Abstract
Microbes in indoor environments are constantly being exposed to antimicrobial surface finishes. Many are rendered non‐viable after spending extended periods of time under low‐moisture, low‐nutrient surface conditions, regardless of whether those surfaces have been amended with antimicrobial chemicals. However, some microorganisms remain viable even after prolonged exposure to these hostile conditions. Work with specific model pathogens makes it difficult to draw general conclusions about how chemical and physical properties of surfaces affect microbes. Here, we explore the survival of a synthetic community of non‐model microorganisms isolated from built environments following exposure to three chemically and physically distinct surface finishes. Our findings demonstrated the differences in bacterial survival associated with three chemically and physically distinct materials. Alkaline clay surfaces select for an alkaliphilic bacterium, Kocuria rosea, whereas acidic mold‐resistant paint favors Bacillus timonensis, a Gram‐negative spore‐forming bacterium that also survives on antimicrobial surfaces after 24 hours of exposure. Additionally, antibiotic‐resistant Pantoea allii did not exhibit prolonged retention on antimicrobial surfaces. Our controlled microcosm experiment integrates measurement of indoor chemistry and microbiology to elucidate the complex biochemical interactions that influence the indoor microbiome.
Keywords: antimicrobial resistance, antimicrobial surface paint, bacterial viability, efficacy of antimicrobial products, indoor microbiome, sporulation
Practical Implications.
The study is one of the few controlled microcosm experiments integrating indoor chemistry and indoor microbiology perspectives, disentangling the complex interactions between surface chemistry and surface microbes.
We applied a defined model community to set a foundation for monitoring the viable surface microbiome.
Our observation adds to the current knowledge on the behavior of spore‐forming bacteria and highlights the importance of these organisms, particularly those which are pathogenic, in indoor environments from a public health perspective.
1. INTRODUCTION
Modern humans spend the majority of their time indoors, surrounded by various microorganisms and material‐associated chemicals. Application of DNA sequencing technologies has revealed the importance of the indoor microbiome in both building operation and occupant health.1 Issues such as human exposure to infectious microorganisms are of significant public health relevance. In 2002, hospital‐acquired infections were associated with 98,987 deaths in the United States alone,2 and so‐called nosocomial infections remain one of the leading causes of mortality.3 Biological exposures associated with mold and dampness also raise concerns regarding occupant exposure to indoor allergens4 as well as building deterioration. To prevent microbial degradation, antimicrobial additives are commonly supplemented into product formulations, ranging from daily‐use personal care products to building materials. Moreover, innovations in engineering “self‐decontaminating” surfaces5 not only aim to prevent biological deterioration of products but also work toward killing microbes on contact with surfaces. Various antimicrobial agents have been used in the fabrication of “self‐decontaminating” surfaces5 for both medical device coatings and latex paint formulations.6, 7, 8, 9 Antimicrobial ingredients such as silver nanoparticles6, 7; crystal violet9; a combination of crystal violet, methylene blue, safranin O, and gold nanoparticles8; and cationic quaternary ammonium salts (QAS)10 have been successfully integrated into latex paints. To assess their antimicrobial performance, researchers often follow the Japanese Industrial Standard (JIS) Z 2801 as a mainstream antimicrobial efficacy testing, utilizing laboratory test strains, mainly Staphylococcus aureus and Escherichia coli, which is a poor representation of the tremendous diversity of Gram‐positive and Gram‐negative bacteria.11, 12, 13 Such assessment heavily relies on enumerating colonies on solidified medium and consequently uses cultivability as a proxy for viability. Tests using populations of these strains on antimicrobial surfaces exhibit significant log reductions in cultivatable colony‐forming units (CFUs). However, the efficacy of these antimicrobials against non‐model microorganisms isolated from indoor environments is largely unproven, and it is unknown whether such activity is necessary or sufficient to influence human health.
Green building materials, for example, clay paints, that are labeled as “eco‐friendly” and “all‐natural” are becoming increasingly market‐popular to meet consumer need for carbon neutrality and sustainable living.14, 15 Green building materials often have characteristics such as “low toxicity, minimal chemical emissions, high recyclability, and long durability”.15 A study done by Mensah‐Attipoe J et al reveals the impacts of different surfaces, including green materials, on fungal communities and experimentally illustrates that different surface materials support the growth of different fungi, but are always dependent on available water.15 While other microbiome and metabolome studies have been performed to explore the growth of bacteria and fungi on different surface materials,16 no studies have explored the impact of “green” surface materials on community dynamics. In addition, some clays have been used in other contexts as antimicrobial agents due to the presence of metal ions that release potent hydroxyl radicals upon being oxidized17, 18; however, the effect of clay surface finishes on the indoor microbiome is unknown.
Our objectives were to evaluate how exposure to natural and synthetic surface finishes, with or without embedded QAS, affects viability of non‐model bacteria isolated from indoor environments. Specifically, the study tested the hypotheses that (a) chemically and physically distinct surfaces will favor the survival of different members of the surface microbiome, and (b) the exposure of microbes to antimicrobial surfaces selects for antimicrobial‐resistant microbes. This is the first study addressing the efficacy of biocidal surface finishes and green building materials using a synthetic community composed of bacteria isolated from indoor environments. This is in contrast with industrialized antimicrobial efficacy standard that uses well‐studied model organisms.
2. MATERIALS AND METHODS
2.1. Test Surface Materials
We selected three commercially available surface paints to characterize their corresponding properties on mold‐resistant drywall surfaces:
a conventional interior acrylic latex paint advertised as including “a VOC‐free formula” that helps improve indoor air quality by reducing VOC levels from potential sources, for example, carpet, cabinets, and fabrics;
an EPA‐registered microbicidal interior latex paint that kills greater than 99.9% of Staphylococcus aureus, MRSA (methicillin‐resistant Staphylococcus aureus), Escherichia coli, VRE (vancomycin‐resistant Enterococcus faecalis), and Enterobacter aerogenes within two hours of exposure on painted surfaces; and
a clay paint that is claimed to be “odorless, non‐toxic and VOC‐free with no chemical biocides.”
To simulate a wall surface environment in a typical indoor environment, mold‐resistant drywall was cut into 2″ × 2″ squares. To minimize the difference in paint surface topography due to uneven paint distribution, each individual coupon was painted consistently twice, including a day of drying in between two coatings.
2.2. Surface pH, conductivity, and moisture content measurements
Following the surface preparations, the pH of each painted surface was measured by adding 1 mL of de‐ionized water on a painted square and allowing the water droplet to equilibrate for one minute. pH readings were obtained from wetted Fisherbrand™ Plastic pH Strips (Thermo Fisher Scientific, Waltham, MA, USA). To measure surface conductivity, five coupons were randomly selected for each paint and 200 µL of DI water was added to each corner (n = 20). Water droplets were allowed to equilibrate for 4 minutes and measured using a HORIBA B‐771 LAQUAtwin Compact Conductivity meter (Kyoto, Japan).
