Abstract
Two well‐characterized carbohydrate epitopes are absent in humans but present in other mammals. These are galactose‐α1,3‐galactose (αGal) and N‐glycolylneuraminic acid (Neu5Gc) which are introduced by the activities of two enzymes including α(1,3) galactosyltransferase (encoded by the GGTA1 gene) and CMP‐Neu5Gc hydroxylase (encoded by the CMAH gene) that are inactive in humans but present in cattle. Hence, bovine‐derived products are antigenic in humans who receive bioprosthetic heart valves (BHVs) or those that suffer from red meat syndrome. Using programmable nucleases, we disrupted (knockout, KO) GGTA1 and CMAH genes encoding for the enzymes that catalyse the synthesis of αGal and Neu5Gc, respectively, in both male and female bovine fibroblasts. The KO in clonally selected fibroblasts was detected by polymerase chain reaction (PCR) and confirmed by Sanger sequencing. Selected fibroblasts colonies were used for somatic cell nuclear transfer (SCNT) to produce cloned embryos that were implanted in surrogate recipient heifers. Fifty‐three embryos were implanted in 33 recipients heifers; 3 pregnancies were carried to term and delivered 3 live calves. Primary cell cultures were established from the 3 calves and following molecular analyses confirmed the genetic deletions. FACS analysis showed the double‐KO phenotype for both antigens confirming the mutated genotypes. Availability of such cattle double‐KO model lacking both αGal and Neu5Gc offers a unique opportunity to study the functionality of BHV manufactured with tissues of potentially lower immunogenicity, as well as a possible new clinical approaches to help patients with red meat allergy syndrome due to the presence of these xenoantigens in the diet.
Keywords: bioprosthetic Heart Valve (BHV), cattle, CMAH, GGTA1, knockout, Neu5Gc, xenotransplantation, αGal
1. INTRODUCTION
Two well‐characterized antigens are absent in humans but present in mammals and include galactose‐α1,3‐galactose (αGal) and N‐glycolylneuraminic acid (Neu5Gc) whose synthesis are catalysed by α(1,3) galactosyltransferase (encoded by the GGTA1 gene)1, 2 and CMP‐Neu5Gc hydroxylase (encoded by the CMAH gene)3, 4, 5 respectively. These have been identified as major antigens in xenotransplantation studies or retrospective clinical findings3. Pigs that carry mutations in both genes, and therefore lack these xenoantigens, have been generated.6 Moreover, porcine kidneys lacking αGal are not hyperacutely rejected.7 It is also expected that such tissues will be less immunogenic for patients being implanted with animal‐derived tissues engineered to lack both antigens.
One of the major clinical applications of xenogenic tissues is for the manufacturing of bioprosthetic heart valves (BHVs), and it has been shown that such tissues carry the same xenoantigens despite the glutaraldehyde treatments used in the manufacturing process8, 9. Almost 300 000 patients are now undergoing BHV replacement each year10 with a growing demand. The sources of BHV are those manufactured from pig or bovine pericardia as compared to mechanical heart valve (MHV) that require lifelong anticoagulation therapy.
Bovine BHVs suffer however premature structural valve degeneration (SVD). The functionality of BHV is maintained for 10‐15 years in older patients. However, in younger (<35 years old) patients, BHVs undergo SVD much earlier.11 It is hypothesized that among various metabolic causes, SVD is also immune‐mediated since both αGal8, 12 and Neu5Gc9, 13 are still present on the BHV used in the clinic.
After BHV replacement, there is an increase of anti‐αGal antibodies14, 15 and it has been reported in an experimental context that implantation of BHV from αGal‐knockout pigs into primates is associated with a reduced anti‐αGal immune response.16 Moreover, valves from αGal/Neu5Gc‐deficient pigs further reduce human IgM/IgG binding when compared to BHV from wild‐type pigs17. A similar situation is likely to occur whether bovine double knockout (DKO) tissue would be used. Seventy per cent of the BHV currently used in the clinic are in fact manufactured with bovine pericardia, that carries non‐negligible amounts of αGal8 and of Neu5Gc9 even after currently used manufacturing treatments.
Pig‐ and cattle‐derived products are also a major source of proteins for human consumption, and particularly, cattle are the major source of dairy products. Such products can become allergenic for some patients or infants consuming baby milk replacers. This allergy, known as the red meat allergy syndrome,18, 19 generally follows a tick bite inducing an isotype shift for IgE against αGal antigen. Neu5Gc is not synthesized by humans, but it can be incorporated through the diet and found in minute amounts in endothelial or epithelial cells of various tissues, likely contributing to inflammation‐related diseases.20, 21 Furthermore, cattle can be used as a “bioreactor” to produce bioactive molecules for nutraceuticals or biomedical use, including r‐human lactoferrin22 in bovine milk. However, the resulting product differs from the human one because of the different glycosylation pattern.23 Similarly, partially “humanized” antibodies24 produced in cattle for various purposes still display Neu5Gc epitopes25, 26 that might be the target of an immune response by the host with clinically relevant side effects.
The scope of the present work was to generate cattle KO for both αGal and Neu5Gc antigens using a genome editing approach.27 A stillborn calf KO for αGal has been reported,28 but to the best of our knowledge, this work has not progressed further. Availability of DKO cattle line offers the opportunity to explore the potential of such animals to provide low immunogenic cattle‐derived products for clinical purposes as well as for the food industry and human consumption.
2. MATERIALS AND METHODS
2.1. Animal experiments and source of animals
All procedures involving the use of animals in this study were approved by the Animal Welfare Committee of Avantea and carried out in accordance with the Italian Law (D.Lgs 26/2014) and EU directive 2010/63/EU regulating animal experimentation after authorization by relevant authorities (Ministry of Health project n 991/2017‐PR). Bovine adult fibroblasts (BAFs) were derived from a skin biopsy of a Holstein bull and a cow with previous successful record of somatic cell nuclear transfer (SCNT). Recipient heifers used as surrogate mothers were also of Holstein breed.
2.2. Chemicals
All chemicals were purchased from Sigma‐Aldrich (Milano, Italy) unless otherwise stated.
2.3. PCR set‐up for identification and validation of target genes
Ensemble database was analysed to obtain the Reference genome sequences for the GGTA1 (ENSBTAG00000012090) and of the CMAH (ENSBTAG00000003892) genes. These sequences were studied in silico to identify possible target sequences, and the selected regions were amplified by PCR and analysed with Sanger sequencing to exclude polymorphisms in male and female fibroblast cell lines selected for the genome editing.
Editing of GGTA1 gene started initially in the male line targeting the exon 9 and because of the paucity of tools available at the time we never found efficient RNA guide. Therefore, we decided to use two guides that targeted the same sequence (Table 2, btGGTA1cr1 and btGGTA1cr2). Years later, when we targeted the female line, we were able to find an efficient guide for exon 4 (Table 3, btGGTA1cr3) used for the pig by Sato et al.29 Editing of the CMAH gene was achieved efficiently in the exon 2 carrying the ATG codon. For the editing of the male, we used one guide (Table 2, btCMAHcr1) and subsequently for the female we found a more efficient guide (Table 3, btCMAHcr2).
