Abstract
Background:
Long noncoding RNAs (lncRNAs) are a new class of cancer regulators. Here, we aimed to investigate the diagnostic and therapeutic values of an lncRNA, differentiation antagonizing noncoding RNA (DANCR), in lung cancer.
Methods:
Real-time polymerase chain reaction was used to compare DANCR levels in normal and cancerous lung tissues as well as lung cancer cells. Lentiviral transduction was used to induce DANCR overexpression or silencing in vitro, followed by monitoring cell proliferation, colony formation, and changes in microRNA-216a (miR-216a) expression. DANCR-specific small hairpin RNA transduction was used to establish cells with stable DANCR knockdown, and silenced cells were used to initiate lung tumor xenografts, followed by monitoring tumor growth.
Results:
DANCR upregulation was seen in lung cancer, particularly in high-grade lung cancer tissues and aggressive cancer cells. Ectopic DANCR expression induced lung cancer cell proliferation and colony formation, whereas DANCR silencing induced opposing effects. The miR-216a level in cancer cells was negatively correlated with DANCR expression. The DANCR knockdown reduced the growth of tumor xenografts in vivo.
Conclusion:
DANCR upregulation is a potential indicator of aggressive lung cancer. Silencing of DANCR has great potential as a potent therapeutic strategy in lung cancer.
Keywords: LncRNA, DANCR, lung cancer, miR-216a, therapeutic strategy
Introduction
Lung cancer is the leading cause of cancer-related death worldwide.1 Particularly, non–small cell lung cancer (NSCLC), which accounts for 85% of newly diagnosed lung cancer cases, is characterized by resistance to chemotherapy and poor prognosis.2 The 5-year survival of patients with lung cancer remains poor, despite tremendous efforts in improving the clinical management of lung cancer. Over the years, studies have unveiled a complex genetic network involved in lung cancer initiation and progression.3 This has led to the development of a large array of therapeutics targeted to these genetic alterations. However, it is still a critical clinical challenge to diagnose NSCLC early and sensitively.4 Treatment of NSCLC at advanced stage is, therefore, problematic due to chemoresistance and recurrence.5 An astonishing number of noncoding RNAs in transcriptome, which were once considered redundant genes, are recently found to be important contributors to carcinogenesis and progression.6 Among the noncoding RNAs, long noncoding RNAs (lncRNAs) are drawing increasing attention due to their role as broad-spectrum regulators in cancer. The LncRNAs are messenger RNA (mRNA)-like transcripts ranging in length from 200 nt to 100 kb. Dysregulated expressions of certain lncRNAs may affect epigenetic information and provide a cellular growth advantage of tumor. Their role in cancer resides in regulating microRNAs (miRNAs, miRs) or mRNAs involved in cancer progression.7 Therefore, lncRNAs are emerging as novel biomarkers for cancer diagnosis and targets of cancer therapy. Regulation of these lncRNAs is an attractive strategy to impede cancer progression and overcome cancer resistance.
DANCR was originally considered an antidifferentiation noncoding RNA (ncRNA) required for the dedifferentiation of epidermal cells.8 The DANCR was found to be higher in a large variety of cancer tissues, including colorectal cancer,8 hepatocellular cancer,9 and prostate cancer.10 These evidences spurred interest in characterizing DANCR as a diagnostic and prognostic marker in cancers. The DANCR upregulates cancer cell stemness and promotes tumor cell dissemination and metastases formation.11 However, the role of DANCR in lung cancer has not been investigated. It is an attractive strategy to diagnose lung cancer based on DANCR levels and suppress lung cancer progression using DANCR interference.
Herein, we analyzed DANCR expression in lung cancer tissues with varying aggressiveness and demonstrated that higher DANCR is linked to cases with lung cancer having higher clinical grade and poorer survival. While DANCR induced lung cancer cell invasion, DANCR interference attenuated tumor progression. Previous evidence suggested direct regulation of microRNA-216a (miR-216a) by DANCR in lung cancer cells.12 Moreover, the tumor suppressing role of miR-216a has been reported previously.13,14 Here we show that miR-216a is suppressed by DANCR in lung cancer. The data reported in this study could potentiate the use of DANCR interference as a novel strategy to impede progression of lung cancer.
