Abstract
Few reports suggested a prognostic impact of Wilms‘Tumor-1 (WT1)-mRNA overexpression in MDS, but translation into clinical routine was hampered by limited patients numbers, differing sample sources, non-standardized methods/cut-offs. To evaluate whether WT1-mRNA expression yields additional prognostic information, we measured peripheral blood (PB) WT1-mRNA expression in 94 MDS using a standardized assay offering a validated cut-off to discriminate between normal and WT1-mRNA overexpression. Overall, 54 patients (57%) showed WT1-mRNA overexpression, while 40 patients (43%) had normal WT1-mRNA expression. This enabled discrimination between MDS and both healthy controls and non-MDS cytopenias. Furthermore, WT1-mRNA expression correlated with WHO 2016 subcategories and IPSS-R as indicated by mean WT1-mRNA expression and frequency of WT1-mRNA overexpressing patients within respective subgroups. Regarding the entire group, PB WT1-mRNA expression was associated with prognosis, as those patients showing WT1-mRNA overexpression had higher risk for disease progression and AML transformation and accordingly shorter progression-free, leukemia-free and overall survival in univariate analysis. In multivariate analysis, prognostic impact of PB WT1-mRNA expression status was independent of IPSS-R and enabled more precise prediction of PFS, but not OS, within IPSS-R very low/low and intermediate risk groups. Overall, measuring PB WT1-mRNA appears valuable to support diagnostics and refine prognostication provided by the IPSS-R.
Subject terms: Myelodysplastic syndrome, Cancer genetics
Introduction
Myelodysplastic syndromes (MDS) are a heterogeneous group of hematopoietic stem cell disorders characterized by hematopoietic insufficiency, variable signs of dysplasia and accumulation of immature precursors in the bone marrow (BM). The course of disease is highly variable and ranges from indolent appearance with almost normal life expectancy to conditions with rapid progression to a more advanced subtype or even acute myeloid leukemia (AML)1–4. To estimate the individual risk for disease progression, AML transformation and survival time scoring systems such as the international prognostic scoring system (IPSS) and its revised version (IPSS-R) have been established and are based on clinical variables such as cytopenias, BM blasts, and cytogenetics5,6. Taking advantage of the knowledge about several gene mutations recently unraveled in MDS the International Working Group for the Prognosis of MDS (IWG-PM) and others are currently trying to incorporate these in order to improve risk assessment7–10. Since this is still work in progress and standardized analyses and reporting of gene mutations is still lacking, we hypothesized that mRNA expression of the Wilms tumor 1 (WT1) gene may offer additional prognostic information in patients with MDS based on the following considerations: (1) Overexpression of WT1-mRNA has been reported in the about 50% of MDS patients and seems to correlate with disease stage and IPSS risk category11,12. (2) A limited number of reports has suggested a prognostic impact of WT1-mRNA expression patients with MDS, but their interpretation and translation into clinical routine are hampered by differing sample sources, non-standardized methods and cut-offs13–16. (3) A standardized, European Leukemia Net (ELN)-certified assay is available and enables measurement of WT1-mRNA expression in peripheral blood (PB) with a reproducible and validated cut-off to distinguish between normal and overexpression of WT1-mRNA17.
We here used this standardized assay to address the prognostic impact of PB WT1-mRNA expression in a cohort of 94 patients covering all common MDS subtypes.