Microcosm experiments were performed in sealed glass jars to maintain a constant humidity. Following the surface preparations, all glass jars and caps were autoclaved and dried. The relative humidity was measured using Fisherbrand™ Traceable™ Humidity‐On‐A‐Card™ Humidity Monitor (Waltham, MA, USA) under three treatment conditions:
dry, 10 g of UV‐sterilized silica gel in a glass jar,
ambient, and
wet, 10 mL of autoclaved milli‐Q water.
Painted drywall coupons were placed in sealed glass jars under each of the three treatment conditions for seven days. The weight of each coupon was recorded after 7 days of treatment as well as after drying at 105°C for 3 days. Gravimetric moisture content was defined as follows.
2.3. Test inoculum
Five isolates, Bacillus timonensis, Enterococcus hirae, Kocuria rosea, Microbacterium oleivorans, and Pantoea allii, cultivated from dust samples collected from athletic facilities in the Pacific Northwest and were selected to build a synthetic community. In preparation of the surface inoculum, individual isolates were grown in tryptic soy broth (TSB) to its late exponential phase. Cells were spun down and resuspended in phosphate‐buffered saline. 100 µL mixed cell suspension was then transferred onto each UV‐sterilized square, and a pipette tip was used to spread liquid culture over the surface. Then, each square was positioned on a small petri dish and transferred into a mason jar (Figure 1) with forceps. Each mason jar can hold up to two 2″ × 2″ squares as technical replicates. All squares were kept as level as possible. Lastly, negative controls were conducted by pipetting 100 µL phosphate‐buffered saline (PBS) onto painted squares. Three humidity levels and three surface finishes were included in this study, resulting in nine total exposure conditions. Moreover, three biological replicates (n = 3) were conducted.
Figure 1.
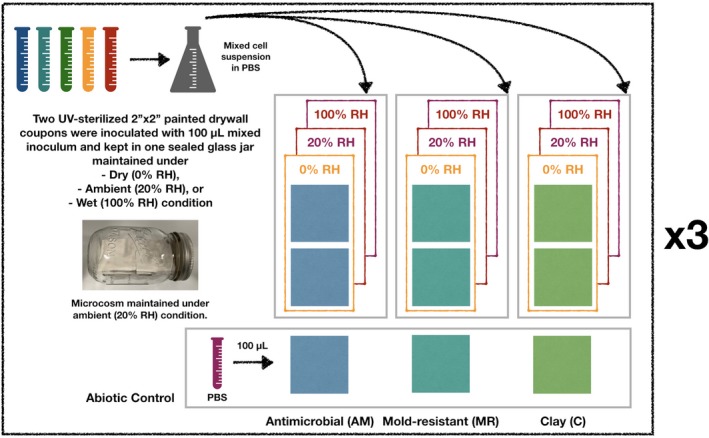
Microcosm setup for three types of surface exposure under dry (0% RH), ambient (20%), and wet (100% RH) conditions (n = 3). Within each biological replicate, two technical replicate surfaces (both AM, both MR, or both Clay) were UV‐sterilized for 30 min, inoculated with 100 µL mixed inoculum, and kept in a single glass jar for up to 7 d. Within each glass jar, we assumed that a uniform and constant humidity level was achieved shortly after the jar was sealed
2.4. Whole‐genome sequencing
To obtain whole‐genome sequences for primer design, isolates were grown for 24‐48 hours in TSB with continuous shaking. Cells were then pelleted by centrifugation at 10 000 g for 3 minutes. DNA was extracted using MasterPure™ Complete DNA and RNA Purification Kit (Lucigen, Middleton, WI, USA), following manufacturer's instructions. DNA quality was measured using a Synergy HTX Multi‐Mode Reader (BioTek, Winooski, VT, USA) and was of acceptable quality if the 260/280 ratio was found to be between 1.8 and 2.0. DNA was quantified using the Quant‐iT™ PicoGreen™ dsDNA Assay Kit (ThermoFisher Scientific, Waltham, MA). Libraries were prepared and indexed using the Nextera XT DNA Library Preparation and Index Kits (Illumina, San Diego, CA) following manufacturer's instructions. Fragment lengths of prepared libraries were measured on an Agilent 2100 Bioanalyzer (Santa Clara, CA). Libraries were then normalized and pooled in preparation for sequencing on an Illumina MiSeq with the MiSeq Reagent Kit v3 for 2x300 bp reads (Illumina, San Diego, CA, USA). The software KneadData (https://bitbucket.org/biobakery/kneaddata/wiki/Home) was then used for raw sequence quality control and contaminant removal using default parameters. Short reads were assembled into contigs using SPAdes,19 and the quality of five draft genomes was assessed using CheckM.20 For each test strain, a primer set was designed using Primer‐BLAST21 and Primer3,22, 23 targeting single‐copy unique functional genes. To assess the genetic potential of each strain, contigs were further annotated with Prokaa24 and the RAST server.25, 26 To further complement Illumina short reads, Bacillus timonensis was also sequenced using Oxford Nanopore MinION. gDNA extraction, library preparation, and flow cell priming were conducted following the ultra‐long read sequencing protocol for RAD004 (https://www.protocols.io/view/ultra-long-read-sequencing-protocol-for-rad004-mrxc57n). Raw sequence reads and five draft genomes were deposited under BioProjectPRJNA524667.
16S rRNA sequences were available for a subset of 436 isolates cultivated from dust samples collected from athletic facilities in the Pacific Northwest. DNA extraction, library preparation, 16S rRNA amplicon sequencing, and taxonomic identification (Data S1) were conducted following the Earth Microbiome Project, targeting the V4 region of the 16S SSU rRNA.27, 28, 29
2.5. Sample recovery and PMA treatment
Swab samples were collected through a combination of dry and wet swabbing. Each surface was first dry swabbed for 5 seconds, and the swab was swirled into an aliquoted PBS buffer. Surfaces were then wet swabbed for a total of 20 seconds, including 5 seconds of rinse and rewet. Finally, the swab was squeezed against the 2‐mL tube to expel any residual liquid. 100 µL swab sample along with its dilutions was spread onto TSA plates, and cell numbers were counted after 5 days.
Diluted solution was also equally divided for propidium monoazide(PMA) treatment and non‐PMA control. 1.25 µL PMA (20 mmol/L, Biotin, Fremont, CA) was added to 1 mL swab suspension. PMA binding was activated using two 500 W halogen lamps facing the samples at a distance of 1 foot. Samples were left on an ice bed for cooling. Tubes were rotated by 180 degrees every 2.5 minutes and vortexed for 5 seconds every 5 minutes for 15 minutes. Treated samples were then stored at −80°C for DNA extraction and qPCR analyses.
