Summary
Aeromonas hydrophila and Aeromonas caviae adapt to saline water environments and are the most predominant Aeromonas species isolated from estuaries. Here, we isolated antimicrobial‐resistant (AMR) Aeromonas strains (A. hydrophila GSH8‐2 and A. caviae GSH8M‐1) carrying the carabapenemase bla KPC‐2 gene from a wastewater treatment plant (WWTP) effluent in Tokyo Bay (Japan) and determined their complete genome sequences. GSH8‐2 and GSH8M‐1 were classified as newly assigned sequence types ST558 and ST13, suggesting no supportive evidence of clonal dissemination. The strains appear to have acquired bla KPC‐2‐positive IncP‐6‐relative plasmids (pGSH8‐2 and pGSH8M‐1‐2) that share a common backbone with plasmids in Aeromonas sp. ASNIH3 isolated from hospital wastewater in the United States, A. hydrophila WCHAH045096 isolated from sewage in China, other clinical isolates (Klebsiella, Enterobacter and Escherichia coli), and wastewater isolates (Citrobacter, Pseudomonas and other Aeromonas spp.). In addition to bla KPC‐2, pGSH8M‐1‐2 carries an IS26‐mediated composite transposon including a macrolide resistance gene, mph(A). Although Aeromonas species are opportunistic pathogens, they could serve as potential environmental reservoir bacteria for carbapenemase and AMR genes. AMR monitoring from WWTP effluents will contribute to the detection of ongoing AMR dissemination in the environment and might provide an early warning of potential dissemination in clinical settings and communities.
Introduction
Antimicrobial resistance (AMR) is a global issue, and carbapenem‐resistant Enterobacteriaceae (CRE) have the potential to facilitate the widespread transmission of AMR. This dissemination is mainly accomplished through mobile genetic elements and the processes of natural competence, transformation and plasmid transconjugation, which can occur in any environment. In addition to clinical settings, AMR genes (ARGs) and AMR bacteria are widely distributed in the environment, particularly in surface waters (Ng et al., 2017), waste water treatment plant (WWTP) effluents (Karkman et al., 2018), soils and animal wastes (Sharma and Reynnells, 2016). In particular, the widespread detection of carbapenemase‐producing organisms (CPOs) in the environment is an emerging environmental issue with potentially serious public health implications.
Klebsiella pneumoniae carbapenemase 2 (KPC‐2) hydrolyzes all class A β‐lactamases, including the most common β‐lactams such as carbapenem, which was initially identified from Salmonella enterica serovar Cubana (Miriagou et al., 2003). In particular, the widespread detection of CPO is an emerging environmental issue with potentially serious public health implications. Notably, Xu et al. (Xu et al., 2018) reported the isolation of KPC‐producing Citrobacter and Aeromonas isolates from sampling sites near a WWTP in China.
Nevertheless, KPC‐2‐type CPOs have rarely been detected, even in clinical settings, in Japan (Saito et al., 2014), implying that Japan is basically free of KPC‐2‐type CPO. However, the monitoring of environmental waters in 2017 in Japan revealed a KPC‐2‐producing K. pneumoniae strain, GSU10‐3, in an effluent from a WWTP in Tokyo Bay (Japan) (Sekizuka et al., 2018). This finding suggested that the Japanese environment, particularly the water in urban areas, may be contaminated with notable levels of such clinically relevant AMR bacteria (Sekizuka et al., 2018), motivating further assessments of the presence of environmental AMR in Japan.
In 2018, we investigated an effluent from a WWTP to identify CPO isolates and isolated Aeromonas strains and determined the complete genomes of two isolates carrying the bla KPC‐2 gene. These genome sequences could suggest the possible role of Aeromonas in the environmental dissemination of AMR. A substantial volume of both treated and untreated sewage is continually discharged into urban bays. Thus, our results may point to the role of coastal waters as reservoirs and vectors for AMR bacteria in cities, which can potentially accelerate the spread of ARGs to other non‐pathogenic reservoir bacteria. These findings thus raise alarms and highlight the importance for adopting measures to mitigate the spread of AMR in the environment, particularly under the ‘One‐Health’ perspective.
Results and discussion
Isolation of CPE from WWTP effluent
The upper effluent flow of an urban WWTP was collected on 6 August 2018 in the canal (N35.631783, E139.7448151) of Tokyo Bay. CPE was isolated as described previously (Sekizuka et al., 2018). Carbapenemase production by bacterial isolates grown on CHROMagar ESBL plates (CHROMagar, Paris, France) was investigated using the Carba NP test as described previously (Nordmann et al., 2012). KPC‐2‐producing K. pneumoniae GSU10‐3 (Sekizuka et al., 2018) and E. coli ATCC 25922 were used as a positive and negative control for the assay respectively. Three isolates exhibited a positive reaction in the Carba NP test among 14 representative colonies with differentiated colour indicator, size and form. These isolates were then subjected to whole‐genome sequencing (WGS) as described previously (Sekizuka et al., 2018), using an Illumina NextSeq 500 platform with a 300‐cycle NextSeq 500 Reagent Kit v2 (2 × 150‐mer). These results confirmed the three clonal isolates of a KPC‐2‐positive A. hydrophila strain (GSH8‐2). In addition, four notable colonies were obtained from the same WWTP effluent under 1 μg/ml meropenem (MER) selection, and a strain that reacted positively in the Carba NP test was obtained and sequenced, revealing a KPC‐2‐positive A. caviae strain (GSH8M‐1).
In general, Aeromonas spp., particularly A. hydrophila, A. caviae and A. veronii, adapt to saline water environments (Janda and Abbott, 2010) and can therefore be isolated from estuaries. Apart from fish, which are widely reported hosts for Aeromonads, other organisms, including insects, crustaceans, reptiles, birds, and mammals, have also been found to harbour Aeromonas species. Aeromonas spp. cause opportunistic systemic disease in immunocompromised patients, diarrheal disease and wound infections both in healthy and diseased states (Pearson et al., 2000; Turutoglu et al., 2005; Evangelista‐Barreto et al., 2006; Ceylan et al., 2009). Although Aeromonas spp. exhibit limited pathogenicity to humans, clinical A. hydrophila isolates producing KPC‐2 were identified from routine perirectal surveillance culture in hospitalized adult patients in the United States (Hughes et al., 2016). Besides KPC‐2, VIM‐producing A. caviae has been isolated in Israeli hospitals (Adler et al., 2014), GES‐24 carbapenemase‐producing A. hydrophila was isolated from an inpatient in Okinawa, Japan (Uechi et al., 2018), and OXA‐181‐carbapenemase was found in A. caviae (Anandan et al., 2017).
