Abstract
Accurate stroke lesion segmentation is a critical step in the neuroimaging processing pipeline for assessing the relationship between poststroke brain structure, function, and behavior. Many multimodal segmentation algorithms have been developed for acute stroke neuroimaging, yet few algorithms are effective with only a single T1‐weighted (T1w) anatomical MRI. This is a critical gap because multimodal MRI is not commonly available due to time and cost constraints in the stroke rehabilitation setting. Although several attempts to automate the segmentation of chronic lesions on single‐channel T1w MRI have been made, these approaches have not been systematically evaluated on a large dataset. We performed an exhaustive review of the literature and identified one semiautomated and three fully automated approaches for segmentation of chronic stroke lesions using T1w MRI within the last 10 years: Clusterize, automated lesion identification (ALI), Gaussian naïve Bayes lesion detection (lesionGnb), and lesion identification with neighborhood data analysis (LINDA). We evaluated each method on a large T1w stroke dataset (N = 181). LINDA was the most computationally expensive approach, but performed best across the three main evaluation metrics (median values: dice coefficient = 0.50, Hausdorff's distance = 36.34 mm, and average symmetric surface distance = 4.97 mm). lesionGnb had the highest recall/least false negatives (median = 0.80). However, across the automated methods, many lesions were either misclassified (ALI: 28, lesionGnb: 39, LINDA: 45) or not identified (ALI: 24, LINDA: 23, lesionGnb: 0). Segmentation accuracy in all automated methods were influenced by size (small: worst) and stroke territory (brainstem, cerebellum: worst) of the lesion. To facilitate reproducible science, our analysis files have been made publicly available online.
Keywords: big data, chronic stroke, lesion segmentation, MRI, stroke
1. INTRODUCTION
Despite intensive research and rehabilitation efforts, stroke remains a leading cause of long‐term disability worldwide (Mozaffarian et al., 2016). Stroke rehabilitation research aims to understand the relationship between brain, behavior, and recovery following a stroke and to use brain changes after a stroke to predict functional outcomes. Neuroimaging, particularly high‐resolution T1‐weighted (T1w) anatomical MRIs, has been used to examine structural brain changes after stroke. Careful investigation of poststroke brain anatomy, using techniques such as voxel‐lesion symptom mapping or calculation of the overlap percentage between the lesion and critical brain structures, have been useful for relating brain changes to behavioral outcomes (e.g., corticospinal tract lesion load; Bates et al., 2003; Lindenberg et al., 2010; Riley et al., 2011; Zhu, Lindenberg, Alexander, & Schlaug, 2010). However, accurate and precise lesion annotation is necessary to conduct and draw valid clinical inferences from these analyses.
To date, manual lesion tracing by an individual with expertise in neuroanatomy remains the gold standard for lesion segmentation. This procedure is a time and labor‐intensive process that requires domain expertise (Fiez, Damasio, & Grabowski, 2000). Consequently, this is not feasible for studies with larger sample sizes, and becomes a limiting factor in large‐scale stroke rehabilitation neuroimaging analyses (Liew et al., 2018). This is especially problematic for stroke rehabilitation research, as compared to acute stroke research, because there are few stroke segmentation algorithms that can be effectively used with only a T1w MRI for lesion segmentation.
Whereas multimodal MRI sequences including diffusion weighted imaging, T2‐FLAIR, and perfusion weighted imaging are commonly acquired within the first few hours to days of stroke onset and used to make clinical decisions about treatment of acute stroke, the primary imaging modality that is commonly acquired for studying chronic stroke anatomy across stroke rehabilitation research sites is a high‐resolution T1w MRI (Albers, 1998; Chalela et al., 2000). This is typically due to patient burden as well as time and financial constraints. Additionally, across different research groups, image modalities that are acquired beyond a T1w MRI vary widely due to varied research interests. For example, some groups may be interested in examining functional changes in the brain, and allocate image acquisition time to acquire task‐ or resting‐state fMRI, while other groups may be more interested in acquiring detailed anatomical scans and therefore acquire a diffusion tractography scan. A wealth of research attention has focused on developing optimal algorithms for quick lesion segmentation and prediction of gross clinical outcomes using multimodal sequences (Maier, Schröder, Forkert, Martinetz, & Handels, 2015). However, fewer lesion segmentation algorithms have focused on only T1w MRIs, on which lesions appear hypointense to the adjacent tissue and sometimes appear as holes in the tissue. Lesion segmentation on a single T1 modality is a challenging task, as such, algorithms have less access to information about the lesion as compared to algorithms that use multiple scans from the same subject (Pustina et al., 2016).
In recent years, a handful of automated and semiautomated lesion segmentation approaches have been developed in response to this problem (see literature search results in Table S1, Supporting Information). Automated segmentation approaches can be divided into two major categories: (a) supervised image classification techniques which use machine learning to train classifiers based on “ground truth” lesion examples (i.e., manually traced lesions), and (b) unsupervised approaches, which use mathematical modeling to first distinguish the lesional tissue characteristics from other tissue types without labeled responses and then separately cluster the voxels belonging to each tissue type. These automated approaches are promising, yet few comparisons between existing T1w lesion segmentation methods have been made, due to (a) the lack of large‐sized, publicly available stroke T1w MRI datasets and (b) the intensive labor necessary to manually segment lesions as the benchmark for comparison.
Systematic evaluations of existing algorithms can be useful for identifying current best solutions, as well as identifying areas where all algorithms could use improvement. An excellent example of a comparative evaluation of lesion segmentation algorithms comes from the multimodal, acute neuroimaging world in the form of the annual ischemic stroke lesion segmentation challenge (ISLES challenge; Maier et al., 2017; http://www.isles-challenge.org/). In the ISLES challenge, teams compete to develop algorithms that accurately segment the lesions and upload their algorithms to the ISLES website, after which the ISLES organizers evaluate automated lesion segmentations and rank participating research teams based on their performance on image metrics, including the dice similarity coefficient (DC), Hausdorff's distance (HD), average symmetric surface distance (ASSD), precision, and recall. However, to our knowledge, no such fair and systematic evaluations have been conducted on T1w MRIs on chronic stroke data.
Here, we evaluated existing approaches for unimodal T1w chronic stroke lesion segmentation by quantitatively measuring the performance of each on a large, publicly available stroke lesion dataset. We also identified areas for improvement across automated lesion segmentation algorithms in the chronic stroke population.
2. METHODS
We first performed a review of the literature to identify existing T1w MRI stroke lesion segmentation approaches. We then implemented the identified lesion segmentation approaches, and compared their performance to a ground‐truth expert segmentation using various image metrics. Finally, we statistically evaluated how each automated segmentation approach performed against one another. All statistical analysis files are publicly available on our GitHub repository to facilitate reproducibility (https://github.com/npnl/elsa).
2.1. Literature search
A computerized search covering the period from April 2007 to April 2017 was conducted on the PubMED online database using the following terms: ([“lesion identification” OR “lesion detection” OR “lesion classification”] OR “lesion” AND [“stroke”[MeSH Terms] OR “brain” [MeSH Terms]]) AND “automated.” Studies were limited to those in the English language.
The initial search yielded 189 results. We then excluded articles that were targeted at lesions not caused by stroke (e.g., multiple sclerosis; n = 144). Any articles that were not specifically on the topic of lesion segmentation methods were also excluded (n = 31). This resulted in 14 remaining results (Table 1, Supporting Information). Finally, to provide a fair evaluation for algorithms for chronic, T1w stroke MRI, we identified only articles on lesion segmentation approaches that were performed on chronic stroke lesions and have been shown to support segmentation on a single T1w (n = 6, Table 1). Of these six different lesion segmentation approaches, two were excluded as the software was either no longer available or not supported (S. Shen, personal communication; Shen, Szameitat, & Sterr, 2010; Guo et al., 2015).
Table 1.
Lesion segmentation approaches meeting literature selection criteria for chronic stroke lesions that support a single T1w image segmentation. Shen et al. and Guo et al. were currently unsupported or unavailable and thus excluded from the current evaluations
| Article | Method name | Availability/support for algorithm | Chronic versus acute | Modalities | Fully automated | Sample size | Healthy data required | Algorithm implemented |
|---|---|---|---|---|---|---|---|---|
| Seghier, Ramlackhansingh, Crinion, Leff, and Price (2008) | ALI toolbox | Available upon request | Chronic | T1 | Y | 10 simulated, 8 stroke T1w MRI | Yes | Fuzzy means clustering |
| Shen et al. (2010) | N/A | Algorithm unavailable | Chronic | T1 | Y | 36 simulated, 29 stroke T1w MRI | No | Fuzzy c‐means clustering |
| Guo et al. (2015) | Automated lesion detection toolbox | Algorithm unsupported | Chronic | T1 | Y | 60 | No | Support vector machine |
| de Haan, Clas, Juenger, Wilke, and Karnath (2015) | Clusterize | Publicly available | Acute (also tested on chronic) | CT, DWI, T2 FLAIR, T1 (any one) | N (semiautomated) | 44 CT/DWI/FLAIR, 11 T1w MRI | No | Iterative region growing (clustering) based on local intensity maxima |
| Griffis, Allendorfer, and Szaflarski (2016) | lesionGnb | Publicly available | Chronic | T1 | Y | 30 | No | GNB classification |
| Pustina et al. (2016) | LINDA | Publicly available | Chronic | T1 | Y | 60 | No | RF |
Abbreviations: ASSD, average symmetric surface distance; GNB, Gaussian naïve Bayes; HD, Hausdorff's distance; lesionGnb, Gaussian naïve Bayes lesion detection; LINDA, lesion identification with neighborhood data analysis; RF, random forest.
