Abstract
Protein target structures for the Critical Assessment of Structure Prediction round 13 (CASP13) were split into evaluation units (EUs) based on their structural domains, the domain organization of available templates, and the performance of servers on whole targets compared to split target domains. 80 targets were split into 112 EUs. The EUs were classified into categories suitable for assessment of high accuracy modeling (or template-based modeling, TBM) and topology (or free modeling, FM) based on target difficulty. Assignment into assessment categories considered the following criteria: 1) the evolutionary relationship of target domains to existing fold space as defined by the Evolutionary Classification of Protein Domains (ECOD) database; 2) the clustering of target domains using eight objective sequence, structure, and performance measures and 3) the placement of target domains in a scatter plot of target difficulty against server performance used in the previous CASP. Generally, target domains with good server predictions had close template homologs and were classified as TBM. Alternately, targets with poor server predictions represent a mixture of fast evolving homologs, structure analogs, and new folds, and were classified as FM or FM/TBM overlap.
Keywords: protein structure, CASP13, classification, fold space, sequence homologs, structure analogs, free modeling, template-based modeling, structure prediction
INTRODUCTION
The Critical Assessment of Structure Prediction (CASP) aims to assess the current state of the art in protein structure modeling methods1, 2. During the prediction timeframe, CASP provides amino acid sequences to participants for modeling targets whose experimental structures are not yet public. Independent assessors evaluate the performance of automated servers and expert groups based on the similarity of their models to experimental structures. In the current round (CASP13), several modeling categories addressed multiple aspects of protein structure prediction; including the detailed positioning of atoms in the high accuracy modeling category, the accuracy of model topologies in the topology category, and the ability to assemble domain and protein complexes.
To more accurately assess tertiary structure predictions, CASP has traditionally split targets into Evaluation Units (EUs) and categorized the resulting EUs based on their difficulty3–5. While the exact criteria used to evaluate such target difficulty has differed slightly throughout the course of CASP experiments, category assignment has largely been dictated by the ability to detect known structure templates from target protein sequence. As such, knowledge of sequence-structure relationships in existing fold space catalogued in databases such as the Evolutionary Classification of Protein Domains (ECOD6) combined with sequence-structure similarity and performance metrics computed by the Prediction Center7 provide a solid basis for target categorization.
The experimental protein structure community contributed 90 single-sequence target structures used in CASP13 (designated T0949-T1022), including subunits of several multi-protein complexes (labeled with subunit captions, e.g. T1022s1 and T1022s2). CASP organizers designated some targets with high sequence similarity to existing templates as “server only” (8 targets), with the rest being released to all groups. Ten targets were cancelled for various reasons, including lack of structure coordinates (8 targets), premature release of the structure paper (1 target), and being identical to a target from CASP12 (1 target). The remaining 80 targets were evaluated. One structure (T0950) originally released for all-group prediction was redefined as “server only” after premature release of paper prior to the expert deadline.
This article describes the procedure used to split CASP13 targets into EUs and assign them into two categories: High Accuracy Modeling (a.k.a. Template Based Modeling, TBM), requiring models of sufficiently high quality to carry out detailed analysis of atom positions, and Topology (a.k.a. Free Modeling, FM), evaluating placement of secondary structure elements (SSEs) in models with lower accuracy. CASP13 target assignment utilized the same semi-automated objective metrics as in CASP123. The category boundary and difficult intermediate cases were decided by clustering CASP12 metrics with additional scores for alignment depth (Neff), which has previously contributed to model accuracy of top performing non-template-based methods1, 37, 38. Table 1 summarizes the evolutionary relationship of CASP13 targets to known folds that guided classification.
TABLE I.
Evolutionary Assignment of CASP13 Targets among Existing Folds
| Target EU | Class | Template | Architecture* | X Group/H Group Name* | Level* |
|---|---|---|---|---|---|
| T0949 | FM/TBM | 3t9wA2 | beta sandwiches | Cupredoxin-related | H-group |
| T0950 | FM | 2nrjA | alpha bundles | Bacterial hemolysins | H-group |
| T0951 | TBM-easy | 5cbkA | a/b three-layered sandwiches | alpha/beta-Hydrolases | F-group |
| T0953s1 | FM | 4ru3A2 | beta duplicates or obligate multimers | Phage tail fiber protein trimerization domain | H-group |
| T0953s2-D1 | FM/TBM | 3v9uD | alpha arrays | LEM/SAP HeH motif-like | X-group |
| T0953s2-D2 | FM | 4pmhA | beta duplicates or obligate multimers | Pectin lyase-like | H-group |
| T0953s2-D3 | FM | 4o47A4 | fibers | N/A | New Fold |
| T0954 | TBM-hard | 2o9kA | beta duplicates or obligate multimers | beta-propeller | F-group |
| T0955 | FM/TBM | 2ikfA4 | a+b two layers | engineered | Analog |
| T0957s1-D1 | FM | 5nwmA/5mjeA1 | a+b two layers | RelE-like | H-group |
| T0957s1-D2 | TBM-hard | 6cud4 | see above | see above | Analog |
| T0957s2 | FM | 5mu7A | alpha superhelices | Repetitive alpha hairpins | X-group |
| T0958 | FM/TBM | 5fd6A | alpha arrays | HTH | F-group |
| T0959 | TBM-hard | 2is5A | a+b complex topology | Lysozyme-like | F-group |
| T0960-D2 | FM | 5m9fA | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | H-group |
| T0960-D3 | TBM-hard | 5efvB | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | H-group |
| T0960-D5 | TBM-easy | 4mtmA | beta sandwiches | Agglutinin HPA-like | H-group |
| 4y9lA2 | alpha arrays | Acyl-CoA dehydrogenase N-terminal domain-like | |||
| T0961 | TBM-easy | 4y9lA3 | beta complex topology | Acyl-CoA dehydrogenase middle domain-like | F-group |
| 4y9lA1 | alpha bundles | Acyl-CoA dehydrogenase C-terminal domain-like | |||
| T0962 | TBM-easy | 4ok7A | a+b complex topology | Lysozyme-like | F-group |
| T0963-D2 | FM | 5m9fA | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | H-group |
| T0963-D3 | TBM-hard | 5efvC | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | H-group |
| T0963-D5 | TBM-easy | 4mtmA | beta sandwiches | Agglutinin HPA-like | H-group |
| T0964 | TBM-hard | 5t7aA | beta sandwiches | Immunoglobulin-related | F-group |
| T0965 | TBM-hard | 1orrA | a/b three-layered sandwiches | Rossmann-related | H-group |
| T0966 | TBM-hard | 2vkdA | alpha bundles | PMT helical bundle domain-like | F-group |
| 2ec5A2 | alpha bundles | PMT central region-like | |||
| a+b two layers | PMT central region-like | ||||
| 2ec5A4 | a/b three-layered sandwiches | EreA/ChaN-like | |||
| T0967 | TBM-easy | 5ho1B | a+b two layers | Cation efflux protein cytoplasmic domain-like | F-group |
| T0968s1 | FM | 3wz3A2/5mjeA1 | a+b two layers | RelE-like | H-group |
| T0968s2 | FM | 5gkfB4 | beta sandwiches | N/A | New Fold |
| T0969 | FM | 2hsjA | a/b three-layered sandwiches | SGNH hydrolase | H-group |
| T0970 | FM/TBM | 1jg5A | a+b two layers | GTP cyclohydrolase I feedback regulatory protein, GFRP | H-group |
| T0971 | TBM-easy | 3ebtA | a+b two layers | NTF2-like | F-group |
| T0973 | TBM-easy | 2vf9C | a+b two layers | RNA bacteriophage capsid protein | H-group |
| T0974s1 | TBM-easy | 5woqA | alpha arrays | HTH | H-group |
| T0975 | FM | 5eaxB6 | a/b three-layered sandwiches | Restriction endonucleaselike | H-group |