Antimicrobial susceptibility testing of strains GSH8‐2 and GSH8M‐1 was performed using the Etest (bioMérieux, Marcy‐l'Étoile, France) and disk diffusion methods according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines v9.0. Strains GSH8‐2 and GSH8M‐1 were resistant to the majority of β‐lactams except for carbapenems [imipenem (IMI) and MER]. GSH8‐2 and GSH8M‐1 showed susceptibility and intermediate susceptibility to MER respectively (Table 1). However, the minimum inhibitory concentration (MIC) breakpoints do not always reliably indicate sensitivity during CPE detection because of possible low expression of carbapenemase by bacterial hosts. Therefore, the EUCAST recommends that detection of potential carbapenemase production be carried out with lower screening cut‐off values (e.g. MER MIC >0.125 mg/l) (EUCAST, 2017) guidelines for detection of resistance mechanisms and specific resistances of clinical and/or epidemiological importance, indicating that molecular (polymerase chain reaction or WGS) or sensitive enzymatic detection (Carba NP test) are more reliable tests for AMR. Opportunistic pathogens appear to be neglected in AMR studies, and this negative attitude towards microbial testing may increase the risk of AMR transfer through non‐pathogenic bacteria behaving as environmental reservoirs (Piotrowska et al., 2017).
Table 1.
Antimicrobial susceptibility test (MIC, mg L−1).
| Electrotransformant in E. coli DH10B T1R | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Antimicrobial agents | A. hydrophila GSH8‐2 | A. caviae GSH8M‐1 | ‐ | pGSH8‐2 | pGSH8M‐1‐2 | |||||
| AMS | 128 | R | 128 | R | 1.5 | S | 128 | R | 48 | R |
| CEP | 4 | I | 16 | R | 0.032 | S | 0.75 | S | 0.75 | S |
| CTA | 32 | R | >32 | R | 0.032 | S | 1.5 | I | 0.75 | I |
| CTZ | 2 | I | 16 | R | 0.032 | S | 2 | I | 1.5 | I |
| IMI | 1 | S | 4 | I | 0.19 | S | 1 | S | 0.75 | S |
| MER | 1 | S | 4 | I | ≤0.125 | S | 0.19 | S | 0.125 | S |
| AZT | 8 | R | 32 | R | 0.064 | S | 12 | R | 12 | R |
| AMI | 2 | S | 16 | I | 1 | S | 1.5 | S | 1.5 | S |
| GEN | 0.5 | S | 1 | S | 0.38 | S | 0.38 | S | 0.5 | S |
| TOB | 2 | S | 8 | R | 0.25 | S | 0.25 | S | 0.38 | S |
| TET | 8 | ‐ | 2 | ‐ | 0.75 | ‐ | 1 | ‐ | 1 | ‐ |
| CIP | 0.5 | I | 1 | R | ≤0.002 | S | ≤0.002 | S | ≤0.002 | S |
| LEV | 2 | R | 2 | R | 0.004 | S | 0.004 | S | 0.004 | S |
| AZI | 64 | ‐ | 32 | ‐ | 2 | ‐ | 3 | ‐ | 96 | ‐ |
| TRS | 0.25 | S | 1 | S | 0.016 | S | 0.023 | S | 0.023 | S |
| CHL | 1 | S | 2 | S | 1.5 | S | 1.5 | S | 1.5 | S |
| COL | 2 | S | 0.5 | S | 0.032 | S | 0.047 | S | 0.047 | S |
AMS, ampicillin‐sulbactam; CEP, cefepime; CTA, cefotaxime; CTZ, ceftazidime; IMI, imipenem; MER, meropenem; AZT, aztreonam; AMI, amikacin; GEN, gentamicin; TOB, tobramycin; TET, tetracycline; CIP, ciprofloxacin; LEV, levofloxacin; AZI, azithromycin; TRS, trimethoprim‐sulfamethoxazole; CHL, chloramphenicol; COL, colistin.
‐, not defined in EUCAST.
Since both strain A. hydrophila GSH8‐2 and A. caviae GSH8M‐1 showed positive reactions in the Carba NP test, we performed WGS, which revealed that both strains carry bla KPC‐2 (Table 2).
Table 2.
Whole genome information of Aeromonas spp. from the WWTP effluent.
| Organism/Strain | Replicon | GC% | Length (bp) | MLSTa | Inc type | Drug Resistance Gene | GenBank ID | BioProject ID | BioSample ID |
|---|---|---|---|---|---|---|---|---|---|
| Aeromonas hydrophila GSH8‐2 | |||||||||
| Chromosome | 61.07 | 4 962 478 | ST558 | ‐ | cepS, cphA2, bla OXA‐ 726 mph(A), tet(E) | AP019193 | PRJDB6962 | SAMD00144877 | |
| pGSH8‐2 | 58.20 | 39 071 | ‐ | IncP‐6 | bla KPC‐ 2 | AP019194 | PRJDB6962 | SAMD00144877 | |
| Aeromonas caviae GSH8M‐1 | |||||||||
| Chromosome | 61.00 | 4 611 279 | ST13 | ‐ | bla MOX‐ 12, bla OXA‐ 780, mcr‐3.18, mcr‐3 variant | AP019195 | PRJDB6962 | SAMD00144878 | |
| pGSH8M‐1‐1 | 54.22 | 153 814 | ‐ | UT | bla OXA‐ 669, aac(6′)‐Ia, aadA2, 3x sul1 | AP019196 | PRJDB6962 | SAMD00144878 | |
| pGSH8M‐1‐2 | 58.76 | 53 629 | ‐ | IncP‐6 | bla KPC‐ 2, mph(A) | AP019197 | PRJDB6962 | SAMD00144878 | |
| pGSH8M‐1‐3 | 52.25 | 8,098 | ‐ | UT | ‐ | AP019198 | PRJDB6962 | SAMD00144878 | |
‐, not available; UT, unknown type.
