Summary
Vitamin B6 (pyridoxine) is vital for key metabolic reactions and reported to have antioxidant properties in planta. Therefore, enhancement of vitamin B6 content has been hypothesized to be a route to improve resistance to biotic and abiotic stresses. Most of the current studies on vitamin B6 in plants are on eudicot species, with monocots remaining largely unexplored. In this study, we investigated vitamin B6 biosynthesis in rice, with a view to examining the feasibility and impact of enhancing vitamin B6 levels. Constitutive expression in rice of two Arabidopsis thaliana genes from the vitamin B6 biosynthesis de novo pathway, AtPDX1.1 and AtPDX2, resulted in a considerable increase in vitamin B6 in leaves (up to 28.3‐fold) and roots (up to 12‐fold), with minimal impact on general growth. Rice lines accumulating high levels of vitamin B6 did not display enhanced tolerance to abiotic stress (salt) or biotic stress (resistance to Xanthomonas oryzae infection). While a significant increase in vitamin B6 content could also be achieved in rice seeds (up to 3.1‐fold), the increase was largely due to its accumulation in seed coat and embryo tissues, with little enhancement observed in the endosperm. However, seed yield was affected in some vitamin B6‐enhanced lines. Notably, expression of the transgenes did not affect the expression of the endogenous rice PDX genes. Intriguingly, despite transgene expression in leaves and seeds, the corresponding proteins were only detectable in leaves and could not be observed in seeds, possibly pointing to a mode of regulation in this organ.
Keywords: PDX proteins, monocot, rice, crop, vitamin B6, stress
Significance Statement
We demonstrate that vitamin B6 can be enhanced in a monocot species (rice), leading to increased accumulation of unphosphorylated B6 vitamers and pyridoxine glucosides, particularly in leaves. Our analysis reveals current limitations and possibly regulatory mechanisms for vitamin B6 biosynthesis in rice endosperm.
Introduction
Vitamin B6 is essential for all kingdoms of life and is composed of a mixture of six different vitamers: pyridoxal (PL), pyridoxine (PN), pyridoxamine (PM), and its 5′‐phosphorylated derivatives: pyridoxal 5′‐phosphate (PLP), pyridoxine 5′‐phosphate (PNP), and pyridoxamine 5′‐phosphate (PMP) (Fitzpatrick et al., 2007; Fitzpatrick, 2011). Vitamin B6 in its form as PLP is required as a coenzyme for more than 200 enzymatic reactions in cells, primarily in amino acid, sugar, and fatty acid metabolism (Percudani and Peracchi, 2003; Colinas et al., 2016). Mammals lack the ability to biosynthesize vitamin B6 de novo, and plants represent one of their major sources of the vitamin (Fitzpatrick, 2011). Acute vitamin B6 deficiency can lead to various chronic diseases in humans, including neurological disorders, cardiovascular disease, and diabetes (Hellmann and Mooney, 2010; Di Salvo et al., 2012; Fudge et al., 2017).
In plants, PLP is biosynthesized de novo via the so‐called ‘DXP independent pathway’ first characterized in Arabidopsis thaliana (Tambasco‐Studart et al., 2005; Fitzpatrick et al., 2007). PLP biosynthesis de novo requires two enzymes, PDX1 and PDX2 (Figure 1), which together act as a glutamine amidotransferase utilizing ribose 5‐phosphate (R5P), glyceraldehyde 3‐phosphate (G3P) and glutamine as substrates (Burns et al., 2005; Raschle et al., 2005, 2007, 2009; Tambasco‐Studart et al., 2005; Moccand et al., 2011). In Arabidopsis, there are three PDX1 homologs, only two of which are catalytically active (PDX1.1 and PDX1.3), and only one gene encoding PDX2 (Tambasco‐Studart et al., 2005). PLP can also be produced by interconversion of the different B6 vitamers through salvage pathways, which are present in all kingdoms of life (Ruiz et al., 2008; Colinas and Fitzpatrick, 2015). Among the known enzymes involved in this pathway, a PMP/PNP/PLP kinase named SALT OVERLY SENSITIVE 4 (SOS4) (Lum et al., 2002; Shi and Zhu, 2002; Shi et al., 2002; Gonzalez et al., 2007b), a PMP/PNP oxidase named PDX3 (Gonzalez et al., 2007b; Sang et al., 2007, 2011; Colinas et al., 2016), and a pyridoxal reductase named PLR1 (Herrero et al., 2011) have been identified in plants (Figure 1). In addition, plants can convert PN to glucosidic derivatives (pyridoxine glucosides, PN‐Glu), for example, pyridoxine‐5′‐β‐d‐glucoside (PNG), using glucosyltransferases (Gregory and Ink, 1987; Gregory, 1998; Ollilainen, 1999).