Patients and methods
Patients and study design
Ninety-four patients (median age: 61.5 years, range 22–84 years) with newly diagnosed, treatment-naive MDS and available information about the peripheral blood (PB) WT1-mRNA expression level at the time diagnosis were included into this retrospective analysis. Among these, three patients (3%) suffered from MDS with single lineage dysplasia (MDS SLD) according to the World Health Organization (WHO) 2016 classification, one patient (1%) from MDS with ring sideroblasts and single lineage dysplasia (MDS RS SLD), 39 patients (41%) from MDS with multilineage dysplasia (MDS MLD), 7 patients (7%) from MDS del(5q), 3 patients (3%) from MDS unclassifiable (3%), 16 patients (17%) from MDS with excess blasts 1 (MDS EB1) and 25 patients (27%) from MDS with excess blasts 2 (MDS EB2). According to IPSS-R 3 (3%), 28 (30%), 28 (30%), 14 (15%), 21 (22%) patients belonged to very low, low, intermediate, high and very high-risk subgroup, respectively. Median follow-up of all patients was 16 months (range, 0.7–142.6 months). Patients proceeding to allogeneic stem cell transplantation (allo-SCT) were censored at transplantation. Excluding those patients, median follow-up was 23 months (range, 0.7–142.6 months). Data lock for this analysis was 15 July 2019. Detailed demographic and clinical characteristics are given in Table 1. All patients gave written informed consent according to Duesseldorf MDS registry and/or MDS biobank which were both approved by the local ethics committee (MDS-registry 3 973 and MDS-biobank 3768). In order to confirm the previously established assay specificity17 and furthermore to discriminate between healthy and malignant hematopoiesis we also analyzed samples of 12 healthy controls (HC) and of 17 non-MDS cytopenias diagnosed at our center with idiopathic cytopenia(s) of undetermined significance (ICUS) (n = 7), idiopathic thrombocytopenia (n = 2), toxic bone marrow failure (n = 2), renal anemia (n = 2), aplastic anemia (n = 2), anemia of chronic disease (n = 1) and cyclic neutropenia (n = 1) (supplementary Table 1).
Table 1.
Patient and clinical characteristics
| No. | % | |
|---|---|---|
| No. | 94 | |
| Median age, years (range) | 61.5 (22–84) | |
| Gender | ||
| Female | 33 | 35 |
| Male | 61 | 65 |
| Bone marrow blasts, median/range | 4 (0–18) | |
| Peripheral blasts, median/range | 0 (0–11) | |
| Leukocytes, median/range (/µl) | 3,3 × 103 (0,7–26 × 103) | |
| Hemoglobin, median range (g/dl) | 9,5 (4,8–14,8 103) | |
| Platelets, median/range (/µl) | 80 × 103 (6–670 × 103) | |
| WHO 2016 classification | ||
| MDS del5q | 7 | 7 |
| MDS-U | 3 | 3 |
| MDS SLD | 3 | 3 |
| MDS RS SLD | 1 | 1 |
| MDS MLD | 39 | 41 |
| MDS EB1 | 16 | 17 |
| MDS EB2 | 25 | 27 |
| Primary MDS | 86 | 91 |
| Therapy related MDS | 8 | 9 |
| IPSS-R | ||
| Very low | 3 | 3 |
| Low | 28 | 31 |
| Intermediate | 28 | 30 |
| High | 14 | 14 |
| Very high | 21 | 22 |
| Karyotype | ||
| Normal | 41 | 44 |
| Abnormal | 52 | 55 |
| Complex | 21 | 22 |
| Missing | 1 | 1 |
| Cytogenetic risk groupa | ||
| Very good | 4 | 4 |
| Good | 52 | 55 |
| Intermediate | 6 | 6 |
| Poor | 22 | 23 |
| Very poor | 9 | 10 |
| Missing | 1 | 1 |
| Presence of certain molecular mutationsb (ASXL1, EZH2, TET2, TP53, DNMT3A, RUNX1) | 24 | 26 |
| Treatment | ||
| Transfusion only | 6 | 7 |
| Growth factors | 22 | 23 |
| Lenalidomide | 7 | 7 |
| HMA | 10 | 11 |
| Intensive chemotherapy | 5 | 5 |
| Allogeneic stem-cell transplantation | 44 | 47 |
No. number, WHO World Health Organization, MDS SLD MDS with single lineage dysplasia, MDS RS SLD MDS with ring sideroblasts and single lineage dysplasia, MDS MLD MDS with multilineage dysplasia, MDS EB1 MDS with excess blasts 1, MDS EB2 MDS with excess of blasts 2, MDS del5q myelodysplastic syndrome with isolated del(5q), MDS-U myelodysplastic syndrome unclassifiable, HMA hypomethylating agents
aAccording to IPSS-R
bInformation based on results from clinical routine, but not on a comprehensive molecular analysis of all patients
Quantitative assessment of PB cell WT1 expression
Quantitative assessment of WT1-mRNA expression in PB mononuclear cells was performed at diagnosis using the Ipsogen® WT1 ProfilQuant® Kit in accordance to manufacturers’ instructions as previously described18. Briefly, after RNA extraction using the RNeasy Mini Kit (Qiagen, Hilden, Deutschland) 1 μg RNA was reversely transcribed. cDNA was then subjected to RT-qPCR reaction using primers and probes in accordance with the manufacturers’ instructions. All experiments were carried out in duplicate on a Rotor-Gene Q 5plex HRM instrument ABL served as control gene and results are expressed as ratio of WT1 copies per 104 ABL copies. In line with the quality control requirements of the manufacturer results from samples containing <4 246 ABL copies were discarded. Based on results from a large cohort of diagnostic AML and healthy samples this ELN certified, standardized, plasmid-based assay uses a validated PB cut-off level of 50 WT1 copies/104 ABL copies to distinguish between normal and overexpression of WT1-mRNA17. Therefore, in our analysis values above this cutoff were judged as overexpression. First, peripheral blood WT1-mRNA expression level was correlated with clinical parameters such as MDS WHO subtype 2016 and IPSS-R risk categories. Next, for the purpose of this analysis patients were dichotomized based on the validated cut-off level into those with normal expression (defined as <50 WT1 copies/104 ABL copies) and those with overexpression of WT1-mRNA (defined as >50 WT1 copies/104 ABL copies) and subsequently compared regarding their outcome in terms of progression-free survival (PFS), leukemia-free survival (LFS) and overall survival (OS).
