performed chromatin immunoprecipitation (ChIP) analysis and showed that polymerase II (Pol II) recruitment to the promoters of MCM6, CDC6, and MAD2L1 was suppressed by TMPO-AS1 knockdown (Fig. 9A and B). We then examined an association of ESR1 expression with regulation of these genes by TMPO-AS1. In cFBS-containing medium, ESR1 knockdown impaired the enhanced proliferation ability mediated by TMPO-AS1 overexpression (Fig. 9C). MCM6, CDC6, and MAD2L1 were downregulated by ESR1 knockdown in MCF-7 cells stably overexpressing TMPO-AS1 (Fig. 9D), indicating that TMPO-AS1-dependent upregulation of ESR1 is important for the proliferation and viability of breast cancer cells.
FIG 9.
TMPO-AS1-dependent ESR1 mRNA stabilization is important for cell cycle progression. (A and B) RNA polymerase II (Pol II) occupancy on MCM6, CDC6, and MAD2L1 promoter regions in MCF7 (A) and T47D (B) cells analyzed by ChIP assay. Data were normalized by input DNA and are presented as means ± SD (n = 3). (C) Viability of MCF-7 cells stably overexpressing TMPO-AS1 treated with the indicated siRNAs in charcoal-stripped FBS (cFBS)-containing medium, analyzed by DNA assay (n = 5). (D) ESR1, MCM6, CDC6, and MAD2L1 mRNA levels in MCF-7 cells stably overexpressing TMPO-AS1 treated with the indicated siRNAs in cFBS-containing medium. Data are presented as mean fold changes ± SD versus levels for siControl (n = 3). (E) Viability of MCF-7 cells stably overexpressing TMPO-AS1, treated with E2 (10−13 M) or left untreated, in cFBS-containing medium after 3 days of pretreatment with cFBS, analyzed by DNA assay (n = 5). (F) Exogenous caERα mRNA levels in MCF-7 cells stably overexpressing caERα treated with the indicated siRNAs. Data are presented as mean fold changes ± SD versus levels for siControl (n = 3). (G) Viability of control and TMPO-AS1 stably overexpressing MCF-7 cells on day 5 after siRNA treatment, analyzed by DNA assay. Values are presented as means ± SD versus levels for siControl in each cell type (n = 5). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
In the next step, we questioned whether the estrogen signaling pathway is the unique target for TMPO-AS1. Control vector-transfected and TMPO-AS1-overexpressing MCF-7 cells were washed and pretreated with cFBS-containing medium for 3 days and again washed thoroughly to exclude the effects of hormone-like components, and then cells were cultured in new cFBS-containing medium with or without a low concentration of E2 (10−13 M) and the proliferative activities of these cells were measured. In the absence of E2 treatment, TMPO-AS1-overexpressing cells could exhibit proliferative activity (Fig. 9E, left). In addition, TMPO-AS1-overexpressing cells respond to low concentrations of E2 (10−13 M) but not control cells (Fig. 9E, right). The results suggest that estrogen signaling activation by TMPO-AS1 is an important event in ER-positive breast cancer cells, although TMPO-AS1 also could promote cell proliferation via an estrogen signaling-independent pathway. To clarify this hypothesis, we evaluated the effect of siTMPO-AS1 on the proliferation of MCF-7 cells overexpressing the caERα coding region without including the 3′UTR of ESR1. We first examined whether or not exogenous caERα is targeted by TMPO-AS1 through detecting the exogenous caERα RNA with a primer set, where one of the pair targets ESR1 and the other targets the vector. Indeed, knockdown of TMPO-AS1 did not change the expression of exogenous caERα RNA (Fig. 9F). TMPO-AS1 knockdown significantly suppressed the proliferation of caERα-overexpressing MCF-7 cells, although the suppression of cell viability was milder in the cells than that in control MCF-7 cells (Fig. 9G). These results suggest that TMPO-AS1 promotes cell proliferation via both ER-dependent and -independent pathways.
Knockdown of TMPO-AS1 inhibits hormone-refractory breast tumor growth in vivo.
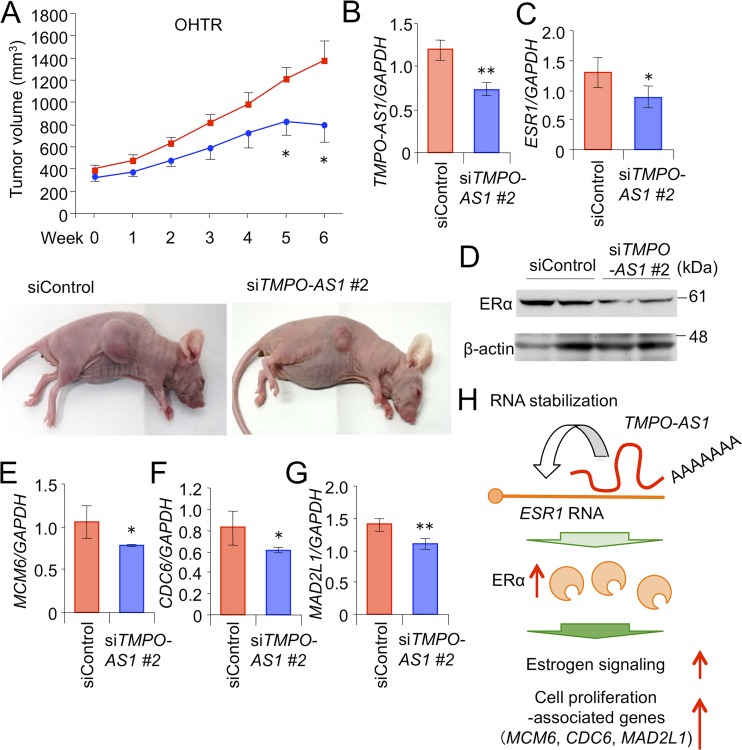
Finally, a pathological role of TMPO-AS1 was further evaluated in OHTR-derived xenograft models. Control or TMPO-AS1-specific siRNAs were injected twice weekly into the flanks of nude mice inoculated with OHTR cells from the time point when the volume of xenografted tumors reached 150 mm3. siTMPO-AS1 injection significantly suppressed the growth of OHTR-derived xenograft tumors (Fig. 10A and B) and repressed the mRNA levels of ESR1 (Fig. 10C), MCM6 (Fig. 10E), CDC6 (Fig. 10F), and MAD2L1 (Fig. 10G), as well as ERα protein levels (Fig. 10D) in OHTR-derived tumors.
