Abstract
Aims
Because the hedgehog signalling pathway plays a major role in many types of cancer and can nowadays be targeted by specific compounds, we aimed to investigate the role of this pathway in squamous cell carcinoma of the head and neck.
Methods and results
Ninety‐eight treatment‐naive head and neck cancer specimens were immunohistologically stained for SMO, GLI‐1, p53 and p16 expression and correlated with clinicopathological factors. Immunoreactivity for SMO and GLI‐1 was found in 20 (20.4%) and 52 (53.1%) cases of tumours, respectively. SMO expression correlated with GLI‐1 expression (ρ = 0.258, P = 0.010) in univariate and multivariate analysis (P = 0.007, t = 2.81). In univariate analysis, high SMO expression was associated with shorter overall survival (HR = 0.56; 95% CI = 0.32–0.98; P = 0.044) and disease‐free survival (HR = 0.53; 95% CI = 0.30–0.95; P = 0.034). In multivariate cox regression analysis SMO expression showed a trend towards an independent predictor for shorter overall survival (HR = 0.57; 95% CI = 0.30–1.05; P = 0.072) and disease‐free survival (HR = 0.53; 95% CI = 0.28–1.02; P = 0.056). In head and neck cancer patients with low tumour p16 expression, SMO expression was an independent factor for overall survival (HR = 0.49; 95% CI = 0.24–0.98; P = 0.043) and disease‐free survival (HR = 0.45; 95% CI = 0.22‐0.96; P = 0.037).
Conclusion
Although it needs to be confirmed in larger cohorts, our results suggest that targeting SMO might be a potentially therapeutic option in patients with head and neck cancer. In line, molecular pathological analyses including mutation analysis in the hedgehog pathway might point to additional therapeutic leads.
Keywords: head and neck cancer, hedgehog, immunohistochemistry, survival
Introduction
Squamous cell carcinoma of the head and neck (HNSCC) is a major cause of cancer‐associated morbidity and mortality, with more than 60 000 newly diagnosed cases per year in the United States and 600 000 cases diagnosed annually worldwide.1, 2 Despite recent advances in cancer therapy only 3% of cases are long‐term survivors.3 This can be explained by two facts: first, the majority of cases are diagnosed in an advanced disease stage with loco‐regional progression, and secondly, therapeutic options are still limited, with a high medical need for new therapeutic options and a deeper understanding of HNSCC biology.4, 5, 6
The Hedgehog Pathway (HhP) is a signal transduction pathway which is mainly known for its role in embryogenesis and wound healing.7 However, in addition to its role in normal human development, it also plays an important role in cancer, including basal cell carcinoma (BCC), where selective inhibitors (i.e. vismodegib and sonidegib) are used successfully.8, 9 Given its clinical efficacy and safety in cancer – in particular in BCC – we aimed to investigate the status of the HhP in treatment‐naive head and neck cancer patients.
Materials and methods
Study Population
The distribution of smoothened, frizzled class receptor (SMO), GLI family zinc finger (GLI)‐1, p53 and p16 expression in 98 HNSCC specimens was studied using tumour tissue archived at the Biobank, Medical University of Graz. This retrospective study included randomly selected patients diagnosed with HNSCC between January 1992 and December 2002 (ethical approval: 24‐236 ex 11/12). For each specimen, both primary tumour tissue and clinicopathological data were available. Patients did not receive neo‐adjuvant chemo‐ or radiotherapy, and all patients underwent a curative resection. Postoperative surveillance was performed on each patient, including routine clinical and laboratory examinations. All clinicopathological as well as laboratory data were retrieved from medical records from the Division of Oncology, Medical University of Graz, as well as from pathology reports from the Institute of Pathology at the same institution. Pathological T‐stage was uniformly adjusted according to the seventh edition of the TNM 2009 classification system. Other clinicopathological parameters included gender, localisation and patients’ age as well as tumour grade, tumour stage and N‐stage.
Immunohistochemistry
Histological sections were deparaffinised in xylene and rehydrated with graded ethanol. For GLI‐1 detection, the sections were subjected to antigen retrieval in a pressure cooker (Pascal; Dako, Santa Clara, CA, USA) in 0.01 m sodium‐citrate buffer, pH 6.0, and subsequently incubated overnight with an antibody to human GLI‐1 (#sc‐20687, 1:200; Santa Cruz, Dallas, TX, USA). For SMO detection, the sections were subjected to antigen retrieval in a microwave in ethylenediamine tetraacetic acid (EDTA) natrium buffer, pH 8.0, for 40 min and subsequently incubated for 1 h with antibody to human SMO (#ab72130; 1:50; Abcam, Cambridge, UK). The reaction was visualised using the UltraVision LP large volume detection system HRP polymer (Thermo Scientific Waltham, MA, USA), and all sections were counterstained with haematoxylin. For the negative control, the primary antibody was omitted. For p53 staining, slides were incubated at 60°C for 1 h and deparaffinised by xylene twice for 10 min and then rehydrated by decreasing ethanol concentrations from 100 to 90%, further to 70% and lastly to 50%, followed by a water bath for 40 min. Immunohistochemistry for p53 (#M7001; 1:100; Dako) was performed on a Dako autostainer (Dako). Dako Real (#K5001; Dako) was used as a detection system. For visualisation, the AEC substrate chromogen ready‐to‐use kit (#K3464; Dako) was utilised.