| T0976-D1 | TBM-easy | 2hhgA | a/b three-layered sandwiches | Flavoproteins/Phospho-tyrosine protein phosphatases-like | F-group |
| T0976-D2 | TBM-easy | 2hhgA | a/b three-layered sandwiches | Flavoproteins/Phospho-tyrosine protein phosphatases-like | F-group |
| T0977-D1 | TBM-easy | 5efvB2 | beta duplicates or obligate multimers | beta-propeller | F-group |
| T0977-D2 | TBM-easy | 5efvB3 | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | F-group |
| 5efvB4 | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | |||
| 1j2bA3 | a/b barrels | TIM barrels | |||
| T0978 | FM/TBM | 1j2bA2 | few secondary structure elements | Zinc binding | F-group |
| T0979 | TBM-hard | 5apzA | fibers | trimeric fiber | Analog |
| T0980s1 | FM | 5mjeA | a+b two layers | RelE-like | H-group |
| T0981-D1 | TBM-hard | 5m9fB | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | H-group |
| T0981-D2 | FM | 5efvA | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | H-group |
| T0981-D3 | FM/TBM | 2zexB | beta sandwiches | Concanavalin A-like | F-group |
| T0981-D4 | TBM-hard | 5m9fA1 | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | F-group |
| T0981-D5 | TBM-hard | 5m9fC2 | beta sandwiches | Immunoglobulin-related | F-group |
| T0982-D1 | TBM-easy | 3q63B | a+b two layers | Bet v1-like | F-group |
| T0982-D2 | TBM-hard | 3tfzB | a+b two layers | Bet v1-like | H-group |
| T0983 | TBM-easy | 4oqdC | a/b three-layered sandwiches | Rossmann-related | F-group |
| T0984-D1 | TBM-easy | 6c9aA1 | alpha complex topology | Voltage-gated ion channels | F-group |
| T0984-D2 | TBM-easy | 6c9aA2 | alpha complex topology | Voltage-gated ion channels | F-group |
| 3afjB2 | beta sandwiches | supersandwich | |||
| 3afjB1 | alpha superhelices | alpha/alpha toroid | |||
| T0985 | TBM-hard | 3afjB3 | beta sandwiches | Glycoside hydrolase family 127 middle domain-related | F-group |
| T0986s1 | FM/TBM | 1tfoA | a+b two layers | Colicin D nuclease domain | H-group |
| T0986s2 | FM | 3al0A12 | a+b two layers | Glutamine synthetase-like | X-group |
| T0987-D1 | FM | 5mcvB | beta sandwiches | Immunoglobulin-like beta-sandwich | X-group |
| T0987-D2 | FM | 5mkdA | beta sandwiches | Immunoglobulin-like beta-sandwich | X-group |
| T0989-D1 | FM | 4uxgK | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | H-group |
| T0989-D2 | FM | 5m9fC1 | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | H-group |
| T0990-D1 | FM | 2rt6A4 | few secondary structure elements | N/A | New Fold |
| T0990-D2 | FM | 5k7vB4 | a+b complex topology | N/A | New Fold |
| T0990-D3 | FM | 5graA2,4 | a+b complex topology | N/A | New Fold |
| T0991 | FM | 5fs4B | a+b two layers | RNA bacteriophage capsid protein | H-group |
| T0992 | FM/TBM | 3uc2C | beta sandwiches | Immunoglobulin-like beta-sandwich | X-group |
| T0993s1 | TBM-easy | 3tuzH2 | a/b three-layered sandwiches | P-loop domains-related | F-group |
| T0993s2 | TBM-easy | 4hylB | a/b three-layered sandwiches | SpoIIaa-like | H-group |
| T0995 | TBM-easy | 3wuyB | a+b four layers | Carbon-nitrogen hydrolase | F-group |
| T0996-D1 | TBM-easy | 5uvnF1 | beta barrels | RIFT-related | F-group |
| T0996-D2 | TBM-easy | 5uvnA2 | beta barrels | RIFT-related | F-group |
| T0996-D3 | TBM-easy | 5uvnB1 | beta barrels | RIFT-related | F-group |
| T0996-D4 | TBM-easy | 5uvnC2 | beta barrels | RIFT-related | F-group |
| T0996-D5 | TBM-easy | 5uvnD2 | beta barrels | RIFT-related | F-group |
| T0996-D6 | TBM-easy | 5uvnB1 | beta barrels | RIFT-related | F-group |
| T0996-D7 | TBM-easy | 5uvnB2 | beta barrels | RIFT-related | F-group |
| T0997 | FM/TBM | 4xzzA2 | beta complex topology | L,D-transpeptidase catalytic domain-like | H-group |
| T0998 | FM | 5tc1H | a+b two layers | RNA bacteriophage capsid protein | H-group |
| T0999-D2 | TBM-hard | 5xwbA1 | a+b two layers | EPT/RTPC-like | F-group |
| 5xwbA2 | a+b two layers | EPT/RTPC-like | |||
| T0999-D3 | TBM-easy | 4bqsA | a/b three-layered sandwiches | P-loop domains-related | F-group |
| T0999-D4 | TBM-easy | 5swuB | a/b barrels | TIM barrels | F-group |
| T0999-D5 | TBM-easy | 1wxdB2 | a/b three-layered sandwiches | Class I glutamine amidotransferase-like | F-group |
| 1wxdB1 | a/b three-layered sandwiches | Rossmann-related | |||
| T1000-D2 | FM | 3edcA2 | a/b three-layered sandwiches | Class I glutamine amidotransferase-like | H-group |
| T1001 | FM | 5nfxA2 | a+b three layers | Sensor domains | H-group |
| T1002-D1 | TBM-easy | 4krtA2 | beta barrels | SH3 | F-group |
| T1002-D2 | TBM-easy | 4krtA3 | beta barrels | SH3 | F-group |
| T1002-D3 | TBM-easy | 4xxtA2 | beta complex topology | L,D-transpeptidase catalytic domain-like | F-group |
| T1003 | TBM-easy | 5txrB1 | a/b three-layered sandwiches | PLP-dependent transferases | F-group |
| 5txrB2 | a+b two layers | C-terminal domain in some PLP-dependent transferases | |||
| T1004-D1 | TBM-easy | 5efvA | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | H-group |
| T1004-D2 | TBM-easy | 6cl6B | a+b two layers | Phi ETA orf 56-like protein C-terminal domains | H-group |
| T1005 | FM/TBM | 4nuzA1 | a/b barrels | TIM barrels | H-group |
| T1006 | TBM-easy | 3w62A | a+b two layers | Cation efflux protein cytoplasmic domain-like | F-group |
| T1008 | FM/TBM | 5hnwK4 | a+b two layers | engineered | Analog |
| 4ba0A1 | beta sandwiches | supersandwich | |||
| T1009 | TBM-hard | 4ba0A2 | a/b barrels | TIM barrels | F-group |
| 4ba0A3 | beta sandwiches | Glycosyl hydrolase domain | |||
| T1010 | FM | 1bxwA/3g7gA3 | beta barrels | Lipocalins/Streptavidin | X-group |
| T1011-D1 | TBM-hard | 5te3A | alpha bundles | Family A G protein-coupled receptor-like | F-group |
| T1013 | TBM-easy | 4rwdA2 | alpha bundles | Family A G protein-coupled receptor-like | F-group |
| T1014-D1 | TBM-easy | 4javB2 | a+b two layers | ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase | F-group |
| T1014-D2 | TBM-easy | 1a2oB1 | a/b three-layered sandwiches | Class I glutamine amidotransferase-like | H-group |
| T1015s1 | FM | 1wkbA1 | few secondary structure elements | Rubredoxin-like | X-group |
| T1015s2 | TBM-hard | 4g6vA | a/b three-layered sandwiches | Restriction endonuclease-like | F-group |
| T1016 | TBM-easy | 4ij5B | a/b three-layered sandwiches | Phosphoglycerate mutase-like | F-group |
| T1017s1 | TBM-easy | 2o5hA | alpha arrays | NMB0513-like | F-group |
| T1017s2 | FM | 4u03A | a+b two layers | Nucleotidyltransferase-like | X-group |
| T1018 | TBM-easy | 1uipA | a/b barrels | TIM barrels | F-group |
| T1019s1 | FM/TBM | 2gjpA/3wwlA3 | few secondary structure elements | Rubredoxin-like | X-group |
| T1019s2 | TBM-easy | 1v74A | a+b two layers | Colicin D nuclease domain | H-group |
| T1020 | TBM-easy | 3m75A | alpha superhelices | Anion channel SLAC1-related | F-group |
| T1021s1 | TBM-hard | 5w5eK | beta barrels | RIFT-related | H-group |
| T1021s2 | TBM-hard | 3j9qm1 | a/b three-layered sandwiches | Domain III of tail sheath protein Gp18 | F-group |
| 3j9qm2 | a+b duplicates or obligate multimers | gpW/gp25-like | |||
| T1021s3-D1 | FM | 3j2mE | a+b two layers | Phage tail protein-like | H-group |
| T1021s3-D2 | FM | 5u0aG/4acvA1 | a+b two layers | Phage tail protein-like | H-group |
| T1022s1-D1 | FM | beta barrels | RIFT-related | H-group | |
| T1022s1-D2 | TBM-hard | 5jceB2 | alpha arrays | LysM domain | H-group |
| 4mtkC4 | beta barrels | RIFT-related | |||
| 4mtkC3 | a+b complex topology | N0 domain in phage tail proteins and secretins | |||
| 4mtkC5 | beta barrels | RIFT-related | |||
| T1022s2 | TBM-hard | 4mtkC7 | a+b complex topology | C-terminal insertion domain in phage tail proteins | F-group |
| 4mtkC2 | beta barrels | Tail-associated lysozyme gp5-N | |||
| 4mtkC6 | beta duplicates or obligate multimers | Phage tail fiber protein trimerization domain |
Domains were assigned to the ECOD hierarchy: Architecture retains similar secondary structure compositions and geometric shapes, X-groups display similar topology but lack justification for homology, and H-groups include homologous folds.