FIG 10.
Knockdown of TMPO-AS1 inhibits hormone-refractory breast tumor growth in vivo. (A) Development of OHTR-derived xenograft tumors treated with siRNAs in nude mice. siControl or siTMPO-AS1 #2 was injected twice weekly into the flanks of mice inoculated with OHTR cells (siControl, n = 7; siTMPO-AS1 #2, n = 6). Tumor volumes are presented as means ± standard errors. Representative photographs of xenografted mice are shown below. (B and C) TMPO-AS1 (B) and ESR1 (C) levels in tumors treated with siControl or siTMPO-AS #2. Tumors were dissected from mice 6 weeks after the beginning of siRNA administration. (D) Immunoblot analysis for ERα and β-actin in tumors dissected from 2 distinct mice for each group treated with either siControl or siTMPO-AS #2. (E to G) mRNA levels of MCM6 (E), CDC6 (F), and MAD2L1 (G) in tumors. Data are presented as mean fold changes ± SD versus levels for siControl (n = 3). (H) Working model of TMPO-AS1 in the proliferation and progression of ER-positive breast cancer cells. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Taking these findings together, we assume that TMPO-AS1 contributes to the pathophysiology of hormone-dependent breast cancer by binding to and stabilizing ESR1 mRNA, leading to the enhanced estrogen signaling that upregulates the proliferation-related gene signature (Fig. 10H).
DISCUSSION
In the present study, we identified TMPO-AS1 as a functional lncRNA that significantly associates with the proliferation signals of invasive breast carcinomas in the TCGA database. Positive ISH signals of TMPO-AS1 in tumor tissues significantly correlated with poorer prognosis of breast cancer patients. The lncRNA expression is substantially elevated in tamoxifen-resistant MCF-7 cells compared with levels of parental cells and is inducible by estrogen treatment. Knockdown and overexpression experiments showed that TMPO-AS1 plays critical roles in the proliferation and viability of ERα-positive breast cancer cells. Intriguingly, knockdown of TMPO-AS1 repressed the in vitro and in vivo proliferation of tamoxifen-resistant breast cancer cells. Pathway analyses based on expression microarray data showed that TMPO-AS1 is strongly associated with estrogen signaling and proliferation-related pathways of ERα-positive breast cancer. The RNA antisense purification method defined that TMPO-AS1 is an ESR1 mRNA-stabilizing lncRNA through a direct interaction with the 3′-UTR of ESR1.
Acquired endocrine resistance is a serious burden for breast cancer patients treated with endocrine therapy. Previous literature proposed multiple molecular mechanisms underlying the development of the pathological state (14–16). Our findings indicate that TMPO-AS1 plays a critical role in the progression of endocrine-resistant as well as hormone-naive breast cancer. In tamoxifen-relapsing breast cancer cases, TMPO-AS1 was also reported as one of the differently expressed genes (31). TMPO-AS1 was originally identified as a target of the E2F signaling pathway (26), an essential signaling pathway for DNA replication and cell cycle progression. Hormone dependency of TMPO-AS1 was recently observed in prostate cancer (32), suggesting that the lncRNA is an important regulator for the proliferation of hormone-dependent cancers.
In particular, we demonstrated that TMPO-AS1 stabilizes ESR1 mRNA and enhances estrogen signaling, suggesting that the lncRNA exerts positive feedback on the estrogen-regulated gene network. lncRNA-mediated alterations of steroid hormone signaling have been reported, although these lncRNAs primarily exert their functions through direct interaction with hormone receptor proteins. HOTAIR was identified as one of the first lncRNAs that associate with breast cancer progression, in this case by interacting with ERα protein and promoting estrogen signaling (28). Steroid receptor RNA activator (SRA) is an lncRNA that binds to steroid hormone receptors and functions as a transcriptional coactivator (33). Our group and others defined several androgen-induced lncRNAs (30, 34–36), some of which enhance androgen signaling (30, 36). Growth arrest-specific 5 (GAS5) is a repressive lncRNA for hormone signaling, inhibiting the DNA binding ability of hormone receptors, including ERα (29). Compared with the lncRNAs that directly interact with hormone receptor proteins, we assume that TMPO-AS1 exerts a unique lncRNA whose posttranscriptional function is mediated through the direct interaction with the 3′-UTR of ESR1, leading to the stabilization of ESR1 mRNA.
The relevance of RNA-RNA interactions has been reported in biological and pathological processes (37, 38). For example, U1 snRNA binds throughout nascent transcripts at the 5′-splice site motif and regulates transcription (39). Antisense lncRNA of beta-secretase 1 (BACE1) interacts with its sense RNA BACE1 and increases the stability of BACE1 mRNA, which may contribute to Alzheimer’s disease (40). Recent advances in methodologies, such as RNA antisense purification, have further enabled the dissection of critical lncRNAs that directly interact with protein-coding RNAs and other lncRNAs. It was reported recently that AR-regulated long noncoding RNA 1 (ARLNC1) interacts with androgen receptor (AR) mRNA and regulates the subcellular localization of AR mRNA (30). In this context, our findings have an impact on cancer research, because TMPO-AS1 might be an ESR1-interacting and -stabilizing lncRNA that contributes to the progression of ERα-positive breast cancer.