For p16 detection the sections were deparaffinised in xylene and rehydrated with graded ethanol. Sections were then subjected to antigen retrieval in the incubator for 1 h at 70°C, after pretreatment in 0.01 m sodium‐citrate buffer, pH 8.0, for 30 min, and incubated with antibody to human p16 (CINtecp16‐Histology, clone E64H, #825‐4713; Ventana Roche, Tucson, AZ, USA) for 32 min. The reaction was visualised using the IVIEW DAB Detection Kit (Ventana Roche) and all sections were counterstained with haematoxylin.
The frequency of p53 (positive nuclear staining), SMO (positive cytoplasmic staining), GLI‐1 (positive nuclear staining) and p16‐positive cells were independently evaluated by A.A. and A.M.A. The percentage of positive tumour cells was quantified by counting 100 cells/section in five randomly chosen high‐power fields (×40) by light microscopy and analysed by Spearman's correlation. For Kaplan–Meier analysis and the multivariate Cox proportional hazard model, samples with at least 5% positive reactivity were considered to be GLI‐1‐ and/or SMO‐positive (high and low expression, respectively). Samples with at least 10% p53 positive nuclear reactivity were considered to be p53‐positive. Samples with at least 1% p16 positive reactivity were considered to be p16‐positive and have been categorized as high (≥1% p16 expression) and low expression (<1% p16 positivity), respectively.
Statistical Analysis
All statistical analyses were performed using the r software package (R Foundation for Statistical Computing, Vienna, Austria; https://www.R-project.org/) and spss version 23.0 software (SPSS, Chicago, IL, USA). Data reported are retrospective in nature. After data closure, all variables passed a plausibility check to detect outliers in the data set. No extreme values have been extracted from the full data set. Spearman's correlation was used to assess the correlation between immunohistochemistry (IHC) staining of GLI‐1, SMO, p16 and p53 with T stage, N stage and grading. Linear regression was used to investigate the impact of different co‐variables on SMO and GLI expression. Overall survival and disease‐free survival of patients was calculated using the Kaplan–Meier method, followed by the log‐rank test and Cox regression analysis. A multivariate Cox proportional hazard model was performed to examine the impact of different predictors on overall and disease‐free survival. Univariate and backward elimination procedures resulted in the same variable selection for the final model. The assumption of proportional hazards was checked by LML plots and residual analyses by Schoenfeld plots. A two‐sided P < 0.05 was considered statistically significant.
Results
Ninety‐eight patients were included in this retrospective immunohistochemical study: 26 women (26.5%) and 72 men (73.5%) (details in Table 1). The cut‐off for staining positivity for GLI‐1 and SMO was set to 5%, for p16 to 1% and for p53 to 10%, due to varying intensities of p53 nuclear staining (strong and weak signals were counted as positive). Samples were divided accordingly into positive and negative (detailed in Figure 1). Using these cut‐offs, 20 samples (20.4%) from the entire cohort were considered positive for SMO, 52 samples (53.1%) for GLI‐1, 41 samples (41.8%) for p53 and 28 samples (28.6%) for p16. Spearman's correlation demonstrated that there was a significant correlation between p53 expression and tumour grading (P < 0.001, ρ = 0.387), and remained consistent when stratified for T stage (P < 0.001, ρ = 0.398).
Table 1.
Clinicopathological features of all patients included in this study
| Clinicopathological parameter | No. of patients (n = 98) (%) |
|---|---|
| Gender | |
| Male | 72 (73.5) |
| Female | 26 (26.5) |
| Tumour stage | |
| T1 | 37 (37.8) |
| T2 | 18 (18.4) |
| T3 | 16 (16.3) |
| T4 | 27 (27.6) |
| N stage | |
| N0 | 63 (64.3) |
| N1 | 19 (19.4) |
| N2 | 16 (16.3) |
| Tumour grade | |
| I | 22 (22.4) |
| II | 49 (50.0) |
| III | 27 (27.6) |
| p53 staining | |
| Negative (<10%) | 48 (49.0) |
| Positive (≥10%) | 41 (41.8) |
| Unrepresentative | 9 (9.2) |
| GLI‐1 staining | |
| Negative (<5%) | 41 (41.8) |
| Positive (≥5%) | 52 (53.1) |
| Unrepresentative | 5 (5.1) |
| SMO staining | |
| Negative (<5%) | 71 (72.4) |
| Positive (≥5%) | 20 (20.4) |
| Unrepresentative | 7 (7.1) |
| p16 staining | |
| Negative (<1%) | 65 (66.3) |
| Positive (≥1%) | 28 (28.6) |
| Unrepresentative | 5 (5.1) |
| Localisation | |
| Palate | 4 (4.1) |
| Lip | 18 (18.4) |
| Base of the mouth | 56 (57.1) |
| Tonsil | 3 (3.1) |
| Buccal | 1 (1.0) |
| Tongue | 16 (16.3) |
Figure 1.