2.6. gDNA extraction and qPCR
Genomic DNA was extracted using the DNeasy Powersoil kit (Qiagen, Venlo, The Netherlands) following the manufacturer's recommended procedures. Each qPCR contained 10 μL 2x PowerUp™ SYBR™ Green Master Mix (Applied Biosystem, Foster City, CA), 0.7 μL forward and reverse primers (350 nmol/L), 4.6 μL nuclease‐free water, and 4 μL DNA template (10‐100 ng). Details of primer sequences and qPCR thermal profile are listed in Table S2. qPCR was conducted using QuantStudio 3 Real‐Time PCR System (Applied Biosystem, Foster City, CA), including a 2‐minute pre‐melt at 95°C and 45 cycles of 15‐second denaturation at 95°C, 15‐second annealing at 52‐54°C, and 1‐minute extension at 72°C. A dissociation curve was generated for each run following manufacturer's recommended conditions with ramp increment of 1.6°C/s from 72°C to 95°C, 1.6°C from 95°C to 60°C, and 0.15°C/s from 60°C to 95°C. Each 96‐well plate contained 24 samples, up to 7 standards, and one no‐template control in triplicates (efficiency > 88% and R 2 > 0.99). Copy numbers obtained from amplifying each functional gene were first normalized to the sum of copy numbers from all five primers and then normalized to the viable copy number of each strain within the mixed inoculum.
2.7. Paint diffusivity assay
The paint diffusivity assay was conducted according to Møretrø et al30 with slight modifications. 2″ × 2″ painted coupons were gently placed onto trypticase soy agar (TSA) plated with individual test strains upside down, allowing painted surfaces to have sufficient contact with TSA (Figure S3). The dimensions of the clearing zone around each coupon were recorded. All five strains were tested individually in triplicate, resulting in 12 measurements for each combination of test strain and surface paint. Individual strains were grown to late exponential phase as described above and inoculum concentrations ranged from 3 × 109/mL to 2 × 1010/mL.
2.8. Visualization of Bacillus spores
Swab suspension samples were first filtered using 5‐µm Thermo Scientific™ Target2™ Syringe Filters (ThermoFisher Scientific, Waltham, MA) to remove dust particles. The filtrate was then concentrated using Vivaspin 10 000 MW concentrator (GE Healthcare Biosciences, Chicago, IL). Following a standard sample preparation for Scanning Electron Microscopy,31 concentrated samples were fixed in 5% glutaraldehyde for 30 minutes and washed in 0.1 mol/L phosphate buffer (pH 7.2) and distilled water. Samples were dehydrated in 35%, 50%, 75%, 95%, and lastly 100% absolute ethanol. Dehydrated samples were dried using hexamethyldisilizane (Electron Microscopy Sciences, Hatfield, PA) and sputter‐coated with 4 mm gold using a Cressington 208HR Sputter Coater (Watford, UK), respectively. Samples were imaged using a Hitachi S‐4800 Field Emission Scanning Electron Microscope (Tokyo, Japan).
2.9. Statistical analysis
Both Welch t test and paired t test were conducted using R (version 3.3.3). Results were considered statistically significantly different with P < 0.05.
3. RESULTS AND DISCUSSION
3.1. Moisture content, pH, and surface conductivity
We tested moisture‐absorbing capacity for three different surfaces under ambient relative humidity ranging from 0% to 100% (Figure 2). Under low relative humidity levels (0% and 20%), all surfaces exhibited similar gravimetric moisture content (P > 0.05), whereas under RH 100%, mold‐resistant surfaces showed the highest moisture content among three types of surfaces. The moisture content of clay surfaces was significantly lower than that of mold‐resistant and antimicrobial surfaces (P < 0.05).
Figure 2.
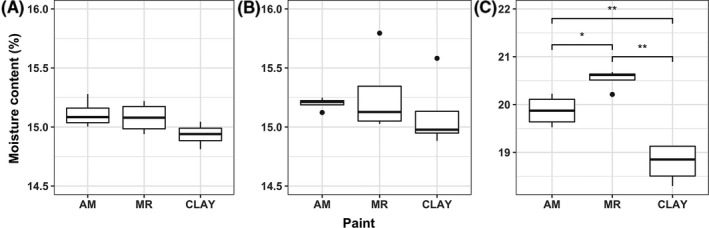
Gravimetric moisture content (%) for Clay (C), Mold‐resistant (MR), and Antimicrobial (AM) surfaces under three ambient relative humidity conditions: (A) 0% RH by adding 10 g desiccant, (B) 20% RH, and (C) 100% RH by supplementing 10 mL sterilized DI water. (*P < 0.05 and **P < 0.01 from Welch's t test)
pH measurements (Table 1) revealed a clear distinction between the surface chemistry of clay and latex paints. Both antimicrobial and mold‐resistant latex paints created slightly acidic environments, in contrast to the alkaline environment on clay surfaces. Surface conductivity (Figure 3) was measured as an indicator for the strength of ion dissociation from painted surfaces. In good agreement with pH measurements, surface conductivities also showed a clear chemistry difference between latex and clay paints. Clay surfaces were significantly different from mold‐resistant and antimicrobial surfaces, but there was no statistically significant difference between the two latex paints, which are commercialized by the same manufacturer. Bacterial membranes serve as the first line of defense from unfavorable environmental conditions. Therefore, ionized molecules leached from surfaces will be either attracted or repelled by the negatively charged membrane at initial contact, potentially leading to disruption of cell functions.
Table 1.
Surface conductivity and average roughness measurements for test surfaces
| Surface | pH | Surface conductivity (mS/cm) | Average roughness (µm) |
|---|---|---|---|
| Clay | 10 | 1.92 ± 0.36 | 19.94 ± 0.36 |
| Mold‐resistant | 4 | 0.49 ± 0.07 | 13.43 ± 1.15 |
| Antimicrobial | 4 | 0.47 ± 0.11 | 11.48 ± 0.79 |
Figure 3.