Currently, it is not clear what kind of factors are involved in ESR1 mRNA stabilization by TMPO-AS1. For example, half-STAU1-binding site RNAs (1/2-sbsRNAs) bind to their target RNAs and recruit staufen 1 protein, leading to a decrease in the RNA stability of target RNAs (41). On the other hand, several lncRNAs, like GAS5, inhibit the interaction between proteins and target molecules (29), suggesting that TMPO-AS1 also recruits RNA-protecting factors to ESR1 mRNA or inhibit the binding between ESR1 mRNA and RNA decay factors. Several microRNAs have been reported to target ESR1 mRNA (42, 43), although none of them targets the ESR1 Spot 1 region for TMPO-AS1 binding as far as we could tell based on the miRNA database. In this study, our findings revealed that TMPO-AS1 could stabilize ESR1 mRNA and activate the estrogen signaling pathway. On the other hand, TMPO-AS1 knockdown also suppressed the proliferation of caERα-overexpressing MCF-7 cells. Microarray analysis showed that TMPO-AS1 is associated with E2F and Myc pathways as well as estrogen signaling pathways (Fig. 4), indicating that TMPO-AS1 also would contribute to E2F or Myc signaling cascades. Future studies will reveal precise mechanisms of TMPO-AS1 in estrogen signaling and breast cancer pathophysiology.
In conclusion, TMPO-AS1 plays a critical role in the proliferation and progression of ERα-positive breast cancer as well as endocrine therapy-resistant breast cancer. We assume that the present results will provide new diagnostic and therapeutic options for advanced states of breast cancer.
MATERIALS AND METHODS
Cell culture and reagents.
The human ERα-positive breast cancer cell lines MCF-7 and T47D were purchased from the ATCC (Manassas, VA) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 50 U/ml penicillin, and 50 μg/ml streptomycin at 37°C in a 5% CO2 humidified atmosphere. The OHTR cells resistant to 4-hydroxytamoxifen (OHT) were established from MCF-7 cells by long-term (>3 months) culture with 1 μM OHT (19). LTED cells were established from MCF-7 cells by long-term (>3 months) culture in cFBS-containing phenol red-free medium, described previously (18). To establish MCF-7 cells stably overexpressing TMPO-AS1, MCF-7 cells were transfected with pCDNA3 empty or TMPO-AS1 plasmids and selected with 800 μg/ml G418 for 2 weeks. At least two clones were picked and used for further experiments. Construction of caERα with Y537S substitution was described previously (27). caERα was introduced into MCF-7 cells by a lentiviral system described previously (44), and transduced cells were selected with 800 μg/ml G418 for 2 weeks. For estrogen treatment experiments, cells were cultured with DMEM (low glucose and no phenol red) (Thermo Fisher Scientific, MA) containing cFBS, 50 U/ml penicillin, and 50 μg/ml streptomycin for 24 to 48 h and were 17β-estradiol (E2) treated. The antibodies used in the present study were anti-ERα (H-184; Santa Cruz Biotechnology, TX), anti-β-actin (A2228; Sigma-Aldrich, MO), and anti-RNA polymerase II (CTD4H8; Merck, Darmstadt, Germany) antibodies.
Bioinformatics.
In screening of proliferation-associated lncRNAs in clinical breast cancer tissues, coexpression genes for proliferative biomarkers MKI67 and PCNA were selected at a threshold Spearman’s correlation value of >0.5, retrieved from The cBioPortal for Cancer Genomics (http://www.cbioportal.org/) (45, 46) based on RNA expression z-scores in RNA-sequencing data sets of breast cancer cohorts in the TCGA database (25). Kaplan-Meier curves of relapse-free survival for breast cancer patients were acquired through the Kaplan-Meier Plotter software (http://kmplot.com/analysis/) (47). TMPO-AS1 target gene expression in breast cancer and normal breast samples were analyzed using Oncomine software (https://www.oncomine.org). Complementary sequences between ESR1 and TMPO-AS1 RNA were acquired through the basic local alignment search tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Collection of human tissue samples and clinical data.
Tissue samples of breast cancer were obtained from 115 Japanese female breast cancer patients who underwent surgical treatment from 2006 to 2013 at Toranomon Hospital, Tokyo, Japan (age range, 31 to 76 years). No patients received chemotherapy or molecular target therapy before surgery. Standard adjuvant treatments were selected according to the clinical practice guidelines of the National Comprehensive Cancer Network (48). Staging was performed according to the TNM Classification of Malignant Tumours (49). The clinical outcome was evaluated by distant disease-free survival in this study, defined as the time span from the date of surgery to the first distant recurrence or last follow-up. The mean follow-up duration was 83 months (range, 8 to 118 months) in the present study. This study was approved by the ethical committee in Toranomon Hospital (approval number 845) and Saitama Medical University (approval number 13-148). All patients provided written informed consent to participate in this study. This study abides by the Declaration of Helsinki principles.
ISH.
