Abstract
Prostatic small cell neuroendocrine carcinoma (SC/NE) is well studied in metastatic castration‐resistant prostate cancer; however, it is not well characterized in the primary setting. Herein, we used gene expression profiling of SC/NE prostate cancer (PCa) to develop a 212 gene signature to identify treatment‐naïve primary prostatic tumors that are molecularly analogous to SC/NE (SC/NE‐like PCa). The 212 gene signature was tested in several cohorts confirming similar molecular profile between prostatic SC/NE and small cell lung carcinoma. The signature was then translated into a genomic score (SCGScore) using modularized logistic regression modeling and validated in four independent cohorts achieving an average AUC >0.95. The signature was evaluated in more than 25,000 primary adenocarcinomas to characterize the biology, prognosis and potential therapeutic response of predicted SC/NE‐like tumors. Assessing SCGScore in a prospective cohort of 17,967 RP and 6,697 biopsy treatment‐naïve primary tumors from the Decipher Genomic Resource Information Database registry, approximately 1% of the patients were found to have a SC/NE‐like transcriptional profile, whereas 0.5 and 3% of GG1 and GG5 patients respectively showed to be SC/NE‐like. More than 80% of these patients are genomically high‐risk based on Decipher score. Interrogating in vitro drug sensitivity analyses, SC/NE‐like prostatic tumors showed higher response to PARP and HDAC inhibitors.
Keywords: neuroendocrine prostate cancer, localized, SC/NE‐like, molecular signature, drug sensitivity
Short abstract
What's new?
While genomic/transcriptomic data analysis has revolutionized cancer biology, this analysis is frequently only available late in the cancer history, often after years of therapy. Here the authors built a single sample genomic classifier to predict primary prostate cancer tumors with early small cell neuroendocrine differentiation. They show in three independent cohorts that small cell neuroendocrine tumors of the prostate are similar to small cell tumors of the lung and predict the specific prostate tumors to be responsive to inhibitors of poly ADP ribose polymerase and histone deacetylases, underscoring the use of these drugs in this subtype of prostate cancer.
Abbreviations
- Adeno
prostatic adenocarcinoma
- ADT
androgen deprivation therapy
- AUC
area under the curve
- CCLE
Cancer Cell Line Encyclopedia
- DRS
drug response score
- FFPE
formalin‐fixed paraffin‐embedded
- GDSC
Genomics of Drug Sensitivity in Cancer
- GG
grade group
- GRID
Genomic Resource Information Database
- HDAC
histone deacetylases
- IC50
half maximal inhibitory concentration
- JHMI
John Hopkins Medical Institute
- mCRPC
metastatic castration resistance prostate cancer
- PARP
poly (ADP‐ribose) polymerase
- PD
poorly differentiated
- RP
radical prostatectomy
- SC/NE
small cell neuroendocrine
- SCGScore
small cell genomic score
- SCLC
small cell lung carcinoma
Introduction
Genomic diversity and clinical relevance of small cell/neuroendocrine (SC/NE) are well established in the castration‐resistant prostate cancer (CRPC) setting,1, 2, 3 but it is less characterized in treatment‐naïve primary tumors due to its rarity. It is well established that some aggressive primary prostate tumors share features with late‐stage therapy‐resistant disease and unsurprisingly confer a poor prognosis.4 Certain genomic alterations that are enriched in SC/NE, including the RB1 and TP53 pathway aberration, can often be found in primary tumors, albeit at a lower overall frequency than metastatic disease.5 It is therefore conceivable that some localized adenocarcinomas are inherently more capable of “transdifferentiating” to SC/NE once under the concerted influence and selective pressure of AR‐targeted therapy.
Most prior studies comparing SC/NE and high‐grade adenocarcinoma that have identified SC/NE signatures and candidates for targeted therapy were based on few SC/NE patients due to disease rareness.6 To accurately identify the wide spectrum of SC/NE and characterize its biology in a large cohort, we leveraged our previously reported meta‐NE signature6 and genome‐wide expression data of more than 25,000 primary tumor samples from the Decipher Genomic Resource Information Database (GRID) registry. The meta‐NE signature was identified to predict histologically SC/NE tumors, but we found genomic heterogeneity within histologically SC/NE tumors. In this work, we refined the meta‐NE signature and modeled it as a single score to predict tumors that are genomically similar to SC/NE tumors. We hypothesize that some primary adenocarcinomas harbor features of SC/NE and that patients with such tumors are at higher risk of progression under the influence of AR‐targeted therapy. Therefore, our objective is to develop a genomic tool to identify and characterize primary tumors with SC/NE‐like features and differentiate them from poorly differentiated (PD) adenocarcinoma with the goal of understanding their biology and identifying potential targeted therapy.