The short‐ and long‐read sequences for DNA‐Seq were deposited in the DNA Data Bank of Japan (BioProject PRJDB6962; BioSample SAMD00144877 for GSH8‐2 and SAMD00144878 for GSH8M‐1).
pubMLST for Aeromonas spp. (https://pubmlst.org/aeromonas/).
WGS analysis of Aeromonas isolates carrying blaKPC‐2
Summary information regarding the complete chromosome and plasmid sequences for Aeromonas isolates GSH8‐2 (Fig. 1) and GSH8M‐1 (Fig. 2) are shown in Table 2. Multilocus sequence typing (MLST) based on the genomic data assigned both strains to novel sequence types (ST), classifying GSH8‐2 and GSH8M‐1 as ST558 and ST13 respectively (Table 2).
Figure 1.
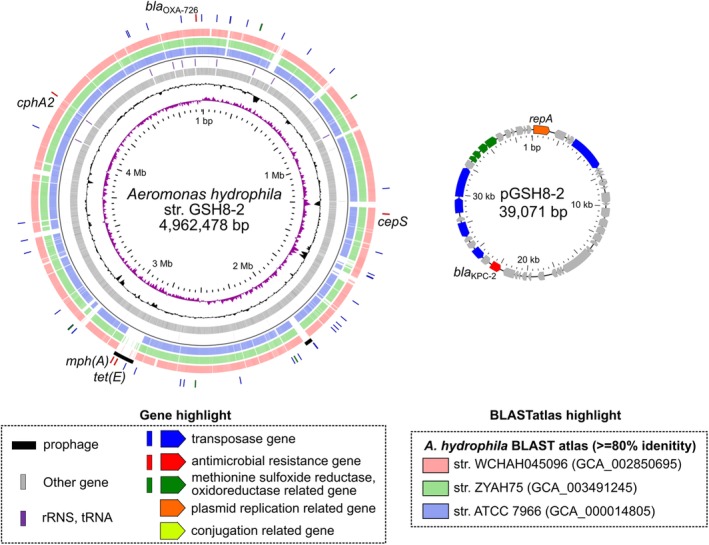
Schematic circular representation of complete genome sequences of KPC‐2‐producing A. hydrophila GSH8‐2. Short paired‐end whole‐genome sequencing was performed using an Illumina NextSeq 500 platform with a 300‐cycle NextSeq 500 Reagent Kit v2 (2 × 150‐mer). The complete genome sequences of the strains were determined using a PacBio Sequel sequencer for long‐read sequencing [Sequel SMRT Cell 1 M v2 (4/tray]; Sequel sequencing kit v2.1; insert size, approximately 10 kb). De novo assembly was performed using Canu version 1.4 (Koren et al., 2017), minimap version 0.2‐r124 (Li, 2016), racon version 1.1.0 (Vaser et al., 2017) and circulator version 1.5.3 (Hunt et al., 2015). Error correction of tentative complete circular sequences was performed using Pilon version 1.18 with Illumina short reads (Walker et al., 2014). Annotation was performed in Prokka version 1.11 (Seemann, 2014), InterPro v49.0 (Finn et al., 2017) and NCBI‐BLASTP/BLASTX. Circular representations of complete genomic sequences were visualized using GView server (Petkau et al., 2010). AMR genes were identified by homology searching against the ResFinder database (Zankari et al., 2012). The class 1 integron was assigned in the INTEGRALL database (http://integrall.bio.ua.pt/) (Moura et al., 2009). Visualization of comparative plasmid ORFs organization was performed using Easyfig (Sullivan et al., 2011). For representation of chromosomal DNA, from the inside: slot 1, GC skew; slot 2, GC content; slot 3, ORFs; slot 4, rRNA/tRNA; slots 5–7, BLASTatlas conserved gene analysis indicating three relative strains (see also Supporting Information Fig. S1); slot 8, prophage; slot 9, notable ARGs or ARG‐related genes (transposase, ARGs and reductases). In representations of circular plasmids, notable ORFs are highlighted as the indicated colour.
Figure 2.
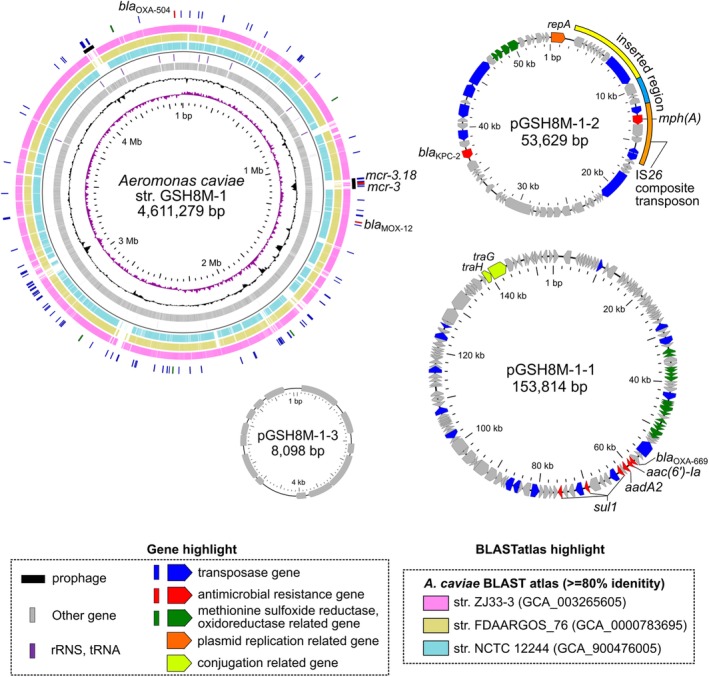
Schematic circular representation of complete genome sequences of KPC‐2‐producing A. caviae GSH8M‐1. See the legend to Fig. 1 for methods and descriptions of representations. Slots 5–7, BLASTatlas conserved gene analysis indicates three relative strains (see also Supporting Information Fig. S2). Marked acquired elements in pGSH8M‐1‐2 compared to pGSH8‐2 are highlighted in the outer slot.