RNA probes for in situ hybridization (ISH) were generated using the digoxigenin (DIG) RNA labeling kit (Roche, Switzerland) in accordance with the manufacturer’s instructions. RNA ISH was performed in breast cancer tissues fixed in 10% formalin and embedded in paraffin wax. Slides were treated with proteinase K (20 mg/ml; Wako Pure Chemical Industries, Ltd., Osaka, Japan) for 10 min at room temperature and then refixed with 10% formalin. Subsequently, the slides were immersed in 0.2 N HCl for 10 min and hybridized with TMPO-AS-1 probe (25 ng per slide) at 63°C for 24 h using G-Hybo-L (Genostaff, Tokyo, Japan) (50). For signal detection, the slides were sequentially labeled with anti-DIG mouse monoclonal antibody (Roche, Switzerland), biotinylated anti-mouse IgG, and alkaline phosphatase-labeled streptavidin (Nichirei Bio, Inc., Tokyo). Finally, chromogenic signals were obtained using nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate solution (Roche, Switzerland) and counterstained by nuclear fast red. The signals obtained from ISH were evaluated by two trained pathologists (T. Suzuki and K. Takagi).
qRT-PCR.
Total RNA was extracted from cells using ISOGEN reagent (Nippon Gene, Toyama, Japan), followed by cDNA synthesis using SuperScript III reverse transcriptase (Invitrogen, MO) with random primers. Quantitative real-time PCR (qRT-PCR) was performed on a StepOnePlus (Thermo Fisher Scientific) using a KAPA SYBR FAST quantitative PCR (qPCR) kit (KAPA Biosystems, MA) and sets of gene-specific primers. RNA expression levels were analyzed by the ΔΔCT method (where CT is threshold cycle) and normalized to the values of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The sequences of primers are the following (forward and reverse, respectively): GAPDH, 5′-GGTGGTCTCCTCTGACTTCAACA-3′ and 5′-GTGGTCGTTGAGGGCAATG-3′; TMPO-AS1, 5′-CTTTTGTGCGCCGTTTCCT-3′ and 5′-CCCAGAGACGAAAGCTGCTT-3′; ESR1, 5′-AGACGGACCAAAGCCACTTG-3′ and 5′-CCCCGTGATGTAATACTTTTG-3′; GREB1, 5′-CCACCCTTTGTGGCGTTTT-3′ and 5′-CGACCATCGGGCTTTAGGTATCTT-3′; WISP2, 5′-CATGCAGAACACCAATATTAAC-3′ and 5′-TAGGCAGTGAGTTAGAGGAAAG-3′; MCM6, 5′-TCGGGCCTTGAAAACATTCGT-3′ and 5′-TGTGTCTGGTAGGCAGGTCTT-3′; CDC6, 5′-TGTTCTCCTCGTGTAAAAGCC-3′ and 5′-GGGGAGTGTTGCATAGGTTGT-3′; MAD2L1, 5′-GGACTCACCTTGCTTGTAACTAC-3′ and 5′-GATCACTGAACGGATTTCATCCT-3′; PGR, 5′-AAGAAATGACTGCATCGTTGATAAAA-3′ and 5′-ATGCCAGCCTGACAGCACTT-3′; ESR1 Spot 1, 5′-GGGACCGTTGCTGTCACTAC-3′ and 5′-GAGGATTTTCTTCCCCAAA-3′; ESR1 Spot2, 5′-TGCCTTTTCTTCAGCCTTGT-3′ and 5′-AATCATCTCCCCATTCACCA-3′; exogenous caERα, 5′-AGACGGACCAAAGCCACTTG-3′ and 5′-CCTCACATTGCCAAAAGACG-3′.
siRNA transfection.
siRNAs against ESR1 and TMPO-AS1 were designed using siDirect and purchased from RNAi Inc. (Tokyo, Japan). A negative-control siRNA (siControl) with no homology to known gene targets in mammalian cells was purchased from RNAi Inc. MCF-7, T47D, and OHTR cells were seeded at 300,000 cells per well in 6-well plates and simultaneously transfected with siRNA at a final concentration of 10 nM using Lipofectamine RNAiMAX (Invitrogen, MO). Forty-eight hours after transfection, cells were collected and used for qRT-PCR, cell cycle analysis, and annexin V and propidium iodide (PI) staining. The sequences of siRNA are the following (forward and reverse, respectively): siESR1 #1, 5′-GCCUGGUCAGAUUACGUAUGC-3′ and 5′-AUACGUAAUCUGACCAGGCCC-3′; siESR1 #2, 5′-GGGAGCGUGAUCUAGAUUACA-3′ and 5′-UAAUCUAGAUCACGCUCCCAA-3′; siTMPO-AS1 #1, 5′-GAAGACUAGUGACCUAUAAUU-3′ and 5′-UUAUAGGUCACUAGUCUUCCU-3′; siTMPO-AS1 #2, 5′-GAGCCGAACUACGAACCAACU-3′ and 5′-UUGGUUCGUAGUUCGGCUCUG-3′.
DNA assay.
Cells were seeded at the indicated densities (1,500 cells per well for MCF-7 and OHTR and 3,000 cells per well for T47D) in 96-well plates with normal FBS- or charcoal-stripped FBS (cFBS)-containing medium. For the stringent washed condition shown in Fig. 9, cells were washed with phosphate-buffered saline without divalent cations [PBS(−)] twice and cultured in phenol red-free cFBS-containing medium for 3 days. Cells then were washed again with PBS(−) twice and reseeded into 96-well plates at 1,000 cells per well with phenol red-free cFBS-containing medium with or without E2 (10−13 M). For the evaluation of OHT sensitivity, cells were seeded at 1,500 cells per well and treated with the indicated concentrations of OHT 24 h after cell seeding. Cells were collected at 1, 3, and 5 days after cell seeding and frozen. Cells were thawed and lysed with TNE buffer (10 mM Tris-HCl [pH 7.5], 2 mM NaCl, and 1 mM EDTA). Extracted DNA samples were stained with Hoechst 33258 pentahydrate (Thermo Fisher Scientific) at a final concentration of 5 μg/ml. The DNA contents in each well were measured on an ARVO5 (Perkin Elmer, MA) at 355 nm for 0.1 s.