Figure 1.
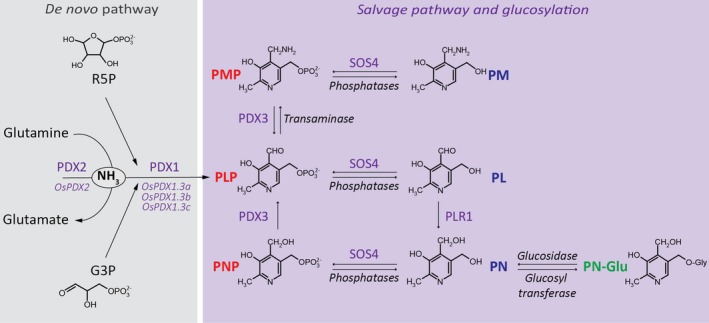
Scheme of the vitamin B6 biosynthesis pathway as described in Arabidopsis and extrapolated to rice.
Vitamin B6 is biosynthesized de novo in the presence of R5P (ribose 5‐phosphate), G3P (glyceraldehyde 3‐phosphate) and glutamine through the combined action of PDX1 (encoded by OsPDX1.3a: LOC_Os07g01020; OsPDX1.3b: LOC_Os10g01080; OsPDX1.3c: LOC_Os11g48080 in rice) and PDX2 (encoded by OsPDX2: LOC_Os02g03740 in rice) to form PLP (pyridoxal 5′‐phosphate). Enzymes from the salvage pathway allow interconversion between the B6 vitamers. Annotation in red depicts the phosphorylated B6 vitamers: PMP (pyridoxamine 5′‐phosphate), PLP and PNP (pyridoxine 5′‐phosphate); annotation in blue depicts the unphosphorylated B6 vitamers: PM (pyridoxamine), PL (pyridoxal) and PN (pyridoxine); annotation in green depicts the glucosylated PN vitamer (PN‐Glu). PDX3 refers to a PN/PM oxidase, SOS4 to a PN/PL/PM kinase and PLR1 to a PL reductase.
B6 vitamers are reported to display antioxidant properties (Bilski et al., 2000; Danon et al., 2005; Denslow et al., 2005) and therefore vitamin B6 has been implicated in abiotic and biotic stress responses in plants (Fitzpatrick, 2011; Hanson et al., 2016). This notion is also supported by the association and susceptibility of Arabidopsis mutants impaired in vitamin B6 biosynthesis de novo or salvage pathways to various abiotic stresses, including osmotic stress (Chen and Xiong, 2005; Titiz et al., 2006), salt stress (Shi et al., 2002; Titiz et al., 2006), oxidative stress (Chen and Xiong, 2005), heat stress (Moccand et al., 2014; Dell'Aglio et al., 2017), as well as high light and photo‐oxidative stress (Titiz et al., 2006; Havaux et al., 2009). Independent studies have also demonstrated that abiotic stresses can modulate the expression of genes of vitamin B6 biosynthesis de novo and salvage pathways (Shi et al., 2002; Savenstrand et al., 2004; Denslow et al., 2005, 2007; Ristila et al., 2011). Coincidently, abiotic stresses also appear to alter vitamin B6 levels in Arabidopsis, with a significant increase in PLP and some changes in PL and PN having been reported (Huang et al., 2013; Moccand et al., 2014). Vitamin B6 is also assumed to play a role in plant responses to pathogens, as evidenced by the enhanced sensitivity of Arabidopsis pdx1.2, pdx1.3 and sos4‐1 mutants to Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea (Zhang et al., 2015). Furthermore, gene expression analyses showed that Arabidopsis pdx3 mutants display a strong upregulation of genes related to plant defense (Colinas and Fitzpatrick, 2016; Colinas et al., 2016). In addition, increased severity of Botrytis cinerea symptoms have been observed in tomato, in which expression of PDX1.2 and PDX1.3 have been knocked‐down (Zhang et al., 2014).
