Abstract
Background
The present study focused on understanding the prognostic value of the methylenetetrahydrofolate reductase (MTHFR) single nucleotide polymorphisms rs1801133 (C667T) and rs1801131 (A1298C) in patients with colorectal cancer (CRC).
Methods
A systematic literature search was conducted in March 2016. Databases, including Medline, EMBASE, Cochrane and Chinese databases (including CNKI, Wanfang and VIP), were searched to identify the relevant articles describing MTHFR polymorphisms in patients with CRC. Data regarding overall survival (OS), progression‐free survival (PFS) and disease‐free survival (DFS) were collected and analysed.
Results
Twenty‐four studies with 5423 patients with CRC were included. Significant differences in OS, PFS and DFS were not observed among the different comparisons of patients carrying different alleles of the MTHFR rs1801133 polymorphism (including TT versus CC, TT versus CT + CC, CT + TT versus CC and CT versus CC). Compared with patients with the rs1801131 CA + AA genotypes, patients with the CC genotype had a shorter OS (hazard ratio = 1.85; 95% confidence interval = 1.30–2.65) and DFS (hazard ratio = 2.16; 95% confidence interval= 1.19–3.93). Significant differences in OS, PFS and DFS were not observed among the other patient groups (including CC versus AA, CC + CA versus AA and CA versus AA). Subgroup analysis of rs1801133 and rs1801131 showed that patients with CRC from Asian regions and Western regions demonstrated similar results.
Conclusions
The MTHFR rs1801133 polymorphism was not associated with the prognosis of patients with CRC; however, rs1801131 may be associated with the prognosis of patients with CRC. Well‐designed prospective studies are necessary to obtain a better understanding of the prognostic value of rs1801133 and rs1801131.
Keywords: colorectal cancer, gene polymorphism, meta‐analysis, methylenetetrahydrofolate reductase, prognosis
1. INTRODUCTION
As the third most commonly diagnosed cancer, colorectal cancer (CRC) has a worldwide incidence of over 1.3 million and a mortality rate of approximately 50%.1, 2, 3 Although the incidence of CRC has decreased in recent years because of improvements in its early diagnosis and treatment,1 the number of CRC cases continues to increase worldwide. Despite recent advances in treatment modalities, the 5‐year survival rate of patients with advanced CRC is not satisfactory as a result of recurrence and drug resistance.4
Methylenetetrahydrofolate reductase (MTHFR) is required for folate metabolism, intracellular homeostasis and DNA synthesis. It converts 5,10‐methylenetetrahydrofolate (5,10‐MTHF) to 5‐methyltetrahydrofolate (5‐MTHF), which is the major circulating form of folate in the blood and provides methyl groups to convert homocysteine into methionine. MTHFR contributes to the imbalance in methylation reactions, leading to genomic DNA hypomethylation, and influences folate metabolism.5, 6
The two most common loci for MTHFR single nucleotide polymorphisms (SNPs) are rs1801133 (C677T) and rs1801131 (A1298C).7 Both are associated with a deficiency in enzymatic activity.8 The MTHFR rs1801133 polymorphism is a point mutation at the position 677C>T, in which alanine is replaced with valine.9 The MTHFR rs1801131 polymorphism is a point mutation at position 1298A>C, in which glutamate is replaced with valine.10 The rs1801133 and rs1801131 polymorphisms reduce the activity of the MTHFR enzyme and increase the homocysteine level in the blood, which may be a risk factor for cancer.11
Recently, some meta‐analyses have reported significant correlations between the MTHFR rs1801133 and rs1801131 polymorphisms and tumour responses to chemoradiotherapy and short‐term clinical benefits.12, 13, 14 For example, two meta‐analyses were performed to investigate the associations between MTHFR polymorphisms and the response of patients with CRC to chemotherapy.13, 14 A meta‐analysis was conducted to investigate the associations between MTHFR polymorphisms and short‐term clinical benefits (complete or partial response, relapse or progression) of chemotherapy in patients with CRC.12 These meta‐analyses only focused on the short‐term prognostic effects of MTHFR polymorphisms on patients with CRC.
No meta‐analysis has been performed investigating the association between these MTHFR polymorphisms and survival (e.g. overall survival [OS], progression‐free survival [PFS] or disease‐free survival [DFS]). By systematically reviewing recent publications, we conducted a meta‐analysis according to the guidelines of the PRISMA statement.15 The aim was to explore whether the MTHFR rs1801133 and rs1801131 polymorphisms might affect the prognosis of patients with CRC and whether these SNPs are potentially useful as predictive biomarkers.
2. MATERIALS AND METHODS
2.1. Literature search strategy
A comprehensive literature search was performed independently by two investigators (XLC and YMW) from the inception of each database up to 14 March 2016. The databases included PubMed, EMBASE, the Cochrane Central Register of Controlled Trials and Chinese databases (including CNKI, Wanfang and VIP). The search terms included the keywords: colorectal cancer (including colorectal cancer, colon cancer, rectal cancer), MTHFR (including MTHFR and methylenetetrahydrofolate reductase) and prognosis (including prognosis, prognoses, predictive, biomarker, marker, survival, log rank, Kaplan–Meier and Cox). The detailed search strategy is documented in the Supporting information (Doc. S1). Google Scholar was also used to search for relevant articles. Systematic reviews and meta‐analyses of MTHFR polymorphisms and CRC were manually screened for potentially eligible articles.
Duplicate articles that were obtained from multiple databases were deleted. The abstract of each article was extracted and screened by two of three investigators (FZ, TGY and GT), and the full texts of potentially eligible articles were reviewed for data analysis. Next, two of three investigators (FZ, TGY and GT) independently reviewed and confirmed the eligibility of the articles. Any disagreement was recorded and resolved by consensus under the guidance of a fourth investigator (XLC). The cross‐referencing strategy was adopted until the two investigators reached a consistent result.
2.2. Inclusion criteria for the studies
This meta‐analysis includes articles reporting the patient's CRC prognosis and MTHFR genotype. The inclusion criteria comprised: (i) a diagnosis of CRC, colon cancer, rectal cancer or metastatic CRC (mCRC); (ii) rs1801133 or rs1801131 polymorphisms identified by polymerase chain reaction (PCR) or polymerase chain reaction restriction fragment length polymorphism (PCR‐RFLP); and (iii) data describing OS, DFS and/or PFS with hazard ratios (HRs), 95% confidence intervals (CIs) or the relevant information (e.g. survival curves) were provided. Articles published in abstract form were included only when sufficient outcome data were presented or when the authors were willing to provide detailed results from the study. If several articles from the same patient population were reported, the most recent or most detailed study was included.