Table 2.
Oligonucleotides synthetized for the assembly of desired CRISPR/Cas9 expression vectors used for the male line and sequence of the ssCMAH‐STOP oligo
| Oligo | Sequence (5′‐3′) | Guide sequence—PAM (5′‐3′) | Target gene (exon) | Expression vector |
|---|---|---|---|---|
| btGGTA1cr1 FW | CACCGGAGACCCTGGGCGAGTCGG | GGAGACCCTGGGCGAGTCGG‐TGG | GGTA1 (9) | pX330‐btGGTA1cr1 |
| btGGTA1cr1 RV | AAACCCGACTCGCCCAGGGTCTCC | |||
| btGGTA1cr2 FW | CACCGCTGGGCCACCGACTCGCCC | GCTGGGCCACCGACTCGCCC‐AGG | GGTA1 (9) | pX330‐btGGTA1cr2 |
| btGGTA1cr2 RV | AAACGGGCGAGTCGGTGGCCCAGC | |||
| btCMAHcr1FW | CACCGACTATGGGCAGGCAAGTGA | GACTATGGGCAGGCAAGTGA‐GGG | CMAH (2) | pX330‐btCMAHcr1 |
| btCMAHcr1 RV | AAACTCACTTGCCTGCCCATAGTC | |||
| ssCMAH‐STOP oligo | GTGACAGCTGCCATTCTTCTGAAATACCCAGGGAGAGGCAACGACAGACTTAAGGCAGGCAAGTGAGGGAGGCATTACTTTGCTGGGAAGGTGGGGTCAA | // | // | // |
Table 3.
Oligonucleotides used for in vitro T7 transcription of sgRNAs used for the female line
| Oligo | Sequence (5′‐3′) | Guide sequence—PAM (5′‐3′) | Target gene (exon) |
|---|---|---|---|
| btGGTA1cr3sgRNA | GAAATTAATACGACTCACTATAGAGAAAATAATGAATGTCAAGTTTTAGAGCTAGAAATAGCAAG | GAGAAAATAATGAATGTCAA‐AGG | GGTA1 (4) |
| btCMAHcr2sgRNA | GAAATTAATACGACTCACTATAGAGAGGCAACGACAGACTATGTTTTAGAGCTAGAAATAGCAAG | GAGAGGCAACGACAGACTAT‐GGG | CMAH (2) |
| sgRNAT7common | AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC | // | // |
Target exons and primers used for PCR analyses and Sanger sequencing of each gene are summarized in Table 1. All the synthetized oligonucleotides and the Sanger sequencing services were purchased from Eurofins Genomics, unless otherwise stated.
Table 1.
Primers used for genotyping of bovine wild‐type cell lines, edited colonies and cloned animals
| Oligo | Sequence (5′‐3′) | Gene | Target exon | Amplicon (bp) |
|---|---|---|---|---|
| FW1 | GGATGCCTTTGATAGAGTTGG | GGTA1 | 9 | 440 |
| RV1 | GCTTTCATCATGCCATTGG | |||
| FW2 | AGCATCTTTCACAACTCAGG | GGTA1 | 4 | 739 |
| RV2 | TGAGACATTAGGAACATGGC | |||
| FW3 | TCAGGAGGAGACATCACCAACGG | CMAH | 2 | 225 |
| RV3 | TGCCCATCCTACTTGTCGAGGG |
2.4. Genomic DNA extraction and PCR conditions
Primary fibroblasts and tissues biopsies were lysed at 55°C for 3 hours using a lysis buffer (100 mmol/L Tris HCl pH 8.3, 5 mmol/L EDTA pH 8.1, 0.2% SDS, 200 mmol/L NaCl) supplemented with Proteinase K (300 µg/mL; Macherey‐Nagel). Genomic DNA was extracted (Sambrook et al, 1989) and resuspended with TE buffer. All the amplifications were performed using Takara La Taq DNA Polymerase (Takara, Japan).
Polymerase chain reaction conditions for GGTA1 in the male line (exon 9) were as follows (FW1 + RV1 = 440 bp): 94°C, 2 minutes; 94°C, 30 seconds, 72°C (−1°C/cycle), 30 seconds; 72°C, 15 seconds for 8 cycles; 94°C, 30 seconds, 58°C, 30 seconds; 72°C, 15 seconds for 35 cycles; and a final extension step of 72°C for 7 minutes. PCR conditions for GGTA1 in the exon 4 of the female line were as follows (FW2 + RV2 = 739 bp): 94°C, 2 minutes; 94°C, 30 seconds, 60°C, 30 seconds, 72°C, 30 seconds for 35 cycles; and a final extension step of 72°C for 7 minutes.
For CMAH, PCR conditions were as follows (FW3 + RV3 = 225 bp): 94°C, 2 minutes; 94°C, 30 seconds; 58°C, 30 seconds; 72°C, 30 seconds for 40 cycles; and a final extension step of 72°C for 7 minutes.
Determination of the absence of any genomic polymorphisms was achieved cloning each resulting PCR products in E coli using the TOPO TA cloning kit (Thermo Fisher Scientific). Resulting purified plasmids (Plasmid Mini kit, Qiagen) were subjected to Sanger sequencing analyses (Eurofins Genomics).
2.5. CRISPR/Cas9 plasmid constructs, single guide RNA synthesis and design of ssCMAH‐STOP oligonucleotide
Editing of the GGTA1 and CMAH genes of the bovine male line was achieved by cloning and expressing the desired single guide RNAs (sgRNAs), into the pX330‐U6‐Chimeric_BB‐CBh‐hSpCas9 expression vector, that was a gift from Feng Zhang (Addgene plasmid # 42230). DNA oligonucleotides for sgRNAs (Table 2) were purchased from Eurofins Genomics. Annealing and molecular cloning of gene‐specific complementary oligos were done following the protocol described by Cong and colleagues.30 The resulting purified expression vectors (Plasmid mini kit, PC‐20, Qiagen) were verified by Sanger sequencing before transfection.
Edited female colonies were obtained transfecting the Cas9 protein/gRNA ribonucleoprotein complexes (Cas9‐RNPs).31, 32 Desired sgRNAs (btGGTA1cr3 and btCMAHcr2) were in vitro synthetized following the CRISPOR guidelines (http://crispor.org/). Briefly, oligonucleotides (Table 3) were annealed, amplified and purified before to use the resulting amplification product as template (1μg) for the following transcription step. Single guide RNAs were finally synthetized using the TranscriptAid T7 High Yield Transcription kit (Thermo Fisher Scientific) and purified on silica membranes columns (MEGAclear Transcription clean‐up kit, Thermo Fisher Scientific) according to the manufacturer's instructions and stored at −80°C.