2.2. Software overview
Based on our literature search, we tested both semiautomated and fully automated approaches to lesion segmentation. In the following sections, a brief overview of each approach is provided.
2.2.1. Semiautomated software
We examined one semiautomated approach, Clusterize (de Haan et al., 2015; Philipp, Groeschel, & Wilke, 2012), as it was the only approach that met our literature search criteria. However, we acknowledge that other tools that may be considered “semiautomated,” such as MRIcron (http://people.cas.sc.edu/rorden/mricron/index.html) and ITKSnap (Yushkevich et al., 2006), are also publicly available and may be used in lesion segmentation, although they were not developed specifically for stroke lesion segmentation. Both MRICron and ITKSnap provide three‐dimensional (3D)‐growth algorithms to fill an initial area, and require a relatively large amount of manual input to guide the automated segmentation mask, thus making them more comparable to manual lesion segmentation methods. For these reasons, we did not include them in our evaluation.
Clusterize toolbox
The Clusterize approach is a semiautomated approach originally developed to identify demyelination load in metachromatic leukodystrophy using T2‐weighted MRIs (Philipp et al., 2012). However, the Clusterize algorithm has been shown to perform comparably on both acute and chronic stroke T1w MRI datasets (de Haan et al., 2015).
The Clusterize approach has an automated preprocessing step followed by a manual cluster selection step. The automated preprocessing step involves identification of the local intensity maxima on each image slice and assignment of each voxel to a single cluster core based on its intensity. This is followed by a manual cluster selection step and an optional freehand correction step to optimize the accuracy of the lesion mask.
2.2.2. Fully automated software
Three fully automated approaches resulted from our literature search and were currently available: the automated lesion identification (ALI) toolbox, a Gaussian naïve Bayes lesion detection method (lesionGnb), and lesion identification with neighborhood data analysis (LINDA; Seghier et al., 2008; Griffis et al., 2016; Pustina et al., 2016).
ALI toolbox
The ALI approach is an unsupervised method that performs outlier detection to segment lesions using a fuzzy c‐means algorithm (Seghier et al., 2008). The outlier detection procedure includes a voxel‐wise comparison between healthy and nonhealthy tissue, using a healthy dataset to define the healthy tissue.
Gaussian naïve Bayes lesion detection
The lesionGnb approach is a supervised method that performs Gaussian naïve Bayes (GNB) classification for the automated delineation of chronic stroke lesions (Griffis et al., 2016). The lesionGnb approach used 30 training cases to create feature maps encoding information about missing and abnormal tissue, obtained from gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) prior probability maps, and tissue probabilistic maps (TPMs). The GNB classifier was trained on ground‐truth manually delineated lesions as well as these feature maps using a leave‐one‐out cross‐validation approach. The trained GNB classifier is provided by the developers of the lesionGnb toolbox.
Lesion identification with neighborhood data analysis
The LINDA approach is a supervised method that relies on feature detection and uses a random forest (RF) algorithm to train and classify lesioned voxels (Pustina et al., 2016). In the LINDA method, features capturing aspects of geometry, subject specific anomalies, and deviation from controls for 60 stroke subjects were passed into a single matrix containing information about a single voxel and its neighboring voxels. The matrix was then used to train the RF algorithm using manually delineated lesions as the ground truth. RF training was repeated two more times with successively hierarchical image resolution. The trained RF classifier is provided by the developers of LINDA.
2.3. Data and implementation of algorithms
2.3.1. Computational platform and software installation
All computations were performed on a Mac OSX Yosemite operating system with a 3.2 GHz Intel Core i5 processor and 8 GB RAM. To run the ALI, lesionGnb, and Clusterize toolboxes, we used MATLAB version R2016b and SPM12. For the LINDA toolkit, we used R version 3.3.3, ANTsR version 0.3.1, ANTsRCore version 0.3.7.4, and ITKR version 0.4.12. See Table 2 for more information.
Table 2.
Processing features of fully automated toolboxes
| ALI | Voxel‐based GNB classification (lesion_gnb) | LINDA | |
|---|---|---|---|
| Compatible operating systems | Windows, Linux, Mac | Windows, Linux, Mac | Windows 10+, Linux, Mac |
| Platform dependencies | MATLAB, SPM5+ | MATLAB 2014b+ (requires statistics and machine learning toolbox), SPM12+ | R v.3.0+, ANTsR package |
| Year developed | 2007 | 2015 | 2016 |
| Open source | No | Yes | Yes |
| Learning type | Unsupervised | Supervised | Supervised |
| Training dataset | Requires user to provide segmented healthy training dataset | Provided (trained on 30 LHS subjects) | Provided (trained on 60 LHS subjects) |
| Amenable to left or right hemisphere lesions | Yes | Yes, provided that the user indicates which hemisphere first | No, right hemisphere lesions must be flipped |
| Template brain space | ICBM152 | ICBM152 | Colin 27 template |
| User‐defined parameters | Sensitivity (tuning factor), fuzziness index in fuzzy means clustering algorithm, threshold probability and size for the extra class prior | Optional smoothing, smoothing kernel, minimum cluster size, implicit masking while smoothing | None |
| Optional postprocessing steps | None | Resegmentation with a tissue prior | None |
Abbreviations: ALI, automated lesion identification; GNB, Gaussian naïve Bayes; lesionGnb, Gaussian naïve Bayes lesion detection; LHS, left hemisphere stroke; LINDA, lesion identification with neighborhood data analysis.
2.3.2. Data
Stroke data
We obtained our stroke dataset from the Anatomical Tracings of Lesions After Stroke (ATLAS) database (Liew et al., 2018). ATLAS is a public database consisting of 304 T1w anatomical MRIs of individuals with chronic stroke collected from research groups worldwide from the ENIGMA Stroke Recovery Working Group consortium. To account for potential confounding factors, we included only MRIs with 1 mm isotropic voxels, and all MRIs were collected from 3‐T scanners. We further excluded any MRIs that highly deviated from the normal range of the standard orientation. We also only included one MRI per individual (no inclusion of longitudinal data). One hundred and eighty‐one T1w anatomical MRIs (100 left hemisphere stroke (LHS), 81 right hemisphere stroke (RHS)) from a total of eight different scanners were included in the current analyses. Average lesion volume for all 181 lesions was 23,387 mm3 (median = 5,584 mm3); for right hemisphere lesions: mean = 31,842 mm3; median = 15,334 mm3; for left hemisphere lesions: mean = 16,539 mm3; median = 4,508 mm3 (see Figure S1, Supporting Information for lesion volume histogram). Further information on image acquisition for stroke data can be found in Liew et al., 2018.
Healthy data
The ALI method required a dataset of healthy controls to perform outlier detection. As the developers of the algorithm did not provide a healthy control dataset, we used images of healthy subjects sampled from the Functional Connectome Project (http://fcon_1000.projects.nitrc.org, n = 100). The developers of ALI did not recommend an optimal number of healthy controls, but rather specified that a larger set of healthy controls would better estimate the normal variability in brain structure (M. Seghier, personal communication). Although we could not match the exact acquisition parameters used in the ATLAS stroke sites, we did include only 3‐T MRIs from seven different research sites with data within the same older adult range of the ATLAS data we used.
2.3.3. Lesion segmentation
Expert segmentation
The ATLAS database included manually segmented lesion masks created by a team of trained individuals. We included only lesions that were designated as the primary stroke, as the original algorithms were developed and tested on a single lesion per brain (Griffis et al., 2016; Pustina et al., 2016; Seghier et al., 2008). Each lesion mask was carefully quality controlled. Briefly, each stroke lesion was segmented using either the coronal or axial view in MRIcron (http://people.cas.sc.edu/rorden/mricron/index.html) with a mouse, track pad, or a tablet (depending on preference) by 1 of 11 trained individuals. Individuals tracing lesions consisted of undergraduate students, graduate students, and postdoctoral fellows. Standardized training included utilization of a detailed protocol and instructional video, and guidance with extensive feedback on lesion tracings from an expert tracer and in consultation with a neuroradiologist. All lesions were checked for accuracy by a separate tracer. Lesion masks were also smoothed using a 2 mm FWHM kernel in order to smooth jagged edges between slices. All lesion locations were reviewed by a neuroradiologist. Interrater and intrarater reliability was computed for five stroke lesions (interrater DC: 0.75 ± 0.18; intrarater DC: 0.83 ± 0.13; Liew et al., 2018). For further information on the labeling and training protocol, see Liew et al., 2018.