2.3. Data extraction and quality assessment
For each article, two of three investigators (FZ, TGY and GT) independently extracted the required data according to a predefined protocol. The extracted data comprised: authors’ names, year of publication, patient characteristics (cancer type, sample size, gender and mean age), therapy (surgery, chemotherapy and radiotherapy), characteristics of MTHFR polymorphisms (rs1801133 or rs1801131, sample source, sample content, test method and cut‐off values) and prognostic outcomes (HRs and their 95% CIs for OS, PFS and DFS). If the data from any of the above categories were unavailable in the text, the corresponding record was marked as “NR (not reported)”. Differences in data extraction were resolved by cross‐checking until a consensus was reached.
2.4. Statistical analysis
Four genetic models existed for rs1801133: TT versus CC (TT/CC, additive model), TT versus CT and CC (TT/CT + CC, recessive model), TT and CT versus CC (TT + CT/CC, dominant model) and CT versus CC (CT/CC, heterozygous model). For rs1801131, the four models included CC versus AA (CC/AA), CC versus CA and AA (CC/CA + AA), CC and CA versus AA (CC + CA/AA) and CA versus AA (CA/AA). None of the included articles reported data about the allele model (wild‐type allele versus mutant‐type allele) for rs1801133 and rs1801131. Therefore, the allele model was not included in our meta‐analysis. OS, PFS and DFS were analysed separately.
The HRs and 95% CIs reflected the effects of rs1801133 and rs1801131 on the prognosis. If these data were available in the collected articles, we extracted these data directly; otherwise, they were calculated from the available numerical data in the articles based on the methods developed by Tierney et al.16
Pooled HRs and their 95% CIs for OS, PFS and DFS between different genetic models were calculated. The heterogeneity of all HRs was calculated using chi‐squared tests. The heterogeneity test with the inconsistency index (I 2) statistic and Q statistic was performed. If the HR was homogeneous, then the fixed‐effects model was employed for analysis; otherwise, a random‐effects model was used. p < 0.05 was considered statistically significant. Additionally, an HR > 1 suggested a poor prognosis. Publication bias was evaluated using the methods described by Begg and Mazumdar.17
Linkage disequilibrium among the variants can vary across populations.18, 19 For example, Haerian and Haerian18 showed that rs1801133 and rs1801131 might be CRC susceptibility variants in Americans and Australians, whereas rs1801133 may be more common in the Brazilian and Japanese populations. Based on these results, patients of different ethnicities may carry different rs1801133 and rs1801131 variants. Therefore, a subgroup analysis based on different regions (e.g. Asia and Western countries) was performed. All calculations were performed using STATA, version 12.0 (StataCorp, College Station, TX, USA).
3. RESULTS
3.1. Article characteristics
Figure 1 shows the process used to screen the included articles. The literature search yielded 539 articles, 152 of which were excluded as a result of duplication. The abstracts of 387 articles were reviewed by the investigators, and the 314 articles that failed to meet the inclusion criteria were excluded. The full texts of the remaining 73 articles were retrieved. Finally, twenty‐four articles were included in the meta‐analysis.20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43
Figure 1.
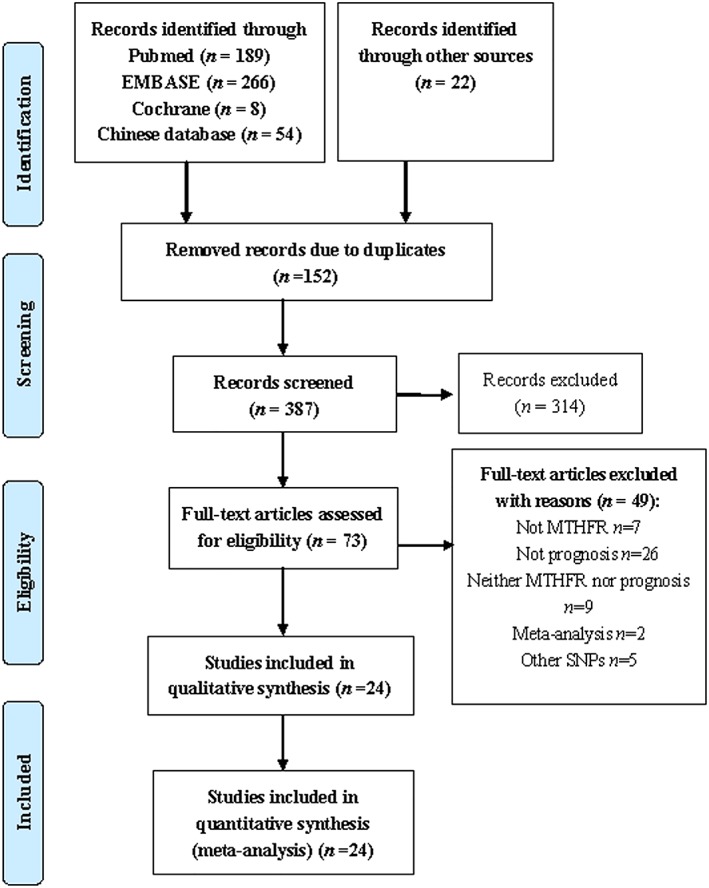
Flow chart of the search strategy
Table 1 summarizes the characteristics of the included articles. Among the 24 included articles, seven were conducted in China or Korea,20, 24, 26, 28, 31, 32, 42 one was conducted in Mexico33 and the remaining articles were conducted in European or North American countries. All but two eligible articles targeted CRC or mCRC: one addressed rectal cancer22 and the other studied colon cancer.28 In total, 5423 patients with CRC were included in our analysis. The sample size of each article ranged from 29 to 784 patients, with a median of 136 patients. All patients received either 5‐fluorouracil (5‐FU) or 5‐FU‐based chemotherapy. Information about the rs1801133 and rs1801131 polymorphisms is provided in Table 2.
Table 1.
