Abstract
Childhood asthma represents a worldwide problem, involving genetic, immune defense and environmental components. MicroRNAs (miRs) are non-coding, single-stranded RNAs involved in immune regulation. The aim was to evaluate clinical potential of plasma miR-21 and miR-146a involved in T helper differentiation in childhood asthma and non-asthmatic controls. Group 1 consisted of 27 asthmatic children receiving inhaled corticosteroids (ICSs), which was compared to group 2 with 21 healthy control children. All patients were assessed by pulmonary function tests. miR-21 and miR-146a expression levels were determined by real-time quantitative PCR, and IL-13 was measured using ELISA. Group 1 showed significant up-regulation of plasma miR-21 and miR-146a levels with mean values 42.6-fold and 4.7-fold higher than average expression, respectively, in group 2. miR-21 levels positively correlated with IL-13 levels and eosinophil percentage, while miR-146a only correlated to eosinophil percentage. There was a linear association between each of miR-21 and miR-146a expression and FEV1 (forced expiratory volume in the first second), miR-21 and miR-146a are up-regulated in asthmatic children. miR-21 served as a better asthma biomarker. Association between both markers and FEV1 points to their role in determining asthma outcome following ICS treatment. miR-21 and miR-146a play a role in eosinophilic endotypic classification of asthma.
Keywords: MicroRNA-21, microRNA-146a, T helper cell, eosinophils, IL-13
Introduction
Asthma is a chronic inflammatory condition of the airways, which is characterized by airway hyper responsiveness (AHR), bronchoconstriction, increased mucus secretion and limited airflow. This is usually associated with elevated serum IgE, eosinophilia with heightened expression of Th2 cytokines, IL-5, IL-9 and IL-13. IL-13 is an essential director of AHR, mucus hyper secretion and inflammation.1
Asthma affects >350 million people worldwide and its incidence continues to increase, thus it represents a serious challenge for any public health system.2 In Egypt, the prevalence of pediatric asthma is nearly 8.2%.3 Its impact is manifested in patients, families and communities as a whole in terms of lost school days, poor quality of life, frequent hospitalizations and deaths.4
Present asthma treatments mainly rely on long-term inhaled corticosteroids (ICSs). While ICSs relieve bronchial asthma, they cannot manage refractory asthma involving hormonal resistance and hormone dependence. Further, ICSs cause side effects such as growth and development inhibition, obesity and osteonecrosis. Therefore, it is urgent that we further understand the pathogenesis of asthma and identify new methods to treat airway inflammation.5
Most of the pathophysiological features of asthma are due to the infiltration of mononuclear cells, resulting in the imbalance of Th1/Th2 paradigm.6 The polarized Th responses could be regulated by multiple microRNAs (miRs) targeting different components of the T-cell polarization pathways.7,8
MiRs are a class of small non-coding RNAs (22 nucleotides in length) that regulate gene expression at the post-transcriptional level.9 miRs base-pair with target mRNAs to induce silencing complexes that degrade mRNAs or block translation, thus affecting protein synthesis. As shown by miR Base, approximately 1900 human miRs have already been identified.5 Over 100 mRNA genes are targeted by each miR, and over 30% of human protein-coding genes are regulated by miRs.10 Therefore, miRs with abnormal expression are closely associated with the occurrence and development of diseases, including asthma.5
It was reported that up-regulation of miR-21 appears to promote Th2 and attenuates Th1.7,8 miR-146a has been found to play a role in allergic diseases by modulating the ability of Treg cells, required to restrain IFN-γ/Th1-dependant pathogenic responses, in a signal transducer and activator transcription 1 (Stat1)-dependent manner.11 It was also reported that the mechanism of miR-146a function is cell-type dependent.12 MiRs were found to be extremely stable even under harsh conditions including heat, extreme pH, long-time storage, and repeated freezing and thawing. The extreme stability of circulating miRs displays a natural advantage as a biomarker.13
This study aimed to clarify the roles of miR-21 and miR-146a as biomarkers for asthma diagnosis in children, and whether they play a role in asthma outcome following ICS. In addition, it sought to explore miR-21 and miR-146a roles in asthma pathogenesis, which may aid endotypic classification related to Th2 augmentation detected by increased IL-13 production.
Patients and methods
Study participants
This case control study was conducted between September and October 2016. The study was carried on group 1 (n = 27) of asthmatic children (17 boys and 10 girls) compared to group 2 (n = 21) of non-asthmatic healthy children (controls). The study was approved by the ethical committee and is in accordance with the current version of the Helsinki Declaration.