We targeted the CMAH gene using as template a synthetized single strand oligonucleotide (ssCMAH‐STOP oligo) specific for the exon 2 and symmetric according to the position of the CMAH‐START codon. Its sequence is characterized by the substitution of the START codon (ATG) with a STOP codon (TAA, in bold Figure 1A), generating a new AflII restriction site (CT TAA G, underlined in Figure 1A), useful for the identification of the knock‐in colonies (152bp + 73 bp) with the AflII‐RFLP analyses (AflII from Thermo Fisher Scientific; 1 hour at 37°C).
Figure 1.
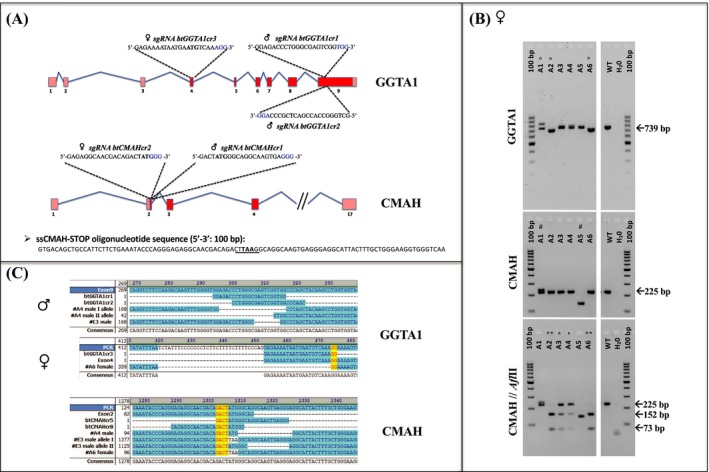
Editing of GGTA1 and CMAH genes in male and female fibroblasts. A, Target sequences for selected sgRNAs and ssCMAH‐STOP oligo sequence. For each bovine gene (GGTA1 and CMAH), target sequences are indicated on the respective exons recognized by the selected sgRNAs. PAM sequences are highlighted in blue. In the ssCMAH‐STOP oligo sequence, the TAA (STOP) codon is highlighted in bold character; the AflII restriction site is underlined. B, PCR analyses of female colonies. The results of the PCR analyses performed for the genomic characterization of the female colonies (A1, A2, A3, A4, A5 and A6) selected after Dynabeads sorting are reported as an example. Each colony was analysed for the GGTA1 gene (739 bp) and for the CMAH gene (225 bp). Resulting electrophoretic patterns determined directly that some colonies were characterized by visible Indels, creating bands different from the WT controls. This situation is clear for colonies A1 (double band), A2 (deletion) and A6 (deletion) in PCR analyses for the GGTA1 gene (°) and for colonies A1 (double band) and A5 (deletion) in PCR analyses for the CMAH gene (#). Resulting CMAH‐PCR products were also digested with the AflII restriction enzyme, detecting the alleles interested by the targeting event. Due to the introduction of a STOP codon (TAA) in the START position (ATG) of the CMAH gene, only the HDR‐CMAH alleles will be cut by the restriction enzyme producing two lower bands (152 + 73 bp). A simple agarose electrophoresis enabled us to identify possible additional edited colonies detecting the STOP codon insertion (**) for colonies A2 and A6 and the single insertion (*) for colonies A3 and A4. In these last ones, the not targeted allele resulted uncut (225 bp) as the WT sample. For this reason, the final determination of the exact Indels, occurred in all the edited colonies, was determined by Sanger sequencing of the resulting TOPO TA E coli clones. 100 = 100 bp ladder (Thermo Fisher Scientific); A1, A2, A3, A4, A5 and A6 = transfected females colonies; WT = wild‐type female line; H20 = Nucleases‐free water. C, Sequences alignments of colonies used for the SCNT. Sanger sequencing outlining the mutations affecting the GGTA1 and the CMAH genes of colonies selected for the SCNT step. For the GGTA1 gene, the exon 9 was used as reference for the male colonies and a PCR product including the exon 4 was used for the female ones. In both cases, deletions of different lengths were obtained (Table S1). For the CMAH gene, all edited alleles of the edited colonies were aligned using as reference a PCR product including the exon 2 sequence. In this case, in both lines, we were able to determine the TAA substitution, as result of the targeting event mediated by the site‐specific cut, produced by the CRISPR/Cas9 system driven by the sgRNA btCMAHcr1
2.6. Culture, transfection and selection of adult fibroblasts
Bovine adult fibroblasts (male and female) were cultured in DMEM + M199 (1:1) +10%FCS in 5%CO2, 5%O2 at 38°C.
Male fibroblasts (2 × 106 cells) were transfected using Nucleofector (V‐024 program, Lonza), two µg of each the 3 CRISPR/Cas9 expressing vectors (pX330‐btGGTA1cr1 and pX330‐btGGTA1cr2—exon9 of GGTA1 gene; pX330‐btCMAHcr1—exon 2 of CMAH gene) and 0.4 nmol of the ssCMAH‐STOP oligo (IDT).
Female fibroblasts (1 × 106 cells) were transfected using Neon system (P‐9 program; Thermo Fisher Scientific) and the Cas9‐RNP complex format of the S pyogenes. Cas9‐RNP was obtained mixing the recombinant Cas9 protein (14.4 μg; Edit‐R Cas9, Dharmacon), with the btGGTA1cr3‐sgRNA (3.6 μg), the btCMAHcr2‐sgRNA (3.6 μg) and 0.4 nmol of the ssCMAH‐STOP oligo.
Transfected cells were plated in a 60‐mm dish and cultured for 3 days when they were passaged 1‐3. Day 7 male (6.5 × 106 cells) and D5 female (2.3 × 106 cells) αGal‐negative cells were selected using biotin‐conjugated IB4 lectin attached to streptavidin‐coated magnetic beads.33, 34, 35 Cells were harvested (6.5 × 106 cells) and suspended in 0.2 mL PBS containing 1 µg biotin‐conjugated IB4 lectin (Sigma) and 0.1 mg Dynabeads M‐280 streptavidin (Life Technologies). The αGal‐positive cells were removed using a magnetic rack. The procedure was repeated three times. The αGal‐negative cells were plated on 150‐mm plates and cultured for 9‐10 days when the largest colonies with good morphology were picked up and expanded.
For each colony, one aliquot was cryopreserved in liquid nitrogen (DMEM/TCM199 1:1, 20% FBS and 10% DMSO) for subsequent SCNT and another was lysed for DNA extraction and molecular analyses (PCR, AflII‐RFLP, TOPO TA cloning and Sanger sequencing).
Only colonies that, during the CMAH molecular screenings, presented detectable Indels in their PCR products and/or that resulted positive for the AflII‐RFLP assay (152 bp + 73 bp) were subjected to the Sanger sequencing analyses for both genes (GGTA1 and CMAH), detecting the occurred Indels and the successful ssCMAH‐STOP oligonucleotide knock‐in events.
