Abstract
Molecular subtypes of urothelial carcinoma may be divided into luminal and nonluminal tumors. Nonluminal tumors are composed of cases with basal/squamous‐like or small cell/neuroendocrine features, with a consensus on the molecular characteristics of the respective subtype. In contrast, luminal tumors are more disparate with three to five suggested subtypes and with definitions that do not always cohere. To resolve some of these disparities we assembled a cohort of 344 luminal tumors classified as urothelial‐like (Uro), with the subtypes UroA, UroAp, UroB, and UroC, or genomically unstable (GU) according to the LundTax system. Cases were systematically analyzed by immunohistochemistry using antibodies for proteins representing important biological processes or cellular states: KRT5, EGFR, and CDH3 for the integrity of a basal cell layer; CCNB1, Ki67, and FOXM1 for proliferation; FGFR3 and ERBB2 for receptor tyrosine kinase status; CCND1, CDKN2A(p16), RB1, and E2F3 for cell cycle regulation; PPARG, GATA3, and TP63 for the differentiation regulatory system; and KRT20 and UPK3 for the differentiation readout. We show that Uro tumors form one, albeit heterogenous, group characterized by FGFR3, CCND1, and RB1 expression, but low or absence of CDKN2A(p16) and ERBB2 expression. The opposite expression pattern is observed in GU cases. Furthermore, Uro tumors are distinguished from GU tumors by showing a high RB1/p16 expression ratio. Class defining characteristics were independent of pathological stage and growth pattern, and thus intrinsic. In Uro tumors, proliferation was limited to a well‐defined single layer of basal‐like cells in UroA tumors but occurred throughout the tumor parenchyma, independent of the basal layer, in the more progressed UroAp and UroC tumors. A similar change in proliferation topology was not observed in GU. We conclude that luminal urothelial carcinomas consist, at the molecular pathology level, of two major subtypes, the larger heterogenous Uro and the biologically distinct GU subtype. © 2019 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Keywords: bladder cancer, molecular subtypes, immunohistochemistry, tumor classification
Introduction
During recent years the molecular classification of urothelial carcinoma (UC) has taken giant steps forward. The first attempt to define clinically relevant subtypes was by Sjödahl et al 1 who described three major molecular subtypes; urothelial‐like (UroA and UroB), genomically unstable (GU), and squamous cell arcinoma‐like. This was followed by three publications in 2014 where Damrauer et al 2 arrived at a two‐group system, Choi et al 3 at a three‐group system, and TCGA 4 at a four‐group system. As several UC classification systems were at hand, a consensus meeting was organized in 2015, at which a basal/squamous‐like subtype (Ba/Sq) was defined 5. Shortly after, Aine et al 6 showed that the four classification systems had a hierarchical relationship. Later, Sjödahl et al 7 extended and refined the classification system to include the UroA‐prog (UroAp), a progressed version of UroA; UroC, a Uro subtype that cluster with GU using genome‐wide gene expression analysis; a small cell/neuroendocrine group (Sc/NE‐like), as well as a minor mesenchymal‐like (Mes‐like) group, the Lund taxonomy (LundTax) 7. This classification system was validated in the extended TCGA data set by combining gene expression, gene mutation, and genomic alteration data 8. Robertson et al published the updated TCGA classification system 9 including the subtypes luminal‐papillary, luminal‐infiltrated, luminal, neuronal, and basal‐squamous, and The Bladder Cancer Molecular Taxonomy Group published the consensus groups luminal papillary, luminal non‐specified, luminal unstable, stroma‐rich, basal/squamous, and neuroendocrine‐like in 2019 10. There are no major inconsistencies between the different classification systems regarding the basal/squamous‐like and the neuroendocrine‐like subtypes. However, on the ‘luminal’ side, the LundTax includes five (UroA, UroAp, UroB, UroC, and GU), TCGA three (luminal‐papillary, luminal infiltrated, and luminal), The Bladder Cancer Molecular Taxonomy three (Luminal Papillary, Luminal Non‐Specified and Luminal Unstable), the UROMOL classification three (classes 1, 2, and 3) 11, and Hurst identified two major groups (GS1 and GS2) 12. Hence, the ‘luminal’ class of UCs seems the most discordant. To shed light on this class of tumors we assembled 344 cases classified by gene expression profiling into the five LundTax luminal subtypes and performed an extensive immunohistochemical investigation using 17 markers. We show that the UroAp, UroB, and UroC represent biologically progressed versions of the canonical UroA subtype, typically low‐grade Ta tumors with features resembling the normal urothelium, and that the GU subtype is biologically distinct from the Uro class of tumors.
Materials and methods
Cohorts
A total of 344 samples classified as UroA, UroAp, UroC, UroB, or GU by global mRNA analysis were selected from the Sjödahl et al 2012 and 2017 cohorts 1, 7. GU cases from the 2012 cohort were re‐evaluated to distinguish between UroC and GU tumors as described in 7. Clinicopathological characteristics are summarized in supplementary material, Table S1. The study was approved by the Lund University Ethical Board (Dnr 2010/5 and 2012/22). Informed consent was obtained from all patients.
Immunohistochemistry
Cases were evaluated using antibodies against CCNB1, CCND1, CDH3, CDKN2A(p16), EGFR, ERBB2, E2F3, FGFR3, FOXM1, GATA3, Ki67, KRT5, KRT20, PPARG, RB1, TP63, UPK3, and ACTA2 (see supplementary material, Table S2). Slides were scanned (Axio Scan.Z1, Zeiss, Germany) and images processed using Xplore (Philips, Amsterdam, The Netherlands). Intensity, percentage and topology of positive cells were assessed in two TMA cores per case. Reference intensity levels were established separately for each cohort. Intensity was evaluated in a 0–3 score range and proportion of positive cells in a 0–5 score range. Tumor cell score was calculated by multiplying intensity with proportion of positive cells. Weak and intermediate staining intensity levels were used as cut‐off to define FGFR3 and ERBB2 positive cases, respectively. Data for clinically HER2 positive cases was retrieved from 13. Histological variants and growth patterns were evaluated according to 14, 15. Urothelial growth pattern was defined as described in 16.
Statistical analysis
Chi‐squared tests or Fisher's exact tests were used for categorical variables, two‐sided t‐test or one‐way ANOVA for comparing immunohistochemistry (IHC) levels between groups, and Spearman's rank correlation to evaluate associations. For hypergeometric test, phyper was used. All statistical analyses were performed in R software (version 3.5.1 R Foundation for Statistical Computing, Vienna, Austria).