Asthmatic children were recruited from the Pediatric Allergy and Pulmonology unit. Controls were selected randomly from healthy children during their routine checkup in the Children's hospital, Cairo University, Egypt. Sample size was calculated by Epi info version 7 according to asthmatic case flow during this period, with 95% confidence level and with % in children assumed to be 8.2%. A systematic random sample technique (one out of two) was adopted from children attending the hospital and the starting point for selection was drawn with a simple random method.
Inclusion criteria were as follows. 1. Patient children aged 5 yr and above, diagnosed with asthma according to the Global Initiative for Asthma (GINA, 2015). Patient criteria are summarized in Table 2. 2. Healthy control children gender- and age-matched to cases were included in the study.
Exclusion criteria were as follows. 1. Cases with systemic immunological disorders, diabetes, malignancy or anatomic abnormalities of the respiratory tract, airway foreign body and respiratory tuberculosis were excluded. Also excluded were asthma patients who received systemic corticosteroids, theophylline, long-acting β2-agonists and/or leukotriene receptor antagonists within at least 4 wk, which may affect levels of expression of miRs in plasma. Lastly, severely ill hospitalized asthma children were not enrolled in our study. 2. Regarding controls, non-asthmatic children with history of allergy, atopy or respiratory tract diseases were excluded.
Table 2.
Disease characteristics among group 1 (asthmatic group).
| Family history of asthma | |
| Yes | 19 (70.4%) |
| No | 8 (29.6%) |
| Age of onset of asthma | |
| < 4 yr | 19 (70.4%) |
| ≥ 4 yr | 8 (29.6%) |
| Frequency of attacks | |
| 3/mo | 11 (40.7%) |
| 1–2/mo | 13 (48.2%) |
| 6/yr | 3 (11.1%) |
| Night symptoms | |
| 2/mo | 16 (59.3%) |
| ≥1/wk | 11 (40.7%) |
| Precipitating factors | |
| No | 2 (7.4%) |
| Yes | 25 (92.6%) |
| Upper respiratory tract symptoms | |
| No | 15 (55.6%) |
| Yes | 12 (44.4%) |
| Chest | |
| Wheeze | 1 (3.7%) |
| Wheeze, cough | 7 (25.9%) |
| Wheeze, cough, dyspnea | 19 (70.4%) |
| Atopic manifestation | |
| No | 21 (77.8%) |
| Yes | 6 (22.2%) |
| FEV1 % of cases, mean ± SD | 83 ± 10 |
FEV1: forced expiratory volume in the first second.
Patient assessment
Structured sheet
Eligible children were subjected to the completion of a designed sheet for full history taking with emphasis on age, sex, family and medical history, and characteristics of asthma regarding cases.
Pulmonary function tests
Spirometry was done for asthmatic children under ICS, detecting forced expiratory volume in the first second (FEV1) (% predicted).
According to disease characteristics, patients were categorized as mild cases (n = 16) characterized with FEV1 > 80%, frequency of attacks ≤ twice/mo and night symptoms <2/mo, while moderate cases (n = 11) were characterized as FEV1 60–80%, frequency of attacks > twice/mo and night symptoms >1/wk (GINA, 2012).
Biochemical and hematological analysis
First, 4 ml blood samples were obtained by venipuncture from each subject and divided into two tubes with EDTA: The first tube for complete blood count and differential blood film analysis, and the second tube centrifuged at 2000 xg for 10 min at room temperature (RT, 23 ± 2℃), then plasma was carefully transferred and divided to two 200 μl aliquots one for isolation of RNA and second for IL-13 detection by ELISA and both were frozen at −20℃ till processing.
Complete blood count was performed using KX21N Hematology analyzer (Sysmex, Kobe, Japan), an automated hematology cell counter.
IL-13 detection in plasma was performed by ELISA using WKEA MED supplies CORP, China. ELISA kits (cat. no. E1243H, Lot 201508).
RNA extraction
Plasma miR-21 and miR-146a were detected by real time quantitative PCR (q-PCR) after reverse transcription using miScript RT kit according to the manufacturer's recommendations.