Materials and Methods
Patient cohorts
Our initial discovery cohort (John Hopkins Medical Institute [JHMI]‐SC) consisted of 33 formalin‐fixed paraffin‐embedded (FFPE) tumors retrieved from John Hopkins Registry.6 This cohort included six morphologically diagnosed “pure” SC/NE specimens (pure SC), 11 high‐grade adenocarcinomas (mostly Grade Group [GG] 5), 1 adenocarcinoma with NE differentiation, as well as tumor foci from 15 specimens harboring concurrent small cell and adenocarcinoma histology. In these 15 specimens, either the predominant adenocarcinoma foci were sampled (termed mixed‐prostatic adenocarcinoma [Adeno], n = 5), or the small cell foci (termed mixed‐SC, n = 10). Additionally, we used 97 FFPE GG5 adenocarcinoma samples from Johns Hopkins natural history cohort7 for model development.
We retrieved external gene expression data from eight publicly available datasets obtained from patients with SC/NE (as well as from more typical AR‐positive metastatic CRPC [mCRPC]): Beltran et al.2 (mCRPC:34, mCRPC‐SC/NE:15), Kumar et al.1 (mCRPC:151, mCRPC‐SC/NE:20), LTL331R system(GSE59984),8 GSE66187 with typical SC/NE and atypical SCC with AR‐positive, GSE43346 with small cell lung carcinoma (SCLC) and normal tissues, Cancer Cell Line Encyclopedia (CCLE) cell lines9 with SCLC or non‐SCLC and Genomics of Drug Sensitivity in Cancer (GDSC) cell lines10 with SCLC or non‐SCLC, and mCRPC cohort (mCRPC:107, NE:12) from Aggarwal et al.3 Finally, we used independent cohorts of low‐grade adenocarcinoma prior to androgen deprivation therapy (ADT) treatment, adenocarcinoma, PD and NE post‐ADT, and de novo SC/NE from the University of Calgary. Tumors slides were reviewed by one of the study pathologists (T.A.B.) to characterize SC/NE features. Tumors were stained with SYP, chromogranin, CD56, PSA, PSAP and AMACR.
To identify primary prostatic tumors that are genomically similar to SC/NE, we used the expression profile of prospective 17,967 radical prostatectomy (RP) and 6,697 biopsies deidentified and anonymized cases from the Decipher Genomic Resource Information Database (GRID) with basic demographic and pathological data obtained through clinical use of the Decipher test. Furthermore, we used 283 neuroblastoma samples (GSE85047) run on the same Human Exon Array technology used for the Decipher GRID comparative analysis. Neuroblastoma samples were used as a positive control for neuroendocrine biomarkers and signatures validation.
Tumor transcriptome profiling
Tumor specimens for the 33 JHMI‐SC, 97 JHMI and GRID samples were obtained from archived paraffin blocks and RNA extraction was performed using the Decipher prostate cancer classifier assay as previously described.11 Briefly, RNA was amplified, labeled and hybridized to Human Exon 1.0 ST microarrays (Affymetrix, Santa Clara, CA) covering 46,050 genes. The SCAN algorithm was used for individual patient profile preprocessing and normalization.12 ComBat13 algorithm was used for adjusting expression for dataset effect. Neuroblastoma cohort was normalized with SCAN algorithm12 as it was profiled on the same Human Exon 1.0ST array.
Development of SC genomic fingerprint and score
We started with 306 neuroendocrine and small cell related genes recently reported from meta‐analysis.6 Since we hypothesized that there is molecular heterogeneity within samples that are not captured by histological annotations, we conducted consensus clustering, using ConsensusClusterPlus R package,14 on the 33 samples with pam clustering algorithm, Pearson correlation distance, ward linkage, 1,000 iterations and 80% sampling rate parameters identifying three clusters (SC/NE, mixed and Adeno). Genes differentially expressed between SC/NE and Adeno clusters were used to define better segregation. For model development, we pooled 97 GG5 samples from natural history cohort with the 33 JHMI‐SC cohorts to define a training set. We grouped genes into modules; genes were ranked based on their expression change in the LTL331R system upon NE differentiation, and grouped into 10 modules based on gene expression fold change (Supporting Information Table S1). For each module of genes, a logistic regression with ridge penalty was fitted and the penalty parameter was selected via 10‐fold cross‐validation, which created 10 models. The prediction probabilities from these 10 models were further averaged with weights proportional to their area under the curve (AUC) on the training data, such that the more powerful model received a higher weight in the final prediction. The final averaged prediction probability is called SCGScore. Flow chart of gene reductions, model developments and overview of model's evaluation are detailed in Supporting Information Methods.
Drug response score
Using in vitro drug sensitivity and microarray data from the GDSC panel, we generated gene signatures predicting lung cancer cell lines (154 cells) sensitivity to 265 drugs from prostate cancer clinical trials. For each drug, we identified gene signature (drug response related genes and their correlations to the half maximal inhibitory concentration [IC50] value). Most significantly correlated genes were selected and the expression of the corresponding genes in the Decipher GRID was extracted for drug response score (DRS) calculations. A patient‐specific DRS was calculated using these correlation coefficients (Cor) as weighting factors of the corresponding gene expression normalized by the sum of Cor. DRSs were calculated for 265 drugs for every patient in the Decipher GRID characterize their associations with SCGScore.
Statistical analysis
Statistical analyses were performed in R version 3.0. All statistical tests were two‐sided using a significant level of 0.05. Chi‐square test was used for statistical associations between categorical variables (GG) and the Wilcoxon test was used for continuous variables (DRSs, Decipher score).