Semiautomated segmentation
Clusterize
We followed the standard procedure (previously described in Section 2.2.1) and manually selected clusters as our lesion mask. We did not perform additional manual correction, as this time‐consuming process would have made the process analogous to a manual labeling procedure.
Automated segmentations
Automated lesion identification
The following automated steps were implemented as part of the ALI toolbox. All adjustable parameters were kept at their default values. First, segmentation and normalization of both healthy and stroke T1w MRI images were performed in SPM12. For stroke T1w MRIs, the ALI toolbox used a modified unified segmentation–normalization algorithm, which included use of an extra lesion tissue class prior (defined as the mean of the standard WM and CSF priors) to inform tissue probability maps for the segmentation of GM, WM, and CSF maps. GM and WM segmentations were then smoothed, and outlier detection comparing both GM and WM segmentations between patients and healthy controls was performed using fuzzy means clustering. Finally, the identified GM and WM outliers were combined into a final lesion mask.
Gaussian naïve Bayes lesion detection
For the lesionGnb approach, the following steps were implemented: we first specified whether the stroke was on the left or right hemisphere, as the program does not automatically detect the stroke hemisphere. Then, probabilistic tissue segmentation on the stroke T1w MRI was carried out using default parameters in the New Segment tool in SPM12. Tissue segmentations were smoothed with an 8 mm FWHM kernel based on default parameters, and feature maps containing information about missing and abnormal tissue were derived from GM/WM/CSF TPMs. The trained and cross‐validated GNB classifier provided by the developers was then used to predict lesion class labels. A minimum cluster size of 100 voxels, again based on default parameters, was specified as the threshold for retention of clusters in the final mask, and the final mask was smoothed using an 8 mm FWHM kernel.
Per personal communication, an additional resegmentation step using the final lesion was recommended for improving normalization performance and outline precision (J. Griffis, personal communication; Sanjuán et al., 2013). However, as we wanted to compare each algorithm based only on the publicly available information, we did not use the additional resegmentation step for this analysis.
Lesion identification with neighborhood data analysis
LINDA requires all strokes to be presented on the left hemisphere. Therefore, using the metadata obtained from ATLAS, as a first step, we automatically flipped the T1w MRIs for subjects with RHS so that the stroke appeared on the left hemisphere (n = 81). The following steps were implemented as a part of the LINDA package: Advanced Normalization Tools (ANTs; Avants et al., 2010) was used to perform two iterations of bias correction and brain extraction, as well as spatial normalization. Six features (deviation of k‐mean segmentation from controls, gradient magnitude, T1 deviation from controls, k‐mean segmentation, deviation of T1 asymmetry from controls, and raw T1 volume) were computed from the preprocessed T1w image. Within the LINDA package, these features were passed to the pretrained RF classifier provided by the developers, and the classifier was then run to detect the lesion using a multiresolution strategy to characterize lesion/perilesional characteristics at different spatial levels. This involves first downsampling the test image to 6 mm, and then using increasing resolutions at 4 and 2 mm, where the predicted lesion mask was then inversely transformed from the template to the subject's image space after each iteration to improve prediction accuracy. For right hemisphere lesions, we flipped the images back to the right hemisphere after lesion segmentation.
2.4. Postprocessing of automated lesion masks
All automated lesion mask outputs were converted back to native space for comparison to expert segmentations. For the ALI and lesionGnb approaches, which relied on SPM for preprocessing, lesion masks were inverse transformed using the transformation matrix resulting from SPM. The LINDA toolbox included two lesion masks, one output in native space and the other in stereotaxic space. Therefore, no further processing was necessary for the LINDA approach.
2.5. Segmentation evaluation
2.5.1. Visual evaluation
As a first step, we manually inspected the quality of the automated lesion mask outputs. To do so, we used an open‐source package (Pipeline for the Analysis of Lesions after Stroke; PALS; http://github.com/NPNL/PALS) to perform a visual evaluation of the automated outputs (Figure 1; Ito et al., 2018). The Visual QC module in PALS facilitates visual inspection of lesion segmentations by creating HTML pages with screenshots of lesion segmentations overlaid on each subject's T1w image, and allows for easy flagging of lesion masks that do not pass inspection.
Figure 1.
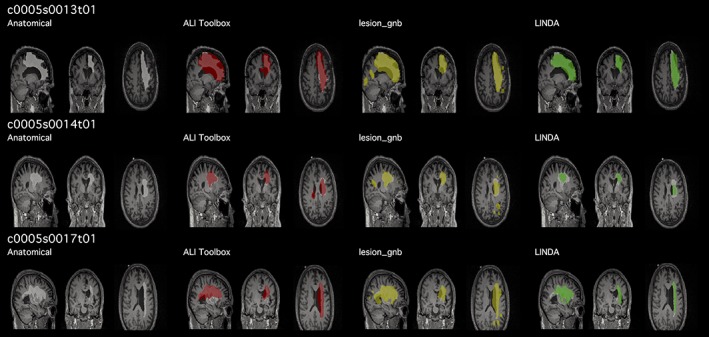
Example of quality control page. Prior to quantitatively evaluating each lesion segmentation performance, we visually assessed the lesion mask for each case. We created a script that automatically output a quality control page (https://github.com/npnl/PALS; Ito, Kumar, Zavaliangos‐Petropulu, Cramer, & Liew, 2018) with each automated lesion mask overlaid (red, yellow, green) on the expert segmentation (white). Subject IDs shown in this figure are kept in the same convention as in the Anatomical Tracings of Lesions After Stroke database [Color figure can be viewed at http://wileyonlinelibrary.com]
Given the nature of our multisite data, we anticipated that there might be cases in which the lesion segmentation algorithms would either (a) produce a lesion mask that has no overlap with the expert segmentation, that is, misclassify the lesion, or (b) identify no lesioned voxels, creating an empty mask file. For evaluating the performance across approaches, we decided to eliminate a case if any automated approach yielded an empty mask file, as this would not yield any comparable distance metric. Additionally, we eliminated cases in which all three automated algorithms produced lesion masks that had no overlap with the expert segmentation as these cases would have the same ranks in the nonparametric evaluation (Table 3; see Section 2.5.3).
Table 3.
Cases with no lesion mask identified, or lesions misclassified. Cases with no lesion mask identified yielded an empty file (containing only 0 value), and cases in which the lesion was misclassified contained lesioned voxels, but had no voxels overlapping with the expert segmentation. For comparisons between algorithms, we removed cases with no lesions identified (24 ALI + 23 LINDA – 8 overlapping = 39), and removed the 10 cases in which all three algorithms misclassified the lesion
| No lesion identified | Lesion misclassified | Total cases | |
|---|---|---|---|
| ALI | 24 | 28 | 52 |
| lesion_gnb | 0 | 39 | 39 |
| LINDA | 23 | 45 | 68 |
| Overlapping cases | 8 (ALI + LINDA only) | 10 across all three approaches | 32 across three approaches |
Abbreviations: ALI, automated lesion identification; lesionGnb, Gaussian naïve Bayes lesion detection; LHS, left hemisphere stroke; LINDA, lesion identification with neighborhood data analysis.
2.5.2. Quantitative evaluation
Evaluation metrics
To evaluate the performance of each automated lesion segmentation approach compared to the expert segmentation, we implemented the following evaluation metrics, which were used to evaluate lesion segmentations in the ISLES challenge: DC, HD and ASSD (Maier et al., 2017). To assess oversegmentaion and undersegmentation, we also obtained values on precision (also known as positive predictive value) and recall (sensitivity). We additionally calculated the lesion volume to assess whether the automated lesion segmentation approaches detected lesions of similar size to the expert segmentation. Finally, algorithmic efficiency was evaluated by obtaining the computational time for each segmentation approach. Evaluation metrics are described in detail below.
Dice similarity coefficient
DC is a measure of segmentation accuracy. DC is calculated with the following equation:
DC ranges from 0 (no overlap) to 1 (complete overlap), and X and Y represent the voxels in the expert segmentation, and those in the automated segmentation respectively.
Hausdorff's distance
HD is a measure of the maximum distance between all surface points of two image volumes. It is defined as:
where x and y are points of lesion segmentations X and Y, respectively, and d(x,y) is a 3D matrix consisting of all Euclidean distances between these points. HD is measured in millimeters and a smaller value indicates higher accuracy.