Materials and Methods
Tissue, Cell Culture, and Viability Assay
Lung cancer tissue of grades II to IV, as well as adjacent normal tissue (>5 cm from tumor), were acquired from Shijiazhuang No.1 Hospital in accordance with protocols approved by ethical committee of Shijiazhuang No.1 Hospital (#SJZ1H-CL-3274).Written consent was acquired from each participant. Detailed pathological information and number of samples are described in Supplementary Table S1. BEAS-2B, NCI-H1299, A549, and NCI-H1975 were acquired from American Type Culture Collection (ATCC; Rockville, MD) and maintained according to ATCC’s recommendations. Cells were incubated in 96-well plates (2 × 104 cells/well) and cultured for 48 hours. For viability assay, Cell Count Kit-8 reagent (Sigma Aldrich, St Louis, Missouri; 10 µL) was added to the medium of cells cultured in 96-well plates, followed by incubation for 4 hours. Absorbance of the solution was measured at 450 nm by an ELx-800 microplate reader (Winooski, Vermont, USA). Kaplan-Meier analysis was performed using an online tool integrated with a public database containing clinical lung cancer data (http://kmplot.com/analysis/).15 Correlation of the survival data of patients and DANCR levels was analyzed using SPSS.
Quantitative Real-Time Polymerase Chain Reaction
RNAs were extracted using the Trizol reagent (Invitrogen, Carlsbad, California), followed by removal of DNA with the TurboDNase kit (Ambion). Quantification of extracted RNA was performed using NanoDrop. Complementary DNA synthesis was performed using PrimeScript real-time (RT) reagent kit (Takara Bio, Japan) using 1000-ng of total RNA. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the SYBR Select Master Mix (Applied Biosystems, Waltham, Massachusetts) on an ABI 7900 system (Applied Biosystems). The level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control. The Ct value was calculated based on the ΔΔCt method. Fold change of gene expression was expressed as 2−ΔΔCt. The primers used in this study were as follows: DANCR sense: 5′-GCCACAGGAGCTAGAGCAGT-3′; DANCR antisense: 5′-GCAGAGTATTCAGGGTAAGGGT-3′; GAPDH sense: 5′-AACGGATTTGGTCGTATTGGG-3′; GAPDH antisense: 5′-CGCTCCTGGAAGATGGTGAT-3′.
Colony Formation Assay
The transfected cells were seeded in 6-well plates with culture medium containing 10% fetal bovine serum and cultured overnight. After 14 days, cells were fixed with methanol and stained with 0.1% Crystal violet. Colonies were manually counted under a light microscope.
Vector Construction and Lentiviral Transfection
The DANCR, siDANCR and small hairpin DANCR (shDANCR) vector, and their noncoding controls were constructed by inserting corresponding DNA to the plenti6.3-MCS-IRES-EGFP vector. Constructed plasmid, along with the packaging plasmids, was cotransfected into 293 T cells. Viral particles were then harvested at 48 hours after transfection with ultracentrifugation. Cells transfected with lentivirus were selected by puromycin for 2 weeks to obtain stable cell lines. The siDANCR sequences used are as follows: sense: 5′-GGCCAAAUAUGCGUACUAAUU-3′, antisense: 3′-UUCCGGUUUAUACGCAUGAUU-5′. The sequences used for shDANCR are as follows: 5′-GGAGCTGAACTGCACTGTTGT-3′.
Ago RNA Immunoprecipitation and RNA Pull-Down Assays
The ThermoFisher RNA immunoprecipitation (RIP) kit (Waltham, MA) was used for Ago RIP assay according to the manufacturer’s recommendations. Ago2 antibody was purchased from Abcam (Cambridge, MA). Immunoglobulin G antibody (Abcam) was used as a negative control. The anti-SNRNP70 antibody (Abcam) was used as the positive control for data normalization. The qRT-PCR analysis was used to analyze purified DANCR RNA. RNA pulldown assay was carried out after DANCR were transcribed using T7 RNA polymerase (Roche, Switzerland). Purified RNAs were then labeled with biotin with the biotin RNA labeling mix (Roche), and magnetic beads were then added to the mixture and incubated at room temperature for 1 hour. Proteins were then eluted from the beads and analyzed by Western blot.
Animal Handling
Cells stably express shDANCR or noncoding shRNA (shNC) were inoculated subcutaneously into the flank of mouse (2 million per mouse). Following tumor inoculation, tumors sizes were monitored by measuring width and length of the tumor. Tumors were measured every 3 days since day 9. Tumor volume is calculated as tumorvolume=width2×length/2. In day 24, tumors were dissected, photographed, and weighted. The miR-216 levels in dissected tumors were also analyzed. Animal study was approved by Ethics Committee of Shijiazhuang No.1 Hospital (#SJZ1H-20161105).