The RNAs were extracted from plasma samples using the miRNeasy Mini Kit (cat. no. 217004, QIAGEN) according to the manufacturer's instruction. Briefly, 1 ml QIAZOL (5 vol) was added to 200 μl of plasma and homogenized by vigorous mixing on a vortexer (15 s), and then incubated at RT for 5 min. Next, 200 μl chloroform was added and mixed on a vortexer for 10 s followed by incubation at RT for 2 min. Samples were then centrifuged at 12000 g at 4℃ for 15 min. The upper (aqueous) phase was transferred to a new RNase-free tube and 1.5 vol ethanol was added. Next, 700 μl of the samples were transferred to a RNeasy Mini Spin Column in a 2 ml collection tube, centrifuged at 8000 g for 15 s at RT and the flow-through was discarded. This step was repeated with the remaining sample and then the column was washed once with 700 μl RWT buffer and two times with 500 μl buffer RPE. Finally, RNA was eluted by adding 30 μl of DNase/RNase-free water to the column and centrifuged at 13000 g for 1 min at RT. Total RNA in 1 μl was evaluated for concentration and purity using a Nano Drop spectrophotometer (ND-1000, Thermo, United States), and 75 ng was used in a reverse transcription reaction.
Reverse transcription
Extracted RNA was reverse transcribed into complementary DNA using the miScript II RT Kit (cat. no. 218161) according to the manufacturer's instructions: miScript reverse transcriptase Mix 2 μl, 10 × miScript Nucleics Mix 2 μl and 5 × miScript Hispec buffer 4 μl. The total volume was 20 μl/reaction. For reverse transcription, template RNA of equal concentration (75 ng) was added to each tube containing the reverse transcription master mix and then the reaction volume was made up to 20 μl using RNase-free water. After gentle mixing and brief centrifugation, samples were incubated for 60 min at 37℃, then incubated for 5 min at 95℃ to inactivate the miScript reverse transcriptase mix.
miR expression analysis
Quantification of miR-21-5p and miR-146a-5p by q-PCR was performed with a PCR ViiA™ 7 System (USA, no 278880908) using human miscript SYBER green Master Mix and the primers for miR-21-5p (cat. no. MS00009079, Lot 20160404121), miR-146a-5p (cat. no. MS00003535, Lot 20160510009s) and SNORD 68 (a housekeeping miR used as the endogenous control) (cat. no. MS00033712, Lot 201601113022s, QIAGEN, Germany). Fluorescence measurements were made in every cycle. The cycling conditions used were as follows: PCR initial active step at 95℃ for 15 min, then 40 cycles (which includes denaturation at 94℃ for 15 s), annealing at 55℃ for 30 s and then extension at 70℃ for 30 s. Melting curve analysis was performed after the thermal profile to ensure specificity in the amplification. Temperature increased very slowly (from 65–95℃) with monitoring of fluorescence signal. Melting curve analysis resulted in the detection of a single sharp peak for each target.
Calculation of q-PCR results: The ΔCT was calculated by subtracting the CT values of SNORD 68 from the CT values of the target miRs. The resulting normalized ΔCT values were used to manually calculate ΔΔCT by subtracting the average normalized ΔCT of control from all different ΔCT values including the control values. Relative expression (fold-change of expression) of each miRs was calculated by the 2−ΔΔCT method.13 Fold-regulation represents fold-change results in a biologically meaningful way. Fold-change values >1 indicate a positive or an up-regulation, and the fold-regulation is equal to the fold-change in this case. Fold-change values < 1 indicate a negative or down-regulation, and the fold-regulation is the negative inverse of the fold-change in cases of fold-change negativity. As miRs in this study are up-regulated, the values of both fold-regulation and fold-change are identical.
Individual miR-21 and miR-146a expression levels for each patient are overlapped in one graph shown in Supplementary figure 1.
Statistical analysis
Data collected was reviewed, coded and statistically analyzed using Statistical Package for the Social Science (SPSS) program version 18 (Inc, Chicago, Illinois, USA). Data was described in terms of mean ± standard deviation ( ± SD) and percentage. The χ2-test was used for the comparison of qualitative data. The Student's t-test was used to compare between quantitative data. Linear regression analysis was done to studying associations. Correlation tests was performed to assess relationships between different biomarkers in asthmatic children. Significance level was taken at P ≤ 0.05 and the results were presented in tables and figures.
miR data was expressed as the means of relative expression (fold-regulation). The P-values were manually calculated based on a Student's t-test of the replicate 2−ΔΔCT values for each miR in studied groups.