Data visualization
The ggplot2 17 package was used to visualize the data and the SPRING algorithm 18 (https://kleintools.hms.harvard.edu/tools/spring.html) for the combined data. The SPRING analysis included the variables: combined basal markers, CCNB1, FGFR3, ERBB2, CCND1, RB1, p16, PPARG, GATA3, TP63, and KRT5.
Results
The experimental cohort
A total of 344 tumor cases of the luminal subtypes UroA (34%), UroAp (12%), UroB (13%), UroC (19%), and GU (22%) were selected 1, 7. The distribution of pathological stage and grade across molecular subtypes are shown in Figure 1A,B. UroA tumors were mainly nonmuscle invasive (Ta, T1) and of lower grade (G1, G2). UroAp, UroC, and GU subtypes showed an almost equal proportion of NMI and MI cases, whereas the majority of UroB tumors were MI. Hence, the luminal tumors show an increasing pathological grade and stage in the order UroA, UroAp, UroB, UroC, and GU.
Figure 1.
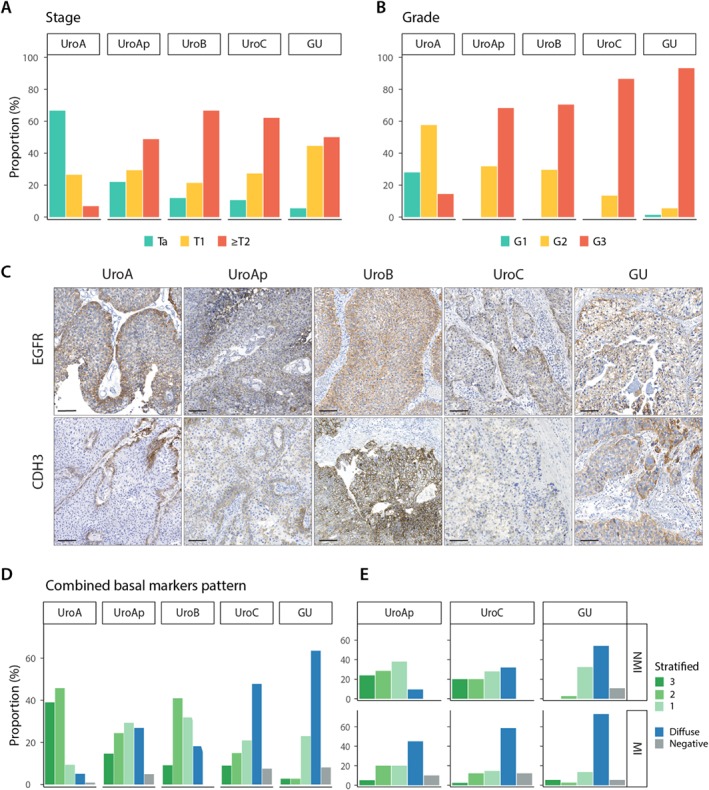
Pathological stage and grade, and expression of EGFR, CDH3, and KRT5 in the luminal subtypes. (A,B) Distribution of pathological stage and grade of the samples within each molecular subtype. (C) Representative IHC staining for EGFR and CDH3 for each molecular subtype. (D) Distribution of the combined basal markers EGFR, CDH3, and KRT5 expression patterns within each molecular subtype. Labels: 3, all markers show stratified expression; 2, two markers show stratified expression; 1, one marker shows stratified expression; diffuse, no marker shows stratified expression and at least one shows diffused staining; 0, all markers are negative. (E) The same data as in D divided into nonmuscle and muscle‐invasive cases for UroAp, UroC, and GU subtypes. UroA and UroB are excluded from this analysis due to low number of cases in one of the groups.
Expression of basal cell markers
Tumor basal cell layer integrity was investigated by expression of the basal epithelial cell markers EGFR, CDH3, and KRT5, evaluated as stratified, diffuse, or negative. A large proportion of the UroA cases showed EGFR and CDH3 expression limited to a single cell layer located along lamina propria (Figure 1C). The proportion of cases with stratified expression pattern for EGFR or CDH3 was reduced in the order UroAp, UroB, UroC, and GU; GU cases showed almost no stratified expression (see supplementary material, Figure S1). Within the UroAp and UroC, the shift to MI growth results in a further loss of stratified expression. KRT5 expression was more complex, showing single‐, multi‐, and noncontinuous layers of positive cells, which defined the stratified pattern; as well as cases with scattered positive or all tumor cells positive, defined as nonstratified expression; and negative (Figure 2A). For KRT5, the dominating pattern in UroA was a single layer of positive cells and hardly any cases negative. UroB tumors showed a similar staining pattern but with overrepresentation of cases with KRT5 positive cells across multiple layers (Figure 2B). UroAp and UroC showed an increasing number of cases negative for KRT5, whereas the majority of GU cases were KRT5 negative. Loss of KRT5 expressing cells was seen in the transition to MI growth for UroAp cases, whereas GU tumors were negative irrespective of stage (see supplementary material, Figure S1). The staining patterns for EGFR, CDH3, and KRT5 were combined, and each case scored as stratified for 3, 2 or 1 marker, diffuse, or all negative (Figure 1D). A majority of the UroA (94%), UroAp (68%), and UroB (87%) showed stratified expression of at least one of these markers. The diffuse pattern was dominating in UroC and GU, seen in 54 and 74% of the cases, respectively. The change to a nonstratified pattern was more pronounced in MI cases of UroAp and UroC (p = 0.014 and p = 0.0007, respectively) (Figure 1E). A similar change was not observed in GU tumors (p = 0.39). Taken together, the seemingly intact layer of basal cells in UroA tumors is sequentially lost in the biologically more progressed variants UroAp, UroB, and UroC, as well as in MI cases. On the contrary, GU tumors show a loss of basal markers expression, irrespective of invasion depth.
Figure 2.
