Abstract
Cell state‐, developmental stage‐, and lineage‐specific combinatorial expression of cluster of differentiation (CD) molecules enables the identification of cellular subsets via multicolor flow cytometry. We describe an exhaustive characterization of neural cell types by surface antigens, exploiting human pluripotent stem cell‐derived neural cell systems. Using multiwell screening approaches followed by detailed validation of expression patterns and dynamics, we exemplify a strategy for resolving cellular heterogeneity in stem cell paradigms. In addition to providing a catalog of surface antigens expressed in the neural lineage, we identified the transferrin receptor‐1 (CD71) to be differentially expressed in neural stem cells and differentiated neurons. In this context, we describe a role for N‐Myc proto‐oncogene (MYCN) in maintaining CD71 expression in proliferating neural cells. We report that in vitro human stem cell‐derived neurons lack CD71 surface expression and that the observed differential expression can be used to identify and enrich CD71− neuronal derivatives from heterogeneous cultures. stem cells 2019;37:1293–1306
Keywords: CD surface markers, Transferrin receptor, CD71, Neural cell selection, Cell sorting, Pluripotent stem cells, Neural stem cells, Cell transplantation, Cell therapy, Neuronal differentiation, CD49b (integrin α‐2)
Comprehensive Cell Surface Antigen Analysis Identifies Transferrin Receptor Protein‐1 (CD71) as a Negative Selection Marker for Human Neuronal Cells.
Significance Statement.
The current article illustrates the strategy to resolve cellular heterogeneity of human stem cell paradigms by conducting comprehensive surface marker screens. The results unequivocally underline the absence of the transferrin receptor protein‐1/CD71 on differentiated neurons, exploiting a broad range of different human neural in vitro differentiation models. This work identifies CD71 as a surface molecule of use for neural cell selection in biomedical stem cell paradigms.
Introduction
In vitro stem cell systems are being extensively used for a wide range of purposes, from studying the basic biology of development to disease modeling and cell therapy 1, 2. The derivation of human embryonic stem cell (hESC) lines 3 and human induced pluripotent stem cells (hiPSCs) 4 has revolutionized concepts in developmental biology and biomedical research. Through directed differentiation, pluripotent sources can be used to generate somatic cell derivatives of all three germ layers 3. Multipotent stem cells derived using such an approach can be further differentiated into defined lineages by providing recombinant factors or small molecule compounds in a time‐dependent manner. Although considerable progress has been made in optimizing differentiation protocols, the inherent heterogeneity of stem cell‐derived products remains an impediment to biomedical application. Stem cell research can greatly benefit from enhanced insights into the microenvironmental cues governing cell proliferation and phenotype establishment. Growth factor signals, cell‐matrix, and cell–cell interactions converge onto surface molecule‐mediated downstream pathways, the specifics of which remain poorly understood 5. Furthermore, characteristic combinations of surface molecules can serve to identify and isolate specific cellular subsets by fluorescence‐activated cell sorting (FACS) or immunomagnetic cell separation 6, 7, 8.
Neural stem/precursor cells (NSCs) derived from hESCs and hiPSCs can be expanded and maintained in their multipotent state in the presence of growth factors 9. By adding specific differentiation cues, NSCs can be differentiated toward neuronal and glial central nervous system cell types 10 as well as neural crest (NCR) derivatives 11, 12. Inadequate in vitro patterning and differentiation can result in cellular heterogeneity, thereby generating considerable variability in downstream applications 8, 13. Unwanted contaminating cells introduce inconsistencies and may affect subsequent scientific interpretation. Although the transplantation of neurons/neuroblasts (NEU) derived from human PSCs is on the verge of becoming a clinical reality for treating neurodegenerative disorders 14, 15, 16, residual proliferating NSCs can form tumors and yield unwanted deleterious progeny in vivo 17, 18. Therefore, eliminating unwanted cells and enriching the cell type of interest will be critical for biomedical in vitro assays, developmental studies and cell therapeutic paradigms alike. The isolation of specific neural cell types using cluster of differentiation (CD) antigens expressed on the cell surface has been established as a proof of principle 8, 19, 20, 21. However, a full characterization of the neural surfaceome is currently not available 22. By exploiting a range of complementary systems ncluding hESCs, long‐term expandable NSCs derived from hiPSCs and immortalized neural precursor cell lines, we performed a comprehensive analysis of neural CD antigen expression.
One of the differentially expressed marker candidates was more extensively investigated to exemplify the validity of our screening strategy. The CD71 epitope representing the transferrin receptor protein 1 (TFR1) is a key regulator of transferrin‐dependent iron uptake in normal development and disease. CD71 (TFR1), a type II membrane glycoprotein, is expressed as a disulfide‐linked homodimer on the cell surface, binds to iron‐bound transferrin (holo‐transferrin) and is internalized through receptor‐mediated endocytosis 23. Most notably, CD71 plays a central role to iron acquisition in erythroid precursors 24, but the protein is expressed on a broad range of cell types at varying levels 25. Rodent model studies indicated the presence of CD71 in neurons and consider transferrin‐ and TFR1‐dependent iron uptake as a major pathway for neural iron homeostasis 26, 27. Our data reveal a significant downregulation of CD71 upon neuronal differentiation in a range of human in vitro systems. Maintenance of CD71 expression in proliferative neural cells was found to be dependent on the basic helix–loop–helix transcription factor N‐Myc proto‐oncogene (MYCN), an indispensable mediator of neural development 28. CD71‐negative (CD71−) cells lacked MYCN and other neural stemness markers. Consequently, we adapted our findings to develop a CD71‐based neuronal selective vulnerability assay as well as a single marker‐based cell isolation strategy, both of which may contribute to optimizing in vitro analytical readout and (therapeutic) cell selection approaches in NSC biology.
Materials and Methods
Maintenance of Cell Lines
For cell lines and reagents, see Supporting Information Tables S7 and S8. iPSC‐NSC (AF22) and Lund human mesencephalic (LUHMES)‐NPC cells were cultured as described previously 9, 11, 29. PSCs (H9 and iPS‐IMR90‐4) were grown on feeder cells and under feeder‐free conditions. SNB‐19 astroglial cells were cultured in Dulbecco's modified Eagle's medium (DMEM) +10% fetal bovine serum 30. Details are provided in Supporting Information Experimental Procedures.
Neural Induction and hESC‐NSC and NCR Expansion
H9 hESC (WA09, WiCell) were enzymatically harvested using TrypLE and replated on Synthemax‐coated 6‐well plates at a density of 2 × 106 cells per well. Cells were grown overnight in E8 medium and switched to E8 containing 1 μM dorsomorphin and 10 μM SB431542 on the next day to initiate neural induction, with a gradual shift (over 4 days) from an E8‐based to DMEM:F12 (1:1) + N2 (1×) supplement‐based medium containing dorsomorphin and SB431542 at the aforementioned concentration. On day 12, cells were TrypLE‐passaged onto poly‐ornithine/laminin (PO/L)‐coated tissue culture flasks and maintained as NSCs in DMEM:F12 with N2 (1×), 20 ng/ml bFGF and 20 ng/ml EGF. Media was changed every day and NSCs were passaged at a seeding density of 500,000 cells per cm2 every 3 to 4 days 11. NCR cells were derived from hESC‐NSC by applying low seeding density over multiple passages as previously published 11.
Neuronal Differentiation
For hiPSC‐ and hESC‐NSC cultures, a differentiation protocol of 14 days was applied. Long‐term expandable hiPSC‐NSC (AF22) were differentiated by removing growth factor‐containing media and replacing it with differentiation media consisting of 50% DMEM:F12‐N2 and 50% Neurobasal‐B27, including 100 U/ml penicillin–streptomycin 11. hESC‐NSC were plated on PO/L‐coated plates at a density of 200,000 cells per cm2. One day after plating, growth factor media was replaced with Neurobasal‐B27 containing 20 ng/ml brain‐derived neurotrophic factor 20 ng/ml glial cell line‐derived neurotrophic factor (GDNF), 200 μM ascorbic acid, 0.5 mM dibutyryl cAMP, and 2 mM l‐glutamine. For terminal differentiation, cells were cultured for 14 days with media being changed every other day. Fresh laminin at a final concentration of 2 μg/ml was added every 4 days. To generate neural cultures enriched for dopamine neurons, established protocols were used 13, 31. LUHMES‐NPC cells were differentiated in DMEM:F12‐N2 medium with 1 μg/ml tetracyclin, 1 mM dibutyryl cAMP, and 2 ng/ml GDNF for 6 days 29.
