Summary
Strigolactones (SLs) are carotenoid‐derived plant hormones that also act in the rhizosphere to stimulate germination of root‐parasitic plants and enhance plant symbiosis with beneficial microbes. Here, the role of SLs was investigated in the interaction of rice (Oryza sativa) roots with the root‐knot nematode Meloidogyne graminicola.
Genetic approaches and chemical sprays were used to manipulate SL signaling in rice before infection with M. graminicola. Then, nematode performance was evaluated and plant defense hormones were quantified.
Meloidogyne graminicola infection induced SL biosynthesis and signaling and suppressed jasmonic acid (JA)‐based defense in rice roots, suggesting a potential role of SLs during nematode infection. Whereas the application of a low dose of the SL analogue GR24 increased nematode infection and decreased jasmonate accumulation, the SL biosynthesis and signaling d mutants were less susceptible to M. graminicola, and constitutively accumulated JA and JA‐isoleucine compared with wild‐type plants. Spraying with 0.1 μM GR24 restored nematode susceptibility in SL‐biosynthesis mutants but not in the signaling mutant. Furthermore, foliar application of the SL biosynthesis inhibitor TIS108 impeded nematode infection and increased jasmonate levels in rice roots.
In conclusion, SL signaling in rice suppresses jasmonate accumulation and promotes root‐knot nematode infection.
Keywords: jasmonic acid, Oryza sativa, phytohormone signaling, plant–nematode interaction, strigolactone
Introduction
Strigolactones (SLs) are carotenoid‐derived plant hormones originally identified as signaling molecules in the rhizosphere (Cook et al., 1966; Akiyama et al., 2005; Umehara et al., 2008). The first known SL, strigol, was isolated from cotton root exudates, and detected as a stimulant of seed germination of the root‐parasitic weed Striga lutea (Cook et al., 1966). To date, > 20 naturally occurring SLs have been identified in different plant species, which have been shown to stimulate seed germination of various root‐parasitic plants (Al‐Babili & Bouwmeester, 2015). The precursor of SL biosynthesis is carlactone, which is derived from trans‐β‐carotene via the action of the isomerase DWARF27 (D27) and then a sequential oxidative cleavage by the carotenoid cleavage dioxygenase enzymes (CCD7 and CCD8) in plastids (Alder et al., 2012). Carlactone is then transported into the cytoplasm where a MAX1‐type monooxygenase transforms it into carlactonic acid that is eventually converted into SLs and SL‐like compounds (Seto et al., 2014). In rice (Oryza sativa), the genes OsD17 and OsD10 encode the enzymes CCD7 and CCD8, respectively. The genes encoding the α/β‐fold hydrolase D14 and the F‐box protein D3 are known as SL‐signaling genes in rice (Cheng et al., 2013). The SL rice mutants are commonly known as dwarf (d) or high‐tillering dwarf (htd) mutants (Gomez‐Roldan et al., 2008; Umehara et al., 2008).
SLs play a pivotal role in various aspects of plant growth and development as well as in plant symbiosis with beneficial microbes (Akiyama et al., 2005; Gomez‐Roldan et al., 2008; Kapulnik et al., 2011). As endogenous signaling molecules, SLs have been shown to modulate plant architecture. For example, SL biosynthesis or signaling mutants of different plant species showed increased numbers of lateral shoot branches (Gomez‐Roldan et al., 2008; Umehara et al., 2008). SLs also promote primary root growth and inhibit the formation of lateral and adventitious roots in Arabidopsis (Kapulnik et al., 2011; Ruyter‐Spira et al., 2011). In rice, SL‐biosynthesis and ‐signaling mutants exhibit shorter crown roots (Arite et al., 2012). Furthermore, SLs negatively regulate mesocotyl elongation in darkness (Hu et al., 2013).
As rhizosphere signaling molecules, SLs promote plant association with mycorrhizas by stimulating hyphal branching (Akiyama et al., 2005) as well as symbiosis with rhizobia in legumes in a complex concentration‐dependent manner (Foo & Davies, 2011; De Cuyper et al., 2014). Recent studies have shown that SLs also play a role in plant responses to various abiotic stresses. For instance, Arabidopsis mutants impaired in SL biosynthesis or signaling are hypersensitive to drought and salinity stress (Bu et al., 2014; Van Ha et al., 2014). Furthermore, SLs are also involved in plant defense responses to biotic stresses. For instance, the SL‐biosynthesis and sensing max mutants of Arabidopsis develop a stronger leafy gall syndrome than wild‐type (WT) controls do as a result of infection by the biotrophic actinomycete Rhodococcus fascians (Stes et al., 2015). Similarly, the SL‐deficient tomato (Solanum lycopersicum) Slccd8 mutant showed an increased susceptibility to the necrotrophic foliar fungal pathogens Botrytis cinerea and Alternaria alternata (Torres‐Vera et al., 2014). This study also demonstrated that Slccd8 mutants contained significantly lower amounts of salicylic acid (SA), jasmonic acid (JA), and abscisic acid (ABA) compared with the WT plants. In pea (Pisum sativum), the SL pathway did not show any effect on colonization by the soil‐borne oomycete Pythium irregulare (Blake et al., 2016).
The root‐knot nematode Meloidogyne graminicola is one of the most damaging plant parasitic nematodes infecting rice roots, especially in aerobic conditions (Bridge et al., 2005; De Waele & Elsen, 2007). It causes yield losses up to 32% (Bridge et al., 2005; De Waele & Elsen, 2007). Meloidogyne graminicola is an obligate biotrophic and sedentary root endoparasite. However, the infective second‐stage juveniles (J2s) that are vermiform in shape are migratory. Upon finding a suitable host plant, the J2s invade roots at the elongation zone and migrate through the root intercellular space until they reach the vascular cylinder to establish a feeding site, containing giant cells (Gheysen & Mitchum, 2011). The nematodes withdraw nutrients from these giant cells throughout their life cycle, which is completed within 2–3 wk. The hyperplasia and hypertrophy of the surrounding cells result in the formation of a root‐knot (gall) on the infected root (Bridge et al., 2005; Kyndt et al., 2014).
Meloidogyne spp. simultaneously influence plant cell differentiation and modulate the defense responses to establish a successful infection via feeding cell formation (Gheysen & Mitchum, 2011; Ji et al., 2013). The plant hormones JA, ethylene, SA, ABA, brassinosteroids (BR), gibberellic acid (GA, and auxin are known to be involved in the interactions between plant and root‐knot nematodes (Karczmarek et al., 2004; Nahar et al., 2011, 2013; Kyndt et al., 2017; Yimer et al., 2018; Gheysen & Mitchum, 2019). For instance, the JA‐based defense pathway plays a vital role in rice defense against M. graminicola infection, whereas SA is also involved in defense but to a lesser extent than JA (Nahar et al., 2011). In tomato, foliar application of methyl jasmonate (MeJA) and JA also induces defense against M. incognita and M. javanica infection in roots (Cooper et al., 2005). In addition, the JA‐precursor cis‐(+)‐12‐oxo‐phytodienoic acid (cis‐OPDA) acts as a signaling molecule in regulating Arabidopsis defense against M. hapla (Gleason et al., 2016). On the other hand, ABA, BR, and GA promote rice susceptibility to M. graminicola infection by interacting antagonistically with the JA pathway (Nahar et al., 2013; Kyndt et al., 2017; Yimer et al., 2018).
Here, we choose to study the interaction between rice as a monocot model plant and its economically important root pathogen M. graminicola to learn more about the function of SL in plant defense. First, we analyzed the expression of some genes related to the SL pathway and quantified defense‐related hormones in rice roots infected with M. graminicola. Upregulation of SL biosynthesis and signaling genes suggested a potential role of SL in rice–M. graminicola interaction. Then, we manipulated SL levels by treating plants with an SL analogue or an SL biosynthetic inhibitor before nematode inoculation, and analyzed the responses of rice d mutants that are deficient in SL biosynthesis and signaling. SL signaling was found to enhance M. graminicola infection and to decrease the levels of jasmonates.
Materials and Methods
Plant materials and growth conditions
Two SL‐biosynthetic rice dwarf (d) mutants (d10 and d27), one SL‐signaling mutant d14 and their corresponding WT (O. sativa cv Shiokari) were used in this study. The seeds of these rice genotypes were provided by Professor Mikio Nakazono, Nagoya University, and Dr Itsuro Takamure, Hokkaido University, Japan. The seeds were first germinated in Petri dishes containing moist tissue paper at 30°C for 4 d in the dark. Each germinated seedling was planted into a specially made polyvinylchloride tube containing sand and absorbent polymer (Reversat et al., 1999). Then, the seedlings were grown in a rice culture room under controlled environmental conditions (26°C : 24°C, day : night temperature, 70% relative humidity, 12 h : 12 h, light : dark cycle). Each plant was fertilized with 10 ml of Hoagland's solution two times per week.
Nematode culture maintenance
The root‐knot nematode M. graminicola was provided by Professor Dirk De Waele (Catholic University Leuven, Belgium) and was originally isolated from rice in the Philippines. The nematode culture was multiplied and maintained using the susceptible rice cv Nipponbare or a grass‐host Echinocloa crusgalli in 2 l plastic pots having soil medium in similar growth conditions as mentioned earlier.