Statistical Analysis
Data were presented as mean (standard deviation). Data were analyzed by Student t test and 1-way or 2-way analysis of variance using SPSS 18.0 software. Difference in data were considered statistically significant if P value is <.05.
Results
Upregulation of DANCR is Indicative of Poor Prognosis in Lung Cancer
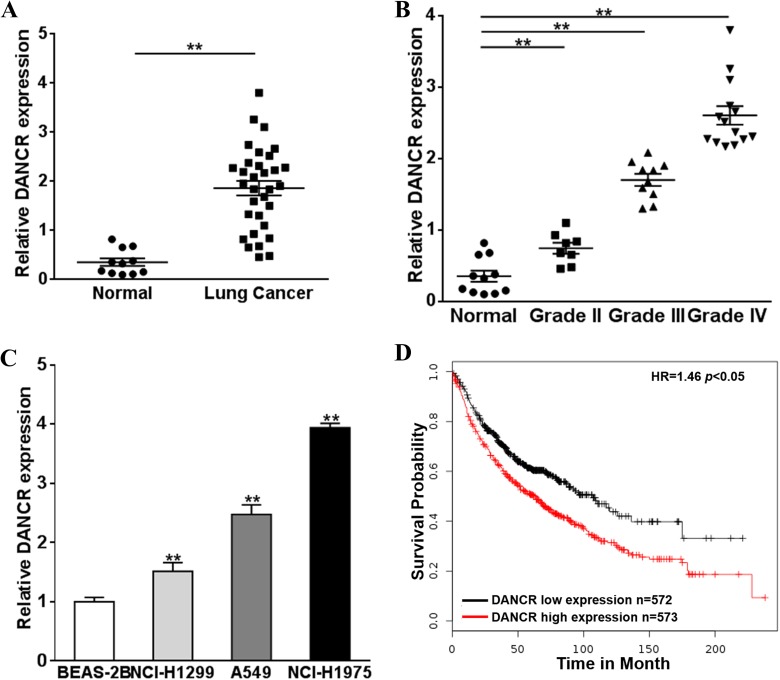
First, to validate DANCR as a prognosis marker of lung cancer, we evaluated DANCR expression in lung cancer and normal lung tissues. As shown in Figure 1A and B, DANCR level was upregulated in lung cancers, and the upregulation was higher in cancers with a higher clinical grade. Further, we evaluated DANCR expression in commonly used lung cell lines, BEAS-2B (epithelial-like lung cell), NCI-H1299, A549, and NCI-H1975 (aggressive lung cancer cells). A higher DANCR expression was seen in lung cancers as shown in Figure 1C. We also validated DANCR expression in correlation with patient survival. Indeed, DANCR overexpression was associated with a poor survival data of patients with lung cancer (Figure 1D). These data suggested that DANCR upregulation was a characteristic of aggressive lung cancer, and DANCR was potentially a valuable diagnostic marker for this high-risk disease.
Figure 1.
DANCR upregulation is indicative of poor prognosis in lung cancer. A, The DANCR expression levels of normal lung tissues (n = 11) and lung cancer specimens (n = 32) were analyzed by qRT-PCR. B, The lung cancer specimens were divided into several grades according to World Health Organization (WHO) standard. C, The DANCR expression levels of normal lung cells BEAS-2B and lung cancer cells (NCI-H1299, A549, and NCI-H1975) were tested by qRT-PCR and were normalized to BEAS-2B group. Data were presented as mean (SD) from 3 independent experiments with triple replicates per experiment. D, The Kaplan-Meier plotter program was employed to detect the survival prognosis of DANCR in lung cancer. ** P < .05. The median value of lncRNA DANCR was used as a cutoff value. DANCR indicates differentiation antagonizing noncoding RNA; qRT-PCR, quantitative real-time polymerase chain reaction; SD, standard deviation.