High‐Throughput Flow Cytometric Screen and Candidate Validation
Human neural cells obtained from proliferation and differentiation conditions were screened for the expression of cell surface molecules using human BD Lyoplate screening panels as recently reported for neuroblastoma cells 32. For flow cytometric analysis, single cell suspensions were dispensed into 96‐well plates at 200,000–500,000 cells per well. Details of sample preparation for flow cytometry, cell sorting, and the antibodies (Supporting Information Table S7) used are provided in Supporting Information Experimental Procedures.
Immunofluorescence and Western Blot Analysis
For details of immunocytochemistry, Western blot protocols and antibodies (Supporting Information Table S8) used, please refer to Supporting Information Experimental Procedures.
Results
Cellular Heterogeneity of Neural Stem Cell Differentiation Systems
The goal of the present study was to identify CD surface marker signatures associated with human PSC neural lineage differentiation that would be generally applicable across cell lines independent of protocols and provenance. To this end, NSCs from two human pluripotent sources were differentiated toward neuronal phenotype (NEU) using defined in vitro protocols 9, 11, 13 (Fig. 1A). hESC‐NSC and hiPSC‐derived long‐term expandable NSC (hiPSC‐NSC; AF22) 9 cultures were predominantly SOX2 (SRY‐Box 2)‐positive (SOX2+), whereas neuronally differentiated cultures derived from either line (hESC‐NEU and hiPSC‐NEU) were largely positive for β‐III‐tubulin (TUJ1+), with few remaining interspersed SOX2+ NSC (Fig. 1B). This was confirmed by Western blot analysis, documenting an increase of TUJ1 and the mature neuronal markers synapsin and microtubule‐associated protein 2 (MAP2; Fig. 1C). Still, quantitative analysis showed a mere ≥65% of TUJ1‐ and ≥35% MAP2‐positive cells in either system (Supporting Information Fig. S1) highlighting the developmental heterogeneity of in vitro neural differentiation paradigms and the need to identify cell surface markers capable of distinguishing the various maturation stages (NSC vs. NEU). In line with previous reports 8, 19, cellular contaminants expressing neural stemness (ZO‐1, MYCN), astroglial glial fibrillary acidic protein (GFAP), and mesenchymal/NCR derivatives (ZEB1, Yes‐associated protein (YAP), peripherin) also contribute to the phenotypic diversity of neuronally differentiated cultures (Fig. 1C). In hiPSC‐NEU, a larger proportion of undifferentiated cells was observed as illustrated by the presence of SOX2+ cells (Fig. 1B) and an enhanced MYCN expression compared with hESC‐NEU (Fig. 1C). To determine subtle, yet functionally relevant variances within and across hESC‐ and hiPSC‐based differentiation systems, a confined analysis of key cell signaling pathways was conducted. Most prominent differences were identified in the activity of MAPK/ERK signaling, yet the expression and activity of other signaling pathways such as AKT and Hippo/YAP also differed between cell systems (Fig. 1C). Taken together, these data illustrate the inherent heterogeneity with respect to cell stage as well as cell state in PSC‐derived neural differentiation systems.
Figure 1.
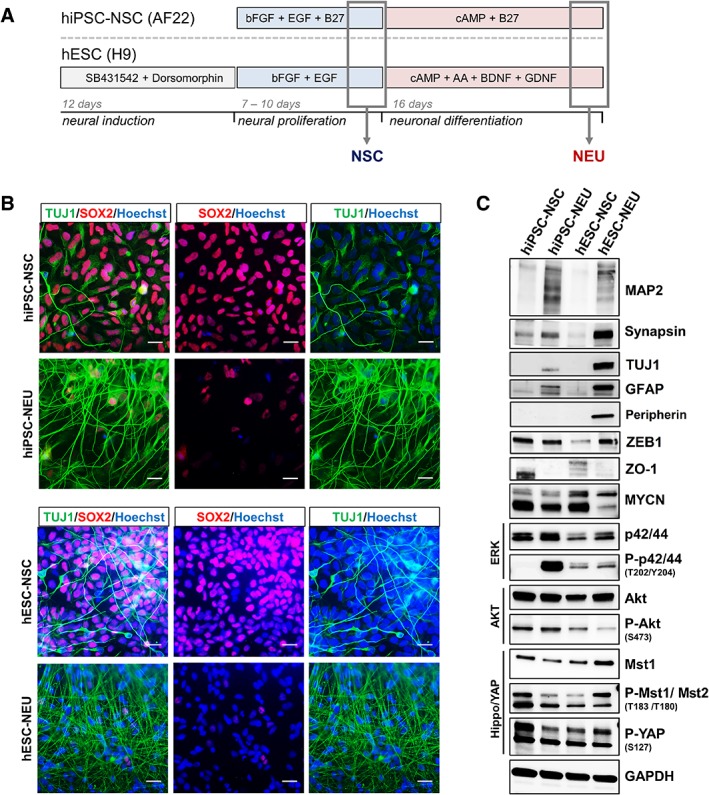
Human pluripotent stem cell‐derived neuronal differentiation systems exhibit cell stage‐ and cell state‐associated phenotypic heterogeneity. (A): Schematic outline of the neural induction and differentiation protocols used to derive proliferating neural stem cells (NSCs) and differentiated neurons (NEU) from pluripotent stem cell (PSC) sources. (B): Immunocytochemistry for the NSC marker SOX2 (red) costained with the neural differentiation marker TUJ1 (green) on human embryonic stem cell‐ as well as hiPSC‐derived proliferating NSC and differentiated NEU. Scale bars: 50 μm. (C): Western blot analysis for expression of key cell signaling pathway proteins (MAPK, AKT, JNK, hippo), neural differentiation (synapsin, MAP2, TUJ1, peripherin, ZEB1, GFAP), and proliferation (MYCN, ZO‐1) markers.
High‐Throughput Cell Surface Antigen Screens of Human Neural Stem Cells and Neurons
Unlike for the hematopoietic lineage, only a limited number of cell surface antigens has previously been assessed on human neural cells 6, 8, 11, 19, 21, 33, 34. To identify novel neural lineage markers, complementing proteomics approaches 22, a comprehensive high‐throughput human cell surface antigen screen was conducted. Most antigens analyzed using the screening panel (BD Lyoplate) are classified under the CD nomenclature 35 and have been frequently used for immunophenotyping. Applying a threshold of ≥5% as the mean percentage of positive cells, hESC‐NSC, hESC‐NEU, hiPSC‐NSC, and hiPSC‐NEU cells were screened for the expression of 242 human cell surface antigens (Supporting Information Fig. S2, Supporting Information Table S3). Of these, a mere 147 were expressed in at least one of the cell systems (Fig. 2A). We further assessed the antigens detected on each of the four cell sources individually and collectively, which helped identify the number of antigens expressed across hESC‐ and hiPSC‐based cell systems (Fig. 2B, marked in green). These 79 antigens were classified as bona fide, globally applicable “neural cell surface antigens.” The remaining 68 antigens expressed in only a subset of the cell sources tested, while not necessarily irrelevant, were neglected for the purpose of the current study. CD numbers simply represent a broadly accepted alternative terminology for known proteins, carbohydrates, and lipid epitopes expressed on the cell surface. Using the HUGO Gene Nomenclature Committee (HGNC) database, CD numbers were converted to their official gene names for subsequent gene ontology (GO) analysis 36. Such processing allowed for the functional annotation of the identified neural surface antigens focusing on their involvement in biological processes. Supporting Information Table S1 provides the complete neural CD antigen list with the corresponding official name, symbol, alternate names, HGNC ID, and Gene ID. Interactions with viruses, cell proliferation, and signal transduction were identified as the most significant biological functions of the identified neural surface antigens (Fig. 2C). As part of our current scope, Supporting Information Figure S3A focuses on the surface molecules known to participate in the process of cell proliferation. To hone in and identify potential stage‐specific markers for neural proliferation versus differentiation (NSC vs. NEU), flow cytometric validation of CD antigen expression patterns was pursued next.
Figure 2.