Eight (four male and four female) confirmed DKO colonies, edited for GGTA1 and CMAH genes, were selected for further screening in SCNT to assess developmental potential. Before SCNT embryos were transferred into recipients, at least 10 cloned embryos of each selected colony were analysed for the absence of wild‐type genotypes. Genomic DNA extraction procedure, PCR amplification and sequencing reactions were done using the same materials and methods described above.
2.7. Somatic cell nuclear transfer (SCNT)
The protocol used is described in Galli et al36 with minor modifications. Briefly, the bovine ovaries were collected at a local abattoir. Follicles larger than 3 mm were aspirated, and cumulus‐oocyte complexes were selected and in vitro matured in TCM 199 supplemented with 10% (v/v) foetal calf serum, 1 mg/mL 17 b‐oestradiol, ITS, 100 mg/mL sodium pyruvate, 90 mg/mL L‐cysteine, 720 mg/mL glycine, 7 nL/mL b‐mercaptoethanol, gonadotropins (0.05 IU/mL FSH and 0.05 IU/mL LH; Meropur 75, Ferring) and growth factors (50 ng/mL long‐EGF and 10 ng/mL bFGF) at 38.5°C in 5% CO2 in humidified air for 22 hours. The day before SCNT, nuclear donor cells were induced into quiescence by serum starvation (0.5% FCS). The day of SCNT, cells were trypsinized and resuspended in H‐SOF37 buffered with 25 mmol/L Hepes (H‐SOF) used for all manipulations. Oocytes with an extruded polar body were stained with Hoechst 33342 (5 μg/mL) and enucleated in the presence of cytochalasin B (5 μg/mL) by the aspiration of polar body and associated metaphase II plate in minimal volume of ooplasm under UV. Donor cells were transferred in the perivitelline space of enucleated oocytes. Donor cell‐cytoplast couplets were washed in 0.3 M mannitol solution and fused by double DC‐pulse (1.5 Kv/cm) 30 µsec long and returned into maturation medium. After one hour, at about 27‐29 hours of maturation, NT embryos were activated with 5 µmol/L ionomycin for 4 minutes followed by 3 hours of incubation in mSOF medium supplemented with 1 mmol/L 6‐DMAP and 5 μg/mL cycloheximide. At the end of the activation, reconstructed embryos were cultured in mSOF supplemented with essential and non‐essential amino acids and 4 mg/mL BSA up to 7‐8 days to the blastocyst stage.
2.8. Recipients synchronization, embryo transfer (ET) and calving
Heifers of 14‐16 months of age were used as recipients. Oestrus was synchronized using the Ovsynch protocol with two injections of a GnRH analogue (Dalmarelin, Fatro, Italy) 8 days apart. Forty‐eight hours after the second injection, animals were observed for oestrus signs and 6 days later those that showed oestrus were ultrasound scanned to detect the presence of a corpus luteum (CL). Those that had a well‐developed CL received 1 or 2 embryos (either fresh or frozen thawed) by non‐surgical ET ipsilateral to the CL. Four weeks after ET, pregnancy diagnosis was performed by ultrasound scanning and then the pregnant animals were checked at monthly interval till the end of the pregnancy. The delivery of the calves was by elective caesarean section at 280 days of gestation.
2.9. Genotyping and phenotyping analyses for αGal and Neu5Gc antigens in DKO cattle‐derived primary cells
Newborn calves were subjected to ear biopsy to establish a primary cell line, to extract the genomic DNA for genotyping by PCR and DNA sequencing as described above. Resulting primary fibroblasts for each calf were expanded and cryopreserved in DMEM:TCM199 1:1 with 10% DMSO and 20% FCS in CBS straws (IMV, Italy). For FACS analysis, bovine fibroblasts were thawed and cultured in DMEM medium (Gibco), with 10% FBS, 1% Peni‐Strepto and bFGF (Sigma, 1 ng/mL). Once confluent, cells were trypsinized and split into two culture dishes, one with the complete medium as above and the other with DMEM medium, 5% human serum (Sigma), 1% Peni/strepto and bFGF (1 ng/mL). For αGal analysis, cells were trypsinized, resuspended and washed in PBS + BSA 0.1%. The cells were pelleted (750g x 1 minute, 4°C) and resuspended in PBS + BSA 0.1% containing the FITC coupled lectin (BS‐I Isolectin B4) diluted 1:50 and incubated at 4°C for 30 minutes. After 3 washes in PBS BSA 0.1%, cells were ready for FACS analysis.
For Neu5Gc analysis, cells were cultured for at least 2 weeks in DMEM + human serum. Foetal calf serum is rich in Neu5Gc that is incorporated by cells in culture. Therefore, to avoid false positives, the cells used for the FACS analysis have to be cultured for at least 2 weeks in culture media without Neu5GC, by replacing FCS with human serum. Cells were seeded in 96‐well plates (106 cells/well), washed once with PBS with 0.5% fish gelatin (PBS‐FG) and then incubated in 200 µL PBS‐FG with anti‐Neu5Gc antibody or control isotype (BioLegend, chicken polyclonal IgY, dilution 1 :1000) for 1 hour at 4°C, washed four times in PBS‐FG, incubated in 100 µL PBS‐FG with Alexa 647‐coupled anti‐IgY antibody (Jackson ImmunoResearch, F(ab′)2 fragment donkey anti‐chicken 1:500) for 1 hour at 4°C, washed four times in PBS‐FG and transferred into FACS tubes. FACS analysis was conducted using a BD Pharmingen LSR‐II flow cytometer and FlowJo software (TreeStar). Despite this culture period, where the cells are also not growing under optimal conditions, sometimes it is not sufficient to clear all the carry‐over of Neu5Gc due to culture conditions and often some background staining remains like the one observed in Figure 4.
Figure 4.