alternate homologous template
alternate increased coverage template
alternate correct topology template
top template analog
METHODS
Defining Evaluation Units
The Prediction Center preprocessed coordinate files of target structures as previously described7. Targets were split into domains using DomainParser29 and Ddomain10 packages. Automatic domain boundaries were inspected manually considering a number of criteria for establishing boundaries; including compactness of secondary structure elements, internal duplications, sequence continuity, and sequence-structure relationships to known folds (using HHpred11 and LGA12 alignments provided by the Prediction Center). The resulting domains were tested for the need to split into EUs based on GDT-TS13 performance of server models using Grishin plots14. CASP targets were not split if servers performed similarly on merged and split domains, and templates with similar domain orientations existed. Traditionally, Grishin plots evaluate the performance of server models designated as “1”. For several CASP13 target splits, model 1 server performance was not linear for plots of whole-target evaluation scores against weighted domain-based scores. As such, Grishin plots for CASP13 targets were re-evaluated using all server models and a few targets were designated in a special category (FM_sp) for consideration of domain interaction. To keep EUs as large as possible and to reward correct inter-domain orientations, other borderline cases with nonlinear plots were merged. All in all, 80 targets were split into 112 EUs.
Mapping EUs to ECOD
Sequences and structures of each defined EU were compared to template domains in the ECOD database. Family level assignments were made using the Conserved Domain search (CD-search15) against the CDD database16. Sequences corresponding to top templates identified by the Prediction Center (HHpred11 or LGA12) were also submitted as queries against the CDD database16. Resulting hits were compared to hits using the target sequences as queries, prioritizing hits from PFAM17, but considering hits from alternate databases where there were no confident PFAM assignments (using default parameters). For cases of EUs without confident family assignments, we compared their topologies to those of top scoring HHpred hits. Topology comparisons were aided by structure superpositions using DaliLite and prioritized 1) consistent alignments made by both HHpred and Dali and 2) similar evolutionary cores defined as the SSEs that are common to all structures belonging to the template H-group. Top scoring LGA templates (according to the LGA_S score) were chosen for Table 1 unless otherwise indicated. For some cases where the top template was a structure analog, a lower scoring homologous LGA template was chosen. Some top scoring LGA templates covered less of the template than lower scoring ones, and the template with higher coverage was chosen. Finally, some top scoring templates did not retain the same overall topology as the target. In these cases, lower scoring templates that retained the same topology were chosen.
Combining Prediction Center Metrics
The Prediction Center provides a number of metrics that facilitate Target Classification7. To maintain consistency between CASP rounds, we utilized the same combination of metrics introduced in CASP123. CASP13 target EU difficulty was established using a CASP12-like plot of the sequence-structure relationship to known folds (average3 of the HHscore and LGA_S score) against server performance (average of top 20 server model 1 GDT_TS13). To determine the HHscore, either the whole target sequences (for unsplit domains) or split EU sequences were used as queries for HHpred11 search against all PDB sequences available prior to the prediction window. The HHscore was calculated as the product of the HHpred probability (HHprob, for either the top hit or for a lower ranked homolog) and the alignment coverage (HHcovg) of the query. To determine the LGA_S score, target EUs were used as query structures to search against the whole PDB using LGA12. For most target EUs, the highest-scoring PDB structure template was chosen. However, for several difficult targets, structure templates with higher coverage were chosen (see table 1).
To help establish the boundary between classification categories on the CASP12-like plot, we clustered targets based on several additional target difficulty scores using the ClustVis web tool18. The following scores were chosen for clustering: GDTtop20 described above, the top GDT_TS among all server models (GDTtop), the average GDT_TS of all server models (GDTall), HHscore, HHprobability, HHcoverage, LGA_S, and Neff (maximum PSI-blast19 or HHblits20 Neff/length). The Neff scores were further transformed using ln(Neff+1) to scale them similarly to the rest. Each of the 8 scores were transformed into Z-scores using the following equation: ZTarget = (Targetscore – Averagescore)/ (St.Dev.score). Z-scores for each measure were uploaded to ClustVis for all target EUs. Uploaded Z-scores were not transformed, centered, or scaled and the singular value decomposition (SVD) with imputation option was used for principal component analysis (PCA) of the scores. The target EUs and measures were clustered using Euclidean distances with complete linkage for display on the heatmap, which was colored using a diverging red-yellow-green scale from low to intermediate to high scores. Clustered targets near the boundary interface of the CASP12-like plot were manually inspected, considering their evolutionary relationship to known folds in the ECOD database6 as a guide for placement in categories.
RESULTS AND DISCUSSION
Defining Evaluation Units from Structure Targets
Domains frequently serve as the basic units of folding and can evolve and function independently21–23. As such, their relative orientations within complete structures as well as their level of prediction difficulty can differ, leading to complications in assessment of whole targets. Traditionally, the tertiary structure prediction categories in CASP have split targets into EUs based on this concept of domains. For domain-based definition of EUs in CASP13, we considered similar criteria as in previous CASPs; including results of domain parsers, self-similarity or internal duplications, sequence continuity, and similarity to known protein sequences and structures. For defined multidomain targets, the decision to split into EUs was based on server performance by inspecting Grishin plots14, with an attempt to balance scoring penalties arising from domain motions with the ability of methods to assemble independent folding units. For a few difficult to define domain boundaries, we generated multiple test splits and considered server performance to establish EUs.
Our domain-based strategy resulted in splitting of 20 targets into 55 EUs, while keeping 13 multidomain targets as single evaluation units. We removed chimeric domains included for protein expression and stabilization from 2 transmembrane targets (T1011 and T1013). Several additional target domains were excluded from the assessment; including extended regions or secondary structure elements that require quaternary interaction for stability (T0960 and T0963, two segments each; T0980s2, T0990, and T0977), and domains or targets with known structures of identical or very close sequences (T0974s2, T0999-D1, T1000-D1, and T1004-D3). To explain the CASP13 procedure for establishing EUs, we highlight a relatively easy example of domain boundary definitions here and more difficult cases in the following section.