Various reports have demonstrated that vitamin B6 content can be increased in planta by genetic engineering strategies (Herrero and Daub, 2007; Chen and Xiong, 2009; Raschke et al., 2011; Li et al., 2015). The combined expression of Arabidopsis PDX1.1 and PDX2 has led to a significant increase in vitamin B6 content in transgenic Arabidopsis leaves and seeds, as well as field‐grown transgenic cassava leaves and roots (Raschke et al., 2011; Li et al., 2015). Recent reports have indicated that constitutive expression of Arabidopsis PDX2 in potato also leads to a significant increase in vitamin B6 content (Bagri et al., 2018). In Arabidopsis, overexpression of PDX1.1 and PDX2 was reported to have pleiotropic effects, including increased size of leaves and embryos (Raschke et al., 2011). Overaccumulation of vitamin B6 in transgenic Arabidopsis and potato was also associated with increased tolerance to oxidative stress, supporting the proposed antioxidant function of vitamin B6 in planta (Raschke et al., 2011; Bagri et al., 2018).
To date, studies investigating vitamin B6 biosynthesis in monocot species have been scarce (Yang et al., 2017). Within this context, it is interesting to note that recent studies have highlighted the almost exclusive presence of the non‐catalytic PDX1 homolog, PDX1.2, to eudicots (Moccand et al., 2014; Dell'Aglio et al., 2017). The absence of PDX1.2 from monocots sets precedence for unraveling further regulatory differences between these two groups of flowering plants. Rice is an important cereal crop for almost half of the global population and its yield is affected by several abiotic and biotic stresses (Seck et al., 2012; Zhao et al., 2016). The impact of vitamin B6 on environmental stress responses in rice has not been studied to date. Furthermore, while rice represents 35–70% of daily caloric intake in several populations (Kennedy et al., 2003; Meng et al., 2005; Bhullar and Gruissem, 2013), most of the micronutrients, which are primarily stored in the husk, aleurone and embryo, are removed during processing (Kennedy et al., 2003; Lucca et al., 2006; Bhullar and Gruissem, 2013). Therefore, these populations suffer from deficiencies in one or several essential micronutrients including vitamin B6 (Thompson and Amoroso, 2011; Vanderschuren et al., 2013; von Grebmer et al., 2014; Singh et al., 2016; Strobbe and Van Der Straeten, 2018). Thus, enhancing vitamin B6 content in rice can provide insight into its impact on rice growth and yield, as well as its tolerance to stress responses, while also providing knowledge on its potential to improve the nutritive value of the consumed grain.
Here, we have studied the effect of heterologous expression of the Arabidopsis PDX1.1 and PDX2 transgenes in rice. We performed a phenotypic assessment as well as stress resistance assays to characterize selected transgenic rice lines. We further characterized the selected transgenic rice lines by profiling the expression of their transgenes as well as the identified rice PDX genes in leaves, unpolished, and polished seeds. We used a high‐performance liquid chromatography (HPLC)‐based method to report an in‐depth profiling of B6 vitamers in wild‐type and transgenic rice lines.