To trace the potential clonal sources of the Aeromonas GSH8‐2 and GSH8M‐1 strains, we performed core‐genome phylogenetic analysis using 108 and 30 publicly available A. hydrophila and A. caviae genome sequences (at 29 October 2018), including the respective draft genomes with at least 40× read coverage (strain phylogenies are presented in the Supporting Information Figs. [Link] and [Link]). Those retrieved sequences were compared using bwaMEM read mapping against either the GSH8‐2 or GSH8M‐1 complete genome sequences as reference respectively. After excluding repeat regions from the whole genome sequences, single nucleotide variants (SNVs) were identified in the core genomes of GSH8‐2 and GSH8M‐1 (see Supporting Information Figs. [Link] and [Link] respectively). Core‐genome MLST (cgMLST) was then performed using these SNVs, and the phylogenies were generated using the maximum‐likelihood phylogenetic method with FastTree v2.1.10.
The core genome of the A. hydrophila strains constituted 48.95% of the total genome based on 491 764 SNVs (Supporting Information Fig. [Link]). The phylogeny placed GSH8‐2 in one of multiple clusters, unrelated to previously identified clinical and environmental isolates (Supporting Information Fig. [Link]). No pathogenic fish clonal cluster was observed among isolates from the United States or China. To date, A. hydrophila has mostly been isolated as a fish pathogen, and from environmental sources such as rivers and sewage treatment plants, as well as from clinical specimens from patients treated with leech therapy. Thus far, only two strains, including GSH8‐2 in this study, have been recovered from sewage treatment plants, making GSH8‐2 useful for characterizing the genetic and phenotypic features of inhabitants of waste processing or decontamination sites.
The core genome of the A. caviae strains constituted 70.53% of the whole genome, and the phylogenetic analysis used 229 493 SNVs (Supporting Information Fig. [Link]). The core‐genome phylogeny of A. caviae strains was variable, and no relevant clinical or environmental isolates related to GSH8M‐1 were found (Supporting Information Fig. [Link]). To date, too few A. caviae genomes have been sequenced to determine the original source or clonal dissemination. In contrast to pathogenic A. hydrophila strains, A. caviae has been more frequently isolated from environmental sources such as sewage treatment plants than from clinical settings. Similar to GSH8‐2, GSH8M‐1 may be used for characterizing the genetic and phenotypic features of waste processing or decontamination site inhabitants.
ARGs in plasmids and chromosomes
Aeromonas hydrophila GSH8‐2
Aeromonas hydrophila GSH8‐2 harbours a 39.1‐kb IncP‐6 replicon plasmid, pGSH8‐2, carrying bla KPC‐2 (Table 2 and Fig. 1). In addition, the strain was found to carry multiple β‐lactamase genes [cepS (FOX/MOX family class C β‐lactamase, 95% identity), cphA2 (CphA family subclass B2 metallo‐β‐lactamase, 98% identity), bla OXA‐726 (class D β‐lactamase)] on its chromosome. These are intrinsic ARGs in A. hydrophila strains. Intriguingly, it also carried chromosomal copies of macrolide 2′‐phosphotransferase [mph(A)] and tetracycline efflux MFS transporter [tet(E)], which were found within a potential prophage (2805–2905 kb, shown in Fig. 1). BLASTatlas analysis suggested that these horizontally acquired phage elements are GSH8‐2 strain‐specific, and are similar to elements from the ATCC 7966 type strain WCHAH045096 (isolated from sewage in China in 2015, bla KPC‐2 positive) and ZYAH75 (isolated from a human wound in China in 2015) (slots 5–7 in Fig. 1). The detection of mph(A) and tet(E) on a prophage in the chromosome of GSH8‐2 could contribute to the reduced susceptibility to azithromycin and tetracycline respectively (Table 1).
Aeromonas caviae GSH8M‐1
Aeromona caviae GSH8M‐1 also harbours the IncP‐6 replicon plasmid pGSH8M‐1‐2 (53.6 kb), which carries bla KPC‐2, as well as additional acquired elements, including mph(A), on the IS26 composite transposon (Table 2 and Fig. 2). pGSH8M‐1‐1 (153.8 kb) is an unknown‐type replicon plasmid, but it shares a partial similar region (47%–51%) with Aeromonas salmonicida plasmids (pS44‐1, pAsa4c, pAsa4b and plasmid 4) (data not shown). pGSH8M‐1‐1 carries an antimicrobial resistance island (3.9–28.0 kb), which includes a class I integron, assigned as In1665 [bla OXA‐669, aac(6′)‐Ia, aadA2, qacEdelta, sul1], and two additional sulfonamide‐resistant dihydropteroate synthase (sul1) genes. pGSH8M‐1‐3 (8.0 kb) is a small and unknown‐type replicon plasmid, sharing a partial similarity region (<25%) with Aeromonas spp. plasmids and does not carry any ARGs. Multiple ARGs intrinsic to A. caviae strains [bla MOX‐12 (CMY‐1/MOX family class C β‐lactamase], as well as bla OXA‐780 (class D β‐lactamase) were identified on the strain's chromosome. Two mcr‐3 variants (mcr‐3.18 and an mcr‐3 variant) are located on a potential prophage in GSH8M‐1 (1112–1155 kb, shown in Fig. 2). BLASTatlas analysis suggested that this horizontally acquired phage similar to GSH8M‐1 strain‐specific elements is comparable to sequences from the NCTC 12244 type strain ZJ33‐3 (from a human rectal swab in China in 2017, mcr‐3.3‐positive) and FDAARGOS_76 (from a human stool in the United States in 2013) (slots 5–7 in Fig. 2). Detection of mcr3 gene variants on a prophage in the chromosome of GSH8M‐1 is of note despite its susceptibility to colistin (Table 1). Indeed, mcr‐3‐positive Aeromonas spp. have been reported and widely isolated from humans, retail meat and environmental water (Shen et al., 2018). Most of these mcr‐3‐positive strains showed moderate susceptibility to colistin and failed in transconjugation of mcr‐3 gene variants to E. coli recipients (Shen et al., 2018), consistent with our findings that mcr‐3 variants are located on a prophage. Thus, Aeromonas spp. and the aquatic environment might be a potential source of mcr genes for exaptation to a hypertonic marine environment (Cabello and Godfrey, 2018).