Characteristics of the included articles
| Reference | Year of publication | Country | Time | Patients | Sample size | Number of males | Mean age (range, years) | Stage | Surgery | Chemotherapy | Radiotherapy | Median (range) follow‐up (months) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Afzal et al.36 | 2009 | Denmark | 1996–2003 | CRC | 331 | 166 | 61 (NR) | II–IV | NR | 5‐FU + LV | NR | 120 (NR) |
| Budai et al.29 | 2012 | Hungary | 2006–2008 | CRC | 85 | NR | NR (NR) | IV | NR | 5‐FU + LV + CPT‐11 + BEV | NR | NR (NR) |
| Castillo‐Fernández et al.33 | 2010 | Mexico | 1998–2004 | mCRC | 29 | 11 | 55.9 (NR) | IV | NR | 5‐FU + FA | NR | NR (NR) |
| Cecchin et al.21 | 2015 | Italy | 2003–2007 | CRC | 112 | 62 | 65 (30–85) | II, III | All | 5‐FU/CAPE | NR | 80 (10–185) |
| Chua et al.37 | 2009 | Britain | 1999–2000 | mCRC | 118 | 80 | 61 (31–75) | IV | NR | 5‐FU + LV + OX | NR | NR (NR) |
| Custodio et al.23 | 2014 | Spain | 2004–2009 | CRC | 202 | 115 | 63.8 (23–85) | II, III | All | 5‐FL + OX | None | 51.4 (7–96) |
| Delgado‐Plasencia et al.27 | 2013 | Spain | 1990–2003 | CRC | 50 | 28 | NR (NR) | NR | All | 5‐FU‐based | NR | NR (NR) |
| Dong et al.20 | 2016 | China | 2012–2013 | CRC | 81 | 44 | 56.2 (27–76) | IV | NR | 5‐FU + LV + OX | NR | 14 (5–20) |
| Etienne et al.43 | 2004 | France | NR | mCRC | 98 | 57 | 64 (40–82) | IV | NR | 5FU + FA | NR | NR (NR) |
| Fernández‐Peralta et al.34 | 2010 | Spain | 1992–1996 | CRC | 143 | 81 | 67.3 (NR) | NR | All | 5‐FU‐based | NR | 44.3 (NR) |
| Gusella et al.35 | 2009 | Britain | 1999–2008 | CRC | 130 | 84 | 64.7 (34–84) | B, C* | NR | 5‐FU + LV | NR | 45.6 (4.8–120.0) |
| Huang et al.31 | 2011 | China | 2005–2009 | mCRC | 157 | 85 | 62.5 (36–82) | IV | NR | 5‐FU + LV + OX | NR | 35 (8–56) |
| Jang et al.24 | 2014 | Korea | 1996–2009 | CRC | 372 | 215 | 62.1 (NR) | I–IV | All | 5‐FU‐based | NR | 34 (4–173) |
| Kim et al.32 | 2010 | Korea | 1995–2004 | CRC | 103 | 49 | 57.0 (NR) | II–IV | All | 5‐FU/5‐FU + OX | NR | 62.2 (18–121) |
| Negandhi et al.25 | 2013 | Canada | 1999–2003 | CRC | 784 | 327 | 61.4 (20.7–75.0) | I–IV | All | 5‐FU (partial) | NR | 76.8 (4.8–130.8) |
| Qiu et al.28 | 2013 | China | 2004–2006 | CRC | 76 | 48 | 57 (21–75) | I–III | All | 5‐FU + OX + LV | NR | NR (37–67) |
| Ruzzo et al.40 | 2007 | Italy | NR | mCRC | 166 | 87 | 66 (NR) | IV | NR | 5‐FU + LV + OX | NR | 24 (NR) |
| Ruzzo et al.39 | 2008 | Italy | NR | mCRC | 146 | 80 | 61 (38–75) | IV | NR | 5‐FU + LV + CPT‐11 | NR | NR (NR) |
| Sharma et al.38 | 2008 | Australia | 2002–2003 | mCRC | 54 | 35 | 72 (42–86) | IV | NR | CAPE | NR | NR (NR) |
| Suh et al.42 | 2006 | Korea | NR | mCRC | 54 | 30 | 57.8 (35–39) | II–IV | NR | 5‐FU + LV + OX | NR | 23.6 (6–35) |
| Taflin et al.30 | 2011 | Sweden | 1999–2006 | CRC | 649 | 88 | 66 (32–82) | III | All | 5‐FU + LV | NR | 70 (NR) |
| Ulrich et al.22 | 2014 | America | 1994–2000 | Rectal cancer | 754 | 482 | 61 (19–86) | II, III | All | 5‐FU + LV | All | NR (NR) |
| Zhang et al.41 | 2007 | America | 1992–2003 | mCRC | 318 | 177 | 58 (25–86) | IV | NR | FU + CPT‐11/FU + OX | NR | 30 (NR) |
| Zhu et al.26 | 2013 | China | 2004–2007 | CRC | 411 | 245 | 60 (NR) | I–IV | All | 5‐FU/5‐FU + OX | NR | 64 (1–88) |
5‐FU, 5‐fluorouracil; BEV, bevacizumab; CAPE, capecitabine; CPT‐11, irinotecan; CRC, colorectal cancer; FA, folinic acid; FU, fluorouracil; LV, leucovorin; mCRC, metastatic CRC; NR, not reported; OX, oxaliplatin;
, Duke's stage.
Table 2.