Results
Development of a prostatic small cell genomic fingerprint
To develop a molecular classifier to identify SC/NE prostate cancer in the localized, treatment‐naïve setting, we first selected 306 genes associated with NE prostate cancer as previously reported by our group.6 Since we hypothesized that there is molecular heterogeneity underlying the histological annotations, the 306 genes were used to guide the consensus clustering of the 33 prostate samples from JHMI‐SC cohort revealing three clusters with distinct biological and histological characteristics. The first cluster was enriched with histologically pure SC and mixed‐SC (SC/NE cluster), the second was enriched with histological adenocarcinoma (Adeno cluster) and the third showed atypical small cell histology with overexpression of AR‐signaling (mixed cluster; Fig. 1 a, Supporting Information Fig. S1). When the cluster memberships were evaluated with respect to histology, we noted a small number of tumors with discordant genotypes relative to their histology (i.e., a mixed‐SC tumor was found in the Adeno cluster).
Figure 1.
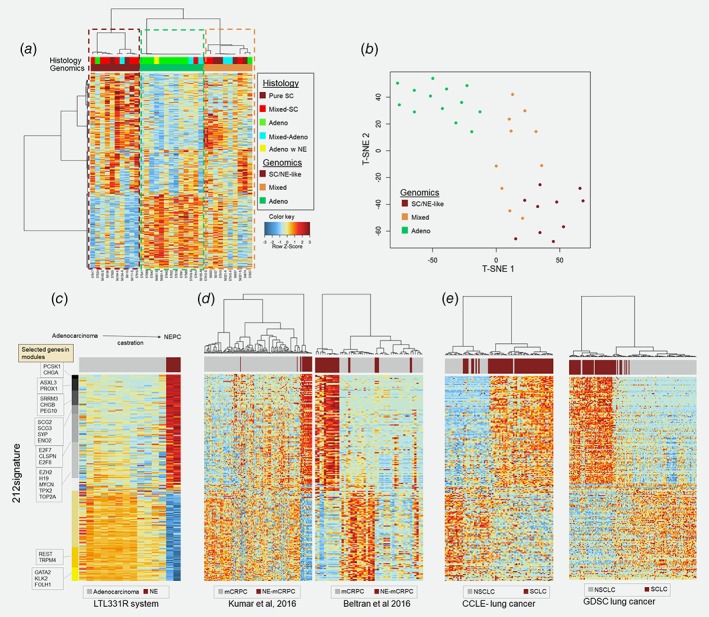
Small cell genomic fingerprint discovery and evaluation. (a) Consensus clustering of the 306 genes revealing three clusters: one enriched with histologically small cell (SC/NE), one enriched with Adeno (Adeno) and one mixed. (b) t‐SNE of the 212 genes (212signature) differentially expressed between SC/NE cluster and adenocluster showing clear discrimination between the groups. (c–e) Identified 212signature is evaluated in four public cohorts with NE. (c) Genes ordered based on their fold change in the LTL331R system. (d) mCRPC‐NE samples were clustered together in mCRPC cohorts with few samples exceptions. (e) 212signature also discriminated small cell lung carcinoma (SCLC) from non‐SCLC in Cancer Cell Line Encyclopedia (CCLE) and Genomics of Drug Sensitivity in Cancer (GDSC). [Color figure can be viewed at http://wileyonlinelibrary.com]
To circumvent some of the challenges posed by the molecular heterogeneity in histological annotation, we compared the SC/NE and Adeno clusters finding 216 genes were differentially expressed (120 upregulated and 96 downregulated) after multiple testing adjustment (adjusted p < 0.05) within the SC cluster (Supporting Information Table S1). These 216 genes represented a set of candidate NE‐specific genes suitable for model development. Only 19 genes were in common with the 69 SC/NE signature presented by Aggarwal et al.3
These 216 genes were found to readily distinguish the three tumor types, with the mixed cluster demonstrating intermediate behavior between the well‐separated SC/NE and Adeno clusters (Fig. 1 b). The SC/NE and mixed clusters had genomic properties consistent with typical small cell biology, including high cell cycle activity, high NE biomarker expression (i.e., PCSK1, SCG2 and CHGB) and expression of low AR, REST and RB1 (Supporting Information Fig. S2). To confirm the association of these 216 genes with neuroendocrine differentiation, we downloaded gene expression data generated from the LTL331R system.8 This is a mouse model where an initial adenocarcinoma tumor, upon host castration, relapses as terminally differentiated NE making this an ideal platform to further refine the 216 gene set. This cross‐platform analysis found the majority (212/216) of genes were covered by the Agilent platform used by the authors of the initial LTL331R study.