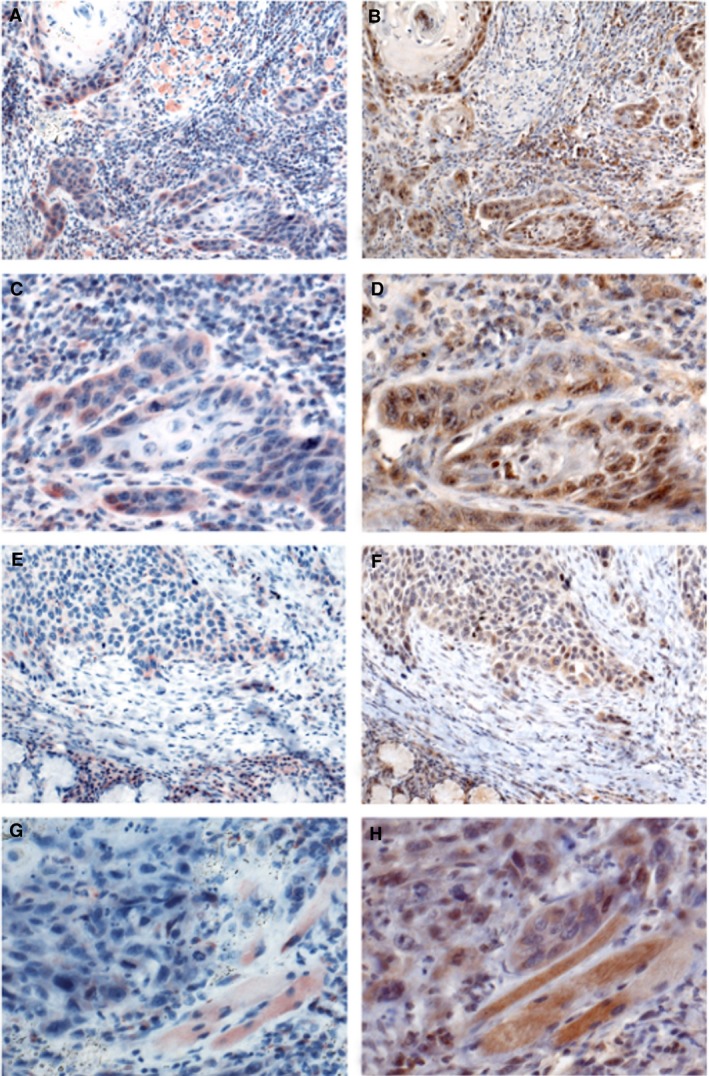
Low and high expression of smoothened, frizzled class receptor (SMO) and GLI family zinc finger (GLI)‐1 in squamous‐cell carcinoma of the head and neck (HNSCC). A–D, HNSCC immunohistochemistry for SMO. A, The majority of tumour cells show strong cytoplasmic staining. Surrounding inflammatory cells are also positive. B, HNSCC, immunohistochemistry for GLI‐1: most tumour cells have a positive nuclear staining. C, Detailed image of A. D, Detailed image of B. E, HNSCC, immunohistochemistry for SMO: the majority of tumour cells are negative. Only a few tumour cells exhibit a positive cytoplasmic staining. Most inflammatory cells at the bottom of the image are also positive. F, Additional case of HNSCC, immunohistochemistry for GLI‐1: only a few cells have a positive nuclear staining. G, Detailed image of E. H, Detailed image of F.
GLI‐1 expression showed a trend towards correlation with lymph node stage (N stage) (P = 0.089, ρ = 0.170), and when stratified for T stage became statistically significant (P = 0.035, ρ = 0.221). SMO expression significantly correlated with its downstream partner GLI (P = 0.010, ρ = 0.258), and remained significant when stratified for T‐stage (P = 0.013, ρ = 0.248).
P16 expression was not associated with any tested parameter in Spearman's correlation analysis, although a trend for negative correlation with SMO expression was observed (P = 0.093, ρ = −0.168; detailed in Table 2) and with male gender (P = 0.054, χ2 test).
Table 2.