Information about and results for the rs1801133 and rs1801131 polymorphisms in the included studies
| Reference | rs1801133 | rs1801131 | Test sample | Test content | Test method | Analytical method | Outcome reported |
|---|---|---|---|---|---|---|---|
| Afzal et al.36 | Yes | Yes | Tumour tissue | DNA | PCR# | Mul | OS*, PFS* |
| Budai et al.29 | Yes | – | Blood | DNA | PCR | Mul | OS^, PFS^ |
| Castillo‐Fernández et al.33 | Yes | – | Tissue | DNA | PCR | Uni | OS^ |
| Cecchin et al.31 | Yes | Yes | Blood or tissue | DNA | PCR# | Mul | DFS* |
| Chua et al.37 | Yes | – | Tissue | DNA | PCR | Uni | OS^, PFS^ |
| Custodio et al.33 | Yes | – | Tissue | DNA | PCR‐RFLP | Uni | DFS^ |
| Delgado‐Plasencia et al.27 | Yes | – | Tumour tissue | DNA | PCR‐RFLP | Uni | OS^ |
| Dong et al.20 | Yes | – | Tissue | DNA | PCR | Uni | DFS^ |
| Etienne et al.43 | Yes | Yes | Tissue | DNA | PCR | Mul | OS* |
| Fernández‐Peralta et al.34 | Yes | Yes | Blood and tissue | DNA | PCR | Mul | OS* |
| Gusella et al.35 | Yes | Yes | Blood | DNA | PCR | Uni | OS*, DFS* |
| Huang et al.31 | Yes | – | Blood | DNA | PCR‐RFLP | Uni | OS^, PFS^ |
| Jang et al.24 | Yes | Yes | Blood | DNA | PCR | Mul | OS*, DFS* |
| Kim et al.32 | Yes | – | Leukocytes | DNA | PCR‐RFLP | Uni | OS^ |
| Negandhi et al.25 | – | Yes | Blood | DNA | PCR# | Mul | OS& |
| Qiu et al.28 | Yes | Yes | Blood | DNA | PCR | Mul | PFS* |
| Ruzzo et al.40 | Yes | Yes | Blood | DNA | PCR | Mul | PFS* |
| Ruzzo et al.39 | Yes | Yes | Blood | DNA | PCR | Mul | PFS* |
| Sharma et al.38 | Yes | – | Blood | DNA | PCR | Uni | OS^ |
| Suh et al.42 | Yes | – | Tissue | DNA | PCR | Uni | OS^ |
| Taflin et al.30 | Yes | – | Blood | DNA | PCR# | Uni | OS^ |
| Ulrich et al.22 | Yes | Yes | Tissue | DNA | PCR# | Mul | OS* |
| Zhang et al.41 | Yes | Yes | Blood and tissue | DNA | PCR# | Mul | OS* |
| Zhu et al.26 | Yes | Yes | Blood | DNA | PCR# | Mul | OS* |
–, Not available;
for both rs1801133 and rs1801131;
for rs1801133 alone;
for rs1801131 alone. PCR, polymerase chain reaction; PCR#, PCR TaqMan; PCR‐RFLP, polymerase chain reaction restriction fragment length polymorphism; Mul, multivariate analysis; Uni, univariate analysis.
3.2. Meta‐analysis of rs1801133
Twenty‐three of the included articles assessed the association between rs1801133 and survival time. According to the heterogeneity analysis, all of the articles were homogeneous and the fixed‐effect model was adopted. Compared with patients carrying the CC genotype, patients carrying the TT genotype did not show an increased HR for OS (HR = 1.17; 95% CI = 0.99–1.40), PFS (HR = 0.90; 95% CI = 0.70–1.15) or DFS (HR = 1.23; 95% CI = 0.93–1.62) (Table 3). Additionally, significant differences in OS (HR = 1.07; 95% CI = 0.76–1.49), PFS (HR = 0.91; 95% CI = 0.53–1.55) and DFS (HR = 1.27; 95% CI = 0.86–1.88) were not observed between patients carrying the TT genotype and patients carrying the CT + CC genotypes (Table 3). In the comparison of patients carrying the TT + CT genotypes with patients carrying the CC genotype, the pooled HRs of OS, PFS and DFS were 1.09 (95% CI = 0.90–1.31), 1.12 (95% CI = 0.85–1.48) and 1.02 (95% CI = 0.69–1.51), respectively (Table 3). Significant differences in OS, PFS and DFS were not observed between patients carrying the CT genotypes and patients carrying the CC genotypes (Table 3).
Table 3.
Results of the meta‐analysis of the MTHFR rs1801133 polymorphism
| Number of articles | Number of patients | HR (95% CI) | Heterogeneity (I 2, p) | |
|---|---|---|---|---|
| TT/CC | ||||
| OS | 11 | 2,526 | 1.17 (0.99–1.40) | 0.0%, 0.957 |
| PFS | 5 | 672 | 0.90 (0.70–1.15) | 0.0%, 0.778 |
| DFS | 3 | 1,256 | 1.23 (0.93–1.62) | 11.1%, 0.325 |
| TT/CT + CC | ||||
| OS | 5 | 840 | 1.07 (0.76–1.49) | 2.1%, 0.395 |
| PFS | 1 | 331 | 0.91 (0.53–1.55) | – |
| DFS | 3 | 686 | 1.27 (0.86–1.88) | 34.4%, 0.218 |
| TT + CT/CC | ||||
| OS | 8 | 1,574 | 1.09 (0.90–1.31) | 40.9%, 0.106 |
| PFS | 3 | 284 | 1.12 (0.85–1.48) | 52.3%, 0.123 |
| DFS | 2 | 448 | 1.02 (0.69–1.51) | 0.0%, 0.387 |
| CT/CC | ||||
| OS | 10 | 2,369 | 1.12 (0.96–1.31) | 0.0%, 0.673 |
| PFS | 2 | 203 | 0.96 (0.68–1.36) | 47.0%, 0.170 |
| DFS | 3 | 1,256 | 1.10 (0.90–1.35) | 0.0%, 0.478 |
TT/CC: TT genotype versus CC genotype; TT/CT + CC: TT genotype versus (CT + CC) genotype; TT + CT/CC: (TT + CT) genotype versus CC genotype; CT/CC: CT genotype versus CC genotype. −, not available.
Subgroup analysis revealed similar results for patients with CRC from Asian regions or Western regions (Table 4). For example, the HR of the TT versus CC genotype for patients from Asian regions was 1.06 (95% CI = 0.75–1.49) and the value for patients from Western regions was 1.22 (95% CI = 0.99–1.50).
Table 4.