We next ranked the 212 genes based on their fold change (FC) upon NE differentiation in the LTL331R system (Fig. 1 c). The most upregulated genes included PCSK1, CHGA, SEZ6L and RNF182 with FC > 9 folds, while AR‐regulated genes like FOLH1 and FOLH1B were the most downregulated (FC < 8 folds). To reduce the bias of the individual genes, we used the fold‐change ranking to categorize the 212 genes into 10 distinct modules, including seven upregulated (total n = 119) and three downregulated (total n = 93) groups (Supporting Information Table S1). The vast majority of these genes were consistent with the established molecular landscape of SC/NE, including notable SC/NE biomarkers (e.g., CHGA) or drivers of SC/NE progression (e.g., PEG10). AR‐regulated genes (i.e., KLK2 and PSMA) were also included, presumably due to their considerable downregulation in AR‐negative SC/NE. The 212 genes also included genes associated with cell proliferation (TOP2A, MCM, TPX2 and EZH2) and those downstream of the E2F1‐RB1 axis (CLSPN). Based on these data, we termed these 212 genes, the “212signature.”
Evaluation of the 212signature in treatment‐induced small cell prostate carcinoma and SCLC
To evaluate the 212signature, we first applied it to eight publicly available transcriptome datasets containing histologically confirmed NE in background of mCRPC and SCLC in a background of non‐SCLC (Figs. 1 c–1 e, Supporting Information Fig. S3). In prostate, SC/NE specimens generally grouped together based on the 212signature, although there were some outliers. Across the three mCRPC cohorts tested (Kumar et al., Beltran et al. and Aggarwal et al.), we found similar patterns of clustering. Even though the 212 genes were selected from de novo SC/NE, the signature remains effective in mCRPC setting differentiating treatment‐induced NE. In a cohort of SCLC and normal samples from 42 tissues (GSE43346), SCLC showed to have unique profiles compared to normal tissues from various sites15 (Supporting Information Fig. S3). However, central nervous system tissues (brain, cerebellum, cerebral cortex, hippocampus and thalamus) clustered with the SCLC. We also selected SCLC and non‐SCLC cell lines from the CCLE database and GDSC finding the 212signature discriminated SCLC and NSCLC cell lines (Fig. 1 e). Here it is worth mentioning that we used clustering based on the 212signatures to assess the feasibility of the genes in the signatures to assess the biological signal. Taken together, these data suggest that the 212signature can be applied to other tumor types with NE features, regardless of the tissue of origin.
Development of a small cell genomic score to identify SC/NE tumors
We used the 212signature to develop a single sample genomic classifier or small cell genomic score (SCGScore) model using a training cohort of 97 GG5 and 33 JHMI‐SC cohorts. When we generated new clusters with this merged cohort, we found the majority of SC/NE and mixed cases clustered together (Fig. 2 a, Supporting Information Fig. S4), but three samples from the natural history cohort also clustered with the SC/NE group. These three cases had developed metastasis within 2 years and had developed CRPC after subsequent hormonal therapy. Upon pathological rereview, one case was found to have small cell morphology. These cases were therefore included in the final SC/NE group (n = 23) for comparison to the Adeno group (n = 107). Using the SCGScore model, we found significantly higher scores in the SC/NE group and mixed group compared to the Adeno group (Fig. 2 b). Additionally, the SC/NE cases had high scores based on prognostic signatures16 indicating that they are very high‐risk tumors, but these signatures had lower sensitivity compared to the SCGScore which had a much higher rate of false positives (Fig. 2 c, Supporting Information Fig. S5). This suggests that prognostic signatures are not suitable for identifying small cell carcinoma.
Figure 2.
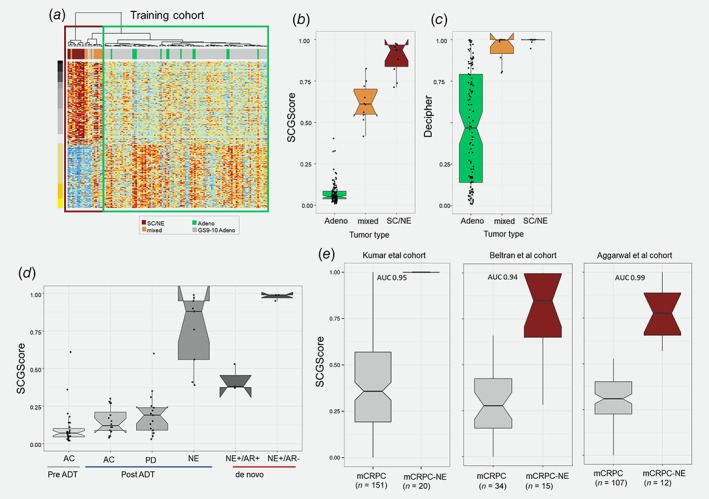
Development and validation of small cell genomic score (SCGScore). (a) Pooling 97 GG5 adenocarcinoma from JHU with the 33 samples to define 23 SC and 107 adenocarcinomas set for modeling. (b) SCGScore is significantly higher in SC/NE compared to mixed and adenocarcinoma. (c) Existing prognosis signature (Decipher) is not ideal for discriminating between SC/NE and adenocarcinoma with low sensitivity. (d) Independent validation of SCGScore in Calgary cohort showing that post‐ADT NE tumors have high scores compared to PD and high‐grade adenocarcinoma. Also, de novo NE with NE biomarker IHC positivity has higher scores that histologically NE tumors with negative IHC. (e) Validating the model in public mCRPC cohorts showing superior performance predicting SC/NE in the setting of mCRPC. [Color figure can be viewed at http://wileyonlinelibrary.com]
Validating SCGScore in four cohorts of de novo NE and treatment‐induced NE
Next, we evaluated the SCGScore model on four independent cohorts from the University of Calgary (Fig. 2 d) with patients presenting with either de novo SC/NE or post‐ADT NE, and Kumar et al., Beltran et al. and Aggarwal et al. cohorts. For comparison, we used post‐ADT PD samples and pre‐ADT adenocarcinoma. The post‐ADT NE group scored higher for SCGScore compared to pre‐ADT Adeno group, post‐ADT Adeno and post‐ADT PD achieving an AUC of 0.98 suggesting that SCGScore could be used to discriminate between histologically challenging cases (i.e., PD vs. small cell). Tumors with de novo NE, tumors with NE+/AR− had higher scores than those with NE+/AR+. The model also achieved superior performance (average AUC 0.95) when applied to mCRPC cohorts distinguishing mCRPC‐NE from mCRPC (Fig. 2 e). Also, among 180 patients with GG1–4 from a case–cohort7 (GG5 were excluded as they were used in model training), two patients had relatively high SCGScore with metastasis‐free survival rate less than 2 years. Finally, we characterized the clinical impact of the SCGScore in retrospective RP tissues (n = 70) treated with adjuvant ADT and have GG5. Patients with the highest SCGScore had a shorter median metastasis‐free survival than patients with low SCGScore (18 vs. 54 months, p = 0.006).