Inoculation of rice seedlings with M. graminicola and susceptibility assessment
The J2s of M. graminicola were extracted from 3‐month‐old infected roots using the modified Baermann method (Luc et al., 2005). Then 2‐wk‐old rice seedlings were each inoculated with c. 200 J2s. The infected root samples were collected at 14 d post‐inoculation (dpi). To visualize the galls and the developmental stages of nematodes inside the galls, the nematode‐infected roots were boiled for 3 min in 0.8% acetic acid and 0.013% acid fuchsin solution. Then the roots were washed with running tap water and incubated in acidified glycerol. This treatment turns the galls and nematodes a purple color, while the rest of the root system is slowly destained in the acidified glycerol. First, the numbers of galls per plant were counted under a stereomicroscope (Leica S8 APO; Leica Microsystems, Wetzlar, Germany). Then each gall was dissected using a needle under the microscope. The developmental stages of all nematodes per plant were counted. The swollen female nematodes containing eggs were counted as egg‐laying females (ELFs), and this criterion was used as a measure of nematode development. The nematode infection experiments were performed at least twice, including at least six individual plants per treatment in each experiment.
RNA isolation and complementary DNA synthesis
The WT rice plants (‘Shiokari’) were grown and infected by the root‐knot nematode M. graminicola as described previously. The whole root systems from both the infected and uninfected control plants were collected 1 d after inoculation. There were four biological replicates per treatment, and each biological replicate consisted of a pool of roots of four to six plants. The samples were immediately frozen in liquid nitrogen (N2) and ground to fine powder using a mortar and pestle. Total RNA was isolated from c. 100 mg finely ground root tissues using the Invitrap Spin Plant RNA Mini Kit (Stratec Biomedical, Birkenfeld, Germany) following the instructions of the manufacturer, except that an additional DNase treatment was included using a Qiagen RNase‐Free DNase Set as described by Ullah et al. (2017). The quantity and quality of the RNA was checked by spectrophotometry (NanoDrop 2000; Thermo Fisher Scientific, Wilmington, DE USA). Reverse transcription of c. 500 ng total RNA into complementary DNA (cDNA) was achieved by using SuperScript III reverse transcriptase (Invitrogen) and 50 pmol Oligo (dT)12‐18 Primer (Invitrogen) in a reaction volume of 20 μl. The cDNA was diluted five times with sterile water.
Quantitative real‐time PCR
The quantitative PCR reactions were performed in a 20 μl volume containing 10 μl Brilliant III Ultra‐Fast SYBR Green QPCR Master Mix (Agilent Technologies, Santa Clara, CA, USA), 10 pmol forward and 10 pmol reverse primers, and 2 μl diluted cDNA (c. 50 ng). The quantitative real‐time (qRT)‐PCR was performed using a CFX Connect Real‐Time PCR Detection System (Bio‐Rad) using a two‐step amplification protocol (cycling parameters: 95°C for 3 min followed by 39 cycles of 95°C for 10 s and 58°C for 30 s). A melt‐curve analysis was performed for each sample run using the Bio‐Rad default parameters (95°C for 10 s, 65–95°C in 0.5°C increments) that yielded one peak. A nontemplate water sample was used as a negative control. Transcript abundance was calculated from five biological replicates for each treatment, with each biological sample consisting of three technical replicates, and was normalized to the abundance of the housekeeping gene OsExp. Relative expression was based on the method (Livak & Schmittgen, 2001). The gene‐specific primers are provided in Supporting Information Table S1.
Extraction of hormones from rice roots
Frozen rice roots were ground to fine powder in liquid N2. Tissues (c. 75 mg) were extracted with 1 ml analytical‐grade methanol containing 2 μl phytohormone standard mix (20 ng of D4‐SA, 20 ng of D6‐JA, and 4 ng D6‐JA–isoleucine (Ile) internal standards). The contents were vortexed vigorously for a few seconds, incubated at 20°C for 25 min at 1000 rpm, and centrifuged at 13 000 g at 4°C for 5 min. Supernatant (c. 950 μl) was transferred to a new microcentrifuge tube. The extracts were directly analyzed using LC–MS/MS for phytohormone quantification.
Quantification of phytohormones by LC–MS/MS
For phytohormone analysis, chromatography was performed on an Agilent 1200 HPLC system (Agilent Technologies). An API 5000 tandem mass spectrometer (AB Sciex, Darmstadt, Germany) equipped with a turbo spray ion source was operated in negative ionization mode. Hormones were separated on a Zorbax Eclipse XDB‐C18 column (1.8 μm, 50 mm × 4.6 mm; Agilent) at 25°C, with two mobile phases consisting of 0.05% formic acid in water and acetonitrile. The flow rate and the elution profile were similar to the method described by Ullah et al. (2019). The parent ion and corresponding fragments of SA and the jasmonates were analyzed by multiple reaction monitoring as described by Vadassery et al. (2012). Mass data were collected and processed using analyst 1.6 software (Applied Biosystems). Linearity in ionization efficiencies was confirmed by analyzing serial dilutions of a standard mixture. The concentrations of cis‐OPDA, JA, JA–Ile, and SA were determined relative to the corresponding internal standard.
Spraying with chemicals followed by nematode inoculation
The SL biosynthesis inhibitor TIS108 and the SL analogue GR24 were purchased from Chiralix (Nijmegen, the Netherlands). The JA biosynthesis inhibitor 5,8,11,14‐eicosatetraynoic acid (ETYA) was purchased from Sigma‐Aldrich. The TIS108 and ETYA were dissolved in ethanol, and GR24 was dissolved in acetone to make a 10 mM stock solution. The final spray concentrations were 3 μM for TIS108, 0.1 and 5 μM for GR24, and 100 μM for ETYA in distilled water containing 0.02% (v/v) Tween 20. Two‐week old rice seedlings were thoroughly sprayed until run‐off with these chemicals. For the mock‐treated plants, 0.02% (v/v) Tween 20 in distilled water containing 0.001% ethanol or acetone for TIS108 and ETYA or GR24 treatment was applied, respectively. Root samples were collected 24 h after spraying to analyze phytohormones. For each treatment, five biological replicates were used, each consisting of a pool with four to six plants. At the same time, 24 h after spraying, another eight plants per treatment were inoculated with M. graminicola to analyze the infection levels. Infected root samples were collected at 14 dpi and stained with acid fuchsin as described in the previous section.
Statistical analysis
All data were analyzed by using the statistical package R v.3.2.0 (R Foundation for Statistical Computing, Vienna, Austria). Normality of data and homogeneity of variances were checked using the Shapiro–Wilk and Levene test, respectively. If test assumptions were not met, data were square root or log transformed. Data were then analyzed by one‐way ANOVA followed by Tukey's post‐hoc test to compare differences among the treatment groups. Comparisons between two means of different treatments were conducted using a two‐tailed Student's t‐test. Percentage data showing nematode development were analyzed using a chi‐squared test.
Results
Meloidogyne graminicola infection in rice activates the SL pathway genes, but inhibits jasmonate accumulation
To investigate the potential role of SL on root‐knot nematode infection in rice, the expression of SL biosynthesis and signaling genes were analyzed in rice roots 1 dpi with M. graminicola and compared with uninfected control roots by qRT‐PCR. Transcript levels of the first biosynthetic gene Osd27 increased in nematode‐infected roots but were not significantly different from those in control roots (Fig. 1a). Meloidogyne graminicola infection in roots significantly induced the transcript levels of Osd17 and Osd10 compared with control roots (Fig. 1a). Furthermore, transcripts of the SL signaling gene Osd14 (Fig. 1b) significantly increased in rice roots infected with M. graminicola compared with control roots (Fig. 1a). To determine whether defense hormones such as jasmonates and SA were changed during the early stage of the rice–M. graminicola interaction, their contents were quantified in infected and uninfected control roots. The levels of the JA precursor, cis‐OPDA, were significantly lower in roots infected by M. graminicola than in control roots (Fig. 1c). The level of JA decreased slightly upon nematode infection; however, the change was not statistically different in comparison with control roots. Interestingly, the level of the bioactive JA–Ile conjugate was three‐fold lower in roots infected by M. graminicola than in control roots (Fig. 1c). The contents of JA and the JA–Ile catabolites, sulfated‐JA (sulfo‐JA) and hydroxyl/carboxy‐JA–Ile (OH/COOH‐JA–Ile), respectively, increased in rice roots infected with M. graminicola compared with control roots (Fig. S1). The level of SA did not change due to nematode infection (Fig. S1). The content of ABA increased slightly in nematode‐infected roots, but was not significantly different from that of control roots (Fig. S1). These results suggest that M. graminicola infection in rice roots induces the SL pathway and suppresses the JA pathway.
Figure 1.