Cell cycle analysis.
siRNA-treated cells and TMPO-AS1 stably transfected cells were harvested and fixed with 70% ethanol for at least 30 min. Fixed cells were treated with RNase A and stained with 5 μg/ml PI. DNA contents were measured using the FACSCalibur platform (Becton, Dickinson, MD). Data were analyzed by CellQuest software (Becton, Dickinson) to determine the percentage of cells in G1, S, and G2/M phases.
Annexin V and PI staining.
After transfection with siRNAs, cells were collected and apoptotic cells were stained using a fluorescein isothiocyanate annexin V apoptosis detection kit (Becton, Dickinson) by following the manufacturer’s instructions. The percentages of apoptotic cells were analyzed on the FACSCalibur platform (Becton, Dickinson).
Microarray and pathway analysis.
For microarray analysis, a human Clariom D array (Thermo Fisher Scientific) was used by following the manufacturer’s instructions. Data were analyzed using Affymetrix Microarray Suite software. Pathway analyses were performed using gene set enrichment analysis (GSEA).
Western blot analysis.
Cells were suspended in Laemmli sample buffer (125 mM Tris-HCl [pH 6.8], 20% glycerol, 4% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, and bromophenol blue) and boiled for 20 min at 100°C. Western blot analysis was described previously (19).
RNA degradation assay.
Twenty-four hours after cell seeding, actinomycin D (Nacalai Tesque, Inc., Kyoto, Japan) was treated at a final concentration of 10 nM. Cells were collected at the indicated times (0, 2, 4, 6, and 8 h after actinomycin D treatment), and RNAs were extracted using ISOGEN (Nippon Gene, Toyama, Japan) reagent.
ChIP assay.
For ChIP assay using Pol II antibody, cells were seeded in 10-cm dishes and transfected with siRNA (10 nM final concentration) using Lipofectamine RNAiMax reagent (Thermo Fisher Scientific) for 48 h. For ChIP assay using ERα antibody, cells were seeded in 10-cm dishes with phenol red-free cFBS-containing medium for 48 h and then treated with 10−13 M E2 for 45 min. Cells were fixed with 1% formaldehyde for 10 min. Cells were collected and lysed in ChIP lysis buffer (10 mM Tris-HCl [pH 7.5], 200 mM NaCl, 10 mM EDTA, 1% SDS) and sonicated. Samples were diluted with a 9× volume of ChIP dilution buffer (16.7 mM Tris-HCl [pH 8.0], 1.2 mM EDTA, 1.1% Triton X-100, 167 mM NaCl, 0.01% SDS). DNA-protein complexes were precipitated with 1 μg of anti-Pol II or anti-ERα antibody overnight at 4°C and captured by protein G-Sepharose 4 Fast Flow beads (GE Healthcare, IL). Beads were washed and incubated at 65°C overnight for decrosslinking. DNA samples were extracted and measured by qPCR. Data were normalized by input DNA samples. The sequences of primers are the following (forward and reverse, respectively): MCM6, 5′-AAGCGACTTGTGGCGGTCGA-3′ and 5′-CCTCCAAGAAGTCCAGGAACAGT-3′; CDC6, 5′-AGTTTGTTCAGGGGCTTGTG-3′ and 5′-CCTCCTCGAGCAATCCTCTTCT-3′; MAD2L1, 5′-GACGTGCTGCGTCGTTACTTTTG-3′ and 5′-CCATGGCCAGGGACACAAACAA-3′; GREB1 ERE, 5′-GAAGGGCAGAGCTGATAACG-3′ and 5′-GACCCAGTTGCCACACTTTT-3′.
RNA antisense purification assay.
RNA antisense purification assay was performed based on techniques previously published by others (51, 52). Briefly, cells were fixed with 1% paraformaldehyde for 10 min. Cells were resuspended in lysis buffer (50 mM Tris-HCl [pH 7.0], 10 mM EDTA, 1% SDS with RNase inhibitor [Nacalai Tesque, Inc., Kyoto, Japan] and cOmplete EDTA-free protease inhibitor cocktail [Sigma-Aldrich]). After sonication on a Bioruptor (COSMO Bio, Tokyo, Japan), lysate was treated with proteinase K for 1 h at 37°C. The lysate was mixed with a 2× volume of hybridization buffer (750 mM NaCl, 1% SDS, 50 mM Tris-HCl [pH 7.0], 1 mM EDTA, 15% formamide) containing pooled 3′-biotinylated probes and incubated at 37°C for 4 h with rotation. Dynabeads MyOne streptavidin C1 (Thermo Fisher Scientific) were mixed and incubated at 37°C for 1 h with rotation. RNA complex and magnet beads were washed with wash buffer five times. Beads were incubated with elution buffer at 65°C for 45 min and then 95°C for 10 min. Eluted samples were resuspended in ISOGEN reagent. Probe sequences for TMPO-AS1 precipitation are the following: 5′-CTACAAAGGCGGGCGTTTGG-3′, 5′-ACTTCTCCAGTGACGAAGAG-3′, 5′-TTTGTGTCCGCGAGTTTTTG-3′, 5′-CGCCTTTTAAACTGCGTTTC-3′, 5′-GCGCACAAAAGCAGTACGAC-3′, 5′-CTACTCTTGGAGCTTCAGTG-3′, 5′-AAAGAAGCGTTCGCGAGGAG-3′, 5′-CCCCAACTATGACACTAAGA-3′, 5′-TAGGTTTAGGATTCTTGCGG-3′, 5′-TTATAGGTCACTAGTCTTCC-3′, 5′-GTGATACTAATTTCCAGGCA-3′, 5′-AGTTTGGAGCTCAGATTCTG-3′, 5′-GAGCTTAATACCATTGCTTA-3′, 5′-AACATTTGCCTATGTGTCCA-3′, 5′-TCAGGCGTATCTAGAATGCA-3′, 5′-TTGGAGCTCTACAGCAGTAA-3′, 5′-GTGTTGCATGGGTCACCTAC-3′, 5′-TTAGGTAAGTGAGAGTACCA-3′, 5′-ATGGTTTAGTCCAAGCAAGG-3′, 5′-AATGGTTAACCCAGAGACTG-3′.