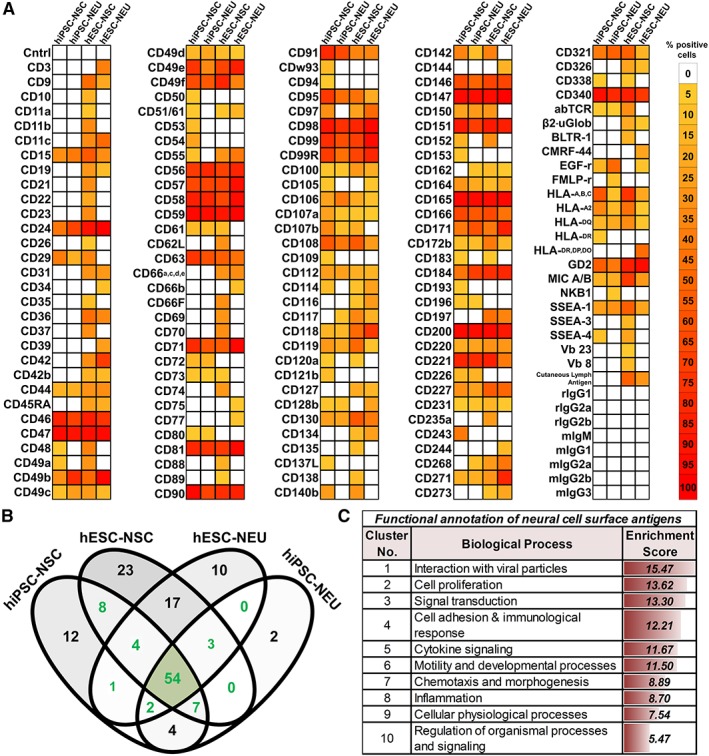
Comprehensive surface molecule screening reveals markers associated with human neural lineage development and functionality. (A): Heatmap depicting the mean percentage of cells positive for each cluster of differentiation (CD) antigen (color‐coded bar). The dataset was generated via high‐throughput flow cytometric screens of 242 human cell surface antigens in human embryonic stem cell‐ as well as hiPSC‐derived proliferating neural stem cell (NSC) versus differentiated neurons, neurons/neuroblasts (NEU; BD Lyoplate; cutoff threshold ≥5% positivity; n ≥ 2). (B): Venn diagram comparing the number of cell surface antigens exclusively expressed and/or shared (intersection) among the four different neural cell samples. Labeled in green are the antigens newly classified as “neural cell surface antigens.” The area shaded in gray highlights generalizable neural surface antigens detected in any of the PSC‐derived neural cell samples analyzed. (C): Biological functional annotation obtained after transformation of neural CD antigen nomenclature into gene names/accession numbers (Supporting Information Table S1), arranged from highest to lowest enrichment score (high enrichment score ~ lowest p‐value/highest significance).
Decrease of CD71 (TFR1) and CD49b (ITGA2) Surface Expression upon Neural Differentiation
High‐throughput screening and subsequent comparative analysis helped determine CD antigen candidates for low‐throughput characterization. A combination of arbitrarily selected 26 candidates and three known lineage‐specific markers was further validated by flow cytometry on neural stem cells (hESC‐NSC; hiPSC‐NSC) and neurons (hESC‐NEU; hiPSC‐NEU; Fig. 3A,3B; Supporting Information Table S4). Nineteen additional antigens, some of which were not part of the initial screen, were also included (Supporting Information Fig. S3B,S3C). Altogether, a systematic expression analysis of 48 cell surface antigens was performed in comparison of human neural proliferation versus neuronal differentiation. This merely represents a starting point for neural surfaceome studies, the confirmatory analysis of 37 antigens identified by the initial screen to be expressed across neural cell systems remains to be conducted.
Figure 3.
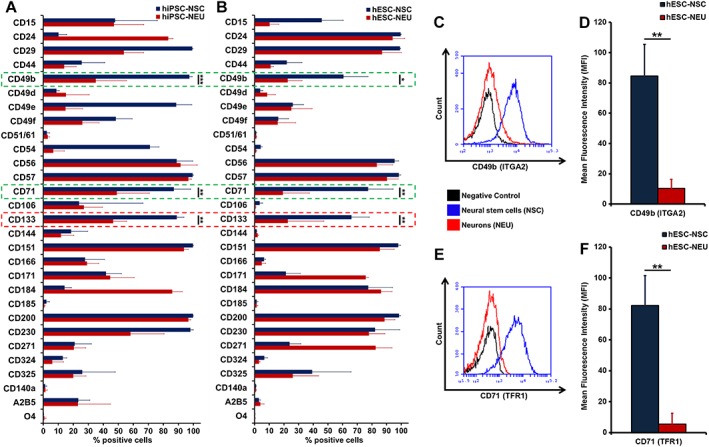
Cell surface expression of the CD71 (TFR1) and CD49b (ITGA2) antigens is downregulated upon neuronal differentiation. (A, B): Flow cytometric validation of cell surface antigen expression on human embryonic stem cell‐ and human induced pluripotent stem cell‐derived neural cells (neural stem cell [NSC]: blue and neurons/neuroblasts [NEU]: red). Bar graphs show the mean percentage of positive cells (X‐axis) for individual CD antigens (Y‐axis; n ≥ 3). (C, E): Univariate histograms representing the number of CD49b‐ and CD71‐expressing cells in NSC (blue) versus NEU (red) plotted against negative controls (black), indicating the absence of CD71 and CD49b surface expression. (D, F): Magnitude of change in cell surface CD49b and CD71 as quantified by mean fluorescence intensity, showing a significant reduction of both markers upon neural differentiation (n ≥ 3; *, p < .05; **, p ≤ .01; ***, p ≤ .001; unpaired Student's t test).
We were able to confirm the presence of known neural lineage CD markers such as CD15 (SSEA‐1), CD24, CD29 (ITGB1), CD44, CD49d (ITGA4), CD171 (L1CAM), CD184 (CXCR4), and CD271 (p75; NGFR) 8, 11, 19. Low frequencies of the oligodendrocyte precursor marker CD140a and the postmitotic oligodendrocyte marker O4 37, suggest minimal oligodendrocyte contamination in all neural cell sources analyzed here. Other markers were only seen in one of the cell systems. The overall higher number of A2B5+ cells in hiPSC‐ versus hESC‐derived neural cultures may reflect the greater proportion of neuroepithelial cells 38. Similarly, presence of CD54 (ICAM1) was only evident in hiPSC, with higher levels at the NSC stage (Fig. 3A,3B). Upon neuronal differentiation, expression levels of CD24 and CD184 increased in hiPSC, while remaining largely stable in hESC neural differentiation. On the other hand, an increase in the number of CD171+ and CD271+ cells was only seen in hESC‐NSC/NEU (Fig. 3B). These observations underline the need for screening approaches exploiting multiple in vitro cell systems when aiming to identify generally applicable neural markers.
The CD133 (Prominin1; PROM1), CD49b (Integrin alpha2; ITGA2), and CD71 (TFR1) antigens showed a significantly reduced surface expression in both systems. CD133 is routinely exploited to isolate multipotent stem cells from normal and cancerous tissue 39, including NSC 40. However, downregulation of CD133 expression (Fig. 3A) has also been observed as NSC enter a dormant/slow‐cycling G0/G1 phase of their cell cycle 41. Therefore, CD133‐negativity alone cannot be used as a hallmark for neural differentiation. The CD71 and CD49b antigens were identified as novel differentially expressed CD antigens (Fig. 3A,3B). Univariate histograms comparing the cell surface expression of CD49b and CD71 show a clear overlap of the NEU peak with the negative control (Fig. 3C,3D). The magnitude of change in cell surface expression for both markers was further quantified by comparing the corresponding mean fluorescence intensities (MFIs). Both CD49b and CD71 showed a significant reduction in MFI upon neuronal differentiation (Fig. 3D,3E; Supporting Information Fig. S4A,S4B). The low MFI levels observed in NEU cultures, we believe, are the cumulative effect of potential undifferentiated NSC or contaminant cells like NCR, astroglial cells in our in vitro NEU‐cell culture system. So far, only resting lymphocytes and mature erythrocytes have been characterized by a lack cell surface CD71 42.