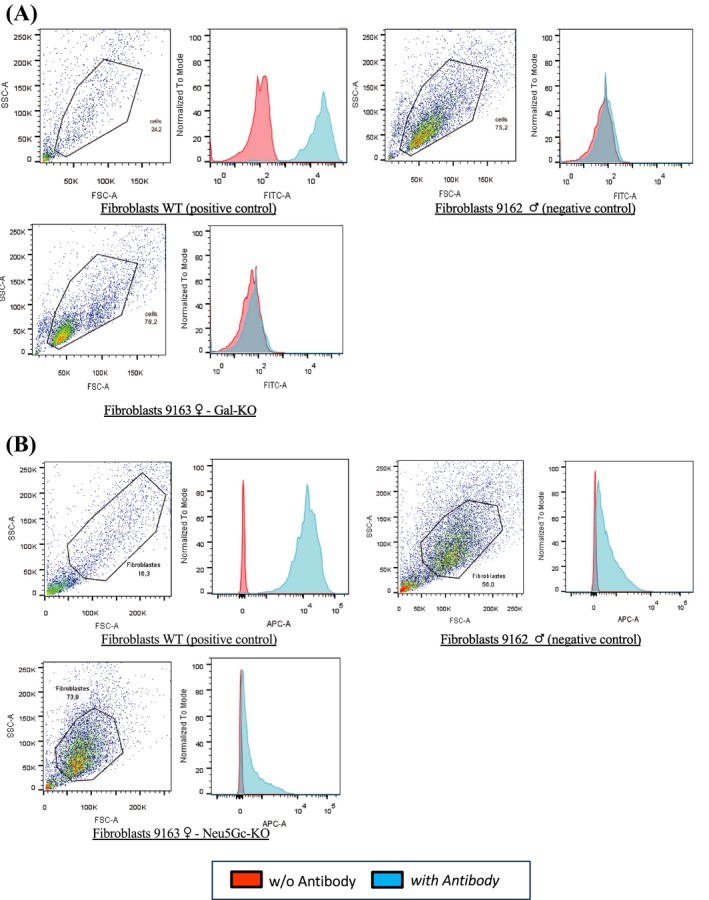
FACS analyses for DKO female 9163 calf. Fibroblasts from wild‐type animal (WT) and from the edited female calf were analysed by FACS. As negative controls, FACS‐validated DKO fibroblasts from 9162 male calf were used. The results demonstrated that the αGal (A) and (B) Neu5Gc antigens were absent from the cell surface of cloned female calf, confirming the genotype analyses for the knocked‐out genes (GGTA1 and CMAH). Fibroblasts WT (positive control): wild‐type primary fibroblasts from the bovine line prior to genetic modification expressing the αGal and the Neu5Gc antigens. Fibroblasts 9162 Gal‐KO and Neu5Gc‐KO (negative control): bovine primary fibroblasts NOT expressing the αGal and the Neu5Gc antigens. Fibroblasts 9163 Gal‐KO and Neu5Gc‐KO: primary fibroblasts derived from cloned DKO female calf
3. RESULTS
3.1. Disruption of GGTA1 and CMAH genes in primary bovine fibroblast lines
Two millions male fibroblasts were nucleofected and expanded for 7 days to 6.5 × 106. After Dynabeads sorting, 4200 αGal‐negative cells (0.065%) were recovered and plated in 20 Petri dishes (∅ = 150 mm) for clonal selection. Ten days after plating, 41 (1%) best growing colonies were picked up for PCR analysis and SCNT. Editing of the female fibroblasts took place a year later, and we used a different system using neon transfection with the Cas9 protein for the first time. From the one million female fibroblasts transfected with Neon and the Cas9‐RNP at D5, 2.3 × 106 cells were used for Dynabeads sorting. The efficiency of transfection was very low compare to the editing of male fibroblasts that was obtained using a plasmid for transfection but all αGal‐negative cells were plated in one 150‐mm dish and after 9 days 6 colonies were picked up. Of the 41 male colonies selected by pick up and analysed, CMAH‐PCR and AflII‐RFLP analyses revealed that 15 appeared to be edited for the CMAH gene and for this reason they were sent for Sanger sequencing analyses of their GGTA1 and CMAH genes (Figure 1C and Table S1). GGTA1‐KO was confirmed in all 15 colonies and 13 (31.7%) resulted also KO for Neu5Gc (Table 4).
Table 4.
Fibroblasts colonies—screening results
| Bovine line | Picked colonies | GGTA1‐KO (beads) | CMAH‐KO (PCR + RFLP) | GGTA1‐KO (sequencing) | CMAH‐KO (sequencing) |
|---|---|---|---|---|---|
| Male | 41 | 41 | 15 | 15 | 13 |
| Female | 6 | 6 | 6 | 6 | 4 |
The female colonies were subjected to the same analysis. The PCR analysis (Figure 1B) followed by Sanger sequencing analyses (Figure 1C and Table S1) on both genes of the six female colonies revealed that two colonies were heterozygotes and four colonies were KO (66.6%) for the CMAH gene and that all six colonies were GGTA1‐KO (Table 4), confirming the high efficiency of Dynabeads selection for the GGTA1 KO.
Male A4 and E3 and female A6 colonies were used for SCNT based on their morphology and growing characteristics and embryo production after SCNT. Ten SCNT embryos of each colony were sequenced to confirm the purity of the selected colonies for the required mutations (Figure 1C) to avoid potential contaminations of WT cells.
3.2. Generation of DKO calves by SCNT
Two male DKO colonies (A4 and E3) were used as nuclear donors for SCNT (Table 5). Seven blastocysts (BLs), derived from colony A4, were transferred in 5 synchronized recipients, 4 (80%) became pregnant and 1 pregnancy (25%) went to term delivering 1 calf (9161, Figure 2A). Fifteen BLs derived from colony E3 were transferred in 12 synchronized recipients; 5 (41.6%) became pregnant, 1 pregnancy (20%) went to term delivering 1 calf (9162, Figure 2A).
Table 5.
Development of cloned embryos after transfer into recipient heifers
| Bovine line | No. of colonies | No. of embryos | No. of recipients | No. of pregnant (%) | No. of born alive at term |
|---|---|---|---|---|---|
| Male | A4 | 7 | 5 | 4 (80) | 1 |
| Male | E3 | 15 | 12 | 5 (41.6) | 1 |
| Female | A6 | 31 | 16 | 6 (37.5) | 1 |
Figure 2.
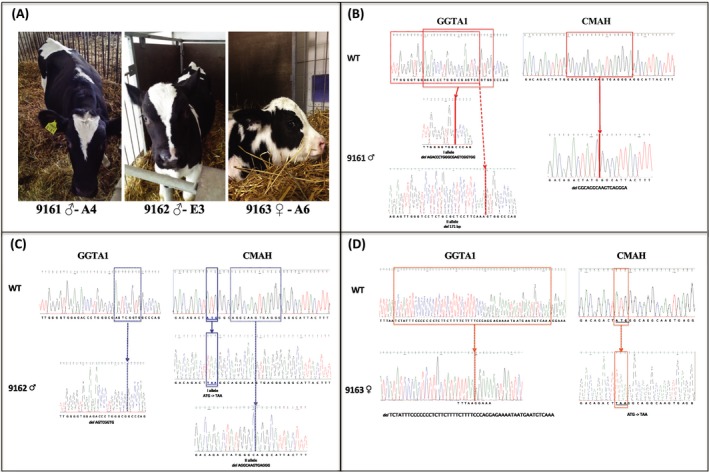
DKO calves and sequencing results. A, Pictures of cloned DKO calves. Two healthy cloned bull calves (9161 and 9162) were generated from two different DKO colonies (A4 and E3). Cloned heifer calf was generated using the colony A6. B, Sequencing results for 9161. For the GGTA1 gene, it was confirmed that this gene is affected by two different deletions (21 and 171 bp), as previously described for the edited colony A4 (Table S1). These data were finally demonstrated by the deletion (17 bp) generated in the CMAH gene. C, Sequencing results for 9162. The GGTA1 gene sequence presented a 8 bp deletion, and the CMAH gene is characterized by the same 2 different mutations (TAA substitution; del 13 bp) detected in colony E3 (Table S1). D, Sequencing results for 9163. The same Indels, characterizing the GGTA1 (del 54 bp) and the CMAH (TAA substitution) genes of A6 colony (Table S1), were confirmed
Female DKO colony A6 was used for SCNT, and 31 BLs were transferred in 16 recipients; 6 (37.5%) became pregnant and 1 pregnancy went to term delivering 1 calf (9163, Figure 2A).