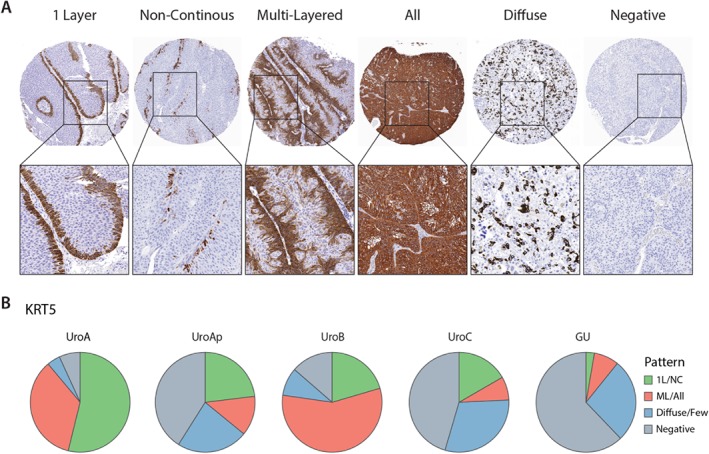
Expression patterns of KRT5 in the luminal subtypes. (A) Representative images of the different types of KRT5 expression patterns. (B) Proportions of the expression patterns within each molecular subtype. Labels: 1 Layer, one continuous layer of positive cells; NC, noncontinuous layer of positive cells; ML, multilayers of positive cells; All, all cells positive; Diffuse, scattered positive cells; Negative.
Proliferation
We used antibodies against CCNB1, Ki67, and FOXM1 to investigate proliferation. The three markers showed a strong concordance with respect to proportion of positive cells and staining pattern, with r > 0.8 in all comparisons. We observed a clear nonrandom distribution of proliferating cells in UroA (Figure 3A), in which positive cells were associated with the basal cell layer. This pattern was reduced in the UroAp, UroB, and UroC cases (Figure 3B), and almost absent in GU cases, where proliferating cells were randomly distributed in the tumor parenchyma. These observations suggest that proliferation in low‐grade UroA tumors is associated with the basal cell layer, an association lost in the more biologically progressed UroAp, UroB, UroC, and GU tumors. To investigate this further, we divided cases into those that showed a stratified expression for EGFR, CDH3, or KRT5, and those that did not, and then overlaid the data for CCNB1 and Ki67 (see supplementary material, Figure S2). An association between presence of stratified basal‐like cell layer and stratified proliferation as determined by CCNB1 was observed in UroA and UroAp (p < 0.02 and <0.02, respectively), but not in UroB, UroC, and GU cases (see supplementary material, Table S3). The overall proliferative activity determined as the proportion of CCNB1 positive cells increased in the order of UroA, UroAp, UroB, UroC, and GU (Figure 3C). The fraction of positive CCNB1 cells was further evaluated regarding the expression pattern of the combined basal markers as stratified or nonstratified (Figure 3D). A strong association between high proliferative activity and absence of a stratified basal‐like cell layer was observed. To investigate the proliferative cell compartment in UroA cases, we zoomed in on CCNB1 positive cells (Figure 3E). Positive cells were frequently attached to the basal‐membrane and dividing cells stretched out perpendicularly to the membrane. This suggests that in low‐grade UroA tumors, proliferation mainly occurs in the basal‐like cells.
Figure 3.
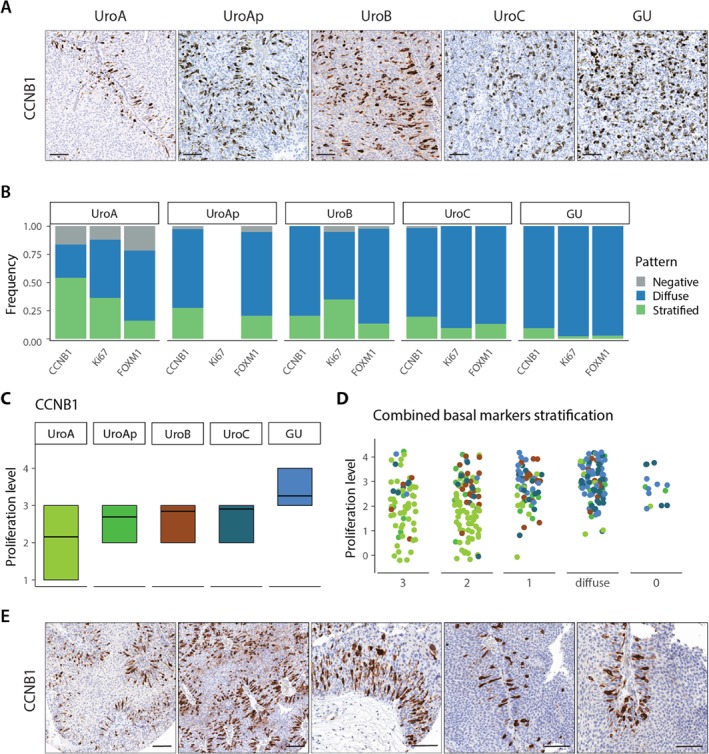
Expression of proliferation makers. (A) Representative images of CCNB1 expression for the respective molecular subtypes. Notice the stratified expression in UroA. (B) Frequency of staining patterns of CCNB1, Ki67, and FOXM1 within each molecular subtype. Ki67 data not available for UroAp. (C) Proliferation level of the tumor samples within each molecular subtype as determined by the fraction of CCNB1 positive tumor cells. Proliferation levels: 1, <10%; 2, 10–20%; 3, 20–50%; 4, 50–80%; 5, >80%. (D) Distribution of the proliferative levels of the samples divided by the combined staining patterns of the basal markers EGFR, CDH3, and KRT5. Labels; 3, all markers show stratified expression; 2, two markers show stratified expression; 1, one marker shows stratified expression; diffuse, no marker shows stratified expression and at least one shows diffused staining; 0, all markers are negative. Samples are colored according to their molecular subtype as in plot C. (E) Examples of CCNB1 staining in UroA tumors showing the association of proliferation with the basal cell layer.
Expression of FGFR3 and ERBB2 receptors
Cases were scored as either positive or negative for FGFR3 and ERBB2 expression, and clinically HER2 positive/negative were obtained from 13(Figure 4A). A large fraction of the Uro tumors was positive for FGFR3, 83% in UroA, 85% in UroAp, 82% in UroB and 72% in UroC, whereas GU showed a significantly lower fraction of positive cases, 27% (p < 2 × 10−7). The fraction of ERBB2 expressing cases increased in the order UroB (9%), UroA (17%), UroAp (30%), and UroC (42%), with the highest proportion of positive cases in GU (57%). The proportion of FGFR3 and ERBB2 positive cases did not differ between NMI and MI variants of UroAp, UroC, and GU tumors (Figure 4B). In each subtype, a part of the cases was positive for both FGFR3 and ERBB2 receptors (see supplementary material, Figure S3). However, there was no enrichment/depletion of double positive cases in any of the molecular subtypes (hypergeometric test, see supplementary material, Table S4), indicating an independent relationship between FGFR3 and ERBB2 expression. Taken together, the Uro tumors are characterized by high expression of FGFR3, and GU by low FGFR3 but high ERBB2 expression. In addition, UroC is similar to UroA, UroAp and UroB with respect to FGFR3 expression, and similar to GU with respect to ERBB2 expression.