Statistical analyses
OS was calculated as time from diagnosis to death from any cause or last follow-up in survivors. PFS was defined as time from diagnosis until (1) progression to a higher IPSS-R risk category, or (2) a higher subgroup according to WHO 2016, e.g., from non-blastic to blastic subgroup, (3) AML transformation or (4) death with those censored at last contact who were alive and had not progressed so far. All patients who underwent allo-SCT were censored at the time of allo-SCT. OS, PFS and LFS were estimated using Kaplan-Meier method using the log-rank test for univariate comparisons. For categorical variables frequencies were displayed and differences were estimated using cross tabulation and Fisher’s exact t-test as well as one-way ANOVA test, while for continuous variables medians (ranges) are given with the Mann-Whitney test employed to detect differences. Multivariate analysis was performed using the proportional hazard regression analysis (multiple Cox regression model). In all analyses, a p-value < 0.05 was considered to be statistically significant. Statistical analyses were performed using GraphPad Prism® 5.01 (GraphPad Software Inc., La Jolla, USA) and SPSS Statistic for Windows (SPSS Inc. Chicago, IL).
Results
Peripheral blood WT1-mRNA expression in patients with MDS according to WHO 2016 and IPSS-R
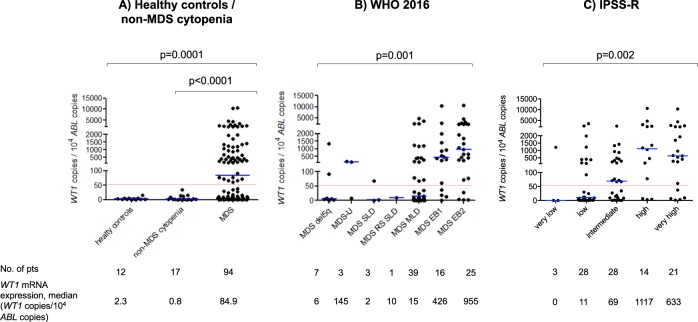
Median PB WT1-mRNA expression level of all patients was 84.9 WT1 copies/104 ABL copies (range, 0 to 10 589 WT1 copies/104 ABL copies) (supplementary Fig. 1). Overall, 40 patients (43%) had normal PB WT1-mRNA expression at diagnosis (median 4.5 WT1 copies/104 ABL copies, range 0–37.5 WT1 copies/104 ABL copies), whereas 54 patients (57%) showed overexpression of WT1-mRNA (median 759 WT1 copies/104 ABL copies, range 61-10589 WT1 copies/104 ABL copies) (supplementary Fig. 2). Thereby, PB WT1-mRNA overexpression enabled significant discrimination between MDS and HC (0/12, 0%, p < 0.0001) as well as between MDS and non-MDS cytopenias (0/17, 0%, p < 0.0001, see also Fig. 1a). In MDS patients, PB WT1-mRNA expression level strongly correlated with disease category according to WHO 2016 classification as indicated both by the frequency of patients with WT1-mRNA overexpression in the respective entities (MDS SLD: 1/3 patients, 33%; MDS MLD: 16/39 patients, 41%; MDS EB1 12/16 patients, 75%; MDS EB2: 21/25 patients, 84%; p = 0.003) and by the median PB WT1-mRNA expression level within each subcategory (MDS SLD: 1.8 WT1 copies/104 ABL copies; MDS MLD: 15 WT1 copies/104 ABL copies; MDS EB1 1: 426.4 WT1 copies/104 ABL copies; MDS EB2: 954.5 WT1 copies/104 ABL copies; p = 0.001, Fig. 1b; supplementary Fig. 3). In addition, PB WT1-mRNA expression significantly correlated with IPSS-R risk categories, again indicated by the frequency of patients with WT1-mRNA overexpression (IPSS-R very low: 1/3 patients, 33%; IPSS-R low: 10/28 patients, 36%; IPSS-R intermediate: 16/28, 57%; IPSS-R high: 11/14 patients, 79%; IPSS-R very high: 16/21, 76%; p = 0.02) as well as by the median WT1-mRNA expression level within the respective subcategories (IPSS-R very low: 0.0 WT1 copies/104 ABL copies; IPSS-R low: 10.8 WT1 copies/104 ABL copies; IPSS-R intermediate: 69.1 copies WT1 copies/104 ABL copies; IPSS-R high: 1117 WT1 copies/104 ABL copies; IPSS-R very high: 632.7 WT1 copies/104 ABL copies; p = 0.002, Fig. 1c; supplementary Fig. 4).
Fig. 1. Peripheral blood WT1-mRNA expression level.
a In patients with non-MDS cytopenia (n = 17) compared to patients with MDS and in 94 patients with MDS according to b WHO 2016 and c IPSS-R