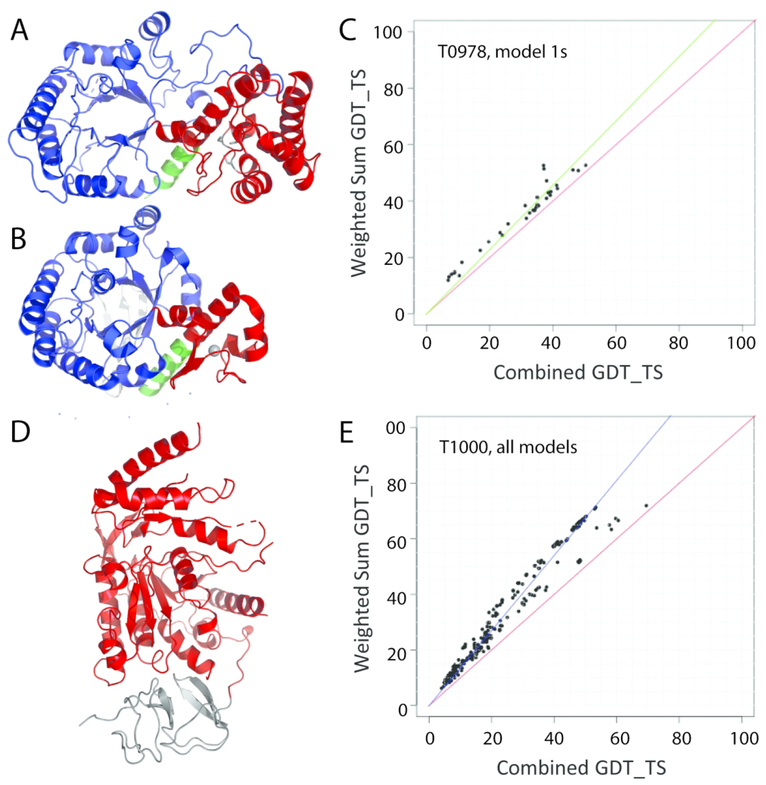
Domain parser split target T0978 into two domains (2–257 and 258–414) that retain sequence continuity. The strict evolutionary definition of T0978 domains would split the TIM barrel domain into a discontinuous sequence range (2–257, and 399–414), with an inserted zinc-binding domain (residues 258–398). Given the relatively short length of the discontinuous C-terminal helix and its extended interactions with the inserted domain (Figure 1A), the domain parser definition that retains sequence continuity provides a reasonable definition for evaluation of splits. The top TIM barrel structure template includes a smaller inserted Zinc-binding domain in a similar orientation (Figure 1B), and the Grishin plot (Figure1C) suggests the server performance on two domains is similar to the combined domain, so T0978 was kept as a single EU.
Figure 1. Evaluation Unit (EU) domain-based definition.
A) T0978 includes a TIM barrel (blue cartoon) with an inserted zinc-binding domain (red cartoon) that separates the last helix (green cartoon) from the rest of the TIM barrel. B) The top structure template 1jtbB (LGA_S 48.09) has a similar domain organization (colored as in A). C) Grishin plot suggests similar server model 1 performance on individual domains (Y-axis) and whole targets (X-axis). D) T1000 includes an N-terminal domain with a previously solved structure (gray cartoon) that was excluded from regular assessment, but was included as a special case (T1000-sp) together with the C-terminal domain (red cartoon). E) Grishin plots for all server models exhibit non-linear performance distribution.
Grishin plots for several CASP13 targets exhibited non-linear server performance when comparing whole-target evaluation scores against weighted domain-based scores. A few targets with ambiguous performance plots were designated in a special category (FM_sp) for consideration of domain interaction (T0984, T1000, and T1002). For example, the target T1000 represents a multidomain protein (Figure 1D), whose sequence is split into an N-terminal SAF domain of known structure (pfam08666) and a C-terminal D-galactarate dehydratase / altronate hydrolase (pfam04295). We initially chose to omit the N-terminal domain from the assessment given its high sequence identity (98%) to a known structure (3lazA), but decided to also keep it as a special case of assessing domain interactions. Grishin plots for the whole target compared to the weighted sum including the omitted N-terminal domain for all server models included two distributions (Figure 1E). While a majority of predictions followed the correlation line above the diagonal, the presence of server predictions in a second distribution along the diagonal (especially for the top predictions) suggested that the domain interactions might need to be evaluated.
Complex Interaction Topologies and Conformation Changes Hamper Domain Definition
CASP13 targets included expected examples of difficult domain definition that have been outlined in classification papers from previous rounds3, 4, 14, 24. For example, extended regions from domain swaps, crystal packing or protein oligomerization that lack interactions with the rest of the domain remain difficult to predict in absence of their presence in existing structure templates. We attempted to exclude some examples of these extended sequence regions in several targets. For example, the R-type pyocin contractile tail fiber structures (T0960 and T0963) form obligate trimeric interactions with two sections of extended segments (Figure 2A). While examples of trimeric tail fibers exist, the CASP13 tail fibers are interspersed with a tandem duplication of more globular Phi ETA orf 56-like protein C-terminal domains (T0960/3-D2 and T0960/3-D3) and an agglutinin HPA-like domain (T0960/3-D5). Given the diversity and fast evolution of the phage tail superfamily, this complex domain interaction topology led us to exclude the extended segments from the assessment.
Figure 2. Complex Interaction Topologies and Conformation Changes.
A) T0960 can be split into 5 sequential domains (left). Globular domains (colored cyan, green, and red) are interspersed between extended segments (gray) whose structure are defined by obligate trimeric interactions (chains colored magenta, cyan and green, right). B) T0953s1 (left) forms an obligate trimer (chains colored magenta, cyan, and green) with a beta-meander and extended segments that are present in the top phage tail fiber protein trimerization domain template (2×3h, below left). T0953s2 (right) adopts 3 domains (blue, green and red). The central domain (green) is defined by similarity to the top single-stranded right-handed beta-helix template (4pmh, below right) and has an inserted compact fiber-like domain (red), with an additional swapped fibrous segment that leads to domain definitions with discontinuous sequence. C) T0950 adopts an extended helical conformation that inserts into membrane that can be split into two domains (blue and red) based on the top template (below), which adopts an alternate soluble conformation (blue and salmon). D) T0999-D2 can be split into two domains (dark and light colors) found in templates with alternate conformations, including an open apo structure (green shades, 5xwb), as well as a closed substrate bound template (yellow shades, 1g6s).
The multi-protein complex target H0953 exhibits obligate interaction topologies in one of the interaction partners (T0953s1). Given the existence of a phage tail fiber protein trimerization domain template for T0953s1, we did not assemble the independent chains into an obligate trimer for evaluation. The second interaction partner (T0953s2) includes a unique compact fiber structure consisting of low complexity sequence, fiber swaps between two domains, and discontinuous sequence resulting from domain insertion (Figure 2B). We split T0953s2 into three evaluation units, with the boundary between T0953s2-D1 and T0953s2-D2 based on domain parser. We manually split the discontinuous and swapped C-terminal domains into T0953s2-D2 (46–114,131–151,229–249) and T0953s2-D3 (115–130,152–228), with the T0953s2-D2 domain having similarity to the top single-stranded right-handed beta helix structure template.
More difficult cases of EU definition included examples of conformation change in target structures. For example, target T0950 exhibits an extended α-helical membrane embedded conformation of the pore forming toxin PaxB (Figure 2C). The top template for this target adopts the soluble conformation of the toxin where the α-helices rearrange to hide the lytic TMH25, 26. Despite the slight outperformance of servers on domains split according to the conformation change, the linear trend of the top performing groups prompted us to keep the target as a single EU. Similarly, one of the largest structures submitted to CASP of the AroM polypeptide (T0999, 1589 residues) contains five central enzymes of the shikimate pathway fused in one chain. We split this target into domains based on known shikimate pathway enzyme structures, some of which are multidomain. Templates for the multidomain enzyme corresponding to the 5-enolpyruvyl shikimate-3-phosphate synthase (EPSP synthase, T0999-D2) component of AroM adopt alternate conformations (Figure 2D), with the structure closing upon substrate binding between its two-domains27. Such flexibility between domains would normally dictate splitting into independent EUs. While most servers outperformed on individual domains in Grishin plots, two predictions outperformed on the domain assembly. Because we expect that the interactions of EPSP synthase with the other enzymes of the pathway present in the target structure should influence the conformation (and not necessarily the presence or absence of substrate), we kept the domain as a single evaluation unit to promote methods that predict domain orientations correctly.
Evolutionary Relationships of Target EUs to Existing Fold Space
Similar to previous CASPs, classifying evaluation units into TBM and FM assessment categories in this round was based largely on prediction difficulty and the ability to detect existing structure templates based on sequence. Accurate assessment of such difficulty requires in depth knowledge of existing fold space so that templates or secondary structure arrangements providing potentially useful information for structure modeling are known. In order to best evaluate prediction difficulty, we assigned each target EU to its evolutionary position in template fold space defined by the ECOD database6 (Table 1). We included three basic levels of ECOD hierarchy for evolutionary assignment; including a close sequence-related family level (F-group), a more distantly related homology level (H-group), and a level of similar topology without evidence for homology (X-group). The remaining target EUs were designated in a special analog category for engineered sequences (T0955, T1008, and T0979) or fragments of another fold (T0957s1-D2); or as new folds when they had novel topologies (discussed below).