Figure 4.
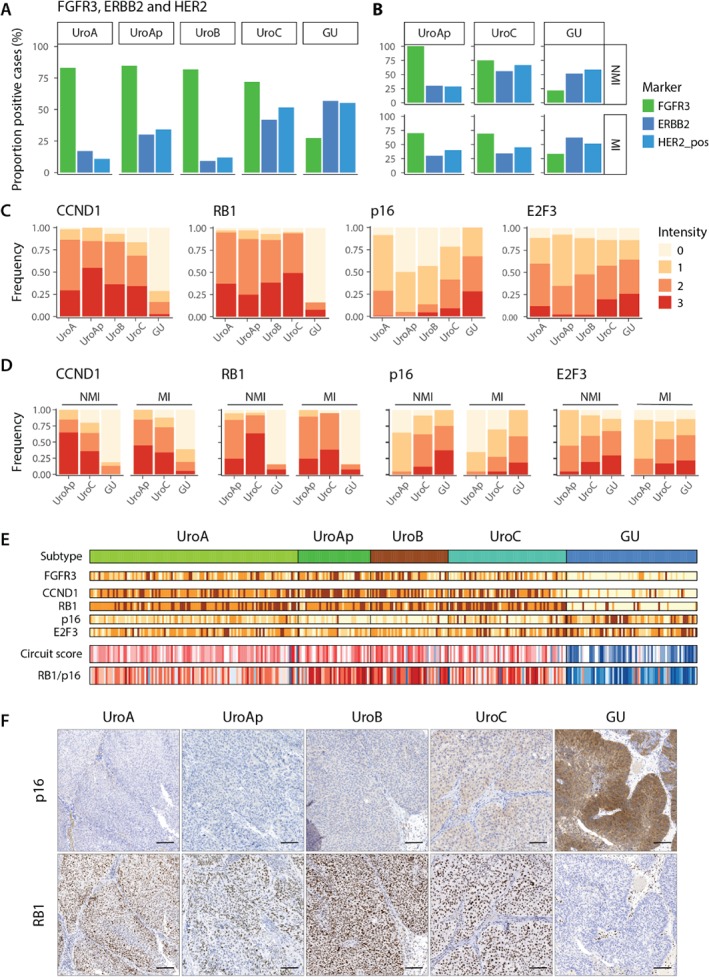
Expression of FGFR3 and ERBB2, and cell cycle regulating proteins. (A) Fraction of FGFR3 and ERBB2 positive and clinically HER2 cases within each molecular subtype. (B) The same data as in A given for UroAp, UroC, and GU divided into nonmuscle and muscle invasive cases. (C) Distribution of staining intensity levels of CCND1, RB1, p16, and E2F3 within each molecular subtype. (D) Same data as in C given for UroAp, UroC, and GU divided into nonmuscle and muscle invasive cases. Data for UroA and UroB not given due to too few samples in the respective groups. (E) Samples within each subtype ordered based on pathological grade and tumor stage. For each individual case a circuit score was calculated as FGFR3+CCND1+RB1‐E2F3 19 as well as a RB1/p16 ratio, using staining intensities. Color codes; yellow, low staining intensity; dark brown, high staining intensity. Red, score or ratio above threshold, blue, score or ratio below threshold. (F) Representative p16 and RB1 staining for the respective molecular subtypes.
Expression of cell cycle regulatory proteins
We determined the expression of the cell cycle regulatory proteins CCND1, RB1, E2F3, and CDKN2A(p16). CCND1 showed high expression in all Uro tumors, ranging from 84 to 100% of positive cases (Figure 4C). In contrast, the majority of GU tumors were negative or showed a low proportion of CCND1 positive cells. The proportion of CCND1 positive cases was not altered between NMI and MI tumors of UroAp, UroC, and GU (Figure 4D). Similarly, RB1 showed high expression in all Uro subtypes, with 95% positive cases on average, whereas the fraction of RB1 positive cases dropped down to 16% in GU cases (Figure 4C). The proportion of RB1 positive cells did not differ between NMI and MI versions of UroAp, UroC, and GU (Figure 4D). CDKN2A(p16), on the other hand, showed a reduced expression in the biologically more progressed UroAp, UroB, and UroC, compared to UroA, particularly evident by the increasing proportion of negative cases (Figure 4C). In contrast, GU showed no negative cases, and a higher overall expression compared to the Uro subtypes. The cell cycle related transcription factor E2F3 showed low expression in the Uro cases, and increased expression in the GU cases (Figure 4C). Detailed data for each tumor expression levels of FGFR3, CCND1, RB1, CDKN2A(p16), and E2F3 are shown in Figure 4E. We then applied a circuit score 7, 19 based on FGFR3, CCND1, RB1, and E2F3 expression that distinguishes Uro from GU tumors (Figure 4E). Noteworthy, the ratio between RB1 and CDKN2A(p16) expression distinguish the two subtypes equally well (AUC = 0.93) (Figure 4E,F).