Evaluating SCGScore in prospective prostate RP and biopsy tissues from the Decipher GRID
To define the prevalence of primary prostate tumors with SC/NE‐like transcriptomic profiles, we measured the SCGScore in 17,967 RP and 6,697 biopsy prospective tissues collected from the clinical use of the Decipher test (Fig. 3 a). Additionally, as neuroblastoma is histologically similar to small cell carcinoma and expresses NE markers, we included a neuroblastoma cohort profiled on the same Human Exon microarray platform as an internal control. The SC/NE samples from discovery cohort and neuroblastoma control samples were found to have very high SCGScores (Fig. 3 a). In both the RP and biopsy settings, 1% of sample (151 in RP, 58 in biopsy) was classified as SC/NE‐like tumors by the model using a cutoff of 0.25 as defined from the discovery cohort. However, only a few tumor samples had SCGScores as high as the SC/NE tumors and these were all GG5. This indicates that the predicted SC/NE‐like tumors based on the model are not fully NE differentiated or that perhaps they are molecularly heterogeneous compared to typical NE. Comparing the SCGScore with previously developed model by Kumar et al., we found good correlation between the two signatures in the 33 SC/NE samples (R 2 = 0.56, p < 0.001) and mCRPC samples (R 2 = 0.66, p < 0.001)1, 2 (Supporting Information Figs. S6A and S6B), with weaker correlation (R 2 = 0.37, p < 0.001) in the primary tumors from prospective RP and biopsy samples (Supporting Information Figs. S6C and S6D) even though there is minimal gene overlap between the two signatures. Similarly, we found stronger correlation between SCGScore and the model developed by Tsai et al. in the 33SC/NE samples (R 2 = 0.85, p < 0.001) but not in the prospective cohort (R 2 = 0.18, p = 0.01) (Supporting Information Fig. 6E).
Figure 3.
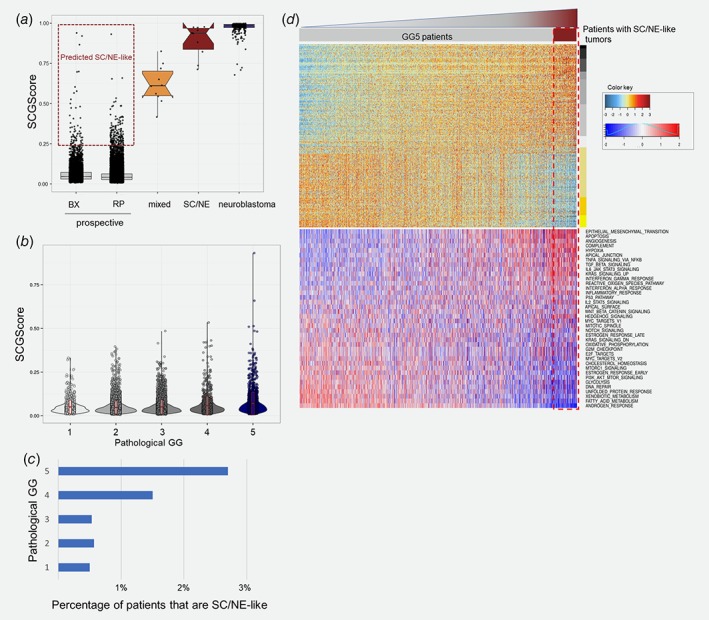
Evaluating SCGScore in a large prospective cohort. (a) Evaluating the SCGScore in prospective prostate RP (n = 17,967) and BX (n = 6,697) and neuroblastoma (n = 283) compared to SC/NE tumors. (b) SCGScore across pathological GG in RP samples. (c) Frequency of predicted SC/NE‐like across GG showing higher frequency in GG5. (d) Predicted SC/NE‐like patients have distinct genomic fingerprint compared to GG5 (n = 1,679) and distinct pathway activity. [Color figure can be viewed at http://wileyonlinelibrary.com]
Characterizing the clinical and molecular features of patients with high SCGS
To gain further insight into the clinical and molecular characteristics of the predicted 151 RP patients with SC/NE‐like features from the prospective cohort, we found the SCGScore tended to trend with GGs (Figs. 3 b and 3 c, Supporting Information Fig. S7A and S7B, Table S2). GG 5 was enriched with SC/NE‐like patients (2.7%) compared to GG 4 (1.5%), GG 3 (0.53%), GG 2 (0.5%) and GG 1 (0.5%; Fig. 3 c), with a similar trend observed for the biopsy samples (Supporting Information Fig. S7B). Interestingly only 31% of the SC/NE‐like cases were GG 5 and about 40% were GG 1–3 suggesting that NE differentiation could begin in early prostate cancer development. In terms of metastatic potential, 80% of the SC/NE‐like patients had high Decipher compared to 31% for Adeno patients. A similar frequency of high Decipher risk scores (74%) was seen in the prospective biopsy SC/NE‐like patients.