Correlation analysis of all immunohistochemically stained proteins with clinicopathological features and all other proteins
| p53 | GLI‐1 | SMO | p16 | |||||
|---|---|---|---|---|---|---|---|---|
| ρ | P | ρ | P | ρ | P | ρ | P | |
| pT | 0.091 | 0.364 | −0.068 | 0.494 | −0.065 | 0.515 | 0.090 | 0.369 |
| pN | 0.099 | 0.324 | 0.170 | 0.089 | 0.060 | 0.551 | 0.063 | 0.528 |
| Grading | 0.434 | <0.001 | 0.120 | 0.231 | −0.033 | 0.740 | 0.006 | 0.951 |
| p53 | NA | NA | 0.038 | 0.702 | 0.056 | 0.575 | 0.010 | 0.922 |
| GLI‐1 | 0.038 | 0.702 | NA | NA | 0.258 | 0.010 | 0.084 | 0.400 |
| SMO | 0.056 | 0.575 | 0.258 | 0.010 | NA | NA | −0.168 | 0.093 |
| p16 | 0.010 | 0.922 | 0.084 | 0.400 | −0.168 | 0.093 | NA | NA |
| pN stratified for pT | 0.062 | 0.536 | 0.211 | 0.035 | 0.104 | 0.297 | 0.003 | 0.973 |
| Grading stratified for pT | 0.426 | <0.001 | 0.155 | 0.121 | −0.010 | 0.921 | −0.015 | 0.879 |
| GLI‐1 stratified for pT | 0.052 | 0.603 | NA | Na | 0.248 | 0.013 | 0.099 | 0.325 |
| SMO stratified for pT | 0.059 | 0.558 | 0.248 | 0.013 | NA | NA | −0.155 | 0.123 |
| P53 stratified for pT | NA | NA | 0.520 | 0.603 | 0.059 | 0.558 | 0.005 | 0.959 |
Data are presented as Spearman's correlation coefficient and P‐value. Significant associations are depicted in bold type. pT, tumour–node–metastasis (TNM) classification of malignant tumours T stage; pN, TNM classification of malignant tumours N stage; GLI‐1, GLI family zinc finger 1; SMO, smoothened, frizzled class receptor; p53, tumour protein p53; P, P‐value; ρ, Spearman's correlation coefficient; NA, not applicable.
In multivariate analysis the association of SMO and GLI‐1 expression (P = 0.007, t = 2.81) remained significant. Furthermore, a trend of SMO expression towards negative p16 expression was observed (P = 0.077, t = −1.80). P53 expression was independently associated with tumour grade (P = 0.004, t = 2.97) (detailed in Table 3).
Table 3.
Multivariate linear regression of SMO, GLI‐1, p53 and p16 correlated with multiple parameters
| SMO | GLI‐1 | p53 | p16 | |||||
|---|---|---|---|---|---|---|---|---|
| t‐value | P‐value | t‐value | P‐value | t‐value | P‐value | t‐value | P‐value | |
| pT | −0.376 | 0.708 | −1.970 | 0.053 | −0.301 | 0.765 | 0.701 | 0.486 |
| pN | 0.795 | 0.430 | 1.249 | 0.216 | −0.420 | 0.676 | 0.328 | 0.744 |
| Grading | −0.391 | 0.697 | 1.024 | 0.309 | 2.969 | 0.004 | −0.536 | 0.594 |
| SMO | NA | NA | 2.810 | 0.007 | 1.084 | 0.282 | −1.798 | 0.077 |
| GLI‐1 | 2.810 | 0.007 | NA | NA | −0.494 | 0.623 | 1.316 | 0.193 |
| p53 | 1.084 | 0.282 | −0.494 | 0.623 | NA | NA | 0.304 | 0.762 |
| p16 | −1.798 | 0.077 | 1.316 | 0.193 | 0.304 | 0.762 | NA | NA |
Significant associations are depicted in bold type. pT, tumour–node–metastasis (TNM) classification of malignant tumours T stage; pN, TNM classification of malignant tumours N stage; GLI‐1, GLI family zinc finger 1; SMO, smoothened, frizzled class receptor; p53, tumour protein p53; NA, not applicable; P16, cyclin‐dependent kinase inhibitor 2A, multiple tumour suppressor 1.
To investigate whether SMO and GLI expression is associated with patients’ clinical outcomes, Kaplan–Meier and Cox proportional hazard models were performed. Kaplan–Meier models showed that patients with low GLI‐1 expression had no longer overall survival (P = 0.264, log‐rank test) (Figure 2A) or longer disease‐free survival (P = 0.364, log‐rank test) (Figure 2B). In contrast to GLI‐1, patients with low SMO expression had a significantly longer overall survival (P = 0.040, log‐rank test) (Figure 2C) and longer disease‐free survival (P = 0.008, log‐rank test) (Figure 2D).
Figure 2.