Results
Our study included 27 asthmatic cases and 21 controls with mean ages 9 ± 2.2 yr and 8.1 ± 3 yr, respectively, and with no statistically significant difference regarding sex and age between them as they were matched (Table 1).
Table 1.
General characteristics and laboratory investigation of studied groups.
| Studied groups | Cases (n = 27) | Control (n = 21) | Test of significance and P-value |
|---|---|---|---|
| Age (yr, mean ± SD) | 9 ± 2.2 | 8.1 ± 3 | t-test = 3.3, P = 0.8 |
| Gender (male/female) n (%) | 17/10 (63/37) | 11/10 (52.4/46.8) | χ2 = 0.5, P = 0.5 |
| Residence n (%) | χ2 = 9, P = 0.007 | ||
| Urban | 17 (63.0) | 21 (100.0%) | |
| Suburban | 6 (22.2) | 0 | |
| Rural | 4 (14.8) | 0 | |
| Eosinophils (%, mean ± SD) | 4 ± 2.8 | 1.5 ± 0.3 | t-test = 3.7, P = 0.001 |
| IL-13 (pg/ml, mean ± SD) | 20.4 ± 6 | 2.3 ± 0.6 | t-test = 13, P < 0.05 |
| FEV1 % of asthma cases Mean ± SD | 83 ± 10 | - - - - - - - | - - - - - - - - - - - |
FEV1: forced expiratory volume in the first second.
The disease characteristics among asthma group are shown in Table 2.
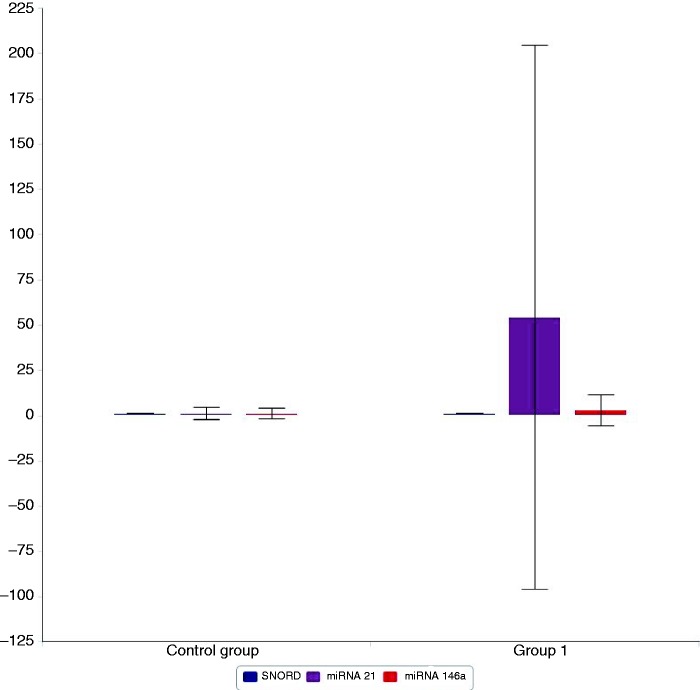
Our results showed that miR-21 and miR-146a were significantly up-regulated in group 1 with 42.6- and 4.7-fold higher than the average expression, respectively (P = 0.02 and 0.04, respectively), as shown in Figure 1.
Figure 1.
Plasma expression of SNORD 68 (in blue), miR-21 (in violet) and miR-146a (in red) in group 1 (asthmatic children) and the control group (group 2). miR-21 and miR-146a were significantly up-regulated in group 1 (42.6- and 4.74-fold higher than the average expression, respectively). P = 0.02 and P = 0.04, respectively.
Comparison of the expression levels of IL-13, miR-21 and miR-146 between mild persistent and moderate persistent cases showed no statistical difference in levels of expression of the three biomarkers among two groups (Table 3).
Table 3.
Expression levels of miR-21, miR-146a and IL-13 according to asthma severity relying on disease characteristics among cases.
| Mild persistent cases n = 16 (59.3%) | Moderate persistent cases n = 11 (40.7%) | Test of significance and P-value | |
|---|---|---|---|
| miR-21 relative expression up-regulationa | 53.55-fold | 55.31-fold | t-test = 0.7, P = 0.47 |
| miR-146a relative expression up-regulationa | 2.3-fold | 3.53-fold | t-test = 0.03, P = 0.9 |
| IL-13 (pg/ml), mean ± SD | 22 ± 7.1 | 18 ± 3.3 | t-test = 1.4, P = 0.2 |
| FEV1 %, mean ± SD | 89.2 ± 7.7 | 73.7 ± 4.5 | t-test = 6, P < 0.05 |
FEV1: forced expiratory volume in the first second.