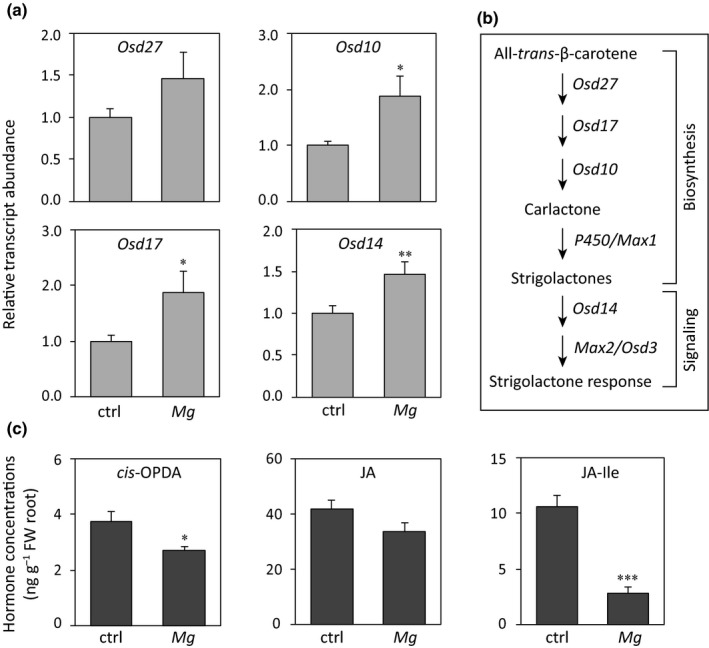
Meloidogyne graminicola infection in rice roots influences the strigolactone (SL) and jasmonic acid (JA) pathways in opposite ways. (a) Relative expression of SL biosynthesis and signaling genes with and without M. graminicola infection 1 d post‐inoculation (dpi). The messenger RNA levels were measured by quantitative real‐time PCR in three technical replicates per sample. Transcripts of each gene were normalized to OsExp transcripts. Data are presented as the mean + SE (n = 4), and each biological replicate was a pool of four to six plants. (b) Biosynthesis and signaling genes of SL pathway in rice. (c) Concentrations of jasmonates in rice roots with and without M. graminicola infection 1 dpi. Hormones were analyzed by LC–MS/MS. Data are presented as the mean + SE (n = 5), and each replicate was a pool of four to six plants. Data were analyzed using a two‐tailed Student's t‐test: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. ctrl, uninfected control; Mg, M. graminicola infected; cis‐OPDA, cis‐(+)‐12‐oxo‐phytodienoic acid; JA–Ile, JA–isoleucine.
Foliar spraying with the SL analogue GR24 increases M. graminicola infection and reduces jasmonate accumulation, but these effects are reversed at high dose of GR24
To investigate the potential role of SL during root‐knot nematode infection in rice, rice shoots were treated with GR24, a functional SL analogue (Besserer et al., 2008). The shoots of WT rice plants were sprayed with two different concentrations of GR24. A subset of the treated plants was harvested to collect root samples 24 h after treatment for hormone analysis. Another subset of plants was inoculated with nematodes at 24 h after treatment. The plant responses to nematode infection were evaluated 14 dpi by counting the number of galls, nematodes, and their different development stages.
Foliar application of GR24 did not affect the number of galls compared with mock‐treated plants (Fig. 2a). However, plants treated with 0.1 μM GR24 contained a higher total number of nematodes as well as ELFs per plant compared with the mock‐treated plants (Fig. 2b,c). By contrast, spraying with 5 μM GR24 decreased the total number of nematodes and ELFs per plant (Fig. 2b,c). The development of nematodes into ELFs increased by 7% in plants treated with 0.1 μM GR24, whereas there was a decrease by 10% in 5 μM GR24‐treated plants in comparison with mock treatment (Fig. 2d,e). Treatments of GR24 followed by nematode infection did not affect shoot and root lengths at 14 dpi compared with mock‐treated plants (Fig. S2).
Figure 2.
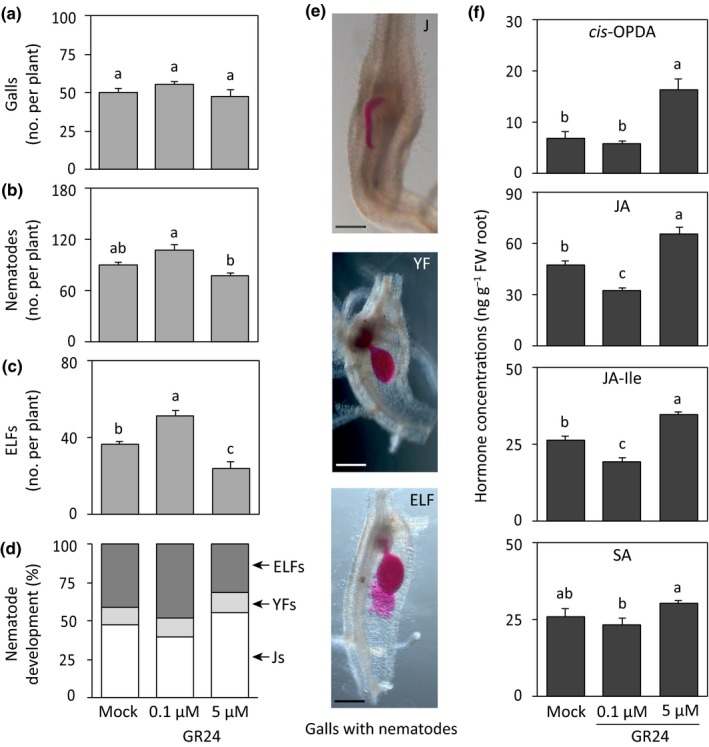
Effect of foliar spraying of the strigolactone analogue GR24 on Meloidogyne graminicola infection and the accumulation of phytohormones in rice roots. Fourteen‐day‐old wild‐type plants were sprayed with GR24 (0.1 or 5 μM) or with solvent only (mock). After 24 h, a subset of plants from each treatment were inoculated with c. 200 second‐stage juveniles of M. graminicola per plant and another subset of plants was sampled for hormone analysis. Plant responses were analyzed at 14 d post‐inoculation. Total numbers of (a) galls, (b) nematodes, (c) egg‐laying females (ELFs), and (d) percentages of the developmental stages of the observed nematodes within the galls per plant. (e) Representative pictures of the three developmental stages (J, juvenile; YF, young female; ELF) of nematodes inside galls observed under stereomicroscope after staining with acid fuchsin. (f) Concentrations of hormones were measured by LC–MS/MS in rice roots 24 h after spraying of GR24. Data were analyzed by one‐way ANOVA followed by Tukey's post‐hoc test. Different letters indicate statistically different means at 95% confidence. For nematode infection, data represent mean + SE (n = 6). For hormones, data represent mean + SE (n = 5), and each replicate was a pool of four to six plants. Percentage data shown in (d) were analyzed by a chi‐squared test (P = 0.042). cis‐OPDA, cis‐(+)‐12‐oxo‐phytodienoic acid; JA, jasmonic acid; JA–Ile, JA–isoleucine; SA, salicylic acid. Bars, 1 mm.
The JA and SA pathways are known to play a pivotal role in rice defense against M. graminicola infection (Nahar et al., 2011; Kyndt et al., 2017). To investigate whether the contrasting effects of 0.1 and 5 μM GR24 on nematode development are also associated with jasmonates and SA, these defense hormones were analyzed in rice roots 24 h after foliar GR24 application. The content of cis‐OPDA was higher in roots of plants treated with 5 μM GR24 than in mock and 0.1 μM GR24‐treated plants (Fig. 2f). The levels of JA and JA–Ile both decreased significantly in 0.1 μM GR24‐treated plants, but their levels were significantly higher in plants treated with 5 μM GR24 than in mock‐treated plants (Fig. 2f). The contents of SA and ABA did not change in GR24‐treated plants compared with mock‐treated plants (Figs. 2f, S3). These results suggest that low doses of GR24 enhance M. graminicola infection and decrease JA accumulation in rice roots, but these effects are reversed at higher concentrations.
SL deficiency in rice reduces the infection of M. graminicola
Next, the effect of reduced SL on the rice–M. graminicola interaction was investigated by inoculating the nematodes into two SL‐biosynthetic rice dwarf (d) mutants, d10 and d27, one SL‐signaling mutant d14 along with their respective WT plants. The nematode infection levels were analyzed by counting the number of galls per plant and the total number of nematodes, including their developmental stages at 14 dpi. The number of galls per plant was similar in d10, d27, and WT plants but was significantly lower in the SL‐signaling d14 mutant (Fig. 3a). Interestingly, the total number of nematodes per plant and females, both young females and ELFs, were significantly lower in all d mutants than in WT plants (Fig. 3b,c). Whereas c. 90% of total nematodes developed into females in WT plants, c. 65–78% of the nematodes developed in the d mutants (Fig. 3d). The difference in the total number of nematodes and females per plant was related to the difference in gall size between d mutants and WT plants (Fig. S4). In the d mutants, only 3–5% galls were (extra‐)large, whereas c. 20% galls were (extra‐)large in the WT plants. The large and extra‐large galls were full with many more developed females and egg masses than the small and medium‐sized galls (Fig. S4). These results indicate that decreased SL levels in rice reduce infection by the root‐knot nematode M. graminicola.
Figure 3.