3.3. Genotyping of cloned calves
Sanger sequencing of TOPO TA‐cloned PCR products of DKO calves confirmed the Indels characterizing the colonies used for cloning. In details, in clone 9161, GGTA1 gene is affected by two different mutations in exon 9 (del AGACCCTGGGCGAGTCGGTGG/del 171bp) and the exon 2 of the CMAH gene carries a deletion (del GGCAGGCAAGTGAGGGA) as it was described for colony A4 (Table S1; Figure 2B). In clone 9162 (Figure 2A), a deletion in GGTA1 gene (del AGTCGGTG) is accompanied by 2 different mutations in the exon 2 of CMAH gene. The first allele was inactivated by the substitution of the ATG codon (START) with the TAA codon (STOP), due to the homology‐directed repair (HDR) event driven by the ssCMAH‐STOP oligo, as described for colony E3 (Table S1, Figure 2C), and the second allele has 13 bp deletion (del AGGCAAGTGAGGG).
Sanger sequencing results of female clone 9163 (Figure 2A) demonstrated that a deletion in the exon 4 of the GGTA1 gene (del 54bp) and the substitution of the START to a STOP codon (ATG TAA) in the exon 2 of the CMAH gene are identical to the Indels described for the donor female colony A6 (Table S1, Figure 2D). PCR analyses on the male calves (data not shown) demonstrated also that the CRISPR/Cas9 expression vectors were not integrated in the genome of the cloned calves.
3.4. Phenotyping of cloned calves
FACS analysis confirmed the genotyping results of the three calves. All primary cell lines derived from biopsies of the cloned male calves do not express αGal (Figure 3A) and Neu5Gc (Figure 3B) as opposed to WT control cells before genetic engineering. As negative controls, pig cells KO for both antigens were used. The female phenotyping was performed in the same way as for the males but in this case the negative control was the male 9162. In this experiment performed a year later with different experimental context, the αGal was completely negative. In the case of Neu5Gc, both the control (9162) and the female (9163) had some background staining. Since the 9162 pictured in Figure 3 is the same as in Figure 4, we can conclude that the tail of Neu5Gc staining is background staining coming from the different experimental setting and culture conditions.
Figure 3.
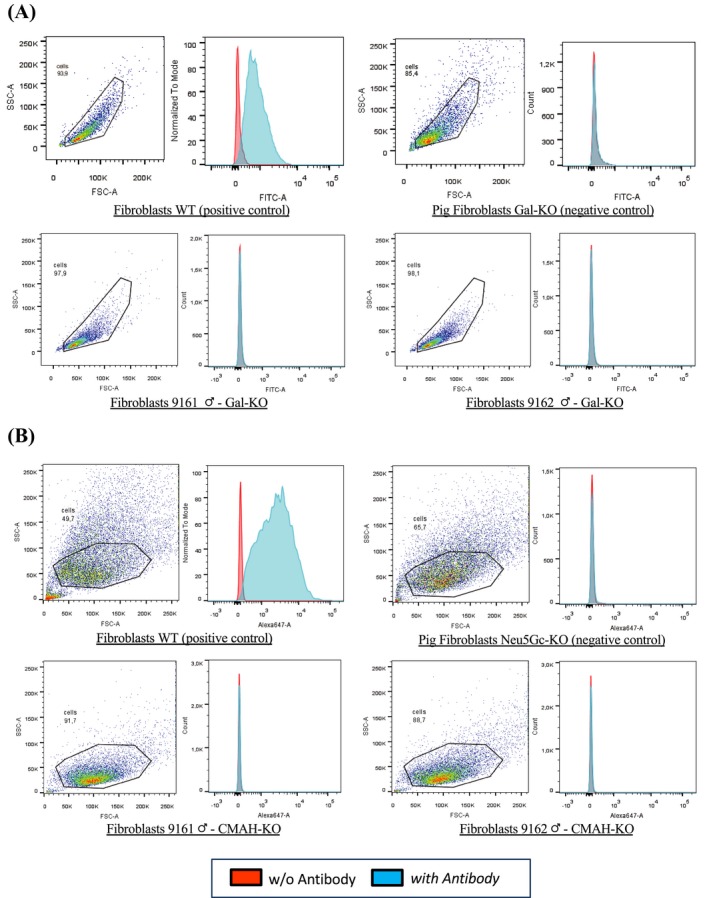
FACS analyses for 9161, 9162 male calves. Fibroblasts from wild‐type animal (WT) and from the edited male calves were analysed by FACS. As negative controls, pig DKO fibroblasts were used as no bovine material was available. The results demonstrated that the αGal (A) and (B) Neu5Gc antigens were absent from the cell surface of cloned calves, confirming the genotype analyses for the knocked‐out genes (GGTA1 and CMAH). Fibroblasts WT (positive control): wild‐type primary fibroblasts from the bovine line prior to genetic modification expressing the αGal and the Neu5Gc antigens. Pig fibroblasts Gal‐KO and Neu5Gc‐KO (negative control): porcine primary fibroblasts NOT expressing the αGal and the Neu5Gc antigens. Fibroblasts 9161/9162 Gal‐KO and Neu5Gc‐KO: primary fibroblasts derived from cloned DKO calves
4. DISCUSSION
In this work, we targeted two well‐known xenoantigens identified as such from pig xenotransplantation studies that are also expressed in cattle. Here, we show that editing bovine fibroblasts are possible using both CRISPR/Cas9 in plasmid and Cas9‐RNP formats and live animals can be generated through SCNT. The advent of programmable nucleases for genome editing in large animals, especially the pig, has greatly increased its efficiency by reducing the number of animals required and the costs involved. The number of genetically modified pigs and the consequent generation of animal models through precise genetic engineering have grown exponentially in the last 10 years. However, genetically modified cattle are still very few due to some constrains for applying this technology to this species such as the long generation interval. Nevertheless, cattle would be more relevant for food production since it is a major source of beef and dairy products. Furthermore, one of the major potential applications for DKO cattle for both GGTA1 and CMAH would be as a source of less immunogenic biological materials (pericadia) to manufacture BHV. In addition, genetically engineered cattle would also allow to produce food to avoid, for example, anaphylactic reaction following the consumption of red meat in some allergic individuals.