Average symmetric surface distance
ASSD is a measure of the average of all Euclidean distances between two image volumes. Given the average surface distance (ASD),
where d(x,y) is a 3D matrix consisting of the Euclidean distances between the two image volumes X and Y, ASSD is given as:
Similar to HD, the ASSD is measured in millimeters, and a smaller value indicates higher accuracy.
Precision and recall
Precision, also called positive predictive value, is the fraction of true positives (i.e., overlapping points between the two images) within the automated segmentation. It is defined as:
where precision ranges from 0 to 1 (1 indicating optimal precision), and TP are the true positives and FP denotes false positives in the automated segmentation.
Recall, also called sensitivity, is the fraction of true positives (overlapping points between the two images) within the expert segmentation. It is calculated with the following equation:
where recall ranges from 0 to 1 (1 indicating optimal recall), and TP denotes true positives and FN (false negative) denotes points that the automated segmentation failed to identify.
2.5.3. Statistical analyses
All statistical analyses were carried out in R version 3.3.3. To prevent any undue influence of extremely easy or extremely difficult cases, we performed nonparametric analyses to use the ranks of each automated approach to determine whether one approach outperformed another. For fair comparison, our statistical analyses were performed only on the fully automated approaches and did not include the lesion masks created in the Clusterize toolbox, which were driven by a degree of manual input. We report segmentation evaluation metrics for Clusterize in Table 4.
Table 4.
Descriptive statistics for each approach. Performance rates for each approach; median (IQR): DC, HD, ASSD, precision, and recall
| Clusterize | ALI | lesion_gnb | LINDA | |
|---|---|---|---|---|
| Image metrics | N = 152 | N = 132 | N = 132 | N = 132 |
| DC | 0.18 (0.31) | 0.4 (0.44) | 0.42 (0.37) | 0.5 (0.61) |
| HD (mm) | 80.89 (36.6) | 62.79 (48.49) | 58.19 (25.22) | 36.34 (42.48) |
| ASSD (mm) | 12.64 (7.68) | 9.58 (13.17) | 8.75 (7.89) | 4.97 (13.98) |
| Precision | 0.11 (0.22) | 0.31 (0.45) | 0.29 (0.33) | 0.6 (0.63) |
| Recall | 0.89 (0.26) | 0.61 (0.51) | 0.8 (0.44) | 0.59 (0.63) |
| Average processing time | 106.43 s for automated clustering + 251.75 s for manual identification | 396.99 + 247.83 s per healthy brain | 246.12 s | 3,843.66 s |
Abbreviations: ALI, automated lesion identification; ASSD, average symmetric surface distance; DC, dice coefficient; HD, Hausdorff's distance; lesionGnb, Gaussian naïve Bayes lesion detection; LINDA, lesion identification with neighborhood data analysis.
A Friedman test, the nonparametric equivalent of a one‐way repeated measures analysis of variance (ANOVA), was carried out to examine whether there was a significant difference in the performance among the fully automated segmentation approaches for each evaluation metric (DC, ASSD, HD, precision, and recall). Post hoc analyses with Wilcoxon signed‐rank tests were carried out using a Bonferroni correction for multiple comparisons. All Type I error rates were set at α < 0.05.
To further evaluate the utility of the automated segmentation approaches on lesion volume, we calculated a Pearson product–moment correlation coefficient for each automated approach to determine the relationship between the lesion volume of the expert segmentation and the lesion volume of the automated segmentation.
2.5.4. Analyses of segmentation performance by lesion characteristics
We assessed whether performance of any of the automated lesion segmentation approaches was associated with any particular lesion characteristics, such as stroke territory (cortical, subcortical, brainstem, cerebellar) and lesion size.
Stroke territory
For each automated approach, we compared DC for each stroke territory (cortical, subcortical, brainstem, cerebellar) using a Kruskal–Wallis rank sum test (a nonparametric equivalent to a one‐way ANOVA) followed by post hoc comparisons with a Bonferroni adjustment to assess whether there were differences in accuracy among the different stroke territories.
Lesion size
We created a lesion size variable by transforming lesion volume based on expert segmentations into three categories using the 33rd and 67th percentiles in the dataset of all lesion volumes as cutoff ranges for small, medium, and large lesions.
Here, again, we compared DC for each category of lesion size using a Kruskal–Wallis rank sum test, followed by post hoc comparisons with a Bonferroni adjustment.
2.5.5. Misclassified cases
Finally, for cases in which lesions were misclassified (i.e., an automated lesion mask was created but the DC yielded 0), we quantified the minimum distance (d min) between the edge of the expert segmentation with the edge of the automated segmentation with the following equation:
where x and y are points of lesion segmentations X and Y, respectively, and d(x,y) is a 3D matrix consisting of all Euclidean distances between these points. Minimum distance is measured in millimeters and a smaller value indicates higher accuracy. We examined this minimum distance measure to better understand, when lesion masks missed the lesion completely, whether they were close or far off from the actual lesioned territory.
3. RESULTS
3.1. Exploratory data analysis
3.1.1. Computational time
We first examined how long it took for each algorithm to run. For this evaluation, we used a subset of n = 100 left hemisphere MRIs (from our total dataset of n = 181). This was because additional steps, albeit automated, were required for processing right hemisphere lesions in the LINDA toolbox, which would have made it difficult to compare total time across toolboxes. The average times to preprocess an image and detect a lesion for 100 LHS MRIs, in order from fastest to slowest, were as follows: Clusterize (106.43 s, but with an additional 251.75 s to manually identify each cluster), lesionGnb (246.12 s), ALI (396.99 s, but with an additional 247.83 s per each healthy brain), and LINDA (3,843.66 s). Notably, for Clusterize, the manual identification time will vary by user and by lesion. For ALI, which requires healthy brains for comparison, we used 100 healthy brains to match the number of stroke MRIs (see Section 2.3.3).
3.1.2. Visual evaluation
We performed a visual evaluation to assess the quality of the automated lesion masks and ascertain that the lesion masks were correctly transformed back to native space.
For the Clusterize toolbox, there were 152 cases which resulted in a lesion mask, and 29 cases (16.02%) in which no cluster was detected as the lesion mask during manual identification.
All fully automated approaches ran completely and produced a lesion mask file for each case. However, we identified a number of cases where either there were no lesioned voxels that resulted from the automated segmentation, creating an empty file (which we refer to as an empty mask; Table 3), or there was a complete mismatch between the automated and expert segmentation (i.e., all voxels in the automated mask were misclassified as the lesion, which we refer to as a misclassification). This had been anticipated as the algorithms were based on supervised learning with a limited number of their own training data for which lesions in some brain regions (e.g., cerebellum) were not included.
Of the 181 cases, ALI successfully generated 129 lesion masks (71%) with at least a single voxel overlapping with the manual label. lesionGnb also detected 142 cases (78%) with at least a single voxel overlap with the manual label, and LINDA detected 113 cases (62%).
In addition, ALI had 28 cases in which the automated segmentation was misclassified (e.g., a lesion was detected, but it did not have any overlapping voxels with the lesion identified by manual segmentation), lesionGnb had 39 misclassified cases, and LINDA had 45 cases (Table 3). Ten of these were the same cases across all three approaches, and we removed these 10 misclassified cases from statistical evaluation of the approaches, as there would be no differences in rank between these cases.
Finally, there were 24 cases in which ALI produced an empty mask; 23 in which LINDA produced an empty mask (eight of these were the same cases as ALI); and zero in lesionGnb (in other words, lesionGnb always created a mask in which it identified what it considered to be lesioned voxels). We subsequently excluded these cases (n = 23 + 24–8 = 39) from the analysis of the evaluation metrics, as they would not yield any measurable metrics. Hence, after exclusion of 39 cases that had empty masks and 10 misclassified cases, 132 total cases remained in our quantitative evaluation.
For a discussion on the implications and possible reasons for misclassification and failed lesion detection, see Section 4.4).
3.2. Quantitative evaluation
The performance of each fully automated toolbox was evaluated across the following metrics: DC, HD, ASSD, precision, and recall (Figures 2 and 3). A summary figure can be found in Figure 4.
Figure 2.
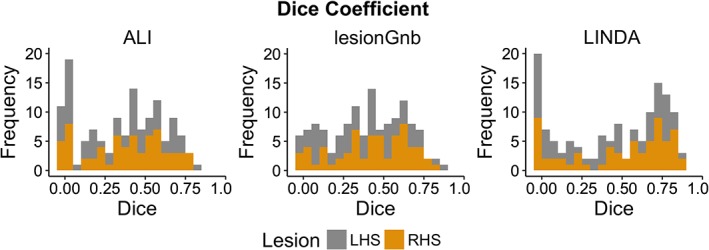
Distribution of dice similarity coefficient values for automated approaches. Histograms of all dice similarity coefficient values (N = 132) for each automated lesion detection approach; left hemisphere stroke (LHS) in gray; right hemisphere stroke (RHS) in gold [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 3.