Differentiation and growth patterns
We applied three markers to evaluate the status of the cellular differentiation process, the nuclear receptor PPARG, the transcription factor GATA3 operating downstream of PPARG, and TP63 that regulate stratified cell growth. The expression of the differentiation genes KRT20 and UPK3 was also evaluated. UroA, UroAp, and UroC showed high expression levels of all three markers PPARG, GATA3, and TP63 (Figure 5A). UroB differed by showing lower expression of PPARG and GATA3, and GU by showing comparable PPARG and GATA3 expression, but reduced TP63 expression. UroAp, UroC, and GU expression levels were similar between NMI and MI, except for a slight reduction of PPARG expression in MI cases (Figure 5B). A large fraction of the UroA, UroAp, UroC, and GU cases showed expression of KRT20, ranging from 60 to 80% (Figure 5C) and with proportions of positive tumor cells exceeding 50%. In contrast, UroB showed an increased number of KRT20 negative cases (Figure 5C) and a lower proportion of positive cells relative UroA. Additionally, KRT20 expression was associated with the less differentiated high‐grade tumors both for staining intensity and proportion of positive cells, G1 versus G3, p < 0.03 and p < 0.001, respectively (Figure 5D). UPK3 showed a similar expression patterns, more frequently expressed in the luminal part of the tumors, particularly in G1 tumors (see supplementary material, Figure S4). KRT20 and UPK3 expression, when positive, were abnormal in the vast majority of cases, either cytoplasmic or nuclear, and expressed in the interior of the tumor parenchyma (Figure 5E). The tumors were analyzed with respect to tumor growth patterns based on cellular orientation and overall organization of tumor cells and surrounding stroma. UroA cases were essentially composed of tumors with a structured, urothelial growth pattern (Figure 5F). An almost even distribution between urothelial and other growth patterns was observed in UroAp, UroB, UroC, and GU tumors (Figure 5F). NMI tumors differed significantly from MI cases; urothelial growth pattern was the far dominating pattern in NMI tumors (Figure 5G). The expression of the class defining genes FGFR3, ERBB2, CCND1, CDKN2A(p16), RB1, and E2F3, as well as the differentiation related genes PPARG, GATA3, TP63, and KRT20 was compared between tumors with urothelial and other growth patterns within UroAp, UroB, UroC, and GU respectively. No significant differences were observed, except for CDKN2A(p16) that showed significant downregulation in UroC tumors (see supplementary material, Table S5). Hence, both class defining and regulatory genes act as intrinsic properties of the tumors, independent of the growth pattern.
Figure 5.
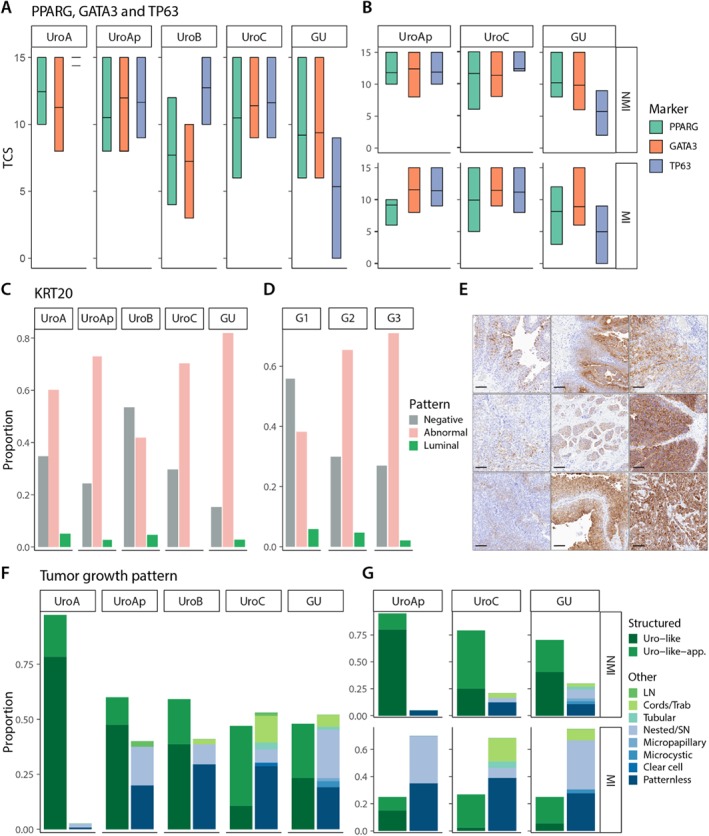
Cellular differentiation and growth patterns. (A) Expression levels of PPARG, GATA3, and TP63 indicated as tumor cell scores for each luminal subtype. (B) Same data as in C given for UroAp, UroC, and GU divided into nonmuscle and muscle invasive cases. (C,D) Frequency of the different KRT20 expression patterns within each molecular subtype and histologic grade. (E) Examples of aberrant KRT20 expression. (F) Distribution of different growth patterns within each molecular subtype. The growth patterns are grouped into urothelial pattern or nonurothelial pattern (other). (G) The same data as in F given for UroAp, UroC, and GU divided into nonmuscle and muscle invasive cases. Data for UroA and UroB not given due to too few samples in the respective groups.
Molecular pathological grouping of luminal UCs
Eleven variables based on molecular pathological data: combined basal markers expression pattern, CCNB1, FGFR3, ERBB2, CCND1, RB1, CDKN2A(p16), PPARG, GATA3, TP63, and KRT5 expression, were used to characterize 318 samples. The SPRING software was used to organize the tumors with respect to similarity between individual cases resulting in three major tumor clusters (Figure 6). One large cluster dominated by UroA and UroB cases, one dominated by UroC, and one distinct cluster composed almost entirely of GU cases (Figure 6A). A good association with tumor grouping and pathological grade (Figure 6B), as well as the integrity of the basal cell layer (Figure 6C) was observed. As expected, cases with a low RB1/p16 ratio were enriched in the GU cluster (Figure 6D). Finally, cases were labeled according to the consensus classification system 10 (see supplementary material, Figure S5). Most of the cases were identified as luminal‐papillary, whereas the GU cluster was mainly assigned to the luminal‐unstable category. The few cases classified as basal/squamous by the consensus were all of the UroB group.
Figure 6.