To gain a deeper understanding of the pathway activity in SC/NE‐like tumors, we compared the 151 SC/NE‐like patients to 1,679 GG5 RP patients. The SC/NE‐like tumors had a distinct pathway activity profile (Fig. 3 d, Supporting Information Figs. S8 and S9). SC/NE‐like tumors had higher activity of KRAS, hypoxia and immune pathways, with lower activity of xenobiotic metabolism, PI3K‐signaling, AR‐signaling and ER‐signaling. This data suggests SC/NE‐like tumors as a biologically distinct, aggressive subclass of PCa with distinctive therapeutic targets.
Characterizing the drug response profile of patients with high SCGScore
As the initial 212signature was able to discriminate between SCLC and NSCLC in both cell lines (CCLE & GDSC) and in tissues (Fig. 1 e, Supporting Information Fig. S3). When we applied SCGScore to the GDSC cohort, we found higher SCGScore values in SCLC compared to squamous, NSCLC (Supporting Information Fig. S10). For prostate cancer cell lines in GDSC, NCIH660 neuroendocrine prostate cancer cell line had a SCGScore of 0.93 (Supporting Information Fig. S10), but, this cell line lacked IC50 values. Next, we correlated the SCGScore with IC50 in 154 lung cancer cell lines from GDSC where a negative correlation would suggest a sensitivity to a given drug, while a positive score would suggest resistance. We found the SCGScore was positively correlated with the IC50 of Erlotinib, Trametinib, Dasatinib, Lapatinib and Afatinib, but was negatively correlated (better response) with Navitoclax, Vorinostat and Belinostat (Supporting Information Table S3).
Unfortunately, none of the six prostate cancer cell lines with combined IC50 and expression data were SC/NE‐like, and therefore we were unable to directly correlate SCGScore to IC50 in the prostate space. As a surrogate for this analysis, we built a drug response signature for each of the 265 drugs from lung carcinoma cell lines. For each drug, we defined a signature that was grouped into a score representing the drug response; high scores are indicative of potential response while low scores would suggest a lack of potential response. We applied these drug response models to our prospective prostate cohort comparing SC/NE‐like to GG5 patients. DRS of histone methyltransferase (SGC0946), histone deacetylases (HDAC) inhibitor (CAY10603, Belinostat) and poly (ADP‐ribose) polymerase (PARP) inhibitor (Rucaparib) showed the highest positive correlation with SCGScore (Figs. 4 a–4 c, Supporting Information Table S4) suggesting potential response. To confirm some of these predictions in neuroendocrine prostate cancer cells, we used drug response profiles from the CCLE experiments that have neuroendocrine prostate cancer cell line NCI‐H660. Drug response profiles showed high sensitivity (low IC50) to Olaparib and Vorinostat supporting predicted DRS results observed in the prospective cohort (Figs. 4 d and 4 e). These data suggest that SC/NE‐like are distinct from GG5 tumors and potentially share common therapeutic targets with SCLC.
Figure 4.