Structural comparison of KPC‐2 plasmids
Both Aeromonas strain GSH8‐2 and GSH8M‐1 carry a bla KPC‐2‐positive plasmid (pGSH8‐2 and pGSH8M‐1‐2 respectively) and share a 39–40‐kb backbone of IncP‐6 replicon plasmids (Fig. 3). pGSH8‐2 shares a common backbone with plasmids in isolates from environmental sources. These include plasmid pKPC‐cd17 in Aeromonas sp. ASNIH3 isolated from hospital wastewater in Bethesda (USA), in 2014, and plasmid pKPC2_045096 in A. hydrophila subsp. hydrophila strain WCHAH045096 isolated from sewage in Sichuan (China) in 2015 (Fig. 3A). pGSH8‐2‐related IncP‐6 plasmids might be fundamental KPC‐2‐positive plasmids in A. hydrophila, which potentially serves as an environmental reservoir for KPC‐2 carbapenemase. Similar IncP‐6 plasmids found in Klebsiella, Citrobacter, Enterobacter, E. coli, Pseudomonas and other Aeromonas spp., have acquired additional ARG elements (aph(6)‐Id and aph(3″)‐Id in pKPC2; qnrS2 in p186‐KPC; mph(A) in pGSH8M‐1‐2) (Fig. 3B). Sequence alignments suggested that IncP‐6 plasmids [equivalent to IncG/U (Haines et al., 2006)] are broadly disseminated in multiple organisms (Fig. 3A), strongly indicating that IncP‐6 replicon shows ability of replication over a broad host range, as previously described (Haines et al., 2005).
Figure 3.
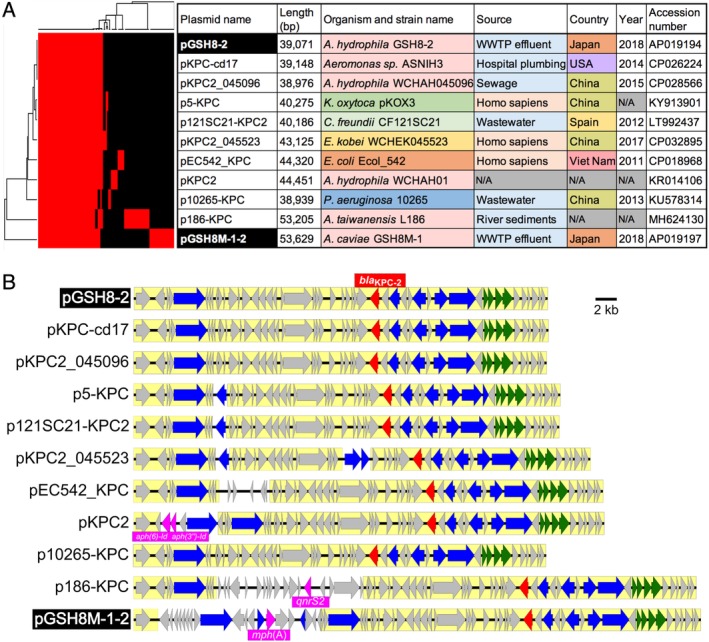
Structural comparison of the bla KPC‐2‐positive plasmids. A. Heat map representation of orthologous gene clustering of bla KPC‐2‐positive IncP‐6 plasmids. B. Comparison of gene structure among plasmids shown in panel (A). Genes for bla KPC‐2, ARGs, transposase, methionine sulfoxide reductase/oxidoreductase, and others are highlighted with red, magenta, blue, green, and grey, respectively, as shown in Fig. 1.
Within this bla KPC‐2‐positive IncP‐6 plasmid family, less pathogenic Aeromonas (Wang et al., 2017), Citrobacter (Yao et al., 2017) and Pseudomonas (Dai et al., 2016) were isolated mostly from environments such as waste water, hospital plumbing (Weingarten et al., 2018), and river sediments (Dai et al., 2016), whereas bacteria with higher pathogenicity, such as Klebsiella oxytoca (Wang et al., 2017), Enterobacter kobei and E. coli, were isolated from clinical specimens, indicating that the isolation source may depend on the pathogenicity of the bacterial host (Fig. 3). In particular, IncP‐6 plasmids [described as IncU in Piotrowska et al. (Piotrowska and Popowska, 2015)] have been characterized as multiple AMR plasmids. In line with the present study, a previous study demonstrated that the IncP‐6 (IncU) replicon plasmid also contributes to the dissemination of the tetracycline resistance gene tetA between Aeromonas species and E. coli isolates from fish farm environments, hospital sewage and a hospital patient (Rhodes et al., 2000)
When compared with pGSH8‐2, pGSH8M‐1‐2 was found to carry an additional elements, the first of which is a Tn3 family transposon (inverted repeats at nt 27 425– 34 564: GGGGATGTAAGCCGGAACCCCAGAAATTTCCGTCACCCTAG –CTGGGCTGACGGAAATTTCTGGGATTCCGGCGTACAACCCC), and the second is the IS26‐mediated composite transposon carrying mph(A) family macrolide 2′‐phosphotransferase (Fig. 1B). A Blastp homology search indicated that this Mph(A) shares 100% identity with those found in 39 E. coli, 14 Klebsiella pneumoniae, 1 Salmonella Typhimurium and 1 Shigella flexneri strains. In addition, IS26 is a ubiquitous IS in Enterobacteriaceae, indicating that mph(A) acquisition could take place through IS26‐mediated recombination with other potential mph(A) resources.
KPC‐2 plasmids reduced antimicrobial susceptibility
To characterize the single effect of each KPC‐2‐positive plasmid (pGSH8‐2 or pGSH8M‐1‐2), electro‐transformants carrying the KPC‐2‐plasmid under an E. coli DH10B T1R strain background were obtained (see detailed experimental methods in the [Link]), and the MIC values were determined (Table 1). These electro‐transformants exhibited significantly reduced susceptibility to all tested β‐lactams but not to other antimicrobials except for azithromycin. pGSH8M‐1‐2 carries mph(A) (Fig. 2), which could be involved in the macrolide resistance in the GSH8M‐1 strain (reduced susceptibility to azithromycin from 2 to 96 mg/l).