Target EUs with close sequence similarity to existing templates as defined by PFAM were assigned to the family (F-group) level. For example, the sequence for T1003 belongs to the aminotransferase class I and II superfamily (pfam00155, E-value 8e–74). The top template (5txrB) belongs to the same pfam superfamily, and both possess identical two domain arrangements with high structure similarity (LGA_S 90.49). Confidently placed targets in the family level (Figure 3A, 52 EUs from 36 Targets) represent a significant portion of CASP13 EUs and span a range of structure similarity to their top template homologs (LGA_S from 48.1 to 99.5). Interestingly, the DpdA sequence for the lowest scoring target T0978 is assigned as a TGT superfamily member (pfam01702) representing queuine tRNA-ribosyltransferases. However, DpdA functions to modify DNA (not tRNA) with queuosine28. As compared to the top template (Figure 1A), this target includes several insertions in TIM barrel loops and a 4-helix insertion in the zinc-binding domain that might explain this substrate shift. So, despite confident sequence relationships, F-level assignments may still represent difficult targets when their structures have diverged.
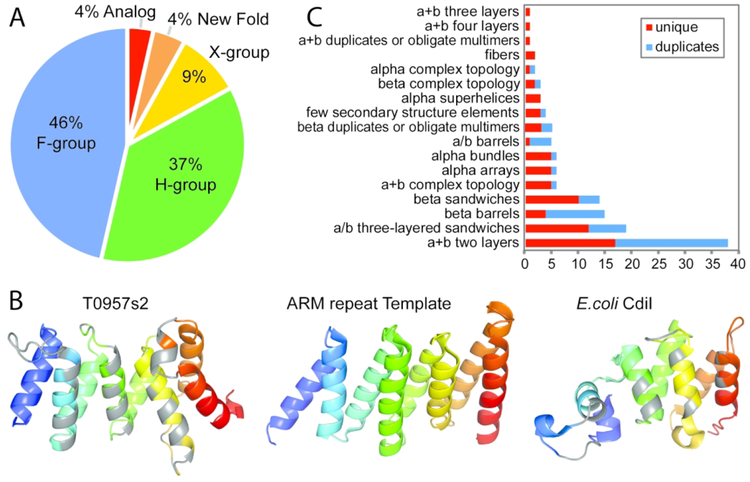
Figure 3. Target EU assignment to fold space hierarchy.
A) Pie chart depicts the proportion of EUs assigned to homologs as F-group (blue) and H-group (green), to potential homologs as X-group (yellow), to New Folds (orange), or to Analogs (red). B) Target T0957s2 immunity protein (left) colored in rainbow from N-terminus (blue) to C-terminus (red) adopts a helical repetitive alpha hairpins topology assigned at the X-group level. Residues within 4Å of the bound toxin (not shown) are colored gray. The best structural template (PDB 5mu7A, LGA_S 51.1) is to half of an ARM repeat (similar elements colored in rainbow), with N-terminal helical repeats that are not shown (center). A functional E. coli CdiI analog with less similarity (PDB 5j5vA, LGA_S 31.2) belongs to a different H-group in the repetitive alpha hairpin X-group (right), with binding residues colored gray. C) Counts of EUs (x axis) assigned to ECOD Architectures.
Because structure tends to be more conserved than sequence in protein evolution29, 30, target EUs without detected family-level sequence similarity can still be homologous to their templates. Assignment of CASP13 EUs as distant homologs required expert curation6, 8 that considered sequence/structure scores, unusual structure features, or shared functional properties as evidence for homology. We assigned 41 EUs from 32 CASP13 targets as being homologous to their templates (Figure 3A, H-groups). These target EUs displayed a diverse range of sequence similarity (13.5–100% HHpred Probability) and structure similarity (23.5–89.4 LGA_S) to known folds. Examples of H-group assignments with low sequence scores include domains from virus or phage that are known for fast evolution31–33, such as the pectin lyase-like single-stranded, right-handed beta-helix domain in T0953s2-D2, the phage tail protein-like domain in T1021s3-D1 and T1021s3-D2, the Phi ETA orf 56-like protein C-terminal domain in T0989-D2, and others, the Phage tail fiber protein trimerization domain in T0953s1, or the RNA bacteriophage capsid protein in T0998. Similarly, fast evolving domains, like RelE-like toxin domains (T0957s1-D1, T0968s1 and T0980s1) or Colicin D nuclease domain (T0986s1) are involved in bacterial resistance or toxicity. Such fast-evolving domains tend to retain a core topology that is common to remote template homologs that is decorated with additional insertions that are difficult to predict.
The remaining CASP13 target EUs lack significant evidence to justify homology to known folds. Ten EUs from nine targets retained similar topologies to existing folds that could be related by either homology or analogy (Figure 3A, X-groups). Target T0957s2 provides a good example of an assignment at the X-group level (Figure 3B left). It adopts a repetitive alpha hairpins fold that resembles the ARM repeat topology of the top identified cotamer subunit template (Figure 3B center). However, T0957s2 functions as an immunity protein that blocks the activity of its bound toxin. The target exhibits a unique twist at the C-terminus that is not found in other ARM repeats and the structure similarity to the top template is relatively low (LGA_S 51.1). Interestingly, another CdiI immunity protein from E.coli (5j5vF) adopts a repetitive alpha hairpin fold placed in its own H-group in ECOD (Figure 3B right). However, the structure similarity of T0957s2 to this immunity protein is much lower than it is to the top template (31.2 LGA_S), with the helices responsible for toxin interaction being shorter in the E.coli CdiI and the number of helical repeats differing between the two. Given the lack of functional similarity to the top template, the lack of structural similarity to a functional analog, and the ease of folding into α-helical repeats exhibited by the large number of existing H-groups that adopt this topology, we chose to assign T0957s2 at the X-group level. In fact, additional examples of immunity proteins (T0986s2, T1015s1, and T1019s1) with vague evolutionary scenarios fall into this category. CASP13 EUs included only five designated new folds (T0953s2-D3, T0968s2, T0990-D1, T0990-D2, and T0990-D3). Thus, the combination of X-groups and new folds that should represent difficult FM targets are limited to only 14 EUs out of a total of 112 (Figure 3A, New Folds, 13%).
Mapping CASP13 targets to fold space provides an added benefit of being able to explore the diversity of fold types provided to the prediction community for assessment of their methods. The architecture level in ECOD represents the top classification category in the hierarchy, grouping domains with similar secondary structure compositions and geometric shapes. As such, enumerating CASP13 domains at this level provides a broad view of fold types provided for prediction. To achieve this enumeration, new fold target EUs, as well as three out of four designated EU analogs, were assigned to ECOD architectures (Table 1) based on the composition and arrangement of their SSEs and multidomain targets were split into their respective ECOD defined domains. All but four architectures (alpha duplicates or obligate multimers, alpha complex topology, beta meanders, and mixed α+β and α/β) were represented in CASP13 targets, with α+βtwo layered sandwiches outnumbering the rest (Figure 3C). Notably, numerous duplications were present among the targets. For example, the beta barrels included 11 rift-related domains and two SH3 domains, leaving only 4 unique folds. After collapsing all evolutionary related duplications among CASP13 target domains, 76 unique folds remain. Among these, all major fold types are represented: 33% α+β, 17% α/β, 18% all α, 25% all β, with 7% being fibers or few secondary structure elements.
Assigning EUs to Tertiary Structure Assessment Categories
Because homology definitions for CASP13 target EUs required manual decisions that were subjective in nature, we decided to impose additional objective criteria for separating targets into assessment categories. Such a strategy has been used in several previous CASP classifications3, 4, 14. However, to maintain consistency with the last round of CASP, we chose to include the same main scores selected previously, including the average performance of the top 20 servers, sequence-related target difficulty measured by HHscore, and structure-related target difficulty measured by LGA_S3. These measures were combined with two additional sequence-related scores (HHpred Probability and HHpred Coverage), two additional performance-based scores (Top server model GDT_TS and Average GDT_TS of all server models), and a score to account for alignment depth (maximum Neff/length from PSI-blast19 or HHblits20) for clustering the target EUs.