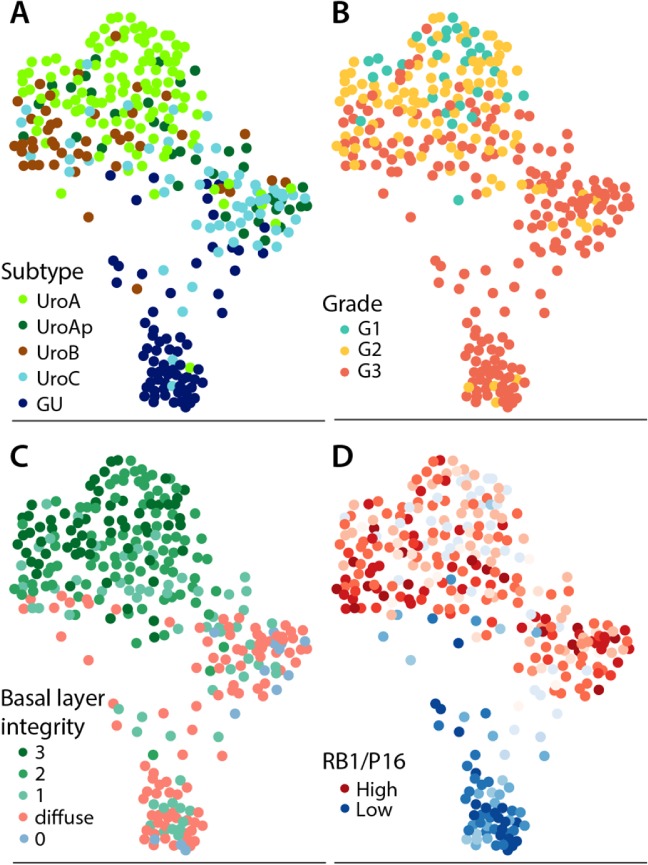
SPRING graph of tumor cases based on similarity of molecular pathological features. Cases were scored for the variables: combined basal markers, CCNB1, FGFR3, HER2, CCND1, RB1, p16, PPARG, GATA3, TP63, and KRT5 expression, and then organized in a k‐nearest neighbor two‐dimensional graph using the SPRING algorithm. Each point represents a tumor sample. The clusters are labeled according to (A) the LundTax, (B) the pathological grade (WHO 1999), (C) combined stratified expression of EGFR, CDH3, and KRT5. Labels: 3, all markers show stratified expression; 2, two markers show stratified expression; 1, one marker shows stratified expression; diffuse, no marker shows stratified expression and at least one shows diffused staining; 0, all markers show negative staining. (D) Cases labeled according to the RB1/p16 ratio. Red: high ratio; blue: low. High ratio indicates expression of p16 but not RB1.
Discussion
Our aim was to clarify discrepancies in the classification of luminal UCs. For this purpose, 344 luminal tumors classified as UroA, UroAp, UroB, UroC, or GU, for which TMAs had been produced, were retrieved from two published studies 1, 7. Tumors were analyzed using IHC with respect to the integrity of a basal cell layer (KRT5, CDH3, and EGFR); proliferation (CCNB1, Ki67, and FOXM1); receptor tyrosine kinase expression (FGFR3 and ERBB2); cell cycle regulation (CCND1, CDKN2A(p16), E2F3, and RB1); regulation of differentiation (PPARG, GATA3, and TP63); and differentiation (KRT20 and UPK3). In addition, we used pathological stage and grade as well as histological evaluation of growth patterns to characterize the tumors.
Tumor cell phenotypes
UroA was originally described in a cohort dominated by nonmuscle invasive cases 1 and the UroAp (UroA‐progressed) identified as UroAs in a cohort of cystectomized patients 7. Regardless, there are no differences between UroA and UroAp with respect to expression of class defining markers such as FGFR3, CCND1, and RB1, as well as PPARG, GATA3, and TP63. In addition, both show low expression of CDKN2A(p16) and ERBB2. The high expression of FGFR3 and low expression of CDKN2A is in accordance with the frequent activating mutations in FGFR3 and homozygous deletions of CDKN2A in this subtype 8. The major differences are seen at the level of proliferation, growth pattern, and pathological grade. Consequently, UroAp is an aggressive version of UroA.
UroB is classified as basal/squamous both by the TCGA and by the Consensus classification systems. However, the UroB shares many characteristics of the UroA tumors, such as high FGFR3, CCND1, and RB1, and low CDKN2A(p16) expression 7, 8. Furthermore, UroB often shows a stratified organization with well‐defined KRT5, CDH3, and EGFR positive cell layers 16. UroB differs from UroA by regularly showing an increased number of cells expressing basal markers, and lower expression of the differentiation factors PPARG and GATA3. In this respect, UroB tumors develop towards a basal/squamous‐like phenotype, however, they do not conform to the consensus Ba/Sq definition 8. In addition, UroB cannot be the major progenitor of Ba/Sq tumors as UroB shows FGFR3 mutations in 50% of the cases whereas the Ba/Sq tumors rarely present with FGFR3 mutations 8. Hence, UroB is a Uro tumor that has acquired some of the molecular features of basal/squamous tumors.
The most distinct feature among the luminal tumors observed in this work was the different characteristics of GU. Even though GU express the differentiation factors PPARG and GATA3, and occasionally the differentiation markers KRT20 and UPK3, and thus belong to the luminal group, they differ radically with respect to the integrity of a basal cell layer, topology of proliferation, expression of receptors, and cell cycle regulatory factors. GU also differs by showing, more frequently, loss of expression of the regulatory factor TP63, involved in epithelial stratification. Furthermore, these characteristics are intrinsic to the GU subtype in as much as NMI cases appeared molecularly identical to the MI cases of the subtype. Apart from frequently loss of KRT5 positive cells and frequent ERBB2 expression, the most distinct difference from the Uro was the inactivation of RB1 and increased expression of CDKN2A(p16). These findings are in line with the frequent genomic alterations of RB1 and almost absence of CDKN2A deletions in the GU subtype 8. In these aspects GU is very similar to carcinoma in situ of the bladder 20, 21. Both the circuit score as well as the RB1/p16 ratio clearly distinguished GU from the remaining Uro cases. Thus, the inverse expression of RB1 and p16 first observed by Benedict et al 22 is related to molecular subtypes of UC. Hence, GU is a biological entity very distinct from the Uro tumors.
UroC is the most ambiguous of the Uro tumors. Originally identified as a group of tumors clustering with GU cases using genome wide gene expression, but which share class defining characteristics with UroA 7. Genomic data demonstrated that UroC express FGFR3 and frequently show CDKN2A homozygous deletions but rarely RB1 alterations 8 indicating a strong similarity to UroA. In the present investigation we show that UroC express FGFR3, CCND1, and RB1, as well as PPARG, GATA3, and TP63, similar to UroA and UroAp. Furthermore, both the circuit score and the RB1/p16 ratio, placed UroC on the Uro side. However, UroC show signs of biological progression away from an UroA‐type as basal cell markers are rarely expressed, proliferation is high, and the majority are of high grade. In addition, UroC frequently express both FGFR3 and ERBB2. Consequently, UroC is an UroA dressed in a GU overcoat.