Absence of CD71 (TFR1) Protein upon Neuronal In Vitro Differentiation
Transferrin/TFR1‐dependent iron uptake plays a key role in iron homeostasis and receptor‐mediated endocytosis, involving in a continuous shuttling of the protein between the cell membrane and the cytoplasm 23. As MFI‐based flow cytometric quantification only takes into consideration the presence of the protein at the cell surface, the observed reduction could be solely due to the internalization of the receptor upon differentiation. Imaging and Western blot analyses were conducted to compare total protein levels. We observed strong CD71 expression on SOX2+ NSC but not on TUJ1+ NEU cells (Fig. 4A,4B; Supporting Information Fig. S4C–S4D). Similarly, total protein analysis showed a prominent decrease in CD71 protein levels between NSCs and NEU (Fig. 4C). The reduction was more profound in hESC‐derived cells than in hiPSC‐derived cells, reflecting the maintained presence of undifferentiated, glial, and NCR cells in the hiPSC‐NEU cultures, as indicated by the higher levels of SOX2 (Figs. 1B, 4C, Supporting Information Fig. S4F), MYCN (Fig. 1C) and CD44 (Supporting Information Fig. S4E). Corroborating our cell surface data, CD49b was also downregulated upon neural differentiation (Fig. 4C). To further confirm our findings, and also in light of potential biomedical utility, we used three independent dopaminergic (DA) in vitro systems: hESC, hiPSC (iPS‐IMR90)‐derived DA neurons (hESC‐DA‐NEU) 31, and the LUHMES‐neural precursor cell line (LUHMES‐NPC) 29. Exploiting LUHMES cells, a drug‐inducible DA differentiation system, we once again observed a significant reduction in the number of both CD71+ and CD49b+ cells upon neuronal differentiation (Fig. 4D). This was corroborated by the magnitude of cell surface expression change of both proteins (MFI; Supporting Information Fig. S5A,S5B). Similarly, at the total protein level, MAP2‐negative LUHMES‐NPC expressed CD71 at high levels whereas MAP2+/tyrosine hydroxylase (TH)+/TUJ1+ LUHMES‐NEU, hESC‐DA‐NEU, and MAP2+ non‐DA hESC‐NEU barely showed any expression (immunoblot, also confirmed by immunocytochemistry; Supporting Information Fig. S5C,S5D). In hiPSC (iPS‐IMR90)‐derived DA neuronal differentiation cultures, a clear reduction of CD71 expression in comparison to the DA precursor cultures was seen (Supporting Information Fig. S5F). Low levels of CD71 expression detected in immunoblots correspond well with the considerable number of CD49b+ and CD71+ cells remaining present at the differentiation stage in PSC‐derived systems (Supporting Information Fig. S5E). From these sets of experiments, we could confidently conclude that in vitro derived human‐NEU do not express CD71 (TFR1).
Figure 4.
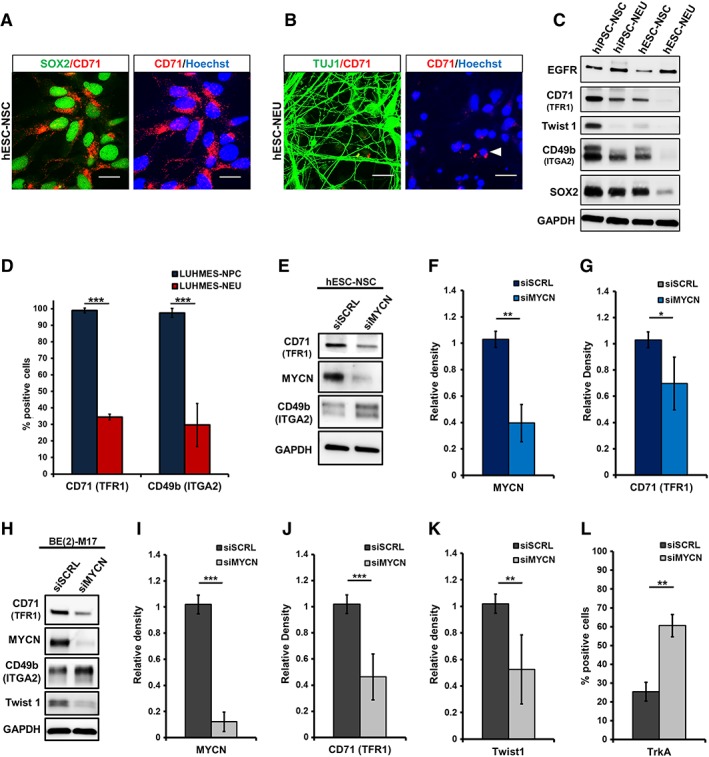
Alteration in total protein expression of CD71 (TFR1) and CD49b (ITGA2) upon neuronal differentiation and MYCN‐knockdown. (A): Immunocytochemistry for CD71 (red), costained with neural stem/precursor cell (NSC)‐marker SOX2 (green) in human embryonic stem cell (hESC)‐NSC. (B): hESC‐neurons/neuroblasts (NEU) stained for CD71 (red) and NEU/NB‐marker TUJ1 (green); Hoechst (blue: nuclei). (C): Western blot analysis of NSC and NEU derived from hESC and hiPSC. Samples were probed for EGFR, CD71, Twist1, CD49b, and SOX2; loading control: GAPDH. (D): Proliferative LUHMES midbrain precursor cells (NPC) and differentiated LUHMES cells (NEU) analyzed by flow cytometry for CD71 and CD49b surface expression. The bar graph represents the mean percentage of positive cells for both markers (n ≥ 4). (E): Western blotting for siRNA‐mediated knockdown (KD) of MYCN in hESC‐NSC. (F, G): Quantification shows a significant downregulation of MYCN and TFR1 (n = 3). (H): MYCN‐KD in MYCN amplified neuroblastoma cell line BE(2)‐M17. Immunoblotting demonstrates the downregulation of CD71, MYCN, and Twist1 upon MYCN knockdown. Scrambled siRNA (siSCRL) and GAPDH were used as controls. (I, J, K): Western blot quantification for MYCN, CD71, and Twist1 post‐MYCN‐KD (n = 6). (L): Flow cytometric quantification of TrKA expression upon MYCN‐KD showed a significant increase in the number of TrKA+ cells (n = 3). Knockdown of MYCN (N‐Myc) significantly downregulated CD71 (TFR1) expression (scale bar: 20 μM; *, p < .05; **, p ≤ .01; ***, p ≤ .001; unpaired Student's t test).
N‐Myc Regulates CD71 (TFR1) Expression on Proliferating Neural Cells
N‐Myc (MYCN) governs one of the key gene networks critical for normal brain development 28 and knockdown of MYCN leads to embryonic lethality 43, 44. Myc proteins regulate the expression of their targets by binding to regulatory elements of the gene, predominantly the E‐box consensus binding sites 45. Studies using B‐lymphocytes have previously identified c‐Myc (MYCC) to bind E‐box sequences upstream of the TFR1 gene 46. However, any potential role of MYCN in regulating CD71 expression on proliferating neural cells has not been addressed. To this end, we conducted a siRNA‐mediated knockdown (KD) of MYCN in hESC‐NSC (Fig. 4E) and a MYCN‐amplified neuroblastoma cell line (Fig. 4H), were we also observe a co‐expression of MYCN with CD71 (Supporting Information Fig. S6B). Unlike neuroblastoma cells, hESC‐NSC cells were prone to cell death when exposed to high doses of MYCN siRNA. Therefore, hESC‐NSCs were exposed to a subthreshold siRNA concentration to achieve a conservative ∼2.9‐fold KD of MYCN (Fig. 4F). As a consequence of MYCN‐KD a significant decrease in CD71 (TFR1) protein was observed (Fig. 4G). A profound KD (∼11.4‐fold) of MYCN protein could be achieved on neuroblastoma cells using the siRNA approach (Fig. 4I), which resulted in a significant downregulation of CD71 (Fig. 4J) and Twist1 (Fig. 4K) total protein expression. Twist1 was used as the positive control for the assay, as it a well‐characterized direct transcriptional target of MYCN 47. In contrast to the effect on CD71 (TFR1), we observed an increase in CD49b (ITGA2) as well as TrkA expression upon MYCN‐KD (Fig. 4E,4H,4L). An increase in TrKA‐positivity as a consequence of MYCN inhibition is well established for neuroblastoma 48. Treatment of NSC's with reversible (Roscovitine) and irreversible (Mitomycin C) proliferation inhibitors (Supporting Information Fig. S7A,S7C) which do not directly target MYCN‐proliferation network were unable to influence CD71 expression (Supporting Information Fig. S7B,S7C). Our data therefore suggest that the MYCN‐gene regulatory network, and not the proliferative network alone of a cell, is critical for the expression of CD71 (TFR1) in proliferating neural cells, yet whether this occurs by directly binding to its upstream E‐box or via an indirect pathway remains to be elucidated.