Overexpression of DANCR Promotes the Proliferation and Growth of Lung Epithelial Cells
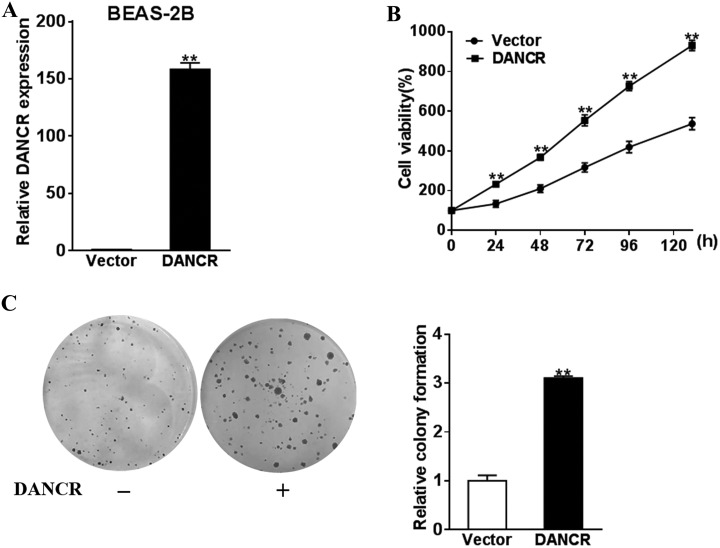
Given that DANCR upregulation is associated with lung cancer, we next examined whether ectopic expression of DANCR in normal lung epithelial cells would increase their survival and colony formation. To this end, we ectopically expressed DANCR in BEAS-2B cells using lentiviral transfection of DANCR expression vector. This induced an upregulation of DANCR level (Figure 2A). Concomitantly, the proliferation (Figure 2B) and colony formation (Figure 2C and D) of BEAS-2B were dramatically increased compared to control cells (P < .01). These evidences indicated that DANCR might contribute to the malignant phenotypes of lung cancer cells.
Figure 2.
Overexpression of DANCR promotes the proliferation and growth of normal lung cells. BEAS-2B cells were transfected with DANCR or control vector. A, The DANCR expression levels were detected by qRT-PCR. B, CCK-8 assay was used to test the cell proliferation. C, The cell growth was analyzed by colony formation assay. ** P < .05. DANCR indicates differentiation antagonizing noncoding RNA; qRT-PCR, quantitative real-time polymerase chain reaction; CCK-8, cell count kit 8.
DANCR Inhibition Suppresses Lung Cancer
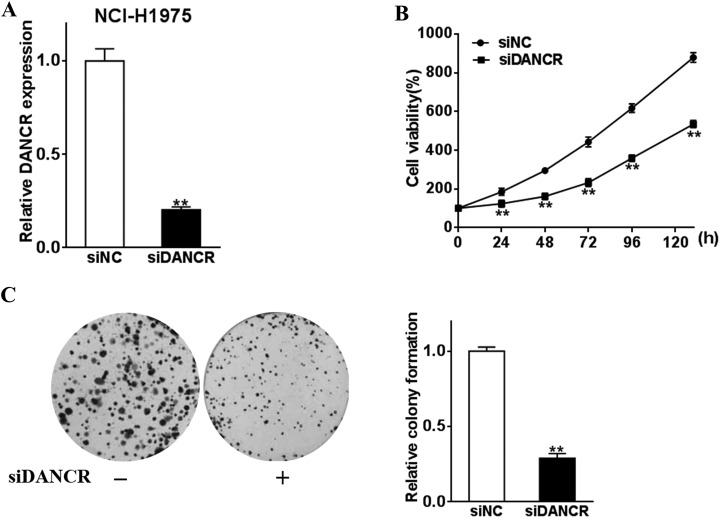
Considering the tumor-promoting role of DANCR, we further reasoned if DANCR silencing would abrogate the proliferation and colony-forming ability in lung cancer cells. Therefore, we transfected DANCR-specific siRNA to NCI-H1925 cancer cells to induce DANCR downregulation (Figure 3A). We found that the proliferation (Figure 3B) and colony formation (Figure 3C) were attenuated by DANCR interference. This further corroborated the tumor-promoting role of DANCR.
Figure 3.
Inhibition of DANCR suppressed lung cancer cellular functions. Lung cancer cells NCI-H1975 were transfected with siDANCR or negative control (siNC). A, The DANCR expression levels were detected by qRT-PCR. B, CCK-8 assay was used to test the cell proliferation. C, The cell growth was analyzed by colony formation assay. ** P < .05. DANCR indicates differentiation antagonizing noncoding RNA; qRT-PCR, quantitative real-time polymerase chain reaction; CCK-8, cell count kit 8.