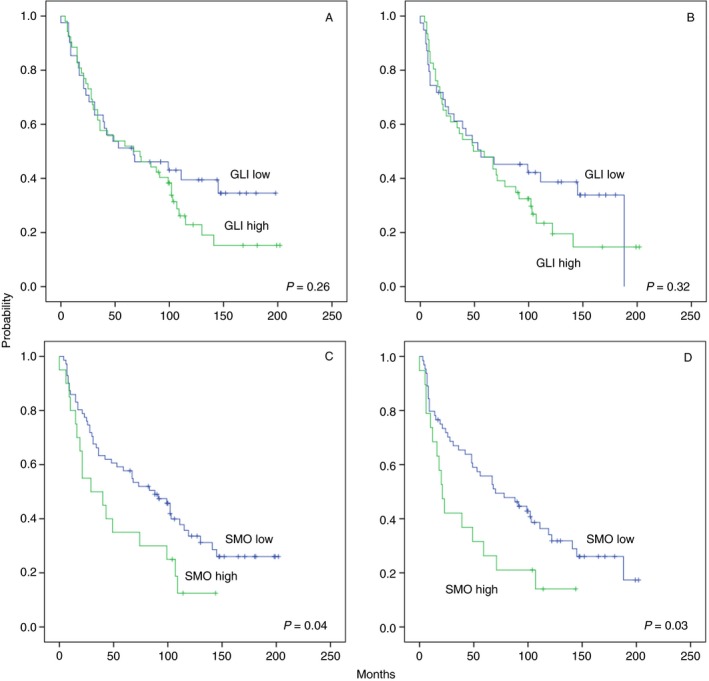
Overall survival (A) and disease‐free survival (B) for all squamous‐cell carcinoma of the head and neck (HNSCC) patients grouped by GLI family zinc finger (GLI)‐1 expression. Overall survival (C) and disease‐free survival (D) for all HNSCC patients grouped by smoothened, frizzled class receptor (SMO) expression.
Low p16 expression was associated with longer overall survival (P = 0.019, log‐rank test) (Figure S1A) and with longer disease‐free survival (P = 0.025, log‐rank test) (Figure S1B).
In addition, univariate Cox proportional hazard analysis revealed that gender [hazard ratio (HR) = 1.23; 95% confidence interval (CI) = 0.71–2.13; P = 0.459)], p53 expression (HR = 1.11; 95% CI = 0.68–1.81; P = 0.267) and GLI expression (HR = 0.75; 95% CI = 0.46–1.24; P = 0.267) had no impact on overall survival in contrast to tumour stage (HR = 0.52; 95% CI = 0.32–0.86; P = 0.011), N‐stage (HR = 0.57; 95% CI = 0.35–0.92; P = 0.020), p16 expression (HR = 0.55; 95% CI = 0.33–0.91; P = 0.021) and SMO expression (HR = 0.56; 95% CI = 0.32–0.98; P = 0.044). For disease‐free survival tumour stage (HR = 0.58; 95% CI = 0.35–0.98; P = 0.040), N‐stage (HR = 0.52; 95% CI = 0.32–0.86; P = 0.011), p16 expression (HR = 0.56; 95% CI = 0.33–0.94; P = 0.028) and SMO expression (HR = 0.53; 95% CI = 0.30–0.95; P = 0.034) have again been identified as significant predictors by univariate analysis. Similar to univariate OS analysis, gender (HR = 1.40; 95% CI = 0.78–2.50; P = 0.257), p53 expression (HR = 1.33; 95% CI = 0.80–2.22; P = 0.273) and GLI expression (HR = 0.77; 95% CI = 0.46–1.29; P = 0.324) had no impact on disease‐free survival.
All significant parameters identified by univariate analysis have been included in multivariate analysis. Using multivariate analyses, N‐stage (overall survival: HR = 0.50; 95% CI = 0.29–0.86; P = 0.012; disease‐free survival: HR = 0.53; 95% CI = 0.30–0.92; P = 0.025) and p16 expression (overall survival: HR = 0.40; 95% CI = 0.23–0.69; P = 0.001; disease‐free survival: HR = 0.41; 95% CI = 0.23–0.72; P = 0.002) were independent predictors of better overall survival and disease‐free survival. SMO expression showed a trend towards better overall survival (HR = 0.57; 95% CI = 0.30–1.05; P = 0.072) and disease‐free survival (HR = 0.53; 95% CI = 0.28–1.02; P = 0.056), although it did not reach statistical significance (detailed in Table 4).
Table 4.