Pluripotent Stem Cells, NCR, and Astroglial Cells Express High Levels of CD71 (TFR1)
Undifferentiated NSCs within neuronally differentiated cultures represent merely one of the contributors to cellular heterogeneity. Other key contaminants include PSC that failed to undergo neural induction as well as NCR and astroglial cells 18, 19. To evaluate the possibility of using negativity for CD71 or CD49b to isolate NEU cells from heterogeneous cultures, we assessed the expression of these marker candidates on neural in vitro culture contaminants. Importantly, both pluripotent stage hESC and hESC‐NCR cells expressed CD71 and CD49b on their surface (≥85% of cells; Fig. 5A). As determined by MFI, human hESC‐NCR exhibited a higher expression level per cell of the migration‐associated integrin subunit CD49b in comparison to pluripotent stage hESC, which was also verified by Western blotting (Fig. 5B,5C). Although vimentin+/Twist1+/SOX2− hESC‐NCR also seemed to have higher CD71 total protein expression, this difference was not significant at the cell surface (Fig. 5B,5D). Hence, the higher cell surface levels of CD49b (ITGA2) could potentially be exploited to distinguish NCR cells (CD71+/CD49bhigh) from hESC, and NSC (both CD71+/CD49b+) as well as NEU (CD71−/CD49b−). Astrocytes generated from NSCs 49 and the glioblastoma cell line SNB‐19 were explored to quantify the expression of CD71 and CD49b in the astroglial lineage. Prominent CD71 and CD49b expression was observed (Fig. 5E) and immunocytochemistry showed coexpression of the glial marker GFAP with CD71 in YAP+/ZO‐1+/vimentin+/MAP2−, PSC‐derived astrocytes as well as SNB‐19 cells (Fig. 5F, Supporting Information Fig. S8A) For CD71, a clear positive correlation with cell proliferation is already known, as it is often overexpressed on a wide range of malignant tissues including glioma, neuroblastoma and colon and pancreatic cancers 50, 51. As a result, various anti‐TFR1 (CD71) tumor‐targeting approaches have been developed 52, 53. Gambogic acid (GA), a naturally occurring, potent anticancer agent 54, is one of the compounds known to trigger apoptosis by binding to TFR1 55. In the context of our study, we treated in vitro neural cultures with GA, which resulted in the induction of apoptosis of in vitro hiPSC‐NSC, but not of CD71− hiPSC‐NEU. GA at a minimum concentration of 2 μM caused a considerable loss of cells in NSC culture (Fig. 5G,5H) within 3 hours, whereas this effect was not observed in neuronally differentiated cells even at a concentration of 4 μM (Fig. 5G, Supporting Information Fig. S8B). Apoptosis was also triggered in other CD71+ multipotent neural cells (hESC‐NSC; hESC‐NCR) and pluripotent stem cells (hESC; Fig. 5I). In contrast, we did not observe cell loss of differentiated neuronal cultures (LUHMES‐NEU, hESC‐NEU, hiPSC‐NEU) when exposed to the identical treatment paradigm (Fig. 5J). Thus, the lack of a toxic response to the known TFR1‐ligand GA further underlines the absence of functional expression of the CD71 antigen on differentiated neuronal cells.
Figure 5.
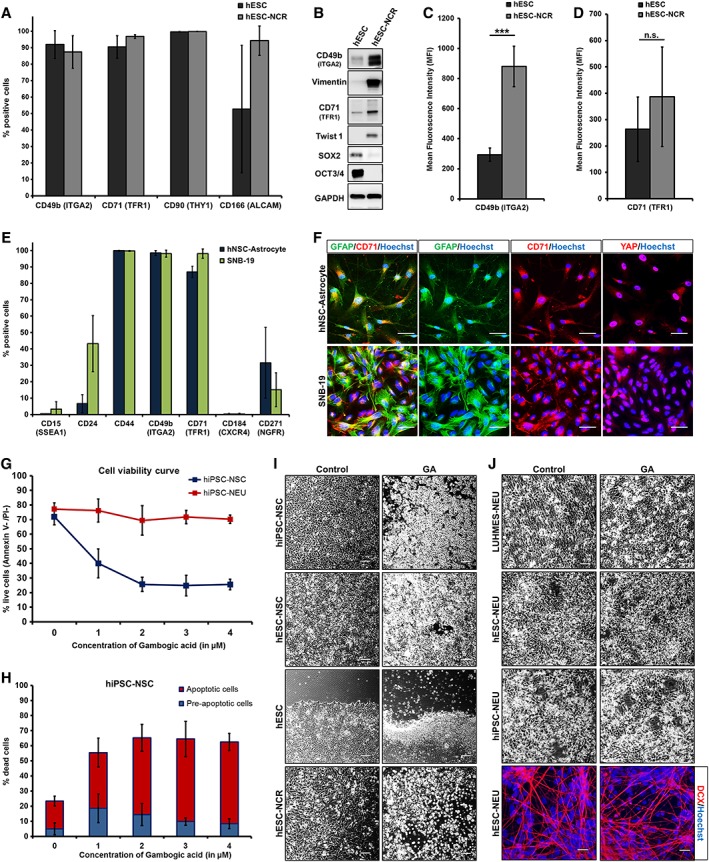
CD71 (TFR1) and CD49b (ITGA2) are expressed on pluripotent stem cells (human embryonic stem cell [hESC]) neural crest (NCR) and astroglial cells. (A): Pluripotent hESC and hESC‐derived NCR cells analyzed by flow cytometry for plasma membrane expression of CD49b, CD71, CD90 (THY1), and CD166 (ALCAM; n ≥ 3). CD71 and CD49b expression is observed in >85% cells. (B): Validation of total protein expression of CD49b, vimentin, CD71, SOX2, OCT3/4, and Twist1. (C): Magnitude of cell surface CD49b expression quantified by mean fluorescence intensity (MFI; n ≥ 4). hESC‐NCR show a significantly higher surface expression of CD49b per cell. (D): CD71 expression quantified by fluorescence intensity (MFI; n ≥ 4). (E): Flow cytometric analysis of hESC‐derived astrocytes and SNB‐19 glioblastoma cells (n = 3). (F): Immunocytochemistry staining of glial cells for GFAP, CD71, and YAP. (G): Gambogic acid (GA) triggers cytotoxicity by targeting cell surface CD71 (TFR1). Dose–response curve for cell viability of hiPSC‐derived neural stem/precursor cell (NSC) and neurons/neuroblasts (NEU) treated with GA for 3 hours. (H): Estimation of live, preapoptotic and apoptotic cells by quantifying annexin‐V/propidium iodide (n = 4). (I): Phase contrast images comparing control (DMSO) versus GA treatment for CD71‐expressing PSC, NSC, and NCR. (J): Phase contrast and immunocytochemistry of NEU cells for control (DMSO) and GA treatments. When applied to CD71− NEU cells, GA treatment was unable to trigger a similar cytotoxic response (scale bar: 50 μM; *, p < .05; **, p ≤ .01; ***, p ≤ .001; unpaired Student's t test).
CD71 (TFR1) as a Marker to Evaluate Cell Type‐Specific Vulnerability for Cytotoxic Compounds
Drug screening platforms heavily rely on human in vitro cell models to evaluate the action of the respective compounds. In order to examine the options of using CD71 as a marker for selective vulnerability studies, we used A23187, a calcium ionophore with defined neurotoxic activity 13. Using a previously established protocol 31, TH+/synapsin+ DA neuronal (hESC‐DA‐NEU) cultures, also containing a small fraction of undifferentiated OTX2+/SOX2+ DA precursors, were derived for the assay (Fig. 6A). Cells were treated with known DA‐NEU toxins A23187, ferrous chloride (FeCl2), and DMSO (vehicle control). Treatment with A23187 resulted in a loss of the neural network, retraction of neurites (Fig. 6B), and significant cell death. After treatment, live cells were analyzed for a combination of surface proteins. A significantly higher number of CD71+ viable cells were identified post‐A23187 treatment (Fig. 6C). Considerable variability in the number of CD49b+ cells was observed under control condition and no significant difference was detected post‐A23187 treatment, pointing to a potential variability of CD49b expression patterns in DA neurons (Fig. 6C). No change was observed with respect to CD166 (ALCAM), a DA precursor/neuron marker 6, 34, which we found to also be expressed by NCR (Fig. 5A). Consistent with our observations, treatment with A23187 resulted in the loss of TH+/synapsin+/MAP2+ DA neurons, represented by the CD71− fraction. Cells remaining in the dish after A23187 treatments were positive for FOXA2, a DA precursor/neuron marker as well as CD71 (Fig. 6D). Thus, using negativity for CD71 as a surrogate marker, the selective cytotoxicity of small molecules (such as A23187) toward mature neurons (DA‐NEU) can be can be assessed in pharmacological screens to rapidly and quantitatively determine neuronal viability, susceptibility and neuroprotection even in heterogeneous neural cell culture systems.
Figure 6.