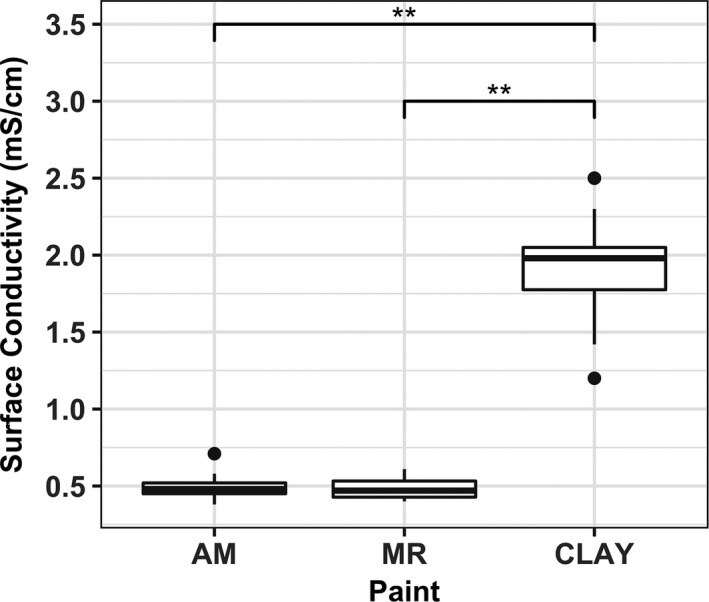
Surface conductivity measurements for Clay (C), Mold‐resistant (MR), and Antimicrobial (AM) surfaces (*P < 0.05 and **P < 0.01 from Welch's t test)
3.2. Selection of test inoculum
We selected five isolates—Bacillus timonensis, Enterococcus hirae, Kocuria rosea, Microbacterium oleivorans, and Pantoea allii, from a collection of 7655 isolates cultivated from over 100 dust samples collected from athletic facilities in the Pacific Northwest.32 All five isolates were recovered from indoor dust, suggesting that they have encountered various stresses in the past, for example, desiccation and oxidation, although the exact duration of exposure is unknown. The morphologies of these strains combined accounted for 37.72% of total morphological diversity (Table 2 and Table S3). Community selection based on morphological abundance, although it did not comprehensively capture the entire diversity of the indoor microbiome, did contain microorganisms that are often abundant indoors, such as Bacillus 33, 34 and Kocuria.34 At the genus level, the five isolates combined represented 21.24% of the diversity captured in 16S rRNA sequences of a subset of 436 isolates for which taxonomic assignments were available (Table 2 and Figure S1). However, the synthetic community covered only 0.8% of the diversity described in a metagenomic analysis (Table 2). Such discrepancies between culture‐independent metagenomic approaches and cultivation approaches have been widely acknowledged.35, 36 As metagenomics is not indicative of viability, our approach using a synthetic community composed of cultivable isolates leads to insights on phenotypic responses rather than deposition of genetic material, laying foundations for future work addressing inter‐species interactions.
Table 2.
Genus‐level abundance comparison among 16S rRNA gene sequences,27, 28, 29 metagenomics,32 and morphological diversity32
|
16S rRNA Genus‐level abundance (%) |
Metagenomics Genus‐level abundance (%) |
Morphological diversity (%) | |
|---|---|---|---|
| Bacillus timonensis | 14.64 | NA | 15.70 |
| Enterococcus hirae | 3.418 | 0.07529 | 12.18 |
| Kocuria rosea | 1.636 | 0.2696 | 6.545 |
| Microbacterium oleivorans | 1.538 | NA | 1.803 |
| Pantoea allii | 0.003148 | 0.8632 | 1.489 |
| Total | 21.34 | 1.2081 | 37.72 |
3.3. Paint diffusivity assay
Indoor microbes are exposed to a variety of continuously occurring stressors in the indoor environment. Despite the fact that culturing comes with certain downfalls, as only a small fraction of bacterial cells can be cultured in the laboratory and culturable bacteria may lose cultivability due to different stressors,37 it remains one of the most efficient and cost‐effective proxies for viability. Here, we used cultivability as a proxy for viability reduction caused specifically by surface exposure by modifying the agar diffusion assay from Møretrø et al30 This assay simulated a scenario where exposure to surface‐available antimicrobial agents becomes dominant over the effects of common indoor stresses such as desiccation and nutrient deprivation. In this scenario, the size of the zone of inhibition was determined by the release rate of antimicrobial agents from the surface finish, as well as the microbial resistance against bioavailable chemicals. Among all surface finishes, the antimicrobial surface had the largest zone of inhibition for all five strains tested (Figure 4). All strains except Pantoea allii showed decreasing sensitivity toward antimicrobial, mold‐resistant, and clay surfaces. It is perhaps not surprising that, although neither clay nor mold‐resistant is explicitly advertised as “bactericidal” surface finishes, both demonstrated inhibition effects against the strains tested (Figure 4).
Figure 4.
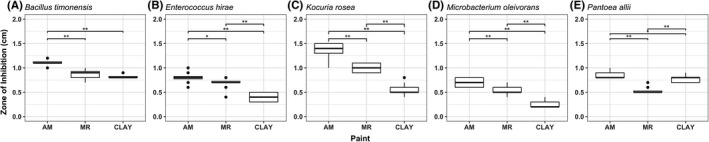
Zone of inhibition (cm) created by Antimicrobial (AM), Mold‐resistant (MR), and Clay (C) surfaces for all five test strains (*P < 0.05 and **P < 0.01). In this paint diffusivity and sensitivity test, thin layers of bacterial culture applied on TSA plates were subjected to various levels of growth inhibition due to the diffusivity of surface‐associated chemicals. The size of the zone of inhibition is determined by the combined effect of antimicrobial release from the surface finish and microbial resistance against bioavailable chemicals
3.4. Microbial composition and viability reduction in relation to surface chemistry
The most significant decrease in viable population occurred between the initial inoculation and 24 hours following the exposure to surfaces (2.41‐5.96 log10 reduction in PMA‐qPCR viable copy numbers; Figure 5C). Viable copy numbers stabilized toward the end of the monitoring period (Day 7) but still had small fluctuations, which may be due to the utilization of cell debris as organic sources. Similarly, Deng et al observed increased levels of rRNA degradation of Salmonella enterica under starvation and desiccation in peanut oil38 as degraded rRNA can serve as a nutrient source.38, 39 Within each viable population, different surfaces favored strains at different survival rates. Antimicrobial and mold‐resistant paint both supported the prolonged survival of B timonensis, whereas the clay surfaces favored the survival of Kocuria rosea (Figure 5A,B).
Figure 5.
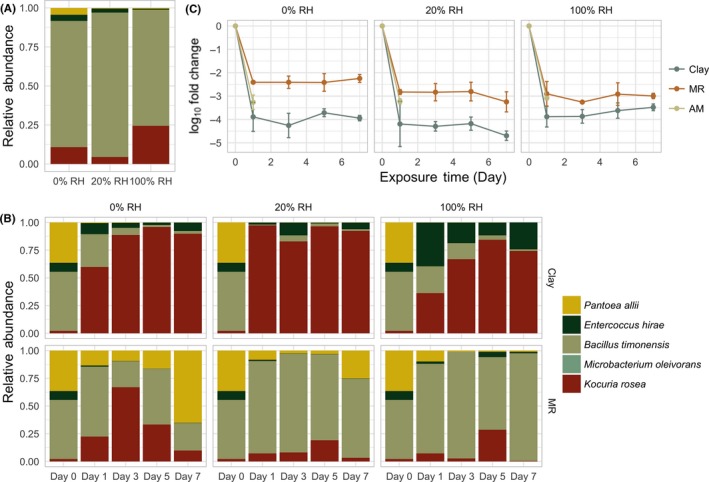
Normalized microbial composition recovered from antimicrobial (A) and other (B) surfaces under 0%, 20%, and 100% relative humidity, and log10 fold changeover time (C). log10 fold changes associated with Clay and MR were significantly different across all humidity levels (paired t test P < 0.05; P‐value 0.01855, 0.01625, and 0.026 for 0%, 20%, and 100% RH, respectively). Swab samples from non‐antimicrobial surfaces (MR and Clay) were collected on Days 1, 3, 5, and 7, whereas samples from antimicrobial surfaces were only collected on the day following the inoculation
Throughout the monitoring period, Microbacterium oleivorans consistently had less than ten copies detected by PMA‐qPCR, likely a consequence of insufficient input in the initial inoculum (Day 0). M.oleivoranson average represented less than 0.05% of the total population (Figure 5B). In subsequent samples, M oleivorans was consistently below the limit of detection for qPCR, so a panel for M oleivorans was not included in Figure 6.
