Figure 1.
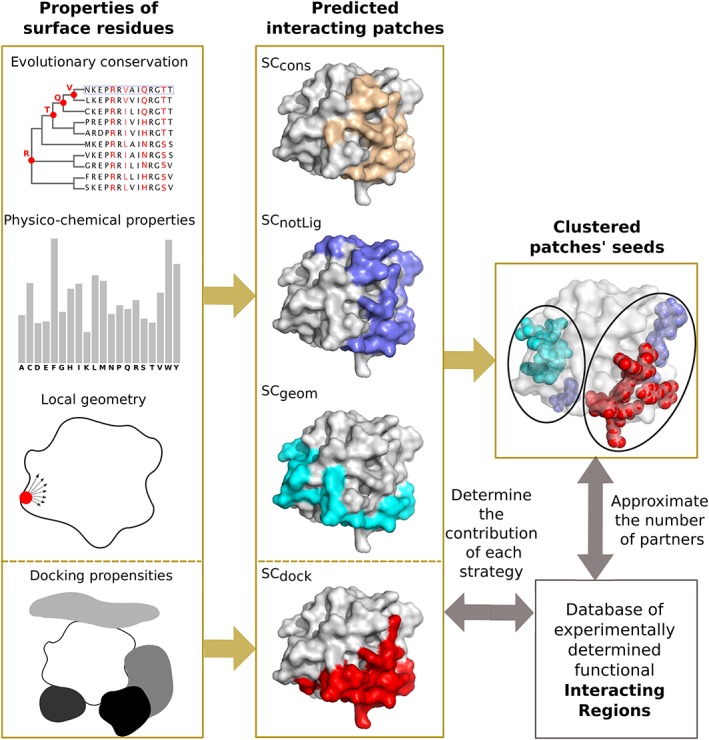
Schematic representation of our workflow. We consider four residue‐based properties (left panel), namely evolutionary conservation, amino acid propensities to be found at an interface, local geometry, and propensities to be found in docked interfaces. We predict interacting patches at the surface of proteins by using four different strategies: SCcons, SCnotLig, and SCgeom combines the first three properties, while SCdock relies exclusively on the fourth property. We compare the predicted patches with a set of experimentally determined functional interacting regions. We analyze and cluster the predicted patches' seeds, from which they were grown, to precisely localize interacting regions and infer the number of partners used by each region [Color figure can be viewed at http://wileyonlinelibrary.com]