In vitro binding assay.
ESR1 Spot 1, ESR1 Spot 2, TMPO-AS1 F1, and TMPO-AS 1 F2, sequences of around 500 to 600 nucleotides, were subcloned into pCDNA3 vector. RNAs were synthesized using RNA polymerase (TaKaRa Bio Inc., Shiga, Japan) with nucleotide triphosphate mix (Thermo Fisher Scientific) or biotin RNA labeling mix (Sigma-Aldrich). Biotinylated RNAs (5 pmol) were incubated with 20 μl of Dynabeads MyOne Streptavidin C1 (Thermo Fisher Scientific) in RNA immunoprecipitation (RIP) buffer (150 mM KCl, 25 mM Tris-HCl [pH 7.4], 5 mM EDTA, 0.5% NP-40) for 1 h at room temperature with rotation. Biotinylated RNA-conjugated magnet beads were washed with RIP buffer and incubated with 2.5 pmol of ESR1 Spot 1 or ESR1 Spot 2 RNA in RIP buffer overnight. RNA-conjugated beads were washed with 1 M NaCl in NT2 buffer (50 mM Tris-HCl [pH 7.5], 1 M NaCl, 1 mM MgCl2, 0.1% NP-40) and resuspended in ISOGEN reagent. Primers for subcloning of ESR1 Spot 1, ESR1 Spot 2, TMPO-AS1 F1, and TMPO-AS1 F2 were the following (forward and reverse, respectively): ESR1 Spot 1, 5′-CTGAGGCACAGCCAGACTTG-3′ and 5′-CACCCAGAGGAAATCAAACA-3′; ESR1 Spot 2, 5′-GTGCCAATTCAAGATGGAAATAGC-3′ and 5′-CTGTATTTCATGATTGCCCCAAAG-3′; TMPO-AS1 F1, 5′-AACCCCAGCCCACACACTAC-3′ and 5′-GAATATGAGTGCCTGCAGAC-3′; TMPO-AS1 F2, 5′-ATCACTGATGACAAATATTT-3′ and 5′-CCCCTTCTGAAGATAAAAAC-3′.
Luciferase assay.
Twenty-four hours after cell seeding at 50,000 cells per well, luciferase vectors (300 ng of psiCHECK2 vector [Promega Corporation, WI] or 300 ng of ERE-luciferase and 10 ng of Renilla-expressing vector per well in a 24-well plate) were transfected with Lipofectamine 2000 reagent (Thermo Fisher Scientific) with the indicated concentrations of OHT. Forty-eight hours after transfection, cells were collected and luciferase activities were measured using the Dual-Luciferase reporter assay system (Promega Corporation, WI) on TriStar2 S LB942 (Berthold, TN).
In vivo tumor formation and siRNA treatment.
Female nude mice were purchased from CREA Japan. MCF-7 and OHTR cells were mixed with equal volumes of Matrigel matrix (Corning, NY) and injected subcutaneously into flanks of 8-week-old female nude mice. When the tumor volume reached 150 mm3, mice were divided in two groups randomly. Five micrograms of siControl or siTMPO-AS #2 was injected with GeneSilencer reagent (Gene Therapy System, CA) into tumors twice a week. Three dimensions of tumors were measured once a week, and tumor volumes were estimated with the following formula: 0.5 × 1st diameter × 2nd diameter × 3rd diameter.
Statistical analysis.
Statistical analyses of in vitro and in vivo experiments were performed by the Student's t test or analysis of variance, respectively.
Data availability.
All microarray data are available in the Gene Expression Omnibus (GEO) database with the accession number GSE129004.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tomoko Suzuki, Miwa Fujitani, and Noriko Sasaki for their technical assistance and Kenichi Takayama for critical discussion and advice.
This work was supported in part by the Support Project of the Strategic Research Centers in Private Universities (to S.I. and K.H.-I.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan; by the Practical Research for Innovative Cancer Control (JP18ck0106194 to K.I.) and the Project for Cancer Research and Therapeutic Evolution (P-CREATE to S.I.) from the Japan Agency for Medical Research and Development, AMED; by Grants in Aid for Scientific Research (B) (17H04205 to K.H.-I.), for Challenging Exploratory Research (16K15496 to K.H.-I.), for Young Scientists (B) (17K18061 to Y.M.), and for a JSPS fellow (18J00252 to Y.M.) from the Japan Society for the Promotion of Science (JSPS), Japan; and by the Takeda Science Foundation (to S.I.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00261-19.