Univariate and multivariate Cox analysis of clinicopathological variables in patients with head and neck cancer (n = 98)
| Overall survival | Disease‐free survival | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||
| HR (CI 95%) | P‐value | HR (CI 95%) | P‐value | HR (CI 95%) | P‐value | HR (CI 95%) | P‐value | |
| Gender | ||||||||
| Female | 1 (Reference) | 0.459 | n.d. | n.d. | 1 (Reference) | 0.257 | n.d. | n.d. |
| Male | 1.23 (0.71–2.13) | 1.40 (0.78–2.50) | ||||||
| pT‐stage | ||||||||
| T1 + T2 | 1 (Reference) | 0.011 | 1 (Reference) | 0.178 | 1 (Reference) | 0.040 | 1 (Reference) | 0.430 |
| T3 + T4 | 0.52 (0.32–0.86) | 0.65 (0.35–1.22) | 0.58 (0.35–0.98) | 0.75 (0.37–1.53) | ||||
| pN‐stage | ||||||||
| N0 | 1 (Reference) | 0.020 | 1 (Reference) | 0.012 | 1 (Reference) | 0.011 | 1 (Reference) | 0.025 |
| N1–3 | 0.57 (0.35–0.92) | 0.50 (0.29–0.86) | 0.52 (0.32–0.86) | 0.53 (0.30–0.92) | ||||
| p53 IHC staining | ||||||||
| <10% | 1 (Reference) | 0.688 | n.d. | n.d. | 1 (Reference) | 0.273 | n.d. | n.d. |
| ≥10% | 1.11 (0.68–1.81) | 1.33 (0.80–2.22) | ||||||
| GLI IHC staining | ||||||||
| <5% | 1 (Reference) | 0.267 | n.d. | n.d. | 1 (Reference) | 0.324 | n.d. | n.d. |
| ≥5% | 0.75 (0.46–1.24) | 0.77 (0.46–1.29) | ||||||
| SMO IHC staining | ||||||||
| <5% | 1 (Reference) | 0.044 | 1 (Reference) | 0.072 | 1 (Reference) | 0.034 | 1 (Reference) | 0.056 |
| ≥5% | 0.56 (0.32–0.98) | 0.57 (0.30–1.05) | 0.53 (0.30–0.95) | 0.53(0.28–1.02) | ||||
| p16 IHC staining | ||||||||
| <1% | 1 (Reference) | 0.021 | 1 (Reference) | 0.001 | 1 (Reference) | 0.028 | 1 (Reference) | 0.002 |
| ≥1% | 0.55 (0.33–0.91) | 0.40 (0.23–0.69) | 0.56 (0.33–0.94) | 0.41 (0.23–0.72) | ||||
All P‐values below a two‐sided alpha of 0.05 have been marked as bold. HR, hazard ratio; CI, confidence interval; n.d., not done in multivariate analysis; IHC, immunohistochemistry; GLI, GLI family zinc finger 1; pT, tumour–node–metastasis (TNM) classification of malignant tumours T stage; pN, TNM classification of malignant tumours N stage; SMO, smoothened, frizzled class receptor.
P16 Expression in Head and Neck Cancer Patients
As pathogenesis is different between patients with and without p16 expression, we further divided our cohort into p16 expression low [n = 65 (66.3%)] and high [n = 28 (28.6%)]. GLI‐1 expression had no impact on disease‐free and overall survival in patients with high p16 expression (disease‐free survival: P = 0.556, log‐rank test; overall survival: P = 0.553, log‐rank test) and low p16 expression (disease‐free survival: P = 0.444, log‐rank test; overall survival: P = 0.534, log‐rank test). In p16 high‐expression head and neck cancer patients, SMO expression had no impact on overall (P = 0.347, log‐rank test) and disease‐free survival (P = 0.325, log‐rank test). In contrast to p16 high‐expression head and neck cancer patients, low SMO expression was associated with longer overall survival (P = 0.019, log‐rank test) and longer disease‐free survival (P = 0.015, log‐rank test) (detailed in Figure S2). In univariate analysis SMO expression was the only statistically significant factor correlating with longer overall (HR = 0.56; 95% CI = 0.32–0.98; P = 0.023) and disease‐free survival (HR = 0.41; 95% CI = 0.19–0.86; P = 0.018) and was confirmed as independent factor in multivariate cox regression analysis (overall survival: HR = 0.49; 95% CI = 0.24–0.98; P = 0.043; disease‐free survival: HR = 0.45; 95% CI = 0.22–0.96; P = 0.037) (detailed in Table 5).
Table 5.
Univariate and multivariate Cox analysis of clinicopathological variables in patients with p16 negative head and neck cancer (n = 65)
| Overall survival | Disease free survival | |||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||
| HR (CI 95%) | P‐value | HR (CI 95%) | P‐value | HR (CI 95%) | P‐value | HR (CI 95%) | P‐value | |
| Gender | ||||||||
| Female | 1 (Reference) | 0.768 | 1 (Reference) | 0.985 | 1 (Reference) | 0.463 | 1 (Reference) | 0.600 |
| Male | 1.10 (0.58–2.10) | 0.99 (0.50–1.99) | 1.30 (0.65–2.58) | 1.21 (0.59–2.50) | ||||
| pT‐stage | ||||||||
| T1 + T2 | 1 (Reference) | 0.233 | 1 (Reference) | 0.512 | 1 (Reference) | 0.495 | 1 (Reference) | 0.921 |
| T3 + T4 | 0.68 (0.35–1.29) | 0.79 (0.40–1.58) | 0.79 (0.40–1.57) | 0.96 (0.46–2.04) | ||||
| pN‐stage | ||||||||
| N0 | 1 (Reference) | 0.202 | 1 (Reference) | 0.790 | 1 (Reference) | 0.127 | 1 (Reference) | 0.999 |
| N1–3 | 0.67 (0.36–1.24) | 0.91 (0.44–1.87) | 0.60 (0.31–1.16) | 1.00 (0.46–2.25) | ||||
| p53 IHC staining | ||||||||
| <10% | 1 (Reference) | 0.935 | 1 (Reference) | 0.880 | 1 (Reference) | 0.267 | 1 (Reference) | 0.216 |
| ≥10% | 1.03 (0.55–1.90) | 1.05 (0.53–2.08) | 1.50 (0.75–2.79) | 1.56 (0.77–3.18) | ||||
| GLI IHC staining | ||||||||
| <5% | 1 (Reference) | 0.537 | n.d. | n.d. | 1 (Reference) | 0.448 | n.d. | n.d. |
| ≥5% | 0.82 (0.46–1.56) | 0.77 (0.39–1.52) | ||||||
| SMO IHC staining | ||||||||
| <5% | 1 (Reference) | 0.023 | 1 (Reference) | 0.043 | 1 (Reference) | 0.018 | 1 (Reference) | 0.037 |
| ≥5% | 0.56 (0.32–0.98) | 0.49 (0.24–0.98) | 0.41 (0.19–0.86) | 0.45 (0.22–0.96) | ||||
All P‐values below a two‐sided alpha of 0.05 have been marked as bold. HR, hazard ratio; CI, confidence interval; n.d., not done in multivariate analysis; IHC, immunohistochemistry; GLI, GLI family zinc finger 1; pT, tumour–node–metastasis (TNM) classification of malignant tumours T stage; pN, TNM classification of malignant tumours N stage; SMO, smoothened, frizzled class receptor.