Mean of relative expression (fold-changes) was calculated according to replicate of 2−ΔΔCT values for miR-21 and miR-146a in each of the mild and moderate groups.
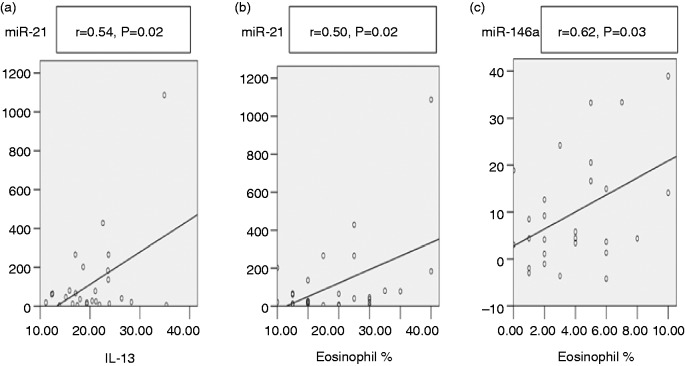
There was a positive correlation between miR-21 levels and both IL-13 levels (P = 0.02) and eosinophil percentage (P = 0.02), while miR-146a was only significantly correlated to eosinophil percentage (P = 0.03) as shown in Figure 2.
Figure 2.
(a) Significant positive correlation between miR-21 and IL-13. (b) Significant positive correlation between miR-21 and eosinophil percentage. (c) Significant positive correlation between miR-146a and eosinophil percentage.
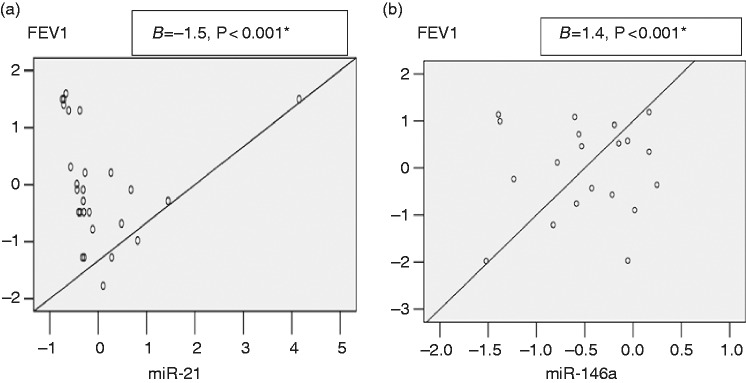
The current study revealed that in the asthma group under ICS treatment, FEV1 (% predicted) of mean ± SD = 83% ± 10. By linear regression, there was a negative association between miR-21 relative expression and FEV1 (B coefficients = 1.5, P < 0.001), while there was a positive association between miR-146a relative expression and FEV1 (B coefficients = 1.4, P < 0.001) (Figure 3).
Figure 3.
(a) Negative linear association between FEV1 (forced expiratory volume in the first second) with miR-21 relative expression. (b) Positive linear association between FEV1 with miR-146a relative expression. Equation for regression: predicted variable = slope × independent variable + intercept. (a) Predicted variable (FEV1) = −0.002 × miR-21 + 2.23. (b) Predicted variable (FEV1) = −0.036 × miR-146a + 2.35.
Discussion
Objective methods of pediatric asthma detection such as spirometry are unavailable for children < 5 yr as they do not show the cooperation needed to perform the test. The lack of detection methods points out the importance of developing asthma biomarkers in pediatrics, which are considered as non-objective methods.