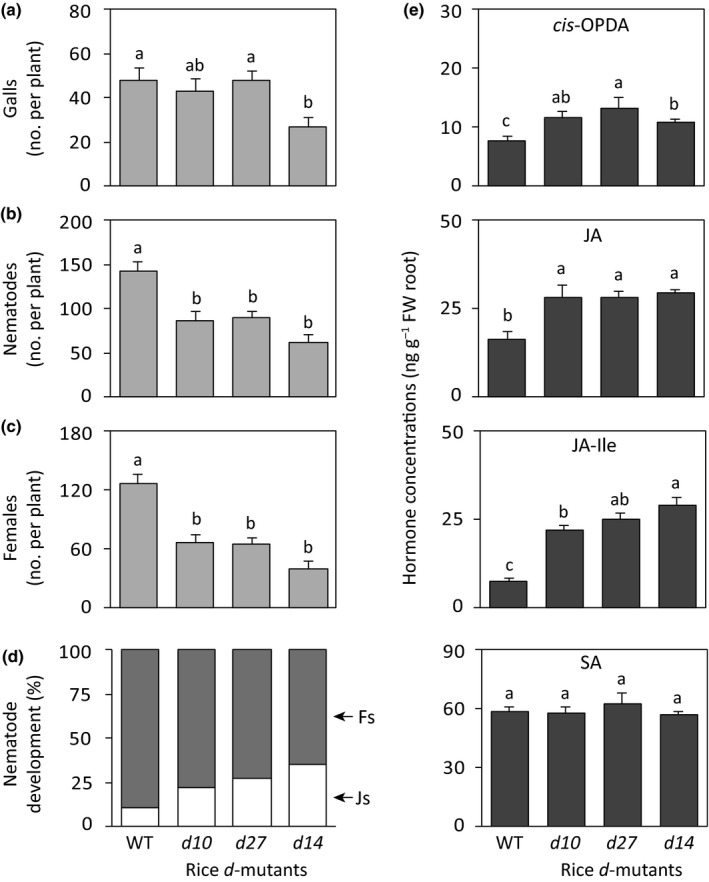
Responses of rice strigolactone (SL)‐biosynthesis and ‐signaling dwarf (d) mutants to Meloidogyne graminicola infection and phytohormone accumulation. The d10 and d27 lines are SL‐biosynthesis mutants, whereas d14 is an SL‐signaling mutant. Each 14‐d‐old rice seedling was inoculated with c. 200 second‐stage juveniles of M. graminicola. Plant responses were analyzed 14 d post‐inoculation. Reported are the total number of (a) galls, (b) nematodes, (c) egg‐laying females, and (d) percentages of the developmental stages of the observed nematodes within the galls of each plant. (e) Enhanced accumulation of jasmonates in rice d mutants. The roots of 14‐d‐old rice seedlings were harvested before nematode inoculation, and hormone metabolites were analyzed by LC–MS/MS. Data were analyzed by one‐way ANOVA followed by Tukey's post‐hoc test. Different letters indicate statistically different means at 95% confidence. For the infection experiment, data represent mean + SE (n = 8). For hormone measurement, data represent mean + SE (n = 5) and each replicate was a pool of four to six plants. Percentage data shown in (d) were analyzed by a chi‐squared test (P < 0.001). Fs, females (young females + egg‐laying females); Js, juveniles; cis‐OPDA, cis‐(+)‐12‐oxo‐phytodienoic acid; JA, jasmonic acid; JA–Ile, JA–isoleucine; SA, salicylic acid.
Plant susceptibility to root‐knot nematode infection might be influenced by the root architecture, especially by the number of root tips, which is depicted by the number of lateral roots. Because the root tips are the entry points of this nematode (Gheysen & Mitchum, 2011), fewer root tips could lead to lower nematode infection. To investigate if root architecture has an influence on the lower nematode susceptibility in the d mutants, the root morphology of the d mutants and WT plants was analyzed. Total root length per plant was lower in all d mutants, but only significantly different in d10 and d14 compared with the WT plants (Fig. S5a). The d mutants also had significantly lower root surface area than WT plants did (Fig. S5b). However, the number of root tips, represented by the number of lateral roots, was comparable in WT and d mutant plants except for d10 (Fig. S5c). The lateral root density was significantly higher in the SL‐signaling d14 mutant than in the WT plants (Fig. S5d). These results suggest that the lower susceptibility of rice d mutants to M. graminicola infection is not associated with the root phenotypes.
SL biosynthesis and signaling rice mutants contain higher jasmonate levels
To investigate whether the lower M. graminicola susceptibility in SL mutants was due to altered JA‐ and SA‐based defense pathways in rice, these hormone metabolites were quantified in roots of SL‐deficient and ‐signaling d mutants and compared with the amounts in WT plants. The level of the JA precursor, cis‐OPDA, JA, and its conjugate JA–Ile were higher in all d mutants than in the WT plants. The concentrations of JA and JA–Ile were up to two‐ and four‐fold higher, respectively, in d mutants in comparison with WT plants. On the other hand, the levels of SA and ABA in d mutants were similar to those in WT plants (Figs 3e, S3). These results suggest that SL deficiency induces the accumulation of jasmonates in rice roots.
GR24 treatment restores M. graminicola susceptibility in SL‐deficient biosynthesis mutants in rice, but not in a signaling mutant
A complementation experiment was performed by treating the SL‐deficient d mutants and WT plants with GR24 at a concentration (0.1 μM) that promotes M. graminicola infection. Both GR24 and mock‐treated plants were inoculated with M. graminicola 24 h after treatment. The number of galls per plant at 14 dpi did not differ due to GR24 treatment compared to the corresponding mock‐treated plants (Fig. 4a). However, GR24 treatment significantly increased the total number of nematodes per plant in SL‐deficient d10 and d27 mutants (Fig. 4b). On the other hand, M. graminicola susceptibility did not increase due to GR24 treatment in d14 plants, which are insensitive to SL (Fig. 4b). The development of nematodes into ELFs was restored in GR24‐treated SL biosynthetic d mutants (d27 and d10) to wild‐type levels, but not in the SL signaling d14 mutant (Fig. 4c,d). The shoot lengths of all rice d‐mutants were significantly lower than corresponding wild‐type plants, irrespective of GR24 and nematode inoculation (Fig. S6). However, the root lengths were similar in all plants at 14 dpi (Fig. S6). This experiment confirmed the role of SL signaling in promoting M. graminicola susceptibility.
Figure 4.
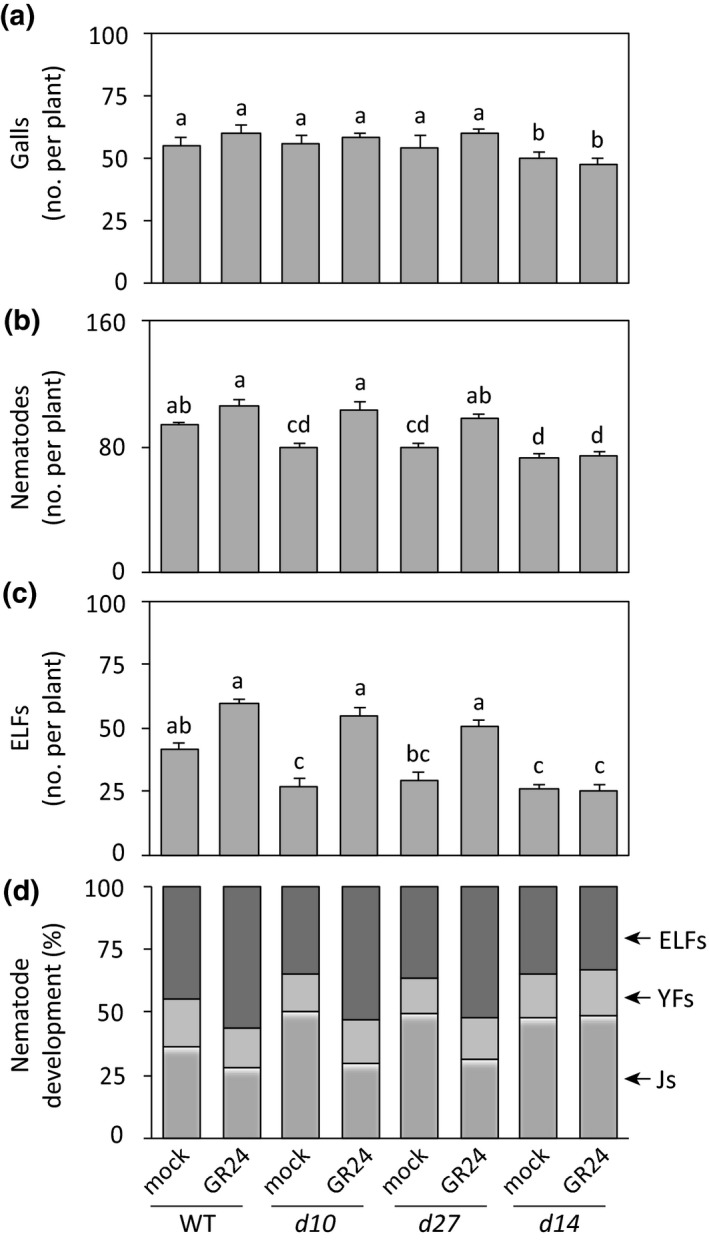
Foliar application of the strigolactone (SL) analogue GR24 restores susceptibility to Meloidogyne graminicola in SL‐deficient biosynthesis mutants (d10 and d27), but not in the SL‐signaling mutant d14. Fourteen‐day‐old wild‐type (WT) and mutant plants were sprayed on the shoots with 0.1 μM GR24 and the respective mock plants were sprayed with solvent only. After 24 h, each plant was inoculated with c. 200 second‐stage juveniles of M. graminicola. Infection levels were analyzed at 14 d post‐inoculation by counting the total number of (a) galls, (b) nematodes, (c) egg‐laying females (ELFs), and (d) percentages of the developmental stages of the observed nematodes within the galls per plant. Data were analyzed by one‐way ANOVA followed by Tukey's post‐hoc test. Different letters indicate statistically different means at 95% confidence. Data represent mean + SE (n = 6). Percentage data (mock vs GR24) for each genotype shown in (d) were analyzed by a chi‐squared test (WT, P = 0.09; d10, P = 0.018; d27, P = 0.04; d10, P = 0.97). YFs, young females; Js, juveniles.
To verify the role of GR24 in suppressing the jasmonate pathway, we conducted another experiment using the SL‐biosynthetic mutants (d10 and d27) and SL‐signaling mutant (d14). Two‐week‐old seedlings were sprayed with GR24 (0.1 μM) and or only solvent (mock). Hormones were measured in whole‐root tissues at 1 d after spraying. In general, the levels of cis‐OPDA, JA and JA‐Ile decreased in SL‐biosynthetic mutants after GR24 application (Fig. S7A). This declining trend was significantly different in d10 (JA, JA‐Ile), d27 (JA‐Ile) mutants when compared between mock and GR24 treated plants. By contrast, the levels of sulfo‐JA, OH‐JA, and COOH‐JA‐Ile slightly increased after GR24 treatment (Fig. S7B). Interestingly, the levels of JA, JA‐Ile and other jasmonates were not reduced after GR24 treatment in the d14 mutant, which is insensitive to SL (Fig. S7). Moreover, the levels of SA and ABA did not change in all d‐mutants after treatment with GR24 (Fig. S7).
Foliar application of the SL biosynthesis inhibitor TIS108 suppresses M. graminicola infection and enhances the accumulation of jasmonates in rice roots
To confirm the ability of SL deficiency in rice to cause lower susceptibility to root‐knot nematode infection and upregulation of the JA pathway, the shoots of 2‐wk‐old WT plants were sprayed with TIS108, an inhibitor of SL biosynthesis (Ito et al., 2011). At 24 h after spraying the plants were inoculated with M. graminicola. The TIS108 treatment did not change the number of galls (Fig. 5a), however, it significantly reduced the total number of nematodes (Fig. 5b) and ELFs (Fig. 5c) per plant compared with the mock‐treated plants at 14 dpi. Approx. 50% of nematodes developed into ELFs in mock‐treated plants at 14 dpi, whereas only 30% developed into this form in TIS108‐treated plants (Fig. 5d). Moreover, the galls in mock‐treated plants contained a higher number of ELFs, whereas a higher number of juveniles was found in the galls in TIS108‐treated plants (Fig. 5e). The average lengths of the shoots and roots were similar in TIS108‐ or mock‐treated plants followed by M. graminicola infection at 14 dpi (Fig. S8). To investigate whether foliar treatment with TIS108 also affects the accumulation of jasmonates, the levels of jasmonates in roots were analyzed 24 h after treatment. While the level of cis‐OPDA increased slightly in TIS108‐treated plants (Fig. 5f), both JA and JA–Ile concentrations increased significantly by approximately two‐fold in roots of the TIS108‐treated plants compared with the mock‐treated plants (Fig. 5b,c). However, the levels of SA and ABA did not change due to the inhibition of the SL biosynthesis by TIS108 (Fig. S9).
Figure 5.
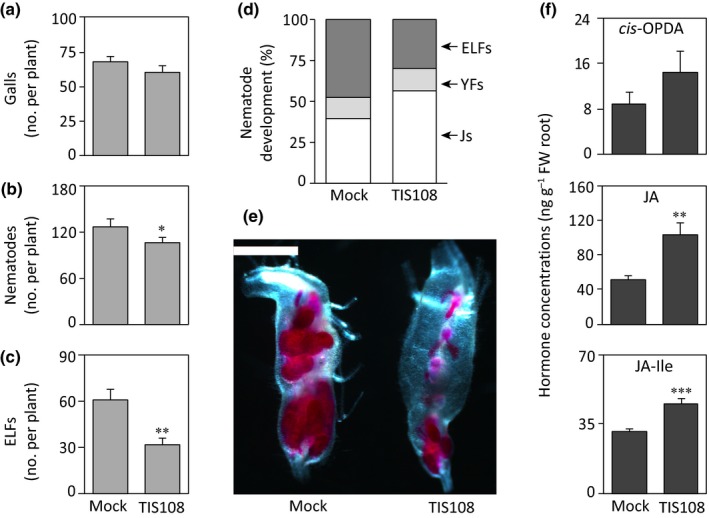
Foliar spraying of the strigolactone (SL) biosynthesis inhibitor TIS108 decreased Meloidogyne graminicola infection in rice roots and increased the contents of jasmonates. The shoots of 14‐d‐old wild‐type plants were sprayed with TIS108 (3 μM) or with solvent only (mock). After 24 h, a subset of plants from each treatment were inoculated with c. 200 second‐stage juveniles of M. graminicola per plant and another subset of plants were sampled for hormone analysis. Nematode infection levels were analyzed at 14 d post‐infection. Total numbers of (a) galls, (b) nematodes, (c) egg‐laying females (ELFs), and (d) percentages of the developmental stages inside the galls per plant. (e) A representative picture of galls in mock‐ and TIS108‐treated plants observed under a stereomicroscope after staining with acid fuchsin. (f) Concentrations of jasmonates were measured by LC–MS/MS in rice roots 24 h after spraying of TIS108. Data were analyzed by a two‐tailed Student's t‐test (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. For nematode infection, data represent mean + SE (n = 8). For hormone measurements, data represent mean + SE (n = 5), and each replicate was a pool of four to six plants. Percentage data shown in (d) were analyzed by a chi‐squared test (P = 0.015). YFs, young females; Js, juveniles; cis‐OPDA, cis‐(+)‐12‐oxo‐phytodienoic acid; JA, jasmonic acid; JA–Ile, JA–isoleucine. Bar, 1 mm.
Reduced M. graminicola infection due to SL deficiency in rice is dependent on the jasmonate pathway
To investigate whether the resistance to M. graminicola infection in rice arising from SL deficiency is dependent on the JA pathway, WT plants were treated with the SL biosynthetic inhibitor TIS108, and the JA biosynthetic inhibitor ETYA, which inhibits the lipoxygenase enzyme (Taheri & Tarighi, 2010). The foliar application of TIS108 or ETYA alone as well as their co‐application 24 h before nematode inoculation did not change the number of galls per plant (Fig. 6a). However, as seen before (Fig. 5b,c), the application of TIS108 alone decreased the total number of nematodes and ELFs per plant compared with the mock‐treated plants (Fig. 6b,c). By contrast, foliar application of ETYA significantly increased the total number of nematodes and ELFs per plant (Fig. 6b,c), consistent with the importance of JA in rice defense against this nematode (Nahar et al., 2011). Interestingly, the co‐application of TIS108 and ETYA also increased susceptibility to M. graminicola infection to the same level as ETYA treatment alone (Fig. 6). These data suggest that the suppression of M. graminicola upon SL deficiency infection in rice depends on jasmonate biosynthesis.
Figure 6.