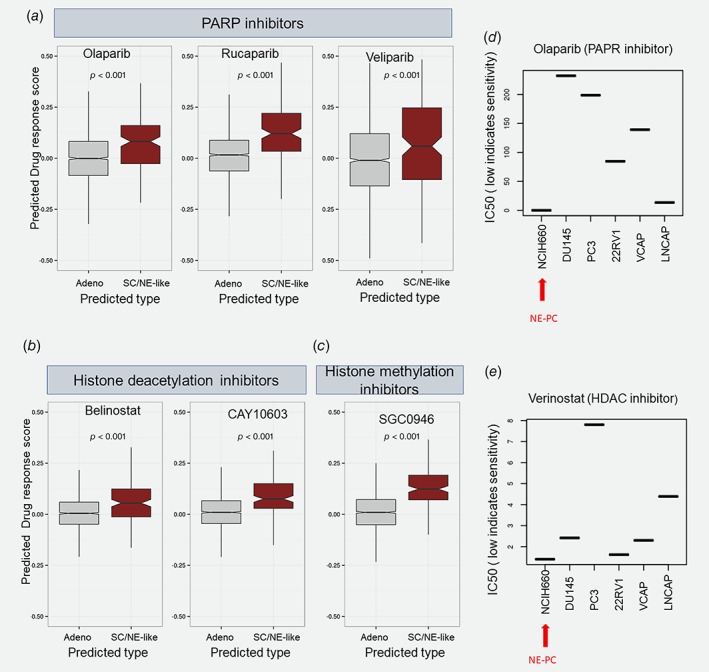
Therapeutic implications of SCGScore. SC/NE‐like are predicted to be more sensitive to (a) PARP inhibitors, (b) HDAC inhibitors and (c) methylation inhibitors. (d–e) NCIH660 (prostatic NE cell line) showed to respond to both PAPR and HDAC inhibitors. [Color figure can be viewed at http://wileyonlinelibrary.com]
Discussion
Herein, we identify a subset of treatment‐naïve pPCa that is molecularly analogous to mCRPC‐NE and SCLC using a 212 gene signature. These genes were grouped these into 10 modules spanning several key pathways including AR‐signaling, NE differentiation, RB‐loss and cell proliferation, with NE differentiation pathway having the largest weight. In what we believe this is the largest ever study that uses transcriptomics to identify SC/NE‐like prostatic tumors.
The data presented here offer several novel insights. First, we demonstrate that prostatic SC/NE are molecularly analogous to SCLC suggesting that prostatic SC/NE suggesting their shared common pathways and potentially common therapeutic targets.
Second, de novo SC/NE and treatment‐induced NE have a similar molecular profile in terms of expressing NE biomarkers and low AR‐activity. Third, about 0.5–1% of GG1 and GG2 have high SCGScore suggesting that NE differentiation can occur in very early stages of cancer. Fourth, the SCGScore might be useful for identifying patients with a NE‐like genomic profile who may not be suitable for AR‐targeted therapy and may be candidates for novel therapeutics. Thus, implementing SCGScore as part of commercially used prognostic test allows for the identification of patients for enrollment in such clinical trials.
Our study did have some noteworthy limitations. Lack of IHC staining of NE markers and lack of outcome in the prospective cohort limited assessing our model in the prospective cases. Also, lack of drug response data in prostatic cell lines prevented us from building prostate specific drug response models.
In conclusion, evaluating SCGScore in very large cohorts of primary tumors provides insights on the early differentiation of neuroendocrine carcinoma. SCGScore can be used together with prognostic models to identify high‐risk tumors that are more likely to develop neuroendocrine disease, and thus not suitable candidates for hormonal therapy.
Supporting information
Appendix S1: Supporting Information
Figure S1: T‐SNE scatter plot of the 33 JHMI‐SC samples based on the 306 meta genes from Tsai et al. 2017.
Figure S2: Genomic‐based groups of Adeno, mixed and SC/NE are representing the expecting biology where SC/NE have high expression of cell cycle, lower AR, REST, and higher neuroendocrine biomarkers.
Figure S3: Evaluating the 212signature in mCRPC cohorts. A. mCRPC cohort (Aggarwal et al.) with NE tumors, B. mCRPC cohort (GSE66187) with NE‐mCRPC samples stained with CHGA, SYP and AR. C. Also, 212signature is able to discriminate SCLC from various normal tissues.
Figure S4: T‐SNE plot of the 33 JHMI‐SC samples with the 97 GG5 from John Hopkins natural history cohort.
Figure S5: Penney signature (prognostic signature) showing high sensitivity to predict SC/NE where more than 50% of the Adeno had high scores similar to SC/NE.
Figure S6: Correlation plots between existing NE signature (Kumar NE) and SCGScore in mCRPC setting (A‐B) and GRID primary data (C‐D). E. Correlation plot between Tsai et al. signature and SCGScore in GRID primary data.
Figure S7: SCGScore distribution across Grade Groups (GG). A. Patients with high SCGScore have GG5 tumors in biopsy. B.GG5 in biopsy showed to be more enriched with SC/NE‐like tumors (3%) compared to GG1‐3 that have only 0.5%.
Figure S8: T‐SNE of the 151 predicted SC/NE‐like from GRID RP and GG5 patients from GRID RP. Results showing that predicted SC/NE‐like tumors are distinct from GG5 based on the 212signature.
Figure S9: Pathway activity associating pathways with SC/NE‐like tumors.
Figure S10: SCGScore across cell lines from GDSC showing SCLC have high scores while other tumors not. Prostate cancer NE cell line (NCIH600) have high score of SCGScore.
Table S1 212genesignature with their p‐value in discovery JHMI‐SC cohort and FC in 331R system
Table S2: Pathological characteristics of SC/NE‐like tumors and Adeno samples from prospective RP cohort
Table S3: Correlation between calculated SCGScore and IC50 for lung cancer sample in GDSC cohort
Table S4: Correlation between calculated Drug response scores and SCGScore in prospective RP patients. Higher correlation indicates higher potential response.