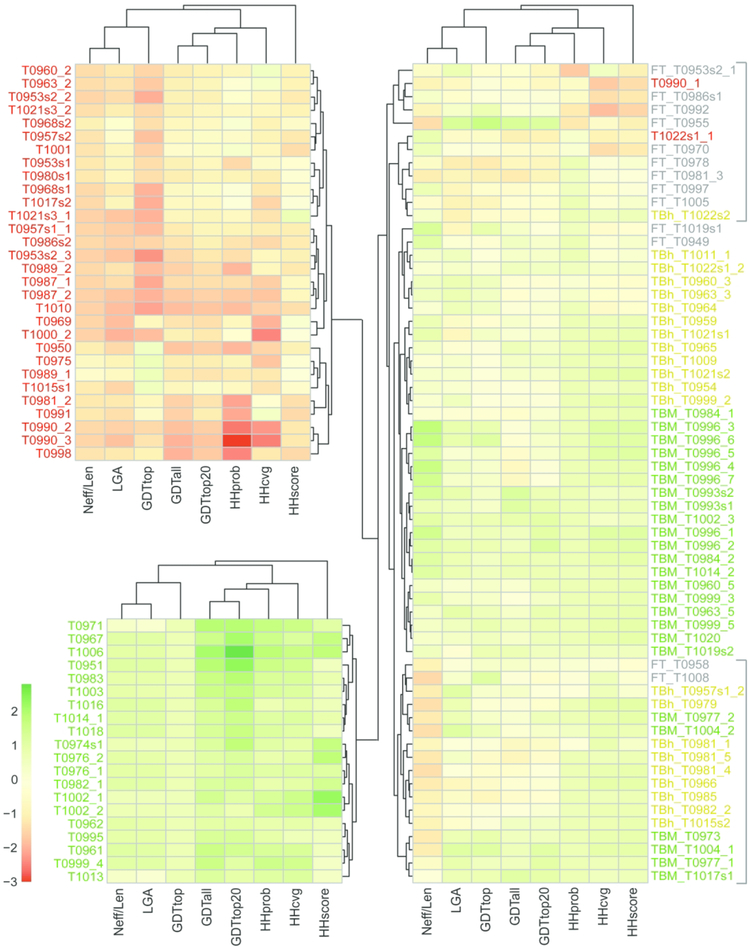
To cluster targets according to difficulty, we converted all chosen measures to Z-scores (see Combining Prediction Center Metrics in Methods). Target EUs were clustered by scaled Z-scores using complete linkage of Euclidean distances, and were visualized with heatmaps colored from low to high on a red-yellow-green scale (Figure 4). Three main clusters correspond to difficult target EUs that should be FM (Figure 4, top left with mainly red blocks), easy target EUs that should be TBM (Figure 4, bottom left, mainly green blocks), and intermediate targets whose clusters required manual inspection (Figure 4, right). Given the number of targets with scores near zero (mainly yellow blocks in the heatmap) whose sequence or structures tend to diverge from their top templates, we ultimately chose to split the TBM assignments into easy and hard subcategories. Interestingly, Neff scores appear to distinguish target difficulty categories in the intermediate cluster, with a central subcluster of relatively high Neff scores being all TBM-easy and two subclusters of lower Neff scores (marked by gray brackets in Figure 4) being mainly TBM-hard and intermediate TBM/FM.
Figure 4. Heatmap clusters target EUs based on objective measures.
Columns include three sequence-based similarity scores (HHprob, HHcovg, and HHscore), three server performance-based scores (GDTtop, GDTall, and GDTtop20), a structure-based score (LGA_S), and a score for alignment depth (Neff/Len), and rows represent target EUs. Rows and columns were clustered (depicted as trees) using Euclidean distance with complete linkage. Scores were colored from low to high using a diverging red yellow green color scheme (depicted on the bottom right). Rows were split into 3 clusters, with the two tightest clusters of clear TBM-easy (EUs labeled green) and clear FM (EUs labeled red) flipped to the left. Intermediate clusters on the right include TBM-hard (EUs labeled yellow) and TBM/FM EUs (labeled gray), with subclusters indicated by gray brackets to the right.
A CASP12-like scatter plot3 of server performance against target difficulty highlights the distribution of EUs assigned to assessment categories (Figure 5A). Similar to the previous round, CASP13 server performance correlated with target difficulty. Furthermore, splitting the performance distribution into quadrants identical to those chosen in the previous round clearly differentiated FM EUs (Figure5A, lower left) from easy TBM EUs (Figure 5A, upper right). Labeled EUs fell in between the two main categories on the difficulty performance scatter or formed clusters of mixed assignments in the heatmap. These borderline cases were manually assigned to assessment categories by considering 1) the scores from the heatmap, 2) the classification of closely related EUs from the heatmap and 3) the evolutionary relationships to existing fold space. Several examples discussed in the following section highlight classification of intermediate EUs based on these criteria.
Figure 5. Target EU difficulty correlates with server performance in CASP12-like plot.
A) Scatter of EU difficulty measured by the average of HHscore and LGA_S similarity to templates and server performance measured by average GDT of the top 20 server models. EUs are colored according to assigned categories: TBM-easy (green), TBM-hard (yellow), TBM/FM (gray), and FM (red). Difficult borderline EUs requiring manual assignment are labeled. B) FM target T0990-D1 (left) with relatively high LGA_S and performance scores does not cluster with other FM EUs. A unique loop includes conserved residues (magenta sphere) that typically bind metal. The metal binding residues are absent from the top unrelated structural template (2rt6, right), which adopts a three-helix bundle. C) TBM-hard target T0960-D3 (left) identified a template homolog (5nxf, right) with a similar overall fold (LGA_S 83.2) with relatively low sequence scores (82%, 0.54 coverage). D) TBM/FM target T0958 identified a template homolog (2kim, right) with relatively low sequence scores (81.4%, 0.59 coverage). The template exhibits SSE shifts (LGA_S 69.2) compared to the target. E) TBM/FM target T1008 represents a designed protein structure that by definition is analogous to its top template (5hnwK, right, LGA_S 73.9).
Manual Assignment of Borderline Target EUs to Assessment Categories
Two FM target EUs (T0990-D1 and T1022s1-D1) did not cluster with the other FM designated targets in heatmaps, although their scores were relatively low. T0990-D1 adopts a bundle of two short helix pairs that interact almost perpendicularly and are joined by a loop with a set of four clustered Zinc-binding residues (Figure 5B). While somewhat similarly arranged four helix bundles exist, the relative positions of the helices and the presence of a potential zinc binding site warrant consideration of this domain as a new fold. The top scoring template (2rt6, LGA_S 56) adopts a three-helix bundle with longer helices, and none of the top ten scoring templates are four helix bundles. Server performance scores for this EU were relatively high (GDTtop20 61.2) due to the simple arrangement of SSEs. We classified T0990-D1 as FM based on the novel topology and lack of evolutionary relationship to existing folds. Several additional boundary targets have low sequence, but relatively high structure scores (T0970, T0992, T0986s1, and T0953s2-D1). Each of these cases were classified as TBM/FM due to 1) the presence of distantly related homologous templates that could potentially be identified by sequence (T0970, T0986s1), 2) the presence of a high scoring template that belongs to a common immunoglobulin-like fold (T0992), or 3) the small size and simplicity of SSEs (T0953s2-D1).
Several intermediate difficulty EUs that border the TBM boundary (i.e. T0960-D3, T0963-D3, T0964, T0919s1, T0949, T1022s2, T0958, T1008, and T0957s1-D2) tend to have relatively low sequence scores when compared to those of confidently assigned TBMs. For example, the R-type pyocin target structure includes a domain duplication near the N-terminus. One of these duplicated domains (T0960-D3, Figure 5C left) adopts a fold that is also found in phage contractile tails. The closest structure domain from the phage 11 host-recognition device would serve as a good template for modeling (5efvB, LGA_S 83.2). However, an alternate domain from T4 proximal long tail fibre protein gp34 was identified by sequence (5nxfC, HHprob 82% with 0.54 coverage). While both templates are homologous to the T0960-D3 target, the one identified by sequence is a worse template (5nxfC, LGA_S 69.7, Figure 5C right). Thus, the diversity of structure template homologs available for modeling this target posed a challenge for servers (GDTtop20 50.93). Ultimately, we classified the target domain as TBM-hard due to the presence of sequence-detectable template homologs. The duplicated phi ETA orf 56-like protein C-terminal domain from the same target (T0960-D2) had much lower scores (Difficulty 42.6, GDTtop20 32.9), was more distantly related by sequence (HHscore 7.8), and was therefore classified as FM, despite the presence of template homologs.