miRs play a role in orchestrating the phenotypic programming of immune and airway epithelial cells to enhance the production of cytokines and mediators that result in the inflammation that characterizes asthma.14 miR-21 achieves its pro-inflammatory role by negatively regulating IL-12, which regulates the Th1/Th2 balance in asthma as decreased IL-12 expression induces an excessive Th2 response.15
Group 1 showed significant up-regulation of plasma miR-21 and miR-146a levels with mean values (Figure 1). This data for miR-21 agreed with that published by Lu et al., who documented miR-21 up-regulation in asthmatic airways and provided the first significant insight into the role of miR-21 in asthma by showing that miR-21 expression could be stimulated by IL-13 and that miR-21 targets IL-12p35 mRNA and lowers IL-12 protein expression.16 Wu et al. concluded that miR-21 expression was significantly up-regulated in bronchial epidermal cells of asthma patients regardless of treatment.5 In addition, several studies have reported that miR-21 is detectable and stable in the sera of asthmatic children, and that it has the potential to be biomarker in diagnosis and response to therapy in asthma.17–19
Regarding plasma miR-146a, the mentioned up-regulation in expression in group 1 indicates its pathogenic role in asthma. This data was in agreement with Jimenez-Morales et al., who revealed that a single nucleotide polymorphism leading to reduced mature miR-146a expression was associated with reduced asthma risk, implicating miR-146a activity in the pathogenesis of allergic diseases.20 Also, Feng et al. suggested that both miR-146a and miR-146b were found to be expressed in spleen CD41 T lymphocytes and appeared to play pro-inflammatory roles in asthma.21 On the other hand, Luo et al. suggested an anti-inflammatory role for miR-146a and that its levels were significantly decreased in children with allergic rhinitis (the most predisposing factor for asthma) compared to controls, but that miR-146a was significantly up-regulated after treatment with immunotherapy.22 This discrepancy could be explained by differences in sample types as they examined miR-146a levels in PBMCs, while we examined its levels in plasma. It was also previously reported that the mechanism of miR-146a function is cell type-dependent, and that miR expression patterns seen in serum are not identical to those seen from miR taken directly from cells.9
This study detected a significant positive correlation between both miR-21 and miR-146a with eosinophil percentage (r = 0.5, P = 0.02) and (r = 0.62, P = 0.03), respectively (Figure 2). This was in agreement with Lu et al., who stated that miR-21 was up-regulated during the differentiation of eosinophils and contributed to eosinophil production, survival and proliferation.11 Also, Rijavec et al. reported that miR-21 expression was associated with eosinophilic Th2-high asthma.14 Moreover, Dougherty et al. found that significant bronchoalveolar lavage eosinophilia was restricted to subjects in the Th2-high group.23 Current data supports previous studies stating that both miR-21 and miR-146a might work additively or synergistically to initiate and/or maintain an exaggerated Th2 response.7,8
This data appears to be of important clinical relevance to the endotypic classification of asthma as it was found that steroid-insensitive asthma was associated with non-eosinophilic endotypes.24 The reason why severe asthma patients were linked to non-eosinophilic types is due to the presence of activated NK cells in severe cases, which were able to promote eosinophil apoptosis.2 This may mark miR-21 and miR-146a as a potential biomarker for eosinophilic Th2 endotypes of asthma.
IL-13, a cytokine secreted by Th2 lymphocytes, critically modulates allergic inflammation and tissue remodeling in allergic asthma.25 The present study revealed a statistically significant difference in IL-13 levels between the asthma and control groups. This data was in agreement with another study that stated that IL-13 has effector functions, and is essential for the induction of mucus production and AHR characterizing asthma.6 Our data were also in line with another study stating that the milder form of asthma is related to Th2 endotype as all patients enrolled in study were mild and moderate cases.25
The current data revealed a positive correlation between miR-21 overexpression and plasma IL-13 levels (Figure 2). This data was in agreement with Wu et al., who stated that miR-21 overexpression in the bronchial epithelia of asthma patients positively correlates with IL-13 due to augmentation of Th2.5 Also, miR-21 overexpression was previously linked to the differentiation of naïve CD4 + T cells into Th2 cells, which were proven to produce IL-13, described as central events in allergy.17