Discussion
We present a cohort of surgically removed, treatment‐naive HNSCC immunohistochemically stained for GLI‐1, SMO and p53 expression. We have demonstrated a correlation between SMO and its downstream target GLI‐1 expression, but not with p53 expression. Previous studies clearly indicated that the HhP is up‐regulated in many types of cancer, including medulloblastoma, pancreatic cancer, prostate cancer and oesophageal cancer.10, 11, 12, 13 It has been further proposed that the HhP plays a crucial role in cancer development and progression.14 In contrast to the findings of Lee et al.,15 we did not find a correlation between GLI‐1 expression and lymph node stage.
However, from a clinical perspective, the HhP in HNSCC can have two important functions: first, as a biomarker for possible therapeutic outcome, and secondly, as a therapeutic drug target.
Yang et al. demonstrated that high GLI‐1 expression was associated with inferior outcome. They further showed that GLI‐1 expression correlated directly with important cancer stem cell markers, including SOX9 and CD44 that are involved in epithelial‐to‐mesenchymal transition (EMT).16, 17 Interestingly, Yang and colleagues found a positive GLI‐1 expression in 28.3% of all cases, which correlated with the expression pattern of SMO (35.6%). In our cohort we have also demonstrated a direct correlation between SMO and GLI‐1 expression, but in contrast to Yang, GLI‐1 expression was no significant indicator of worse overall or disease‐free survival. This may be due to the fact that, although GLI‐1 is the downstream target of SMO, GLI‐1 has several possible upstream targets.18
One additional factor suggested by Enzenhofer and colleagues was human papillomavirus (HPV) status. They demonstrated that in HPV‐negative head and neck cancer high pan‐GLI expression was associated with better outcome.19
One approach in targeted therapy of HNSCC is the use of the EGFR inhibitor cetuximab, although a predictive biomarker for resistance is not available.20 An up‐regulation of the HhP – in particular GLI expression – has been suggested as one acquired resistance mechanism to anti‐EGFR therapy.21 Although we did not test for EGFR in our cohort, it would be of clinical interest if patients with tumour that positively stained for SMO would profit from vismodegib monotherapy or in combination with anti‐EGFR therapy.
In a recently conducted Phase I study of the HhP inhibitor IPI‐926, together with the EGFR inhibitor cetuximab, it has been shown that patients with no prior anti‐EGFR therapy, in particular, could benefit from this therapeutic approach.22
Our study is not without limitations due to its retrospective nature, unicentre experience, heterogeneity in anatomical locations and small sample size.
Conclusion
This study has demonstrated that in therapy‐naive HNSCC patients, SMO expression correlated directly with its downstream partner GLI‐1. In addition, we have demonstrated that SMO expression is an independent parameter for reduced overall and disease‐free survival in patients with low p16 expression head and neck cancer. Further studies should focus on the possibility to pharmacologically target SMO expression by a HhP inhibitor in SMO expression‐positive tumour patients.
Conflicts of interest
None declared.
Supporting information
Figure S1. Overall survival (A) and disease free survival (B) for all HNSCC patients grouped by p16 expression. Overall survival (C) and disease free survival (D) for all HNSCC patients grouped by SMO expression.
Figure S2. Disease free survival in HNSCC patients with low p16 expression (A) and high p16 expression (B) grouped by SMO expression. Overall survival in HNSCC patients with low p16 expression (C) and high p16 expression (D) grouped by SMO expression.
Acknowledgements
We gratefully thank Jessica Damberger for linguistic help.