Results
Enhancement of vitamin B6 content in rice using the Arabidopsis de novo pathway
In order to enhance the vitamin B6 content in rice, we transformed rice variety Taipei 309 (TP309) with binary vectors carrying both the Arabidopsis PDX1.1 and PDX2 genes under the constitutive CaMV 35S promoter (p35S::AtPDX1.1‐p35S::AtPDX2) (Figure 2a). We selected Arabidopsis genes because they were previously demonstrated to enhance vitamin B6 content in cassava (Li et al., 2015). The use of Arabidopsis gene sequences also aimed at avoiding unintentional silencing of endogenous PDX rice genes. A molecular characterization of the transgenic rice lines was performed at the T0 generation assessing 26 lines for the presence of the p35S::AtPDX1.1‐p35S::AtPDX2 construct (hereafter called 35S lines). Polymerase chain reaction (PCR) genotyping indicated that 21 of the 35S lines harbored both of the Arabidopsis PDX transgenes (Figure S1a). The number of T‐DNA integration events was determined by Southern blot and showed that the 21 35S lines carrying both transgenes were genetically independent and included seven single insertion lines (Figure S1b). Of these, 14 35S lines (seven single insertion lines and seven multiple insertion lines) were selected for further evaluation. The segregating T1 generation was germinated on media containing hygromycin and resistant seedlings were transferred to soil for growth under greenhouse conditions. A TP309 wild‐type control and a transgenic line transformed with the empty vector, pCAMBIA1300 (p1300), were simultaneously grown under these conditions. To select the best performing transgenic lines, vitamin B6 content was evaluated in leaves of 45‐day‐old plants (vegetative stage) using a yeast bioassay for total vitamin B6 content (Figure S2a). All selected 35S lines accumulated more vitamin B6 than the controls, with higher levels detected in single insertion lines in general, compared to multiple insertion lines (Figure S2b). Six single insertion 35S lines (35S‐12a, 35S‐12b, 35S‐30, 35S‐31, 35S‐32, 35S‐35) were selected for further characterization in the T2 generation. The measurement of total vitamin B6 content by yeast bioassay confirmed enhanced vitamin B6 levels in leaves of these lines, when compared with wild‐type (TP309) and empty vector control plants, as seen in the previous generation (Figure 2b, c). We also measured the vitamin B6 content in the roots of these lines and observed significant enhancement in five out of six of the 35S lines (Figure 2d).
Figure 2.
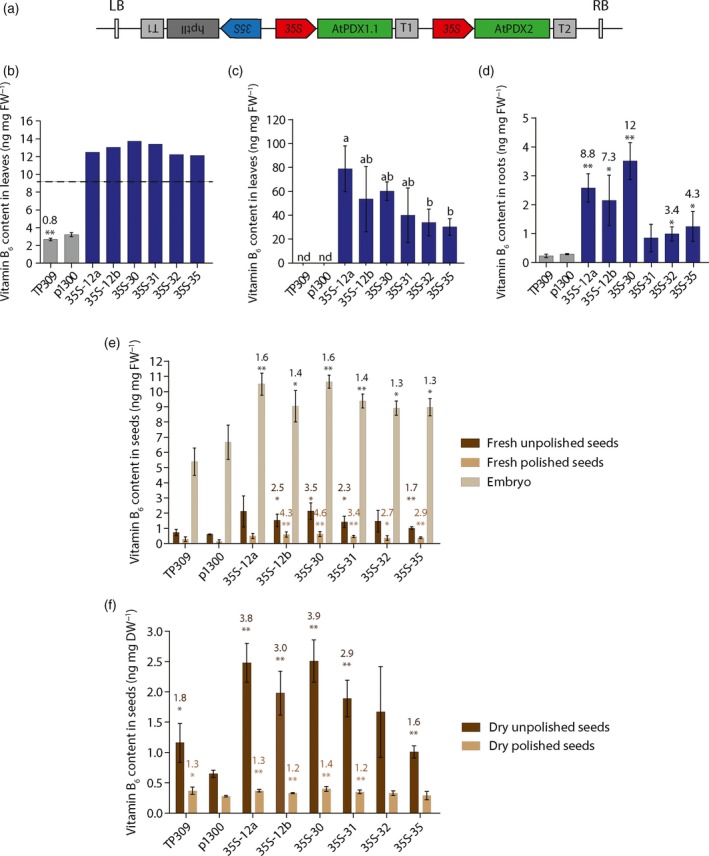
Enhancement of vitamin B6 content in rice by heterologous expression of Arabidopsis PDX1/PDX2.