Figure 6.
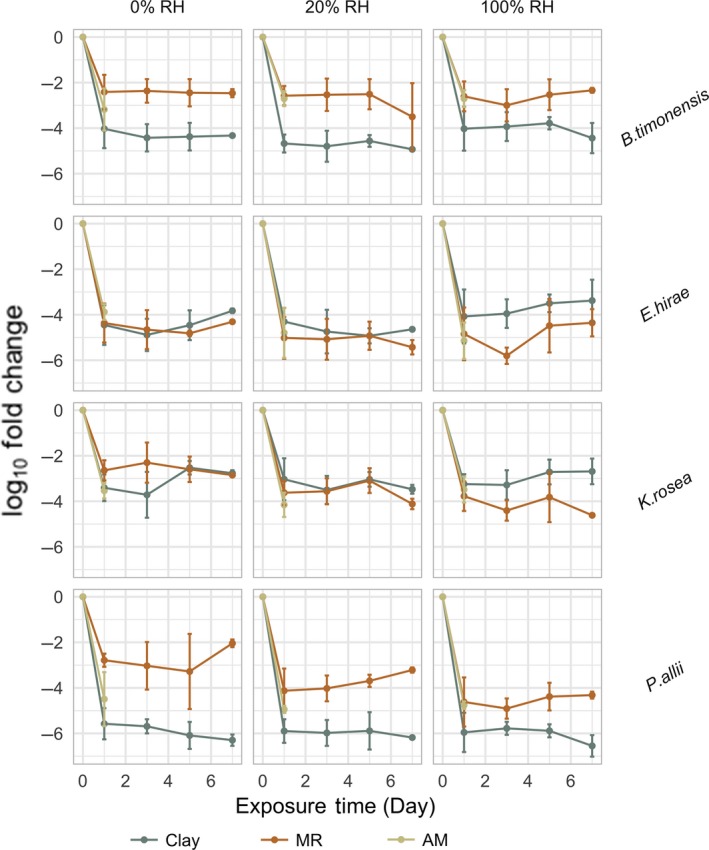
PMA‐qPCR log10 fold change normalized to the starting abundance of each species. Both B timonensis and P allii demonstrated statistically significant differences in surface survival on MR and Clay surfaces for all humidity levels (0%, 20%, and 100%), whereas the survival patterns for E hirae and K rosea were only significantly different under 100% RH (Table S2). Swab samples from MR and Clay surfaces were collected on Days 1, 3, 5, and 7, whereas samples from the antimicrobial surfaces were only collected on the day following the inoculation
Both B timonensis and P allii demonstrated statistically significant differences between mold‐resistant(MR) and clay surface survival for all humidity levels (0%, 20%, and 100%), whereas the survival patterns for E hirae and K rosea were only significantly different under 100% RH (Table S2 and Figure 6). Despite the observation that both B timonensis and K rosea demonstrated relatively successful retention on surfaces, they exhibited different survival patterns. We consistently recovered more B timonensis from MR than clay, whereas K rosea exhibited the opposite pattern under 100% RH.
To determine whether the pH of the different paints was contributing to selection within the microbial community, we tested the tolerance of each individual community member to variations in pH in liquid media. Specifically, we tested pH 4 and 10, which correspond to the pH of latex and clay surfaces, respectively (Figure S2). In agreement with a previous study on Kocuria rosea,40 growth of K rosea was observed at pH 10 (Figure S2C); this strain's alkaline tolerance potentially explains its persistence on alkaline clay surfaces compared to other community members. However, the microbial community composition on acidic mold‐resistant surfaces is not consistent with bacterial growth at pH 4, whereas B timonensis did not grow under pH 4 or pH 10, highlighting the complex nature of biological and physical processes associated with surface survival.
3.5. Survival on antimicrobial surfaces and benzalkonium chloride susceptibility
All five community members show significant log reduction (2.70‐5.15) following inoculation on antimicrobial surfaces. This biocidal effect can be largely attributed to the use of quaternary ammonia salts (QAS) in the paint formulation,10 although the contribution of desiccation and lack of nutrients cannot be overlooked. QAS irreversibly attach to the negatively charged cell membrane, leading to deformation of the membrane and subsequent leakage of intracellular materials.41, 42 Studies have further shown that QAS have different bactericidal activities against Gram‐positive and Gram‐negative bacteria. Gram‐positive bacteria are generally more susceptible to QAS than Gram‐negative bacteria43 due to the presence of both inner and outer cellular membranes.44 Despite the use of quaternary ammonia additives in paint formulation, B timonensis demonstrated relatively minor log reduction (2.70‐3.18) compared to others. We consistently observed cultivable Bacillus timonensis, a Gram‐negative spore‐forming bacteria,45 24 hours after exposure to antimicrobial surfaces across all humidity levels (Table S1). A subpopulation of B timonensis remained detectable after PMA treatment and was able to grow on trypticase soy agar (TSA). We speculate that the copy numbers obtained from PMA‐treated samples are dominated by live spores because bacterial spores are intrinsically resistant to various disinfectants including QAS,46 owing to the structure of spore coats and cortex. However, these results could also be influenced by the contribution of vegetative cells and non‐viable spores.47
B timonensis showed significant resistance against the presence of QAS on surfaces, although genetic determinants (eg, QacA, QacB, QacC, and smr)48 were not identified from its genome. Therefore, sporulation induced by common stresses encountered on indoor surfaces was likely to be one of the mechanisms contributing to the survival of B timonensis in the presence of QAS. Using Scanning Electron Microscopy, we confirmed the presence of Bacillus spores (Figure 7C) in addition to vegetative cells (Figure 7D) in swab samples. Owing to the structure of spore coats and cortex, bacterial spores are intrinsically resistant to various disinfectants including QAS.46 Silver‐ and zinc‐containing zeolite stainless steel surfaces also yield satisfactory inactivation of vegetative cells of B anthracis, B cereus, and B subtilis, but have limited effects on spores.49 Although live spores on surfaces can be metabolically inactive, they are constantly chemically sensing their environments to determine an optimal time to return to a vegetative state, and therefore continue to pose potential risk of exposure to occupants.
Figure 7.