REFERENCES
- 1.Wang KC, Chang HY. 2011. Molecular mechanisms of long noncoding RNAs. Mol Cell 43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopp F, Mendell JT. 2018. Functional classification and experimental dissection of long noncoding RNAs. Cell 172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt AM, Chang HY. 2016. Long noncoding RNAs in cancer pathways. Cancer Cell 29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitobe Y, Takayama KI, Horie-Inoue K, Inoue S. 2018. Prostate cancer-associated lncRNAs. Cancer Lett 418:159–166. doi: 10.1016/j.canlet.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Sun M, Gadad SS, Kim DS, Kraus WL. 2015. Discovery, annotation, and functional analysis of long noncoding RNAs controlling cell-cycle gene expression and proliferation in breast cancer cells. Mol Cell 59:698–711. doi: 10.1016/j.molcel.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y, Wei Y, Ma G, Park PK, Zhou J, Zhou Y, Hu Z, Zhou Y, Marks JR, Liang H, Hung M-C, Lin C, Yang L. 2016. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol 18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghoncheh M, Pournamdar Z, Salehiniya H. 2016. Incidence and mortality and epidemiology of breast cancer in the world. Asian Pac J Cancer Prev 17:43–46. doi: 10.7314/apjcp.2016.17.s3.43. [DOI] [PubMed] [Google Scholar]
- 11.Arnedos M, Vicier C, Loi S, Lefebvre C, Michiels S, Bonnefoi H, Andre F. 2015. Precision medicine for metastatic breast cancer–limitations and solutions. Nat Rev Clin Oncol 12:693–704. doi: 10.1038/nrclinonc.2015.123. [DOI] [PubMed] [Google Scholar]
- 12.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. 2011. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart CD, Migliaccio I, Malorni L, Guarducci C, Biganzoli L, Di Leo A. 2015. Challenges in the management of advanced, ER-positive, HER2-negative breast cancer. Nat Rev Clin Oncol 12:541–552. doi: 10.1038/nrclinonc.2015.99. [DOI] [PubMed] [Google Scholar]
- 14.Jeselsohn R, Bergholz JS, Pun M, Cornwell M, Liu W, Nardone A, Xiao T, Li W, Qiu X, Buchwalter G, Feiglin A, Abell-Hart K, Fei T, Rao P, Long H, Kwiatkowski N, Zhang T, Gray N, Melchers D, Houtman R, Liu XS, Cohen O, Wagle N, Winer EP, Zhao J, Brown M. 2018. Allele-specific chromatin recruitment and therapeutic vulnerabilities of ESR1 activating mutations. Cancer Cell 33:173–186. doi: 10.1016/j.ccell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musgrove EA, Sutherland RL. 2009. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 16.Stender JD, Nwachukwu JC, Kastrati I, Kim Y, Strid T, Yakir M, Srinivasan S, Nowak J, Izard T, Rangarajan ES, Carlson KE, Katzenellenbogen JA, Yao XQ, Grant BJ, Leong HS, Lin CY, Frasor J, Nettles KW, Glass CK. 2017. Structural and molecular mechanisms of cytokine-mediated endocrine resistance in human breast cancer cells. Mol Cell 65:1122–1135. doi: 10.1016/j.molcel.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oyama M, Nagashima T, Suzuki T, Kozuka-Hata H, Yumoto N, Shiraishi Y, Ikeda K, Kuroki Y, Gotoh N, Ishida T, Inoue S, Kitano H, Okada-Hatakeyama M. 2011. Integrated quantitative analysis of the phosphoproteome and transcriptome in tamoxifen-resistant breast cancer. J Biol Chem 286:818–829. doi: 10.1074/jbc.M110.156877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda K, Horie-Inoue K, Ueno T, Suzuki T, Sato W, Shigekawa T, Osaki A, Saeki T, Berezikov E, Mano H, Inoue S. 2015. miR-378a-3p modulates tamoxifen sensitivity in breast cancer MCF-7 cells through targeting GOLT1A. Sci Rep 5:13170. doi: 10.1038/srep13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ujihira T, Ikeda K, Suzuki T, Yamaga R, Sato W, Horie-Inoue K, Shigekawa T, Osaki A, Saeki T, Okamoto K, Takeda S, Inoue S. 2015. MicroRNA-574-3p, identified by microRNA library-based functional screening, modulates tamoxifen response in breast cancer. Sci Rep 5:7641. doi: 10.1038/srep07641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urano T, Saito T, Tsukui T, Fujita M, Hosoi T, Muramatsu M, Ouchi Y, Inoue S. 2002. Efp targets 14–3-3 sigma for proteolysis and promotes breast tumour growth. Nature 417:871–875. doi: 10.1038/nature00826. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda K, Orimo A, Higashi Y, Muramatsu M, Inoue S. 2000. Efp as a primary estrogen-responsive gene in human breast cancer. FEBS Lett 472:9–13. doi: 10.1016/s0014-5793(00)01421-6. [DOI] [PubMed] [Google Scholar]
- 22.Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, Li Z, Gala K, Fanning S, King TA, Hudis C, Chen D, Taran T, Hortobagyi G, Greene G, Berger M, Baselga J, Chandarlapaty S. 2013. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 45:1439–1445. doi: 10.1038/ng.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhan A, Mandal SS. 2016. Estradiol-induced transcriptional regulation of long non-coding RNA, HOTAIR. Methods Mol Biol 1366:395–412. doi: 10.1007/978-1-4939-3127-9_31. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. 2013. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, Akbani R, Bowlby R, Wong CK, Wiznerowicz M, Sanchez-Vega F, Robertson AG, Schneider BG, Lawrence MS, Noushmehr H, Malta TM, Stuart JM, Benz CC, Laird PW, Caesar-Johnson SJ, Demchok JA, Felau I, Kasapi M, Ferguson ML, Hutter CM, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Zhang JJ, Chudamani S, Liu J, Lolla L, Naresh R, Pihl T, Sun Q, Wan Y, Wu Y, Cho J, DeFreitas T, Frazer S, Gehlenborg N, Getz G, Heiman DI, Kim J, Lawrence MS, Lin P, Meier SN. 2018. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173:291–304. doi: 10.1016/J.Cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parise P, Finocchiaro G, Masciadri B, Quarto M, Francois S, Mancuso F, Muller H. 2006. Lap2alpha expression is controlled by E2f and deregulated in various human tumors. Cell Cycle 5:1331–1341. doi: 10.4161/cc.5.12.2833. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda K, Tsukui T, Horie-Inoue K, Inoue S. 2011. Conditional expression of constitutively active estrogen receptor α in osteoblasts increases bone mineral density in mice. FEBS Lett 585:1303–1309. doi: 10.1016/J.Febslet.2011.03.038. [DOI] [PubMed] [Google Scholar]