Results for the subgroup analysis of the MTHFR rs1801133 polymorphism in different geographic regions
| Subgroup | Number of articles | Number of patients | HR (95% CI) | |
|---|---|---|---|---|
| TT/CC | ||||
| OS | Asian | 4 | 994 | 1.06 (0.75–1.49) |
| Western | 7 | 1,532 | 1.22 (0.99–1.50) | |
| PFS | Asian | 1 | 157 | 1.53 (0.36–6.51) |
| Western | 4 | 515 | 0.88 (0.68–1.14) | |
| DFS | Asian | 1 | 372 | 0.71 (0.33–1.53) |
| Western | 2 | 884 | 1.33 (0.99–1.79) | |
| TT/CT + CC | ||||
| OS | Asian | 2 | 426 | 0.94 (0.59–1.51) |
| Western | 3 | 414 | 1.19 (0.78–1.81) | |
| PFS | Asian | – | – | – |
| Western | 1 | 331 | 0.91 (0.53–1.55) | |
| DFS | Asian | 1 | 372 | 0.86 (0.45–1.64) |
| Western | 2 | 314 | 1.59 (0.98–2.58) | |
| TT + CT/CC | ||||
| OS | Asian | 3 | 426 | 1.13 (0.76–1.69) |
| Western | 5 | 995 | 1.07 (0.87–1.33) | |
| PFS | Asian | 1 | 81 | 1.81 (0.99–3.32) |
| Western | 2 | 203 | 0.98 (0.72–1.34) | |
| DFS | Asian | 2 | 448 | 1.02 (0.69–1.51) |
| Western | – | – | – | |
| CT/CC | ||||
| OS | Asian | 3 | 837 | 1.22 (0.89–1.67) |
| Western | 7 | 1,532 | 1.09 (0.92–1.30) | |
| PFS | Asian | – | – | – |
| Western | 2 | 203 | 0.96 (0.68–1.36) | |
| DFS | Asian | 1 | 372 | 0.89 (0.53–1.50) |
| Western | 2 | 884 | 1.14 (0.92–1.43) | |
–, Not available.
3.3. Meta‐analysis of rs1801131
Thirteen articles assessed the association between rs1801131 and survival time. Significant differences in OS, PFS and DFS were not observed between patients carrying the CC genotype and patients carrying the AA genotype (Table 5). Compared with patients with the CA + AA genotypes, patients with the CC genotype had a shorter OS (HR = 1.85; 95% CI = 1.30–2.65) and DFS (HR = 2.16; 95% CI = 1.19–3.93) (Figure 2 and Table 5). Significant differences in OS, PFS and DFS were not observed between patients with the CC + CA genotypes and patients with the AA genotype (Table 5). Significant differences in OS, PFS and DFS were not observed between patients with the CA genotype and patients with the AA genotype (Table 5).
Table 5.
Results of the meta‐analysis of the MTHFR rs1801131 polymorphism
| Number of articles | Number of patients | HR (95% CI) | Heterogeneity (I 2, p) | |
|---|---|---|---|---|
| CC/AA | ||||
| OS | 7 | 2,867 | 1.13 (0.81–1.59)* | 50.9%, 0.057 |
| PFS | 2 | 312 | 0.89 (0.58–1.37) | 0.0%, 0.660 |
| DFS | 3 | 1,256 | 0.78 (0.53–1.13) | 0.0%, 0.738 |
| CC/CA + AA | ||||
| OS | 3 | 1,254 | 1.85 (1.30–2.65) | 0.0%, 0.584 |
| PFS | – | – | – | – |
| DFS | 2 | 484 | 2.16 (1.19–3.93) | 0.0%, 0.337 |
| CC + CA/AA | ||||
| OS | 5 | 1,948 | 1.11 (0.85–1.45)* | 62.3%, 0.031 |
| PFS | 2 | 412 | 0.79 (0.55–1.14) | 0.0%, 0.547 |
| DFS | 1 | 372 | 0.92 (0.55–1.54) | – |
| CA/AA | ||||
| OS | 6 | 2,549 | 0.97 (0.84–1.12) | 0.0%, 0.507 |
| PFS | 2 | 312 | 0.97 (0.60–1.57) | 0.0%, 0.933 |
| DFS | 3 | 1,256 | 0.88 (0.73–1.07) | 0.0%, 0.974 |
CC/AA: CC genotype versus AA genotype; CC/CA + AA: CC genotype versus (CA + AA) genotype; CC + CA/AA: (CC + CA) genotype versus AA genotype; CA/AA: CA genotype versus AA genotype. −, not available.
Results from the random‐effects model.
Figure 2.
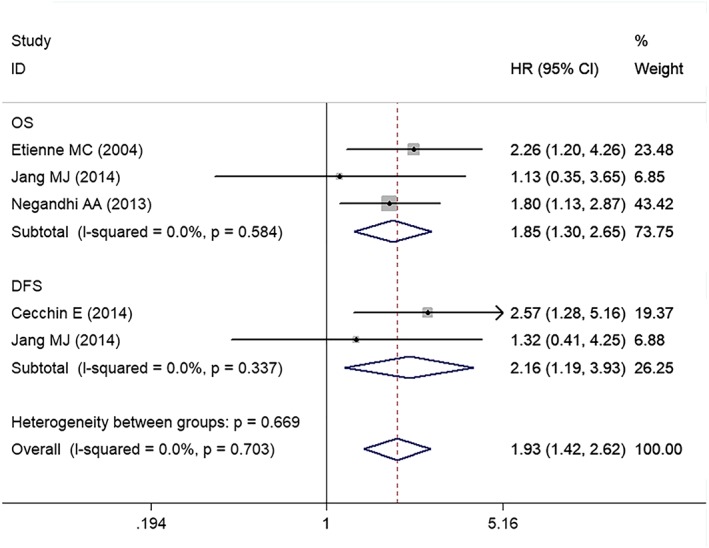
Meta‐analysis plots of the HRs for survival in the comparison of patients with the CC genotype and patients with the AA + CA genotypes of rs1801131. OS, overall survival; PFS, progression‐free survival; DFS, disease‐free survival
Subgroup analysis revealed similar results for patients with CRC from Asian regions and Western regions (Table 6). For example, the HR of the CC versus AA genotype in patients from Asian regions was 0.81 (95% CI = 0.51–1.29) and the HR for this same comparison of patients from Western regions was 1.25 (95% CI = 0.82–1.91).
Table 6.