Correlation of PB WT1-mRNA expression level with hematologic parameters
Next, we correlated PB WT1 expression with several hematologic parameters (Table 2). WT1-mRNA overexpression was significantly associated with decreased platelet and WBC count as well as with an increase of BM blast count (p = 0.04, 0.03 and 0.001, respectively), while no correlation with hemoglobin level was found (p = 0.46). Additionally, the presence of blasts in PB (p = 0.002) as well as an aberrant or complex karyotype (p = 0.02 and p = 0.03) were significantly linked to PB WT1-mRNA overexpression.
Table 2.
Correlation of WT1-mRNA expression and hematologic parameters
| WBC (x109/L) | Hb (g/dl) | PLT (×109/L) | PB blast count (%) | BM blast count (%) | Age (years) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT1 < 50* | 3.85 (0.7–17) | 0.0345 | 9.8 (4.8–14.8) | 0.4616 | 103 (9–540) | 0.0425 | 0 (0–5) | 0.0025 | 3 (0–17) | 0.0013 | 63.5 (45–84) | 0.0089 |
| WT1 > 50* | 2.7 (0.7–26) | 9.3 (6–13) | 67 (6–670) | 0 (0–11) | 6 (1–18) | 59 (22–77) |
| Gender (female/male) | Karyotype (normal/abnormal) | Karyotype (normal/komplex) | PB blasts (presence/absence) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT1 < 50a | 12 | 28 | 0.3923 | 23 | 16 | 0.0198 | 23 | 5 | 0.0296 | 1 | 37 | 0.0023 |
| WT1 > 50a | 21 | 33 | 18 | 36 | 18 | 15 | 13 | 34 | ||||
aCopies/104 ABL copies; values are presented as medians with ranges
p-value < 0.05 was considered to be statistically significant
No. number, WBC white blood cell count, Hb hemoglobin, PLT platelet count, PB peripheral blood, BM bone marrow
Prognostic impact of PB WT1-mRNA expression level in patients with MDS
Median PFS, LFS and OS for all patients were 28.9 months (range, 0.8–142.6 months; estimated 5-year PFS 27%, 95% CI: 14–39%), 30.8 months (range, 0.8–60.8 months; estimated 5-year LFS 44%, 95% CI: 27–64%) and 79.1 months (range, 0.8–142.6 months; estimated 5-year OS 58%, 95% CI: 44–74%) (supplementary Fig. 5). By comparing patients based on the WT1-mRNA expression status (overexpression, >50 WT1 copies/104 ABL copies vs. normal expression, <50 WT1 copies/104 ABL copies), we found that patients with WT1-mRNA overexpression had significantly higher frequency of disease progression compared to those with normal WT1-mRNA expression (WT1-mRNA overexpressing patients: 37 of 54 patients = 68.5% vs. patients with normal WT1-mRNA expression: 13 of 40 patients = 32.5%; p = 0.001). Accordingly, PFS was significantly lower in patients with WT1-mRNA overexpression in comparison to those with normal WT1-mRNA expression (median PFS: 18.2 months vs. not reached, p < 0.0001). Also the rate of patients who transformed into AML was higher in those exhibiting a WT1-mRNA overexpression compared to patients with normal pB WT1-mRNA expression (patients with normal WT1-mRNA expression: 2 of 13 patients, 15% vs. WT1-mRNA overexpressing patients: 18 of 37 patients, 48.6%; p = 0.0498). Consequently, LFS was significantly lower in patients with pB WT1-mRNA overexpressing compared to those with normal WT1-mRNA expression (median LFS: 18.2 months vs. not reached, p = 0.045). Regarding OS, we saw a similar trend in favor for patients with normal WT1-mRNA expression compared to those exhibiting pB WT1-mRNA overexpression, but without reaching statistical significance (median OS not reached vs. 79.10, p = 0.06) (Fig. 2). The differences regarding PFS and OS were statistically significant after excluding patients who underwent allo-SCT during the course of disease (Fig. 3). PB WT1-mRNA expression level on PFS retained its independent prognostic value also in multivariate analysis (p = 0.0001, Table 3).