DANCR Sequesters Tumor Suppresser miR-216a
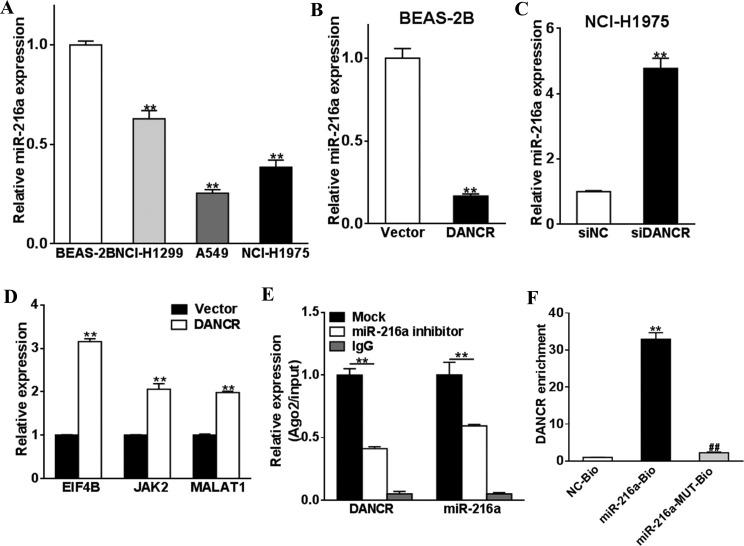
The LncRNAs usually serve as molecular sponges that sequester the expression of other RNAs. Due to the cancer-promoting function DANCR, we examined whether miR-216a could be sequestered by DANCR. As shown in Figure 4A, miR-216 expression was lower in lung cancer cells (NCI-H1299, A549, and NCI-1975 cells) compared to lung epithelial cells (BEAS-2B cells). DANCR overexpression, which was shown to promote cell proliferation and colony formation, also downregulated the level of miR-216a (Figure 4B). On the other hand, silencing of DANCR with siRNA increased the expression of miR-216a in NCI-H1299 cells (Figure 4C). Further, ELF4B, JAK2, and MALAT1, which are suppressed by miR-216a, demonstrated an upregulation under DANCR overexpression (Figure 4D). Therefore, DANCR overexpression abrogated the suppression of these oncogenes by miR-216a, inducing tumorigenesis and invasion. To further validate the antagonizing effects of DANCR on miR-216, we showed that cotransfection of siDANCR and anti-miR-216a abrogated the inhibitory effect of siDANCR on lung cancer proliferation and colony formation (Supplementary Figure S1A and B). In addition, DANCR and miR-216a demonstrated a negative correlation in patient samples (Supplementary Figure S2). Ago RIP (Figure 4E) and RNA (Figure 4F) pull-down assays were used to detect the association between DANCR and miR-216a in HEK293 cells. We showed that miR-216a inhibitor significantly blocked the pulled RNA level of DANCR and miR-216a (Figure 4E). DANCR enrichment in RNA pull-down assay was also reduced using biotin-labeled mutant miR-216a when compared to unmodified miR-216a (Figure 4F). Similar results were observed in NCI-H1975 cells (Supplementary Figure S4). Taken together, these data suggested a close interaction between DANCR and miR-216a in lung cancer. Additionally, we tested whether miR-216b and miR-217 were regulated by DANCR. However, we did not see changes in the levels of these 2 miRNAs after siDANCR transfection (Supplementary Figure S3).
Figure 4.
DANCR sequesters tumor suppresser miR-216a. A, The miR-216a expression levels of normal lung cells BEAS-2B and lung cancer cells (NCI-H1299, A549, and NCI-H1975) were tested by qRT-PCR and were normalized to BEAS-2B group. B-D, BEAS-2B cells were transfected with DANCR or control vector. NCI-H1975 was transfected with siDANCR or siNC. The miR-216a and the identified targets (EIF4B, JAK2, and MALAT1) of miR-216a expression levels were detected by qRT-PCR. E and F, RIP and RNA pull-down assays were used to detect the association between DANCR and miR-216a in HEK293 cells. ** P < .05. DANCR indicates differentiation antagonizing noncoding RNA; qRT-PCR, quantitative real-time polymerase chain reaction; miR-216a; microRNA-216a; RIP, RNA immunoprecipitation.