Results from the subgroup analysis of the MTHFR rs1801131 polymorphism in different geographic regions
| Subgroup | Number of articles | Number of patients | HR (95% CI) | |
|---|---|---|---|---|
| CC/AA | ||||
| OS | Asian | 2 | 783 | 0.81 (0.51–1.29) |
| Western | 5 | 2,084 | 1.25 (0.82–1.91)* | |
| PFS | Asian | – | – | – |
| Western | 2 | 203 | 0.89 (0.58–1.37) | |
| DFS | Asian | 1 | 372 | 1.20 (0.37–3.89) |
| Western | 2 | 884 | 0.74 (0.50–1.10) | |
| CC/CA + AA | ||||
| OS | Asian | 1 | 372 | 1.13 (0.35–3.65) |
| Western | 2 | 882 | 1.95 (1.34–2.84) | |
| PFS | Asian | – | – | – |
| Western | – | – | – | |
| DFS | Asian | 1 | 372 | 1.32 (0.41–4.25) |
| Western | 1 | 112 | 2.57 (1.28–5.16) | |
| CC + CA/AA | ||||
| OS | Asian | 1 | 372 | 0.70 (0.41–1.19) |
| Western | 4 | 1,576 | 1.21 (0.94–1.56)* | |
| PFS | Asian | 1 | 81 | 0.69 (0.39–1.23) |
| Western | 1 | 331 | 0.87 (0.54–1.41) | |
| DFS | Asian | 1 | 372 | 0.92 (0.55–1.54) |
| Western | – | – | – | |
| CA/AA | ||||
| OS | Asian | 2 | 783 | 0.94 (0.68–1.28) |
| Western | 4 | 1,766 | 0.98 (0.83–1.16) | |
| PFS | Asian | – | – | – |
| Western | 2 | 312 | 0.97 (0.60–1.57) | |
| DFS | Asian | 1 | 372 | 0.89 (0.52–1.53) |
| Western | 2 | 884 | 0.88 (0.72–1.09) | |
–, Not available.
Results from the random‐effects model.
4. DISCUSSION
Our meta‐analysis highlighted the long‐term prognostic effects (including OS, PFS and DFS) of MTHFR polymorphisms on patients with CRC. The rs1801131 polymorphism may predict the prognosis. Compared with patients with the CA + AA genotypes, patients with the CC genotype had a shorter OS (HR = 1.85) and DFS (HR = 2.15). However, significant differences were not observed among the other comparisons (CC versus AA, CC + CA versus AA and CA versus AA). Other researchers also reported similar results. For example, rs1801131 appears to be a potential prognostic factor for patients with gastric cancer.13, 44, 45
The MTHFR rs1801131 polymorphism may predict the prognosis; the possible explanations are described below. As a crucial enzyme in metabolism, MTHFR catalyses the transformation of 5,10‐MTHF into 5‐MTHF.25, 46, 47, 48 Notably, 5,10‐MTHF mainly synthesizes purines and thymidine. Furthermore, 5‐MTHF participates in the synthesis of S‐adenosyl‐methionine, which is an important mediator of methylation reactions.47, 48 Regarding rs1801131, its mutation is linked to reduced MTHFR enzyme activity, although the decrease is less pronounced than the change induced by 677CNT.49 Therefore, the reduction in MTHFR enzyme activity as a result of the rs1801131 polymorphism may lead to a higher level of the precursor 5,10‐MTHF and a correspondingly lower level of 5‐MTHF, given the relatively low catalytic activity of the enzyme. The accumulation of 5,10‐MTHF would provide a greater pool of nucleotides for DNA synthesis, thus prompting tumour cell proliferation, which requires an abundant supply of nucleic acids. Once CRC has developed, folate supplementation might enhance its growth and progression,42, 48, 50, 51 presumably by providing large amounts of nucleotide precursors for tumour growth.42, 48, 51 Folate supplementation is associated with a higher risk of CRC.52 These findings indicate a negative effect of high levels of MTHFR on patients with CRC. Therefore, the association between MTHFR polymorphisms and a worse prognosis of CRC may be ascribed to decreased MTHFR activity.
In the present study, data heterogeneity was not observed; therefore, all of the data were analysed using fixed‐effects models. Subgroup analysis was performed according to the patient's nationality and revealed that rs1801133 and rs1801131 exerted the same effects on patients from Asian regions and patients from Western regions.
The MTHFR rs1801131 polymorphism may be associated with the prognosis of patients with CRC. In the future, additional high‐quality prospective studies should be conducted to obtain a better understanding of the prognostic value of the MTHFR polymorphisms. The MTHFR rs1801131 polymorphism may be regarded as a target for drugs that are widely used to treat cancer and inflammatory diseases.12 This polymorphism may better predict the prognosis of patients with CRC and facilitate the administration of individualized treatments.
Some limitations exist in this meta‐analysis. (i) The eligible articles included in our meta‐analysis were restricted to studies published in English and Chinese, which likely caused selection bias. Articles published in other languages were excluded, which might cause selection bias as a result of low reporting qualities. (ii) The therapy method substantially affected the survival of patients with CRC. Although all of the included patients with CRC were treated with 5‐FU chemotherapy, the use of specific therapies differed among the included articles. Thus, the confounding effects of different therapies remain unclear. (ii) HRs calculated from the data or extracted from survival curves may be less reliable than HRs directly calculated with an analysis of variance.
In summary, the MTHFR rs1801133 polymorphism was not associated with the OS, PFS or DFS of patients with CRC. However, the MTHFR rs1801131 polymorphism was associated with a shorter OS and DFS in patients with CRC (CC + CA versus AA), although the other genotypes of MTHFR rs1801131 did not produce significant differences. Both rs1801133 and rs1801131 produced similar results among patients with CRC from Asian regions and Western regions. These results might provide guidance and prognostic predictive power for physicians during the clinical treatment of patients with CRC who are undergoing 5‐FU chemotherapy. Well‐designed prospective studies are necessary to further investigate the precise prognostic value of the MTHFR rs1801133 and rs1801131 polymorphisms.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflicts of interest.
Supporting information
Doc. S1. The search strategy
ACKNOWLEDGEMENTS
This study was funded by grants from the National Natural Science Foundation of China (81774451), the Outstanding Youth Foundation of Guangdong Province Colleges and Universities (YQ2015041), the Young Talents Foundation of Guangzhou University of Chinese Medicine (QNYC20140101), and the Natural Science Foundation of Guangdong Province (2017A030313827).
Chen X‐L, Wang Y‐M, Zhao F, et al. Methylenetetrahydrofolate reductase polymorphisms and colorectal cancer prognosis: A meta‐analysis. J Gene Med. 2019;21:e3114 10.1002/jgm.3114
Contributor Information
Xin‐Lin Chen, Email: chenxlsums@126.com.
Feng‐Bin Liu, Email: liufb163@163.com.
