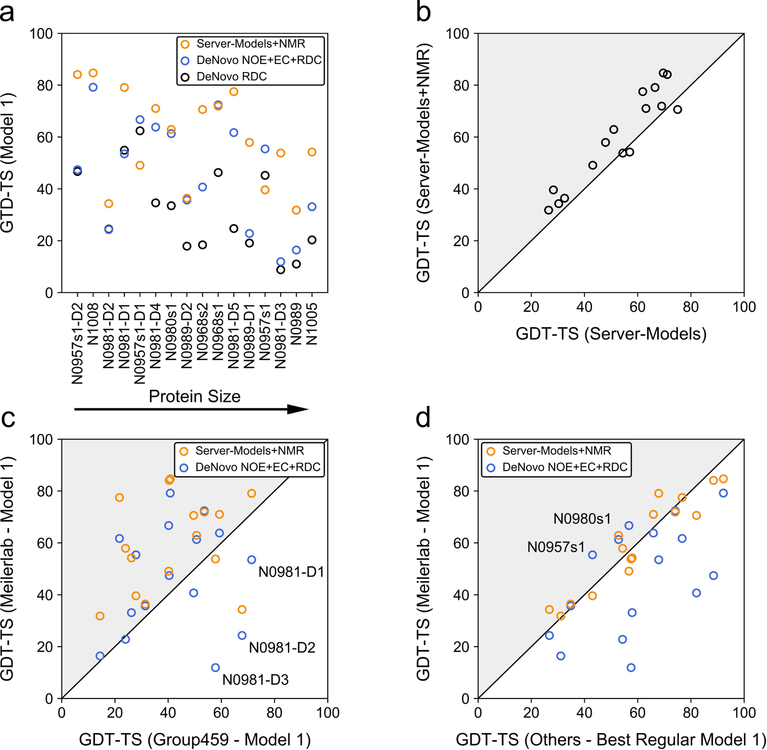
Figure 2. Overview of model GDT-TS of Meilerlab models obtained with different restraint sets and comparison to other structure predictions in the NMR-assisted and free modeling category.
(a) GDT-TS of de novo predicted models created in CASP13 using only RDCs or NOEs, ECs and RDCs, respectively. In addition, the GDT-TS obtained when modeling started from server-models and employed an iterative refinement with NOEs, ECs, and RDCs, which was investigated in our post-CASP13 analysis, is shown. (b) Improvement of server-models through refinement with NMR data. The GDT-TS of the best model among all submissions of five different servers (Robetta35, I-TASSER36,37, QUARK38, RaptorX-Contact39, RaptorX-TBM40) (x-axis) is compared to the GDT-TS of our best-scoring model after hybridization and NMR-refinement of the respective server-models (y-axis). (c) Comparison of GDT-TS of the Meilerlab submitted Model 1 and NMR-refined server-models to Model 1 created by the ‘baseline’ group (Group 459) in the NMR-assisted category. Gray triangles indicate an improvement of the GDT-TS. Targets for which the Meilerlab Model 1 was significantly less accurate (GDT-TS difference > 10) then Model 1 from Group 459 are labeled. (d) Comparison of GDT-TS of the Meilerlab submitted Model 1 and NMR-refined server-models to the best Model 1 in the regular unassisted modeling category.