- 28.Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, Li S, Zhao JC, Yu J. 2016. LncRNA Hotair enhances Er signaling and confers tamoxifen resistance in breast cancer. Oncogene 35:2746–2755. doi: 10.1038/Onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hudson WH, Pickard MR, de Vera IM, Kuiper EG, Mourtada-Maarabouni M, Conn GL, Kojetin DJ, Williams GT, Ortlund EA. 2014. Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate. Nat Commun 5:5395. doi: 10.1038/Ncomms6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Pitchiaya S, Cieślik M, Niknafs YS, Tien JC, Hosono Y, Iyer MK, Yazdani S, Subramaniam S, Shukla SK, Jiang X, Wang L, Liu TY, Uhl M, Gawronski AR, Qiao Y, Xiao L, Dhanasekaran SM, Juckette KM, Kunju LP, Cao X, Patel U, Batish M, Shukla GC, Paulsen MT, Ljungman M, Jiang H, Mehra R, Backofen R, Sahinalp CS, Freier SM, Watt AT, Guo S, Wei JT, Feng FY, Malik R, Chinnaiyan AM. 2018. Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for Arlnc1 in prostate cancer progression. Nat Genet 50:814–824. doi: 10.1038/S41588-018-0120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Notas G, Pelekanou V, Kampa M, Alexakis K, Sfakianakis S, Laliotis A, Askoxilakis J, Tsentelierou E, Tzardi M, Tsapis A, Castanas E. 2015. Tamoxifen induces a pluripotency signature in breast cancer cells and human tumors. Mol Oncol 9:1744–1759. doi: 10.1016/J.Molonc.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang W, Su X, Yan W, Kong Z, Wang D, Huang Y, Zhai Q, Zhang X, Wu H, Li Y, Li T, Wan X. 2018. Overexpression of Ar-regulated lncRNA Tmpo-As1 correlates with tumor progression and poor prognosis in prostate cancer. Prostate 78:1248–1261. doi: 10.1002/Pros.23700. [DOI] [PubMed] [Google Scholar]
- 33.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW. 1999. A steroid receptor coactivator, Sra, functions As an RNA and is present in an Src-1 complex. Cell 97:17–27. doi: 10.1016/S0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 34.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, Debruyne FM, Ru N, Isaacs WB. 1999. Dd3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res 59:5975–5979. [PubMed] [Google Scholar]
- 35.Misawa A, Takayama K, Urano T, Inoue S. 2016. Androgen-induced long noncoding RNA (lncRNA) Socs2-As1 promotes cell growth and inhibits apoptosis in prostate cancer cells. J Biol Chem 291:17861–17880. doi: 10.1074/Jbc.M116.718536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takayama K, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, Ikeda K, Urano T, Fujimura T, Takagi K, Takahashi S, Homma Y, Ouchi Y, Aburatani H, Hayashizaki Y, Inoue S. 2013. Androgen-responsive long noncoding RNA Ctbp1-As promotes prostate cancer. EMBO J 32:1665–1680. doi: 10.1038/Emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guil S, Esteller M. 2015. RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem Sci 40:248–256. doi: 10.1016/J.Tibs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Sharma E, Sterne-Weiler T, O'Hanlon D, Blencowe BJ. 2016. Global mapping of human RNA-RNA interactions. Mol Cell 62:618–626. doi: 10.1016/j.molcel.2016.04.030. [DOI] [PubMed] [Google Scholar]
- 39.Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. 2014. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell 159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, Kenny PJ, Wahlestedt C. 2008. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong C, Maquat LE. 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. 2008. miR-206 expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res 68:5004–5008. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 43.Leivonen SK, Mäkelä R, Ostling P, Kohonen P, Haapa-Paananen S, Kleivi K, Enerly E, Aakula A, Hellström K, Sahlberg N, Kristensen VN, Børresen-Dale AL, Saviranta P, Perälä M, Kallioniemi O. 2009. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene 28:3926–3936. doi: 10.1038/onc.2009.241. [DOI] [PubMed] [Google Scholar]
- 44.Namekawa T, Ikeda K, Horie-Inoue K, Suzuki T, Okamoto K, Ichikawa T, Yano A, Kawakami S, Inoue S. 12 June 2019. ALDH1A1 in patient-derived bladder cancer spheroids activates retinoic acid signaling leading to TUBB3 overexpression and tumor progression. Int J Cancer. doi: 10.1002/ijc.32505. [DOI] [PubMed] [Google Scholar]
- 45.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. 2010. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 48.National Comprehensive Cancer Network. 2017. National comprehensive cancer network guidelines of treatment of cancer by site. https://www.nccn.org/professionals/physician_gls/f_guidelines.asp. Accessed 25 July 2017.
- 49.Sobin LH, Gospodarowicsz MK, Wittekind C. 2009. TNM classification of malignant tumors, 7th ed John Wiley & Sons, New York, NY. [Google Scholar]
- 50.Suzuki Y, Kitahara S, Suematsu T, Oshima M, Sato Y. 2017. Requisite role of vasohibin-2 in spontaneous gastric cancer formation and accumulation of cancer-associated fibroblasts. Cancer Sci 108:2342–2351. doi: 10.1111/cas.13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torres M, Becquet D, Guillen S, Boyer B, Moreno M, Blanchard MP, Franc JL, François-Bellan AM. 2018. RNA pull-down procedure to identify RNA targets of a long non-coding RNA. J Vis Exp 134:57379. doi: 10.3791/57379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All microarray data are available in the Gene Expression Omnibus (GEO) database with the accession number GSE129004.