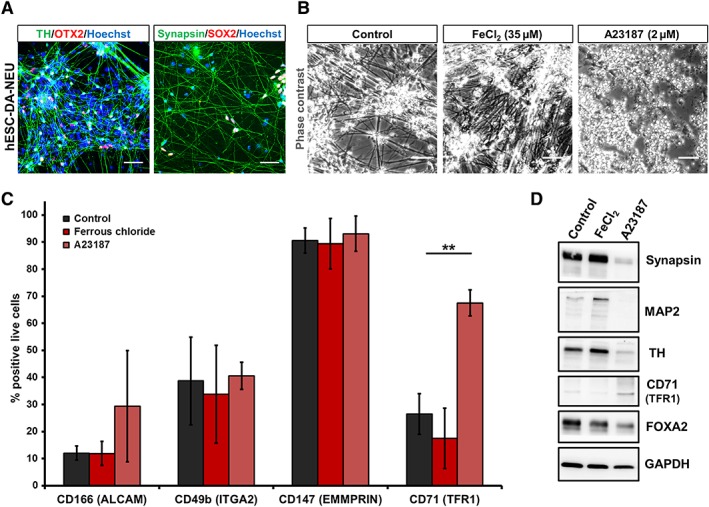
Exploiting negativity for CD71 (TFR1) to assess selective vulnerability of neurons to cytotoxic small molecules. (A): Immunocytochemistry staining of DA neurons (human embryonic stem cell [hESC]‐dopaminergic [DA]‐neurons/neuroblasts [NEU]) for TH, OTX2, SOX2, and synapsin. (B): Phase contrast image of DA‐NEU treated with vehicle control, FeCl2 (35 μM), and A23187 (2 μM) for 24 hours. Cell death and disruption of neural network was observed upon A23187 treatment. (C): Post‐treatment flow cytometry‐based analysis for the expression of CD166, CD49b, CD147, and CD71. Post‐A23187 treatment, a significant number of viable cells was CD71 and CD166 positive (n = 4). (D): Western blotting analysis post‐treatment (scale bar: 50 μM; *, p < .05; **, p ≤ .01; ***, p ≤ .001; one‐way analysis of variance with post hoc Tukey test).
Isolation of NEU Fraction from Heterogeneous In Vitro Neural Cultures Using Fluorescence‐Based and Magnetic Immunoseparation
Following the confirmation that CD71 and CD49b are expressed on PSC, NSC, and NCR but not on NEU, we exploited the identified difference to develop novel cell selection strategies. Current NEU cell isolation either requires a combination of cell surface markers or multiple FACS experiments to enrich the target cell 8, 13, 19. We isolated CD71+/CD49b+ and CD71−/CD49b− subpopulations from hESC‐NEU cultures at 10 days of differentiation using FACS. The CD71−/CD49b− population was enriched for DCX+ and TUJ1+ NEU cells (Fig. 7A). DCX+/TUJ1+ were nearly absent in the CD71+/CD49b+ subset. To simplify the sorting method and given the availability of CD71 antibody for clinically approved magnetic immunoseparation, we next pursued single‐marker CD71‐negative selection for NEU enrichment. Having demonstrated the neuronal identity of CD71−/CD49b− cells, we further assessed the coexpression of CD71 and CD49b in order to evaluate the possibility of using CD49b‐negative selection alone as an additional strategy to enrich neurons. To this end, CD49b expression was analyzed on the CD71+ and CD71− subpopulations (Supporting Information Fig. S8C). Although the majority (≥90%) of CD71+ cells were CD49b+, CD71− cells showed heterogeneous CD49b expression on their cell surface (Supporting Information Fig. S8D). Furthermore, only a fraction (≤50%) of CD49b‐postive cells expressed CD71 (Supporting Information Fig. S8E), thereby highlighting the inability of CD49b+ versus CD49b− selection to provide optimal resolution for cell sorting. Based on these findings and to simplify the sorting method, hiPSC‐NEU cells were subjected to CD71‐negative versus positive selection by FACS (Sort purity in Supporting Information Fig. S7D). Similar to our previous observation the CD71− cell fraction exhibited a TUJ1+/DCX+ neuronal phenotype (Fig. 7B). In contrast, the CD71+ fraction comprised a mixture of Ki67+ proliferative cells also positive for GFAP and nestin while being negative for TUJ1 and DCX (Fig. 7B,7C). Five days post‐CD71− sorting, the viability and functionality of the isolated mature neurons were evaluated by whole‐cell current clamp (Supporting Information Fig. S9). All recorded cells exhibited a clear neuronal phenotype in which action potential could be evoked after current injection (8/8 cells) or occurred spontaneously (5/8; Supporting Information Fig. S9A), and showed clear neurites extending from the soma (Supporting Information Fig. S9B). This CD71− selection strategy was then replicated using immunomagnetic cell separation (MACS) on a heterogeneous mixture of hESC‐derived neural cells. The CD71− subset was Ki67−/GFR1alpha+/high/MAP2+/synapsin+/TH+ and immunoreactive for DCX as well as MAP2 highlighting the presence of NEU cells (Fig. 7D,7E). These data illustrate that the selection of CD71− cells represents a viable option to enrich for neuronally differentiated cells in a range of neural differentiation systems. As previously stated, CD71+ expression alone cannot distinguish between NSC, NCR, and PSC. Hence, for better resolution between NCR versus NSC and PSC, high CD49b expression could be exploited (Fig. 5C).
Figure 7.
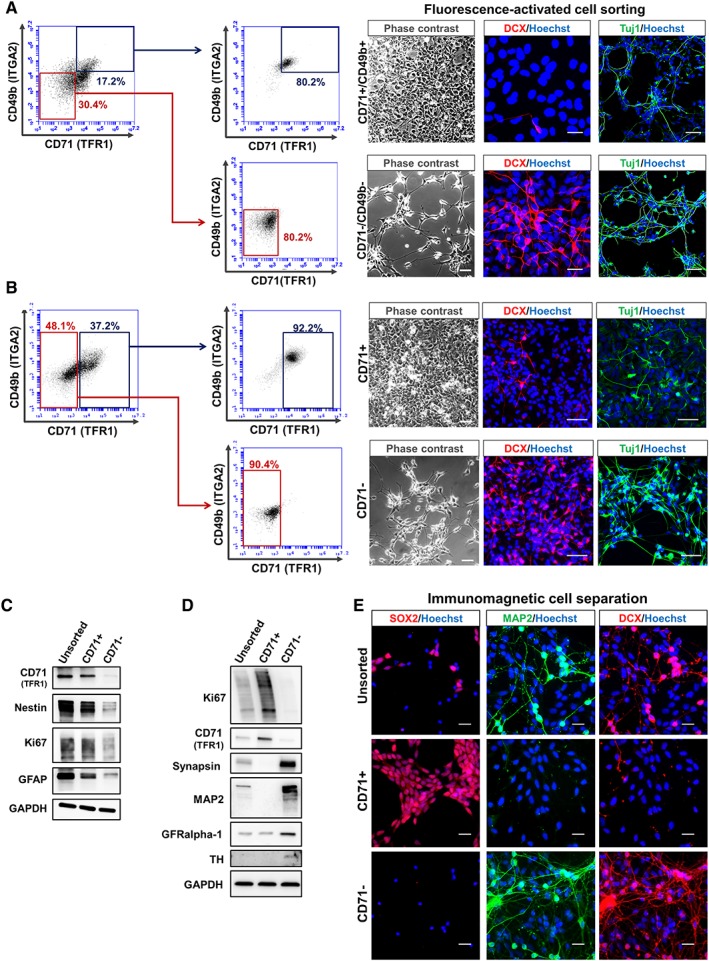
Using CD71− selection to isolate neuronal cells (neurons/neuroblasts [NEU]) from heterogeneous PSC‐derived neural in vitro cultures. (A): Immunocytochemistry and phase contrast images of CD49b+/CD71+ and CD49b−/CD71− subpopulations isolated by fluorescence‐activated cell sorting (FACS). (B): FACS sorting CD71+ and CD71− cells from hiPSC‐NEU cultures. DCX+ (Doublecortin) and TUJ1+ NEU were enriched in the CD49b−/CD71− and CD71− only fractions. (C): Immunomagnetic separation of CD71+ and CD71− subpopulations from heterogeneous neural cell cultures containing proliferating (neural stem/precursor cell/neural precursor cell) and differentiated cells (NEU/NB). Separation was followed by Western blot analysis for the isolated subsets versus unsorted. The CD71− fraction was Ki67−/synapsin+/MAP2+/TH+. (D): Immunocytochemical analysis of cellular subsets isolated via immunomagnetic cell separation and the unsorted fraction. SOX2−/DCX+/MAP2+ cells were predominantly isolated in the CD71− subset (scale bar: 50 μM).
Discussion
Cells express a plethora of antigens on their plasma membrane. Intracellular transmission of external stimuli, cell adhesion, and transport of cargo across the cell membrane are among the common functions associated with cell surface molecules. Characteristic combinations of these antigens can define the identity of a cell. Hematopoietic and immunological research has greatly benefited from capitalizing on the extensive number of lineage‐defining cell surface molecules 56, 57. In contrast, for the human neural lineage only a limited number of antigens have been explored 7, 8, 19, 21. The comprehensive in vitro screening approach on multiple cell sources including postmitotic neurons provided here represents a foundation for cataloging CD marker expression dynamics on proliferating as well as differentiated human neural cells.