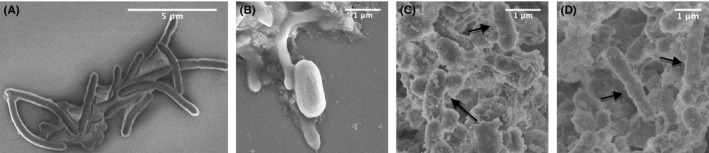
Scanning electron microscopy images of B timonensis in vegetative state (A), sporulation state (B), and antimicrobial surface swab samples (C and D). Both spores (C) and vegetative cells (D) can be identified in swab samples. Figure 7A was captured under 1 kV acceleration voltage, 8.1 mm working distance, and ×9.0 k magnification. Figure 7B was obtained under 4 kV acceleration voltage, 8.6 mm working distance, and ×22.0 k magnification. Figure 7C was obtained under 3 kV acceleration voltage, 8.8 mm working distance, and ×20.0 k magnification. Figure 7D was captured under 3 kV, 8.8 mm working distance, and ×15.0 k magnification
Many daily‐use consumer products with embedded antimicrobial chemicals have been studied so far, and in general, the antimicrobial effectiveness is dependent on the context in which the product is used. Triclosan‐containing cutting boards were shown to have an antibacterial effect that depends upon humidity as well as the type and load of bacteria.30 Furthermore, repeated washing of triclosan‐containing cutting boards reduced antimicrobial activity, suggesting a reduction in the bioavailability of surface triclosan over time. Despite the fact that the added concentration of QAS in the antimicrobial paint is reported between 0.25% and 3%,10 the bioavailable fraction of QAS on the surfaces is context‐dependent as well.
We further tested the susceptibility of the five strains against one of the most commonly used QAS disinfectants—benzalkonium chloride (BKC). Pantoea allii had the greatest resistance with a minimum inhibitory concentration (MIC) of 0.0156% by weight (equivalent to 156.25 μg/mL), while others were highly susceptible, with MICs < 0.001%. Despite this phenotypic resistance, P allii did not present with a strong survival pattern on antimicrobial surfaces, suggesting that the concentrations of surface‐available antimicrobials are well‐above corresponding lethal levels. Alternatively, we can infer that this strain is particularly sensitive to other environmental conditions. In this particular case, the exposure of microbes to antimicrobial surfaces did not explicitly select for antimicrobial‐resistant microbes, but rather spore‐forming bacteria.
3.6. Potential functions associated with stress tolerance
Indoor surface environments have been considered as a “microbial wasteland”50 because of water and nutrient scarcity. The majority of surface‐associated microbes are likely to be dormant for an extended period of time under desiccation and starvation, and only revive when a desirable water activity is achieved.51 Survival in this “wasteland” requires a diverse range of adaptive strategies such as dormancy, for example, sporulation and biofilm formation. We characterized the genetic potential for stress tolerance in each of the cultured community members. All five successfully coped with the various stresses encountered in dust samples and remained viable, although their exact exposure time varied. Desiccation is not only one of the most commonly encountered stresses in indoor environments, but also one of the significant relevance for transmission of pathogenic microorganisms in healthcare and daycare facilities.52, 53 Desiccation‐tolerant cells are typically equipped with physiological, structural, and molecular mechanisms to withstand water scarcity.54 Osmotic regulation through aquaporins, accumulation of osmoprotectants, and synthesis and uptake of biomolecules such potassium, trehalose, glutamate, glutamine, proline, glycine betaine, and glucosylglycerol have been related to desiccation tolerance in prokaryotic cells.40, 55, 56 Osmoprotectant transporters (eg, YehX, YehZ, YehW, and YehY) and osmoprotectant import permease proteins (OsmW and OsmY) could be identified based on sequence homology only from the Pantoea allii genome (Figure 8B). Trehalose transport system permease proteins (SugA, SugB) and trehalose import ATP‐binding protein (SugC) were identified in the genomes of B timonensis, P allii, and K rosea (Figure 8).
Figure 8.
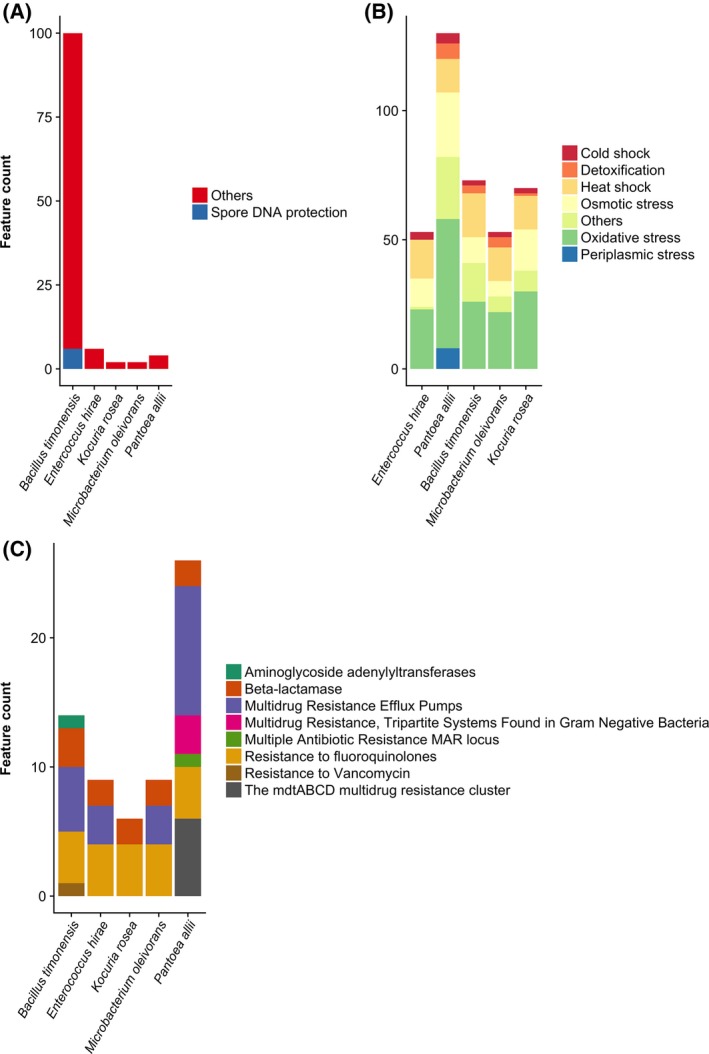
Genetic potential of surface survival for each community member represented by number of features associated with (A) dormancy and sporulation, (B) stress response, and (C) antibiotic resistance. The complete functional annotation results can be found in Data S2
4. CONCLUSION
We demonstrated the differences in survival for a synthetic community comprised of 5 non‐model bacteria on three chemically and physically distinct materials (Figures 5 and 6). The bacterium carrying antibiotic resistance genes (which also displayed the corresponding phenotypes) failed to exhibit a strong retention pattern on antimicrobial surfaces, indicating that the possession of antibiotic resistance genes alone is not predictive of survival on antimicrobial surfaces. In this particular case, the exposure to antimicrobial surfaces did not explicitly select for antimicrobial‐resistant microbes, but rather spore‐forming bacteria, potentially due to the confounding effect of having multiple stressors (ie, desiccation and nutrient limitation).