Fig. 2. Outcome of patients with MDS based on WT1-mRNA expression.
a Progression free survival (PFS), b overall survival (OS), and c leukemia free survival (LFS)
Fig. 3. Outcome of patients with MDS based on WT1-mRNA expression excluding those patients proceeded to transplant.
a Progression free survival (PFS) and b overall survival (OS)
Table 3.
Prognostic impact of WT1-mRNA expression on outcome of patients with MDS in multivariate analysis
| Progression-free survival HR (95% CI) | p-value | Overall survival HR (95% CI) | p-value | |
|---|---|---|---|---|
| WT1-mRNA expression status (<50 vs. >50a) | 0.239 (0.152–0.566) | 0.0001 | 0.486 (0.208–1.136) | 0.096 |
| IPSS-R (very low/low vs. int/high/very high) | 0.640 (0.325–1.258) | 0.196 | 1.222 (0.875–1.705) | 0.240 |
aCopies/104 ABL copies
PB WT1-mRNA expression status can predict PFS in low and intermediate risk MDS
Having demonstrated that WT1-mRNA expression status correlated with IPSS-R category as well as survival and progression in the entire cohort, we then examined whether WT1-mRNA expression status may also refine the prediction of prognosis (OS and PFS) within each IPSS-R risk categories separately. Due to limitations regarding patient numbers in the very low and high-risk subgroups patients within these categories were summarized with patients of low and very high-risk group, respectively. Hereby, it became apparent that in the very low/low and intermediate risk subcategories PFS significantly differed between patients showing normal PB WT1-mRNA expression (IPSS-R very low/low, median PFS: not reached; IPSS-R intermediate, median PFS: 59.4 months) compared to patients exhibiting PB WT1-mRNA overexpression (IPSS-R very low/low, median PFS: 30.8 months; IPSS-R intermediate, median PFS: 7.8 months) (p = 0.047 for IPSS-R very low/low and p = 0.01 for IPSS-R intermediate, respectively; Fig. 4a, b). In contrast, no impact of WT1-mRNA expression status on PFS was found in patients within the IPSS-R high/very high-risk categories (Fig. 4c). Furthermore, no impact on OS was observed within any IPSS-R defined risk group. Overall, these data implied that in addition to a general prognostic impact in patients with MDS, WT1-mRNA expression seemed to refine the prediction of disease progression particularly in patients with IPSS-R very low/low and intermediate risk.
Fig. 4. Progression free survival (PFS) of IPSS-R subgroups based on WT1-mRNA expression.
a IPSS-R very low (n = 3) and low (n = 28) risk MDS, b intermediate risk MDS (n = 28), and c high (n = 14) and very high (n = 21) risk MDS
Discussion
Using a standardized ELN-certified assay we here showed that the transcription factor WT1 was overexpressed on mRNA level in 57% of patients with MDS and that PB WT1-mRNA overexpression strongly correlated with disease categories and risk stages according to WHO 2016 classification and IPSS-R respectively. Furthermore, our data indicated that PB WT1-mRNA expression status significantly correlated with prognosis of MDS patients with those patients showing WT1-mRNA overexpression having a higher risk for disease progression and AML transformation and accordingly shorter progression-free, leukemia-free and overall survival. This prognostic impact of PB WT1-mRNA expression was independent of the IPSS-R as confirmed by multivariate analysis. In further support of this, WT1-mRNA expression status enabled a more precise prediction of prognosis in terms of PFS in patients within the IPSS-R very low/low and intermediate risk groups.