Richtig G, Aigelsreiter A M, Asslaber M, Weiland T, Pichler M, Eberhard K, Sygulla S, Schauer S, Hoefler G & Aigelsreiter A (2019) Histopathology. 75, 118–127 10.1111/his.13860 Hedgehog pathway proteins SMO and GLI expression as prognostic markers in head and neck squamous cell carcinoma
References
- 1. Ferlay J, Soerjomataram I, Dikshit R et al Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015; 136; E359–E386. [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018; 68; 7–30. [DOI] [PubMed] [Google Scholar]
- 3. Cohen EEW, LaMonte SJ, Erb NL et al American Cancer Society head and neck cancer survivorship care guideline. CA Cancer J. Clin. 2016; 66; 203–239. [DOI] [PubMed] [Google Scholar]
- 4. Ferris RL, Blumenschein G, Fayette J et al Nivolumab for recurrent squamous‐cell carcinoma of the head and neck. N. Engl. J. Med. 2016; 375; 1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aigelsreiter AM, Aigelsreiter A, Wehrschuetz M et al Loss of the putative tumor suppressor protein spinophilin is associated with poor prognosis in head and neck cancer. Hum. Pathol. 2014; 45; 683–690. [DOI] [PubMed] [Google Scholar]
- 6. Bauernhofer T, Pichler M, Wieckowski E et al Prolactin receptor is a negative prognostic factor in patients with squamous cell carcinoma of the head and neck. Br. J. Cancer 2011; 104; 1641–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gan GN, Jimeno A. Emerging from their burrow: hedgehog pathway inhibitors for cancer. Expert Opin. Investig. Drugs 2016; 25; 1153–1166. [DOI] [PubMed] [Google Scholar]
- 8. Li T, Liao X, Lochhead P et al SMO expression in colorectal cancer: associations with clinical, pathological, and molecular features. Ann. Surg. Oncol. 2014; 21; 4164–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jacobsen AA, Aldahan AS, Hughes OB et al Hedgehog pathway inhibitor therapy for locally advanced and metastatic basal cell carcinoma. JAMA Dermatol. 2016; 152; 816. [DOI] [PubMed] [Google Scholar]
- 10. Wadhwa R, Wang X, Baladandayuthapani V et al Nuclear expression of Gli‐1 is predictive of pathologic complete response to chemoradiation in trimodality treated oesophageal cancer patients. Br. J. Cancer 2017; 117; 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. di Magliano MP, Hebrok M. Hedgehog signalling in cancer formation and maintenance. Nat. Rev. Cancer 2003; 3; 903–911. [DOI] [PubMed] [Google Scholar]
- 12. Nagai S, Nakamura M, Yanai K et al Gli1 contributes to the invasiveness of pancreatic cancer through matrix metalloproteinase‐9 activation. Cancer Sci. 2008; 99; 1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karhadkar SS, Steven Bova G, Abdallah N et al Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature 2004; 431; 707–712. [DOI] [PubMed] [Google Scholar]
- 14. Katoh Y, Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr. Mol. Med. 2009; 9; 873–886. [DOI] [PubMed] [Google Scholar]
- 15. Lee D‐F, Kuo H‐P, Chen C‐T et al IKKβ suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell 2007; 130; 440–455. [DOI] [PubMed] [Google Scholar]
- 16. Gonzalez AC, Ferreira M, Ariel T et al Immunohistochemical evaluation of hedgehog signalling in epithelial/mesenchymal interactions in squamous cell carcinoma transformation: a pilot study. J. Oral Pathol. Med. 2016; 45; 173–179. [DOI] [PubMed] [Google Scholar]
- 17. Yang Z, Cui Y, Ni W et al Gli1, a potential regulator of esophageal cancer stem cell, is identified as an independent adverse prognostic factor in esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2017; 143; 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rimkus T, Carpenter R, Qasem S et al Targeting the sonic hedgehog signaling pathway: review of smoothened and GLI inhibitors. Cancers (Basel) 2016; 8; 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Enzenhofer E, Parzefall T, Haymerle G et al Impact of sonic hedgehog pathway expression on outcome in HPV negative head and neck carcinoma patients after surgery and adjuvant radiotherapy. PLoS One 2016; 11; e0167665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blasco MA, Svider PF, Raza SN et al Systemic therapy for head and neck squamous cell carcinoma: historical perspectives and recent breakthroughs. Laryngoscope 2017; 127; 2565–2569. [DOI] [PubMed] [Google Scholar]
- 21. Keysar SB, Le PN, Anderson RT et al Hedgehog signaling alters reliance on EGF receptor signaling and mediates anti‐EGFR therapeutic resistance in head and neck cancer. Cancer Res. 2013; 73; 3381–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bowles DW, Keysar SB, Eagles JR et al A pilot study of cetuximab and the hedgehog inhibitor IPI‐926 in recurrent/metastatic head and neck squamous cell carcinoma. Oral Oncol. 2016; 53; 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Overall survival (A) and disease free survival (B) for all HNSCC patients grouped by p16 expression. Overall survival (C) and disease free survival (D) for all HNSCC patients grouped by SMO expression.
Figure S2. Disease free survival in HNSCC patients with low p16 expression (A) and high p16 expression (B) grouped by SMO expression. Overall survival in HNSCC patients with low p16 expression (C) and high p16 expression (D) grouped by SMO expression.