Understanding the interaction between surface chemistry and surface microbiology is central to understanding the viable indoor microbiome. The use of surface finishes with different chemical and physical properties might not be the most significant driving force for microbial community composition in the long term due to frequent occupant disturbances.51 Nevertheless, selection for dormant and persistent microbes on surfaces is constantly impacting the transmission of surface microbes either through air or human contact. Dormant cells, although not highly active on surfaces, may still serve as a sink for pathogenic microbes such as Acinetobacter baumannii 57 or Staphylococcus aureus,11 both of which have been previously related to one or more outbreaks in healthcare systems. Neither the use of antimicrobial surfaces nor rigorous cleaning procedures eradicate all surface microbes along with their associated risks, which draws attention to the potential persistence of spore‐forming pathogens indoors, for example, Bacillus 53 and Clostridium. Lastly, microbial adaptation to extreme environments and mechanisms to cope with environmental stresses are still not fully understood. Such complex dynamics might not be comprehensively captured by whole‐genome sequences, indicating the need for studies that investigate the viable portion of indoor microbiome as well as their phenotypic responses, and the need to develop tractable models for gene knockout‐knockdown mutants for these organisms so as to determine the importance of specific cellular functions in desiccation tolerance, carbon storage, and oxidative resistance.
CONFLICT OF INTEREST
The authors declare no competing interests.
Supporting information
ACKNOWLEDGMENTS
The authors are grateful for the financial support from the Alfred P. Sloan Foundation (G‐2016‐7291), the Northwestern University Office of Undergraduate Research (999URAP139596), and the Searle Leadership Fund. The authors would like to thank Nicolas Martinez Prieto for his training in surface roughness measurement as well as Reiner Bleher, Charlene D. Wilke, Tirzah Abbott, and Karl W. Hagglund from Northwestern University Atomic and Nanoscale Characterization Experimental Center (NUANCE) for SEM sample preparation and image generation. We are further grateful to Julius Lucks and Katherine Berman for the use of their MiSeq. Last but not least, we would like to thank Daniela Ruiz and Huseyin Demir for their help in experiment preparation. We acknowledge Vlad Tchompalov for setting up a useful photo booth.
Hu J, Ben Maamar S, Glawe AJ, Gottel N, Gilbert JA, Hartmann EM. Impacts of indoor surface finishes on bacterial viability. Indoor Air. 2019;29:551–562. 10.1111/ina.12558
REFERENCES
- 1. Stephens B. What have we learned about the microbiomes of indoor environments? mSystems. 2016;1(4):e00083‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klevens RM, Edwards JR, Richards CL, et al. Estimating Health Care‐Associated Infections and Deaths in U.S. Hospitals, 2002. Public Health Rep. 2007;122(2):160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lax S, Gilbert JA. Hospital‐associated microbiota and implications for nosocomial infections. Trends Mol Med. 2015;21(7):427‐432. [DOI] [PubMed] [Google Scholar]
- 4. Mendell MJ, Mirer AG, Cheung K, Tong M, Douwes J. Respiratory and allergic health effects of dampness, mold, and dampness‐related agents: a review of the epidemiologic evidence. Environ Health Perspect. 2011;119(6):748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fulmer PA, Wynne JH. Development of broad‐spectrum antimicrobial latex paint surfaces employing active amphiphilic compounds. ACS Appl Mater Interfaces. 2011;3(8):2878‐2884. [DOI] [PubMed] [Google Scholar]
- 6. Kumar A, Vemula PK, Ajayan PM, John G. Silver‐nanoparticle‐embedded antimicrobial paints based on vegetable oil. Nat Mater. 2008;7:236. [DOI] [PubMed] [Google Scholar]
- 7. Sahoo PC, Kausar F, Lee JH, Han JI. Facile fabrication of silver nanoparticle embedded CaCO3 microspheres via microalgae‐templated CO2 biomineralization: application in antimicrobial paint development. RSC Advances. 2014;4(61):32562‐32569. [Google Scholar]
- 8. Beech SJ, Noimark S, Page K, Noor N, Allan E, Parkin IP. Incorporation of crystal violet, methylene blue and safranin O into a copolymer emulsion; the development of a novel antimicrobial paint. RSC Advances. 2015;5(33):26364‐26375. [Google Scholar]
- 9. Hwang GB, Allan E, Parkin IP. White light‐activated antimicrobial paint using crystal violet. ACS Appl Mater Interfaces. 2016;8(24):15033‐15039. [DOI] [PubMed] [Google Scholar]
- 10. Gisser KR, Sibbald MS, Smith WJ, Dreshar JK, Prochazka DA. High quality antimicrobial paint composition. In: oogle Patents; 2015.
- 11. Michels H, Noyce J, Keevil CW. Effects of temperature and humidity on the efficacy of methicillin‐resistant Staphylococcus aureus challenged antimicrobial materials containing silver and copper. Lett Appl Microbiol. 2009;49(2):191‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawakami H, Yoshida K, Nishida Y, Kikuchi Y, Sato Y. Antibacterial properties of metallic elements for alloying evaluated with application of JIS Z 2801: 2000. ISIJ Int. 2008;48(9):1299‐1304. [Google Scholar]
- 13. Egger S, Lehmann RP, Height MJ, Loessner MJ, Schuppler M. Antimicrobial properties of a novel silver‐silica nanocomposite material. Appl Environ Microbiol. 2009;75(9):2973‐2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoang CP, Kinney KA, Corsi RL, Szaniszlo PJ. Resistance of green building materials to fungal growth. Int Biodeterior Biodegradation. 2010;64(2):104‐113. [Google Scholar]
- 15. Mensah‐Attipoe J, Reponen T, Salmela A, Veijalainen A‐M, Pasanen P. Susceptibility of green and conventional building materials to microbial growth. Indoor Air. 2015;25(3):273‐284. [DOI] [PubMed] [Google Scholar]
- 16. Adams RI, Lymperopoulou DS, Misztal PK, et al. Microbes and associated soluble and volatile chemicals on periodically wet household surfaces. Microbiome. 2017;5(1):128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Otto CC, Cunningham TM, Hansen MR, Haydel SE. Effects of antibacterial mineral leachates on the cellular ultrastructure, morphology, and membrane integrity of Escherichia coli and methicillin‐resistant Staphylococcus aureus . Ann Clin Microbiol Antimicrob. 2010;9(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Williams LB, Metge DW, Eberl DD, et al. What makes a natural clay antibacterial? Environ Sci Technol. 2011;45(8):3768‐3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single‐cell sequencing. J Comput Biol. 2012;19(5):455‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043‐1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer‐BLAST: a tool to design target‐specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13(1):134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23(10):1289‐1291. [DOI] [PubMed] [Google Scholar]
- 23. Untergasser A, Cutcutache I, Koressaar T, et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115‐e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068‐2069.24642063 [Google Scholar]
- 25. Aziz RK, Bartels D, Best AA, et al. The RAST server: rapid annotations using subsystems technology. BMC Genom. 2008;9(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wattam AR, Parrello B, Xia F, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2013;42(D1):D206‐D214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gilbert JA, Jansson JK, Knight R. The Earth Microbiome project: successes and aspirations. BMC Biol. 2014;12(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thompson LR, Sanders JG, McDonald D, et al. A communal catalogue reveals Earth's multiscale microbial diversity. Nature. 2017;551:457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marotz C, Amir A, Humphrey G, Gaffney J, Gogul G, Knight R. DNA extraction for streamlined metagenomics of diverse environmental samples. Biotechniques. 2017;62(6):290‐293. [DOI] [PubMed] [Google Scholar]
- 30. Møretrø T, Høiby‐Pettersen GS, Habimana O, Heir E, Langsrud S. Assessment of the antibacterial activity of a triclosan‐containing cutting board. Int J Food Microbiol. 2011;146(2):157‐162. [DOI] [PubMed] [Google Scholar]
- 31. Murtey MD, Ramasamy P. Sample preparations for scanning electron microscopy–life sciences. In: Modern Electron Microscopy in Physical and Life Sciences. InTech; 2016.