REFERENCES
- 1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11‐30. [DOI] [PubMed] [Google Scholar]
- 2. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490‐1502. [DOI] [PubMed] [Google Scholar]
- 3. Torres Stone RA, Waring ME, Cutrona SL, Kiefe CI, Allison J, Doubeni CA. The association of dietary quality with colorectal cancer among normal weight, overweight and obese men and women: a prospective longitudinal study in the USA. BMJ Open. 2017;7:e015619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Asteria CR, Pucciarelli S, Gerard L, et al. The impact of colorectal screening program on the detection of right‐sided colorectal cancer. A 5‐year cohort study in the Mantua District. Int J Colorectal Dis. 2015;30:1627‐1637. [DOI] [PubMed] [Google Scholar]
- 5. Stern LL, Mason JB, Selhub J, Choi SW. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol Biomarkers Prev. 2000;9:849‐853. [PubMed] [Google Scholar]
- 6. Jeon YJ, Kim JW, Park HM, et al. Genetic variants in 3'‐UTRs of methylenetetrahydrofolate reductase (MTHFR) predict colorectal cancer susceptibility in Koreans. Sci Rep. 2015;5:11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shiao SP, Yu CH. Meta‐prediction of MTHFR gene polymorphism mutations and associated risk for colorectal cancer. Biol Res Nurs. 2016;18:357‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liew SC, Gupta ED. Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism: epidemiology, metabolism and the associated diseases. Eur J Med Genet. 2015;58:1‐10. [DOI] [PubMed] [Google Scholar]
- 9. Yeh CC, Lai CY, Chang SN, et al. Polymorphisms of MTHFR C677T and A1298C associated with survival in patients with colorectal cancer treated with 5‐fluorouracil‐based chemotherapy. Int J Clin Oncol. 2017;22:484‐493. [DOI] [PubMed] [Google Scholar]
- 10. Zhu XL, Liu ZZ, Yan SX, et al. Association between the MTHFR A1298C polymorphism and risk of cancer: evidence from 265 case‐control studies. Mol Genet Genomics. 2016;291:51‐63. [DOI] [PubMed] [Google Scholar]
- 11. Wu NC, Su SM, Lin TJ, et al. Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and fluorouracil‐based treatment in Taiwan colorectal cancer. Anticancer Drugs. 2015;26:888‐893. [DOI] [PubMed] [Google Scholar]
- 12. Jennings BA, Kwok CS, Willis G, Matthews V, Wawruch P, Loke YK. Functional polymorphisms of folate metabolism and response to chemotherapy for colorectal cancer, a systematic review and meta‐analysis. Pharmacogenet Genomics. 2012;22:290‐304. [DOI] [PubMed] [Google Scholar]
- 13. Zhao T, Gu D, Xu Z, et al. Polymorphism in one‐carbon metabolism pathway affects survival of gastric cancer patients: large and comprehensive study. Oncotarget. 2015;6:9564‐9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zintzaras E, Ziogas DC, Kitsios GD, Papathanasiou AA, Lau J, Raman G. MTHFR gene polymorphisms and response to chemotherapy in colorectal cancer: a meta‐analysis. Pharmacogenomics. 2009;10:1285‐1294. [DOI] [PubMed] [Google Scholar]
- 15. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. J Clin Epidemiol. 2009;62:1006‐1012. [DOI] [PubMed] [Google Scholar]
- 16. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time‐to‐event data into meta‐analysis. Trials. 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088‐1101. [PubMed] [Google Scholar]
- 18. Haerian BS, Haerian MS. Evaluation of association studies and meta‐analyses of MTHFR gene polymorphisms in colorectal cancer. Pharmacogenomics. 2015;16:413‐425. [DOI] [PubMed] [Google Scholar]
- 19. Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE‐1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734‐1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dong QM, Li Y, Huang SH. Prognostic impact of MTHFR polymorphism on colorectai cancer patients ttreated by chemotherapy. J Sun Yat‐Sen Univ (Med Sci). 2016;37:95‐99. [Google Scholar]
- 21. Cecchin E, Perrone G, Nobili S, et al. MTHFR‐1298 a>C (rs1801131) is a predictor of survival in two cohorts of stage II/III colorectal cancer patients treated with adjuvant fluoropyrimidine chemotherapy with or without oxaliplatin. Pharmacogenomics J. 2015;15:219‐225. [DOI] [PubMed] [Google Scholar]
- 22. Ulrich CM, Rankin C, Toriola AT, et al. Polymorphisms in folate‐metabolizing enzymes and response to 5‐fluorouracil among patients with stage II or III rectal cancer (INT‐0144; SWOG 9304). Cancer. 2014;120:3329‐3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Custodio A, Moreno‐Rubio J, Aparicio J, et al. Pharmacogenetic predictors of outcome in patients with stage II and III colon cancer treated with oxaliplatin and fluoropyrimidine‐based adjuvant chemotherapy. Mol Cancer Ther. 2014;13:2226‐2237. [DOI] [PubMed] [Google Scholar]
- 24. Jang MJ, Kim JW, Jeon YJ, et al. Polymorphisms of folate metabolism‐related genes and survival of patients with colorectal cancer in the Korean population. Gene. 2014;533:558‐564. [DOI] [PubMed] [Google Scholar]
- 25. Negandhi AA, Hyde A, Dicks E, et al. MTHFR Glu429Ala and ERCC5 His46His polymorphisms are associated with prognosis in colorectal cancer patients: analysis of two independent cohorts from Newfoundland. PLoS ONE. 2013;8:e61469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu L, Wang F, Hu F, et al. Association between MTHFR polymorphisms and overall survival of colorectal cancer patients in Northeast China. Med Oncol. 2013;30:467. [DOI] [PubMed] [Google Scholar]
- 27. Delgado‐Plasencia L, Medina‐Arana V, Bravo‐Gutierrez A, et al. Impact of the MTHFR C677T polymorphism on colorectal cancer in a population with low genetic variability. Int J Colorectal Dis. 2013;28:1187‐1193. [DOI] [PubMed] [Google Scholar]