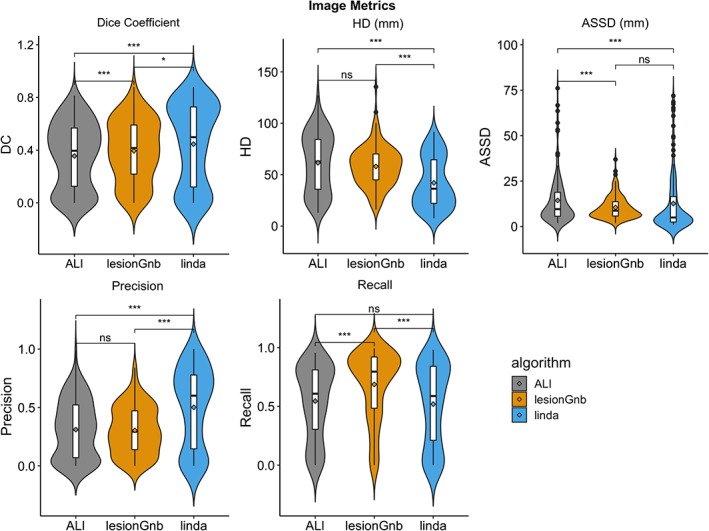
Violin plots of evaluation metrics for automated approaches. For dice coefficient, precision, and recall, range is from 0 to 1, where 0 = worst and 1 = best; Hausdorff's distance and average symmetric surface distance are measured in millimeters, and smaller values indicate better performance. *p < .05, **p < .01, ***p < .001, ns, not significant; Bonferroni corrected [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 4.
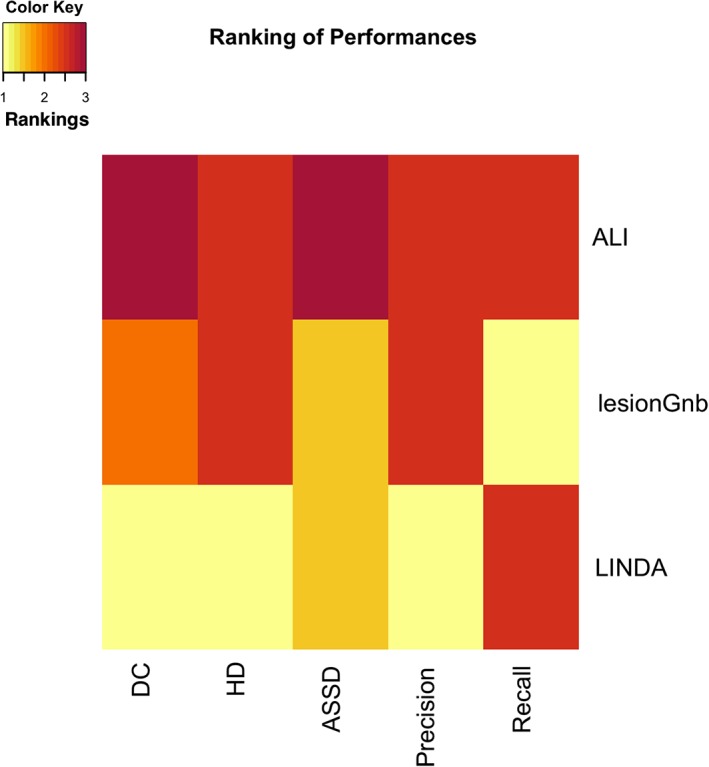
Heat map summary of the performances of algorithms. Each of the automated lesion algorithms were assigned a value (1–3) based on their median dice coefficient, Hausdorff's distance, average symmetric surface distance, precision, and recall scores. If pairwise comparisons (Section 3.2) showed they were not significantly different, the algorithms were marked as tied [Color figure can be viewed at http://wileyonlinelibrary.com]
3.2.1. Dice similarity coefficient
Using a Friedman test, we found a statistically significant difference in DC among the three fully automated lesion segmentation approaches, χ 2(2) = 27.10, p < .0001; corrected using the Bonferroni adjustment which was applied to all the following tests. Median (IQR) DC values for ALI, lesionGnb, and LINDA approaches were 0.40 (0.00–0.81), 0.42 (0.00–0.88), and 0.50 (0.00–0.88), respectively. Post hoc analyses using Wilcoxon signed‐rank tests on DC showed that LINDA outperformed lesionGnb and ALI (sum of positive ranks, lesionGnb: V = 5,359, p = .01; ALI: V = 1944, p < .0001), and lesionGnb outperformed ALI (V = 2,601, p < .0001).
3.2.2. Hausdorff's distance
A statistically significant difference in ranks for HD was found among the three fully automated segmentation approaches, χ 2(2) = 43.09, p < .0001. Median (IQR) HD values for ALI, lesionGnb, and LINDA are as follows: 62.79 mm (12.81–127.10), 58.19 mm (16.06–135.50), and 36.34 mm (7.55–91.75), where smaller values indicate better performance. Wilcoxon signed‐rank tests showed that LINDA performed better than ALI and lesionGnb (ALI: V = 7,156, p < .0001; lesionGnb: V = 1,915, p < .0001). There were no significant differences between ALI and lesionGnb (V = 5,085, p = .34).
3.2.3. Average symmetric surface distance
We also found a statistically significant difference in ASSD among the three fully automated segmentation approaches, χ 2(2) = 42.97, p < .0001. Median (IQR) ASSD values for ALI, lesionGnb, and LINDA approaches were 9.58 mm (1.94–76.11), 8.75 mm (1.88–36.94), and 4.97 (1.11–71.95), respectively. Again, smaller values indicate better performance. Pairwise comparisons showed that LINDA and lesionGnb both performed better than ALI (LINDA: V = 6,832, p < .0001; lesionGnb: V = 5,990, p = .0008), but there were no significance differences between LINDA and lesionGnb (V = 3,766, p = .47).
3.2.4. Precision and recall
We found a statistically significant difference in median precision among the three fully automated approaches, χ 2(2) = 41.59, p < .0001. Median (IQR) precision values for ALI, lesionGnb, and LINDA approaches were 0.31 (0.00–0.99), 0.29 (0.00–0.84), and 0.60 (0.00–1.00), respectively. Here, higher values indicate better performance. Wilcoxon signed‐rank tests showed that LINDA had higher precision rates than both ALI (V = 1,423, p < .0001) and lesionGnb (V = 6,961, p < .0001), and there were no significant differences between ALI and lesionGnb (V = 3,998, p = 1.00).
We also found a statistically significant difference in recall among the three fully automated lesion segmentation approaches, χ 2(2) = 97.86, p < .0001. Median (IQR) recall values for ALI, lesionGnb, and LINDA approaches were 0.61 (0.00–0.96), 0.80 (0.00–0.99), and 0.59 (0.00–0.98), respectively, again with higher values indicating better performance. We found that lesionGnb performed better than both LINDA (V = 1,084, p < .0001) and ALI (V = 1,171, p < .0001), and there were no significant differences between LINDA and ALI (V = 4,479, p = .55).
3.3. Volume correlation
We found a significant positive correlation between lesion volumes of automated segmentations and manual lesion segmentations for each approach: ALI (r = .69, p < .0001), lesionGnb (r = .61, p < .0001), and LINDA (r = .59, p < .0001). Root mean square error (RMSE) is as follows: ALI (58,275.71 mm3); lesionGnb (73,931.39 mm3); and LINDA (47,421.61 mm3). However, we detected two outliers, defined as having a Cook's distance >1 for LINDA and one outlier for ALI and lesionGnb each. After outlier removal, we found a statistically significant positive correlation between the lesion volumes of the expert segmentations and the lesion volumes of each of the automated segmentations: ALI (r = .75, p < .0001), lesionGnb (r = .90, p < .0001), and LINDA (r = .84, p < .0001). RMSE is as follows: ALI (57,427.09 mm3); lesionGnb (42,619.39 mm3); and LINDA (25, 092.11 mm3).
3.4. Analyses of segmentation performance by lesion characteristics
3.4.1. Analyses of cases by stroke territory
We found significant differences in DC between different stroke territories for each automated approach (ALI: χ 2 = 76.78, p < .0001; lesionGnb: χ 2 = 97.22, p < .0001; LINDA: χ 2 = 77.61, p < .0001; Figure 5). Pairwise comparisons for each of the approaches followed a similar pattern, where cortical lesions had significantly higher DC than all other stroke territories (p < .0001), subcortical lesions had higher DC than brainstem and cerebellar lesions (p < .05), and there were significant no differences in DC between brainstem and cerebellar lesions.
Figure 5.