CASP13 targets included two engineered proteins (T1008 and T0955) with topologies present in the PDB. By definition, the top structure templates for these engineered proteins are analogs. Target T1008 adopts an α+β two-layered sandwich (Figure 5D left). While the same topology is present in the top structure analog (5hnwK, LGA_S 73.9, Figure 5D right), templates identified by sequence were incorrect. Compared to previous examples of engineered protein structures in CASP4, target difficulty estimates (67.4 for T1008 and 56.4 for T0955) and performance measured by GDTtop20 (47.44 for T1008 and 78.0 for T0955) on these engineered protein targets was surprisingly good, causing us to classify them as FM/TBM.
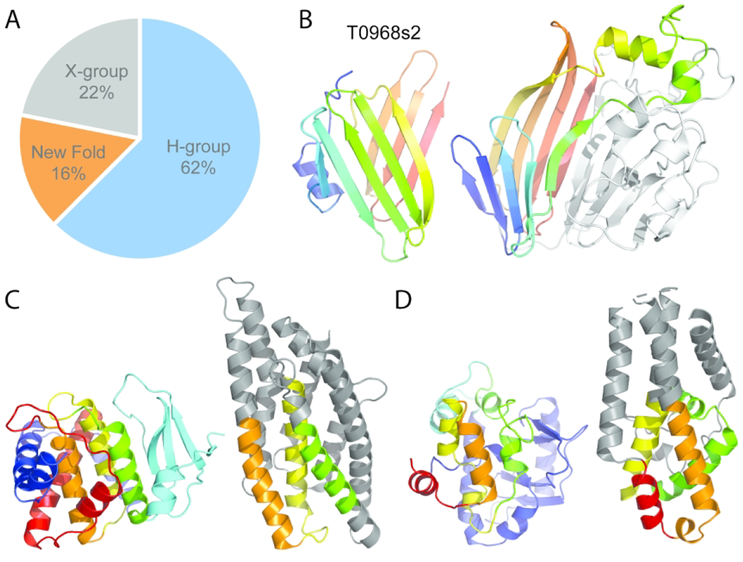
FM Targets Represent Fast Evolving Homologs, Potential Analogs, and New Folds
The majority of CASP13 target EUs that were classified as FM had distantly related structure template homologs (Figure 6A, H-group, 62% of FM EUs). As previously discussed with difficult target EU assignments (i.e. T0960-D2 and T0950), these targets represent rapidly evolving sequences whose functions require rapid adaptation, such as in phage host recognition or bacterial warfare. Such fast evolution results in the presence of multiple divergent templates (i.e. T0960-D2). Thus for these examples, the ever-increasing size of the protein structure database serves as a double-edge sword. On the positive side, the expanding structure database can add novel topologies to fold space that fill in voids. However, new structures can also adopt alternate conformations, acquire alternate topologies, or gain/lose SSEs. Thus, satisfactory structure modeling in the era of large databases relies on correct selection of templates (or fragments). Those targets with diverse templates (T0960-D2) or conformation changes (T0950) remain difficult to predict.
Figure 6. FM Targets are Mainly Fast-Evolving Homologs with Few New Folds.
A) Pie chart depicts evolutionary relationships of FM target domains to existing folds. B) Potential new β-sandwich fold in T0968s2 (left) is colored in rainbow from the N-terminus (blue) to the C-terminus (red). A structure template with much lower similarity (4ttgA, LGA_S 16.0) than the top β-meander template (not shown) has similar topology as a subcomponent of a larger supersandwich fold, with similar elements colored in rainbow (right). C) T0990-D2 (left) includes N-terminal helices (blue), followed by a β-meander (cyan), a central 3-helix bundle (green, yellow, and orange), and C-terminal helices (red). A top template (3eb7A, right) includes analogous helices (colored like the target) arranged like the three-helix bundle. D) T0990-D3 (left) has an N-terminal α+β subdomain (blue), followed by a connecting α-helix (cyan), and 4 broken helices in a bundle (green, yellow, orange, and red). The top template (4alyB, left) has four analogous helices (colored like the target) as a subcomponent of the overall fold.
In contrast to the large number of FM targets with template homologs, examples of potential analogs and new folds are more limited (22% and 16%, respectively, Figure 6A). The topology of target T0953s2-D1 represents one domain with questionable evolutionary relationship to known folds (designated as X-group). The top structure template RnaseT (3v9uD) includes a helical insertion to the core Ribonuclease H-like topology common to homologous structures. This insertion resembles the topology of T0953s2-D1, with each possessing three α-helices arranged in parallel. The parallel arrangement of α-helices requires an extended connecting segment that, together with the two helices, resembles a HEH (Helix-Extension-Helix) motif found in various proteins involved in nucleic acid metabolism34. However, the HEH motif domains share a distinct fold, with the first short helix (not present in T0953s2 or RnaseT) leading almost immediately into the second helix. The following extended-region positions the third helix parallel to the second to adopt the characteristic HEH structure. In addition to missing the first helix found in HEH domains, the extended-region and third helix are shorter in the target domain, bringing into question any potential evolutionary relationship.
Five potential new folds were represented in three CASP13 targets. First, the Salmonella phage S16 gp38 adhesin includes a low complexity polyglycine rich sequence (T0953s2-D3) that folds into a compact fiber of packed type II helices35 (Figure 2B, red domain). While type II helices are present in the structure database, their arrangement into this topology is new. Second, the immunity component (T0968s2) of the contact-dependent growth inhibition toxin - immunity complex from Klebsiella pneumoniae forms a β-sandwich using an N-terminal 5-stranded β-meander and a C-terminal 4-stranded β-meander (Figure 6B left) that pack against each other at an unusual angle, half way in between parallel and perpendicular. The simple β-meander SSE components that make up this fold exist in many structures (i.e. in the ECOD beta meanders architecture) and some structures could serve as relatively good templates. The top scoring template (5gkf LGA_S 50.2) adopts a PH domain-like β-barrel fold that includes an analogous 4-stranded β-meander. A more distantly related template (4ttgA4, LGA_S 16.0) with a glycosyl hydrolase domain-like supersandwich fold includes a subdomain with similar topology as the target, but with more parallel β-sheets (figure 6B right). The unique orientation of the T0968s2 β-strands, together with low structure similarity to existing folds suggest the target to be a new fold.
The translation regulatory protein NS1 of bluetongue virus target provides the remaining three domains (T0990-D1, T0990-D2, and T0990-D3) designated as new folds. The N-terminal domain was previously discussed as a borderline classification EU (Figure 5B). The two C-terminal domains each adopt a complex α+β topology, with T0990-D3 (Figure 6C left) inserted into T0990-D2 (figure 6D left). T0990-D2 includes a twisted 4-stranded β-meander followed by an adjacent central three helix bundle. The bundle is surrounded by five additional α-helices from the N-terminus and the C-terminus of the domain. The top scoring templates, which include a designed helical repeat (5k7vB, LGA_S 26.4) and a toxin membrane translocation domain (3eb7B, LGA_S 24.5, Figure 6C right), have analogous 3-helix bundles as subcomponents of their folds. T0990-D3 includes an N-terminal α+β subdomain followed by a connecting helix and then four broken helices arranged as a bundle. The top template (4alyB, LGA_S 23.8) includes a similar arrangement of analogous helices as a subcomponent of the overall fold (figure 6D right).