Multiple sulfo‐related reductase enzymes
Intriguingly, A. caviae GSH8M‐1 has acquired a genomic island including multiple sulfo‐related reductase enzymes such as glutathione S‐transferase (GST), two peptide methionine sulfoxide reductases (MsrA/MsrB), two glutaredoxins, two zinc‐type alcohol dehydrogenase‐like proteins, two 3‐ketoacyl‐CoA thiolases and other oxidoreductases (green highlighted ORFs in Fig. 2). In total, four sets of redundant MsrA/B were identified in A. caviae GSH8M‐1 (one on its chromosome, two on pGSH8M‐1‐1 and one on pGSH8M‐1‐2). Likewise, A. hydrophila GSH8‐2 plasmid pGSH8M‐1‐2 also carries GST and MsrA/B reductase (Fig. 1). Such multiple sulfo‐related reductase enzymes might contribute to adaptation to oxidative stresses encountered in WWTP environments or during antiseptic treatment.
Conclusions and prospects
We isolated a potential KPC‐2 carbapenemase reservoir in environmental Aeromonas isolates from a WWTP effluent in Tokyo Bay. KPC‐type CPOs have rarely been detected, even in clinical settings, in Japan. Thus, although Aeromonas species are opportunistic pathogens, such rather neglected pathogens might play a key role as environmental antimicrobial resistance reservoirs.
In conclusion, AMR monitoring of WWTP effluent contributes to the detection of ongoing bacterial AMR dissemination in the environment, and can provide early warning of the potential dissemination in clinical settings and communities. CPO‐colonized and infected patients may disseminate AMR bacteria in excreta, which encounter active broad‐spectrum antimicrobials in hospital sewage systems, leading to the perfect selection pressure for the exchange of resistance genes between clinical pathogens and environmental bacteria (Taylor et al., 2011; Picao et al., 2013). Indeed, KPC‐type CPOs have been found in hospital effluents in Brazil, China, and the United States (Chagas et al., 2011; Zhang et al., 2012; Picao et al., 2013; Woodford et al., 2014; Weingarten et al., 2018). Subsequently, identification of the presence of CPO in polluted aquatic environments such as recreational waters indicated a potential threat to beach frequenters (de Araujo et al., 2016; Paschoal et al., 2017; Leonard et al., 2018).
Because contamination in the environment can serve as AMR reservoirs, potentially causing infectious diseases or infecting healthy carriers through consumption of contaminated water or food, or through recreational activities (Schijven et al., 2015; Leonard et al., 2018), a comprehensive approach will be required to uncover environmental contamination through WWTP effluents, and more attention should be focused on non‐ or opportunistic pathogens as environmental AMR reservoirs.
Authors' contributions
TS and MK contributed to the isolation of the KPC‐2‐positive Aeromonas strains. TS and MH performed antimicrobial susceptibility testing. YI performed the genome sequencing. TS and KY performed the comparative genome analysis. MK wrote the manuscript.
Funding
This work was supported by the Research Program on Emerging and Re‐emerging Infectious Diseases from the Japan Agency for Medical Research and Development, AMED (grant number: JP17fk0108121 and JP18fk0108048). This work was also supported by a grant for Research on Emerging and Re‐emerging Infectious Diseases and Immunization (H30 Shinkogyosei‐Ippan‐002) from the Ministry of Health, Labour and Welfare, Japan. The funding agencies had no role in the study design, data collection and analysis, decision to publish, or manuscript preparation.
Supporting information
Appendix S1. Supplementary Information
Supplemental Figure S1 Core genome phylogeny of Aeromonas hydrophila GSH8‐2 among publicly available 108 strains (updated at 2018/10/29).
All publicly available draft genome sequences of Aeromonas strains were retrieved. Analysis of conserved plasmid gene sequences was performed with BLASTn searches (≥99% nt identity), followed by visualization using FriPan (http://drpowell.github.io/FriPan/). Using the complete genome sequence of GSH8‐2 (4,962,478 bp), bwa‐read mapping analysis identified 48.95% of the full genome as core‐genome sequence, and a total of 491,764 single nucleotide variations (SNVs) were identified, excluding prophages and repeated regions.
Supplemental Figure S2 Core genome phylogeny of Aeromonas caviae GSH8M‐1 among publicly available 30 strains (updated at 2018/10/29).
Using the complete genome sequence of GSH8M‐1 (4,611,279 bp), bwa‐read mapping analysis identified 70.53% of the full genome as core‐genome sequence, and a total of 229,493 single nucleotide variations (SNVs) were identified, excluding prophages and repeated regions.
Acknowledgements
The authors would like to thank Miho Nishio and Norimi Kishi for technical assistance with the NGS experiments, and Dr. Satowa Suzuki for providing E. coli ATCC 25922 strain. We would like to thank Editage (http://www.editage.com) for English language editing.
References
- Adler, A. , Assous, M.V. , Paikin, S. , Shulman, A. , Miller‐Roll, T. , Hillel, S. , et al (2014) Emergence of VIM‐producing Aeromonas caviae in Israeli hospitals. J Antimicrob Chemother 69: 1211–1214. [DOI] [PubMed] [Google Scholar]
- Anandan, S. , Gopi, R. , Devanga Ragupathi, N.K. , Muthuirulandi Sethuvel, D.P. , Gunasekaran, P. , Walia, K. , and Veeraraghavan, B. (2017) First report of blaOXA‐181‐mediated carbapenem resistance in Aeromonas caviae in association with pKP3‐a: threat for rapid dissemination. J Glob Antimicrob Resist 10: 310–314. [DOI] [PubMed] [Google Scholar]
- Cabello, F.C. , and Godfrey, H.P. (2018) Aquaculture, exaptation, and the origin of mcr‐positive Colistin resistance. Antimicrob Agents Chemother 62: e01903–e01918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceylan, E. , Berktas, M. , and Agaoglu, Z. (2009) The occurrence and antibiotic resistance of motile Aeromonas in livestock. Tropl Anim Health Prod 41: 199–204. [DOI] [PubMed] [Google Scholar]
- Chagas, T.P. , Seki, L.M. , da Silva, D.M. , and Asensi, M.D. (2011) Occurrence of KPC‐2‐producing Klebsiella pneumoniae strains in hospital wastewater. J Hosp Infect 77: 281. [DOI] [PubMed] [Google Scholar]
- Dai, X. , Zhou, D. , Xiong, W. , Feng, J. , Luo, W. , Luo, G. , et al (2016) The IncP‐6 plasmid p10265‐KPC from Pseudomonas aeruginosa carries a novel DeltaISEc33‐associated Bla KPC‐2 gene cluster. Front Microbiol 7: 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Araujo, C.F. , Silva, D.M. , Carneiro, M.T. , Ribeiro, S. , Fontana‐Maurell, M. , Alvarez, P. , et al (2016) Detection of Carbapenemase genes in aquatic environments in Rio de Janeiro, Brazil. Antimicrob Agents Chemother 60: 4380–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST (2017) In: EUCAST [Internet]. URL http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf.