Persisting cytopenias and signs of dysplasia in the BM are prerequisites to establish the diagnosis of MDS. Still, in particular if dysplastic features are subtle it is sometimes difficult even for trained hematologists to distinguish between MDS and reactive cytopenias or cytopenias related other bone marrow syndromes such as aplastic anemia19. Furthermore, even the detection of gene mutations such as DNMT3A, ASXL1, and TET2 may not be sufficient enough to accurately diagnose MDS, since these mutation can be found in approximately 10% of healthy individuals older than 65 years without evidence for a hematological malignancy summarized as “clonal hematopoiesis of indeterminate potential” (CHIP) as well as in patients with aplastic anemia20,21. In our analysis PB WT1-mRNA overexpression was found in 57% of patients with MDS, while the remaining 43% had normal WT1-mRNA expression. Still, WT1-mRNA overexpression nicely enabled discrimination between MDS and and non-MDS cytopenias, and this effect applied when looking at all MDS, but also when focusing on those with a BM blast count <5% (WT1-mRNA overexpression in non-MDS cytopenia 0/17 and WT1-mRNA overexpression in MDS < 5% BM blast count 26/53, p = 0.0001, see also supplementary Fig. 6). These data are in line with two previous reports22,23 and indicate that measurement of PB WT1-mRNA expression may serve as an easy accessible marker and helpful tool to differentiate between MDS and non-MDS cytopenias. In those patients with a definitive diagnosis of MDS it has been previously shown using an in-house assay that WT1-mRNA expression level, which was determined in BM in the majority of patients, correlated with disease category according French American Britain (FAB) classification and IPSS risk categories11. In our analysis, we confirm and expand this strong correlation of WT1-mRNA expression level measured in the PB to the WHO 2016 classification and to IPSS-R. For instance, in our cohort PB WT1-mRNA overexpression was detected at diagnosis in 68% of the patients with intermediate-, high- or very high risk according to IPSS-R, who generally represent potential candidates for allo-SCT. In contrast to this, individual mutations of the most frequently affected genes such as TET2 or ASXL1, which may be accessible for tracking by mutation-specific techniques, are present in only about 20 to 35% of patients. These findings suggested a greater informativeness and applicability of WT1-mRNA expression for MRD monitoring after allo-SCT. Indeed, supporting its clinical usefulness we and others have recently demonstrated that monitoring of WT1-mRNA expression levels in PB allows sensitive and specific detection of MRD after allo-SCT in a large proportion of MDS patients17,18,24. Furthermore, monitoring of disease and response kinetics by PB WT1-mRNA expression to guide therapeutic decisions may also be used in MDS patients treated with other therapies such as hypomethylating agents25 or proteasome inhibitors11,26.
Besides its value as diagnostic tool and MRD marker a limited number of reports suggested a prognostic impact of WT1-mRNA expression level on outcome of patients with MDS. However, interpretation and translation of these results into clinical practice has been hampered by a limited number of patient samples, varying sample sources (PB vs. BM) and the use of different assays without comparable cut-off levels13–16. Still, it would be of interest to correlate PB and BM WT1-mRNA expression levels in paired samples, a point we were not able to perform in our analysis due to insufficient number of paired BM samples. As a specific strength we here used a commercially available, ELN-certified assay, which offered a validated cut-off level to discriminate between normal and WT1-mRNA overexpression and thereby enabled reproducible and comparable analysis of WT1-mRNA expression in a standardized manner across different laboratories. Another advantage of this assay is that it facilitates measurement of WT1-mRNA in with a greater sensitivity and specificity than in BM thereby offering patient comfort.