- 32. Fahimipour AK, Ben Maamar S, McFarland AG, et al. Antimicrobial chemicals associate with microbial function and antibiotic resistance indoors. mSystems. 2018;3(6):e00200‐00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Okten S, Asan A. Airborne fungi and bacteria in indoor and outdoor environment of the Pediatric Unit of Edirne Government Hospital. Environ Monit Assess. 2012;184(3):1739‐1751. [DOI] [PubMed] [Google Scholar]
- 34. Park HK, Han JH, Joung Y, Cho SH, Kim SA, Kim SB. Bacterial diversity in the indoor air of pharmaceutical environment. J Appl Microbiol. 2013;116(3):718‐727. [DOI] [PubMed] [Google Scholar]
- 35. Dickson RP, Erb‐Downward JR, Prescott HC, et al. Analysis of culture‐dependent versus culture‐independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid. J Clin Microbiol. 2014;52(10):3605‐3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stefani F, Bell TH, Marchand C, et al. Culture‐dependent and ‐independent methods capture different microbial community fractions in hydrocarbon‐contaminated soils. PLoS ONE. 2015;10(6):e0128272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li L, Mendis N, Trigui H, Oliver JD, Faucher SP. The importance of the viable but non‐culturable state in human bacterial pathogens. Front Microbiol. 2014;5:258‐258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deng X, Li Z, Zhang W. Transcriptome sequencing of Salmonella enterica serovar Enteritidis under desiccation and starvation stress in peanut oil. Food Microbiol. 2012;30(1):311‐315. [DOI] [PubMed] [Google Scholar]
- 39. Deutscher MP. Degradation of stable RNA in bacteria. J Biol Chem. 2003; 278 (46): 45041-45044. [DOI] [PubMed] [Google Scholar]
- 40. Gholami M, Etemadifar Z, Bouzari M. Isolation a new strain of Kocuria rosea capable of tolerating extreme conditions. J Environ Radioact. 2015;144:113‐119. [DOI] [PubMed] [Google Scholar]
- 41. Maris P. Modes of action of disinfectants. Revue Scientifique et Technique‐Office International des Epizooties. 1995;14:47‐47. [DOI] [PubMed] [Google Scholar]
- 42. Hamilton WA. The mechanism of the bacteriostatic action of tetrachlorosalicylanilide: a membrane‐active antibacterial compound. Microbiology. 1968;50(3):441‐458. [DOI] [PubMed] [Google Scholar]
- 43. Fazlara A, Ekhtelat M. The disinfectant effects of benzalkonium chloride on some important foodborne pathogens. Am‐Eurasian J Agric Environ Sci. 2012;12(1):23‐29. [Google Scholar]
- 44. Jennings MC, Minbiole K, Wuest WM. Quaternary ammonium compounds: an antimicrobial mainstay and platform for innovation to address bacterial resistance. ACS Infectious Diseases. 2015;1(7):288‐303. [DOI] [PubMed] [Google Scholar]
- 45. Kokcha S, Mishra AK, Lagier J‐C, et al. Non contiguous‐finished genome sequence and description of Bacillus timonensis sp. nov. Stand Genomic Sci. 2012;6(3);346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Louie A, VanScoy BD, Brown DL, Kulawy RW, Heine HS, Drusano GL. Impact of spores on the comparative efficacies of five antibiotics for treatment of Bacillus anthracis in an in vitro hollow fiber pharmacodynamic model. Antimicrob Agents Chemother. 2012;56(3):1229‐1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rawsthorne H, Dock CN, Jaykus LA. PCR‐based method using propidium monoazide to distinguish viable from nonviable Bacillus subtilis spores. Appl Environ Microbiol. 2009;75(9):2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sidhu MS, Heir E, Sørum H, Holck A. Genetic linkage between resistance to quaternary ammonium compounds and β‐lactam antibiotics in food‐related Staphylococcus spp. Microb Drug Resist. 2001;7(4):363‐371. [DOI] [PubMed] [Google Scholar]
- 49. Galeano B, Korff E, Nicholson WL. Inactivation of vegetative cells, but not spores, of Bacillus anthracis, B. cereus, and B. subtilis on stainless steel surfaces coated with an antimicrobial silver‐ and zinc‐containing zeolite formulation. Appl Environ Microbiol. 2003;69(7):4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gibbons SM. The built environment is a microbial wasteland. mSystems. 2016;1(2):e00033‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chase J, Fouquier J, Zare M, et al. Geography and location are the primary drivers of office microbiome composition. mSystems. 2016;1(2):e00022‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ramos JL, Ma‐T G, Marqués S, Ramos‐González M‐I, Espinosa‐Urgel M, Segura A. Responses of Gram‐negative bacteria to certain environmental stressors. Curr Opin Microbiol. 2001;4(2):166‐171. [DOI] [PubMed] [Google Scholar]
- 53. Lee L, Tin S, Kelley ST. Culture‐independent analysis of bacterial diversity in a child‐care facility. BMC Microbiol. 2007;7(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Potts M. Desiccation tolerance: a simple process? Trends Microbiol. 2001;9(11):553‐559. [DOI] [PubMed] [Google Scholar]
- 55. Billi D, Potts M. Life and death of dried prokaryotes. Res Microbiol. 2002;153(1):7‐562. [DOI] [PubMed] [Google Scholar]
- 56. Welsh DT. Ecological significance of compatible solute accumulation by micro‐organisms: from single cells to global climate. FEMS Microbiol Rev. 2006;24(3):263‐290. [DOI] [PubMed] [Google Scholar]
- 57. Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol. 1998;36(7):1938‐1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