- Evangelista‐Barreto, N.S. , Vieira, R.H. , Carvalho, F.C. , Torres, R.C. , Sant'Anna, E.S. , Rodrigues, D.P. , and Reis, C.M. (2006) Aeromonas spp. isolated from oysters (Crassostrea rhizophorea) from a natural oyster bed, Ceara, Brazil. Rev Inst Med Trop Sao Paulo 48: 129–133. [DOI] [PubMed] [Google Scholar]
- Finn, R.D. , Attwood, T.K. , Babbitt, P.C. , Bateman, A. , Bork, P. , Bridge, A.J. , et al (2017) InterPro in 2017‐beyond protein family and domain annotations. Nucleic Acids Res 45: D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines, A.S. , Cheung, M. , and Thomas, C.M. (2006) Evidence that IncG (IncP‐6) and IncU plasmids form a single incompatibility group. Plasmid 55: 210–215. [DOI] [PubMed] [Google Scholar]
- Haines, A.S. , Jones, K. , Cheung, M. , and Thomas, C.M. (2005) The IncP‐6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J Bacteriol 187: 4728–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, H.Y. , Conlan, S.P. , Lau, A.F. , Dekker, J.P. , Michelin, A.V. , Youn, J.H. , et al (2016) Detection and whole‐genome sequencing of Carbapenemase‐producing Aeromonas hydrophila isolates from routine perirectal surveillance culture. J Clin Microbiol 54: 1167–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, M. , Silva, N.D. , Otto, T.D. , Parkhill, J. , Keane, J.A. , and Harris, S.R. (2015) Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol 16: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda, J.M. , and Abbott, S.L. (2010) The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev 23: 35–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkman, A. , Do, T.T. , Walsh, F. , and Virta, M.P.J. (2018) Antibiotic‐resistance genes in waste water. Trends Microbiol 26: 220–228. [DOI] [PubMed] [Google Scholar]
- Koren, S. , Walenz, B.P. , Berlin, K. , Miller, J.R. , Bergman, N.H. , and Phillippy, A.M. (2017) Canu: scalable and accurate long‐read assembly via adaptive k‐mer weighting and repeat separation. Genome Res 27: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, A.F.C. , Zhang, L. , Balfour, A.J. , Garside, R. , Hawkey, P.M. , Murray, A.K. , et al (2018) Exposure to and colonisation by antibiotic‐resistant E. coli in UKcoastal water users: environmental surveillance, exposure assessment, and epidemiological study (beach bum survey). Environ Int 114: 326–333. [DOI] [PubMed] [Google Scholar]
- Li, H. (2016) Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32: 2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miriagou, V. , Tzouvelekis, L.S. , Rossiter, S. , Tzelepi, E. , Angulo, F.J. , and Whichard, J.M. (2003) Imipenem resistance in a Salmonella clinical strain due to plasmid‐mediated class A carbapenemase KPC‐2. Antimicrob Agents Chemother 47: 1297–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura, A. , Soares, M. , Pereira, C. , Leitao, N. , Henriques, I. , and Correia, A. (2009) INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25: 1096–1098. [DOI] [PubMed] [Google Scholar]
- Ng, C. , Tay, M. , Tan, B. , Le, T.H. , Haller, L. , Chen, H. , et al (2017) Characterization of metagenomes in urban aquatic compartments reveals high prevalence of clinically relevant antibiotic resistance genes in wastewaters. Front Microbiol 8: 2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann, P. , Poirel, L. , and Dortet, L. (2012) Rapid detection of carbapenemase‐producing Enterobacteriaceae. Emerg Infect Dis 18: 1503–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschoal, R.P. , Campana, E.H. , Correa, L.L. , Montezzi, L.F. , Barrueto, L.R.L. , da Silva, I.R. , et al (2017) Concentration and variety of Carbapenemase producers in recreational coastal waters showing distinct levels of pollution. Antimicrob Agents Chemother 61: e01963–e01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, M.D. , Hirono, I. , Aoki, T. , Miranda, R. , and Inglis, V. (2000) Virulence properties of motile aeromonads isolated from farmed frogs Rana tigerina and R. rugulosa . Dis Aquat Organ 40: 185–193. [DOI] [PubMed] [Google Scholar]
- Petkau, A. , Stuart‐Edwards, M. , Stothard, P. , and Van Domselaar, G. (2010) Interactive microbial genome visualization with GView. Bioinformatics 26: 3125–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picao, R.C. , Cardoso, J.P. , Campana, E.H. , Nicoletti, A.G. , Petrolini, F.V. , Assis, D.M. , et al (2013) The route of antimicrobial resistance from the hospital effluent to the environment: focus on the occurrence of KPC‐producing Aeromonas spp. and Enterobacteriaceae in sewage. Diagn Microbiol Infect Dis 76: 80–85. [DOI] [PubMed] [Google Scholar]
- Piotrowska, M. , and Popowska, M. (2015) Insight into the mobilome of Aeromonas strains. Front Microbiol 6: 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska, M. , Przygodzinska, D. , Matyjewicz, K. , and Popowska, M. (2017) Occurrence and variety of beta‐lactamase genes among Aeromonas spp. isolated from urban wastewater treatment plant. Front Microbiol 8: 863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes, G. , Huys, G. , Swings, J. , McGann, P. , Hiney, M. , Smith, P. , and Pickup, R.W. (2000) Distribution of oxytetracycline resistance plasmids between aeromonads in hospital and aquaculture environments: implication of Tn1721 in dissemination of the tetracycline resistance determinant tet a. Appl Environ Microbiol 66: 3883–3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, R. , Takahashi, R. , Sawabe, E. , Koyano, S. , Takahashi, Y. , Shima, M. , et al (2014) First report of KPC‐2 Carbapenemase‐producing Klebsiella pneumoniae in Japan. Antimicrob Agents Chemother 58: 2961–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schijven, J.F. , Blaak, H. , Schets, F.M. , and de Roda Husman, A.M. (2015) Fate of extended‐Spectrum beta‐lactamase‐producing Escherichia coli from Faecal sources in surface water and probability of human exposure through swimming. Environ Sci Technol 49: 11825–11833. [DOI] [PubMed] [Google Scholar]