In addition, our results showed that PB WT1-mRNA expression at diagnosis represented a new prognostic factor in MDS patients which was independent from the IPSS-R and exhibited additional prognostic information. This was true when analysing the entire MDS cohort (PFS, LFS), but also enabled us to more precisely predict PFS in patients with IPSS-R very low/low and intermediate risk. Even though patients may fall in one of the same of these categories their course of disease may vary individually ranging from indolent conditions with near normal life expectancy to those with rapid progression to AML. Thus, refinement of the prognosis by the addition of WT1-mRNA expression status may help to better stratify patients and tailor surveillance strategies or even support individual treatment decisions, for example to consider allo-SCT at an earlier time point.
A similar approach like ours to improve IPSS-R-based risk stratification by integrating molecular markers is the subject of an on-going initiative of the IWG-PM which tries to incorporate the prognostic information of somatic mutations into the IPSS-R. Results reported so far showed that inclusion of gene mutations improved assessment of prognosis in MDS7–10. However, they also indicated that screening procedures for somatic mutations is a complex issue, integrating numerous mutations, e.g., up to 13 in the analysis of the IWG-PM27, and is thereby time- and resource-intensive. In addition, the limited access to molecular diagnostics at least in some regions, lack of standardized analyses and reporting, the need of BM as optimal sample source and the limited frequency of individual mutations also hampers the broad application of gene mutation testing in the entire MDS population. Given the intrinsic limitation of a retrospective analysis information on molecular aberrations at primary diagnosis was only available for a limited proportion of patients impeding inclusion of molecular data into uni- and/or multivariate outcome analyses.
In summary, measurement of PB WT1-mRNA expression as single molecular marker using a standardized, ELN-certified assay with reproducible cut-offs represented a clinically valuable, patient friendly and resource-effective supplementation in diagnostic work up and refinement of prognostic information provided by the IPSS-R.
Supplementary information
Acknowledgements
T.S., C.R., M.L., S.P. and C.F. performed the research. T.S. and G.K. designed the research study. C.R., U.G., M.L., S.P., C.F., P.J., S.G., G.K. and T.S. contributed essential data. C.R., U.G., G.K. and T.S. analyzed the data. C.R., U.G., G.K. and T.S. wrote the paper. All authors critically revised the manuscript.
Conflict of interest
A Rotorgene PCR cycler was provided free of charge by Qiagen for routine use in this study. No other financial or logistic support was provided by the company for this retrospective analysis.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41408-019-0248-y).
References
- 1.Montalban‐Bravo G, Garcia‐Manero G. Myelodysplastic syndromes: 2018 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2018;93:129–147. doi: 10.1002/ajh.24930. [DOI] [PubMed] [Google Scholar]
- 2.Gangat N, Patnaik MM, Tefferi A. Myelodysplastic syndromes: contemporary review and how we treat. Am. J. Hematol. 2016;91:76–89. doi: 10.1002/ajh.24253. [DOI] [PubMed] [Google Scholar]
- 3.Germing U, Kobbe G, Haas R, Gattermann N. Myelodysplastic syndromes: diagnosis, prognosis, and treatment. Dtsch Arztebl Int. 2013;110:783–790. doi: 10.3238/arztebl.2013.0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Germing U, Kündgen A. Prognostic scoring systems in MDS. Leuk. Res. 2012;36:1463–1469. doi: 10.1016/j.leukres.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Greenberg P, et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. doi: 10.1182/blood.V89.6.2079. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg PL, et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120:2454–2465. doi: 10.1182/blood-2012-03-420489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bejar R, et al. Clinical effect of point mutations in myelodysplastic syndromes. New England J. Medi. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bejar R, et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J. Clin. Oncol. 2014;32:2691–2698. doi: 10.1200/JCO.2013.52.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papaemmanuil E, et al. Genomic classification and prognosis in acute myeloid leukemia. New England J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nazha A, et al. A personalized prediction model to risk stratify patients with myelodysplastic syndromes. Blood. 2018;132:793. doi: 10.1182/blood-2018-99-114774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cilloni D, et al. Significant correlation between the degree of WT1 expression and the International Prognostic Scoring System Score in patients with myelodysplastic syndromes. J. Clin. Oncol. 2003;21:1988–1995. doi: 10.1200/JCO.2003.10.503. [DOI] [PubMed] [Google Scholar]