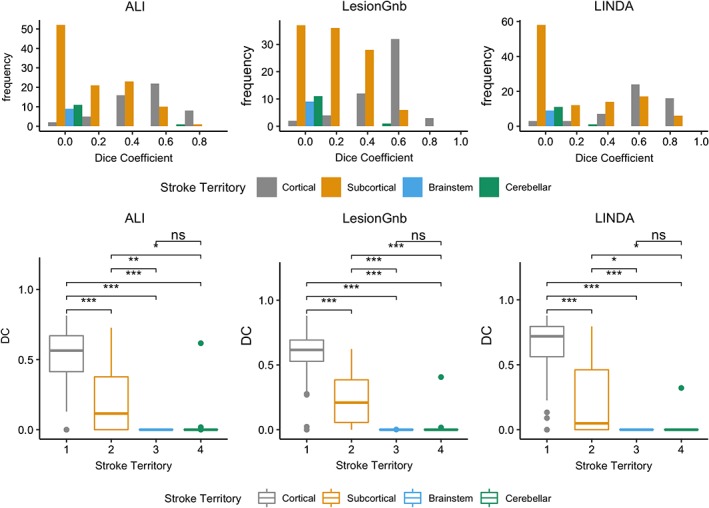
Analysis of the dice coefficient by stroke territory. (above) histograms of dice coefficient values for each stroke territory. (below) box plots showing pairwise comparisons of DC values for each stroke territory. *p < .05, **p < .01, ***p < .001, ns, not significant; Bonferroni corrected [Color figure can be viewed at http://wileyonlinelibrary.com]
3.4.2. Analyses of cases by lesion size
Lesion volume cutoffs for lesion size category as determined by the 33rd and 67th percentiles of all manual lesion volumes were as follows: small: 12–2,510 mm3; medium: 2,794–21,352 mm3; and large: 21,623–164,300 mm3.
We found significant differences in DC between different lesion volumes for each automated approach (ALI: χ 2 = 116.29, p < .0001; lesionGnb: χ 2 = 126.88, p < .0001; LINDA: χ 2 = 121.91, p < .0001; Figure 6). Pairwise comparisons for each of the approaches showed that large lesions had significantly higher DC than both medium and small lesions (p < .0001), and medium‐sized lesions also had higher DC than small lesions (p < .0001).
Figure 6.
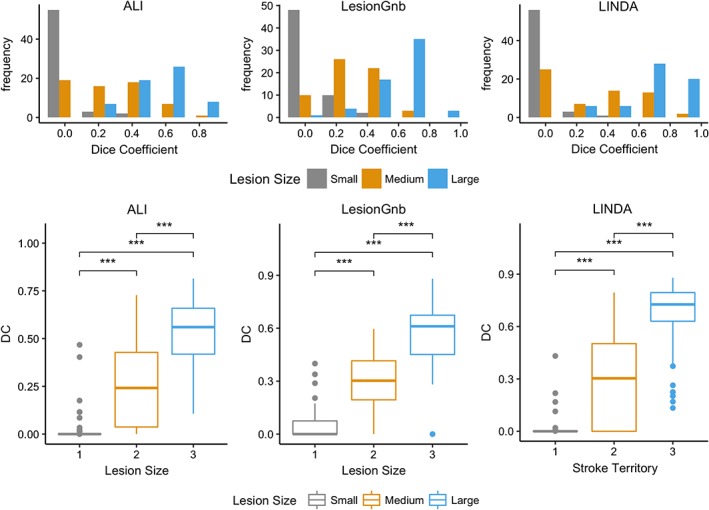
Analysis of the dice coefficient (DC) by lesion size. (Above) Histograms of DC values for each lesion size category. (Below) Box plots showing pairwise comparisons of DC values for each lesion size category. *p < .05, **p < .01, ***p < .001, ns, not significant; Bonferroni corrected [Color figure can be viewed at http://wileyonlinelibrary.com]
3.5. Misclassified cases
To quantify how far off misclassified lesions were from the expert mask, we calculated the minimum distance between the edge of the automated mask and the edge of the expert mask for cases in which there were no overlapping voxels. Average d min is as follows: ALI: 36.10 ± 21.72 mm (range: 2.24–93.25 mm); lesionGnb: 19.31 ± 13.73 mm (range: 1.41–42.91 mm); and LINDA: 29.67 ± 19.82 mm (range: 1.00–83.36 mm). A density plot of minimum distances to the manual segmentation is shown in Figure 7. As the plot shows, masks from lesionGnb were closest to the lesion, followed by ALI and then LINDA. However, due to the low precision of lesionGnb (i.e., high false positive rate), it is likely that lesionGnb creates multiple false positive labels, some of which may have been in closer proximity to the true lesion.
Figure 7.
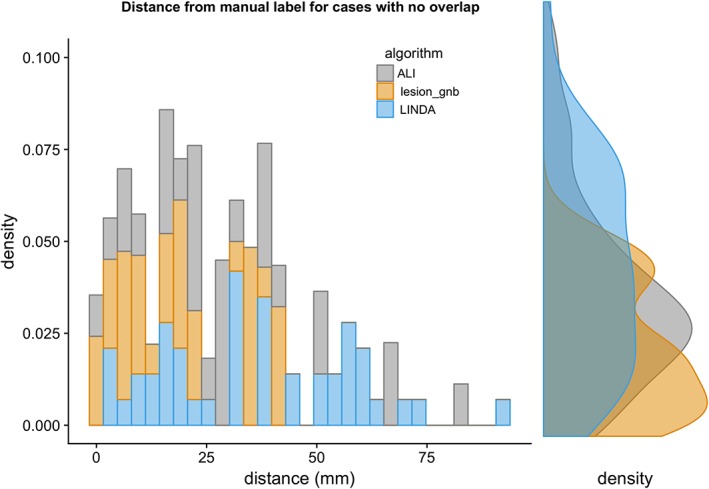
Density plot of minimum distances to manual segmentations for lesions with no overlap [Color figure can be viewed at http://wileyonlinelibrary.com]
4. DISCUSSION
In the present paper, we systematically evaluated the performance of existing stroke lesion segmentation approaches for chronic T1w MRI on a large common dataset. Overall, we found that LINDA performed the best out of the fully automated lesion segmentation methods. In addition, all methods performed the worst on small lesions, as well as lesions in the brainstem and cerebellum. These findings provide implications for how to improve existing lesion segmentation algorithms for T1w MRIs.
4.1. Fully automated software
Our findings showed that each of the fully automated approaches resulted in different patterns in the various evaluation metrics used in the current study, indicating that each approach had its own benefits and drawbacks. Specifically, we found that lesionGnb yielded the least number of cases (in fact, 0) in which no lesion mask was detected, compared to both LINDA and ALI. However, LINDA consistently performed the best out of the evaluation metrics (DC, HD, ASSD).
A closer examination of precision and recall provides insight to the results obtained from the DC and distance metrics: LINDA resulted in higher precision (positive predictive value) rates than both ALI and lesionGnb, while recall values were highest in lesionGnb and similar in ALI and LINDA. Moreover, LINDA had roughly equivalent median precision and recall values (0.60 and 0.59, respectively), whereas both ALI and lesionGnb had relatively better recall compared to precision (ALI precision: 0.31, recall: 0.61; lesionGnb precision: 0.29, recall: 0.80). This suggests that both the lesionGnb and ALI approaches tended to oversegment lesions (high false positives). These findings were confirmed by our visual evaluation.
Our findings suggest that LINDA was the approach that consistently performed best across all metrics—but only when it successfully identified a lesion. However, LINDA was also the most computationally expensive approach: the average time to process a single image on LINDA took roughly 16 times as long as lesionGnb, and six times as long as ALI. Additionally, in cases in which automated segmentations were misclassified, the misclassified lesion was in closer proximity to the expert segmentation for lesionGnb as compared to LINDA.
The three fully automated segmentation approaches implemented distinct machine learning algorithms in their approach to lesion segmentation. ALI used an unsupervised approach with fuzzy means clustering to detect outliers in gray and WM segmentations, lesionGnb used a supervised naïve Bayesian classification algorithm to estimate the probability of a lesion class, and LINDA used a supervised RF approach with a multi‐resolution framework to classify voxels and their neighbors as lesional tissue. We expected that the supervised learning algorithms would have higher performance than an unsupervised approach, given that supervised approaches are trained with ground‐truth lesions. Indeed, we found that both LINDA and lesionGnb had higher values than ALI on the DC. However, this was not consistently the case for the distance metrics (ASSD and HD).
Notably, both lesionGnb and ALI have adjustable parameters. As mentioned above, in order to systematically evaluate performance without bias from expert feedback, we implemented the approaches with their default settings. This may have caused a drop in accuracy in these two approaches. Related, additional preprocessing steps, such as excluding voxels that fall within the CSF mask, may have improved results.