No correlation was found between miR-146a and IL-13 levels. This disagreed with a previous study that reported that increased miR-146a expression contributed to IL-13 production due to Th2 augmentation.26 This discrepancy could be explained by the action of miR-146a being directed toward Th1 mainly as up-regulation of miR-146a, potentially enhancing the Treg cell-mediated suppression of Th1 responses and resulting in unopposed Th2 activation.7 miR-146a has no direct effect on Th2 responses.8
Phenotypically, asthma can be classified as mild, moderate or severe by measures of lung function, particularly FEV1. FEV1 is essential for categorizing asthma severity as it reflects the grade of airway obstruction.27 The current study revealed that in the asthma group under ICS treatment, FEV1 (% predicted) was mean ± SD = 83% ± 10, while a normal value is considered to be > 79% of the predicted value,22 indicating that collectively those patients were categorized as mild asthma patients due to normal FEV1. Genome-wide association studies have revealed that genes involved in airway structure/remodeling are associated with lung function and that those genes might affect asthma severity.27
The current study revealed a negative linear association between miR-21 relative expression and FEV1 post-ICS treatment (Figure 3), which points out their role in ICS treatment outcome as FEV1 reflects the grade of airway obstruction after ICS treatment. This was in agreement with the study by Elbehidy et al., which reported that miR-21 expression level was negatively correlated with FEV1 and added that expression levels of serum miR-21 were significantly higher in asthmatic steroid-resistant compared to steroid-sensitive patients, and that serum miR-21 had a high predictive value in differentiating those patients.19 Another study by Kim et al. identified a role for the miR-21/phosphatidylinositol-3 kinase (PI3K) axis in severe steroid-insensitive allergic airway disease, which highlights miR-21 as a novel therapeutic target for the treatment of steroid-insensitive asthma.24
Our study revealed a positive linear association between miR-146a relative expression and FEV1 (Figure 3). This was in agreement with Tsitsiou et al., who reported that miR-146a is reduced in T cells in patients with severe asthma with decreased FEV1,28 and is in line with another study that demonstrated that many loci regulated by miR-146a and related to the Th1 pathway are associated with FEV1.27 On the other hand, Luo et al. reported that miR-146a level is negatively associated with disease severity. This discrepancy could be explained by differences in sample types as they examined miR-146a levels in PBMCs while we examined its levels in plasma, as mentioned before, and could also be due to differences in the groups studied as in Luo et al. the study cases were children with allergic rhinitis.22
Perry et al. proposed that both miR-21 and miR-146a may be epithelial biomarkers of asthma but their relationship to asthma severity was attributed to their possible role regarding Th2.29 Severe asthma patients were reported to have low IL-13 levels and low Th2.30
Conclusion
We conclude that miR-21 and miR-146a are both up-regulated in the plasma of asthmatic children, but that miR-21 has better potential to serve as asthma biomarker. Also, the association of miR-21 and miR-146a with FEV1 points out their role in ICS treatment outcome as FEV1 reflects the grade of airway obstruction after treatment. miR-21 and miR-146a both play a role in the eosinophilic endotypic classification of asthma. Also, the role of miR-21 in Th2 differentiation may serve to direct the application of therapies like Th2 cytokine blockers in patients comprising the Th2 asthma endotype.
Expanding miR-21 studies on a larger scale across the normal population, to study its usefulness in screening for asthma, particularly asthmatic children < 5 yr, to confirm its role as a biomarker for diagnosis, is recommended. Also, studying miR-21 and miR-146a expression in severe asthmatic children with precautions is recommended to avoid factors affecting their plasma levels, which will add to the assessment of their role in asthma severity, monitoring of response to treatment and the personalization of patient therapy.
Supplemental Material
Supplementary figure
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1.Marashian SM, Mortaz E, Jamaati HR, et al. Role of innate lymphoid Cells in lung disease. Iran J Allergy Asthma 2015; 14: 346–360. [PubMed] [Google Scholar]
- 2.To T, Stanojevic S, Moores G, et al. Global asthma prevalence in adults: findings from the cross-sectional world health survey. BMC Public Health 2012; 12: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Mashad GM, Mahmoud AA, Abdel Hafez AA. The prevalence of bronchial asthma among primary school children in Menoufiya Governorate (El-Bagour Center). Menoufiya Med J 2016; 29: 89–94. [Google Scholar]