Perspectives for future CASPs
Assignment of CASP targets into EUs and classifying the EUs according to difficulty has been an important task for CASP assessment over the years. The task relies on relating targets to existing fold space in a meaningful way, which has been successfully accomplished in the past by providing an evolutionary-based domain assignment. The number of existing templates in the PDB is growing (14,622 PDB structures in 2018), and includes structures with increased size and complexity36. This ever-increasing database of templates makes such evolutionary-based domain classification difficult, as highlighted by the examples of complex topologies, conformation changes and fast evolution outlined in this paper. The decision to split targets into domains for evaluation requires considerations that are not necessarily reflected in traditional Grishin plots. Splits are also not easily discerned when the number of related templates extends into the hundreds. When conformation changes are large, templates can be quite distant from the target and can easily be missed. Domain interactions are influenced by bound ligands, interaction partners, or even crystal contacts. Such information is not always provided to CASP participants (or assessors), but might need to be a required component of future target submissions. Given the importance of conformational state on protein function and the relatively good success of prediction methods, we chose to keep domains together wherever possible. Finally, assignment of difficulty based on prediction performance and traditional evolutionary considerations might be becoming antiquated. Potentially, similarity of the target structure to any existing template, regardless of their evolutionary relationship, might provide a better indication of its difficulty. Our CASP13 classification relied heavily on sequence relationships, with HHpred-related scores contributing to three out of eight scores in the Figure 4 heatmap used to cluster EUs and half of the difficulty component of the scatterplot in Figure 5 used to assign EUs into assessment categories. While this choice of scores allowed a classification consistent with the previous CASP, perhaps the future should rely more heavily on the presence of existing structures, or substructures in the PDB. Future classifications should also consider Neff, which tends to distinguish targets of intermediate difficulty, and not necessarily targets that are classified as FM. Neff provides a good indication of the evolutionary information contained in a target sequence that complements template-based sequence and structure scores.
ACKNOWLEDGEMENTS
We thank the CASP organizers for their invitation to participate in CASP13. We thank the authors of two unpublished targets (as of the release of this paper) for allowing us to publish their images: Manal A. Swairjo and Joshua Savage (San Diego State University, T0978) and George Minasov (Northwestern University, T1000). This research was supported by the National Institutes of Health (grants GM094575 and GM127390 to NVG; and GM100482 to AK and BM), and the Welch Foundation (I-1505 to NVG).
REFERENCES
- 1.Moult J, Fidelis K, Kryshtafovych A, Schwede T, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)-Round XII. Proteins. 2018;86 Suppl 1:7–15. doi: 10.1002/prot.25415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moult J A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr Opin Struct Biol. 2005;15(3):285–9. doi: 10.1016/j.sbi.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 3.Abriata LA, Kinch LN, Tamo GE, Monastyrskyy B, Kryshtafovych A, Dal Peraro M. Definition and classification of evaluation units for tertiary structure prediction in CASP12 facilitated through semi-automated metrics. Proteins. 2018;86 Suppl 1:16–26. doi: 10.1002/prot.25403. [DOI] [PubMed] [Google Scholar]
- 4.Kinch LN, Li W, Schaeffer RD, Dunbrack RL, Monastyrskyy B, Kryshtafovych A, Grishin NV. CASP 11 target classification. Proteins. 2016;84 Suppl 1:20–33. doi: 10.1002/prot.24982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kryshtafovych A, Fidelis K, Moult J. CASP10 results compared to those of previous CASP experiments. Proteins. 2014;82 Suppl 2:164–74. doi: 10.1002/prot.24448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng H, Schaeffer RD, Liao Y, Kinch LN, Pei J, Shi S, Kim BH, Grishin NV. ECOD: an evolutionary classification of protein domains. PLoS Comput Biol. 2014;10(12):e1003926. doi: 10.1371/journal.pcbi.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kryshtafovych A, Monastyrskyy B, Fidelis K. CASP11 statistics and the prediction center evaluation system. Proteins. 2016;84 Suppl 1:15–9. doi: 10.1002/prot.25005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng H, Liao Y, Schaeffer RD, Grishin NV. Manual classification strategies in the ECOD database. Proteins. 2015;83(7):1238–51. doi: 10.1002/prot.24818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo JT, Xu D, Kim D, Xu Y. Improving the performance of DomainParser for structural domain partition using neural network. Nucleic Acids Res. 2003;31(3):944–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, Xue B, Zhou Y. DDOMAIN: Dividing structures into domains using a normalized domain-domain interaction profile. Protein Sci. 2007;16(5):947–55. doi: 10.1110/ps.062597307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244–8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zemla A LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31(13):3370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zemla A, Venclovas C, Moult J, Fidelis K. Processing and analysis of CASP3 protein structure predictions. Proteins. 1999;Suppl 3:22–9. [DOI] [PubMed] [Google Scholar]
- 14.Kinch LN, Shi S, Cheng H, Cong Q, Pei J, Mariani V, Schwede T, Grishin NV. CASP9 target classification. Proteins. 2011;79 Suppl 10:21–36. doi: 10.1002/prot.23190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32(Web Server issue):W327–31. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Geer LY, Bryant SH. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45(D1):D200–D3. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–D32. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43(W1):W566–70. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Remmert M, Biegert A, Hauser A, Soding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9(2):173–5. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 21.Bork P Shuffled domains in extracellular proteins. FEBS Lett. 1991;286(1–2):47–54. [DOI] [PubMed] [Google Scholar]
- 22.Richardson JS. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. [DOI] [PubMed] [Google Scholar]
- 23.Wetlaufer DB. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc Natl Acad Sci U S A. 1973;70(3):697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinch LN, Qi Y, Hubbard TJ, Grishin NV. CASP5 target classification. Proteins. 2003;53 Suppl 6:340–51. doi: 10.1002/prot.10555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brauning B, Bertosin E, Praetorius F, Ihling C, Schatt A, Adler A, Richter K, Sinz A, Dietz H, Groll M. Structure and mechanism of the two-component alpha-helical pore-forming toxin YaxAB. Nat Commun. 2018;9(1):1806. doi: 10.1038/s41467-018-04139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganash M, Phung D, Sedelnikova SE, Lindback T, Granum PE, Artymiuk PJ. Structure of the NheA component of the Nhe toxin from Bacillus cereus: implications for function. PLoS One. 2013;8(9):e74748. doi: 10.1371/journal.pone.0074748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park H, Hilsenbeck JL, Kim HJ, Shuttleworth WA, Park YH, Evans JN, Kang C. Structural studies of Streptococcus pneumoniae EPSP synthase in unliganded state, tetrahedral intermediate-bound state and S3P-GLP-bound state. Mol Microbiol. 2004;51(4):963–71. [DOI] [PubMed] [Google Scholar]
- 28.Hutinet G, Swarjo MA, de Crecy-Lagard V. Deazaguanine derivatives, examples of crosstalk between RNA and DNA modification pathways. RNA Biol. 2017;14(9):1175–84. doi: 10.1080/15476286.2016.1265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26(1):316–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sadreyev RI, Grishin NV. Exploring dynamics of protein structure determination and homology-based prediction to estimate the number of superfamilies and folds. BMC Struct Biol. 2006;6:6. doi: 10.1186/1472-6807-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krupovic M, Cvirkaite-Krupovic V, Iranzo J, Prangishvili D, Koonin EV. Viruses of archaea: Structural, functional, environmental and evolutionary genomics. Virus Res. 2018;244:181–93. doi: 10.1016/j.virusres.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanjuan R, Domingo-Calap P. Mechanisms of viral mutation. Cell Mol Life Sci. 2016;73(23):4433–48. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stern A, Sorek R. The phage-host arms race: shaping the evolution of microbes. Bioessays. 2011;33(1):43–51. doi: 10.1002/bies.201000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11(8):1365–74. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunne M, Denyes JM, Arndt H, Loessner MJ, Leiman PG, Klumpp J. Salmonella Phage S16 Tail Fiber Adhesin Features a Rare Polyglycine Rich Domain for Host Recognition. Structure. 2018;26(12):1573–82 e4. doi: 10.1016/j.str.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 36.Burley SK, Berman HM, Christie C, Duarte JM, Feng Z, Westbrook J, Young J, Zardecki C. RCSB Protein Data Bank: Sustaining a living digital data resource that enables breakthroughs in scientific research and biomedical education. Protein Sci. 2018;27(1):316–30. doi: 10.1002/pro.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaarschmidt J, Monastyrskyy B, Kryshtafovych A, Bonvin AMJJ. Assessment of contact predictions in CASP12: Co-evolution and deep learning coming of age. Proteins: Structure. Function, and Bioinformatics 2017;86:51–66. 10.1002/prot.25407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abriata LA, Tamò GE, Monastyrskyy B, Kryshtafovych A, Dal Peraro M. Assessment of hard target modeling in CASP12 reveals an emerging role of alignment-based contact prediction methods. Proteins: Structure. Function, and Bioinformatics 2017;86:97–112. 10.1002/prot.25423. [DOI] [PubMed] [Google Scholar]