Proliferation
We used antibodies for CCNB1, Ki67, and FOXM1 to evaluate proliferation. In contrast to Ki67 and FOXM1, CCNB1 only labels cells in late cell cycle, G2, and not after cell division has occurred. Thus, CCNB1 identifies cells in active cell division and, as the staining is cytoplasmic, the extension/polarity of dividing cells is traceable. In UroA almost all CCNB1 positive cells were attached to the basal membrane and extended, perpendicularly, into the tumor parenchyma. As these cells were consistently positive for KRT5, CDH3, or EGFR, markers for basal cell identity, this indicates that, for UroA, proliferation only occurs in the basal cell compartment. In the more progressed UroAp two changes occur, first, the nature of the basal cells dwindles by being positive for fewer basal cell markers, and second, CCNB1 positive cells are detected at some distance from the basal layer. Consequently, proliferation in UroAp is not limited to the basal cells nor to the proximity of a basal membrane. A similar pattern was seen in UroC. Hence, the transition from UroA to UroAp and to UroC, involves the release of proliferation from basal cells linked to a basal membrane. The UroB seems to take a slightly different route. In UroB an increased number of cells become positive for KRT5, indicating the existence of basal‐like cells not in contact with a basal membrane. Hence, UroB seem to achieve increased proliferation by expansion of the basal cell compartment. The topology of proliferation in GU was in stark contrast to the Uro‐subtypes. First, proliferation occurred independent of a basal membrane, second, the polarity of cell divisions was randomly oriented, and third, there was no topological difference between NMI and MI cases. This suggests that decoupling of proliferation from the ‘basal cell compartment’ occurs earlier in GU compared to Uro tumors. This difference may be linked to key dissimilarities in the regulatory make‐up of the two tumor classes, e.g. Uro‐cases show FGFR3 expression and an accelerating inactivation of CDKN2A(p16), whereas GU show ERBB2 expression and inactivation of RB1.
Differentiation and growth patterns
All tumors subtypes, with the possible exception of UroB, expressed PPARG and GATA3, indicating that parts of the normal differentiation pathway are present 23, 24, 25. Consequently, the expression of the differentiation markers KRT20 and UPK3 in a subpopulation of tumors may not come as a surprise. Intriguingly, KRT20 and UPK3 expression was increased in tumors of high pathological grade. Furthermore, expression was abnormal, either by cellular or topological location, in almost all cases. The most pertinent interpretation of these observations is that pseudo‐differentiation occurs in response to a general dysregulation in high‐grade tumors, rather than a sign of cellular origin. Additionally, tumors expressing KRT20/UPK3 did not differ from negative tumors concerning class‐defining markers. This also held true for tumors of the same subtype showing different growth patterns. Hence, tumors maintain their molecular identity when transformed into a more aggressive growing pattern. As nonurothelial growth patterns almost invariably occurred in MI cases, one may speculate that the change in growth pattern is determined, at least partly, by cues in the tumor microenvironment, or by the cellular composition of the stromal compartment surrounding the tumor. This points to the possible importance of tumor associated stromal cells for a proper (clinical) classification of tumors.
We conclude that luminal UCs consist of two major classes, Uro and GU. Of these two, the Uro‐group is the larger and more heterogeneous. This became particularly evident by the SPRING clustering of the tumors using molecular pathological features. The Uro tumors formed a large cluster with discernable subgroupings whereas the GU cases formed a tight single group at some distance from the Uro cases. Although clearly distinct from GU, the large heterogeneous group of Uro‐tumors exists in a spectrum, which may explain why investigators arrive at different number of luminal‐like subtypes. Furthermore, most classification systems are based on mRNA profiling of whole biopsy specimens, an approach affected by infiltrating stromal and/or immunological cells. Consequently, both the TCGA and the Consensus classification systems have subtypes assigned as luminal‐infiltrated or stroma‐rich masking the underlying tumor cell phenotypes. In contrast, the LundTax mRNA‐based classifier 8 is independent of signals from nontumors cells, and therefore, based on features of the tumor cell phenotypes only.
Author contributions statement
CB performed the IHC reevaluations, statistical analyses, and drafted the manuscript. PE performed gene expression classifications. NM performed statistical analyses and global analyses, FL provided samples, pathology, and clinical follow up. GS performed the initial IHC analyses. MH conceived the study and drafted the manuscript.
SUPPLEMENTARY MATERIAL ONLINE.
Figure S1. Expression pattern of CDH3, EGFR, and KRT5
Figure S2. Proliferation pattern as determined by CCNB1 and Ki67 expression grouped by the combined basal markers EGFR, CDH3, and KRT5 expression pattern as stratified (Strat) or diffuse (Diff) for each molecular subtype
Figure S3. Tumor expression of FGFR3 and ERBB2
Figure S4. UPK3 tumor expression pattern stratified by molecular subtype and pathological grade
Figure S5. SPRING graph of the tumor cases based on similarity of molecular pathological features
Table S1. Clinicopathological characteristics of the cohort (Grade according to WHO 1999)
Table S2. Antibodies used for immunohistochemistry
Table S3. Number of cases per stratification status for CCNB1 and combined basal markers (at least one versus diffuse)
Table S4. Number of positive cases and proportion by marker and subtype
Table S5. Statistical analysis of the proportion of positive and negative cases for each marker divided according to morphology (Uro‐Like or other), FGFR3/ERBB2: pos/neg; CCND1, p16, RB1, E2F3: intensity; PPARG, GATA3, P63: tumor cell score
Supporting information
Figure S1. Expression pattern of CDH3, EGFR, and KRT5
Figure S2. Proliferation pattern as determined by CCNB1 and Ki67 expression grouped by the combined basal markers EGFR, CDH3, and KRT5 expression pattern as stratified (Strat) or diffuse (Diff) for each molecular subtype
Figure S3. Tumor expression of FGFR3 and ERBB2
Figure S4. UPK3 tumor expression pattern stratified by molecular subtype and pathological grade
Figure S5. SPRING graph of the tumor cases based on similarity of molecular pathological features
Table S1. Clinicopathological characteristics of the cohort (Grade according to WHO 1999)
Table S2. Antibodies used for immunohistochemistry
Table S3. Number of cases per stratification status for CCNB1 and combined basal markers (at least one versus diffuse)
Table S4. Number of positive cases and proportion by marker and subtype
Table S5. Statistical analysis of the proportion of positive and negative cases for each marker divided according to morphology (Uro‐Like or other), FGFR3/ERBB2: pos/neg; CCND1, p16, RB1, E2F3: intensity; PPARG, GATA3, P63: tumor cell score
Acknowledgements
This work was supported by the Swedish Research Council, The Swedish Cancer Foundation, The Nilsson Cancer Foundation, The Kamprad Foundation, and by BioCare.