- 4.Al-Moamary MS, Alhaider SA, Idrees MM, et al. The Saudi Initiative for Asthma - 2016 update: guidelines for the diagnosis and management of asthma in adults and children. Ann Thorac Med 2016; 11: 3–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu XB, Wang MY, Zhu HY, et al. Overexpression of microRNA-21 and microRNA-126 in the patients of bronchial asthma. Int J Clin Exp Med 2014; 7: 1307–1312. [PMC free article] [PubMed] [Google Scholar]
- 6.Gosh B. Involvement of microRNA in asthma: new perspective in respiratory biology. Indian J Allerg Asthma Immunol 2013; 27: 3–8. [Google Scholar]
- 7.Lu TX, Rothenberg ME. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J Allergy Clin Immunol 2013; 132: 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebane A. MicroRNA and allergy. Adv Exp Med Biol 2015; 888: 331–352. [DOI] [PubMed] [Google Scholar]
- 9.Moret-Tatay I, Iborra M, Cerrillo E, et al. Possible biomarkers in blood for Crohn's disease: oxidative stress and microRNAs-current evidences and further aspects to unravel. Oxid Med Cell Longev 2016; 2016: 2325162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol 2013; 425: 3582–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu TX, Lim EJ, Itskovich S, et al. Targeted ablation of mir-21 decreases murine eosinophil progenitor cell growth. PLoS One 2013; 8: e59397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larner-Svensson HM, Williams AE, Tsitsiou E, et al. Pharmacological studies of the mechanism and function of interleukin-1beta-induced miRNA-146a expression in primary human airway smooth muscle. Respir Res 2010; 11: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS One 2008; 3: e3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rijavec M, Korošec P, Žavbi M, et al. Let-7a is differentially expressed in bronchial biopsies of patients with severe asthma. Sci Rep 2014; 4: 6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greene CM, Gaughan KP. MicroRNAs in asthma: potential therapeutic targets. Curr Opin Pulm Med 2013; 19: 66–72. [DOI] [PubMed] [Google Scholar]
- 16.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol 2009; 182: 4994–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawant DV, Yao W, Wright Z, et al. Serum microRNA-21 as a biomarker for allergic inflammatory disease in children. Microrna 2015; 4: 36–40. [DOI] [PubMed] [Google Scholar]
- 18.Jiang C, Yu H, Sun QW, et al. Extracellular microRNA-21 and microRNA-26a increase in body fluids from rats with antigen induced pulmonary inflammation and children with recurrent wheezing. BMC Pulm Med 2016; 16: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elbehidy RM, Youssef DM, El-Shal AS, et al. MicroRNA-21 as a novel biomarker in diagnosis and response to therapy in asthmatic children. Mol Immunol 2016; 71: 107–114. [DOI] [PubMed] [Google Scholar]
- 20.Jimenez-Morales S, Gamboa-Becerra R, Baca V, et al. MiR-146a polymorphism is associated with asthma but not with systemic lupus erythematosus and juvenile rheumatoid arthritis in Mexican patients. Tissue Antigens 2012; 80: 317–321. [DOI] [PubMed] [Google Scholar]
- 21.Feng MJ, Shi F, Qiu C, et al. MicroRNA-181a, -146a and -146b in spleen CD41 T lymphocytes play proinflammatory roles in a murine model of asthma. Int Immunopharmacol 2012; 13: 347–353. [DOI] [PubMed] [Google Scholar]
- 22.Luo X, Hong H, Tang J, et al. Increased expression of miR-146a in children with allergic rhinitis after allergen-specific immunotherapy. Allergy Asthma Immunol Res 2016; 2: 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dougherty RH, Sidhu SS, Raman K, et al. Accumulation of intraepithelial mast cells with a unique protease phenotype in TH2-high asthma. J Allergy Clin Immunol 2010; 125: 1046–1053.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim RY, Horvat JC, Pinkerton JW, et al. MicroRNA-21 drives severe, steroid-insensitive experimental asthma by amplifying phosphoinositide 3-kinase-mediated suppression of histone deacetylase 2. J Allergy Clin Immunol 2017; 139: 519–532. [DOI] [PubMed] [Google Scholar]
- 25.Kumar M, Ahmad T, Sharma A, et al. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy Clin Immunol 2011; 128: 1077–1085.e1–10. [DOI] [PubMed] [Google Scholar]
- 26.Koch MA, Tucker-Heard G, Perdue NR, et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol 2009; 10: 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Hawkins GA, Ampleford EJ, et al. Genome-wide association study identifies TH1 pathway genes associated with lung function in asthmatic patients. J Allergy Clin Immunol 2013; 132: 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsitsiou E, Williams AE, Moschos SA, et al. Transcriptome analysis shows activation of circulating CD8 + T cells in patients with severe asthma. J Allergy Clin Immunol 2012; 129: 95–103. [DOI] [PubMed] [Google Scholar]
- 29.Perry MM, Adcock IM, Chung KF. Role of microRNAs in allergic asthma: present and future. Curr Opin Allergy Clin Immunol 2015; 15: 156–162. [DOI] [PubMed] [Google Scholar]
- 30.Woodruff PG, Modrek B, Choy DF, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med 2009; 180: 388–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure