Abstract
Genome integrity is essential for proper cell function such that genetic instability can result in cellular dysfunction and disease. Mutations in the human genome are not random, and occur more frequently at “hotspot” regions that often co-localize with sequences that have the capacity to adopt alternative (i.e. non-B) DNA structures. Non-B DNA-forming sequences are mutagenic, can stimulate the formation of DNA double-strand breaks, and are highly enriched at mutation hotspots in human cancer genomes. Thus, small molecules that can modulate the conformations of these structure-forming sequences may prove beneficial in the prevention and/or treatment of genetic diseases. Further, the development of molecular probes to interrogate the roles of non-B DNA structures in modulating DNA function, such as genetic instability in cancer etiology are warranted. Here, we discuss reported non-B DNA stabilizers, destabilizers, and probes, recent assays to identify ligands, and the potential biological applications of these DNA structure-modulating molecules.
Keywords: non-B DNA, genetic instability, small molecule ligands, small molecule DNA probes, DNA stabilizers, DNA destabilizers
There has been an explosive growth in structure-based drug design in the past decade, in part due to the progress in molecular and structural biology techniques, making 3D structures of macromolecular targets at an atomic-level readily available, and the rapid advancement in computer-aided drug design [1–3]. Based on the detailed information of the interactions between the targets and ligands, this approach has resulted in the identification of new therapeutic agents and/or the optimization of existing drugs.
The vast majority of these efforts have been focused on targeting proteins because their structural features and biological activities can often be recognized and targeted in a specific fashion. Nucleic acids have received less attention as potential targets, though they have clearly defined structural features and biological functions [4]. This is reflected in the dwindling number of FDA-approved drugs that target DNA. Before 1982, ~31% (12 of 38) of FDA-approved drugs were associated with DNA damage mechanisms, which has dropped to ~2% of new drugs (1 out of 47) during 2010–2015 [5].
DNA arguably remains to be an effective therapeutic target for genetic-instability-related diseases such as cancer. Recent investigations have shown that certain repetitive DNA sequences with the capacity to adopt alternatively-structured DNA (i.e. non-B DNA) often co-localize with “mutation hotspots” implicated in genetic instability-related diseases [6–9]. Naturally occurring non-B DNA structure-forming sequences can stimulate genetic instability [10–16], although the fundamental mechanisms are still unclear.
Beyond the Watson-Crick B-DNA helix
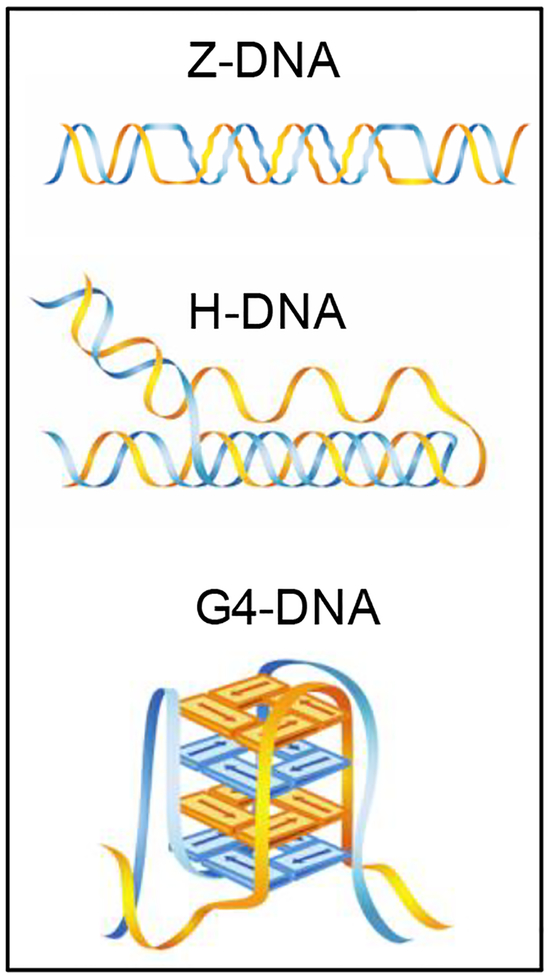
Watson and Crick first described the canonical B-form structure of the DNA duplex structure over 60 years ago. However, since this first description, more than a dozen alternative DNA structures have been characterized. These include left-handed Z-DNA, three-stranded triplex or H-DNA, 4-way junction-containing cruciforms, self-annealing hairpins, slipped DNA, guanine quartet-containing G4-DNA, A-DNA, cytosine-rich i-motifs, etc. [17–19].
These structures require certain sequence elements for their formation. For example, inverted repeat sequences can adopt hairpin or cruciform structures, where intra-strand annealing occurs on one or both strands due to the self-complementary feature of both strands. Simple repeats can form loop-out structures if the two strands are not perfectly re-aligned after unwinding [20]. If the looped-out regions are self-complementary, then duplex stem regions can be formed, such as in CNG triplet repeats [11, 21, 22]. Alternating purine-pyrimidine tracks can form left-handed Z-DNA structures (Figure 1), where the alternating stacking of bases in the syn- and anti- conformations results in a zigzag pattern of the sugar-phosphate backbone [23]. The Z-DNA conformation results in a double-stranded helix with ~12 base pairs (bp) per turn, compared to the ~10 bp per helical turn in canonical B-form DNA. A polypurine/pyrimidine region with mirror symmetry can form an intramolecular triplex structure (H-DNA, Figure 1) where the purine-rich strand in one half of the symmetry-containing duplex folds back and binds to the duplex in the other half of the mirror repeat via Hoogsteen hydrogen bonding in the major groove [24, 25]. Because initial studies showed that protonation of cytosine is required for Hoogsteen hydrogen bonding to the guanine in a GC Watson-Crick bp, the structure is named H-DNA. G4-DNA or G-quadruplex DNA can form at tandem repeats of guanine (G) sequences with a contiguous run of three or more guanine bases [26–28]. Four Gs associate in a square-planar cyclic array, stabilized by Hoogsteen H-bonds to form a G-tetrad that can stack and thereby form a G-quadruplex (G4-DNA, Figure 1) structure stabilized by monovalent cations, such as K+ or Na+. Because the formation of non-B DNA structures requires specific sequence elements, we and others have designed algorithms to search for such structure-forming sequences in genomes/databases of interest [29–31].
Figure 1.
Non-B DNA structures discussed in this review. Figure adapted with permission from Zhao, et al., Nature Springer, CMLS, 2010 [7].
In addition, RNA can also form alternative structures, either intramolecularly or intermolecularly with other RNA strands or DNA strands. For example, RNA G-quadruplexes have been reported to impact transcription, translation, RNA localization and splicing. Intermolecular G-quadruplex structures formed between two copies of HIV-1 genomic RNA at their 5′ end dimerization-linked sequence domains, can anchor the virus for dimerization, which is important for viral recombination, translation, and encapsidation (for reviews see [32, 33]). Both RNA*DNA-DNA triplexes and RNA*RNA-RNA triplexes can alter transcription and splicing [34]. Further, RNA can also form a left-handed alternative conformation, i.e., Z-RNA, which can interact with Zα domain family proteins and regulate immunity in vivo [35, 36]. However, the focus of this review is on non-B DNA structures.
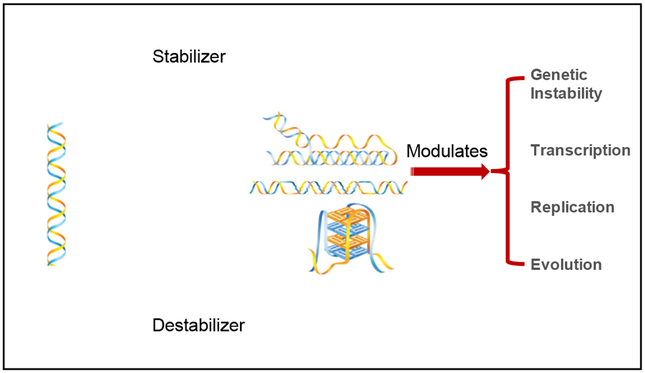
Although most non-B DNA structures exist in higher energy states compared to the B-DNA structure, negative supercoiling generated by the unwinding of DNA from histone cores behind the replication and transcription machinery, or other chromatin remodeling processes, can drive the B-DNA to non-B DNA transition and maintain non-B structures. Importantly, non-B DNA is involved in wide range of biological processes, such as DNA replication, transcription, recombination and telomere maintenance [7, 19, 37, 38], such that modulation of and/or alterations in DNA conformation could lead to significant biological outcomes (Figure 2).
Figure 2.
Modulating DNA structure-related metabolism using small molecules.
DNA structure-induced genetic instability
While triplet repeat-forming hairpins have been implicated in a number of neurological disorders, largely via expansion of the repeats, here we will focus on Z-DNA, H-DNA and G4-DNA structures (Figure 1) that have been implicated in cancer etiology.
In human genomes, G4-DNA, H-DNA, and Z-DNA-forming sequences often occur near, and are enriched at chromosomal mutation hotspots in disease-related genes, implicating them in genomic instability and disease [6, 39, 40]. For example, in c-MYC translocation-induced lymphomas and leukemias, a major breakage hotspot region in the promoter overlaps with sequences capable of adopting H-DNA, Z-DNA, and G-4 DNA [41–49]. We have discovered that an H-DNA-forming sequence within this region is susceptible to genetic instability and stimulates the formation of DNA double-strand breaks (DSBs), deletions, and point mutations in mammalian cells both in vitro and in vivo [10, 13, 16]. Taking advantage of the accumulating sequencing data of mutations in human cancer genomes, and search engines used to identify sequences that have the potential to adopt non-B DNA structures, we and others have found that many types of non-B DNA-forming sequences are significantly enriched at cancer mutation hotspots, suggesting a biological role of DNA structures in cancer etiology [6, 10, 11, 50]. Z-DNA-forming sequences map near breakpoints in c-MYC [51, 52], and a 5’ breakage hotspot in the BCL-2 gene [53, 54]. Z-DNA-forming sequences are also mutagenic and lead to DSBs, deletions, rearrangements and point mutations in mammalian cells in vitro and in vivo [13, 15]. In yeast, the introduction of G4-DNA-forming sequences leads to genome instability [55–58], and in human epidermal keratinocytes, intrinsic DSB sites are enriched within G4-forming sequences [59]. G4-forming sequences are also found near sites of mutation hotspots, such as the mutationally activated hTERT promoter in melanoma [60–62]. Thus, naturally occurring Z-DNA, H-DNA, and G4-DNA-forming sequences can stimulate mutations in mammalian cells and in mouse tissues supporting a role for non-B DNA structure in genetic instability in the form of point mutations, chromosomal deletions, translocations, and rearrangements, leading to disease etiology [7, 11, 15, 16, 39, 63–69].
Interestingly, studies have suggested that some single nucleotide polymorphisms (SNPs) within non-B DNA-forming sequences can affect their structure-forming potential, and thereby impact biological outcomes. For example, replacement of an A with a G in a short palindromic sequence can reduce its hairpin-forming potential, and has been associated with a higher risk of ß-thalassemia in Indian populations [70]. Another example includes the promoter of the GRIN1M gene that has been shown to adopt G-quadruplex structures in vitro, where a one base change (−855 G to C) can alter the G-quadruplex structure formation and has been associated with schizophrenic patients [71]. A +2985 (T to C) polymorphism in the APOE gene has been associated with Alzheimer’s disease, and such a change can also convert a hairpin structure of a “T” allele to a G-quadruplex in vitro [72]. Further, a bioinformatics study showed that SNPs in quadruplex-forming sequences were significantly associated with the expression of the corresponding genes [73]. Because a one base-pair alteration can affect non-D DNA formation and impact the associated phenotypes, modulating DNA conformation by small molecules may have a wide variety of applications (Figure 2).
Based on many studies of the roles of DNA structure in DNA transcription, replication, chromatin structure, and genetic instability, it has become evident that DNA structures are functional genomic elements. Thus, DNA is more than just an inert source of genetic information; it has conformational features that modulate genetic processes, some of which have been implicated in disease etiology, genome plasticity, and evolution. Moreover, the specific structural features of non-B DNA could overcome the presumed lack of specificity in targeting DNA, potentially providing selective targets for drug design. The structural formation of non-B DNA can be predicted given that the required sequence elements are known for the formation of various types of such alternative DNA structures [74, 75]. Thus, DNA structural features represent important targets of investigation, with non-B-DNA structure formation and stability as site-specific key targets of DNA functional regulation and/or disease intervention.
Modulation of DNA structures in vitro and in vivo
Non-B DNA-forming sequences are dynamic in nature. The propensity of these structures to form in genomic DNA is dependent upon many variables, including the stabilizing effect of super-helical stress in the form of negative supercoiling, protein interactions, chromatin structure, and intracellular conditions (i.e. cations, and polyamines such as spermine and spermidine). An example of structure-altering cellular conditions and protein interactions include the presence of helicases such as the RecQ family members, some of which can unwind these mutagenic structures to preserve genomic integrity [76–78]. There are also exogenous small molecule DNA-interacting ligands that can stabilize or destabilize these structures, and can be used as tools in the study of non-B-DNA structure formation and processing, which is the focus of this review.
Small molecules as tools to study DNA structure and function
Because of their size, small molecules can often reach their targets in vivo more efficiently than larger compounds. Thus, the use of small molecules has provided an approach to advance studies of biological mechanisms, evaluate therapeutic targets underlying diseases, and reveal novel therapeutic avenues [79–81]. As such, many small molecules have been found to interact with non-B DNA in a conformation-specific manner (see below). Many of the challenges associated with studies of non-B DNA structure-induced genetic instability could also benefit by the use of small molecules that have the potential to stabilize or destabilize these mutagenic structures. By using structure-specific recognition agents, precise temporal and spatial modulation of non-B DNA structure formation could be performed leading to a better understanding of the recognition and processing of a particular non-B DNA structure. In addition, fluorescent probes that can detect specific non-B DNA structures may be used to visualize non-B DNA structures, and therefore assist in revealing their cellular loci and possible biological functions. While there have been antibodies generated to visualize a number of non-B-DNA structures [82–90], there have been concerns regarding their use. For example, permeabilization of cells prior to antibody probing could potentially alter the conditions that are critical for DNA conformation.
There are many examples of the use of DNA structure-specific ligands and probes in the literature. Here, we review recent findings regarding small molecule non-B-DNA structure modulators (stabilizers and destabilizers) as well as probes, focusing on Z-DNA, H-DNA, and G4-DNA structures. We refer to previous or current reviews where applicable, and highlight salient points of known ligands relevant to discussion (e.g. studies with a focus on biological outcomes). We also present a brief overview on the recent techniques and analyses developed to enable successful identification of DNA structure-specific ligands.
Z-DNA
Among the known non-B DNA structures, Z-DNA is the only entirely left-handed form. Its nucleobases alternate between the syn (purines) and anti (pyrimidines) positions, resulting in the phosphate groups being closer together (compared to the canonical B-DNA form) in a zigzag fashion. With the close proximity of the phosphate groups in this structure, steric hindrance and anionic repulsion is generated [91, 92]. This also leads to a deepening and narrowing of the minor groove [93]. Thus, while non-B-DNA structures are generally transient and in high-energy states, Z-DNA is arguably the structure most heavily dependent on extrinsic factors for its formation and stability in vivo and in vitro [92, 93]. This is reflected in the literature where most Z-DNA-related small molecule studies describe inducers (i.e. promote the formation) of Z-DNA structures as opposed to ligands that only recognize the structure [91, 92, 94, 95].
Z-DNA stabilizers –
Pivotal to Z-DNA recognition is ‘handedness’ and the ability to counteract repulsive factors (phosphate-phosphate proximity and steric hindrance), which are largely provided for by chiral molecules and/or metal complexes. For example, the ruthenium complexes developed by the Barton group, e.g., Ru(phen)32+, Ru(DIP)32+, and Ru(bpy)2dppz2+ (phen, 1, 10-phenanthroline; DIP, 4,7-diphenylphenanthroline; dppz, dipyridophenazine) whose left-handed lambda (˄) enantiomers bind preferentially to the left-handed Z-DNA by intercalation [96–98]. It was previously thought that intercalation through the rigid Z-DNA backbone was not permissive until these complexes indicated otherwise [99, 100]. Moreover, their binding to Z-DNA was accompanied with marked enhancement in their visible and luminescence spectra, thus also serving as probes for Z-DNA structure [96–98]. This chiral recognition of Z-DNA by a left-handed enantiomer (left-handed recognizing left-handed) is again demonstrated in more recent publications where an (M)-chiral, 1,14-dimethyl[5]helicene-spermine ligand selectively recognized and stabilized Z-DNA over B-DNA [101, 102]. A zinc (II) porphyrin (ZnTMPyP4) complex also selectively bound Z-DNA, and prevented the Z-DNA to B-DNA transition, as demonstrated by several independent studies [103–106]. The metal complex-Z-DNA interaction was indicated by a strong induced circular dichroism (CD) spectrum representative of a Z-DNA signature, and a positive wavelength-dependent linear dichroism (LD) signal. Parsing the kinetics of Z-DNA binding revealed two coexisting binding modes: one from the electrostatic interaction between the Z-DNA phosphate groups and the porphyrin positive charges, and another from the axial coordination of the central Zn(II) of the complex to the exposed N7 of guanine in the Z-DNA [105]. A sulfonated nickel porphyrin from the same group also demonstrated spectroscopic discrimination between the B-DNA and the spermine-induced Z-DNA structures [107]. The strong induced CD signal from binding with the spermine-induced Z-DNA structure, was not observed in the presence of the B-DNA, nor spermine alone [107].
Z-DNA destabilizers –
Many of the studied Z-DNA destabilizers have shown preferred binding to the canonical B-DNA structure, and were able to revert Z-DNA spectroscopic signals back to those indicative of the B-DNA form. Among them are known intercalators, initially thought not to be easily accommodated in the rigid structure of Z-DNA [99, 100]. For example, netropsin [108], distamycin, daunomycin [109, 110], adriamycin [111], ethidium bromide (EB) [112–115], dipyrandium [112], actinomycin D [113,116] tilorone [117] and thiazole orange (TO) [118] facilitate the Z-DNA to BDNA conversion when added to poly(dG-dC) or poly(dG-m5dC) in the Z-DNA form. The anticancer drug (+)daunorubicin, binds selectively to B-DNA and was found to be an allosteric effector. It converted a [poly(dGdC)]2 in the Z-DNA conformation to its intercalated B-DNA form [119, 120]. More recently, quinacrine and 9-amino-acridine were observed to bind strongly to B-DNA and decrease the rate of transition from BDNA to Z-DNA, in addition to converting the left handed poly(dGm5-dC) back to the right-handed B-DNA form [121]. The tetrapeptide, lysyl tryptophenyl glycyl lysine O-tert butyl ester (KWGK) was also shown to convert poly(dG-m5dC) Z-DNA to its B-DNA form under low salt conditions as determined by CD spectroscopy. Although, this tetrapeptide seemed to have a stronger affinity for the Z-DNA compared to the B-form, and for alternating (over non-alternating) GC sequences, due in part to the intercalation of the tryptophan moiety to the DNA [122].
As for metal complexes, a similar induction of the poly(dGm5-dC) in the Z-form to the B-form was seen for both the inversion-labile Fe(phen)32+ and inversion-stable Ru(phen)32+ complexes, even when no binding specificity was detected [123]. Unlike its zinc counterpart, the copper (II) porphyrin (CuTMPyP4) was able to convert the Z-poly(dG-dC) to the B-form [124].
The antibiotic elsamicin A was found to inhibit both the rate and extent of the B- to Z-transition of poly(dG-dC) or poly(dG-m5dC) DNA. It was also shown to convert Z-DNA to the intercalated B-form even under conditions that otherwise favored Z-DNA formation [125].
Z-DNA probes –
While there are many examples of the use of spectroscopy to monitor Z-DNA binding (almost exclusively changes in CD patterns), probes and techniques that can be used to visualize the cellular loci of Z-DNA formation are still lacking. The use of fluorescence resonance energy transfer (FRET) technology provides an alternative approach at a molecular level that allows for the monitoring and visualization of Z-DNA formation and binding that may be more sensitive and applicable to high-throughput screens. This was first demonstrated by using a tethered proto-Z-DNA-forming sequence [d(m5CG)10] flanked by a fluorescein (donor) and a Cy3 (acceptor) containing rigid DNA sequence motifs [126]. The swivel of the proto-Z-DNA-forming sequence upon its Z-DNA formation resulted in a concomitant change in the distance between the donor and acceptor and hence, FRET efficiency. Several laboratories have used the Cy3 (donor) and Cy5 (acceptor) pairs in similar successive studies [127, 128]. Examples include a study in combination with magnetic tweezers, in real-time monitoring of Z-DNA formation under controlled tension and superhelical density [127], and another study that observed the B-DNA to Z-DNA transition under low-salt conditions and Z-DNA-binding protein association and dissociation events [128].
It is notable that because the DNA conformations are dynamic and can transition between B-DNA and non-B forms, using probes to detect/monitor Z-DNA (and other types of non-B DNA) can potentially affect such equilibrium. Thus, biases can arise from the probe-induced or stabilized non-B-DNA formation.
Z-DNA ligands and biological activity –
Studies concerning biological outcomes as a result of modulating Z-DNA formation through ligands are limited. In a study investigating the mechanism by which EB exerts its anti-trypanosomal effect, Chowdhury, et al. (2010) reported that low concentrations of EB blocked replication initiation of the trypanosomic minicircle DNA (and at higher drug concentrations, nuclear replication) [129]. Interestingly, 16–24 hour exposure of trypanosomic minicircle DNA to EB produced highly supertwisted minicircle fractions containing Z-DNA (probed with an anti-Z DNA antibody), which were otherwise fully relaxed in the absence of EB [129]. Whether the replication blockage was due to Z-DNA structure formation, however, was not explored.
H-DNA
In a polypurine-polypyrimidine-containing mirror-repeat sequence, an intramolecular triplex serves as a major structural element of H-DNA resulting from the association of the DNA strand (serving as the 3rd strand) from the symmetric half of the mirror-repeat with the major groove of the purine-rich strand of the underlying duplex [130, 131]. The triplex can be of the divalent cation-dependent R*RY type (R: purine, Y: pyrimidine, *Hoogsteen/reverse Hoogsteen H-bonds) if the purine-rich tract serves as the third strand, or an acidic pH-dependent Y*RY type if the pyrimidine-rich strand serves as the third strand. This triad/triplex association is mediated either by Hoogsteen (Y third strand, e.g. T*AT or C+*GC) or reverse Hoogsteen (R third strand, e.g. A*AT, G*GC) H-bonding. Hoogsteen and reverse Hoogsteen interactions also run in parallel and antiparallel directions, respectively, with respect to the purine-rich strand of the underlying duplex. RNA strands or combinations of RNA and DNA strands can also form triplex structures [25, 34]. Thermodynamic studies in vitro have demonstrated that RNA*DNA:DNA triplexes are as stable as three DNA strands [132].
Another mode of triplex formation is through an intermolecular mechanism where the third strand is provided by an exogenous short DNA strand, i.e., a ‘triplex-forming oligonucleotide’ (TFO). TFOs have been studied for their specific DNA duplex targeting characteristics as potential gene regulatory and therapeutic agents, for example, in the regulation of gene expression, stimulation of DNA repair, site-specific mutation, etc. [131]. Thus, most studies on TFO-derived intermolecular triplexes have been focused on inducing the formation of triplex DNA. Many modifications have been made to TFOs to enhance their duplex binding specificity and affinity, including alterations to the base, sugar, 5’- and/or 3’- ends, and the phosphate backbone. These have been extensively reviewed elsewhere [131, 133–136]. However, small molecule ligands, utilized to stabilize or destabilize the triads of the intermolecular triplex are considered herein.
Triplex formation occurs with inherent instabilities, which small molecule ligands can help alleviate. For example, the interaction of a DNA third strand with a DNA duplex results in high local negative charge density, leading to unfavorable electrostatic repulsion that must be overcome to enable and maintain stable triplex formation. As such, charge-neutralizing factors that counteract this repulsion such as multivalent cations (e.g. Mg2+) and polyamines, have been well-documented [131, 132, 137, 138]. Additionally, the hydrogen donor and acceptor groups from the third strand must be available to maintain two stabilizing Hoogsteen H-bonds between the 3rd strand nucleobase and the duplex purine; thus, the formation of a Y*RY triplex such as those involving C+*GC triads, requires acidic pH for stabilization [138]. These Hoogsteen-duplex interactions are also critical to allow for base stacking among the triads [25]. Thus, triplex-specific recognition can be modulated by small molecule ligands through electrostatic interactions, Hoogsteen hydrogen bonding, third strand binding, base stacking and therefore the shape of the polyaromatic ring system.
Triplex stabilizers –
Triplex stabilizers often consist of an extended aromatic ring system and surface areas analogous (and preferably overlapping) with that of the triad, enabling strong stacking interactions. This renders specificity to the triplex structure as binding to the duplex could be disfavored due to the presence of the larger surface area. The polyaromatic ring system can be fused and planar, typical of intercalators, or unfused and non-planar linked by a torsional bond that allows flexibility for matching the propeller twist of the base triplet [25, 137]. The torsional freedom of the latter can also allow for a twist in the molecule that enables groove binding in the triplex region. Lastly, cationic moieties, either as protonated ring heteroatoms or side chains that mediate electrostatic binding to the grooves tend to be triplex-stabilizing [25, 95, 137–140], but may not always be applicable for C+*GC-containing triads due to electrostatic repulsion. Aliphatic nitrogen atoms, generally protonated below pH 8.0, have been widely used as side chain functional groups of some intercalating agents [141].
Triplex intercalators –
An early report of a molecule binding to a poly(dA)•2poly(dT) triplex, albeit weakly, was ethidium bromide [142]. However, its binding with C+*GC-containing triplexes was found to be destabilizing. This is presumably due to electrostatic repulsion to the protonated cytosines of the triad, providing evidence for its T*AT specificity [136, 142]. The characterization of benzo[e]pyridoindole (BePI) and its derivatives followed, which may be the first compound shown to interact preferentially with triplex DNA over duplex DNA [143–146]. Other intercalators that have been shown to have triplex-stabilizing properties include the unfused aromatic napthylquinolines [146, 147], LS-08 and MHQ-12 [148–150] and their self-dimers [151]. Although favoring T*AT triads, the napthylquinolines can also stabilize a triplex with a GA strand at acidic pH due to the protonation of the chromophore, which is advantageous for electrostatic binding [146, 149]. Other triplex stabilizers include: midazothioxanthones [152]; coralyne, which has a very high affinity for T*AT triplexes such that it can disproportionate a poly(dA)•poly(dT) duplex into a poly(dA)•2poly(dT) triplex and a free poly(dA) strand [153–155]; dibenzophenanthroline derivatives [156, 157]; 2,6-disubstituted anthraquinones whose neutral scaffold may preclude the requirement of T*AT triads for binding [146, 158]; acridines [159, 160]; SN-18071, a bis-quaternary ammonium (BQA) heterocycle that binds via electrostatics in the T*AT minor groove, reportedly more effectively than spermine [161, 162]; benzoindoloquinolone derivatives whose electron-withdrawing substituents can improve triplex formation and selectivity [163]; quinacridines, which have been shown to have photocleaving properties [157]; NB-506 [164]; and 3,3’-diethyloxadicarbocyanine (DODC) [165].
Diazoniapolycyclic salts, because of their extended aromatic system, have been investigated for triplex binding [137, 166, 167]. They have been shown to afford a marked 10–30°C stabilization for the T*AT triplex-to-duplex transition with minimal effect on the duplex-to-single strand transition (1–2°C stabilization) [166]. However, they have also been shown to increase the thermal melting of AT duplexes as well as calf thymus DNA. This highlights the importance of having separate structural and or sequence controls, rather than relying on the effects of the ligands on changes only in the transitions of the triplex structures.
U*AU triplex binding by the coralyne family of isoquinoline alkaloids [154], such as berberine [168] and palmatine has been recently explored [169, 170]. Berberine and palmatine showed triplex stabilization via thermal melting (by monitoring the triplex to duplex transition, without the use of separate duplex controls) but not to the same extent as coralyne [169, 170]. This was further reflected in their binding affinities, which were an order of magnitude lower compared to coralyne for triplex DNA substrates [170]. Based on fluorescence quenching and viscosity measurements, the authors inferred that coralyne bound via full intercalation, whereas berberine and palmatine only partially intercalated into the triplex DNA structures [170]. Similarly-shaped naturally occurring indoquinoline alkaloids, cryptolepine and neocryptolepine, were tested against an extended panel of substrates [171]. Cryptolepine showed a 10°C stabilization for a poly(dA)•2poly(dT) triplex. However, for the shorter (T,C), (G,A) and (G,T)-containing triplexes, no change in the thermal melting profiles were detected in the presence of 6 μM cryptolepine. In addition, competition dialysis revealed that cryptolepine had modest affinity for GC-rich structures (e.g. duplex DNA and G4-DNA) [171].
Further, 4,9-dimethoxy-11-phenyl-substituted indoloquinoline analogs have been synthesized and characterized for their interactions with intramolecular triplex substrates containing varying lengths of T*AT tracts, as well as an antiparallel GT-containing triplex (Tap), duplex, and single-strand controls [172]. Interestingly, the analog with a singly-charged aminoethylamine side chain demonstrated Tap preference with a pH-dependent stabilization and affinity, despite the modest T*AT affinity. The stronger stabilization and binding of the protonated ligand at lower pH was associated with a more exothermic binding process [172]. A similar requirement for protonated amines (aside from the napthylquinolines) to afford triplex stabilization was also observed with the recently studied benzimidazoles [173]. To determine the number and type of interactions of triplex-selective indoloquinolines with parallel triplexes, NMR studies were performed using short intermolecular triplexes specifically labeled with 3’−15N thymidine probes as substrates [174]. This study, which may be the only reported ligand-triplex NMR structural determination to date, reported that several co-existing species were observed. But a general observation was that the 5’-triplex-duplex junction was the most favorable intercalation site, in particular when flanked by a T*AT base triad. The triplex-duplex junction, as a strong site of intercalation, has been demonstrated by other intercalators such as acridine, phenanthroline, ellipticine, and ethidium bromide [175–178].
Extending the studies on substituted quinolines, the phenanthroline-containing derivatives were able to stabilize a T*AT triplex by as much as 28°C without any effect detected on the AT duplex [147, 179]. Other 2-arylquinolin-4-amines were studied in which a 4-hexamethylenediamine substituent resulted in a 44°C increase in triplex thermal melting with no effect detected on the duplex melting [137].
The importance of the complementary shape between the triads and an intercalator to promote binding efficiency was highlighted in the development of benzo[f]quino[3,4-b]quinoxaline (BQQ), which is possibly one of the most triplex-selective and effective ligands to date. BQQ, a product of rational design after molecular modeling of benzo[f]pyrido[3,4-b]quinoxaline (BfPQ), showed that better overlap (i.e. better stacking interactions) with the purine nucleobase/strand of a T*AT triplex can be achieved if the aromatic system can be extended by adding a new ring [180]. From a functional perspective, the use of BQQ and its conjugates to direct triplex-directed double-strand cleavage of plasmid DNA has been demonstrated [181].
Triplex-stabilizing groove binders –
Even in the absence of the aromaticity that provides base stacking stabilization, the polyamine/carbohydrate structure of aminoglycosides, neomycin in particular [182, 183], proved to be effective in specifically stabilizing a T*AT triplex DNA structure. The change in thermal melting by as much as 25°C, at a neomycin/base triplex ratio of ~2, without any effect detected on the DNA duplex substrate, may be the highest stabilization among triplex groove binders [184]. Neomycin has a novel and specific “Watson-Crick groove recognition” (see discussion below) that precludes duplex binding even in the presence of salt. Also, though it preferentially stabilized T*AT triads, it was also able to accommodate C+*GC triads [182, 185], despite its polycationic feature. However, it was also shown to induce the formation of hybrid DNA-RNA-DNA and DNA-RNA-RNA triplexes [184].
Combining the triplex-selective intercalator (BQQ) with the triplex-selective groove binder (neomycin), a BQQ-neomycin conjugate was synthesized, which proved to be more effective in stabilizing poly(dA)•2poly(dT) and poly(rA)•2poly(rU) than either BQQ or neomycin alone [186–188]. Its 2.7×108 M binding affinity to poly(dA)•2poly(dT) was nearly 1000-fold greater than that of neomycin alone, increasing the melting temperature by 46°C [188]. Other intercalators (e.g. naphthalene diimide, anthraquinone, pyrene) were also conjugated to neomycin, and similarly increased the stability of the T*AT triads over that of neomycin alone, but were still second to the BQQ-neomycin conjugate [188]. Whether these intercalator-neomycin conjugates are specific and can induce the formation of hybrid triplexes similarly to neomycin alone is still an open question. Other ligands that bind triple helices via groove binding include Hoechst 33258, berenil, DAPI, distamycin A and its analogs [189–191].
Some of the triplex stabilizers have been shown to induce the formation of hybrid triplex structures that otherwise would not have formed in the absence of the ligand. In the presence of poly(rA) and poly(dT) strands and at low ionic strength (~18 mM Na+), berenil, DAPI, netropsin, and ethidium bromide induced the formation of dT*rAdT triplexes, whereas the structurally similar berenil and DAPI induced the formation of rA*rAdT triplexes [192, 193]. For reference, a dT*rAdT triplex was shown to form in the presence of high Na+ (2.5 M) [194]. There is also an intercalator, oxazine 170, which can induce the formation of a hybrid poly rA:(poly dT)2 triplex [195].
It should be noted that most of these studies utilized poly(dA)•2poly(dT) triplex substrates and other Y*RY triplets [146]. The presence of the positive charge on the aromatic portion of the ligand can prevent interactions near the C+*GC triads. This was shown by the broader affinity of a neutral 2,7-disubstituted anthraquinone to (TC)n, (CCT)n, (TTC)n and (CCTT)n-containing third strands, whereas the positively-charged napthylquinoline only stabilized (TTC)n and (CCTT)n [146]. Some triplex-stabilizing ligands can bind to triplexes containing C+*GC triads but with lower affinities than to poly(dA)•2poly(dT) triplexes [136]. On the other hand, the use of R*RY triplexes as substrates of ligand interactions have been limited [149, 160, 172, 196, 197]. Some factors that may have contributed to this limitation include observations that antiparallel R*RY triplexes often possess G-rich sequences that may form competing G4-DNA structures, complicating the structural studies [198, 199]. Also, in our studies, an intramolecular R*RY triplex-forming sequence from a chromosomal breakpoint hotspot in the human c-MYC gene [200], formed a very thermally stable intramolecular triplex structure, such that further thermal stabilization effected by a potential stabilizing ligand would be difficult to observe by the currently available techniques.
Triplex destabilizers –
Historically, some groove-binding agents have been shown to destabilize triplexes, largely due to their higher affinities for the B-DNA/duplex structure. This then either prevented the binding of the third strand to the major groove of the duplex or resulted in the displacement of the bound third strand. The binding of a third strand to form the triplex creates new groove structures. Using the denotations used by Escude and Sun [136] for the triplex-forming strands, the oligopyrimidine- and oligopurine-rich strands of the duplex are referred to as the Watson and Crick strands, respectively, and the third, Hoogsteen strand. The three new grooves are referred to as the Watson-Crick (highly similar to the B-DNA/duplex groove [201, 202]), Watson-Hoogsteen, and the Crick-Hoogsteen grooves. With exceptions, most classical duplex minor groove binders have an affinity for the Watson-Crick groove (i.e. B-DNA/duplex structure) even in the presence of the third strand in the major groove. This Watson-Crick minor groove-preferred binding generates an intergroove modulation that can result in the displacement of the third strand [140, 159, 203, 204]. Thus, H-bond mediated binding to the Watson-Crick minor groove can destabilize triplexes. Conversely, while the Crick-Hoogsteen groove is very narrow, the Watson-Hoogsteen groove can accommodate the binding of ligands, for example, neomycin, which is triplex stabilizing [182, 183].
There are only a handful of triplex destabilizers reported in the literature [138, 139, 189, 205]. Although most are minor groove binders, the details of destabilization are quite varied and can be condition specific. Mithramycin binding was shown to inhibit the formation of an R*RY triplex in the human c-Ki-ras promoter. Its binding in the minor groove also resulted in the displacement of the major groove-bound TFO [206, 207]. Using the same intermolecular triplex in the human c-Ki-ras promoter, the anti-trypanosomal agent, berenil, destabilized the triplex but did not cause TFO binding inhibition or TFO displacement like that seen with mithramycin [206]. In addition, prior investigation showed that although berenil bound both poly(dA):2poly(dT) DNA triplexes and poly(rA):2poly(rU) RNA triplexes without displacing the third strand, it thermally destabilized a T*AT triplex at [Na+] ≥ 0.125 M, whereas it was thermally stabilizing at [Na+] ≤ 0.08 M. Berenil also affected the thermal stability of the RNA triplex to duplex equilibrium depending on the [base triplet]/[total berenil] ratio, with weakly destabilizing effects at [base triplet]/[total berenil] ratios >5, while thermally stabilizing this equilibrium at [base triplet]/[total berenil] ratios <5 [204, 208]. Netropsin has been shown to have a higher affinity for duplex DNA [209, 210], it bound to T*AT triplets in the minor groove, and destabilized them relative to duplexes, and yet did not displace the major-groove bound pyrimidine-rich third strand [203]. Molecular dynamics simulations suggested that near saturation of the minor groove with ligand is needed to completely dissociate the third strand [211].
As predicted by molecular modeling, 2,6-diamidoanthraquinones (see stabilizers above) can stabilize T*AT triplexes due to the extended planarity of anthraquinone that can significantly stack with the triads and side chains that are well-oriented along the grooves. The 1,4-diamidoanthraquinones, on the other hand, have steric requirements that precluded an effective triad overlap and groove accessibility by the side chains [158, 212]. Effectively competing for the third strand, the 1,4-diamidoanthraquinones showed preferential binding to duplex DNA and prevented the formation of triplex structures as evidenced by the absence of a triplex footprint [158, 212]. SN-6999, another BQA derivative, is unlike SN-18071 (see stabilizers above) in its ability to form H-bonds in the poly(dA):2poly(dT) minor groove, which can destabilize the triplex structure [161]. Other destabilizing groove binders include Hoechst 33258 [190, 213], and distamycin A [207, 214].
A more recent report, which investigated the interactions of poly(rA)·2poly(rU) triple strands with proflavine (PR) and its proflavine cis-platinum derivative (PRPt) showed that they both thermally destabilized the RNA triplex structure [215]. The destabilizing effect of PRPt was greater than that of PR (as much as 22°C destabilization versus only 8°C for PR) since its occupation of the major groove prevented the full intercalation of PR and also destabilized the binding of the third strand.
Triplex probes –
The most well-characterized triplex ligands, BePI [144] and coralyne [154], lose their fluorescence upon binding to their triplex substrates [139]. Further, the fluorescent staining of a rationally-designed H-DNA/triplex intercalator, BQQ, showed non-specific interactions with DNA [180, 216]. Thus, ligands that can specifically probe for the presence of triplex formation in real-time and provide an approach to visualize H-DNA loci in vivo are still lacking. While most triplex stabilizers are made up of extended aromatic ring systems, very few, if any, reports have fully addressed and investigated the spectroscopic changes that accompany binding of ligands to triplexes/H-DNA structures. The family of ruthenium complexes was first tested against Z-DNA, of which the derivatives Ru(phen)2dppz2+ and Ru(bpy)2dppz2+ were noted for their “light switch” effect upon nucleic acid binding [217]. However, they were found to attain their highest level of luminescent emission upon interacting with T*AT triplexes [217]. Luminescence data and modeling studies supported an intercalative model where substantial overlap and stacking occurred between the dppz ligand and the triplex bases [217]. Subsequent studies incorporating a larger intercalative ‘wing’, Ru(phen)2bdppz2+ [218], showed similar spectral properties between duplex or triplex DNA substrates confirming the triplex intercalative model, and further indicated that the binding occurred from the minor groove. More recent Ru2+-based triplex studies have been tested against a U*AU RNA triplex substrate, and with varying complexities of coordinating aromatic intercalators. The intercalative mode of interaction and subsequent stabilization of the RNA triplex still holds, albeit without demonstration of specificity (e.g. duplex versus triplex, DNA triplex versus RNA triplex) [219–221].
Ligands that can stabilize intermolecular triplexes, a number of which have reported increases in fluorescence intensities, have been characterized. A bisintercalator YOYO was used in the recognition of a mixed DNA sequence by a homologous single-strand oligonucleotide [222]. The free and bound YOYO absorbed at 458 and 490 nm, respectively, which precluded signal interference from a number of biological molecules. YOYO also increased its fluorescence ~40-fold when intercalated in DNA, compared to the fluorescent free YOYO. However, when its relative quantum yield was estimated, it was found to be similar when it interacted with duplex or triplex DNA [223]. Similarly, a pyrene moiety tethered to the 3’-end of a triplex‐ forming homopyrimidine oligonucleotide exhibited strong fluorescence when triplex formation occurred [224]. The probe alone was spectroscopically silent but was increased to as much as 45-fold in fluorescence quantum yield depending on the thermal stability of the triplex substrate. Thiazole orange (TO), a synthetic cyanine dye, which has shown a >1000-fold increase in fluorescence quantum yield when intercalated into duplex DNA [225], may have higher affinity and selectivity for G*GC triplexes and G4-DNA over duplex DNA [226]. A TO displacement assay has been developed based on its affinity to G4 DNA structures. The assay relies on the displacement of G4-bound TO to test the affinity and selectivity of putative G4-specific ligands [227–230]. A related cyanine dye, Cyan 40, which absorbs at ~435 nm as a free dye, was found to be specific for G*GC triplexes relative to G4-DNA [231]. DMT (methyl-2,6-[2-(4-methyl-sulfanyl-phenyl)-vinyl]-pyridine) is one of the more recently reported triplex-probing synthetic small molecules; however, it showed only moderate fluorescence enhancement and little structure selectivity [232].
Wang et al. (2015) tested a panel of flavonoids for their interactions with DNA triplex structures, and fisetin (7,3′,4′-flavon-3-ol), a plant polyphenol, was identified as a T*AT triplex-specific probe [233]. When bound to a triplex substrate, it exhibited a bright green fluorescence emission that was observable by the naked eye under UV illumination. This emission was reported to be the result of a process termed ‘excited state intramolecular proton transfer’ (ESIPT) between its exocyclic 3-hydroxyl and 4-carbonyl groups. Fisetin was shown to stabilize the triplex structure by ~14°C, without affecting the duplex control. With ESIPT requiring aprotic solvents, the use of fisetin may be limited to low pH-requiring C+*GC triplexes. It will be interesting to see whether the strong emission also occurs in the presence of the R*RY triplexes. Another natural product, chelerythrine, was isolated from a panel of natural isoquinoline alkaloids, and found to be fluorescent (~120-fold) upon its interaction with T*AT triplexes, increasing the triplex to duplex thermal transition by ~7°C [234].
Triplex ligands and biological activity –
Despite the development of small molecules as modulators of triplex/H-DNA structures, cellular studies that demonstrate biological effects of such modulation are still underexplored. BePI was used in one of the first few studies that demonstrated that a ligand binding to H-DNA resulted in a biological outcome [235]. Here, a 55-bp polypurine-polypyrimidine sequence able to adopt a Y*RY H-DNA triplex structure, was inserted into a plasmid (pIbla69) between the bla promoter and the coding start site of the β-lactamase gene. In the presence of BePI, a stabilizer of H-DNA, chloroacetaldehyde modification showed increased hyperreactivity of the adenines on the 3’-side of the R strand consistent with the folding of the 5’ half of the Y strand to form H-DNA. H-DNA structures can cause replication arrests [236–238] during in vitro replication, and in the presence of increasing BePI concentration, dose-dependent arrests (to as much as 90% in the presence of 2 μM BePI) of the elongation products of E.coli DNA polymerase I were observed. Using Taq polymerase at high temperature, the presence of BePI resulted in 55% and 40% inhibition of replication at 52°C and 62°C, respectively. In contrast, in the absence of BePI, very few replication arrests were detected at 52°C, and no arrests were observed >52°C.
H-DNA-forming sequences have also been shown to cause transcription arrest [238]. Thus, using the same plasmid system described above, a decade later, the effect of the BQQ ligand on the H-DNA insert and subsequent β-lactamase expression (via β-lactamase activity) was assessed [239]. The expectation was that an H-DNA-forming sequence positioned upstream of the open reading frame would reduce β-lactamase gene expression. Indeed, there was decreased β-lactamase activity in the cells with the H-DNA-containing plasmid (pIbla69). Addition of increasing concentrations of BQQ (0.5 – 4 μM) led to a further dose-dependent reduction of β-lactamase activity with as much as 48% reduction in the presence of 4 μM BQQ. Conversely, the presence or absence of BQQ did not affect the enzymatic activity of the cells harboring the parental control plasmid (pBR322). The authors did not detect a marked difference in the quantity of plasmid DNA in the presence or absence of BQQ. From this, they eliminated the possibility of inhibition of plasmid DNA replication, and attributed the repression of transcription due to BQQ stabilization of H-DNA as the cause of reduced β-lactamase activity.
(GAA•TTC)n repeats implicated in Friedreich’s ataxia (FRDA) were shown to form ‘sticky DNA’ arising from the association of two R*RY triplexes in negatively supercoiled plasmids at neutral pH [240]. These triplexes can stall RNA polymerases, leading to the inhibition of transcription and subsequent reduced frataxin (FXN) protein levels in FRDA patients [205, 241, 242]. Low FXN levels led to progressive neurodegeneration and cardiomyopathy, characteristic of FRDA [243]; thus, destabilizing the formation of the triple helical structure may restore FXN levels [205, 244–246]. One approach has been to use a short, low molecular weight oligonucleotide that can bind to the ‘third’ strand of the triplex structure. The oligonucleotide competed with the formation of the triplex structure, which led to a specific and concentration-dependent increase in the full transcript of the FXN gene [244]. Another approach was to use a linear β-alanine-linked polyamide FA1 (ImPyβImPyβImβDp, where Py=pyrrole, Im-imidazole, β=β-alanine, Dp=dimethylaminopropylamine), which is a duplex minor groove binder. By stabilizing the duplex, strand separation and subsequent triplex formation was disfavored [245]. Its use also increased FXN transcription by ~3-fold at both the mRNA and protein levels in an FRDA lymphoid cell line [245]. Pentamidine, another minor groove binding agent identified via a competition dialysis method, increased the levels of FXN by 2-fold in patient cells [246].
G4-DNA
G4-DNA structures can form at tandem repeats of guanine (G)-rich DNA or RNA sequences [32, 33, 247]. The four guanines associate with each other via Hoogsteen hydrogen bonding in a planar, cyclic array (i.e. a G-quartet or G-tetrad), either intramolecularly (single-strand folding upon itself) or intermolecularly (two or more strands) with parallel or antiparallel strand directionality. The large planar surface of two or more G-tetrad, in part due to strong Van der Waals attraction, can stack on top of each other to form the G4-DNA structure. G4-DNA structures can be stabilized by monovalent cations in the order of K+ > Na+ > NH4+ > Li+ [248], although some divalent cations have also been reported to stabilize G4 structures [249]. The cations can counterbalance the high electronegative potential created by the carbonyl groups of the guanines, which are directed toward the interior of the G-tetrad [28]. This ‘central’ coordinating role of cations was calculated to be an important stabilizing factor in addition to the Hoogsteen H-bonding and stacking interactions of the G-tetrads [249, 250]. This is in addition to the known roles of cations in neutralizing phosphate backbones that are also in close proximity in G4-DNA. As dictated by their ionic radii, smaller Na+ ions can either be in-plane of a G-tetrad or equidistant from two successive G-tetrads, while K+ ions are exclusively found between two adjacent G-quartets [251].
G4-DNA structures have four grooves with dimensions that vary with the phosphodiester groups, their dimensions dictated by topology and the nature of the loops. The topology and loop conformation of the G4-DNA structures are known to be highly polymorphic [reviewed in [28, 247, 252–257]]. They are regulated by the nucleic acid sequence (length and composition; e.g., the number of loop nucleotides) [258], orientation (parallel versus antiparallel) and stoichiometry (one to four) of strands, the glycosidic conformation of the G nucleosides (syn versus anti), and solution conditions (e.g. cations, co-solutes) [249, 259, 260].
Thus, the structural features of G4-DNA amenable for ligand modulation include the G-quartets, the central channel of the G-quadruplex typically occupied by metal ions, the phosphate groups, the loop nucleobases, and the grooves at the surface of the G4-DNA stack.
G4-DNA stabilizers –
Much work has been published on G4-DNA ligand interactions, with the majority of the studies focusing on G4-DNA stabilization. We would like to direct the readers to reviews generated from the last ~5 years covering organic compounds and metal complexes as G4-DNA ligands [252–254, 261–267].
Studies on G4-DNA binding showed that an intercalative mode of binding in between G-tetrads is highly unfavorable because it may require displacing a cation within a very strong stacking environment [252, 263]. Only a few reports have indicated that intercalation may have taken place between G-quartets, and these were performed in the absence of K+ and with TMPyP4 as a ligand [268–270]. Thus, the most common modes of stabilizing G4-DNA interactions are via: 1) stacking into one of the end G-quartets of G4-DNA (end-stacking); 2) groove/loop; and 3) combined end-stacking and groove/loop binding.
Small molecules that can end-stack with G4-DNA include planar aromatic surfaces that mimic the large planar surface of the G-quadruplex. Fused aromatic polycyclic systems, macrocycles, and non-fused aromatic systems have been cited as three major families of G4-DNA interactive molecules that target the external G-tetrads [263, 271]. Some of these include anthraquinones, cationic porphyrins, acridines [272] and others, which are derived from duplex DNA binders, and therefore often do not offer high specificity for G-quadruplexes. TMPyP4, for example, is a porphyrin-based macrocycle, that also binds duplex and triplex DNA [273, 274]. However, it was one of the first ligands to have been studied for its potential to stabilize a G4-DNA-forming sequence in the c-MYC promoter (via partial end-stacking, as assessed by footprinting studies) with associated suppression of transcriptional activation [275]. A possible exception is Phen-DC3, a bisquinolinium compound, which despite its interaction with a c-MYC intramolecular G4-DNA-forming sequence via extensive π-π end stacking [276], has also displayed exceptional selectivity for G4-DNA relative to duplex DNA [228, 277]. It is also very potent in that it can enhance G4-DNA thermal stability (ΔTm) up to ~30°C [228, 277]. Thus, it has been used in several assays to probe genetic instability from stabilized G4-DNA formation in yeast and mammalian cells [56, 278, 279]. Further, as end-stacking only requires a G-tetrad (i.e. a planar aromatic surface) to stack with, the binding of these molecules may not require a specific G4-DNA topology, and thus, may lack discrimination among the various G4-DNA conformations, and may even promote ligand-mediated G4-DNA structure formation [266].
Alternatively, the groove and loop regions of G4-DNA differ from the canonical DNA duplex and other non-B DNA structures, and even among G4-DNA structures, and thus may provide a better approach for structural selectivity in binding [reviewed in [280]; [252, 254, 266]]. Finally, recent reviews have suggested that the most thermally stabilizing and selective G4-DNA ligands often have the combined features of end-stacking and groove/loop binding. Specifically, these include aromatic ligands with a U-(or V) shape, exemplified by fused aromatic polycyclic and macrocyclic ligands, with electron withdrawing atoms/groups (e.g. protonable nitrogens, halogens) that can be locked in a planar conformation (i.e. with constrained flexibility) for effective G-tetrad stacking, and with basic side chains that target the loops and grooves for better selective G4-DNA binding and stabilization [263]. Pyridostatin (PDS) is such an example. PDS is a G4-selective stabilizing ligand whose design was intuited from previous studies on a potent macrocyclic oligoamide derivative [281]. Müller, et al. (2012), designed PDS as a flexible molecule, yet able to adopt a flat conformation such that it can participate in π-π interactions with the G-tetrad. It also contains amino groups as side chains to participate in electrostatic interactions [281]. As discussions below indicate, PDS has been useful in studies of co-localization [282] and in mapping the genome-wide distribution of potential G4-DNA-forming sequences [283].
G4-DNA destabilizers –
In contrast to the number of G4-DNA stabilizers that have been characterized, there have only been a limited number of reports that pertain to G4-DNA destabilizers. Bioavailable molecules such as polyamines at >1 mM concentration can destabilize G4-DNA structures [284], whereas 20 wt% urea resulted in an ~11°C Tm decrease for a human telomeric G4-DNA sequence in the presence of K+ [285]. Many of the reported G4-DNA destabilizing ligands include cationic porphyrins and their derivatives. While TMPyP4 has stabilizing effects toward telomeric G4-DNA structures [286], it was found to be destabilizing toward a bimolecular d(CGG)n and intramolecular r(CGG)n G4-DNA-forming trinucleotide repeats [286, 287]. The formation of secondary structures in these trinucleotide repeats has been implicated in the silencing the FMRI gene in Fragile X syndrome [241, 243]. Excess TMPyP4 in the presence of d(CGG)7 and 20 mM K+ resulted in a 15°C decrease in thermal melting [286], whereas TMPyP4 slowed the electrophoretic migration of a r(CGG)33-containing plasmid and enhanced the translation efficiency in vitro of (CGG)99 firefly luciferase mRNA [287]. In a complex with platinum, PtTMPyP4 was found to be weakly destabilizing (Tm decrease of 3°C) to a parallel bimolecular G4-DNA from the d(TAGGG)2 sequence in the presence of 0.1 M K+[288]. While the TMPyP3 isomer was reported to have destabilizing effects on the antiparallel human telomeric G4-DNA structure-forming sequence, d(TTAGGG)4, decreasing its thermostability by 8°C [289]. A derivative bearing spermine pendants in the four meso positions of the porphyrin scaffold, H2TCPPSpm4, led to a 30°C decrease in melting temperature of the tetramolecular G4-DNA-forming DNA aptamer (TGGGAG)4 when added at a 2 μM concentration in a 1:1 molar ratio [290].
Lanthanide metallacrown complexes (MCs) have been shown to decrease the melting temperature of human telomeric G4-DNA (22Htel) by 16–20°C at a 5:1 complex:DNA ratio [291]. The destabilizing effect was also seen via CD spectroscopy studies where addition of the MCs led to the disappearance of the signature G4-DNA CD bands without the appearance of new bands. The destabilizing effect was ascribed to either the large MC plane that caused unfavorable steric interactions with the G4-DNA, or to the presence of Cu2+ ions, which also comprise the MCs, which can interact/coordinate with N7 of guanines [291].
A triarylpyridine with three side chains (TAP1) has been shown to have a moderate affinity (Kd ~11 μM) and a high stabilizing potential (ΔTm ~23°C) for a G4-DNA-forming sequence from the promoter of the c-kit proto-oncogene [292]. Yet CD and NMR studies have revealed that TAP1 can significantly disrupt and weaken the G-quartets. Further support came from gene expression studies where human HGC-27 cancer cells overexpressing c-kit were incubated with different concentrations of TAP1. Previous studies showed that stabilization of G4-DNA led to reduced c-kit gene expression [275, 293–295], and treatment with TAP1 resulted in a dose-dependent increase (~90% increase with 5 μM treatment) in c-kit expression relative to control genes, as measured by quantitative real-time PCR [296].
A synthetic pyrrole-inosine nucleoside was designed to form a specific extended three-point Hoogsteen H-bonding interaction with guanines [297, 298]. From 1H NMR (CDCl3, CD2Cl2 as solvents) and ESI-mass spectrometry studies, this competitive interaction was shown to disrupt guanosine dimerization and G4-DNA formation from the parallel tetramolecular [dTG4T]4 and the intramolecular d(T2G4)4 sequences [297]. While this represents a promising approach to destabilize G4-DNA, it may not be G4-DNA structure-specific and thus, may also disrupt other guanine-rich sequences in the genome.
G4-DNA ligands and biological outcomes –
There have been a large number of studies published utilizing ligand-mediated G4-DNA structure modulation to interrogate biological activities, and potential therapeutic outcomes. These include (but are not limited to) telomeric-end processing, effects on gene expression via transcriptional control, and genetic instability. These reflect telomeres and gene promoters as major loci of G4-DNA-forming sequences, the former being currently exclusive to G4-DNAs. As a result, some G4-DNA-specific ligands have entered human clinical trials. Quarfloxin/CX3543 [299] completed Phase I/II clinical trials for neuroendocrine and carcinoid tumors, whereas CX5461 [300, 301] is currently in late Phase II clinical trials for breast cancer patients with BRCA1/2 or homologous recombination deficiency (HRD) germline aberrations.
This rapid growth of ligand-G4-DNA investigation was, in part, stimulated by initial findings that sequences from the human telomere can form G4-DNA [302–304] and inhibit telomerase activity [305, 306]. Human telomeres are comprised of nucleoprotein complexes that can be found at the end of chromosomes, the extended maintenance of which has been implicated in cancer etiology [26]. The telomeric DNA is composed of a double-stranded region terminating into a single-stranded, G-rich 3’-overhang with a unit sequence of (TTAGGG)n whose G4-DNA formation [302–304] was shown to inhibit telomerase activity [305–307] by restricting access of the telomerase RNA template to the overhang. It has also been suggested that G4-DNA folding protects the overhang from replication protein A (RPA) binding, and thereby suppressing DNA damage signals [308]. Telomerase has been shown to be upregulated by as much as 85% in some human cancer cells [309], and is thought to promote the lifespan of the cancer cells by further synthesizing telomeric DNA [310, 311]. This gave rise to a biochemically informative assay to probe ligand interactions with telomeric G4-DNA, known as the telomeric repeat amplification protocol (TRAP) assay [312]. In the TRAP assay, telomerase activity appears as extension products, but in the presence of a G4-DNA stabilizing ligand, telomerase activity will be inhibited and prevent the formation of the extension products. Thus, ligand-mediated G4-DNA stabilization has become a potential anticancer approach through its interference with telomere function and/or inhibition of telomerase (and other telomere-related proteins), providing an impetus to identify telomeric G4-DNA interactive agents [261, 263, 264, 266, 267].
G4-DNA-forming sequences have been found to be overrepresented in biologically active regions of the genome, including gene regulatory and promoter regions [313]. Interestingly, while estimates predict that ~40% of human gene promoters contain G4-DNA-forming sequences [314], proto-oncogenes are reportedly more enriched in these sequences compared to tumor suppressor and housekeeping genes [315, 316]. Thus, ligand-mediated structure modulation of G4-DNA in gene promoter regions may represent an approach to control deleterious gene expression. Studies to stimulate G4-DNA formation and stabilization have demonstrated potential for transcriptional control, either gene expression upregulation or downregulation, depending on the interactions of G4-DNA with proteins involved in the regulation of transcription [263, 264, 267, 317]. A number of reviews are available related to ligand targeting of promoter-specific G4-DNA formation [261, 266, 317]. On the other hand, G4-DNA structure destabilization, while relatively underexplored, is another means to control gene expression. This was exemplified by the increase in translation efficiency in vitro of (CGG)99 firefly luciferase mRNA through treatment with the TMPyP4 ligand [287].
G4-DNA probes –
The abundance of characterized G4-DNA-stabilizing ligands provides ligands that can be used as potential probes of G4-DNA structure, from organic fluorophores to metal complexes [263, 264, 318–322]. In addition to the “light up” and “light off” probes [321, 323], which show gain or loss of spectroscopic/fluorescent signals upon G4-DNA binding, high-affinity ligands ‘tagged’ with fluorophores have also been investigated [323, 324]. Some ligands with promising biophysical characteristics have advanced to testing by imaging in cells at the chromosome level [321, 325, 326]. While selective staining in the nucleus or nucleoli was observed in a few studies, the results may not able to be unequivocally interpreted as the occurrence of G4-DNA structures in vivo, as there could be non-specific binding to other biomolecules (e.g. RNA with other 3D topologies, etc.). Further, cellular localization of some ligands were noted to vary, whether in living or fixed cells, or depending on the mode of crosslinking or fixation used. Only by the use of orthogonal assays, such as crosslinking, pulldown, or sequencing (or a combination), using live cells, would such staining be confirmed as an actual visualization of G4-DNA structures in vivo.
Methods to identify small-molecule modulators of DNA structure
With increasing evidence of the involvement of non-B DNA structures in regulating biological processes, this review aims to highlight the current status of the recognition of non-B DNA (Z-DNA, H-DNA, and G4-DNA) by structure-specific ligands. This approach takes advantage of distinct structural features of a particular non-B DNA structure compared to the canonical B-form DNA (and other non-B DNA structures) to obtain small-molecule ligand recognition. Using methods that span from biophysical and biochemical assays to functional cellular investigations - the binding selectivity, binding affinity, and subsequent biological outcomes of structure-specific binding can be defined. The methods that have been employed to investigate non-B DNA-ligand interactions are essentially similar to those used to identify B-DNA-ligand interactions [327–330], but under experimental conditions (e.g. buffer, pH, wavelength, signatures, etc.) that are suited for the formation/detection of a particular non-B DNA structure. For example, to probe optical thermal melting for short sequences of G4-DNA structures, the temperature-dependent absorbance at 295 nm (A295) characterized by a distinctive hypochromic shift, is monitored [331, 332], rather than absorbance at 260 nm (A260), which is used to detect canonical B-DNA and H-DNA structures [333–335]. Examples of methods to detect and evaluate non-B DNA-ligand interactions are summarized in Table 1 and have been previously reviewed [91, 138, 336–341].
Table 1.
Various methods to detect and evaluate non-B DNA-ligand interactions.
| Analysis of in vitro binding | Information on ligand-DNA binding interactions |
|---|---|
| Spectroscopy | |
| UV-Vis | Concentration dependence/titration: affinity, stoichiometry Temperature dependence/thermal melting: stabilizing/destabilzing effect |
| Fluorescence | Concentration dependence/titration: affinity, stoichiometry Temperature dependence/thermal melting: stabilizing/destabilzing effect Competition/fluorescent intercalator displacement (FID): relative affinity, structural specificity (vs. competing non-B DNA structures), structure-related sequence preference (vs. other structure-forming sequences) Förster resonance energy transfer (FRET) with thermal melting: stability Quenching experiments: mode of binding |
| Circular/Linear Dichroism (CD/LD) | Comparison of CD/LD signatures: structural/topological changes, mode of binding Temperature dependence/thermal melting: stability |
| Nuclear Magnetic Resonance (NMR) | 3D Structure, structural/topological changes, mode of binding |
| Surface Plasmon Resonance (SPR) | Concentration dependence/titration: affinity, stoichiometry, kinetics of binding |
| Electrospray ionization-mass spectrometry (ESI-MS) | Stoichiometry, relative binding affinity, relative abundance of ligand-DNA complexes |
| Viscosity/hydrodynamic measurement | Mode of binding |
| Isothermal titration calorimetry (ITC) | Binding energetics, stoichiometry |
| Electrophoretic mobility-shift assay (EMSA) | Stabilizing effect on a destabilized structure, binding affinity, specificity, kinetics of binding |
| Footprinting assay | Structural/topological changes, mode of binding |
| Competition equilibrium dialysis | Structural specificity (vs. competing non-B DNA structures), structure-related sequence preference (vs. other structure-forming sequences) |
| Capillary electrophoresis | Relative binding affinity |
| X-ray crystallography | 3D Structure, structural/topological changes, mode of binding |
| Atomic force microscopy/optical tweezers | Stability, structure |
| In vitro functional assays | Information on ligand-DNA binding interactions |
| DNA polymerase stop assay | Efficacy of ligand stabilizing (or destabilizing) effect that stalls (or improves) enzymatic processing of substrate |
| Reverse transcriptase stalling (RTS) stop assay | Efficacy of ligand stabilizing (or destabilizing) effect that stalls (or improves) enzymatic processing of substrate |
| G4 TRAP assay | Efficacy of ligand stabilizing (or destabilizing) effect that stalls (or improves) enzymatic processing of substrate |
| DNA polymerase extension assay | Efficacy of ligand stabilizing (or destabilizing) effect that stalls (or improves) enzymatic processing of substrate |
| Replication/transcription arrest assay | Efficacy of ligand stabilizing (or destabilizing) effect that stalls (or improves) enzymatic processing of substrate |
| Cell/genome-based functional assays | Information on ligand-DNA binding interactions |
| Footprinting assay | Structural/topological changes, binding loci |
| Co-localization assay | Visualization of non-B DNA-ligand loci, specificity of loci |
To better define the mechanistic details of non-B DNA and ligand binding interactions, in vitro analyses employing short non-B DNA structure-forming sequences have often been used [329, 330]. Such in vitro analyses can be used to address many questions, including: 1) how is the ligand binding to the non-B DNA-forming sequence (e.g. binding mode, binding affinity, kinetics, stoichiometry, energetics); 2) where is the ligand binding the non-B DNA-forming sequence (nucleotide-level binding site); 3) how specific is the ligand binding to the non-B DNA structure of interest (versus duplex DNA or other non-B DNA structures or structure-related sequence preferences); and 4) what is the effect of the ligand binding to the non-B DNA-forming sequence (stability, i.e., stabilizing or destabilizing). Consequently, the answers to these questions are better defined from combined results using different techniques, rather than from a single type of experiment.
On the other hand, in vitro biochemical- and molecular biology-based assays, able to accommodate longer non-B DNA-forming sequences (e.g. incorporated into plasmid DNA), can further define the functional effects of ligand binding to non-B DNA. By demonstrating that ligand-modulated structure formation affects the function and/or enzymatic processing of the DNA substrate, the putative ligand-non-B DNA interaction and its stabilizing or destabilizing effect(s), can be further validated.
Lastly, cell/genome-based investigations using small molecule ligands have been employed to identify biologically-relevant functions of non-B DNA-forming sequences and to determine their loci in cells [338]. For example, footprinting-based studies have been used to detect non-B DNA structures in the genome [316, 342], and have revealed the presence of non-B DNA in promoter regions of developmental regulatory genes and oncogenes [316], and in human precursor mRNA [342]. Co-localization studies that involved treatment of live cells with G4-DNA-stabilizing ligands such as PDS, TMPyP4, and telomestatin showed increased numbers of G4-DNA-antibody foci, suggesting that either the ligands trapped the G4-DNA structures when they formed [86, 89], or that they induced the formation of G4-DNA. Similarly, telomestatin treatment of Fanconi anemia group J (FANCJ)-depleted chicken DT40 cells further increased the numbers of G4-DNA-antibody foci, which the authors claimed to be supportive of the existence of G4-DNA structures in mammalian cells and their processing by helicases, such as FANCJ [89].
While not intended to investigate mechanistic or functional effects of ligand binding, next generation sequencing (NGS) has been utilized with G4-DNA-specific ligands to investigate the genome-wide distribution of G4-DNA loci [343, 344]. A biotin-tagged Schiff-base catechol derivative was used to crosslink G4-DNA structures in murine melanoma cells [345], after which the G4-DNA-forming sequences were isolated using streptavidin magnetic beads and subjected to NGS. However, out of the 1,294 sequences isolated (mostly from gene promoter regions), only 120 (~10%) were putative G4-DNA forming sequences [345]. Another approach, G4-seq, combined the G4-DNA-stabilizing effects of PDS or PhenDC3 ligands with the polymerase stop assay followed by Illumina NGS [346] to profile G4-DNA structures in purified human genomic DNA [248]. The technique relied on ligand-mediated, G4-induced polymerase stalling, which introduced sequencing errors and mismatches at sequences of G4-DNA start sites. In comparison to the sequencing data obtained under conditions that disfavor G4-DNA formation, the exact position, as well as the putative G4-DNA-forming sequences can be elucidated [346]. G4-seq identified ~700,000 putative G4-DNA-forming sequences in the human genome, which is twice that estimated by standard G4-DNA predictive algorithms [344, 346] and orders of magnitude more than the sites revealed by chromatin immunoprecipitation (ChIP)-sequencing methodologies [283, 347]. A model study involving a cyclic hepta-oxazole compound with a biotin-affinity tag was reported to have isolated G4-DNA forming sequences from a mixture of G4-DNA and non G4-DNA forming sequences [348].
Recent developments in experimental methods of ligand-non-B DNA interactions have involved the use of high-throughput screens (HTS), alone or in combination with other techniques. Screening for triplex ligands from natural plant extracts using peak area-fading ultra high-performance liquid chromatography (HPLC) coupled with Orbitrap mass spectrometry (MS) [349], and a combination of HPLC-MS (ESI) [350] has been reported. An evaluation that involved ESI-MS, tandem mass spectrometry (MS/MS) and molecular modeling was performed on benzopyridoindole and benzopyridoquinoxaline triplex ligands [351]. Rosu et al. (2007) utilized molecular modeling data in combination with tandem mass spectrometry data [which calculated the energy to dissociate 50% (CE50) of the bound ligand], to parse binding interactions of ligands with duplex or triplex DNA [351]. The combined analyses indicated that the ligands intercalated via the minor groove of the Watson-Crick duplex, and the benzopyridoindoles, in particular, interacted with the triplex via the duplex [351].
Thermal melting measurements [333–335] have been a standard method to determine stabilizing or destabilizing effects of ligands on nucleic acids. The low-throughput sampling of this approach has been alleviated through melting mixtures of poly- or oligonucleotides containing the structure-forming sequences of interest [352]. In this approach, addition of the ligand at low molar ratios will shift the thermal melting temperature (Tm) of the preferred structure or sequence [352]. High-throughput FRET-based thermal melting assays using fluorophore-quencher (FQ) combinations [128, 353], have also been described [354–356]. An in-depth testing of the ‘two-state’ model [334, 357], often assumed during thermal melting experiments, was performed by analysis of multidimensional ‘3D’ melting curves for G4-DNA [358]. This 3D-melting involves the recording of the entire spectra (absorbance, CD, and fluorescence) as a function of temperature, as opposed to the common practice of recording temperature-dependent spectral responses at a single wavelength. The 3D melting data was then subjected to analysis using singular value decomposition that led to better characterization of the unfolding of G4-DNA by revealing intermediate states previously unknown [358].
The use of FRET in the context of molecular beacon (MB)-based methodology has been explored for triplex ligand screening with T*AT triads [359]. Inspired by the MB technology and other published assays utilizing fluorophore-quencher (FQ) combinations [353–356, 360, 361], our group has developed a FRET-based assay that can identify both triplex stabilizers and destabilizers, simultaneously, in a facile HTS-compatible approach using a biologically-relevant triplex-forming sequence [362].
Due to their high extinction coefficients, absorption in the 500–700 nm range, and unique aggregate optical properties [363], gold nanoparticles (AuNPs) and their interactions with DNA have been utilized for facile colorimetric assays to screen for triplex-binding ligands. Whether the AuNP was functionalized with DNA [364, 365] or was label-free [363], the assay depended on the visible color change of the AuNP as a result of the ‘aggregation’ concomitant with triplex formation in the presence of a binding ligand.
Recently, the use of chemometric approaches through the application of principal component analysis (PCA) and hierarchical cluster analysis (HCA) on large data sets derived from G4-DNA fluorescent intercalator displacement (FID) assays was reported [366]. PCA and HCA were used as tools to provide information on the often complex relationship between binding affinity and binding selectivity [366].
Lastly, computational studies concerning non-B DNA structures continue to advance, with regards to molecular dynamic simulations of structures, and their thermodynamic stability [367–369]. And as virtual ligand screening still serves as an economical tool to identify possible ligand candidates, the best approaches and platforms have been reviewed [370], and used in conjunction with an oligo affinity support assay for G4-DNA structures [371].
Challenges and perspectives
Challenges still remain to be addressed in optimizing ligands as tools to study non-B DNA structure formation and function. There is a need to study ligand-binding interactions using biologically-relevant structure-forming sequences as model targets to increase therapeutic relevance and increase the likelihood of an effective biological outcome. Also, there has been considerable progress made in identifying G4-DNA-interacting ligands as exemplified by the development of cell-based and genome-based functional methodologies that employ G4-DNA-interacting molecules in an effort to locate genome-wide G4-DNA loci, and the advance of G4-DNA-specific ligands, such as Quarfloxin/CX3543 [299] and CX5461 [300, 301] to Phase II clinical trials. However, there are fewer examples of Z-DNA- and H-DNA-specific ligands, and additional studies to identify non-B DNA-specific ligands are warranted. With regards to use of experimental methods, there is advantage in incorporating biochemical or in vitro functional assays, and when possible, cell and genome-based functional assays in studies to determine non-B DNA-ligand interactions. Consequently, facile and accessible assays to monitor biochemical or cellular responses (e.g. interference with processing of helicases or DNA damage responses) upon treatment with non-B DNA-interacting ligands are warranted. The development of such assays will likely improve as more information regarding the mechanistic processing of non-B DNA is obtained.
Because non-B DNA can stimulate genetic instability, the precise temporal and spatial modulation of a non-B DNA structure and its mutagenic outcome might be achieved by the use of DNA structure-specific small-molecule ligands. Though the majority of non-B DNA-specific small molecule ligands identified to date stabilize the DNA structures, it is also of importance to identify non-B DNA destabilizers. For example, non-B DNA destabilizers may prove useful in preventing genetic instability induced by these mutagenic structures. However, both non-B DNA stabilizers and destabilizers can serve as molecular tools to interrogate mechanisms involved in DNA structure-induced genetic instability, to probe structure formation in vivo and thus, have potential toward diagnostic and/or therapeutic applications.
Structure-destabilizing ligands are predicted to be a synthetic approach to decrease or even prevent the genetic instability associated with mutagenic non-B DNA structures. Thus, they may be impactful toward therapeutic approaches to prevent and/or treat disorders arising from DNA structure-induced genetic instability. An example is the recovery of frataxin (FXN) protein levels by destabilization of the triplex structure that inhibited the transcription of the FXN gene, characteristic of Friedreich’s ataxia [205, 241, 242] as cited in the ‘Triplex ligands and biological outcome’ section of this review. Destabilization of the triplex was achieved through the use of an oligonucleotide [244], polyamide [245] or a pentamidine ligand [246]. Only a handful of non-B DNA-destabilizing ligands are known and this bottleneck is, in part, due to the shortage of efficient methods to assay for structure-destabilizing molecules. To effectively identify ligands that can destabilize mutagenic non-B DNA structures, the development of additional facile assays or screens is needed.
On the other hand, the use of effective and selective non-B DNA stabilizers will prove useful for mechanistic or diagnostic investigations, by allowing a better understanding of structural features, structure formation, location, and the biological functions of non-B DNA in vivo. Further, there are genomic regions that contain multiple and overlapping potential non-B DNA-forming sequences [19], or more than one topology of a particular non-B DNA structure [267]. Direct and conclusive methods to detect specific DNA structures within a region of putative non-B-DNA forming sequences in vivo are still lacking. Confounding such complexity is the accessibility limits posed by chromatin organization within the context of chromatin in a cell. Thus, there is a need for tools such as structure-specific fluorescent probes derived from stabilizers that can visualize and report on the conformational dynamics, cellular loci, and processing of non-B DNA structures. However, it should be noted that super resolution microscopy, which can provide better monitoring of biomolecular processes in live cells (compared to other forms of microscopy) [372], is fluorescent-label dependent, and has not yet achieved the resolution to visualize individual/monomeric non-B DNA structures [323]. Its current resolution is at 10–70 nm [373], which still needs improvement to resolve single-molecule non-B DNA structures. Thus, even in the case of a specific non-B DNA structure ligand, only large domains with a high density of non-B DNA structures can be imaged by microscopy [323]. Under what conditions such domains exist in the context of chromatin in a cell remain to be elucidated. As such, ligand-induced staining, which has been attempted for G4-DNA structures (see G4-DNA Probes discussion above), needs independent validation for the sequences and loci of non-B DNA structure formation [323]. To observe conformational dynamics of non-B DNA formation in vivo, parallel developments in super-resolution imaging, in conjunction with improved small molecule fluorophores, is required. Thus, in the search for non-B DNA ligands, consideration should be given to the features of ligand/fluorophores that would be compatible with or further improve the current limit of super-resolution imaging. For example, bright, photoswitchable (or photoactivatable), photon-emissive, photostable fluorophores [373–375], should be considered, in addition to the minimum ligand requirements such as non-B DNA selectivity (versus canonical B-DNA and other biomolecules), high affinity, and biocompatibility.
Another concern with the use of ligands as biological probes is their propensity to induce (versus detect) DNA structure-formation. Until new methods become available to monitor DNA conformation without impacting the DNA structure, researchers should carry this potential bias in mind to avoid over-interpretation of the results obtained.
With the involvement of non-B DNA structures in various biological processes and genetic instability, they may be considered as potential druggable targets, but how can non-B DNA structures be validated as such targets? Unlike proteins, non-B DNA structures do not have defined ‘active sites’ nor particular enzymatic functions. Instead, they contain unique structural features arising from known structure-forming sequences, as well as mutagenic potential as a result of error-prone processing. More information is being obtained with respect to their recognition and processing. As the field moves toward unraveling the mechanistic processing of non-B DNA, as well as their in vivo loci, the information provided will be invaluable in exploring non-B DNA structures as valid targets for the prevention and/or treatment of disease.
Highlights:
Genomic mutations co-localize with sequences that can form non-B DNA structures.
Non-B DNA impacts transcription, replication, and genetic instability.
Small molecules can be used as tools to study non-B DNA structure formation.
Stabilizers/destabilizers of DNA structure, assays and applications are discussed.
Acknowledgements
Funding: This work was supported by the National Institutes of Health [NIH/NCI CA093729] to K.M.V.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement: None declared
References
- [1].Baig MH, Ahmad K, Roy S, Ashraf JM, Adil M, Siddiqui MH, Khan S, Kamal MA, Provaznik I, Choi I, Computer Aided Drug Design: Success and Limitations, Curr Pharm Des, 22 (2016) 572–581. [DOI] [PubMed] [Google Scholar]
- [2].Abdolmaleki A, Ghasemi JB, Ghasemi F, Computer Aided Drug Design for Multi-Target Drug Design: SAR/QSAR, Molecular Docking and Pharmacophore Methods, Curr Drug Targets, 18 (2017) 556–575. [DOI] [PubMed] [Google Scholar]
- [3].van Montfort RLM, Workman P, Structure-based drug design: aiming for a perfect fit, Essays Biochem, 61 (2017) 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Navia MA, Peattie DA, Structure-based drug design: applications in immunopharmacology and immunosuppression, Immunol Today, 14 (1993) 296–302. [DOI] [PubMed] [Google Scholar]
- [5].Coussens NP, Braisted JC, Peryea T, Sittampalam GS, Simeonov A, Hall MD, Small-Molecule Screens: A Gateway to Cancer Therapeutic Agents with Case Studies of Food and Drug Administration-Approved Drugs, Pharmacol Rev, 69 (2017) 479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bacolla A, Tainer JA, Vasquez KM, Cooper DN, Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences, Nucleic Acids Res, 44 (2016) 5673–5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhao J, Bacolla A, Wang G, Vasquez KM, Non-B DNA structure-induced genetic instability and evolution, Cell Mol Life Sci, 67 (2010) 43–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Javadekar SM, Raghavan SC, Snaps and mends: DNA breaks and chromosomal translocations, FEBS J, 282 (2015) 2627–2645. [DOI] [PubMed] [Google Scholar]
- [9].Lopez Castel A, Cleary JD, Pearson CE, Repeat instability as the basis for human diseases and as a potential target for therapy, Nat Rev Mol Cell Biol, 11 (2010) 165–170. [DOI] [PubMed] [Google Scholar]
- [10].Zhao J, Wang G, Del Mundo IM, McKinney JA, Lu X, Bacolla A, Boulware SB, Zhang C, Zhang H, Ren P, Freudenreich CH, Vasquez KM, Distinct Mechanisms of Nuclease-Directed DNA-Structure-Induced Genetic Instability in Cancer Genomes, Cell Rep, 22 (2018) 1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lu S, Wang G, Bacolla A, Zhao J, Spitser S, Vasquez KM, Short Inverted Repeats Are Hotspots for Genetic Instability: Relevance to Cancer Genomes, Cell Rep, 10 (2015) 1674–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Inagaki H, Ohye T, Kogo H, Kato T, Bolor H, Taniguchi M, Shaikh TH, Emanuel BS, Kurahashi H, Chromosomal instability mediated by non-B DNA: cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans, Genome Res, 19 (2009) 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang G, Carbajal S, Vijg J, DiGiovanni J, Vasquez KM, DNA structure-induced genomic instability in vivo, J Natl Cancer Inst, 100 (2008) 1815–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wang G, Vasquez KM, Non-B DNA structure-induced genetic instability, Mutat Res, 598 (2006) 103–119. [DOI] [PubMed] [Google Scholar]
- [15].Wang G, Christensen LA, Vasquez KM, Z-DNA-forming sequences generate large-scale deletions in mammalian cells, Proc Natl Acad Sci U S A, 103 (2006) 2677–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang G, Vasquez KM, Naturally occurring H-DNA-forming sequences are mutagenic in mammalian cells, Proc Natl Acad Sci U S A, 101 (2004) 13448–13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Choi J, Majima T, Conformational changes of non-B DNA, Chemical Society Reviews, 40 (2011) 5893–5909. [DOI] [PubMed] [Google Scholar]
- [18].Mirkin SM, Discovery of alternative DNA structures: a heroic decade (1979–1989), Frontiers in Bioscience, 13 (2008) 1064–1071. [DOI] [PubMed] [Google Scholar]
- [19].Wang G, Vasquez KM, Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability, DNA Repair (Amst), 19 (2014) 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mirkin SM, Expandable DNA repeats and human disease, Nature, 447 (2007) 932–940. [DOI] [PubMed] [Google Scholar]
- [21].Voineagu I, Narayanan V, Lobachev KS, Mirkin SM, Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins, Proc Natl Acad Sci U S A, 105 (2008) 9936–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sinden RR, Zheng GX, Brankamp RG, Allen KN, On the deletion of inverted repeated DNA in Escherichia coli: effects of length, thermal stability, and cruciform formation in vivo, Genetics, 129 (1991) 991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rich A, Zhang S, Timeline: Z-DNA: the long road to biological function, Nat Rev Genet, 4 (2003) 566–572. [DOI] [PubMed] [Google Scholar]
- [24].Voloshin ON, Mirkin SM, Lyamichev VI, Belotserkovskii BP, Frank-Kamenetskii MD, Chemical probing of homopurine-homopyrimidine mirror repeats in supercoiled DNA, Nature, 333 (1988) 475–476. [DOI] [PubMed] [Google Scholar]
- [25].Frank-Kamenetskii MD, Mirkin SM, Triplex DNA structures, Annu Rev Biochem, 64 (1995) 65–95. [DOI] [PubMed] [Google Scholar]
- [26].Bochman ML, Paeschke K, Zakian VA, DNA secondary structures: stability and function of G-quadruplex structures, Nat Rev Genet, 13 (2012) 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lane AN, Chaires JB, Gray RD, Trent JO, Stability and kinetics of G-quadruplex structures, Nucleic Acids Research, 36 (2008) 5482–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S, Quadruplex DNA: sequence, topology and structure, Nucleic Acids Res, 34 (2006) 5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang G, Gaddis S, Vasquez KM, Methods to detect replication-dependent and replication-independent DNA structure-induced genetic instability, Methods, 64 (2013) 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cer RZ, Donohue DE, Mudunuri US, Temiz NA, Loss MA, Starner NJ, Halusa GN, Volfovsky N, Yi M, Luke BT, Bacolla A, Collins JR, Stephens RM, Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools, Nucleic Acids Res, 41 (2013) D94–D100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kikin O, Antonio L, Bagga PS, QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences, Nucleic Acids Research, 34 (2006) W676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Agarwala P, Pandey S, Maiti S, The tale of RNA G-quadruplex, Org Biomol Chem, 13 (2015) 5570–5585. [DOI] [PubMed] [Google Scholar]
- [33].Malgowska M, Czajczynska K, Gudanis D, Tworak A, Gdaniec Z, Overview of the RNA G-quadruplex structures, Acta Biochim Pol, 63 (2016) 609–621. [DOI] [PubMed] [Google Scholar]
- [34].Bacolla A, Wang G, Vasquez KM, New Perspectives on DNA and RNA Triplexes As Effectors of Biological Activity, PLoS Genet, 11 (2015) e1005696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Herbert A, Z-DNA and Z-RNA in human disease, Commun Biol, 2 (2019) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hall K, Cruz P, Tinoco I Jr., Jovin TM, van de Sande JH, ‘Z-RNA’--a left-handed RNA double helix, Nature, 311 (1984) 584–586. [DOI] [PubMed] [Google Scholar]
- [37].Saini N, Zhang Y, Usdin K, Lobachev KS, When secondary comes first--the importance of non-canonical DNA structures, Biochimie, 95 (2013) 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kaushik M, Kaushik S, Roy K, Singh A, Mahendru S, Kumar M, Chaudhary S, Ahmed S, Kukreti S, A bouquet of DNA structures: Emerging diversity, Biochem Biophys Rep, 5 (2016) 388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kamat MA, Bacolla A, Cooper DN, Chuzhanova N, A Role for Non-B DNA Forming Sequences in Mediating Microlesions Causing Human Inherited Disease, Hum Mutat, 37 (2016) 65–73. [DOI] [PubMed] [Google Scholar]
- [40].Dong DW, Pereira F, Barrett SP, Kolesar JE, Cao K, Damas J, Yatsunyk LA, Johnson FB, Kaufman BA, Association of G-quadruplex forming sequences with human mtDNA deletion breakpoints, BMC Genomics, 15 (2014) 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kinniburgh AJ, A cis-acting transcription element of the c-myc gene can assume an H-DNA conformation, Nucleic Acids Res, 17 (1989) 7771–7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wiener F, Ohno S, Babonits M, Sumegi J, Wirschubsky Z, Klein G, Mushinski JF, Potter M, Hemizygous interstitial deletion of chromosome 15 (band D) in three translocation-negative murine plasmacytomas, Proc Natl Acad Sci U S A, 81 (1984) 1159–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Akasaka T, Akasaka H, Ueda C, Yonetani N, Maesako Y, Shimizu A, Yamabe H, Fukuhara S, Uchiyama T, Ohno H, Molecular and clinical features of non-Burkitt’s, diffuse large-cell lymphoma of B-cell type associated with the c-MYC/immunoglobulin heavy-chain fusion gene, J Clin Oncol, 18 (2000) 510–518. [DOI] [PubMed] [Google Scholar]
- [44].Kovalchuk AL, Muller JR, Janz S, Deletional remodeling of c-myc-deregulating chromosomal translocations, Oncogene, 15 (1997) 2369–2377. [DOI] [PubMed] [Google Scholar]
- [45].Joos S, Haluska FG, Falk MH, Henglein B, Hameister H, Croce CM, Bornkamm GW, Mapping chromosomal breakpoints of Burkitt’s t(8;14) translocations far upstream of c-myc, Cancer Res, 52 (1992) 6547–6552. [PubMed] [Google Scholar]
- [46].Haluska FG, Tsujimoto Y, Croce CM, The t(8;14) breakpoint of the EW 36 undifferentiated lymphoma cell line lies 5’ of MYC in a region prone to involvement in endemic Burkitt’s lymphomas, Nucleic Acids Res, 16 (1988) 2077–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Saglio G, Grazia Borrello M, Guerrasio A, Sozzi G, Serra A, di Celle PF, Foa R, Ferrarini M, Roncella S, Borgna Pignatti C, et al. , Preferential clustering of chromosomal breakpoints in Burkitt’s lymphomas and L3 type acute lymphoblastic leukemias with a t(8;14) translocation, Genes Chromosomes Cancer, 8 (1993) 1–7. [DOI] [PubMed] [Google Scholar]
- [48].Care A, Cianetti L, Giampaolo A, Sposi NM, Zappavigna V, Mavilio F, Alimena G, Amadori S, Mandelli F, Peschle C, Translocation of c-myc into the immunoglobulin heavy-chain locus in human acute B-cell leukemia. A molecular analysis, EMBO J, 5 (1986) 905–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wilda M, Busch K, Klose I, Keller T, Woessmann W, Kreuder J, Harbott J, Borkhardt A, Level of MYC overexpression in pediatric Burkitt’s lymphoma is strongly dependent on genomic breakpoint location within the MYC locus, Genes Chromosomes Cancer, 41 (2004) 178–182. [DOI] [PubMed] [Google Scholar]
- [50].De S, Michor F, DNA secondary structures and epigenetic determinants of cancer genome evolution, Nature Structural & Molecular Biology, 18 (2011) 950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rimokh R, Rouault JP, Wahbi K, Gadoux M, Lafage M, Archimbaud E, Charrin C, Gentilhomme O, Germain D, Samarut J, et al. , A chromosome 12 coding region is juxtaposed to the MYC protooncogene locus in a t(8;12)(q24;q22) translocation in a case of B-cell chronic lymphocytic leukemia, Genes Chromosomes Cancer, 3 (1991) 24–36. [DOI] [PubMed] [Google Scholar]
- [52].Wolfl S, Wittig B, Rich A, Identification of transcriptionally induced Z-DNA segments in the human c-myc gene, Biochim Biophys Acta, 1264 (1995) 294–302. [DOI] [PubMed] [Google Scholar]
- [53].Adachi M, Tsujimoto Y, Potential Z-DNA elements surround the breakpoints of chromosome translocation within the 5’ flanking region of bcl-2 gene, Oncogene, 5 (1990) 1653–1657. [PubMed] [Google Scholar]
- [54].Seite P, Leroux D, Hillion J, Monteil M, Berger R, Mathieu-Mahul D, Larsen CJ, Molecular analysis of a variant 18;22 translocation in a case of lymphocytic lymphoma, Genes Chromosomes Cancer, 6 (1993) 39–44. [DOI] [PubMed] [Google Scholar]
- [55].Kim N, Jinks-Robertson S, Guanine repeat-containing sequences confer transcription-dependent instability in an orientation-specific manner in yeast, DNA Repair (Amst), 10 (2011) 953–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Piazza A, Serero A, Boule JB, Legoix-Ne P, Lopes J, Nicolas A, Stimulation of gross chromosomal rearrangements by the human CEB1 and CEB25 minisatellites in Saccharomyces cerevisiae depends on G-quadruplexes or Cdc13, PLoS Genet, 8 (2012) e1003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Aksenova AY, Greenwell PW, Dominska M, Shishkin AA, Kim JC, Petes TD, Mirkin SM, Genome rearrangements caused by interstitial telomeric sequences in yeast, Proc Natl Acad Sci U S A, 110 (2013) 19866–19871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA, Pif1 family helicases suppress genome instability at G-quadruplex motifs, Nature, 497 (2013) 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lensing SV, Marsico G, Hansel-Hertsch R, Lam EY, Tannahill D, Balasubramanian S, DSBCapture: in situ capture and sequencing of DNA breaks, Nat Methods, 13 (2016) 855–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA, Highly recurrent TERT promoter mutations in human melanoma, Science, 339 (2013) 957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Liu T, Yuan X, Xu D, Cancer-Specific Telomerase Reverse Transcriptase (TERT) Promoter Mutations: Biological and Clinical Implications, Genes, 7 (2016) 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kang H-J, Cui Y, Yin H, Scheid A, Hendricks WPD, Schmidt J, Sekulic A, Kong D, Trent JM, Gokhale V, Mao H, Hurley LH, A Pharmacological Chaperone Molecule Induces Cancer Cell Death by Restoring Tertiary DNA Structures in Mutant hTERT Promoters, Journal of the American Chemical Society, 138 (2016) 13673–13692. [DOI] [PubMed] [Google Scholar]
- [63].Wang G, Vasquez KM, Non-B DNA structure-induced genetic instability, Mutat Res, 598 (2006) 103–119. [DOI] [PubMed] [Google Scholar]
- [64].Wells RD, Dere R, Hebert ML, Napierala M, Son LS, Advances in mechanisms of genetic instability related to hereditary neurological diseases, Nucleic Acids Res, 33 (2005) 3785–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Du X, Gertz EM, Wojtowicz D, Zhabinskaya D, Levens D, Benham CJ, Schaffer AA, Przytycka TM, Potential non-B DNA regions in the human genome are associated with higher rates of nucleotide mutation and expression variation, Nucleic Acids Res, 42 (2014) 12367–12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Choi J, Majima T, Conformational changes of non-B DNA, Chemical Society reviews, 40 (2011) 5893–5909. [DOI] [PubMed] [Google Scholar]
- [67].Chen X, Shen Y, Zhang F, Chiang C, Pillalamarri V, Blumenthal I, Talkowski M, Wu BL, Gusella JF, Molecular analysis of a deletion hotspot in the NRXN1 region reveals the involvement of short inverted repeats in deletion CNVs, Am J Hum Genet, 92 (2013) 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bacolla A, Jaworski A, Larson JE, Jakupciak JP, Chuzhanova N, Abeysinghe SS, O’Connell CD, Cooper DN, Wells RD, Breakpoints of gross deletions coincide with non-B DNA conformations, Proc Natl Acad Sci U S A, 101 (2004) 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Bacolla A, Wells RD, Non-B DNA conformations, genomic rearrangements, and human disease, J Biol Chem, 279 (2004) 47411–47414. [DOI] [PubMed] [Google Scholar]
- [70].Kukreti R, C BR, Das SK, De M, Talukder G, Vaz F, Verma IC, Brahmachari SK, Study of the single nucleotide polymorphism (SNP) at the palindromic sequence of hypersensitive site (HS)4 of the human beta-globin locus control region (LCR) in Indian population, Am J Hematol, 69 (2002) 77–79. [DOI] [PubMed] [Google Scholar]
- [71].Chaudhary S, Kaushik M, Kukreti R, Kukreti S, Structural switch from a multistranded G-quadruplex to single strands as a consequence of point mutation in the promoter of the human GRIN1 gene, Mol Biosyst, 13 (2017) 1805–1816. [DOI] [PubMed] [Google Scholar]
- [72].Chaudhary S, Kaushik M, Ahmed S, Kukreti R, Kukreti S, Structural Switch from Hairpin to Duplex/Antiparallel G-Quadruplex at Single-Nucleotide Polymorphism (SNP) Site of Human Apolipoprotein E (APOE) Gene Coding Region, ACS Omega, 3 (2018) 3173–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Baral A, Kumar P, Halder R, Mani P, Yadav VK, Singh A, Das SK, Chowdhury S, Quadruplex-single nucleotide polymorphisms (Quad-SNP) influence gene expression difference among individuals, Nucleic Acids Res, 40 (2012) 3800–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wells RD, Non-B DNA conformations, mutagenesis and disease, Trends Biochem Sci, 32 (2007) 271–278. [DOI] [PubMed] [Google Scholar]
- [75].Bacolla A, Cooper DN, Vasquez KM, Tainer JA, Non-B DNA Structure and Mutations Causing Human Genetic Disease, eLS, (2018) 1–15. [Google Scholar]
- [76].Urban V, Dobrovolna J, Janscak P, Distinct functions of human RecQ helicases during DNA replication, Biophys Chem, 225 (2017) 20–26. [DOI] [PubMed] [Google Scholar]
- [77].Mendoza O, Bourdoncle A, Boule JB, Brosh RM Jr., Mergny JL, G-quadruplexes and helicases, Nucleic Acids Res, 44 (2016) 1989–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bochman ML, Roles of DNA helicases in the maintenance of genome integrity, Mol Cell Oncol, 1 (2014) e963429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Blagg J, Workman P, Choose and Use Your Chemical Probe Wisely to Explore Cancer Biology, Cancer Cell, 32 (2017) 9–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Schreiber SL, Kotz JD, Li M, Aube J, Austin CP, Reed JC, Rosen H, White EL, Sklar LA, Lindsley CW, Alexander BR, Bittker JA, Clemons PA, de Souza A, Foley MA, Palmer M, Shamji AF, Wawer MJ, McManus O, Wu M, Zou B, Yu H, Golden JE, Schoenen FJ, Simeonov A, Jadhav A, Jackson MR, Pinkerton AB, Chung TD, Griffin PR, Cravatt BF, Hodder PS, Roush WR, Roberts E, Chung DH, Jonsson CB, Noah JW, Severson WE, Ananthan S, Edwards B, Oprea TI, Conn PJ, Hopkins CR, Wood MR, Stauffer SR, Emmitte KA, N.I.H.M.L.P. Team, Advancing Biological Understanding and Therapeutics Discovery with Small-Molecule Probes, Cell, 161 (2015) 1252–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lee JS, Lee JW, Kang N, Ha HH, Chang YT, Diversity-oriented approach for chemical biology, Chem Rec, 15 (2015) 495–510. [DOI] [PubMed] [Google Scholar]
- [82].Ohno M, Fukagawa T, Lee JS, Ikemura T, Triplex-forming DNAs in the human interphase nucleus visualized in situ by polypurine/polypyrimidine DNA probes and antitriplex antibodies, Chromosoma, 111 (2002) 201–213. [DOI] [PubMed] [Google Scholar]
- [83].Agazie YM, Burkholder GD, Lee JS, Triplex DNA in the nucleus: direct binding of triplex-specific antibodies and their effect on transcription, replication and cell growth, Biochem J, 316 (Pt 2) (1996) 461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lee JS, Latimer LJ, Haug BL, Pulleyblank DE, Skinner DM, Burkholder GD, Triplex DNA in plasmids and chromosomes, Gene, 82 (1989) 191–199. [DOI] [PubMed] [Google Scholar]
- [85].Artusi S, Perrone R, Lago S, Raffa P, Di Iorio E, Palu G, Richter SN, Visualization of DNA G-quadruplexes in herpes simplex virus 1-infected cells, Nucleic Acids Res, 44 (2016) 10343–10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Biffi G, Tannahill D, McCafferty J, Balasubramanian S, Quantitative visualization of DNA G-quadruplex structures in human cells, Nat Chem, 5 (2013) 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Liu HY, Zhao Q, Zhang TP, Wu Y, Xiong YX, Wang SK, Ge YL, He JH, Lv P, Ou TM, Tan JH, Li D, Gu LQ, Ren J, Zhao Y, Huang ZS, Conformation Selective Antibody Enables Genome Profiling and Leads to Discovery of Parallel G-Quadruplex in Human Telomeres, Cell Chem Biol, 23 (2016) 1261–1270. [DOI] [PubMed] [Google Scholar]
- [88].Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A, In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei, Proc Natl Acad Sci U S A, 98 (2001) 8572–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Henderson A, Wu Y, Huang YC, Chavez EA, Platt J, Johnson FB, Brosh RM Jr., Sen D, Lansdorp PM, Detection of G-quadruplex DNA in mammalian cells, Nucleic Acids Res, 42 (2014) 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Polymenis M, Brigido MM, Sanford DG, Stollar BD, The targets and genes for antibodies to Z-DNA, Biotechnology and Applied Biochemistry, 18 (1993) 175–183. [PubMed] [Google Scholar]
- [91].Rich A, Nordheim A, Wang AH, The chemistry and biology of left-handed Z-DNA, Annu Rev Biochem, 53 (1984) 791–846. [DOI] [PubMed] [Google Scholar]
- [92].Jovin TM, Soumpasis DM, McIntosh LP, The Transition Between B-DNA and Z-DNA, Annual Review of Physical Chemistry, 38 (1987) 521–558. [Google Scholar]
- [93].Wang G, Vasquez KM, Z-DNA, an active element in the genome, Front Biosci, 12 (2007) 4424–4438. [DOI] [PubMed] [Google Scholar]
- [94].Yang L, Wang S, Tian T, Zhou X, Advancements in Z-DNA: development of inducers and stabilizers for B to Z transition, Curr Med Chem, 19 (2012) 557–568. [DOI] [PubMed] [Google Scholar]
- [95].Song G, Ren J, Recognition and regulation of unique nucleic acid structures by small molecules, Chem Commun (Camb), 46 (2010) 7283–7294. [DOI] [PubMed] [Google Scholar]
- [96].Barton JK, Basile LA, Danishefsky A, Alexandrescu A, Chiral probes for the handedness of DNA helices: enantiomers of tris(4,7-diphenylphenanthroline)ruthenium(II), Proceedings of the National Academy of Sciences of the United States of America, 81 (1984) 1961–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Barton JK, Tris (phenanthroline) Metal Complexes: Probes for DNA Helicity, Journal of Biomolecular Structure and Dynamics, 1 (1983) 621–632. [DOI] [PubMed] [Google Scholar]
- [98].Friedman AE, Kumar CV, Turro NJ, Barton JK, Luminescence of ruthenium(II) polypyridyls: evidence for intercalative binding to Z-DNA, Nucleic acids research, 19 (1991) 2595–2602. [PMC free article] [PubMed] [Google Scholar]
- [99].Thomas TJ, Bloomfield VA, Chain flexibility and hydrodynamics of the B and Z forms of poly(dG-dC).poly(dG-dC), Nucleic Acids Res, 11 (1983) 1919–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gupta G, Dhingra MM, Sarma RH, Left-handed intercalated DNA double helix: rendezvous of ethidium and actinomycin D in the Z-helical conformation space, J Biomol Struct Dyn, 1 (1983) 97–113. [DOI] [PubMed] [Google Scholar]
- [101].Kawara K, Tsuji G, Taniguchi Y, Sasaki S, Synchronized Chiral Induction between [5]Helicene-Spermine Ligand and B-Z DNA Transition, Chemistry, 23 (2017) 1763–1769. [DOI] [PubMed] [Google Scholar]
- [102].Tsuji G, Kawakami K, Sasaki S, Enantioselective binding of chiral 1,14-dimethyl[5]helicene–spermine ligands with B- and Z-DNA, Bioorganic & Medicinal Chemistry, 21 (2013) 6063–6068. [DOI] [PubMed] [Google Scholar]
- [103].Balaz M, De Napoli M, Holmes AE, Mammana A, Nakanishi K, Berova N, Purrello R, A Cationic Zinc Porphyrin as a Chiroptical Probe for Z-DNA, Angewandte Chemie International Edition, 44 (2005) 4006–4009. [DOI] [PubMed] [Google Scholar]
- [104].Gong L, Jang YJ, Kim J, Kim SK, Z-Form DNA Specific Binding Geometry of Zn(II) meso-Tetrakis(N-methylpyridinium-4-yl)porphyrin Probed by Linear Dichroism Spectroscopy, The Journal of Physical Chemistry B, 116 (2012) 9619–9626. [DOI] [PubMed] [Google Scholar]
- [105].Qin T, Liu K, Song D, Yang C, Zhao H, Su H, Binding Interactions of Zinc Cationic Porphyrin with Duplex DNA: From B-DNA to Z-DNA, International Journal of Molecular Sciences, 19 (2018) 1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Choi JK, Reed A, Balaz M, Chiroptical properties, binding affinity, and photostability of a conjugated zinc porphyrin dimer complexed with left-handed Z-DNA and right-handed B-DNA, Dalton Trans, 43 (2014) 563–567. [DOI] [PubMed] [Google Scholar]
- [107].D’Urso A, Mammana A, Balaz M, Holmes AE, Berova N, Lauceri R, Purrello R, Interactions of a Tetraanionic Porphyrin with DNA: from a Z-DNA Sensor to a Versatile Supramolecular Device, Journal of the American Chemical Society, 131 (2009) 2046–2047. [DOI] [PubMed] [Google Scholar]
- [108].Zimmer C, Marck C, Guschlbauer W, Z-DNA and other non-B-DNA structures are reversed to B-DNA by interaction with netropsin, FEBS Lett, 154 (1983) 156–160. [DOI] [PubMed] [Google Scholar]
- [109].Chaires JB, Long-range allosteric effects on the B to Z equilibrium by daunomycin, Biochemistry, 24 (1985) 7479–7486. [DOI] [PubMed] [Google Scholar]
- [110].Chaires JB, Daunomycin inhibits the B leads to Z transition in poly d(G-C), Nucleic acids research, 11 (1983) 8485–8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Chen C, Knop RH, Cohen JS, Adriamycin inhibits the B to Z transition of poly(dGm5dC).cntdot.poly(dGm5dC), Biochemistry, 22 (1983) 5468–5471. [Google Scholar]
- [112].Kypr J, Vorlickova M, Conformations of alternating purine-pyrimidine DNAs in high-CsF solutions and their reversal by dipyrandium, ethidium and high temperature, Biochim Biophys Acta, 838 (1985) 244–251. [DOI] [PubMed] [Google Scholar]
- [113].Walker GT, Stone MP, Krugh TR, Interaction of drugs with Z-DNA: cooperative binding of actinomycin D or actinomine to the left-handed forms of poly(dG-dC).poly(dG-dC) and poly(dG-m5dC).poly(dG-m5dC) reverses the conformation of the helix, Biochemistry, 24 (1985) 7471–7479. [DOI] [PubMed] [Google Scholar]
- [114].Walker GT, Stone MP, Krugh TR, Ethidium binding to left-handed (Z) DNAs results in regions of right-handed DNA at the intercalation site, Biochemistry, 24 (1985) 7462–7471. [DOI] [PubMed] [Google Scholar]
- [115].Shafer RH, Brown SC, Delbarre A, Wade D, Binding of ethidium and bis(methidium)spermine to Z DNA by intercalation, Nucleic Acids Res, 12 (1984) 4679–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Mirau PA, Kearns DR, The effect of intercalating drugs on the kinetics of the B to Z transition of poly(dG-dC), Nucleic Acids Res, 11 (1983) 1931–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Nishimura T, Takeda Y, Shimada N, Sakurai K, DNA Conformational Switching by Use of an Intercalator and Its Receptor, Chemistry Letters, 36 (2007) 388–389. [Google Scholar]
- [118].Biver T, Garcia B, Leal JM, Secco F, Turriani E, Left-handed DNA: intercalation of the cyanine thiazole orange and structural changes. A kinetic and thermodynamic approach, Phys Chem Chem Phys, 12 (2010) 13309–13317. [DOI] [PubMed] [Google Scholar]
- [119].Qu X, Trent JO, Fokt I, Priebe W, Chaires JB, Allosteric, chiral-selective drug binding to DNA, Proceedings of the National Academy of Sciences, 97 (2000) 12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Chaires JB, Allosteric conversion of Z DNA to an intercalated right-handed conformation by daunomycin, J Biol Chem, 261 (1986) 8899–8907. [PubMed] [Google Scholar]
- [121].Das S, Kundu S, Suresh Kumar G, Quinacrine and 9-Amino Acridine Inhibit B-Z and B-HL Form DNA Conformational Transitions, DNA and Cell Biology, 30 (2011) 525–535. [DOI] [PubMed] [Google Scholar]
- [122].Rajeswari MR, Tryptophan intercalation in G, C containing polynucleotides: Z to B conversion of poly [d(G-5M C)] in low salt induced by a tetra peptide, J Biomol Struct Dyn, 14 (1996) 25–30. [DOI] [PubMed] [Google Scholar]
- [123].Härd T, Hiort C, Nordén B, On the Use of Chiral Compounds for Probing the DNA Handedness: Z to B Conversion in Poly(dGm5dC) Upon Binding of Fe(phen)3 2+ and Ru(phen)3 2+, Journal of Biomolecular Structure and Dynamics, 5 (1987) 89–96. [DOI] [PubMed] [Google Scholar]
- [124].Pasternack RF, Sidney D, Hunt PA, Snowden EA, Gibbs EJ, Interactions of water soluble porphyrins with Z-poly(dG-dC), Nucleic acids research, 14 (1986) 3927–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Jimenez-Garcia E, Portugal J, Elsamicin A can convert the Z-form of poly[d(G-C)] and poly[(G-m5C)] back to B-form DNA, Biochemistry, 31 (1992) 11641–11646. [DOI] [PubMed] [Google Scholar]
- [126].Mao C, Sun W, Shen Z, Seeman NC, A nanomechanical device based on the B-Z transition of DNA, Nature, 397 (1999) 144–146. [DOI] [PubMed] [Google Scholar]
- [127].Lee M, Kim SH, Hong SC, Minute negative superhelicity is sufficient to induce the B-Z transition in the presence of low tension, Proc Natl Acad Sci U S A, 107 (2010) 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Bae S, Kim D, Kim KK, Kim YG, Hohng S, Intrinsic Z-DNA is stabilized by the conformational selection mechanism of Z-DNA-binding proteins, J Am Chem Soc, 133 (2011) 668–671. [DOI] [PubMed] [Google Scholar]
- [129].Roy Chowdhury A, Bakshi R, Wang J, Yildirir G, Liu B, Pappas-Brown V, Tolun G, Griffith JD, Shapiro TA, Jensen RE, Englund PT, The killing of African trypanosomes by ethidium bromide, PLoS Pathog, 6 (2010) e1001226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Mirkin SM, Frank-Kamenetskii MD, H-DNA and related structures, Annu. Rev. Biophys. Biomol. Struct, 23 (1994) 541–576. [DOI] [PubMed] [Google Scholar]
- [131].Jain A, Wang G, Vasquez KM, DNA triple helices: biological consequences and therapeutic potential, Biochimie, 90 (2008) 1117–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Buske FA, Mattick JS, Bailey TL, Potential in vivo roles of nucleic acid triple-helices, RNA Biol, 8 (2011) 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Smith CIE, Zain R, Therapeutic Oligonucleotides: State of the Art, Annu Rev Pharmacol Toxicol, 59 (2019) 605–630. [DOI] [PubMed] [Google Scholar]
- [134].Mukherjee A, Vasquez KM, Triplex technology in studies of DNA damage, DNA repair, and mutagenesis, Biochimie, 93 (2011) 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB, The triple helix: 50 years later, the outcome, Nucleic Acids Res, 36 (2008) 5123–5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Escudé C, Sun J-S, DNA Major Groove Binders: Triple Helix-Forming Oligonucleotides, Triple Helix-Specific DNA Ligands and Cleaving Agents, in: Waring MJ, Chaires JB (Eds.) DNA Binders and Related Subjects: −/−, Springer; Berlin Heidelberg, 2005, pp. 109–148. [Google Scholar]
- [137].Strekowski L, Wolinska E, Mojzych M, DNA triple-helix stabilizing agents, in: Lee M, Strekowski L (Eds.) Synthetic and biophysical studies of DNA-binding compounds, Transworld Research Network, 2007, pp. 263–278. [Google Scholar]
- [138].Soyfer VN, Potaman VN, Triple-Helical Nucleic Acids, Springer-Verlag, 1995. [Google Scholar]
- [139].Escude C, Garestier T, Sun JS, Drug interaction with triple-helical nucleic acids, Methods Enzymol, 340 (2001) 340–357. [DOI] [PubMed] [Google Scholar]
- [140].Jenkins TC, Targeting multi-stranded DNA structures, Curr Med Chem, 7 (2000) 99–115. [DOI] [PubMed] [Google Scholar]
- [141].Escude C, Garestier T, Sun JS, Drug interaction with triple-helical nucleic acids, Methods in enzymology, 340 (2001) 340–357. [DOI] [PubMed] [Google Scholar]
- [142].Scaria PV, Shafer RH, Binding of ethidium bromide to a DNA triple helix. Evidence for intercalation, J Biol Chem, 266 (1991) 5417–5423. [PubMed] [Google Scholar]
- [143].Mergny JL, Duval-Valentin G, Nguyen CH, Perrouault L, Faucon B, Rougee M, Montenay-Garestier T, Bisagni E, Helene C, Triple helix-specific ligands, Science, 256 (1992) 1681–1684. [DOI] [PubMed] [Google Scholar]
- [144].Pilch DS, Waring MJ, Sun JS, Rougee M, Nguyen CH, Bisagni E, Garestier T, Helene C, Characterization of a triple helix-specific ligand. BePI (3-methoxy-7H-8-methyl-11- [(3′-amino)propylamino]-benzo[e]pyrido[4,3-b]indole) intercalates into both double-helical and triple-helical DNA, J Mol Biol, 232 (1993) 926–946. [DOI] [PubMed] [Google Scholar]
- [145].Pilch DS, Martin MT, Nguyen CH, Sun JS, Bisagni E, Garestier T, Helene C, Self-association and DNA-binding properties of two triple helix-specific ligands: comparison of a benzo[e]pyridoindole and a benzo[g]pyridoindole, Journal of the American Chemical Society, 115 (1993) 9942–9951. [Google Scholar]
- [146].Keppler MD, James PL, Neidle S, Brown T, Fox KR, DNA sequence specificity of triplex-binding ligands, European Journal of Biochemistry, 270 (2003) 4982–4992. [DOI] [PubMed] [Google Scholar]
- [147].Strekowski L, Gulevich Y, Baranowski TC, Parker AN, Kiselyov AS, Lin SY, Tanious FA, Wilson WD, Synthesis and structure-DNA binding relationship analysis of DNA triple-helix specific intercalators, J Med Chem, 39 (1996) 3980–3983. [DOI] [PubMed] [Google Scholar]
- [148].Chaires JB, Ren J, Henary M, Zegrocka O, Bishop GR, Strekowski L, Triplex selective 2-(2-naphthyl)quinoline compounds: origins of affinity and new design principles, J Am Chem Soc, 125 (2003) 7272–7283. [DOI] [PubMed] [Google Scholar]
- [149].Cassidy SA, Strekowski L, Fox KR, DNA sequence specificity of a naphthylquinoline triple helix-binding ligand, Nucleic Acids Res, 24 (1996) 4133–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Holt PA, Ragazzon P, Strekowski L, Chaires JB, Trent JO, Discovery of novel triple helical DNA intercalators by an integrated virtual and actual screening platform, Nucleic acids research, 37 (2009) 1280–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Keppler M, Zegrocka O, Strekowski L, Fox KR, DNA triple helix stabilisation by a naphthylquinoline dimer, FEBS Lett, 447 (1999) 223–226. [DOI] [PubMed] [Google Scholar]
- [152].Fox KR, Thurston DE, Jenkins TC, Varvaresou A, Tsotinis A, Siatra-Papastaikoudi T, A novel series of DNA triple helix-binding ligands, Biochem Biophys Res Commun, 224 (1996) 717–720. [DOI] [PubMed] [Google Scholar]
- [153].Moraru-Allen AA, Cassidy S, Asensio Alvarez JL, Fox KR, Brown T, Lane AN, Coralyne has a preference for intercalation between TA.T triples in intramolecular DNA triple helices, Nucleic Acids Res, 25 (1997) 1890–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Lee JS, Latimer LJ, Hampel KJ, Coralyne binds tightly to both T.A.T- and C.G.C(+)-containing DNA triplexes, Biochemistry, 32 (1993) 5591–5597. [DOI] [PubMed] [Google Scholar]
- [155].Jain SS, Polak M, Hud NV, Controlling nucleic acid secondary structure by intercalation: effects of DNA strand length on coralyne-driven duplex disproportionation, Nucleic Acids Res, 31 (2003) 4608–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Baudoin O, Marchand C, Teulade-Fichou M-P, Vigneron J-P, Sun J-S, Garestier T, Hélène C, Lehn J-M, Stabilization of DNA Triple Helices by Crescent-Shaped Dibenzophenanthrolines, Chemistry – A European Journal, 4 (1998) 1504–1508. [Google Scholar]
- [157].Teulade-Fichou MP, Perrin D, Boutorine A, Polverari D, Vigneron JP, Lehn JM, Sun JS, Garestier T, Helene C, Direct photocleavage of HIV-DNA by quinacridine derivatives triggered by triplex formation, J Am Chem Soc, 123 (2001) 9283–9292. [DOI] [PubMed] [Google Scholar]
- [158].Fox KR, Polucci P, Jenkins TC, Neidle S, A molecular anchor for stabilizing triplehelical DNA, Proc Natl Acad Sci U S A, 92 (1995) 7887–7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Kim HK, Kim JM, Kim SK, Rodger A, Norden B, Interactions of intercalative and minor groove binding ligands with triplex poly(dA).[poly(dT)]2 and with duplex poly(dA).poly(dT) and poly[d(A-T)]2 studied by CD, LD, and normal absorption, Biochemistry, 35 (1996) 1187–1194. [DOI] [PubMed] [Google Scholar]
- [160].Missailidis S, Modi C, Trapani V, Laughton CA, Stevens MF, Antitumor polycyclic acridines. Part 16. Triplex DNA as a target for DNA-binding polycyclic acridine derivatives, Oncol Res, 15 (2005) 95–105. [PubMed] [Google Scholar]
- [161].Fortsch I, Birch-Hirschfeld E, Schutz H, Zimmer C, Different effects of nonintercalative antitumor drugs on DNA triple helix stability: SN-18071 promotes triple helix formation, J Biomol Struct Dyn, 14 (1996) 317–329. [DOI] [PubMed] [Google Scholar]
- [162].Fortsch I, Baguley B, Zimmer C, Structure-dependent effects of minor groove binders on the DNA triple helix motif poly(dA).2poly(dT): influence of antitumoractive nonintercalative bisquaternary ammonium heterocycles, Anticancer Drug Des, 13 (1998) 417–429. [PubMed] [Google Scholar]
- [163].Schmitt P, Nguyen CH, Sun J-S, Grierson DS, Bisagni E, Garestier T, Hélène C, 13H-benzo[6–7]indolo[3,2-c]quinolines (B[6,7]IQ): optimization of their DNA triplex-specific stabilization properties, Chemical Communications, (2000) 763–764. [Google Scholar]
- [164].Ren J, Bailly C, Chaires JB, NB-506, an indolocarbazole topoisomerase I inhibitor, binds preferentially to triplex DNA, FEBS Letters, 470 (2000) 355–359. [DOI] [PubMed] [Google Scholar]
- [165].Ren J, Chaires JB, Preferential Binding of 3,3’-Diethyloxadicarbocyanine to Triplex DNA, Journal of the American Chemical Society, 122 (2000) 424–425. [Google Scholar]
- [166].Granzhan A, Ihmels H, Selective stabilization of triple-helical DNA by diazoniapolycyclic intercalators, Chembiochem, 7 (2006) 1031–1033. [DOI] [PubMed] [Google Scholar]
- [167].Basili S, Bergen A, Dall’Acqua F, Faccio A, Granzhan A, Ihmels H, Moro S, Viola G, Relationship between the structure and the DNA binding properties of diazoniapolycyclic duplex- and triplex-DNA binders: efficiency, selectivity, and binding mode, Biochemistry, 46 (2007) 12721–12736. [DOI] [PubMed] [Google Scholar]
- [168].Pal S, Saha C, Hossain M, Dey SK, Kumar GS, Influence of galloyl moiety in interaction of epicatechin with bovine serum albumin: a spectroscopic and thermodynamic characterization, PLoS One, 7 (2012) e43321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Bhowmik D, Das S, Hossain M, Haq L, Suresh Kumar G, Biophysical characterization of the strong stabilization of the RNA triplex poly(U)*poly(A)*poly(U) by 9-O-(omega-amino) alkyl ether berberine analogs, PLoS One, 7 (2012) e37939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Sinha R, Kumar GS, Interaction of Isoquinoline Alkaloids with an RNA Triplex: Structural and Thermodynamic Studies of Berberine, Palmatine, and Coralyne Binding to Poly(U).Poly(A)*Poly(U), The Journal of Physical Chemistry B, 113 (2009) 13410–13420. [DOI] [PubMed] [Google Scholar]
- [171].Guittat L, Alberti P, Rosu F, Van Miert S, Thetiot E, Pieters L, Gabelica V, De Pauw E, Ottaviani A, Riou JF, Mergny JL, Interactions of cryptolepine and neocryptolepine with unusual DNA structures, Biochimie, 85 (2003) 535–547. [DOI] [PubMed] [Google Scholar]
- [172].Riechert-Krause F, Autenrieth K, Eick A, Weisz K, Spectroscopic and calorimetric studies on the binding of an indoloquinoline drug to parallel and antiparallel DNA triplexes, Biochemistry, 52 (2013) 41–52. [DOI] [PubMed] [Google Scholar]
- [173].Jain AK, Gupta SK, Tawar U, Dogra SK, Tandon V, Benzimidazoles: a minor groove-binding ligand-induced stabilization of triple helix, Oligonucleotides, 19 (2009) 53–62. [DOI] [PubMed] [Google Scholar]
- [174].Dickerhoff J, Riechert-Krause F, Seifert J, Weisz K, Exploring multiple binding sites of an indoloquinoline in triple-helical DNA: a paradigm for DNA triplex-selective intercalators, Biochimie, 107 Pt B (2014) 327–337. [DOI] [PubMed] [Google Scholar]
- [175].Perrouault L, Asseline U, Rivalle C, Thuong NT, Bisagni E, Giovannangeli C, Le Doan T, Helene C, Sequence-specific artificial photo-induced endonucleases based on triple helix-forming oligonucleotides, Nature, 344 (1990) 358–360. [DOI] [PubMed] [Google Scholar]
- [176].Collier DA, Mergny JL, Thuong NT, Helene C, Site-specific intercalation at the triplex-duplex junction induces a conformational change which is detectable by hypersensitivity to diethylpyrocarbonate, Nucleic Acids Res, 19 (1991) 4219–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Sun JS, Francois JC, Montenay-Garestier T, Saison-Behmoaras T, Roig V, Thuong NT, Helene C, Sequence-specific intercalating agents: intercalation at specific sequences on duplex DNA via major groove recognition by oligonucleotide-intercalator conjugates, Proc Natl Acad Sci U S A, 86 (1989) 9198–9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Sun J.-s., Lavery R, Chomilier J, Zakrzewska K, Montenay-Garestier T, Hélène C, Theoretical Study of Ethidium Intercalation in Triple-Stranded DNA and at Triplex-Duplex Junctions, Journal of Biomolecular Structure and Dynamics, 9 (1991) 425–436. [DOI] [PubMed] [Google Scholar]
- [179].Strekowski L, Hojjat M, Wolinska E, Parker AN, Paliakov E, Gorecki T, Tanious FA, Wilson WD, New triple-helix DNA stabilizing agents, Bioorg Med Chem Lett, 15 (2005) 1097–1100. [DOI] [PubMed] [Google Scholar]
- [180].Escude C, Nguyen CH, Kukreti S, Janin Y, Sun JS, Bisagni E, Garestier T, Helene C, Rational design of a triple helix-specific intercalating ligand, Proc Natl Acad Sci U S A, 95 (1998) 3591–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Zain R, Polverari D, Nguyen CH, Blouquit Y, Bisagni E, Garestier T, Grierson DS, Sun JS, Optimization of triple-helix-directed DNA cleavage by benzoquinoquinoxalineethylenediaminetetraacetic acid conjugates, Chembiochem, 4 (2003) 856–862. [DOI] [PubMed] [Google Scholar]
- [182].Arya DP, New approaches toward recognition of nucleic acid triple helices, Acc Chem Res, 44 (2011) 134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Arya DP, Coffee RL Jr., Willis B, Abramovitch AI, Aminoglycoside-nucleic acid interactions: remarkable stabilization of DNA and RNA triple helices by neomycin, J Am Chem Soc, 123 (2001) 5385–5395. [DOI] [PubMed] [Google Scholar]
- [184].Arya DP, Coffee RL Jr., Charles I, Neomycin-induced hybrid triplex formation, J Am Chem Soc, 123 (2001) 11093–11094. [DOI] [PubMed] [Google Scholar]
- [185].Arya DP, Aminoglycoside–Nucleic Acid Interactions: The Case for Neomycin, in: Waring MJ, Chaires JB (Eds.) DNA Binders and Related Subjects: −/−, Springer; Berlin Heidelberg, 2005, pp. 149–178. [Google Scholar]
- [186].Arya DP, Xue L, Tennant P, Combining the best in triplex recognition: synthesis and nucleic acid binding of a BQQ-neomycin conjugate, J Am Chem Soc, 125 (2003) 8070–8071. [DOI] [PubMed] [Google Scholar]
- [187].Xi H, Arya DP, Recognition of triple helical nucleic acids by aminoglycosides, Curr Med Chem Anticancer Agents, 5 (2005) 327–338. [DOI] [PubMed] [Google Scholar]
- [188].Xue L, Xi H, Kumar S, Gray D, Davis E, Hamilton P, Skriba M, Arya DP, Probing the recognition surface of a DNA triplex: binding studies with intercalator-neomycin conjugates, Biochemistry, 49 (2010) 5540–5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Jain AK, Bhattacharya S, Groove binding ligands for the interaction with parallel-stranded ps-duplex DNA and triplex DNA, Bioconjug Chem, 21 (2010) 1389–1403. [DOI] [PubMed] [Google Scholar]
- [190].Wilson WD, Mizan S, Tanious FA, Yao S, Zon G, The interaction of intercalators and groove-binding agents with DNA triple-helical structures: the influence of ligand structure, DNA backbone modifications and sequence, J Mol Recognit, 7 (1994) 89–98. [DOI] [PubMed] [Google Scholar]
- [191].Umemoto K, Sarma MH, Gupta G, Luo J, Sarma RH, Structure and stability of a DNA triple helix in solution: NMR studies on d(T)6.d(A)6.d(T)6 and its complex with a minor groove binding drug, Journal of the American Chemical Society, 112 (1990) 4539–4545. [Google Scholar]
- [192].Pilch DS, Breslauer KJ, Ligand-induced formation of nucleic acid triple helices, Proc Natl Acad Sci U S A, 91 (1994) 9332–9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Xu Z, Pilch DS, Srinivasan AR, Olson WK, Geacintov NE, Breslauer KJ, Modulation of nucleic acid structure by ligand binding: induction of a DNA.RNA.DNA hybrid triplex by DAPI intercalation, Bioorg Med Chem, 5 (1997) 1137–1147. [DOI] [PubMed] [Google Scholar]
- [194].Riley M, Maling B, Physical and chemical characterization of two- and three-stranded adenine-thymine and adenine-uracil homopolymer complexes, J Mol Biol, 20 (1966) 359–389. [DOI] [PubMed] [Google Scholar]
- [195].Song G, Xing F, Qu X, Chaires JB, Ren J, Oxazine 170 Induces DNA:RNA:DNA Triplex Formation, Journal of Medicinal Chemistry, 48 (2005) 3471–3473. [DOI] [PubMed] [Google Scholar]
- [196].Escude C, Sun JS, Nguyen CH, Bisagni E, Garestier T, Helene C, Ligand-induced formation of triple helices with antiparallel third strands containing G and T, Biochemistry, 35 (1996) 5735–5740. [DOI] [PubMed] [Google Scholar]
- [197].Keppler MD, Neidle S, Fox KR, Stabilisation of TG- and AG-containing antiparallel DNA triplexes by triplex-binding ligands, Nucleic Acids Res, 29 (2001) 1935–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Olivas WM, Maher LJ 3rd, Overcoming potassium-mediated triplex inhibition, Nucleic Acids Res, 23 (1995) 1936–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Cheng AJ, Van Dyke MW, Monovalent cation effects on intermolecular purine-purine-pyrimidine triple-helix formation, Nucleic Acids Res, 21 (1993) 5630–5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Del Mundo IMA, Zewail-Foote M, Kerwin SM, Vasquez KM, Alternative DNA structure formation in the mutagenic human c-MYC promoter, Nucleic Acids Res, 45 (2017) 4929–4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Radhakrishnan I, Patel DJ, DNA triplexes: solution structures, hydration sites, energetics, interactions, and function, Biochemistry, 33 (1994) 11405–11416. [DOI] [PubMed] [Google Scholar]
- [202].Radhakrishnan I, Patel DJ, Solution structure of a purine.purine.pyrimidine DNA triplex containing G.GC and T.AT triples, Structure, 1 (1993) 135–152. [DOI] [PubMed] [Google Scholar]
- [203].Park YW, Breslauer KJ, Drug binding to higher ordered DNA structures: netropsin complexation with a nucleic acid triple helix, Proc Natl Acad Sci U S A, 89 (1992) 6653–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Pilch DS, Kirolos MA, Breslauer KJ, Berenil binding to higher ordered nucleic acid structures: complexation with a DNA and RNA triple helix, Biochemistry, 34 (1995) 16107–16124. [DOI] [PubMed] [Google Scholar]
- [205].Rajeswari MR, DNA triplex structures in neurodegenerative disorder, Friedreich’s ataxia, J Biosci, 37 (2012) 519–532. [DOI] [PubMed] [Google Scholar]
- [206].Vigneswaran N, Mayfield CA, Rodu B, James R, Kim HG, Miller DM, Influence of GC and AT specific DNA minor groove binding drugs on intermolecular triplex formation in the human c-Ki-ras promoter, Biochemistry, 35 (1996) 1106–1114. [DOI] [PubMed] [Google Scholar]
- [207].Stonehouse TJ, Fox KR, DNase I footprinting of triple helix formation at polypurine tracts by acridine-linked oligopyrimidines: stringency, structural changes and interaction with minor groove binding ligands, Biochim Biophys Acta, 1218 (1994) 322–330. [DOI] [PubMed] [Google Scholar]
- [208].Durand M, Thuong NT, Maurizot JC, Berenil complexation with a nucleic acid triple helix, J Biomol Struct Dyn, 11 (1994) 1191–1202. [DOI] [PubMed] [Google Scholar]
- [209].Durand M, Thuong NT, Maurizot JC, Binding of netropsin to a DNA triple helix, J Biol Chem, 267 (1992) 24394–24399. [PubMed] [Google Scholar]
- [210].Rentzeperis D, Marky LA, Ligand Binding to the Hoogsteen-WC Groove of TAT Base Triplets. Thermodynamic Contribution of the Thymine Methyl Groups, Journal of the American Chemical Society, 117 (1995) 5423–5424. [Google Scholar]
- [211].Boehm BJ, Whidborne C, Button AL, Pukala TL, Huang DM, DNA triplex structure, thermodynamics, and destabilisation: insight from molecular simulations, Phys Chem Chem Phys, 20 (2018) 14013–14023. [DOI] [PubMed] [Google Scholar]
- [212].Haq I, Ladbury JE, Chowdhry BZ, Jenkins TC, Molecular Anchoring of Duplex and Triplex DNA by Disubstituted Anthracene-9,10-diones: Calorimetric, UV Melting, and Competition Dialysis Studies, Journal of the American Chemical Society, 118 (1996) 10693–10701. [Google Scholar]
- [213].Durand M, Thuong NT, Maurizot JC, Interaction of Hoechst 33258 with a DNA triple helix, Biochimie, 76 (1994) 181–186. [DOI] [PubMed] [Google Scholar]
- [214].Howard FB, Miles HT, Liu K, Frazier J, Raghunathan G, Sasisekharan V, Structure of d(T)n.d(A)n.d(T)n: the DNA triple helix has B-form geometry with C2’-endo sugar pucker, Biochemistry, 31 (1992) 10671–10677. [DOI] [PubMed] [Google Scholar]
- [215].García B, Leal JM, Paiotta V, Ruiz R, Secco F, Venturini M, Role of the Third Strand in the Binding of Proflavine and Pt-Proflavine to Poly(rA)·2poly(rU): A Thermodynamic and Kinetic Study, The Journal of Physical Chemistry B, 112 (2008) 7132–7139. [DOI] [PubMed] [Google Scholar]
- [216].Amiri H, Nekhotiaeva N, Sun JS, Nguyen CH, Grierson DS, Good L, Zain R, Benzoquinoquinoxaline derivatives stabilize and cleave H-DNA and repress transcription downstream of a triplex-forming sequence, J Mol Biol, 351 (2005) 776–783. [DOI] [PubMed] [Google Scholar]
- [217].Jenkins Y, Friedman AE, Turro NJ, Barton JK, Characterization of dipyridophenazine complexes of ruthenium(II): the light switch effect as a function of nucleic acid sequence and conformation, Biochemistry, 31 (1992) 10809–10816. [DOI] [PubMed] [Google Scholar]
- [218].Choi SD, Kim MS, Kim SK, Lincoln P, Tuite E, Norden B, Binding mode of [ruthenium(II) (1,10-phenanthroline)2L]2+ with poly (dT*dA-dT) triplex. Ligand size effect on third-strand stabilization, Biochemistry, 36 (1997) 214–223. [DOI] [PubMed] [Google Scholar]
- [219].Li J, Sun Y, Xie L, He X, Tan L, Effect of ancillary ligands on the interaction of ruthenium(II) complexes with the triplex RNA poly(U)·poly(A)*poly(U), Journal of Inorganic Biochemistry, 143 (2015) 56–63. [DOI] [PubMed] [Google Scholar]
- [220].Li J, Sun Y, Zhu Z, Zhao H, Tan L, Binding properties of ruthenium(II) complexes [Ru(bpy)2(ppn)]2+ and [Ru(phen)2(ppn)]2+ with triplex RNA: As molecular “light switches” and stabilizers for poly(U)·poly(A)*poly(U) triplex, Journal of Inorganic Biochemistry, 161 (2016) 128–133. [DOI] [PubMed] [Google Scholar]
- [221].Peng M, Ni W, Tan L, Effects of the fluorine substituent positions of the intercalating ligands on the binding behavior and third-strand stabilization of two Ru(II) complexes toward poly(U)•poly(A)*poly(U) triplex RNA, Journal of Inorganic Biochemistry, 175 (2017) 276–283. [DOI] [PubMed] [Google Scholar]
- [222].Daksis JI, Erikson GH, Specific triplex binding capacity of mixed base sequence duplex nucleic acids used for single-nucleotide polymorphism detection, Genet Test, 9 (2005) 111–120. [DOI] [PubMed] [Google Scholar]
- [223].Kaluzhny DN, Timoshin VV, Borisova OF, Zhurkin VB, Florentiev VL, Shchyolkina AK, Intramolecular recombination R-triplex in solution: stabilization by bis-intercalator YOYO, J Biomol Struct Dyn, 26 (2008) 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Van Daele I, Bomholt N, Filichev VV, Van Calenbergh S, Pedersen EB, Triplex Formation by Pyrene-Labelled Probes for Nucleic Acid Detection in Fluorescence Assays, ChemBioChem, 9 (2008) 791–801. [DOI] [PubMed] [Google Scholar]
- [225].Nygren J, Svanvik N, Kubista M, The interactions between the fluorescent dye thiazole orange and DNA, Biopolymers, 46 (1998) 39–51. [DOI] [PubMed] [Google Scholar]
- [226].Lubitz I, Zikich D, Kotlyar A, Specific high-affinity binding of thiazole orange to triplex and G-quadruplex DNA, Biochemistry, 49 (2010) 3567–3574. [DOI] [PubMed] [Google Scholar]
- [227].Monchaud D, Allain C, Teulade-Fichou MP, Thiazole orange: a useful probe for fluorescence sensing of G-quadruplex-ligand interactions, Nucleosides Nucleotides Nucleic Acids, 26 (2007) 1585–1588. [DOI] [PubMed] [Google Scholar]
- [228].Monchaud D, Allain C, Bertrand H, Smargiasso N, Rosu F, Gabelica V, De Cian A, Mergny JL, Teulade-Fichou MP, Ligands playing musical chairs with G-quadruplex DNA: a rapid and simple displacement assay for identifying selective G-quadruplex binders, Biochimie, 90 (2008) 1207–1223. [DOI] [PubMed] [Google Scholar]
- [229].Yang P, De Cian A, Teulade-Fichou MP, Mergny JL, Monchaud D, Engineering bisquinolinium/thiazole orange conjugates for fluorescent sensing of G-quadruplex DNA, Angew Chem Int Ed Engl, 48 (2009) 2188–2191. [DOI] [PubMed] [Google Scholar]
- [230].Monchaud D, Allain C, Teulade-Fichou MP, Development of a fluorescent intercalator displacement assay (G4-FID) for establishing quadruplex-DNA affinity and selectivity of putative ligands, Bioorg Med Chem Lett, 16 (2006) 4842–4845. [DOI] [PubMed] [Google Scholar]
- [231].Kovalska VB, Losytskyy MY, Yarmoluk SM, Lubitz I, Kotlyar AB, Mono and trimethine cyanines Cyan 40 and Cyan 2 as probes for highly selective fluorescent detection of non-canonical DNA structures, J Fluoresc, 21 (2011) 223–230. [DOI] [PubMed] [Google Scholar]
- [232].Chen Z, Zhang H, Ma X, Lin Z, Zhang L, Chen G, A novel fluorescent reagent for recognition of triplex DNA with high specificity and selectivity, Analyst, 140 (2015) 7742–7747. [DOI] [PubMed] [Google Scholar]
- [233].Wang Y, Hu Y, Wu T, Zhou X, Shao Y, Triggered Excited-State Intramolecular Proton Transfer Fluorescence for Selective Triplex DNA Recognition, Anal Chem, 87 (2015) 11620–11624. [DOI] [PubMed] [Google Scholar]
- [234].Hu Y, Lin F, Wu T, Wang Y, Zhou X-S, Shao Y, Fluorescently Sensing of DNA Triplex Assembly Using an Isoquinoline Alkaloid as Selector, Stabilizer, Inducer, and Switch-On Emitter, Chemistry – An Asian Journal, 11 (2016) 2041–2048. [DOI] [PubMed] [Google Scholar]
- [235].Duval-Valentin G, de Bizemont T, Takasugi M, Mergny JL, Bisagni E, Helene C, Triple-helix specific ligands stabilize H-DNA conformation, J Mol Biol, 247 (1995) 847–858. [DOI] [PubMed] [Google Scholar]
- [236].Baran N, Lapidot A, Manor H, Formation of DNA triplexes accounts for arrests of DNA synthesis at d(TC)n and d(GA)n tracts, Proc Natl Acad Sci U S A, 88 (1991) 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Wang G, Zhao J, Vasquez KM, Detection of cis- and trans-acting Factors in DNA Structure-Induced Genetic Instability Using In silico and Cellular Approaches, Frontiers in Genetics, 7 (2016) 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Belotserkovskii BP, De Silva E, Tornaletti S, Wang G, Vasquez KM, Hanawalt PC, A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription, J Biol Chem, 282 (2007) 32433–32441. [DOI] [PubMed] [Google Scholar]
- [239].Amiri H, Nekhotiaeva N, Sun JS, Nguyen CH, Grierson DS, Good L, Zain R, Benzoquinoquinoxaline derivatives stabilize and cleave H-DNA and repress transcription downstream of a triplex-forming sequence, Journal of molecular biology, 351 (2005) 776–783. [DOI] [PubMed] [Google Scholar]
- [240].Sakamoto N, Chastain PD, Parniewski P, Ohshima K, Pandolfo M, Griffith JD, Wells RD, Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich’s ataxia, Mol Cell, 3 (1999) 465–475. [DOI] [PubMed] [Google Scholar]
- [241].Ohshima K, Montermini L, Wells RD, Pandolfo M, Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo, J Biol Chem, 273 (1998) 14588–14595. [DOI] [PubMed] [Google Scholar]
- [242].Ohshima K, Kang S, Larson JE, Wells RD, Cloning, characterization, and properties of seven triplet repeat DNA sequences, J Biol Chem, 271 (1996) 16773–16783. [DOI] [PubMed] [Google Scholar]
- [243].Campuzano V, Montermini L, Lutz Y, Cova L, Hindelang C, Jiralerspong S, Trottier Y, Kish SJ, Faucheux B, Trouillas P, Authier FJ, Durr A, Mandel JL, Vescovi A, Pandolfo M, Koenig M, Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes, Hum Mol Genet, 6 (1997) 1771–1780. [DOI] [PubMed] [Google Scholar]
- [244].Grabczyk E, Usdin K, Alleviating transcript insufficiency caused by Friedreich’s ataxia triplet repeats, Nucleic Acids Res, 28 (2000) 4930–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Burnett R, Melander C, Puckett JW, Son LS, Wells RD, Dervan PB, Gottesfeld JM, DNA sequence-specific polyamides alleviate transcription inhibition associated with long GAA.TTC repeats in Friedreich’s ataxia, Proc Natl Acad Sci U S A, 103 (2006) 11497–11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Grant L, Sun J, Xu H, Subramony SH, Chaires JB, Hebert MD, Rational selection of small molecules that increase transcription through the GAA repeats found in Friedreich’s ataxia, FEBS Lett, 580 (2006) 5399–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Neidle S, Balasubramanian S, Quadruplex Nucleic Acids, Royal Society of Chemistry, 2007. [Google Scholar]
- [248].Hansel-Hertsch R, Di Antonio M, Balasubramanian S, DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential, Nat Rev Mol Cell Biol, 18 (2017) 279–284. [DOI] [PubMed] [Google Scholar]
- [249].Bhattacharyya D, Mirihana Arachchilage G, Basu S, Metal Cations in G-Quadruplex Folding and Stability, Frontiers in Chemistry, 4 (2016) 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Gu J, Leszczynski J, Bansal M, A new insight into the structure and stability of Hoogsteen hydrogen-bonded G-tetrad: an ab initio SCF study, Chemical Physics Letters, 311 (1999) 209–214. [Google Scholar]
- [251].Hud NV, Plavec J, The role of cations in determining quadruplex structure and stability, Quadruplex Nucleic Acids, (2006) 100–130. [Google Scholar]
- [252].Neidle S, Quadruplex Nucleic Acids as Novel Therapeutic Targets, Journal of Medicinal Chemistry, 59 (2016) 5987–6011. [DOI] [PubMed] [Google Scholar]
- [253].Parotta L, Ortuso F, Moraca F, Rocca R, Costa G, Alcaro S, Artese A, Targeting unimolecular G-quadruplex nucleic acids: a new paradigm for the drug discovery? AU - Parrotta, Lucia, Expert Opinion on Drug Discovery, 9 (2014) 1167–1187. [DOI] [PubMed] [Google Scholar]
- [254].Zhang S, Wu Y, Zhang W, G-Quadruplex Structures and Their Interaction Diversity with Ligands, ChemMedChem, 9 (2014) 899–911. [DOI] [PubMed] [Google Scholar]
- [255].Sannohe Y, Sugiyama H, Overview of formation of G-quadruplex structures, Curr Protoc Nucleic Acid Chem, Chapter 17 (2010) Unit 17 12 11–17. [DOI] [PubMed] [Google Scholar]
- [256].Lane AN, Chaires JB, Gray RD, Trent JO, Stability and kinetics of G-quadruplex structures, Nucleic Acids Res, 36 (2008) 5482–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Kerwin SM, G-Quadruplex DNA as a target for drug design, Curr Pharm Des, 6 (2000) 441–478. [DOI] [PubMed] [Google Scholar]
- [258].Sattin G, Artese A, Nadai M, Costa G, Parrotta L, Alcaro S, Palumbo M, Richter SN, Conformation and stability of intramolecular telomeric G-quadruplexes: sequence effects in the loops, PLoS One, 8 (2013) e84113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Fujii T, Podbevšek P, Plavec J, Sugimoto N, Effects of metal ions and cosolutes on G-quadruplex topology, Journal of Inorganic Biochemistry, 166 (2017) 190–198. [DOI] [PubMed] [Google Scholar]
- [260].You J, Li H, Lu X-M, Li W, Wang P-Y, Dou S-X, Xi X-G, Effects of monovalent cations on folding kinetics of G-quadruplexes, Bioscience Reports, 37 (2017) BSR20170771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Asamitsu S, Obata S, Yu Z, Bando T, Sugiyama H, Recent Progress of Targeted G-Quadruplex-Preferred Ligands Toward Cancer Therapy, Molecules, 24 (2019) 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Asamitsu S, Bando T, Sugiyama H, Ligand Design to Acquire Specificity to Intended G-Quadruplex Structures, Chemistry, 25 (2019) 417–430. [DOI] [PubMed] [Google Scholar]
- [263].Duarte AR, Cadoni E, Ressurreicao AS, Moreira R, Paulo A, Design of Modular G-quadruplex Ligands, ChemMedChem, 13 (2018) 869–893. [DOI] [PubMed] [Google Scholar]
- [264].Vilar R, Nucleic Acid Quadruplexes and Metallo-Drugs, in: Sigel A, Sigel H, Freisinger E, Sigel RKO (Eds.) Metallo-Drugs: Development and Action of Anticancer Agents, de Guyter, 2018, pp. 325–349. [Google Scholar]
- [265].Francisco AP, Paulo A, Oncogene Expression Modulation in Cancer Cell Lines by DNA G-Quadruplex-Interactive Small Molecules, Curr Med Chem, 24 (2017) 4873–4904. [DOI] [PubMed] [Google Scholar]
- [266].Cimino-Reale G, Zaffaroni N, Folini M, Emerging Role of G-quadruplex DNA as Target in Anticancer Therapy, Curr Pharm Des, 22 (2016) 6612–6624. [DOI] [PubMed] [Google Scholar]
- [267].Muller S, Rodriguez R, G-quadruplex interacting small molecules and drugs: from bench toward bedside, Expert Rev Clin Pharmacol, 7 (2014) 663–679. [DOI] [PubMed] [Google Scholar]
- [268].Lubitz I, Borovok N, Kotlyar A, Interaction of Monomolecular G4-DNA Nanowires with TMPyP: Evidence for Intercalation, Biochemistry, 46 (2007) 12925–12929. [DOI] [PubMed] [Google Scholar]
- [269].Haq I, Trent JO, Chowdhry BZ, Jenkins TC, Intercalative G-Tetraplex Stabilization of Telomeric DNA by a Cationic Porphyrin1, Journal of the American Chemical Society, 121 (1999) 1768–1779. [Google Scholar]
- [270].Anantha NV, Azam M, Sheardy RD, Porphyrin Binding to Quadruplexed T4G4, Biochemistry, 37 (1998) 2709–2714. [DOI] [PubMed] [Google Scholar]
- [271].Phan AT, Kuryavyi V, Gaw HY, Patel DJ, Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter, Nature Chemical Biology, 1 (2005) 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Mergny J-L, Gros J, De Cian A, Bourdoncle A, Rosu F, Sacca B, Guittat L, Amrane S, Mills M, Alberti P, Takasugi M, Lacroix L, Energetics, kinetics and dynamics of quadruplex folding, Quadruplex Nucleic Acids, (2006) 31–80. [Google Scholar]
- [273].Guliaev AB, Leontis NB, Cationic 5,10,15,20-tetrakis(N-methylpyridinium-4-yl)porphyrin fully intercalates at 5’-CG-3’ steps of duplex DNA in solution, Biochemistry, 38 (1999) 15425–15437. [DOI] [PubMed] [Google Scholar]
- [274].Lee YA, Kim JO, Cho TS, Song R, Kim SK, Binding of meso-tetrakis(N-methylpyridium-4-yl)porphyrin to triplex oligonucleotides: evidence for the porphyrin stacking in the major groove, J Am Chem Soc, 125 (2003) 8106–8107. [DOI] [PubMed] [Google Scholar]
- [275].Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH, Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription, Proc Natl Acad Sci U S A, 99 (2002) 11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Chung WJ, Heddi B, Hamon F, Teulade-Fichou MP, Phan AT, Solution structure of a G-quadruplex bound to the bisquinolinium compound Phen-DC(3), Angew Chem Int Ed Engl, 53 (2014) 999–1002. [DOI] [PubMed] [Google Scholar]
- [277].De Cian A, Delemos E, Mergny JL, Teulade-Fichou MP, Monchaud D, Highly efficient G-quadruplex recognition by bisquinolinium compounds, J Am Chem Soc, 129 (2007) 1856–1857. [DOI] [PubMed] [Google Scholar]
- [278].Piazza A, Boule JB, Lopes J, Mingo K, Largy E, Teulade-Fichou MP, Nicolas A, Genetic instability triggered by G-quadruplex interacting Phen-DC compounds in Saccharomyces cerevisiae, Nucleic Acids Res, 38 (2010) 4337–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Lopes J, Piazza A, Bermejo R, Kriegsman B, Colosio A, Teulade-Fichou MP, Foiani M, Nicolas A, G-quadruplex-induced instability during leading-strand replication, EMBO J, 30 (2011) 4033–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Jain AK, Bhattacharya S, Recent developments in the chemistry and biology of G-quadruplexes with reference to the DNA groove binders, Curr Pharm Des, 18 (2012) 1917–1933. [DOI] [PubMed] [Google Scholar]
- [281].Muller S, Sanders DA, Di Antonio M, Matsis S, Riou JF, Rodriguez R, Balasubramanian S, Pyridostatin analogues promote telomere dysfunction and long-term growth inhibition in human cancer cells, Org Biomol Chem, 10 (2012) 6537–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Muller S, Kumari S, Rodriguez R, Balasubramanian S, Small-molecule-mediated G-quadruplex isolation from human cells, Nat Chem, 2 (2010) 1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, Oelschlaegel T, Xhemalce B, Balasubramanian S, Jackson SP, Small-molecule-induced DNA damage identifies alternative DNA structures in human genes, Nat Chem Biol, 8 (2012) 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Sun H, Xiang J, Liu Y, Li L, Li Q, Xu G, Tang Y, A stabilizing and denaturing dual-effect for natural polyamines interacting with G-quadruplexes depending on concentration, Biochimie, 93 (2011) 1351–1356. [DOI] [PubMed] [Google Scholar]
- [285].Ueda YM, Zouzumi YK, Maruyama A, Nakano SI, Sugimoto N, Miyoshi D, Effects of trimethylamine N-oxide and urea on DNA duplex and G-quadruplex, Sci Technol Adv Mater, 17 (2016) 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Weisman-Shomer P, Cohen E, Hershco I, Khateb S, Wolfovitz-Barchad O, Hurley LH, Fry M, The cationic porphyrin TMPyP4 destabilizes the tetraplex form of the fragile X syndrome expanded sequence d(CGG)n, Nucleic acids research, 31 (2003) 3963–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Ofer N, Weisman-Shomer P, Shklover J, Fry M, The quadruplex r(CGG)n destabilizing cationic porphyrin TMPyP4 cooperates with hnRNPs to increase the translation efficiency of fragile X premutation mRNA, Nucleic Acids Res, 37 (2009) 2712–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Bhattacharjee AJ, Ahluwalia K, Taylor S, Jin O, Nicoludis JM, Buscaglia R, Brad Chaires J, Kornfilt DJ, Marquardt DG, Yatsunyk LA, Induction of G-quadruplex DNA structure by Zn(II) 5,10,15,20-tetrakis(N-methyl-4-pyridyl)porphyrin, Biochimie, 93 (2011) 1297–1309. [DOI] [PubMed] [Google Scholar]
- [289].Dutikova Iu V, Borisova OF, Shchelkina AK, Lin J, Huang S, Shtil AA, Kaliuzhnyi DN, [5,10,15,20-Tetra-(N-methyl-3-pyridyl)porphyrin destabilizes the anti-parallel telomeric quadruplex d(TTAGGG)4], Mol Biol (Mosk), 44 (2010) 929–937. [PubMed] [Google Scholar]
- [290].D’Urso A, Randazzo R, Rizzo V, Gangemi CMA, Romanucci V, Zarrelli A, Tomaselli G, Milardi D, Borbone N, Purrello R, Piccialli G, Di Fabio G, Oliviero G, Stabilization vs. destabilization of G-quadruplex superstructures: the role of the porphyrin derivative having spermine arms, Physical Chemistry Chemical Physics, 19 (2017) 17404–17410. [DOI] [PubMed] [Google Scholar]
- [291].Rajczak E, Gluszynska A, Juskowiak B, Interaction of metallacrown complexes with G-quadruplex DNA, J Inorg Biochem, 155 (2016) 105–114. [DOI] [PubMed] [Google Scholar]
- [292].Waller ZA, Shirude PS, Rodriguez R, Balasubramanian S, Triarylpyridines: a versatile small molecule scaffold for G-quadruplex recognition, Chem Commun (Camb), (2008) 1467–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Bejugam M, Sewitz S, Shirude PS, Rodriguez R, Shahid R, Balasubramanian S, Trisubstituted isoalloxazines as a new class of G-quadruplex binding ligands: small molecule regulation of c-kit oncogene expression, J Am Chem Soc, 129 (2007) 12926–12927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Cogoi S, Xodo LE, G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription, Nucleic Acids Res, 34 (2006) 2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Ou TM, Lu YJ, Zhang C, Huang ZS, Wang XD, Tan JH, Chen Y, Ma DL, Wong KY, Tang JC, Chan AS, Gu LQ, Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc by quindoline derivatives, J Med Chem, 50 (2007) 1465–1474. [DOI] [PubMed] [Google Scholar]
- [296].Waller ZA, Sewitz SA, Hsu ST, Balasubramanian S, A small molecule that disrupts G-quadruplex DNA structure and enhances gene expression, J Am Chem Soc, 131 (2009) 12628–12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Pierce SE, Sherman CL, Jayawickramarajah J, Lawrence CM, Sessler JL, Brodbelt JS, ESI-MS characterization of a novel pyrrole-inosine nucleoside that interacts with guanine bases, Anal Chim Acta, 627 (2008) 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [298].Sessler JL, Jayawickramarajah J, Sherman CL, Brodbelt JS, Enhancing Hoogsteen interactions: a pyrrole-containing purine nucleoside that competes with guanosine self-assembly, J Am Chem Soc, 126 (2004) 11460–11461. [DOI] [PubMed] [Google Scholar]
- [299].Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JK, Von Hoff D, Anderes K, Rice WG, Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis, Cancer Res, 69 (2009) 7653–7661. [DOI] [PubMed] [Google Scholar]
- [300].Xu H, Di Antonio M, McKinney S, Mathew V, Ho B, O’Neil NJ, Santos ND, Silvester J, Wei V, Garcia J, Kabeer F, Lai D, Soriano P, Banáth J, Chiu DS, Yap D, Le DD, Ye FB, Zhang A, Thu K, Soong J, Lin S.-c., Tsai AHC, Osako T, Algara T, Saunders DN, Wong J, Xian J, Bally MB, Brenton JD, Brown GW, Shah SP, Cescon D, Mak TW, Caldas C, Stirling PC, Hieter P, Balasubramanian S, Aparicio S, CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours, Nature Communications, 8 (2017) 14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, Huser N, Proffitt C, Bliesath J, Haddach M, Schwaebe MK, Ryckman DM, Rice WG, Schmitt C, Lowe SW, Johnstone RW, Pearson RB, McArthur GA, Hannan RD, Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53, Cancer Cell, 22 (2012) 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Wang Y, Patel DJ, Guanine residues in d(T2AG3) and d(T2G4) form parallel-stranded potassium cation stabilized G-quadruplexes with anti glycosidic torsion angles in solution, Biochemistry, 31 (1992) 8112–8119. [DOI] [PubMed] [Google Scholar]
- [303].Henderson E, Hardin CC, Walk SK, Tinoco I Jr., Blackburn EH, Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs, Cell, 51 (1987) 899–908. [DOI] [PubMed] [Google Scholar]
- [304].Sundquist WI, Klug A, Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops, Nature, 342 (1989) 825–829. [DOI] [PubMed] [Google Scholar]
- [305].Zahler AM, Williamson JR, Cech TR, Prescott DM, Inhibition of telomerase by G-quartet DNA structures, Nature, 350 (1991) 718–720. [DOI] [PubMed] [Google Scholar]
- [306].Williamson JR, Raghuraman MK, Cech TR, Monovalent cation-induced structure of telomeric DNA: the G-quartet model, Cell, 59 (1989) 871–880. [DOI] [PubMed] [Google Scholar]
- [307].Neidle S, Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer, FEBS J, 277 (2010) 1118–1125. [DOI] [PubMed] [Google Scholar]
- [308].Ray S, Bandaria JN, Qureshi MH, Yildiz A, Balci H, G-quadruplex formation in telomeres enhances POT1/TPP1 protection against RPA binding, Proc Natl Acad Sci U S A, 111 (2014) 2990–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW, Specific association of human telomerase activity with immortal cells and cancer, Science, 266 (1994) 2011–2015. [DOI] [PubMed] [Google Scholar]
- [310].Hanahan D, Weinberg RA, The hallmarks of cancer, Cell, 100 (2000) 57–70. [DOI] [PubMed] [Google Scholar]
- [311].Shay JW, Reddel RR, Wright WE, Cancer. Cancer and telomeres--an ALTernative to telomerase, Science, 336 (2012) 1388–1390. [DOI] [PubMed] [Google Scholar]
- [312].Gomez D, Mergny J-L, Riou J-F, Detection of telomerase inhibitors based on g-quadruplex ligands by a modified telomeric repeat amplification protocol assay, Cancer research, 62 (2002) 3365–3368. [PubMed] [Google Scholar]
- [313].Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation, Cell, 144 (2011) 646–674. [DOI] [PubMed] [Google Scholar]
- [314].Huppert JL, Balasubramanian S, G-quadruplexes in promoters throughout the human genome, Nucleic Acids Res, 35 (2007) 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Eddy J, Maizels N, Gene function correlates with potential for G4 DNA formation in the human genome, Nucleic Acids Res, 34 (2006) 3887–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Kouzine F, Wojtowicz D, Baranello L, Yamane A, Nelson S, Resch W, Kieffer-Kwon KR, Benham CJ, Casellas R, Przytycka TM, Levens D, Permanganate/S1 Nuclease Footprinting Reveals Non-B DNA Structures with Regulatory Potential across a Mammalian Genome, Cell Syst, 4 (2017) 344–356 e347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Morgan RK, Brooks TA, Chapter 7 Targeting Promoter G-Quadruplexes for Transcriptional Control, Small-molecule Transcription Factor Inhibitors in Oncology, The Royal Society of Chemistry, 2019, pp. 169–193. [Google Scholar]
- [318].Suseela YV, Narayanaswamy N, Pratihar S, Govindaraju T, Far-red fluorescent probes for canonical and non-canonical nucleic acid structures: current progress and future implications, Chemical Society Reviews, 47 (2018) 1098–1131. [DOI] [PubMed] [Google Scholar]
- [319].Ma D-L, Zhang Z, Wang M, Lu L, Zhong H-J, Leung C-H, Recent Developments in G-Quadruplex Probes, Chemistry & Biology, 22 (2015) 812–828. [DOI] [PubMed] [Google Scholar]
- [320].Bhasikuttan AC, Mohanty J, Targeting G-quadruplex structures with extrinsic fluorogenic dyes: promising fluorescence sensors, Chem Commun (Camb), 51 (2015) 7581–7597. [DOI] [PubMed] [Google Scholar]
- [321].Vummidi BR, Alzeer J, Luedtke NW, Fluorescent Probes for G-Quadruplex Structures, ChemBioChem, 14 (2013) 540–558. [DOI] [PubMed] [Google Scholar]
- [322].Chilka P, Desai N, Datta B, Small Molecule Fluorescent Probes for G- Quadruplex Visualization as Potential Cancer Theranostic Agents, Molecules, 24 (2019) 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].Largy E, Granzhan A, Hamon F, Verga D, Teulade-Fichou M-P, Visualizing the Quadruplex: From Fluorescent Ligands to Light-Up Probes, in: Chaires JB, Graves D (Eds.) Quadruplex Nucleic Acids, Springer; Berlin Heidelberg, 2013, pp. 111–177. [DOI] [PubMed] [Google Scholar]
- [324].Chen S-B, Hu M-H, Liu G-C, Wang J, Ou T-M, Gu L-Q, Huang Z-S, Tan J-H, Visualization of NRAS RNA G-Quadruplex Structures in Cells with an Engineered Fluorogenic Hybridization Probe, Journal of the American Chemical Society, 138 (2016) 10382–10385. [DOI] [PubMed] [Google Scholar]
- [325].Amor S, Yang SY, Wong JMY, Monchaud D, Cellular Detection of G-Quadruplexes by Optical Imaging Methods, Curr Protoc Cell Biol, 76 (2017) 4.33.31–34.33.19. [DOI] [PubMed] [Google Scholar]
- [326].Dik-Lung M, Modi W, Sheng L, Quan-Bin H, Chung-Hang L, Recent Development of G-Quadruplex Probes for Cellular Imaging, Current Topics in Medicinal Chemistry, 15 (2015) 1957–1963. [DOI] [PubMed] [Google Scholar]
- [327].Kellett A, Molphy Z, Slator C, McKee V, Farrell NP, Molecular methods for assessment of non-covalent metallodrug-DNA interactions, Chem Soc Rev, 48 (2019) 971–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [328].Majus M, Francesco M, Giovanni LB, Methods for Elucidation of DNA-Anticancer Drug Interactions and their Applications in the Development of New Drugs, Current Pharmaceutical Design, 22 (2016) 6596–6611. [DOI] [PubMed] [Google Scholar]
- [329].Rehman SU, Sarwar T, Husain MA, Ishqi HM, Tabish M, Studying non-covalent drug-DNA interactions, Arch Biochem Biophys, 576 (2015) 49–60. [DOI] [PubMed] [Google Scholar]
- [330].Palchaudhuri R, Hergenrother PJ, DNA as a target for anticancer compounds: methods to determine the mode of binding and the mechanism of action, Curr Opin Biotechnol, 18 (2007) 497–503. [DOI] [PubMed] [Google Scholar]
- [331].Mergny JL, Phan AT, Lacroix L, Following G-quartet formation by UV-spectroscopy, FEBS Lett, 435 (1998) 74–78. [DOI] [PubMed] [Google Scholar]
- [332].Mergny JL, Lacroix L, UV Melting of G-Quadruplexes, Curr Protoc Nucleic Acid Chem, Chapter 17 (2009) Unit 17 11. [DOI] [PubMed] [Google Scholar]
- [333].Wilson WD, Tanious FA, Fernandez-Saiz M, Rigl CT, Evaluation of drug-nucleic acid interactions by thermal melting curves, Methods Mol Biol, 90 (1997) 219–240. [DOI] [PubMed] [Google Scholar]
- [334].Schroeder SJ, Turner DH, Optical melting measurements of nucleic acid thermodynamics, Methods Enzymol, 468 (2009) 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [335].Mergny JL, Lacroix L, Analysis of thermal melting curves, Oligonucleotides, 13 (2003) 515–537. [DOI] [PubMed] [Google Scholar]
- [336].Kumar GS, Basu A, The use of calorimetry in the biophysical characterization of small molecule alkaloids binding to RNA structures, Biochim Biophys Acta, 1860 (2016) 930–944. [DOI] [PubMed] [Google Scholar]
- [337].Halder K, Benzler M, Hartig JS, Reporter assays for studying quadruplex nucleic acids, Methods, 57 (2012) 115–121. [DOI] [PubMed] [Google Scholar]
- [338].Di Antonio M, Rodriguez R, Balasubramanian S, Experimental approaches to identify cellular G-quadruplex structures and functions, Methods, 57 (2012) 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [339].Shafer RH, Stability and Structure of Model DNA Triplexes and Quadruplexes and Their Interactions with Small Ligands, in: Moldave K (Ed.) Progress in Nucleic Acid Research and Molecular Biology, Academic Press, 1997, pp. 55–94. [DOI] [PubMed] [Google Scholar]
- [340].Jaumot J, Gargallo R, Experimental methods for studying the interactions between G-quadruplex structures and ligands, Curr Pharm Des, 18 (2012) 1900–1916. [DOI] [PubMed] [Google Scholar]
- [341].Murat P, Singh Y, Defrancq E, Methods for investigating G-quadruplex DNA/ligand interactions, Chem Soc Rev, 40 (2011) 5293–5307. [DOI] [PubMed] [Google Scholar]
- [342].Kwok CK, Sahakyan AB, Balasubramanian S, Structural Analysis using SHALiPE to Reveal RNA G-Quadruplex Formation in Human Precursor MicroRNA, Angew Chem Int Ed Engl, 55 (2016) 8958–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [343].Anandhakumar C, Kizaki S, Bando T, Pandian GN, Sugiyama H, Advancing small-molecule-based chemical biology with next-generation sequencing technologies, Chembiochem, 16 (2015) 20–38. [DOI] [PubMed] [Google Scholar]
- [344].Kwok CK, Merrick CJ, G-Quadruplexes: Prediction, Characterization, and Biological Application, Trends Biotechnol, 35 (2017) 997–1013. [DOI] [PubMed] [Google Scholar]
- [345].Yuan L, Tian T, Chen Y, Yan S, Xing X, Zhang Z, Zhai Q, Xu L, Wang S, Weng X, Yuan B, Feng Y, Zhou X, Existence of G-quadruplex structures in promoter region of oncogenes confirmed by G-quadruplex DNA cross-linking strategy, Sci Rep, 3 (2013) 1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [346].Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S, High-throughput sequencing of DNA G-quadruplex structures in the human genome, Nat Biotechnol, 33 (2015) 877–881. [DOI] [PubMed] [Google Scholar]
- [347].Hansel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, Tannahill D, Balasubramanian S, G-quadruplex structures mark human regulatory chromatin, Nat Genet, 48 (2016) 1267–1272. [DOI] [PubMed] [Google Scholar]
- [348].Iida K, Tsushima Y, Ma Y, Sedghi Masoud S, Sakuma M, Yokoyama T, Yoshida W, Ikebukuro K, Nagasawa K, Model studies for isolation of G-quadruplex-forming DNA sequences through a pull-down strategy with macrocyclic polyoxazole, Bioorg Med Chem, 27 (2019) 1742–1746. [DOI] [PubMed] [Google Scholar]
- [349].Yang H, Wang Y, Yu W, Shi L, Wang H, Su R, Chen C, Liu S, Screening and investigation of triplex DNA binders from Stephania tetrandra S. Moore by a combination of peak area-fading ultra high-performance liquid chromatography with orbitrap mass spectrometry and optical spectroscopies, Journal of Separation Science, 41 (2018) 2878–2885. [DOI] [PubMed] [Google Scholar]
- [350].Xu N, Yang H, Cui M, Wan C, Liu S, High-Performance Liquid Chromatography–Electrospray Ionization-Mass Spectrometry Ligand Fishing Assay: A Method for Screening Triplex DNA Binders from Natural Plant Extracts, Analytical Chemistry, 84 (2012) 2562–2568. [DOI] [PubMed] [Google Scholar]
- [351].Rosu F, Nguyen CH, De Pauw E, Gabelica V, Ligand binding mode to duplex and triplex DNA assessed by combining electrospray tandem mass spectrometry and molecular modeling, J Am Soc Mass Spectrom, 18 (2007) 1052–1062. [DOI] [PubMed] [Google Scholar]
- [352].Shi X, Chaires JB, Sequence- and structural-selective nucleic acid binding revealed by the melting of mixtures, Nucleic Acids Res, 34 (2006) e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [353].Mendoza O, Gueddouda NM, Boule JB, Bourdoncle A, Mergny JL, A fluorescence-based helicase assay: application to the screening of G-quadruplex ligands, Nucleic Acids Res, 43 (2015) e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [354].Darby RA, Sollogoub M, McKeen C, Brown L, Risitano A, Brown N, Barton C, Brown T, Fox KR, High throughput measurement of duplex, triplex and quadruplex melting curves using molecular beacons and a LightCycler, Nucleic Acids Res, 30 (2002) e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [355].Schneider UV, Severinsen JK, Geci I, Okkels LM, Johnk N, Mikkelsen ND, Klinge T, Pedersen EB, Westh H, Lisby G, A novel FRET pair for detection of parallel DNA triplexes by the LightCycler, BMC Biotechnol, 10 (2010) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [356].Renciuk D, Zhou J, Beaurepaire L, Guedin A, Bourdoncle A, Mergny JL, A FRET-based screening assay for nucleic acid ligands, Methods, 57 (2012) 122–128. [DOI] [PubMed] [Google Scholar]
- [357].Benight AS, Pančoška P, Owczarzy R, Vallone PM, Nešetřil J, Riccelli PV, Calculating sequence-dependent melting stability of duplex DNA oligomers and multiplex sequence analysis by graphs, Methods in Enzymology, Academic Press, 2001, pp. 165–192. [DOI] [PubMed] [Google Scholar]
- [358].Gray RD, Chaires JB, Analysis of multidimensional G-quadruplex melting curves, Curr Protoc Nucleic Acid Chem, Chapter 17 (2011) Unit17 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [359].Lin K-C, Kuo C-Y, Nieh C-C, Tseng W-L, Molecular beacon-based NAND logic gate for sensing triplex DNA binders, RSC Advances, 4 (2014) 38389–38392. [Google Scholar]
- [360].You Y, Tataurov AV, Owczarzy R, Measuring thermodynamic details of DNA hybridization using fluorescence, Biopolymers, 95 (2011) 472–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [361].James PL, Brown T, Fox KR, Thermodynamic and kinetic stability of intermolecular triple helices containing different proportions of C+*GC and T*AT triplets, Nucleic Acids Res, 31 (2003) 5598–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [362].Del Mundo IMA, Cho EJ, Dalby KN, Vasquez KM, A tunable assay for modulators of genome-destabilizing DNA structures, Nucleic Acids Res, 47 (2019) e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [363].Chen C, Song G, Yang X, Ren J, Qu X, A gold nanoparticle-based strategy for label-free and colorimetric screening of DNA triplex binders, Biochimie, 92 (2010) 1416–1421. [DOI] [PubMed] [Google Scholar]
- [364].Han MS, Lytton-Jean AK, Mirkin CA, A gold nanoparticle based approach for screening triplex DNA binders, J Am Chem Soc, 128 (2006) 4954–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [365].Lytton-Jean AK, Han MS, Mirkin CA, Microarray detection of duplex and triplex DNA binders with DNA-modified gold nanoparticles, Anal Chem, 79 (2007) 6037–6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [366].Del Villar-Guerra R, Gray RD, Trent JO, Chaires JB, A rapid fluorescent indicator displacement assay and principal component/cluster data analysis for determination of ligand-nucleic acid structural selectivity, Nucleic Acids Res, 46 (2018) e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [367].Bachurin SS, Kletskii ME, Burov ON, Kurbatov SV, Non-canonical DNA structures: Comparative quantum mechanical study, Biophys Chem, 235 (2018) 19–28. [DOI] [PubMed] [Google Scholar]
- [368].Moraca F, Amato J, Ortuso F, Artese A, Pagano B, Novellino E, Alcaro S, Parrinello M, Limongelli V, Ligand binding to telomeric G-quadruplex DNA investigated by funnel-metadynamics simulations, Proceedings of the National Academy of Sciences, 114 (2017) E2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [369].Stadlbauer P, Krepl M, Cheatham TE 3rd, Koca J, Sponer J, Structural dynamics of possible late-stage intermediates in folding of quadruplex DNA studied by molecular simulations, Nucleic Acids Res, 41 (2013) 7128–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [370].Monsen RC, Trent JO, G-quadruplex virtual drug screening: A review, Biochimie, 152 (2018) 134–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [371].Musumeci D, Amato J, Zizza P, Platella C, Cosconati S, Cingolani C, Biroccio A, Novellino E, Randazzo A, Giancola C, Pagano B, Montesarchio D, Tandem application of ligand-based virtual screening and G4-OAS assay to identify novel G-quadruplex-targeting chemotypes, Biochim Biophys Acta Gen Subj, 1861 (2017) 1341–1352. [DOI] [PubMed] [Google Scholar]
- [372].Deng X, Xiong F, Li X, Xiang B, Li Z, Wu X, Guo C, Li X, Li Y, Li G, Xiong W, Zeng Z, Application of atomic force microscopy in cancer research, J Nanobiotechnology, 16 (2018) 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [373].Sigal YM, Zhou R, Zhuang X, Visualizing and discovering cellular structures with super-resolution microscopy, Science, 361 (2018) 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [374].van de Linde S, Aufmkolk S, Franke C, Holm T, Klein T, Loschberger A, Proppert S, Wolter S, Sauer M, Investigating cellular structures at the nanoscale with organic fluorophores, Chem Biol, 20 (2013) 8–18. [DOI] [PubMed] [Google Scholar]
- [375].Huang B, Babcock H, Zhuang X, Breaking the diffraction barrier: super-resolution imaging of cells, Cell, 143 (2010) 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]