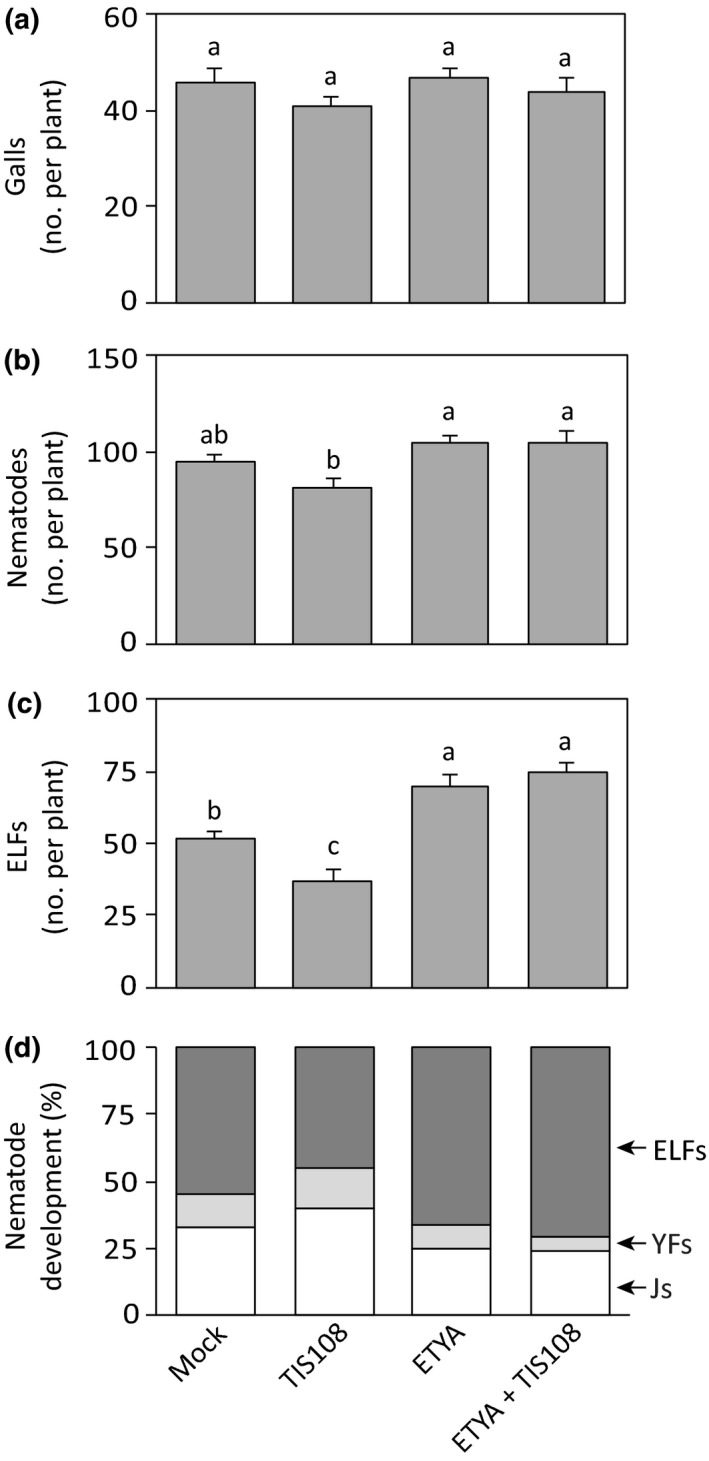
Effect of the strigolactone biosynthesis inhibitor TIS108 and the jasmonic acid (JA) biosynthesis inhibitor 5,8,11,14‐eicosatetraynoic acid (ETYA) on nematode infection. Inhibitors were sprayed on the shoots alone or in combination 24 h before Meloidogyne graminicola inoculation. Nematode infection was assessed 14 d post‐infection by counting total number of (a) galls, (b) nematodes, (c) egg‐laying females (ELFs), and (d) percentages of the developmental stages of the observed nematodes within the galls per plant. Data were analyzed by one‐way ANOVA followed by Tukey's post‐hoc test. Different letters indicate means were statistically different at 95% confidence. Data represent mean + SE (n = 6). Percentage data shown in (d) were analyzed by a chi‐squared test (P = 0.01). YFs, young females; Js, juveniles.
Discussion
SLs, a relatively recently described group of plant hormones, have been extensively studied in the last decade. These hormones are known to play an important role in various aspects of plant developmental and physiological processes, including shoot branching (Gomez‐Roldan et al., 2008; Umehara et al., 2008), leaf senescence (Ueda & Kusaba, 2015), and root development (Kapulnik et al., 2011; Ruyter‐Spira et al., 2011; Arite et al., 2012). They also stimulate seed germination of root‐parasitic weeds (Cook et al., 1966), promote arbuscular mycorrhizal association with their host plants (Akiyama et al., 2005), and affect nodulation in legumes in a dose‐dependent manner (De Cuyper et al., 2014). A few studies also demonstrated that SLs play a role in plant responses to abiotic stress; for example, drought tolerance (Bu et al., 2014; Van Ha et al., 2014), salt tolerance (Van Ha et al., 2014), and root development under nutrient deficiencies (Sun et al., 2014). Whether SLs play a role in plant–pathogen interactions is still under investigation. The rice root‐knot nematode M. graminicola causes substantial yield losses in rice production, especially under aerobic conditions. This nematode is able to manipulate developmental processes in the host cell and suppress innate immunity during infection (Kyndt et al., 2012). Our previous work described the importance of plant hormones such as JA, SA, ABA, ethylene, BR and auxin during the rice–M. graminicola interaction (Nahar et al., 2011, 2013; Kyndt et al., 2017; Yimer et al., 2018). Here, we focus on the role of SLs in the interaction between rice and M. graminicola.
In our study, M. graminicola infection in rice roots transcriptionally activated SL biosynthesis and signaling at 1 d after inoculation, a time when root‐knot nematodes initiate feeding site induction. We also measured defense hormones and found that the jasmonate levels were strongly reduced in nematode‐infected roots. Accumulation of hydroxylated‐ and carboxylated‐JA–Ile, which seem to be inactive forms (Wasternack & Strnad, 2018), in nematode‐infected roots also correlated with the reduction of bioactive JA–Ile. Upregulation of the SL signaling pathway and suppression of the JA pathway raised the question of whether SLs play a direct role in rice defense against M. graminicola infection. To address this question, SL biosynthesis and signaling mutants were analyzed in a nematode infection experiment. The infection and development of M. graminicola were significantly lower in SL‐deficient (d27 and d10) and ‐responsive (d14) rice d mutants, suggesting that SLs act as negative regulators of rice defense against root‐knot nematode infection. Inhibition of SL biosynthesis by spraying TIS108 also resulted in lower nematode susceptibility in rice roots. In contrast to our result, a previous study reported an increased susceptibility to the necrotrophic foliar fungi B. cinerea and A. alternata in the SL‐deficient tomato Slccd8 mutant (equivalent to the d10 rice mutant) (Torres‐Vera et al., 2014). Similarly, in Arabidopsis, SL deficiency promoted plant susceptibility to the biotrophic actinomycete R. fascians, causing leafy gall syndrome (Stes et al., 2015). However, the SL pathway did not show any effect on pea defense during the soil‐borne oomycete P. irregulare infection (Blake et al., 2016). Therefore, the role of SLs in plant defense against pathogen infections appears to vary in different plants, different tissues, and with different types of attackers.
On the other hand, treatment with a low dose of GR24 (a functional analogue of SL; Besserer et al., 2008) on rice shoots 24 h before nematode inoculation increased susceptibility to M. graminicola infection. A foliar spray with 0.1 μM GR24 significantly increased the number of ELFs at 14 dpi, but treatment with 5 μM GR24 decreased infection levels. In agreement with our observations, the number of nodules increased in Medicago truncatula treated with 0.1 μM GR24, whereas they were strongly reduced after treatment with 5 μM GR24 (De Cuyper et al., 2014). Application of GR24 at low concentration restored the nematode susceptibility in the SL‐biosynthesis mutants (d27 and d10), but not in the SL signaling mutant (d14), which is deficient in SL/GR24 perception. These findings suggest that SL signaling in rice is required for enhanced root‐knot nematode susceptibility. In rice, phosphate and nitrate deficiency‐induced reduction in lateral root density was rescued by GR24 treatment in WT as well as in SL‐biosynthesis mutants but not in the SL‐signaling mutant (Sun et al., 2014). SL signaling plays an important role in determining rice root architecture. Thus, one could argue that SL deficiency in the d mutants might have negatively influenced nematode infection levels by decreasing lateral root density, as the root tips are the sites of M. graminicola penetration into the host. However, we found a higher lateral root density in SL‐deficient and ‐signaling d mutants, suggesting that the possibility of nematode host entrance is not decreased in these mutant lines even though these plants were less susceptible to M. graminicola infection.
Since the root phenotypes of rice d mutants do not seem to be at the basis of their increased M. graminicola resistance, we hypothesized that altered defense hormones might be involved. Plant hormones, especially JA‐mediated defense signaling, play an important role in both monocot and dicot plant species to protect against root‐knot nematode infections (Cooper et al., 2005; Nahar et al., 2011; Gheysen & Mitchum, 2019). It was shown that the JA‐precursor cis‐OPDA also acts as a key signaling molecule in the regulation of Arabidopsis defense against the root‐knot nematode M. hapla (Gleason et al., 2016). On the other hand, SA signaling also mediates defense in rice against M. graminicola infection, but to a lesser extent than JA (Nahar et al., 2011).
In this study, we could show a negative association between active SL biosynthesis and signaling and JA accumulation in rice roots. The contents of cis‐OPDA, JA, and JA–Ile all significantly increased in the roots of SL biosynthesis and signaling rice d mutants in comparison with the WT plants. In line with these results, chemical inhibition of SL biosynthesis by TIS108 (Ito et al., 2011) also increased the levels of jasmonates in rice roots, whereas no effects on SA were observed. Therefore, decreased root‐knot nematode susceptibility in SL‐deficient rice mutants appears to be due to the activation of the JA pathway, which is known as a positive regulator of rice defense against M. graminicola infection (Nahar et al., 2011). In contrast to our results, the tomato Slccd8 mutant showed reduced levels of defense hormones, including JA and SA, which consequently increased susceptibility to the foliar necrotrophic fungi B. cinerea and A. alternata (Torres‐Vera et al., 2014). However, JA accumulation was not affected in SL‐deficient (max1) or ‐responsive mutant (max2) of Arabidopsis thaliana (Rozpądek et al., 2018). By contrast, the max1 mutant accumulated a higher amount of SA than WT plants did. In Arabidopsis, GR24 treatment at 1 μM increased the production of SA in roots (Rozpądek et al., 2018). However, GR24 treatment in rice did not change the contents of SA in our study. It is possible that the interplay between SL and defense hormones such as JA and SA is different in monocot and dicot plants, or that their interaction is concentration dependent. Indeed, in our study, the levels of JA and JA–Ile decreased in systemic roots treated with 0.1 μM GR24 on the foliage, but increased after treatment with 5 μM GR24, and nematode infection correlated inversely with the jasmonate level. Furthermore, we could show that foliar application of GR24 on d10 and d27 lines (SL‐biosynthetic mutants) reduced the levels of JA and JA–Ile but not in d14 signaling mutant that is unable to perceive exogenous SL or GR24. These findings indicate that increased root‐knot nematode susceptibility after GR24 application in SL‐biosynthetic mutants might be due the suppression of active JA signaling. In addition, we showed that chemical inhibition of the JA pathway by ETYA increased M. graminicola susceptibility in rice, whereas treatment with the SL biosynthetic inhibitor TIS108 decreased susceptibility. The co‐application of both of these inhibitors resulted in nematode susceptibility to a similar level as ETYA treatment alone, suggesting that increased nematode resistance due to SL deficiency is dependent on an intact JA pathway.
Because ABA and SLs share the same biosynthetic precursor, β‐carotene, we also investigated whether ABA plays a role in SL‐induced M. graminicola susceptibility. It is known that ABA plays a complex role in the interaction between rice and M. graminicola (Kyndt et al., 2017). In our study, neither the exogenous GR24 treatment nor the TIS108 treatment or the d‐mutants changed the contents of ABA in rice roots. In agreement with our findings, chemical inhibition of SL biosynthesis in tomato did not affect the ABA content in roots (López‐Ráez et al., 2010). However, this study also proposed that endogenous ABA might be important for SL synthesis because inhibition of the ABA pathway compromised SL production. Our findings do not support any direct relation between SLs and ABA in rice roots.
In conclusion, this study shows that SLs play an important role in the interaction between rice and the root‐knot nematode M. graminicola. Inhibition of SL biosynthesis in rice either chemically or genetically resulted in enhanced JA accumulation, which led to a lower nematode infection. During the early stage of M. graminicola infection in rice roots, SL biosynthesis is increased, and this is also associated with a strong downregulation of JA‐induced defense response. We conclude that SLs enhance susceptibility of rice to M. graminicola by antagonizing the JA biosynthesis pathway (Fig. 7).
Figure 7.
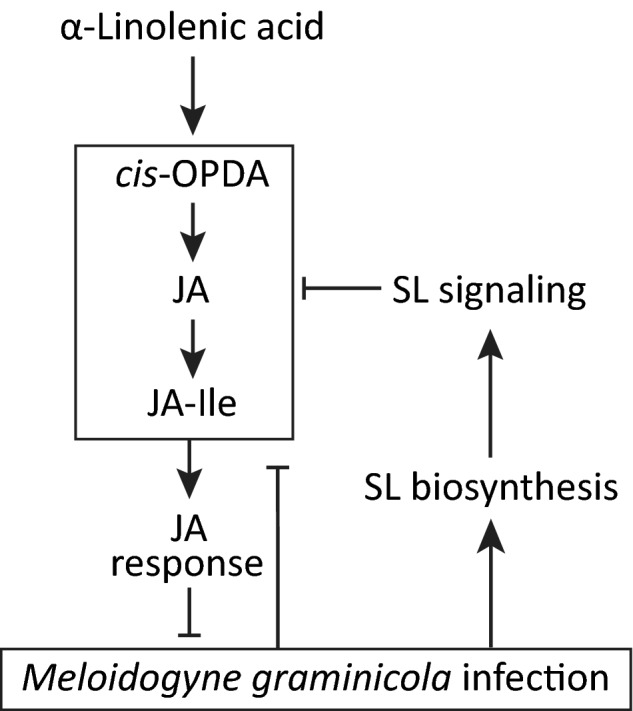
A model depicting the antagonism between the strigolactone (SL) and the jasmonic acid (JA) pathway in rice roots. Meloidogyne graminicola infection in rice roots induces the SL pathway and represses the JA pathway. On the other hand, the JA pathway reduces M. graminicola infection and the SL pathway represses the JA pathway to promote infection. cis‐OPDA, cis‐(+)‐12‐oxo‐phytodienoic acid; JA–Ile, JA–isoleucine.
Author contributions
ZL, CU, TK and GG designed the study. ZL and CU carried out experiments and analyzed data. ZL prepared the draft manuscript. CU, TK, JG and GG helped in critical discussion and revised the manuscript.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Changes in salicylic acid, abscisic acid and JA‐catabolites in rice roots upon Meloidogyne graminicola infection.
Fig. S2 Effect of foliar GR24 application followed by M. graminicola infection on shoot and root lengths of rice plants.
Fig. S3 Abscisic acid concentrations in rice roots after foliar application of GR24 and in rice d‐mutants.
Fig. S4 Percentages of M. graminicola induced galls of different sizes in rice d mutants.
Fig. S5 Root system architectures of rice d‐mutants and the corresponding wild‐type plants.
Fig. S6 Effect of foliar GR24 application followed by M. graminicola infection on shoot and root lengths of rice d‐mutants and wild‐type plants.
Fig. S7 Hormone metabolites in roots of the rice d‐mutants after GR24 application.
Fig. S8 Effect of foliar TIS108 application followed by M. graminicola infection on shoot and root lengths of rice plants.
Fig. S9 Effect of foliar application of strigolactone biosynthesis inhibitor (TIS108) on salicylic acid and abscisic acid contents.
Table S1 Primer sequences used in this study for qRT‐PCR.
Acknowledgements
We thank Prof. Mikio Nakazono, Nagoya University, and Dr Itsuro Takamure, Hokkaido University, Japan, for providing SL d‐mutant rice seeds. We are grateful to Professor Aska Goverse, Casper van Schaik, and Sonja Warmerdam for support during the root morphology measurements. We thank Dr Michael Reichelt for his assistance in the analytical lab. We also thank Sofie Goormachtig of the VIB‐UGent Center for Plant Systems Biology for helpful discussions on this project. We would like to acknowledge the Research Foundation Flanders FWO for financial support (FWO grant 3G008718W) and the Special Research Fund of Ghent University for financial support via the BOF13/GOA/030 project.
References
- Akiyama K, Matsuzaki K‐i, Hayashi H. 2005. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827. [DOI] [PubMed] [Google Scholar]
- Al‐Babili S, Bouwmeester HJ. 2015. Strigolactones, a novel carotenoid‐derived plant hormone. Annual Review of Plant Biology 66: 161–186. [DOI] [PubMed] [Google Scholar]
- Alder A, Jamil M, Marzorati M, Bruno M, Vermathen M, Bigler P, Ghisla S, Bouwmeester H, Beyer P, Al‐Babili S. 2012. The path from β‐carotene to carlactone, a strigolactone‐like plant hormone. Science 335: 1348–1351. [DOI] [PubMed] [Google Scholar]
- Arite T, Kameoka H, Kyozuka J. 2012. Strigolactone positively controls crown root elongation in rice. Journal of Plant Growth Regulation 31: 165–172. [Google Scholar]
- Besserer A, Bécard G, Jauneau A, Roux C, Séjalon‐Delmas N. 2008. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiology 148: 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake SN, Barry KM, Gill WM, Reid JB, Foo E. 2016. The role of strigolactones and ethylene in disease caused by Pythium irregulare . Molecular Plant Pathology 17: 680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridge J, Plowright RA, Peng D. 2005. Plant parasitic nematodes in subtropical and tropical agriculture In: Luc M, Sikora RA, Bridge J, eds. Nematode parasites of rice. Wallingford, UK: CABI, 87–130. [Google Scholar]
- Bu Q, Lv T, Shen H, Luong P, Wang J, Wang Z, Huang Z, Xiao L, Engineer C, Kim TH. 2014. Regulation of drought tolerance by the F‐box protein MAX2 in Arabidopsis. Plant Physiology 164: 424–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Ruyter‐Spira C, Bouwmeester H. 2013. The interaction between strigolactones and other plant hormones in the regulation of plant development. Frontiers in Plant Science 4: e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook C, Whichard LP, Turner B, Wall ME, Egley GH. 1966. Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154: 1189–1190. [DOI] [PubMed] [Google Scholar]
- Cooper W, Jia L, Goggin L. 2005. Effects of jasmonate‐induced defenses on root‐knot nematode infection of resistant and susceptible tomato cultivars. Journal of Chemical Ecology 31: 1953–1967. [DOI] [PubMed] [Google Scholar]
- De Cuyper C, Fromentin J, Yocgo RE, De Keyser A, Guillotin B, Kunert K, Boyer F‐D, Goormachtig S. 2014. From lateral root density to nodule number, the strigolactone analogue GR24 shapes the root architecture of Medicago truncatula . Journal of Experimental Botany 66: 137–146. [DOI] [PubMed] [Google Scholar]
- De Waele D, Elsen A. 2007. Challenges in tropical plant nematology. Annual Review of Phytopathology 45: 457–485. [DOI] [PubMed] [Google Scholar]
- Foo E, Davies NW. 2011. Strigolactones promote nodulation in pea. Planta 234: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Gheysen G, Mitchum MG. 2011. How nematodes manipulate plant development pathways for infection. Current Opinion in Plant Biology 14: 415–421. [DOI] [PubMed] [Google Scholar]
- Gheysen G, Mitchum MG. 2019. Phytoparasitic nematode control of plant hormone pathways. Plant Physiology 179: 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C, Leelarasamee N, Meldau D, Feussner I. 2016. OPDA has key role in regulating plant susceptibility to the root‐knot nematode Meloidogyne hapla in Arabidopsis . Frontiers in Plant Science 7: e1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Roldan V, Fermas S, Brewer PB, Puech‐Pagès V, Dun EA, Pillot J‐P, Letisse F, Matusova R, Danoun S, Portais J‐C. 2008. Strigolactone inhibition of shoot branching. Nature 455: 189. [DOI] [PubMed] [Google Scholar]
- Hu Z, Yamauchi T, Yang J, Jikumaru Y, Tsuchida‐Mayama T, Ichikawa H, Takamure I, Nagamura Y, Tsutsumi N, Yamaguchi S. 2013. Strigolactone and cytokinin act antagonistically in regulating rice mesocotyl elongation in darkness. Plant and Cell Physiology 55: 30–41. [DOI] [PubMed] [Google Scholar]
- Ito S, Umehara M, Hanada A, Kitahata N, Hayase H, Yamaguchi S, Asami T. 2011. Effects of triazole derivatives on strigolactone levels and growth retardation in rice. PLoS ONE 6: e21723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Gheysen G, Denil S, Lindsey K, Topping JF, Nahar K, Haegeman A, De Vos WH, Trooskens G, Van Criekinge W. 2013. Transcriptional analysis through RNA sequencing of giant cells induced by Meloidogyne graminicola in rice roots. Journal of Experimental Botany 64: 3885–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y, Delaux P‐M, Resnick N, Mayzlish‐Gati E, Wininger S, Bhattacharya C, Séjalon‐Delmas N, Combier J‐P, Bécard G, Belausov E. 2011. Strigolactones affect lateral root formation and root‐hair elongation in Arabidopsis . Planta 233: 209–216. [DOI] [PubMed] [Google Scholar]
- Karczmarek A, Overmars H, Helder J, Goverse A. 2004. Feeding cell development by cyst and root‐knot nematodes involves a similar early, local and transient activation of a specific auxin‐inducible promoter element. Molecular Plant Pathology 5: 343–346. [DOI] [PubMed] [Google Scholar]
- Kyndt Tina, Denil Simon, Haegeman Annelies, Trooskens Geert, Bauters Lander, Van Criekinge Wim, De Meyer Tim, Gheysen Godelieve. 2012. Transcriptional reprogramming by root knot and migratory nematode infection in rice. New Phytologist 196: 887–900. [DOI] [PubMed] [Google Scholar]
- Kyndt T, Fernandez D, Gheysen G. 2014. Plant‐parasitic nematode infections in rice: molecular and cellular insights. Annual Review of Phytopathology 52: 135–153. [DOI] [PubMed] [Google Scholar]
- Kyndt T, Nahar K, Haeck A, Verbeek R, Demeestere K, Gheysen G. 2017. Interplay between carotenoids, abscisic acid and jasmonate guides the compatible rice–Meloidogyne graminicola interaction. Frontiers in Plant Science 8: e951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real‐time quantitative PCR and the method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- López‐Ráez JA, Kohlen W, Charnikhova T, Mulder P, Undas AK, Sergeant MJ, Verstappen F, Bugg TD, Thompson AJ, Ruyter‐Spira C et al 2010. Does abscisic acid affect strigolactone biosynthesis? New Phytologist 187: 343–354. [DOI] [PubMed] [Google Scholar]
- Luc M, Sikora RA, Bridge J. 2005. Plant Parasitic Nematodes in Subtropical and Tropical Agriculture, 2nd edn. Wallingford, UK: CABI. [Google Scholar]
- Nahar K, Kyndt T, De Vleesschauwer D, Höfte M, Gheysen G. 2011. The jasmonate pathway is a key player in systemically induced defense against root knot nematodes in rice. Plant Physiology 157: 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahar K, Kyndt T, Hause B, Höfte M, Gheysen G. 2013. Brassinosteroids suppress rice defense against root‐knot nematodes through antagonism with the jasmonate pathway. Molecular Plant–Microbe Interactions 26: 106–115. [DOI] [PubMed] [Google Scholar]
- Reversat G, Boyer J, Sannier C, Pando‐Bahuon A. 1999. Use of a mixture of sand and water‐absorbent synthetic polymer as substrate for the xenic culturing of plant‐parasitic nematodes in the laboratory. Nematology 1: 209–212. [Google Scholar]
- Rozpądek P, Domka AM, Nosek M, Ważny R, Jędrzejczyk RJ, Wiciarz M, Turnau K. 2018. The role of strigolactone in the cross‐talk between Arabidopsis thaliana and the endophytic fungus Mucor sp. Frontiers in Microbiology 19: e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter‐Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez‐Raez JA, Matusova R, Bours R. 2011. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiology 155: 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y, Sado A, Asami K, Hanada A, Umehara M, Akiyama K, Yamaguchi S. 2014. Carlactone is an endogenous biosynthetic precursor for strigolactones. Proceedings of the National Academy of Sciences, USA 111: 1640–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stes E, Depuydt S, De Keyser A, Matthys C, Audenaert K, Yoneyama K, Werbrouck S, Goormachtig S, Vereecke D. 2015. Strigolactones as an auxiliary hormonal defence mechanism against leafy gall syndrome in Arabidopsis thaliana . Journal of Experimental Botany 66: 5123–5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Tao J, Liu S, Huang S, Chen S, Xie X, Yoneyama K, Zhang Y, Xu G. 2014. Strigolactones are involved in phosphate‐ and nitrate‐deficiency‐induced root development and auxin transport in rice. Journal of Experimental Botany 65: 6735–6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri P, Tarighi S. 2010. Riboflavin induces resistance in rice against Rhizoctonia solani via jasmonate‐mediated priming of phenylpropanoid pathway. Journal of Plant Physiology 167: 201–208. [DOI] [PubMed] [Google Scholar]
- Torres‐Vera R, García JM, Pozo MJ, López‐Ráez JA. 2014. Do strigolactones contribute to plant defence? Molecular Plant Pathology 15: 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Kusaba M. 2015. Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiology 169: 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah C, Tsai C‐J, Unsicker SB, Xue L, Reichelt M, Gershenzon J, Hammerbacher A. 2019. Salicylic acid activates poplar defense against the biotrophic rust fungus Melampsora larici‐populina via increased biosynthesis of catechin and proanthocyanidins. New Phytologist 221: 960–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah C, Unsicker SB, Fellenberg C, Constabel CP, Schmidt A, Gershenzon J, Hammerbacher A. 2017. Flavan‐3‐ols are an effective chemical defense against rust infection. Plant Physiology 175: 1560–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda‐Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K. 2008. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195. [DOI] [PubMed] [Google Scholar]
- Vadassery J, Reichelt M, Hause B, Gershenzon J, Boland W, Mithöfer A. 2012. CML42‐mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis . Plant Physiology 159: 1159–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ha C, Leyva‐González MA, Osakabe Y, Tran UT, Nishiyama R, Watanabe Y, Tanaka M, Seki M, Yamaguchi S, Van Dong N. 2014. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proceedings of the National Academy of Sciences, USA 111: 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Strnad M. 2018. Jasmonates: news on occurrence, biosynthesis, metabolism and action of an ancient group of signaling compounds. International Journal of Molecular Sciences 19: e2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yimer HZ, Nahar K, Kyndt T, Haeck A, Van Meulebroek L, Vanhaecke L, Demeestere K, Höfte M, Gheysen G. 2018. Gibberellin antagonizes jasmonate‐induced defense against Meloidogyne graminicola in rice. New Phytologist 218: 646–660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Changes in salicylic acid, abscisic acid and JA‐catabolites in rice roots upon Meloidogyne graminicola infection.
Fig. S2 Effect of foliar GR24 application followed by M. graminicola infection on shoot and root lengths of rice plants.
Fig. S3 Abscisic acid concentrations in rice roots after foliar application of GR24 and in rice d‐mutants.
Fig. S4 Percentages of M. graminicola induced galls of different sizes in rice d mutants.
Fig. S5 Root system architectures of rice d‐mutants and the corresponding wild‐type plants.
Fig. S6 Effect of foliar GR24 application followed by M. graminicola infection on shoot and root lengths of rice d‐mutants and wild‐type plants.
Fig. S7 Hormone metabolites in roots of the rice d‐mutants after GR24 application.
Fig. S8 Effect of foliar TIS108 application followed by M. graminicola infection on shoot and root lengths of rice plants.
Fig. S9 Effect of foliar application of strigolactone biosynthesis inhibitor (TIS108) on salicylic acid and abscisic acid contents.
Table S1 Primer sequences used in this study for qRT‐PCR.