Despite the low transfection efficiency in bovine fibroblasts that affected the total number of edited colonies, because of the thorough screening of the few colonies selected and the combination with SCNT, we were able to generate DKO male and female calves. All the bovine genome editing work was undertaken to disrupt simultaneously the GGTA1 and the CMAH genes without the need of a selectable marker, choosing primary cell lines whose genomic sequences were not affected by polymorphisms. We started the bovine genome editing work in the male line transfecting the plasmid format of the S pyogenes CRISPR/Cas9 system, while later its Cas9‐RNP format was tested in the female line.
We selected to target exon 9 of GGTA1 gene in the male line using together two different sgRNAs (btGGTA1cr1 and btGGTA1cr2—Figure 1A). In contrast to the female line, we targeted the exon 4, using the protein (Cas9‐RNP), designing a sgRNA specific for the START codon (btGGTA1cr3 Figure 1A). The use of Dynabeads and IB4 lectins greatly compensated for the low transfection efficiency very effectively since all the analysed colonies derived from cells that did not bind IB4 were all KO for the GGTA1 (Table 4). As a consequence, also the KO rate for the CMAH gene was very high indicating that when these nucleases enter the cells, they are very effective on all the targets. This event was also described for the pig by Li et al.38 The use of ssODN‐mediated KI with CRISPR/Cas9 system for KO purposes was also possible in cattle and facilitated the PCR screening because of the insertion of an AflII restriction site. The use of the plasmid to introduce and express all the machinery required was in our experiments more efficient than the use of the protein but because we required only a few cell clones for SCNT, it did not affect the success at the end since we had far more cell clone that we could need for SCNT. The reason for preferring the Cas9 protein to the plasmid is to avoid the risk of integration of the plasmid. Luckily in this case, we did not detect any integration of the CRISPR/Cas9 expressing plasmids in the genome of the male calves. CRISPR/Cas9‐mediated genome editing procedures are compatible with SCNT, and the efficiency is comparable when WT cells are used. Three live calves were delivered by caesarean section and the first born (9161) is about to reach puberty while the female is only a few days old. The genotype of the three claves born alive exactly matched the genotype of the three cell clones selected for SCNT (Table S1). To further validate the genotyping findings with the phenotype, FACS analysis was performed on fibroblasts derived from the three newborn animals. The absence of αGal and Neu5Gc was clearly confirmed on the two bull calves. To perform the FACS analysis, primary cells were grown from biopsy taken in the first days of life of the calves that were gestated by WT surrogate mother and fed after birth with milk from WT cows; moreover, the culture of primary cells was performed with FCS supplementation to culture media from WT source. All these conditions favour incorporation of Neu5Gc into the cells that before the analysis requires 2‐3 week in culture with serum lacking Neu5Gc. We used human serum in this period to allow the cells to shed the incorporated Neu5Gc but this time is variable depending on culture conditions, not ideal with human serum for bovine fibroblasts and the reagents used. The phenotypical characterization of the female calf by FACS was not yet extensively completed (only one experiment was performed). There is some background noise due to Neu5Gc remnants of the culture conditions; on the other end, also the male cells used as negative control that was completely clear in a previous experiment (Figure 3B) had the same right shift for Neu5Gc (Figure 4B). We can conclude that the generation of DKO cattle is possible using the latest genome editing technologies combined with SCNT. This will offer the opportunity to use novel biological materials of bovine origin for medical and industrial application as well as for human consumption in the form of beef or dairy products for allergic individuals.
CONFLICT OF INTERESTS
The authors declare no conflict of interests.
AUTHOR CONTRIBUTION
AP, EC, JPS and CG conceived the project and designed the experiments. AP, IL, RD, EZ, GL, JPJ and SC performed the experiments, collected and analysed the data. JMB, EC, JPS, VPK, CC, GG, MG and TB provided advice for experimental design discussion of results and review of the manuscript. EC, JPS and CG secured the funding. CG, AP and IL wrote the manuscript. All authors reviewed and approved the manuscript.
Supporting information
ACKNOWLEDGEMENTS
The authors acknowledge the technical support of Gabriella Crotti, Paola Turini, Silvia Colleoni, Gaia Fiorini from Avantea. Luigi Scordamaglia CEO of INALCA Spa for the access to bovine ovaries. Yasuhiro Takeuchi, Michael Breimer, Jan Holgersson, Rafael Manez Mendiluce and Xi Chen of the Translink consortium for discussion during the course of the work and reading the manuscript.
Perota A, Lagutina I, Duchi R, et al. Generation of cattle knockout for galactose‐α1,3‐galactose and N‐glycolylneuraminic acid antigens. Xenotransplantation. 2019;26:e12524 10.1111/xen.12524
Funding information
This work was supported by the European Union Seventh Framework Program (FP7/2007/2013) under the Grant agreement 603049 for Translink consortium (http://www.translinkproject.com/)
REFERENCES
- 1. Kobayashi T, Cooper DK. Anti‐Gal, alpha‐Gal epitopes, and xenotransplantation. Subcell Biochem. 1999;32:229‐257. [DOI] [PubMed] [Google Scholar]
- 2. Bouhours D, Pourcel C, Bouhours JE. Simultaneous expression by porcine aorta endothelial cells of glycosphingolipids bearing the major epitope for human xenoreactive antibodies (Gal alpha 1–3Gal), blood group H determinant and N‐glycolylneuraminic acid. Glycoconj J. 1996;13(6):947‐953. [DOI] [PubMed] [Google Scholar]
- 3. Scobie L, Padler‐Karavani V, Le Bas‐Bernardet S, et al. Long‐term IgG response to porcine Neu5Gc antigens without transmission of PERV in burn patients treated with porcine skin xenografts. J Immunol (Baltimore, Md: 1950). 2013;191(6):2907‐2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu A, Hurst R. Anti‐N‐glycolylneuraminic acid antibodies identified in healthy human serum. Xenotransplantation. 2002;9(6):376‐381. [DOI] [PubMed] [Google Scholar]
- 5. Salama A, Evanno G, Harb J, Soulillou JP. Potential deleterious role of anti‐Neu5Gc antibodies in xenotransplantation. Xenotransplantation. 2015;22(2):85‐94. [DOI] [PubMed] [Google Scholar]
- 6. Lutz AJ, Li P, Estrada JL, et al. Double knockout pigs deficient in N‐glycolylneuraminic acid and Galactose α‐1,3‐Galactose reduce the humoral barrier to xenotransplantation. Xenotransplantation. 2013;20(1):27‐35. [DOI] [PubMed] [Google Scholar]