Acknowledgements
This work was supported in part by the Prostate Cancer Foundation Young Investigator Award (to T.A.B). This work was also supported by Prostate Cancer Canada and is proudly funded by the Movember Foundation Grant #B2013‐01.
Conflict of interests: MA, YL, EG, NE, JL, MT, ED are employees of GenomeDx Biosciences. Dr. Lotan has received research funding for other projects from GenomeDx. Other authors declare no potential conflicts of interest.
References
- 1. Kumar A, Coleman I, Morrissey C, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 2016;22:369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beltran H, Prandi D, Mosquera JM, et al. Divergent clonal evolution of castration‐resistant neuroendocrine prostate cancer. Nat Med 2016;22:298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aggarwal R, Huang J, Alumkal JJ, et al. Clinical and genomic characterization of treatment emergent small‐cell neuroendocrine prostate cancer: a multi‐institutional prospective study. J Clin Oncol 2018;36:2492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cancer Genome Atlas Research Network . Electronic address: schultz@cbio.Mskcc.Org, cancer genome atlas research network. The molecular taxonomy of primary prostate cancer. Cell 2015;163:1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsai H, Lehrer J, Alshalalfa M, et al. Gene expression signatures of neuroendocrine prostate cancer and primary small cell prostatic carcinoma. BMC Cancer 2017;17:759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ross AE, Johnson MH, Yousefi K, et al. Tissue‐based genomics augments post‐prostatectomy risk stratification in a natural history cohort of intermediate‐ and high‐risk men. Eur. Urol. 2015;69:157–65. [DOI] [PubMed] [Google Scholar]
- 8. Akamatsu S, Wyatt AW, Lin D, et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep 2015;12:922–36. [DOI] [PubMed] [Google Scholar]
- 9. Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iorio F, Knijnenburg TA, Vis DJ, et al. A landscape of Pharmacogenomic interactions in cancer. Cell 2016;166:740–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Glass AG, Leo MC, Haddad Z, et al. Validation of a genomic classifier for predicting post‐prostatectomy recurrence in a community based health care setting. J Urol 2015;195:1748–53. [DOI] [PubMed] [Google Scholar]
- 12. Piccolo SR, Sun Y, Campbell JD, et al. A single‐sample microarray normalization method to facilitate personalized‐medicine workflows. Genomics 2012;100:337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27. [DOI] [PubMed] [Google Scholar]
- 14. Wilkerson M, Hayes D. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sato T, Kaneda A, Tsuji S, et al. PRC2 overexpression and PRC2‐target gene repression relating to poorer prognosis in small cell lung cancer. Sci Rep 2013;3:1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Penney KL, Sinnott JA, Fall K, et al. mRNA expression signature of Gleason grade predicts lethal prostate cancer. J Clin Oncol 2011;29:2391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information
Figure S1: T‐SNE scatter plot of the 33 JHMI‐SC samples based on the 306 meta genes from Tsai et al. 2017.
Figure S2: Genomic‐based groups of Adeno, mixed and SC/NE are representing the expecting biology where SC/NE have high expression of cell cycle, lower AR, REST, and higher neuroendocrine biomarkers.
Figure S3: Evaluating the 212signature in mCRPC cohorts. A. mCRPC cohort (Aggarwal et al.) with NE tumors, B. mCRPC cohort (GSE66187) with NE‐mCRPC samples stained with CHGA, SYP and AR. C. Also, 212signature is able to discriminate SCLC from various normal tissues.
Figure S4: T‐SNE plot of the 33 JHMI‐SC samples with the 97 GG5 from John Hopkins natural history cohort.
Figure S5: Penney signature (prognostic signature) showing high sensitivity to predict SC/NE where more than 50% of the Adeno had high scores similar to SC/NE.
Figure S6: Correlation plots between existing NE signature (Kumar NE) and SCGScore in mCRPC setting (A‐B) and GRID primary data (C‐D). E. Correlation plot between Tsai et al. signature and SCGScore in GRID primary data.
Figure S7: SCGScore distribution across Grade Groups (GG). A. Patients with high SCGScore have GG5 tumors in biopsy. B.GG5 in biopsy showed to be more enriched with SC/NE‐like tumors (3%) compared to GG1‐3 that have only 0.5%.
Figure S8: T‐SNE of the 151 predicted SC/NE‐like from GRID RP and GG5 patients from GRID RP. Results showing that predicted SC/NE‐like tumors are distinct from GG5 based on the 212signature.
Figure S9: Pathway activity associating pathways with SC/NE‐like tumors.
Figure S10: SCGScore across cell lines from GDSC showing SCLC have high scores while other tumors not. Prostate cancer NE cell line (NCIH600) have high score of SCGScore.
Table S1 212genesignature with their p‐value in discovery JHMI‐SC cohort and FC in 331R system
Table S2: Pathological characteristics of SC/NE‐like tumors and Adeno samples from prospective RP cohort
Table S3: Correlation between calculated SCGScore and IC50 for lung cancer sample in GDSC cohort
Table S4: Correlation between calculated Drug response scores and SCGScore in prospective RP patients. Higher correlation indicates higher potential response.