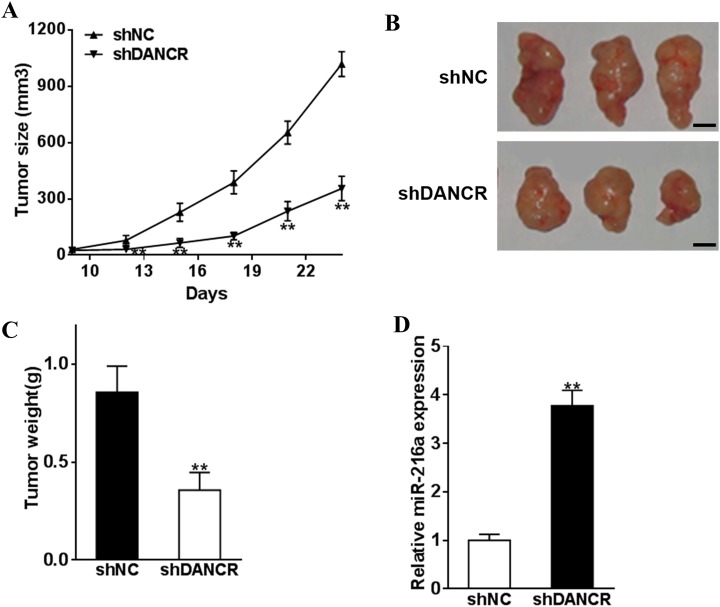
Silence of DANCR Inhibits Lung Cancer Tumor Growth In Vivo
To demonstrate DANCR as a therapeutic target with great clinical potential, we performed in vivo evaluation of the effect of DANCR silencing on tumor growth. We showed that inoculated cancer cells with DANCR silencing had a considerably reduced tumor burden as indicated by tumor growth rate (Figure 5A), tumor size (Figure 5B), and tumor weight (Figure 5C; P < .01). Further, we also found that miR-216a levels in tumors with DANCR silencing were also higher (Figure 5D). These data indicate that DANCR interference is a promising strategy to impede lung cancer progression.
Figure 5.
Silence of DANCR inhibits lung cancer tumor growth in vivo. NCI-H1975 cells were infected by lentivirus to establish NCI-H1975-shDANCR or NCI-H1975-shNC stable overexpression cells and were injected into each side of the nude mice posterior flanks (8 tumors of each group). A, Tumors were measured every 3 days since day 9, and the volumes were calculated using the following formula: volume = 0.5 × Length × Width2. B, C, In day 24, the mice were sacrificed, weighed with representative pictures of tumors shown (bar = 10 mm). D, The miR-216a expression levels of the tumors were analyzed by qRT-PCR. ** P < .05. DANCR indicates differentiation antagonizing noncoding RNA; shDANCR, small hairpin DANCR; shNC, negative control; miR-216a; microRNA-216a; qRT-PCR, quantitative real-time polymerase chain reaction.
Discussions
The development of aggressive lung cancer requires a series of oncogenic stimuli to maintain their tumor-initiating capacity. Here we have identified DANCR as a potential inducer of aggressive lung cancers and demonstrated a close correlation between DANCR expression and lung cancer malignancy. In our study, poorer survival was found to be associated with cases with lung cancer having higher DANCR expressions. This potentiates the use of DANCR as a biomarker for risk prognostication of lung cancer. Our study is juxtaposed with a number of researches in characterizing DANCR as a diagnostic marker for other cancers8,9,11,16 and broadens the clinical implication of DANCR. Currently, biopsy remains the primary approach for risk stratification of lung cancer.17 However, biopsy examines only limited sample of the disease region and is highly susceptible to false-negative results. Although recent advances in imaging technologies have potentiated evaluation of lung cancer risk by examining the entire lung noninvasively, the high cost of imaging examinations limits their clinical use. Still, accurate imaging biomarkers are difficult to establish. Our data provide an opportunity to evaluate lung cancer aggressiveness by assessing DANCR levels. Although our study only investigated the correlation of tissue or intracellular DANCR levels with lung cancer risk, circulating DANCR levels may also indicate cancer aggressiveness.18 A facile testing based on plasma DANCR level could be developed for lung cancer diagnosis. For example, Ma et al demonstrated the correlation of plasma DANCR with the aggressiveness of hepatocellular carcinoma.19 Further studies are warranted to evaluate the correlation between plasma DANCR levels and lung cancer malignancy to validate the clinical utility of DANCR as a lung cancer marker.