Among the neural cell systems tested, 95 antigens of the 242 tested in the original screen were found to exhibit little to no expression (<5% positive cells). These include known T cell markers (CD4, CD5, CD6, CD8a/b, CD45) as well as antigen presenting proteins (CD1a, CD1b, CD1d, CD83), CD62P (P‐selectin), CD274 (PD‐L1), and CD279 (PD‐1). Of the markers in fact identified as present on neural cells, the majority is prominently involved in the interaction with viruses, cell proliferation, and signal transduction (Fig. 2C). Of note, the deleterious effects of infection by Zika and other neurotropic viruses on neural precursor cells have recently sparked an interest in studying the participation of cell surface proteins in viral entry and susceptibility of neural cells to such pathogens 58. Many of the candidate surface antigens identified in this study, including the CD71 (TFR1) have previous reported interactions with viruses 59, and may therefore provide new leads toward future research in these fields.
Expression of CD49b (integrin α‐2; ITGA2) has previously been observed on human NSCs 60, but follow‐up analysis regarding how levels may vary upon exiting the stem cell stage has been limited. Although the current study focused on enrichment of neurons, the expression pattern of CD49b (ITGA2) could potentially be exploited as a marker for the enrichment of NCR progeny. NCR cells were found to exhibit a significantly higher CD49b expression on their cell surface compared with NSC, NEU, and PSC. Consequently, a CD71+/CD49bhigh cell selection strategy could be used to isolate this cell type. To discriminate between NSC and PSC in vitro, the CD71/CD49b code would have to be combined with pluripotency‐associated surface markers such as SSEA‐4, SSEA‐3, and CD9 61. Thereby, one could define NEU (CD71−/CD49b−), NCR (CD71+/CD49bhigh), NSC (CD71+/CD49b+; SSEA‐3/−4/CD9‐negative), and PSC (CD71+/CD49b+; SSEA‐3/−4/CD9‐positive).
Across multiple in vitro human neural cell systems, a significant reduction of CD71+ cells was observed upon neuronal differentiation, the CD71 (TFR1) protein neither being expressed on the cell surface nor intracellularly. Compared with CD49b, CD71 showed a less variable expression pattern (Supporting Information Figs. S5E, S8C–S8E), a clear association with stemness (Fig. 4E–4G) and a greater sensitivity for detecting neuronal cell loss (Fig. 6C). As a consequence of its pivotal role in iron homeostasis, nearly all cell types are expected to express CD71 on their cell surface, although at varying levels 25. Therefore, the observation that in vitro human neurons are CD71− is unique from a biological perspective. In proliferating neural cells, the expression of CD71 was identified to be dependent on MYCN activity, yet whether the absence of CD71 in NEU is directly due to the downregulation of the neural cell proliferation protein MYCN remains open.
Exploiting the differences in CD71 expression in the neural lineage, we could highlight a simplified method to enrich neurons and use CD71 as a marker for selective vulnerability assays, in PSC‐derived neural cultures. Cellular heterogeneity of these in vitro cell systems remains a common concern in disease modeling and cell therapy and developing strategies to resolve this heterogeneity is an important necessity for the advancement toward NSC‐derived therapeutics 16, 20, 21. The negative selection method toward elimination of Ki67+ proliferating cells and enrichment of neurons (CD71−) as well as the described CD71‐based cytotoxicity read‐out can facilitate biomedical advancement in cell therapy and drug discovery. Tissue level comparison of TFR1 transcript expression in humans using Harmonizome database 62 further supports the low presence of CD71 in CNS‐neurons (Supporting Information Fig. S6A).
Taken together, this study provides an exhaustive analysis of neural CD surface antigen expression and describes CD71 as a novel, differentially expressed surface marker in the neural lineage. Beyond the utility of CD71 for cell selection, CD71‐mediated contribution to neural iron metabolism and differential susceptibility to virus in human neurons lend themselves to future in‐depth investigation. Ultimately, the extended list of markers of neuropoiesis may further aid in elucidating cell–cell interactions involved in the regulation of growth and differentiation in neural development, stemness, and cancer.
Author Contributions
V.M.: performed majority of the experiments, experimental design, data analysis and interpretation, wrote the manuscript, critically reviewed and approved the final manuscript; R.T.: performed human pluripotent stem cell‐based derivation of NCR cells, critically reviewed and approved the final manuscript; C.E., M.B.: planned and executed electrophysiological testing, critically reviewed and approved the final manuscript; M.H.: assisted with stem cell culture and flow cytometric analysis, critically reviewed and approved the final manuscript; T.O.: contributed to dopaminergic differentiation, critically reviewed and approved the final manuscript; P.J.H., O.I.: experimental design, data analysis and interpretation, critically reviewed and approved the final manuscript; J.P.: experimental design, data analysis and interpretation, wrote the manuscript, critically reviewed and approved the final manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
Supporting information
Appendix S1: Supporting Information
Acknowledgments
We thank M. Follo, D. Herchenbach, and K. Geiger of the Lighthouse Core Facility, Medical Center—University of Freiburg, for cell sorting and A. Falk (Karolinska Institute) for providing the hiPSC‐NSC (AF22) cell line. This study was supported by the Emmy Noether‐Program of the German Research Foundation (DFG; PR1132/3‐1; J.P.) and supported in part by the DFG Excellence Initiative (GSC‐4, Spemann Graduate School of Biology and Medicine), and scholarship funds from the State Graduate Funding Program of Baden‐Württemberg (V.M.). V.M. is currently affiliated with the Wellcome Trust/CRUK Gurdon Institute, Wellcome Trust/MRC Stem Cell Institute, Department of Pathology, University of Cambridge, Cambridge, United Kingdom; J.P. is currently affiliated with the Institute of Anatomy and Cell Biology, Paracelsus Medical University, Salzburg, Austria.
Contributor Information
Vishal Menon, Email: v.menon@gurdon.cam.ac.uk.
Jan Pruszak, Email: jan.pruszak@pmu.ac.at.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Zhu Z, Huangfu D. Human pluripotent stem cells: An emerging model in developmental biology. Development 2013;140:705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol 2011;13:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thomson JA, Itskovitz‐Eldor J, Shapiro SS et al. Embryonic stem cell lines derived from human blastocysts. Science 1998;80:282. [DOI] [PubMed] [Google Scholar]
- 4. Takahashi K, Tanabe K, Ohnuki M et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861–872. [DOI] [PubMed] [Google Scholar]
- 5. Pruszak J. Neural Surface Antigens: From Basic Biology Towards Biomedical Applications. London, Oxford: Elsevier, Inc., 2015. [Google Scholar]
- 6. Doi D, Samata B, Katsukawa M et al. Isolation of human induced pluripotent stem cell‐derived dopaminergic progenitors by cell sorting for successful transplantation. Stem Cell Rep 2014;2:337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Panchision DM, Chen H‐L, Pistollato F et al. Optimized flow cytometric analysis of central nervous system tissue reveals novel functional relationships among cells expressing CD133, CD15, and CD24. Stem Cells 2007;25:1560–1570. [DOI] [PubMed] [Google Scholar]
- 8. Pruszak J, Ludwig W, Blak A et al. CD15, CD24, and CD29 define a surface biomarker code for neural lineage differentiation of stem cells. Stem Cells 2009;27:2928–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Falk A, Koch P, Kesavan J et al. Capture of neuroepithelial‐like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS One 2012;7:e29597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reubinoff BE, Itsykson P, Turetsky T et al. Neural progenitors from human embryonic stem cells. Nat Biotechnol 2001;19:1134–1140. [DOI] [PubMed] [Google Scholar]
- 11. Hindley CJ, Condurat AL, Menon V et al. The Hippo pathway member YAP enhances human neural crest cell fate and migration. Sci Rep 2016;6:23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee G, Kim H, Elkabetz Y et al. Isolation and directed differentiation of neural crest stem cells derived from human embryonic stem cells. Nat Biotechnol 2007;25:1468–1475. [DOI] [PubMed] [Google Scholar]
- 13. Schöndorf DC, Aureli M, McAllister FE et al. iPSC‐derived neurons from GBA1‐associated Parkinson's disease patients show autophagic defects and impaired calcium homeostasis. Nat Commun 2014;5:4028. [DOI] [PubMed] [Google Scholar]