- 12.Ueda Y, et al. Clinical evaluation of WT1 mRNA expression levels in peripheral blood and bone marrow in patients with myelodysplastic syndromes. Leuk. Lymphoma. 2013;54:1450–1458. doi: 10.3109/10428194.2012.745074. [DOI] [PubMed] [Google Scholar]
- 13.Tamaki H, et al. The Wilms’ tumor gene WT1 is a good marker for diagnosis of disease progression of myelodysplastic syndromes. Leukemia. 1999;13:393–399. doi: 10.1038/sj.leu.2401341. [DOI] [PubMed] [Google Scholar]
- 14.Tamura H, et al. Prognostic significance of WT1 mRNA and anti-WT1 antibody levels in peripheral blood in patients with myelodysplastic syndromes. Leuk. Res. 2010;34:986–990. doi: 10.1016/j.leukres.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi S, et al. Prognostic significance of Wilms tumor 1 mRNA expression levels in peripheral blood and bone marrow in patients with myelodysplastic syndromes. Cancer Biomark. 2016;17:21–32. doi: 10.3233/CBM-160612. [DOI] [PubMed] [Google Scholar]
- 16.Nagasaki J, et al. Wilms Tumor 1 (WT1) mRNA expression level at diagnosis is a significant prognostic marker in elderly patients with Myelodysplastic Syndrome. AHA. 2017;137:32–39. doi: 10.1159/000452732. [DOI] [PubMed] [Google Scholar]
- 17.Cilloni D, et al. Real-time quantitative polymerase chain reaction detection of minimal residual disease by standardized WT1 assay to enhance risk stratification in acute myeloid leukemia: a European LeukemiaNet study. J. Clin. Oncol. 2009;27:5195–5201. doi: 10.1200/JCO.2009.22.4865. [DOI] [PubMed] [Google Scholar]
- 18.Rautenberg C, et al. Wilms’ Tumor 1 gene expression using a standardized european leukemianet-certified assay compared to other methods for detection of minimal residual disease in myelodysplastic syndrome and acute myelogenous leukemia after allogeneic blood stem cell transplantation. Biol. Blood Marrow Transplant. 2018;24:2337–2343. doi: 10.1016/j.bbmt.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Valent P, et al. Proposed minimal diagnostic criteria for myelodysplastic syndromes (MDS) and potential pre-MDS conditions. Oncotarget. 2017;8:73483–73500. doi: 10.18632/oncotarget.19008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malcovati L, et al. Clinical significance of somatic mutation in unexplained blood cytopenia. Blood. 2017;129:3371–3378. doi: 10.1182/blood-2017-01-763425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valent P. ICUS, IDUS, CHIP and CCUS: Diagnostic Criteria, Separation from MDS and Clinical Implications. Pathobiology. 2019;86:30–38. doi: 10.1159/000489042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamauchi T, et al. Wilms’ tumor-1 transcript in peripheral blood helps diagnose acute myeloid leukemia and myelodysplastic syndrome in patients with pancytopenia. Anticancer Res. 2012;32:4479–4483. [PubMed] [Google Scholar]
- 23.Baba M, et al. The level of bone marrow WT1 message is a useful marker to differentiate myelodysplastic syndromes with low blast percentage from cytopenia due to other reasons. Intern. Med. 2015;54:445–451. doi: 10.2169/internalmedicine.54.3123. [DOI] [PubMed] [Google Scholar]
- 24.Lange T, et al. Monitoring of WT1 expression in PB and CD34(+) donor chimerism of BM predicts early relapse in AML and MDS patients after hematopoietic cell transplantation with reduced-intensity conditioning. Leukemia. 2011;25:498–505. doi: 10.1038/leu.2010.283. [DOI] [PubMed] [Google Scholar]
- 25.Du X, et al. WT1 mRNA level reflects disease changes and progression of myelodysplastic syndromes patients with ‘stable disease’. Br. J. Haematol. 2019;184:447–450. doi: 10.1111/bjh.15098. [DOI] [PubMed] [Google Scholar]
- 26.Alimena G, et al. Erythroid response and decrease of WT1 expression after proteasome inhibition by bortezomib in myelodysplastic syndromes. Leuk. Res. 2011;35:504–507. doi: 10.1016/j.leukres.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 27.Bejar R, et al. Somatic Mutations in MDS Patients Are Associated with Clinical Features and Predict Prognosis Independent of the IPSS-R: Analysis of Combined Datasets from the International Working Group for Prognosis in MDS-Molecular Committee. Blood. 2015;126:907. doi: 10.1182/blood.V126.23.907.907. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.