Meanwhile, a new therapeutic strategy for lung cancer is revealed which is based on the modulation of DANCR in lung cancers. First, we validated that overexpression of DANCR is sufficient in promoting the proliferation and migration of BEAS-2B cells, which is an epithelial type of lung cells. Although we did not investigate the mesenchymal markers associated with cancer stemness, it is likely that DANCR upregulation promoted the epithelial to mesenchymal transition in BEAS-2B cells. This is consistent with the role of DANCR as an oncogene. Further, DANCR interference with siRNA was shown to decrease the proliferation and colony formation of NCI-H1975 cells, which lays the foundation of lung cancer therapy using DANCR interference. To elucidate the mechanism of DANCR in regulating lung cancer progression, we investigated the correlation between DANCR expression and miR-216a. The DANCR is a putative regulator of a number of tumor suppressor miRNAs.20,21 Thus far, the interaction between DANCR and miR-216a has not been demonstrated. Dysregulated miR-216a expression is thought of as a significant contributor to hepatocellular carcinoma.22 We found that miR-216a expression is lower in high-risk lung cancers, and DANCR suppresses miR-216a. The targets of miR-216a, EIF4B, JAK2, and MALAT1 were therefore upregulated by DANCR. It should be noted that miR-216a is only one of the targets mediated by DANCR. Other cancer-related molecules, such as CTNNB19 and miR-33a-5p,23 were also demonstrated to be regulated by DANCR. Further studies are still warranted to unravel additional molecular pathways regulated by DANCR. On the other hand, DANCR interference with siRNA induced an opposite effect. This encouraged us to test whether DANCR silencing would successfully reduce lung cancer progression.
We have demonstrated that DANCR interference is an effective strategy to suppress lung cancer. Cells stably expressing shDANCR demonstrated attenuated tumor growth. This antitumor effect was also accompanied by the upregulation of miR-216a. Therefore, our study offered a novel antitumor strategy by transfecting shDANCR in lung cancer cells. Our study also provides an alternative route for miR-216a restoration, which has been employed in a number of studies for cancer therapy.24,25 It should be noted that tumors expressing shDANCR still demonstrated increased rate of tumor growth after 20 days of treatment, and no tumor regression can be observed. The suboptimal tumor suppression with shDANCR can potentially be improved by combinatory therapy with other chemotherapy drugs, which has been demonstrated in other similar studies.26 Additionally, we are aware that our approach does not represent the clinical practice in lung cancer treatment since the cells were transfected with shRNA prior to inoculation. It can be envisioned that with the development of efficient gene delivery systems,27,28 administration of shDANCR or other interfering genes in vivo would be possible. In fact, the lentiviral system for shDANCR transfection could be readily tested for DANCR silencing.29 Other nonviral system for DANCR interference can also be employed.
In conclusion, we have demonstrated that DANCR upregulation is linked to poor survival of patients with lung cancer. The DANCR overexpression promotes cell proliferation and colony formation in vitro, and DANCR interference effectively suppresses lung cancer progression both in vitro and in vivo. MiR-216a is downregulated by DANCR. DANCR interference has great potential to attenuate the progression of aggressive lung cancers. The diagnostic and therapeutic value of DANCR is worthy of being tested in clinical arena to improve the clinical management of lung cancer.
Supplemental Material
Supplementary_material for LncRNA DANCR Promotes Lung Cancer by Sequestering miR-216a by Qiang Zhen, Li-na Gao, Ren-feng Wang, Wei-wei Chu, Ya-xiao Zhang, Xiao-jian Zhao, Bao-lei Lv, and Jia-bao Liu in Cancer Control
Acknowledgments
The authors thank the Center Lab of Shijiazhuang No.1 Hospital and Research Center of Hebei General Hospital for the selfless helps and supports on their experimental technology and experimental.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Supplemental Material: Supplementary material for this article is available online
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. [DOI] [PubMed] [Google Scholar]
- 2. Tannenbaum SL, Koru-Sengul T, Zhao W, Miao F, Byrne MM. Survival disparities in non–small cell lung cancer by race, ethnicity, and socioeconomic status. Cancer J. 2014;20(4):237–245. [DOI] [PubMed] [Google Scholar]
- 3. Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382(9893):720–731. [DOI] [PubMed] [Google Scholar]