The three approaches also implemented different image processing tools (e.g., ANTs, SPM) that include various preprocessing steps, such as brain extraction, registration, and tissue classification. It is likely that performance variability in these preprocessing steps may have had downstream effects on lesion segmentation accuracy. Manual quality control and inspection of preprocessing steps could enhance the lesion segmentation process, and would ideally be a part of any neuroimaging analysis pipeline.
4.2. Comparison to other evaluations
Previously reported results from the developers of each automated algorithm provide a useful tool for comparison and evaluation of the results we obtained from the current study. Our DC values were approximately 0.20–0.24 lower than those reported by the developers in their original papers (ALI: original = 0.64, ATLAS = 0.40; lesionGnb: 0.66, 0.42; LINDA: 0.70, 0.50; Griffis et al., 2016; Pustina et al., 2016; Seghier et al., 2008). There are several likely explanations for this.
First, each of these automated algorithms was originally tested on single site, single scanner‐acquired data. This makes these algorithms vulnerable to over‐fitting to their own data. In particular, the supervised methods (LINDA, lesionGnb) were dependent on machine learning classifiers that were pretrained using data acquired from a single scanner from the original study (see, Supporting Information for more information on training data and on analysis of site effects). Here, we implemented a large data evaluation and tested each pretrained algorithm on multisite data. Not surprisingly, we found a significant drop in segmentation accuracy, as variability in machine characteristics was likely not addressed using the initial training set. Whereas the training data from the developers of the algorithms included only unilateral lesions, the dataset we obtained from ATLAS included bilateral lesions. However, the inclusion of MRIs with bilateral lesions is unlikely to have contributed to the drop in DC values in the current data as compared to those reported in the original papers, as we did not find significant differences in DC between bilateral and unilateral lesions (see, Supporting Information).
Second, to provide an equal comparison across toolboxes, we implemented the fully automated approaches as they were without modifications to the parameters selected using the original training dataset. As previously stated, this could have resulted in a decrease in accuracy for detecting lesions in this dataset. For example, keeping the standard thresholding parameters suggested by the developers of lesion_gnb at 100 voxels and 8 mm FWHM may have created a bias against small lesions. We also kept built‐in preprocessing steps prior to lesion detection. While performance of the approaches may have been improved by fine‐tuning the default parameters to our dataset, the current results obtained without modifications provide valuable baseline information to researchers and clinicians who may be interested in using any of the tested algorithms and wish to bypass the time‐intensive and computationally intensive training procedure.
4.3. Small brainstem and cerebellar lesions perform worst
We also assessed whether automated lesion segmentation performance was related to specific lesion characteristics. Overall, we found that the fully automated approaches were less likely to detect small lesions, with half of all total small lesion cases (30/60) failing to be detected by all three fully automated approaches. This is consistent with the literature, which has shown that automated and semiautomated approaches for T1w lesion segmentation to be biased for detection of large lesions (Wilke et al., 2011; Griffis et al., 2016). Users of these algorithms should thus manually inspect lesion segmentation quality, and pay specific attention to small lesions. However, as these are typically the fastest to manually segment, they should also be the fastest to correct. Using an automated segmentation algorithm may therefore still save considerable time, even with manual inspection and corrections for smaller lesions.
Regarding lesion location, fully automated approaches displayed significantly higher segmentation accuracy on cortical lesions than subcortical, brainstem, and cerebellar lesions, and subcortical lesions generally displayed higher segmentation accuracy than brainstem and cerebellar lesions. As brainstem and cerebellar strokes occur less frequently, brainstem and cerebellar lesions were likely not included in the original training set for the automated algorithms (Chua & Kong, 1996; Datar & Rabinstein, 2014; Kase, Norrving, Levine, & Babikian, 1993; Teasell, Foley, Doherty, & Finestone, 2002). Moreover, features implemented in the algorithms to classify lesions may not be sensitive to brainstem or cerebellar strokes. Finally, subcortical, brainstem, and cerebellar lesions are often smaller than cortical lesions, suggesting a potential additive effect on accuracy. Users with datasets containing multiple brainstem or cerebellar strokes may need to retrain the algorithm with a training dataset that contains more of these types of lesions to increase algorithm sensitivity.
4.4. Dropped cases
In our evaluation, we found that there were 39 cases in which lesions were not detected for the ALI and LINDA algorithms (ALI: 24, LINDA: 23, with eight same cases between approaches). Importantly, we note that dropping these cases may have inflated the results in favor of LINDA and ALI.
One potential explanation for the failed detection is that the thresholds that were used to classify lesioned versus nonlesioned voxels was too high. We had opted to use default values in implementing the algorithms, since we wanted to be systematic in our evaluation, and because ALI had user‐adjustable parameters but LINDA did not. However, this meant that the thresholds that were used might not have been optimally tuned for this dataset. Although here the focus was to fairly evaluate the various algorithms using a common set of parameters, an actual user who is trying to generate lesion masks for his or her data could try to better optimize a specific method for a specific dataset.
Additionally, we found that there were a number of cases in which lesions were misclassified (ALI: 28, lesionGnb: 39, LINDA: 45, with 10 same cases between all three, which are shown in, Supporting Information). These are cases where the automated algorithm generated a lesion mask that did not overlap at all with the manual segmentations. Of the misclassified cases, lesionGnb produced lesion masks that were closest to the manual segmentation. However, we note that this may be due to the increased false positive rate of the lesionGnb algorithm. Notably, across all of the segmentation algorithms, most of these cases were small and brainstem/cerebellar lesions, which may not be reflective of the cases that were used in the training datasets of the algorithms. Finally, the number of cases with suboptimal segmentations may also have been inflated in due to our use of secondary, combined multisite data.
4.5. Semiautomated software
We tested one semiautomated software, the Clusterize toolbox, for lesion segmentation. The Clusterize toolbox was designed for use with manual input and corrections. We only performed the initial manual step of cluster selection (identifying the lesioned region), but did not perform the subsequent manual correction, as this would have made the segmentation analogous to an expert segmentation. However, because a manual correction was expected as a part of the procedure for the Clusterize toolbox, we expected the Clusterize toolbox to have a less favorable performance compared to the other methods that did not require additional manual input (i.e., the automated approaches). We therefore did not evaluate performance on Clusterize against the automated segmentations (see Table 4).
The automated preprocessing plus manual cluster selection resulted in a relatively low DC value (M = 0.18, IQR: 0.06, 0.37), but a fairly high recall value (sensitivity; M = 0.89, IQR: 0.71, 0.96). The high recall was likely driven by the manual selection of clusters: due to expert feedback, a cluster corresponding to the true lesion was accurately selected for most cases. However, Clusterize tended to overestimate the lesioned region, which led to lower precision in the lesion segmentation. In particular, we found that the cluster corresponding to the true lesion often included the ventricle as part of the lesion when the lesion was adjacent to the ventricle. These lower precision values may also have been partly driven by the fact that Clusterize was originally designed for the detection of a different type of the brain lesion (i.e., metachromatic leukodystrophy) on T2‐weighted images, suggesting a need for additional feature modeling or parameter optimization. The creation of a mask for the ventricles and exclusion of any voxels within that mask could also enhance this method.
4.6. Limitations
Here, we strived to evaluate existing T1w lesion segmentation algorithms on a large, naturalistic dataset. However, we note a few limitations in this evaluation.
First, the evaluation of lesion segmentation algorithms—or any machine‐learning algorithm—is by nature, highly dependent on the dataset that is used for testing. Typically, the greater the similarity between lesions in the testing dataset (i.e., ATLAS) and lesions used by the original developers to train and develop the algorithms, the better the performance will be. It is possible that our findings could be related to dataset‐specific effects that may have biased one algorithm over another, if there was greater similarity between ATLAS and the specific training data. Interestingly, however, the average lesion volume of lesionGnb (30,314 mm3) was closer to the average lesion volume of the ATLAS dataset (23,387 mm3) than LINDA (68,000 mm3), yet LINDA had higher DC values than lesionGnb (lesion volumes for developing ALI was unavailable; see Table 2, Supporting Information).
Second, as previously noted, small lesions and brainstem and cerebellar lesions performed worst across the algorithms. This could also be due to a discrepancy in lesion size and representation of the type of stroke lesions in the training and testing datasets. However, we believe that this evaluation is useful precisely for this reason: the ideal lesion segmentation algorithm should be robust to new data, regardless of stroke size and territory.
Finally, ALI required a healthy dataset to define healthy tissue from nonhealthy tissue. While we attempted to match parameters of the healthy data from the Functional Connectome Project (e.g., scanner strength, age range of individuals, number of scanners), we were still limited in our ability match the acquisition parameters of the datasets due to our use of retrospective data. This may have biased performance against the ALI method.