No conflicts of interest were declared.
References
- 1. Sjödahl G, Lauss M, Lovgren K, et al A molecular taxonomy for urothelial carcinoma. Clin Cancer Res 2012; 18: 3377–3386. [DOI] [PubMed] [Google Scholar]
- 2. Damrauer JS, Hoadley KA, Chism DD, et al Intrinsic subtypes of high‐grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci USA 2014; 111: 3110–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choi W, Porten S, Kim S, et al Identification of distinct basal and luminal subtypes of muscle‐invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014; 25: 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cancer Genome Atlas Research Network . Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014; 507: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lerner SP, McConkey DJ, Hoadley KA, et al Bladder cancer molecular taxonomy: summary from a consensus meeting. Bladder Cancer 2016; 2: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aine M, Eriksson P, Liedberg F, et al Biological determinants of bladder cancer gene expression subtypes. Sci Rep 2015; 5: 10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sjödahl G, Eriksson P, Liedberg F, et al Molecular classification of urothelial carcinoma: global mRNA classification versus tumour‐cell phenotype classification. J Pathol 2017; 242: 113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marzouka N, Eriksson P, Rovira C, et al A validation and extended description of the Lund taxonomy for urothelial carcinoma using the TCGA cohort. Sci Rep 2018; 8: 3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Robertson AG, Kim J, Al‐Ahmadie H, et al Comprehensive molecular characterization of muscle‐invasive bladder cancer. Cell 2017; 171: 540–556.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kamoun A, de Reynies A, Allory Y, et al The consensus molecular classification of muscle‐invasive bladder cancer. bioRxiv 2018. 10.1101/488460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hedegaard J, Lamy P, Nordentoft I, et al Comprehensive transcriptional analysis of early‐stage urothelial carcinoma. Cancer Cell 2016; 30: 27–42. [DOI] [PubMed] [Google Scholar]
- 12. Hurst CD, Alder O, Platt FM, et al Genomic subtypes of non‐invasive bladder cancer with distinct metabolic profile and female gender bias in KDM6A mutation frequency. Cancer Cell 2017; 32: 701–715.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eriksson P, Sjödahl G, Chebil G, et al HER2 and EGFR amplification and expression in urothelial carcinoma occurs in distinct biological and molecular contexts. Oncotarget 2017; 8: 48905–48914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amin MB. Histological variants of urothelial carcinoma: diagnostic, therapeutic and prognostic implications. Mod Pathol 2009; 22: S96–S118. [DOI] [PubMed] [Google Scholar]
- 15. Humphrey PA, Moch H, Cubilla AL, et al The 2016 WHO classification of tumours of the urinary system and male genital organs. Part B: prostate and bladder tumours. Eur Urol 2016; 70: 106–119. [DOI] [PubMed] [Google Scholar]
- 16. Sjödahl G, Lövgren K, Lauss M, et al Toward a molecular pathologic classification of urothelial carcinoma. Am J Pathol 2013; 183: 681–691. [DOI] [PubMed] [Google Scholar]
- 17. Scanlon DO, Morgan BJ, Watson GW. Modeling the polaronic nature of p‐type defects in Cu2O: the failure of GGA and GGA+U. J Chem Phys 2009; 131: 124703. [DOI] [PubMed] [Google Scholar]
- 18. Weinreb C, Wolock S, Klein AM. SPRING: a kinetic interface for visualizing high dimensional single‐cell expression data. Bioinformatics 2018; 34: 1246–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindgren D, Sjödahl G, Lauss M, et al Integrated genomic and gene expression profiling identifies two major genomic circuits in urothelial carcinoma. PLoS One 2012; 7: e38863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edgecombe A, Nguyen BN, Djordjevic B, et al Utility of cytokeratin 5/6, cytokeratin 20, and p16 in the diagnosis of reactive urothelial atypia and noninvasive component of urothelial neoplasia. Appl Immunohistochem Mol Morphol 2012; 20: 264–271. [DOI] [PubMed] [Google Scholar]
- 21. Barth I, Schneider U, Grimm T, et al Progression of urothelial carcinoma in situ of the urinary bladder: a switch from luminal to basal phenotype and related therapeutic implications. Virchows Arch 2018; 472: 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Benedict WF, Lerner SP, Zhou J, et al Level of retinoblastoma protein expression correlates with p16 (MTS‐1/INK4A/CDKN2) status in bladder cancer. Oncogene 1999; 18: 1197–1203. [DOI] [PubMed] [Google Scholar]
- 23. Varley CL. Role of PPAR and EGFR signalling in the urothelial terminal differentiation programme. J Cell Sci 2004; 117: 2029–2036. [DOI] [PubMed] [Google Scholar]
- 24. Eriksson P, Aine M, Veerla S, et al Molecular subtypes of urothelial carcinoma are defined by specific gene regulatory systems. BMC Med Genomics 2015; 8: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Warrick JI, Walter V, Yamashita H, et al FOXA1, GATA3 and PPARɣ cooperate to drive luminal subtype in bladder cancer: a molecular analysis of established human cell lines. Sci Rep 2016; 6: 38531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Expression pattern of CDH3, EGFR, and KRT5
Figure S2. Proliferation pattern as determined by CCNB1 and Ki67 expression grouped by the combined basal markers EGFR, CDH3, and KRT5 expression pattern as stratified (Strat) or diffuse (Diff) for each molecular subtype
Figure S3. Tumor expression of FGFR3 and ERBB2
Figure S4. UPK3 tumor expression pattern stratified by molecular subtype and pathological grade
Figure S5. SPRING graph of the tumor cases based on similarity of molecular pathological features
Table S1. Clinicopathological characteristics of the cohort (Grade according to WHO 1999)
Table S2. Antibodies used for immunohistochemistry
Table S3. Number of cases per stratification status for CCNB1 and combined basal markers (at least one versus diffuse)
Table S4. Number of positive cases and proportion by marker and subtype
Table S5. Statistical analysis of the proportion of positive and negative cases for each marker divided according to morphology (Uro‐Like or other), FGFR3/ERBB2: pos/neg; CCND1, p16, RB1, E2F3: intensity; PPARG, GATA3, P63: tumor cell score