- 14. Hallett PJ, Cooper O, Sadi D et al. Long‐term health of dopaminergic neuron transplants in Parkinson's disease patients. Cell Rep 2014;7:1755–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hallett PJ, Deleidi M, Astradsson A et al. Successful function of autologous iPSC‐derived dopamine neurons following transplantation in a non‐human primate model of Parkinson's disease. Cell Stem Cell 2015;16:269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kikuchi T, Morizane A, Doi D et al. Human iPS cell‐derived dopaminergic neurons function in a primate Parkinson's disease model. Nature 2017;548:592–596. [DOI] [PubMed] [Google Scholar]
- 17. Roy NS, Cleren C, Singh SK et al. Functional engraftment of human ES cell‐derived dopaminergic neurons enriched by coculture with telomerase‐immortalized midbrain astrocytes. Nat Med 2006;12:1259–1268. [DOI] [PubMed] [Google Scholar]
- 18. Brederlau A, Correia AS, Anisimov SV et al. Transplantation of human embryonic stem cell‐derived cells to a rat model of Parkinson's disease: Effect of in vitro differentiation on graft survival and teratoma formation. Stem Cells 2006;24:1433–1440. [DOI] [PubMed] [Google Scholar]
- 19. Yuan SH, Martin J, Elia J et al. Cell‐surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS One 2011;6:e17540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Isacson O. Sorting the wheat from the chaff in dopamine neuron‐based cell therapies. Proc Natl Acad Sci USA 2015;112:4512–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lehnen D, Barral S, Cardoso T et al. IAP‐based cell sorting results in homogeneous transplantable dopaminergic precursor cells derived from human pluripotent stem cells. Stem Cell Rep 2017;9:1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tyleckova J, Valekova I, Zizkova M et al. Surface N‐glycoproteome patterns reveal key proteins of neuronal differentiation. J Proteomics 2016;132:13–20. [DOI] [PubMed] [Google Scholar]
- 23. Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: Molecular control of mammalian iron metabolism. Cell 2004;117:285–297. [DOI] [PubMed] [Google Scholar]
- 24. Andrews N, Levy JE, Jin O et al. Transferrin receptor is necessary for development of erythrocytes and the nervous system. Nat Genet 1999;21:396–399. [DOI] [PubMed] [Google Scholar]
- 25. Ponka P, Lok CN. The transferrin receptor: Role in health and disease. Int J Biochem Cell Biol 1999;31:1111–1137. [DOI] [PubMed] [Google Scholar]
- 26. Leitner DF, Connor JR. Functional roles of transferrin in the brain. Biochim Biophys Acta 2012;1820:393–402. [DOI] [PubMed] [Google Scholar]
- 27. Matak P, Matak A, Moustafa S et al. Disrupted iron homeostasis causes dopaminergic neurodegeneration in mice. Proc Natl Acad Sci USA 2016;113:3428–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Knoepfler PS, Cheng PF, Eisenman RN. N‐myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev 2002;16:2699–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lotharius J, Barg S, Wiekop P et al. Effect of mutant alpha‐synuclein on dopamine homeostasis in a new human mesencephalic cell line. J Biol Chem 2002;277:38884–38894. [DOI] [PubMed] [Google Scholar]
- 30. Turaç G, Hindley CJ, Thomas R et al. Combined flow cytometric analysis of surface and intracellular antigens reveals surface molecule markers of human neuropoiesis. PLoS One 2013;8:e68519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kriks S, Shim J‐W, Piao J et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature 2011;480:547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ferlemann FC, Menon V, Condurat AL et al. Surface marker profiling of SH‐SY5Y cells enables small molecule screens identifying BMP4 as a modulator of neuroblastoma differentiation. Sci Rep 2017;7:13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gennet N, Tamburini C, Nan X et al. FolR1: A novel cell surface marker for isolating midbrain dopamine neural progenitors and nascent dopamine neurons. Sci Rep 2016;6:32488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bye CR, Jönsson ME, Björklund A et al. Transcriptome analysis reveals transmembrane targets on transplantable midbrain dopamine progenitors. Proc Natl Acad Sci USA 2015;112:E1946–E1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Engel P, Boumsell L, Balderas R et al. CD nomenclature 2015: Human leukocyte differentiation antigen workshops as a driving force in immunology. J Immunol 2015;195:4555–4563. [DOI] [PubMed] [Google Scholar]
- 36. Gray KA, Yates B, Seal RL et al. Genenames.org: The HGNC resources in 2015. Nucleic Acids Res 2015;43:D1079–D1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cahoy JD, Emery B, Kaushal A et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci 2008;28:264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rao MS, Mayer‐Proschel M. Glial‐restricted precursors are derived from multipotent neuroepithelial stem cells. Dev Biol 1997;188:48–63. [DOI] [PubMed] [Google Scholar]
- 39. Wu Y, Wu PY. CD133 as a marker for cancer stem cells: Progresses and concerns. Stem Cells Dev 2009;18:1127–1134. [DOI] [PubMed] [Google Scholar]
- 40. Peh GS‐L, Lang RJ, Pera MF et al. CD133 expression by neural progenitors derived from human embryonic stem cells and its use for their prospective isolation. Stem Cells Dev 2009;18:269–282. [DOI] [PubMed] [Google Scholar]
- 41. Sun Y, Kong W, Falk A et al. CD133 (Prominin) negative human neural stem cells are clonogenic and tripotent. PLoS One 2009;4:e5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Judd W, Poodry CA, Strominger JL. Novel surface antigen expressed on dividing cells but absent from nondividing cells. J Exp Med 1980;152:1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stanton BR, Perkins AS, Tessarollo L et al. Loss of N‐myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev 1992;6:2235–2247. [DOI] [PubMed] [Google Scholar]
- 44. Charron J, Malynn BA, Fisher P et al. Embryonic lethality in mice homozygous for a targeted disruption of the N‐myc gene. Genes Dev 1992;6:2248–2257. [DOI] [PubMed] [Google Scholar]
- 45. Zeller KI, Zhao X, Lee CWH et al. Global mapping of c‐Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA 2006;103:17834–17839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. O'Donnell KA, Yu D, Zeller KI et al. Activation of transferrin receptor 1 by c‐Myc enhances cellular proliferation and tumorigenesis. Mol Cell Biol 2006;26:2373–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Selmi A, de Saint‐Jean M, Jallas A‐C et al. TWIST1 is a direct transcriptional target of MYCN and MYC in neuroblastoma. Cancer Lett 2015;357:412–418. [DOI] [PubMed] [Google Scholar]
- 48. Brodeur GM, Minturn JE, Ho R et al. Trk receptor expression and inhibition in neuroblastomas. Clin Cancer Res 2009;15:3244–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shaltouki A, Peng J, Liu Q et al. Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells 2013;31:941–952. [DOI] [PubMed] [Google Scholar]
- 50. Ryschich E, Huszty G, Knaebel HP et al. Transferrin receptor is a marker of malignant phenotype in human pancreatic cancer and in neuroendocrine carcinoma of the pancreas. Eur J Cancer 2004;40:1418–1422. [DOI] [PubMed] [Google Scholar]
- 51. Szekeres T, Sedlak J, Novotny L. Benzamide riboside, a recent inhibitor of inosine 5‐monophosphate dehydrogenase induces transferrin receptors in cancer cells. Curr Med Chem 2002;9:759–764. [DOI] [PubMed] [Google Scholar]
- 52. Daniels TR, Bernabeu E, Rodríguez JA et al. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim Biophys Acta 2012;1820:291–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Martell LA, Agrawal A, Ross DA et al. Efficacy of transferrin receptor‐targeted immunotoxins in brain tumor cell lines and pediatric brain tumors. Cancer Res 1993;53:1348–1353. [PubMed] [Google Scholar]
- 54. Guo Q‐L, You Q‐D, Wu Z‐Q et al. General gambogic acids inhibited growth of human hepatoma SMMC‐7721 cells in vitro and in nude mice. Acta Pharmacol Sin 2004;25:769–774. [PubMed] [Google Scholar]
- 55. Kasibhatla S, Jessen KA, Maliartchouk S et al. A role for transferrin receptor in triggering apoptosis when targeted with gambogic acid. Proc Natl Acad Sci USA 2005;102:12095–12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Seita J, Weissman IL. Hematopoietic stem cell: Self‐renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med 2010;2:640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lv F‐J, Tuan RS, Cheung KMC et al. Concise review: The surface markers and identity of human mesenchymal stem cells. Stem Cells 2014;32:1408–1419. [DOI] [PubMed] [Google Scholar]
- 58. Wen Z, Song H, Ming G‐L. How does Zika virus cause microcephaly? Genes Dev 2017;31:849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci USA 2013;110:10777–10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sundberg M, Jansson L, Ketolainen J et al. CD marker expression profiles of human embryonic stem cells and their neural derivatives, determined using flow‐cytometric analysis, reveal a novel CD marker for exclusion of pluripotent stem cells. Stem Cell Res 2009;2:113–124. [DOI] [PubMed] [Google Scholar]
- 61. Adewumi O, Aflatoonian B, Ahrlund‐Richter L et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol 2007;25:803–816. [DOI] [PubMed] [Google Scholar]
- 62. Rouillard AD, Gundersen GW, Fernandez NF et al. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016;2016:baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