5. CONCLUSION
Our systematic evaluation facilitates and informs future use and development of automated approaches. Notably, we found that the supervised algorithms performed best, but there was a high failure rate across all approaches. We found systematic differences in segmentation accuracy depending on stroke territory and size. Based on these findings, we recommend two primary areas for improvement in the future development of automated lesion detection algorithms: (a) that algorithms be trained on larger and more diverse datasets, allowing for interscanner variability from multisite, multiscanner data, and (b) that prior knowledge about lesion size and territory be integrated into algorithms to increase segmentation performance. For clinicians and researchers who wish to use currently available lesion detection approaches, we suggest selection of an automated lesion detection approach most suitable for their purposes and performance of a thorough visual inspection of the automated segmentations to ensure the accuracy of each mask. We strongly recommend manual quality control following any of these approaches. By facilitating and informing the use and development of automated segmentation approaches, we hope that this systematic review will advance the discovery of clinically meaningful findings about stroke recovery.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
6.
Supporting information
Appendix S1 Supporting Information
ACKNOWLEDGMENTS
The authors would like to thank Allie Schmiesing for her contribution to the literature search and performing lesion segmentations on the Clusterize Toolbox and Jennifer Chan for performing segmentations on the Clusterize Toolbox and her comments on this article. The authors would also like to thank all ATLAS contributors and contributors to the Functional Connectome Project healthy dataset. This work was supported by the National Institutes of Health NCMRR (grant number 1K01HD091283 to S.‐L.L.). H.K. was supported by Baxter Foundation Fellowship Award.
Ito KL, Kim H, Liew S‐L. A comparison of automated lesion segmentation approaches for chronic stroke T1‐weighted MRI data. Hum Brain Mapp. 2019;40:4669–4685. 10.1002/hbm.24729
Funding information Baxter Foundation Fellowship; NIH NCMRR, Grant/Award Number: 1K01HD091283
DATA AVAILABILITY
The raw dataset used for this study is openly available in ICPSR at https://doi.org/10.3886/ICPSR36684.v3[doi].
REFERENCES
- Albers, G. W. (1998). Diffusion‐weighted MRI for evaluation of acute stroke. Neurology, 51(3 Suppl. 3), S47–S49. [DOI] [PubMed] [Google Scholar]
- Avants, B. B. , Yushkevich, P. , Pluta, J. , Minkoff, D. , Korczykowski, M. , Detre, J. , & Gee, J. C. (2010). The optimal template effect in hippocampus studies of diseased populations. NeuroImage, 49(3), 2457–2466. 10.1016/j.neuroimage.2009.09.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, E. , Wilson, S. M. , Saygin, A. P. , Dick, F. , Sereno, M. I. , Knight, R. T. , & Dronkers, N. F. (2003). Voxel‐based lesion–symptom mapping. Nature Neuroscience, 6(5), 448–450. 10.1038/nn1050 [DOI] [PubMed] [Google Scholar]
- Chalela, J. A. , Alsop, D. C. , Gonzalez‐Atavales, J. B. , Maldjian, J. A. , Kasner, S. E. , & Detre, J. A. (2000). Magnetic resonance perfusion imaging in acute ischemic stroke using continuous arterial spin labeling. Stroke, 31(3), 680–687. [DOI] [PubMed] [Google Scholar]
- Chua, K. S. G. , & Kong, K. H. (1996). Functional outcome in brain stem stroke patients after rehabilitation. Archives of Physical Medicine and Rehabilitation, 77(2), 194–197. 10.1016/S0003-9993(96)90167-7 [DOI] [PubMed] [Google Scholar]
- Datar, S. , & Rabinstein, A. A. (2014). Cerebellar infarction. Neurologic Clinics, 32(4), 979–991. https://doi.org/S0733-8619(14)00061-9 [pii]. [DOI] [PubMed] [Google Scholar]
- de Haan, B. , Clas, P. , Juenger, H. , Wilke, M. , & Karnath, H.‐O. (2015). Fast semi‐automated lesion demarcation in stroke. NeuroImage: Clinical, 9, 69–74. 10.1016/j.nicl.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiez, J. A. , Damasio, H. , & Grabowski, T. J. (2000). Lesion segmentation and manual warping to a reference brain: Intra‐ and interobserver reliability. Human Brain Mapping, 9(4), 192–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis, J. C. , Allendorfer, J. B. , & Szaflarski, J. P. (2016). Voxel‐based Gaussian naive Bayes classification of ischemic stroke lesions in individual T1‐weighted MRI scans. Journal of Neuroscience Methods, 257, 97–108. 10.1016/j.jneumeth.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K. L. , Kumar, A. , Zavaliangos‐Petropulu, A. , Cramer, S. C. , & Liew, S.‐L. (2018). Pipeline for analyzing lesions after stroke (PALS). Frontiers in Neuroinformatics, 12, 63. Retrieved from https://www.frontiersin.org/article/10.3389/fninf.2018.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kase, C. S. , Norrving, B. , Levine, S. R. , & Babikian, V. L. (1993). Cerebellar infarction: Clinical and anatomic observations in 66 cases. Stroke, 24, 76–83. 10.1161/01.STR.24.1.76 [DOI] [PubMed] [Google Scholar]
- Liew, S.‐L. , Anglin, J. M. , Banks, N. W. , Sondag, M. , Ito, K. L. , Kim, H. , … Stroud, A. (2018). A large, open source dataset of stroke anatomical brain images and manual lesion segmentations. Scientific Data, 5, 180011 10.1038/sdata.2018.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenberg, R. , Renga, V. , Zhu, L. L. , Betzler, F. , Alsop, D. , & Schlaug, G. (2010). Structural integrity of corticospinal motor fibers predicts motor impairment in chronic stroke. Neurology, 74(4), 280–287. 10.1212/WNL.0b013e3181ccc6d9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, O. , Menze, B. H. , Von Der Gablentz, J. , Häni, L. , Heinrich, M. P. , Liebrand, M. , … Reyes, M. (2017). ISLES 2015 — A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Medical Image Analysis, 35, 250–269. 10.1016/j.media.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, O. , Schröder, C. , Forkert, N. D. , Martinetz, T. , & Handels, H. (2015). Classifiers for ischemic stroke lesion segmentation: A comparison study. PLoS One, 10(12), 1–16. 10.1371/journal.pone.0145118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffarian, D. , Benjamin, E. J. , Go, A. S. , Arnett, D. K. , Blaha, M. J. , Cushman, M. , … Turner, M. B. (2016). Heart disease and stroke statistics‐2016 update a report from the American Heart Association. Circulation, 133, e38–e360. 10.1161/CIR.0000000000000350 [DOI] [PubMed] [Google Scholar]
- Philipp, C. , Groeschel, S. , & Wilke, M. (2012). A semi‐automatic algorithm for determining the demyelination load in metachromatic leukodystrophy. Academic Radiology, 19(1), 26–34. 10.1016/j.acra.2011.09.008 [DOI] [PubMed] [Google Scholar]
- Pustina, D. , Coslett, H. B. , Turkeltaub, P. E. , Tustison, N. , Schwartz, M. F. , & Avants, B. (2016). Automated segmentation of chronic stroke lesions using LINDA: Lesion identification with neighborhood data analysis. Human Brain Mapping, 37(4), 1405–1421. 10.1002/hbm.23110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, J. D. , Le, V. , Der‐Yeghiaian, L. , See, J. , Newton, J. M. , Ward, N. S. , & Cramer, S. C. (2011). Anatomy of stroke injury predicts gains from therapy. Stroke, 42(2), 421–426. 10.1161/STROKEAHA.110.599340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seghier, M. L. , Ramlackhansingh, A. , Crinion, J. , Leff, A. P. , & Price, C. J. (2008). Lesion identification using unified segmentation‐normalisation models and fuzzy clustering. NeuroImage, 41(4), 1253–1266. 10.1016/j.neuroimage.2008.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasell, R. , Foley, N. , Doherty, T. , & Finestone, H. (2002). Clinical characteristics of patients with brainstem strokes admitted to a rehabilitation unit. Archives of Physical Medicine and Rehabilitation, 83(7), 1013–1016. 10.1053/apmr.2002.33102 [DOI] [PubMed] [Google Scholar]
- Yushkevich, P. A. , Piven, J. , Hazlett, C. , Smith, G. , Ho, S. , Gee, J. C. , & Gerig, G. (2006). User‐guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage, 31, 1116–1128. 10.1016/j.neuroimage.2006.01.015 [DOI] [PubMed] [Google Scholar]
- Zhu, L. L. , Lindenberg, R. , Alexander, M. P. , & Schlaug, G. (2010). Lesion load of the corticospinal tract predicts motor impairment in chronic stroke. Stroke, 41(5), 910–915. 10.1161/STROKEAHA.109.577023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Supporting Information
Data Availability Statement
The raw dataset used for this study is openly available in ICPSR at https://doi.org/10.3886/ICPSR36684.v3[doi].