- 28. Qiu Y, Zhou C, Yang N, Li K, Wang J, Liu X. Relationship between polymorphism of MTHFR and curative effect of FOLFOX4 adjuvant chemotherapy in postoperative colon cancer patients. Cancer Res Prev Treat. 2013;40:595‐598. [Google Scholar]
- 29. Budai B, Komlosi V, Adleff V, et al. Impact of SHMT1 polymorphism on the clinical outcome of patients with metastatic colorectal cancer treated with first‐line FOLFIRI+bevacizumab. Pharmacogenet Genomics. 2012;22:69‐72. [DOI] [PubMed] [Google Scholar]
- 30. Taflin H, Wettergren Y, Odin E, Carlsson G, Derwinger K. Gene polymorphisms MTHFRC677T and MTRA2756G as predictive factors in adjuvant chemotherapy for stage III colorectal cancer. Anticancer Res. 2011;31:3057‐3062. [PubMed] [Google Scholar]
- 31. Huang MY, Huang ML, Chen MJ, et al. Multiple genetic polymorphisms in the prediction of clinical outcome of metastatic colorectal cancer patients treated with first‐line FOLFOX‐4 chemotherapy. Pharmacogenet Genomics. 2011;21:18‐25. [DOI] [PubMed] [Google Scholar]
- 32. Kim JW, Lee JH, Hong SH, et al. Combination of polymorphisms between MTHFR and TS gene modulates survival after 5‐fluorouracil‐based therapy in colorectal cancer patients. Genes Genomics. 2010;32:469‐475. [Google Scholar]
- 33. Castillo‐Fernandez O, Santibanez M, Bauza A, et al. Methylenetetrahydrofolate reductase polymorphism (677 C>T) predicts long time to progression in metastatic colon cancer treated with 5‐fluorouracil and folinic acid. Arch Med Res. 2010;41:430‐435. [DOI] [PubMed] [Google Scholar]
- 34. Fernández‐Peralta AM, Daimiel L, Nejda N, Iglesias D, Medina Arana V, Gonzalez‐Aguilera JJ. Association of polymorphisms MTHFR C677T and A1298C with risk of colorectal cancer, genetic and epigenetic characteristic of tumors, and response to chemotherapy. Int J Colorectal Dis. 2010;25:141‐151. [DOI] [PubMed] [Google Scholar]
- 35. Gusella M, Frigo AC, Bolzonella C, et al. Predictors of survival and toxicity in patients on adjuvant therapy with 5‐fluorouracil for colorectal cancer. Br J Cancer. 2009;100:1549‐1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Afzal S, Jensen SA, Vainer B, et al. MTHFR polymorphisms and 5‐FU‐based adjuvant chemotherapy in colorectal cancer. Ann Oncol. 2009;20:1660‐1666. [DOI] [PubMed] [Google Scholar]
- 37. Chua W, Goldstein D, Lee CK, et al. Molecular markers of response and toxicity to FOLFOX chemotherapy in metastatic colorectal cancer. Br J Cancer. 2009;101:998‐1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sharma R, Hoskins JM, Rivory LP, et al. Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphisms and toxicity to capecitabine in advanced colorectal cancer patients. Clin Cancer Res. 2008;14:817‐825. [DOI] [PubMed] [Google Scholar]
- 39. Ruzzo A, Graziano F, Loupakis F, et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first‐line FOLFIRI chemotherapy. Pharmacogenomics J. 2008;8:278‐288. [DOI] [PubMed] [Google Scholar]
- 40. Ruzzo A, Graziano F, Loupakis F, et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first‐line FOLFOX‐4 chemotherapy. J Clin Oncol. 2007;25:1247‐1254. [DOI] [PubMed] [Google Scholar]
- 41. Zhang W, Press OA, Haiman CA, et al. Association of methylenetetrahydrofolate reductase gene polymorphisms and sex‐specific survival in patients with metastatic colon cancer. J Clin Oncol. 2007;25:3726‐3731. [DOI] [PubMed] [Google Scholar]
- 42. Suh KW, Kim JH, Kim DY, Kim YB, Lee C, Choi S. Which gene is a dominant predictor of response during FOLFOX chemotherapy for the treatment of metastatic colorectal cancer, the MTHFR or XRCC1 gene? Ann Surg Oncol. 2006;13:1379‐1385. [DOI] [PubMed] [Google Scholar]
- 43. Etienne MC, Formento JL, Chazal M, et al. Methylenetetrahydrofolate reductase gene polymorphisms and response to fluorouracil‐based treatment in advanced colorectal cancer patients. Pharmacogenetics. 2004;14:785‐792. [DOI] [PubMed] [Google Scholar]
- 44. Ott K, Rachakonda PS, Panzram B, et al. DNA repair gene and MTHFR gene polymorphisms as prognostic markers in locally advanced adenocarcinoma of the esophagus or stomach treated with cisplatin and 5‐fluorouracil‐based neoadjuvant chemotherapy. Ann Surg Oncol. 2011;18:2688‐2698. [DOI] [PubMed] [Google Scholar]
- 45. Blank S, Rachakonda S, Keller G, et al. A retrospective comparative exploratory study on two methylentetrahydrofolate reductase (MTHFR) polymorphisms in esophagogastric cancer: the A1298C MTHFR polymorphism is an independent prognostic factor only in neoadjuvantly treated gastric cancer patients. BMC Cancer. 2014;14:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jennings BA, Willis G. How folate metabolism affects colorectal cancer development and treatment; a story of heterogeneity and pleiotropy. Cancer Lett. 2015;356:224‐230. [DOI] [PubMed] [Google Scholar]
- 47. Kim YI. Folate and colorectal cancer: an evidence‐based critical review. Mol Nutr Food Res. 2007;51:267‐292. [DOI] [PubMed] [Google Scholar]
- 48. Williams EA. Folate, colorectal cancer and the involvement of DNA methylation. Proc Nutr Soc. 2012;71:592‐597. [DOI] [PubMed] [Google Scholar]
- 49. Weisberg I, Tran P, Christensen B, Sibani S, Rozen R. A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab. 1998;64:169‐172. [DOI] [PubMed] [Google Scholar]
- 50. Ulrich CM, Potter JD. Folate and cancer – timing is everything. JAMA. 2007;297:2408‐2409. [DOI] [PubMed] [Google Scholar]
- 51. Holmes RS, Zheng Y, Baron JA, et al. Use of folic acid‐containing supplements after a diagnosis of colorectal cancer in the Colon Cancer Family Registry. Cancer Epidemiol Biomarkers Prev. 2010;19:2023‐2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ma J, Stampfer MJ, Giovannucci E, et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res. 1997;57:1098‐1102. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Doc. S1. The search strategy