- 7. Chen G, Qian H, Starzl T, et al. Acute rejection is associated with antibodies to non‐Gal antigens in baboons using Gal‐knockout pig kidneys. Nat Med. 2005;11(12):1295‐1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Naso F, Gandaglia A, Bottio T, et al. First quantification of alpha‐Gal epitope in current glutaraldehyde‐fixed heart valve bioprostheses. Xenotransplantation. 2013;20(4):252‐261. [DOI] [PubMed] [Google Scholar]
- 9. Reuven EM, Leviatan Ben‐Arye S, Marshanski T, et al. Characterization of immunogenic Neu5Gc in bioprosthetic heart valves. Xenotransplantation. 2016;23(5):381‐392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Naso F, Gandaglia A. Different approaches to heart valve decellularization: A comprehensive overview of the past 30 years. Xenotransplantation. 2018;25(1). 10.1111/xen.12354 [DOI] [PubMed] [Google Scholar]
- 11. Schoen FJ, Levy RJ. Founder's Award, 25th Annual Meeting of the Society for Biomaterials, perspectives. Providence, RI, April 28‐May 2, 1999. Tissue heart valves: current challenges and future research perspectives. J Biomed Mater Res . 1999;47(4):439‐465. [DOI] [PubMed] [Google Scholar]
- 12. Park CS, Park SS, Choi SY, Yoon SH, Kim WH, Kim YJ. Anti alpha‐gal immune response following porcine bioprosthesis implantation in children. J Heart Valve Dis. 2010;19(1):124‐130. [PubMed] [Google Scholar]
- 13. Barone A, Benktander J, Whiddon C, et al. Glycosphingolipids of porcine, bovine, and equine pericardia as potential immune targets in bioprosthetic heart valve grafts. Xenotransplantation. 2018;25(5):e12406. [DOI] [PubMed] [Google Scholar]
- 14. Bloch O, Golde P, Dohmen PM, Posner S, Konertz W, Erdbrügger W. Immune response in patients receiving a bioprosthetic heart valve: lack of response with decellularized valves. Tissue Eng Part A. 2011;17(19–20):2399‐2405. [DOI] [PubMed] [Google Scholar]
- 15. McGregor CG, Carpentier A, Lila N, Logan JS, Byrne GW. Cardiac xenotransplantation technology provides materials for improved bioprosthetic heart valves. J Thorac Cardiovasc Surg. 2011;141(1):269‐275. [DOI] [PubMed] [Google Scholar]
- 16. McGregor CG, Kogelberg H, Vlasin M, Byrne GW. Gal‐knockout bioprostheses exhibit less immune stimulation compared to standard biological heart valves. J Heart Valve Dis. 2013;22(3):383‐390. [PubMed] [Google Scholar]
- 17. Zhang R, Wang Y, Chen L, et al. Reducing immunoreactivity of porcine bioprosthetic heart valves by genetically‐deleting three major glycan antigens, GGTA1/beta4GalNT2/CMAH. Acta Biomater. 2018;72:196‐205. [DOI] [PubMed] [Google Scholar]
- 18. Steinke JW, Platts‐Mills TA, Commins SP. The alpha‐gal story: lessons learned from connecting the dots. J Allergy Clin Immunol. 2015;135(3):589–596; quiz 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Apostolovic D, Tran TA, Starkhammar M, Sanchez‐Vidaurre S, Hamsten C, Van Hage M. The red meat allergy syndrome in Sweden. Allergo J Int. 2016;25(2):49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alisson‐Silva F, Kawanishi K, Varki A. Human risk of diseases associated with red meat intake: Analysis of current theories and proposed role for metabolic incorporation of a non‐human sialic acid. Mol Aspects Med. 2016;51:16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Samraj AN, Pearce OM, Laubli H, et al. A red meat‐derived glycan promotes inflammation and cancer progression. Proc Natl Acad Sci USA. 2015;112(2):542–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang M, Sun Z, Yu T, et al. Large‐scale production of recombinant human lactoferrin from high‐expression, marker‐free transgenic cloned cows. Sci Rep. 2017;7(1):10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parc AL, Karav S, Rouquie C, Maga EA, Bunyatratchata A, Barile D. Characterization of recombinant human lactoferrin N‐glycans expressed in the milk of transgenic cows. PLoS ONE. 2017;12(2):e0171477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sano A, Matsushita H, Wu H, et al. Physiological level production of antigen‐specific human immunoglobulin in cloned transchromosomic cattle. PLoS ONE. 2013;8(10):e78119–e78119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buist M, Komatsu E, Lopez P, et al. Features of N‐Glycosylation of Immunoglobulins from Knockout Pig Models. J Anal Bioanal Tech. 2016;7:5. [Google Scholar]
- 26. Ghaderi D, Zhang M, Hurtado‐Ziola N, Varki A. Production platforms for biotherapeutic glycoproteins. Occurrence, impact, and challenges of non‐human sialylation. Biotechnol Genet Eng Rev. 2012;28:147–175. [DOI] [PubMed] [Google Scholar]
- 27. Tan W, Carlson DF, Lancto CA, et al. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Natl Acad Sci. 2013;110(41):16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sendai Y, Sawada T, Urakawa M, et al. α1,3‐Galactosyltransferase‐Gene knockout in cattle using a single targeting vector with loxP sequences and cre‐expressing adenovirus. Transplantation. 2006;81(5):760‐6. [DOI] [PubMed] [Google Scholar]
- 29. Sato M, Miyoshi K, Nagao Y, et al. The combinational use of CRISPR/Cas9‐based gene editing and targeted toxin technology enables efficient biallelic knockout of the alpha‐1,3‐galactosyltransferase gene in porcine embryonic fibroblasts. Xenotransplantation. 2014;21(3):291–300. [DOI] [PubMed] [Google Scholar]
- 30. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, NY). 2013;339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA‐guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liang X, Potter J, Kumar S, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol. 2015;208:44–53. [DOI] [PubMed] [Google Scholar]
- 33. Fujimura T, Takahagi Y, Shigehisa T, et al. Production of alpha 1,3‐galactosyltransferase gene‐deficient pigs by somatic cell nuclear transfer: a novel selection method for gal alpha 1,3‐Gal antigen‐deficient cells. Mol Reprod Develop. 2008;75(9):1372–1378. [DOI] [PubMed] [Google Scholar]
- 34. Hauschild J, Petersen B, Santiago Y, et al. Efficient generation of a biallelic knockout in pigs using zinc‐finger nucleases. Proc Natl Acad Sci USA. 2011;108(29):12013–12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tucker BA, Rahimtula M, Mearow KM. A procedure for selecting and culturing subpopulations of neurons from rat dorsal root ganglia using magnetic beads. Brain Res Brain Res Protoc. 2005;16(1–3):50–57. [DOI] [PubMed] [Google Scholar]
- 36. Galli C, Lagutina I, Vassiliev I, Duchi R, Lazzari G. Comparison of microinjection (piezo‐electric) and cell fusion for nuclear transfer success with different cell types in cattle. Cloning Stem Cells. 2002;4(3):189–196. [DOI] [PubMed] [Google Scholar]
- 37. Tervit HR, Whittingham DG, Rowson LE. Successful culture in vitro of sheep and cattle ova. J Reprod Fertil. 1972;30(3):493–497. [DOI] [PubMed] [Google Scholar]
- 38. Li P, Estrada JL, Burlak C, et al. Efficient generation of genetically distinct pigs in a single pregnancy using multiplexed single‐guide RNA and carbohydrate selection. Xenotransplantation. 2015;22(1):20–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