- 4. Moyer VA. Screening for lung cancer: US Preventive Services Task Force recommendation statement. Ann Intern Med. 2014;160(5):330–338. [DOI] [PubMed] [Google Scholar]
- 5. Stewart EL, Tan SZ, Liu G, Tsao MS. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations—a review. Transl Lung Cancer Res. 2015;4(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. [DOI] [PubMed] [Google Scholar]
- 7. Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kretz M, Webster DE, Flockhart RJ, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26(4):338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63(2):499–511. [DOI] [PubMed] [Google Scholar]
- 10. Jia J, Li F, Tang XS, et al. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget. 2016;7(25):37868–37881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jia J, Li F, Tang XS, et al. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget. 2016;7(25):37868–37881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu QC, Rui ZH, Guo ZL, Xie W, Shan S, Ren T. LncRNA-DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mTOR expression. J Cell Mol Med. 2017;22(3):1527–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hou BH, Jian ZX, Cui P, Li SJ, Tian RQ, Ou JR. miR-216a may inhibit pancreatic tumor growth by targeting JAK2. Febs Lett. 2015;589(17):2224–2232. [DOI] [PubMed] [Google Scholar]
- 14. Wang SL, Chen XD, Tang MY. MicroRNA-216a inhibits pancreatic cancer by directly targeting Janus kinase 2. Oncol Rep. 2014;32(6):2824–2830. [DOI] [PubMed] [Google Scholar]
- 15. Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLos One. 2014;9(10): e111842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Y, Zhang M, Liang L, Li J, Chen YX. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8(9):11480–11484. [PMC free article] [PubMed] [Google Scholar]
- 17. Rivera MP, Mehta AC, Wahidi MM. Establishing the diagnosis of lung cancer: diagnosis and management of lung cancer: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest. 2013;143(5):e142S–e165S. [DOI] [PubMed] [Google Scholar]
- 18. Kohls K, Schmidt D, Holdenrieder S, Müller S, Ellinger J. Detection of cell-free lncRNA in serum of cancer patients. Der Urologe Ausg A. 2015;54(6):819–825. [DOI] [PubMed] [Google Scholar]
- 19. Ma X, Wang X, Yang C, et al. DANCR acts as a diagnostic biomarker and promotes tumor growth and metastasis in hepatocellular carcinoma. Anticancer Res. 2016;36(12):6389–6398. [DOI] [PubMed] [Google Scholar]
- 20. Yuan SX, Wang J, Yang F, et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63(2):499–511. [DOI] [PubMed] [Google Scholar]
- 21. Zhang L, Sun X, Chen S, et al. Long noncoding RNA DANCR regulates miR-1305-Smad 4 axis to promote chondrogenic differentiation of human synovium-derived mesenchymal stem cells. Biosci Rep. 2017;37(4):BSR20170347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen PJ, Yeh SH, Liu WH, et al. Androgen pathway stimulates microRNA-216a transcription to suppress the tumor suppressor in lung cancer-1 gene in early hepatocarcinogenesis. Hepatology. 2012;56(2):632–643. [DOI] [PubMed] [Google Scholar]
- 23. Jiang N, Wang X, Xie X, et al. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. [DOI] [PubMed] [Google Scholar]
- 24. Hou BH, Jian ZX, Cui P, Li SJ, Tian RQ, Ou JR. miR-216a may inhibit pancreatic tumor growth by targeting JAK2. FEBS Lett. 2015;589(17):2224–2232. [DOI] [PubMed] [Google Scholar]
- 25. Zhang X, Shi H, Lin S, Ba M, Cui S. MicroRNA-216a enhances the radiosensitivity of pancreatic cancer cells by inhibiting beclin-1-mediated autophagy. Oncol Rep. 2015;34(3):1557–1564. [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y, Tang X, Shi M, Wen C, Shen B. Mir-216a decreases malat1 expression, induces g2/m arrest and apoptosis in pancreatic cancer cells. Biochem Biophys Res Commun. 2017;483(2):816–822. [DOI] [PubMed] [Google Scholar]
- 27. Chen Z, Zhang L, He Y, Shen Y, Li Y. Enhanced shRNA delivery and ABCG2 silencing by charge-reversible layered nanocarriers. Small. 2015;11(8):952–962. [DOI] [PubMed] [Google Scholar]
- 28. Che J, Tao A, Chen S, Li X, Zhao Y, Yuan W. Biologically responsive carrier-mediated anti-angiogenesis shRNA delivery for tumor treatment. Sci Rep. 2016;6:35661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singer O, Verma IM. Applications of lentiviral vectors for shRNA delivery and transgenesis. Curr Gene Ther. 2008;8(6):483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_material for LncRNA DANCR Promotes Lung Cancer by Sequestering miR-216a by Qiang Zhen, Li-na Gao, Ren-feng Wang, Wei-wei Chu, Ya-xiao Zhang, Xiao-jian Zhao, Bao-lei Lv, and Jia-bao Liu in Cancer Control