- Seemann, T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069.24642063 [Google Scholar]
- Sekizuka, T. , Yatsu, K. , Inamine, Y. , Segawa, T. , Nishio, M. , Kishi, N. , and Kuroda, M. (2018) Complete genome sequence of a blaKPC‐2‐positive Klebsiella pneumoniae strain isolated from the effluent of an urban sewage treatment Plant in Japan. mSphere 3: e00314–e00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, M. , and Reynnells, R. (2016) Importance of soil amendments: survival of bacterial pathogens in manure and compost used as organic fertilizers. Microbiol Spectr 4: 1–13. [DOI] [PubMed] [Google Scholar]
- Shen, Y. , Xu, C. , Sun, Q. , Schwarz, S. , Ou, Y. , Yang, L. , et al (2018) Prevalence and genetic analysis of mcr‐3‐positive Aeromonas species from humans, retail meat, and environmental water samples. Antimicrob Agents Chemother 62: e00404–e00418. 10.1128/AAC.00404-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, M.J. , Petty, N.K. , and Beatson, S.A. (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27: 1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G. , Verner‐Jeffreys, D.W. , and Baker‐Austin, C. (2011) Aquatic systems: maintaining, mixing and mobilising antimicrobial resistance? Trends Ecol Evol 26: 278–284. [DOI] [PubMed] [Google Scholar]
- Turutoglu, H. , Ercelik, S. , and Corlu, M. (2005) Aeromonas hydrophila‐associated skin lesions and septicaemia in a Nile crocodile (Crocodylus niloticus). J S Afr Vet Assoc 76: 40–42. [DOI] [PubMed] [Google Scholar]
- Uechi, K. , Tada, T. , Sawachi, Y. , Hishinuma, T. , Takaesu, R. , Nakama, M. , et al (2018) A carbapenem‐resistant clinical isolate of Aeromonas hydrophila in Japan harbouring an acquired gene encoding GES‐24 beta‐lactamase. J Med Microbiol 67: 1535–1537. [DOI] [PubMed] [Google Scholar]
- Vaser, R. , Sovic, I. , Nagarajan, N. , and Sikic, M. (2017) Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27: 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, B.J. , Abeel, T. , Shea, T. , Priest, M. , Abouelliel, A. , Sakthikumar, S. , et al (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Yuan, M. , Chen, H. , Chen, X. , Jia, Y. , Zhu, X. , et al (2017) First report of Klebsiella oxytoca strain simultaneously producing NDM‐1, IMP‐4, and KPC‐2 Carbapenemases. Antimicrob Agents Chemother 61: e00877–e00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten, R.A. , Johnson, R.C. , Conlan, S. , Ramsburg, A.M. , Dekker, J.P. , Lau, A.F. , et al (2018) Genomic analysis of hospital plumbing reveals diverse reservoir of bacterial plasmids conferring Carbapenem resistance. MBio 9: e02011–e02017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford, N. , Wareham, D.W. , Guerra, B. , and Teale, C. (2014) Carbapenemase‐producing Enterobacteriaceae and non‐Enterobacteriaceae from animals and the environment: an emerging public health risk of our own making? J Antimicrob Chemother 69: 287–291. [DOI] [PubMed] [Google Scholar]
- Xu, H. , Wang, X. , Yu, X. , Zhang, J. , Guo, L. , Huang, C. , et al (2018) First detection and genomics analysis of KPC‐2‐producing Citrobacter isolates from river sediments. Environ Pollut 235: 931–937. [DOI] [PubMed] [Google Scholar]
- Yao, Y. , Lazaro‐Perona, F. , Falgenhauer, L. , Valverde, A. , Imirzalioglu, C. , Dominguez, L. , et al (2017) Insights into a novel blaKPC‐2‐encoding IncP‐6 plasmid reveal Carbapenem‐resistance circulation in several Enterobacteriaceae species from wastewater and a hospital source in Spain. Front Microbiol 8: 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari, E. , Hasman, H. , Cosentino, S. , Vestergaard, M. , Rasmussen, S. , Lund, O. , et al (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67: 2640–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Lu, X. , and Zong, Z. (2012) Enterobacteriaceae producing the KPC‐2 carbapenemase from hospital sewage. Diagn Microbiol Infect Dis 73: 204–206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supplementary Information
Supplemental Figure S1 Core genome phylogeny of Aeromonas hydrophila GSH8‐2 among publicly available 108 strains (updated at 2018/10/29).
All publicly available draft genome sequences of Aeromonas strains were retrieved. Analysis of conserved plasmid gene sequences was performed with BLASTn searches (≥99% nt identity), followed by visualization using FriPan (http://drpowell.github.io/FriPan/). Using the complete genome sequence of GSH8‐2 (4,962,478 bp), bwa‐read mapping analysis identified 48.95% of the full genome as core‐genome sequence, and a total of 491,764 single nucleotide variations (SNVs) were identified, excluding prophages and repeated regions.
Supplemental Figure S2 Core genome phylogeny of Aeromonas caviae GSH8M‐1 among publicly available 30 strains (updated at 2018/10/29).
Using the complete genome sequence of GSH8M‐1 (4,611,279 bp), bwa‐read mapping analysis identified 70.53% of the full genome as core‐genome sequence, and a total of 229,493 single nucleotide variations (SNVs) were identified, excluding prophages and repeated regions.