(a) Schematic representation of the T‐DNA regions of the transformation vector used to generate the 35S lines. LB, Left Border; RB, Right Border; 35S, cauliflower mosaic virus 35S (CaMV 35S) promoter; AtPDX1.1, At2g38230 coding sequence; AtPDX2, At5g60540 coding sequence; hptII, hygromycin phosphotransferase gene; T1, CaMV 35S terminator; T2, Octopine synthase terminator.
(b) Vitamin B6 content in leaf sample extracts of 35S lines in the T2 generation compared to wild‐type (TP309) and empty vector control (p1300). Average ± SD of four biological replicates. Student's t‐test (p1300 versus transgenic lines and TP309), *P < 0.05, **P < 0.01. Standard deviation is not indicated for samples with at least one O.D. (Optical Density) value above the linear range of the standard curve indicated by the area above the dotted line.
(c) Vitamin B6 content as in (b) in 15‐fold diluted leaf sample extracts. Average ± SD of four biological replicates. Tukey's multiple comparison test (P < 0.05). nd, not detected.
(d) Vitamin B6 content in root sample extracts from mature plants in the T2 generation grown under greenhouse conditions. Average ± SD of four biological replicates. Student's t‐test (p1300 versus transgenic lines and TP309), *P < 0.05, **P < 0.01. Values above the bars represent the fold increase compared to p1300.
(e) Vitamin B6 content in fresh mature unpolished and polished seeds and in the embryo from plants in the T2 generation grown in the greenhouse. Average ± SD of four biological replicates. Student's t‐test (p1300 versus transgenic lines and TP309), *P < 0.05, **P < 0.01.
(f) Vitamin B6 content in dry mature unpolished and polished seeds of the samples as in (e). Average ± SD of four biological replicates. Student's t‐test (p1300 versus transgenic lines and TP309), *P < 0.05, **P < 0.01. Values above the bars represent the fold increase compared to p1300. For panels (b) to (f), the vitamin B6 content was determined by a yeast bioassay.
We next measured the vitamin B6 content in seeds of the transgenic rice lines. In the first instance, the content of mature fresh whole seeds (unpolished), polished seeds (removal of outer hull and aleurone layers) and isolated embryo were compared. In all cases, the highest vitamin B6 content was observed in the isolated embryos per unit fresh weight, and considerably less in polished versus unpolished seeds (Figure 2e). A statistically relevant increase (albeit modest) in vitamin B6 content could be observed in the embryos of all lines examined (Figure 2e). Similar increases were also observed in fresh unpolished seeds of most of the lines, and substantial vitamin B6 enhancement (up to 6.1‐fold) was observed in most of the fresh polished seeds compared with wild‐type and the empty vector control lines (Figure 2e). The content of dry unpolished and polished seeds was also determined. In all cases, the highest vitamin B6 content was observed in the mature dry unpolished seeds per unit dry weight (Figure 2f). Notably, a statistically significant increase in vitamin B6 levels was also observed in dry mature unpolished seeds of several of the 35S lines (Figure 2f). However, the vitamin B6 content was not enhanced in the mature dry polished seeds from the transgenic lines compared to wild‐type (Figure 2f).
Enhanced vitamin B6 content has only limited impacts on rice growth and development
The ability to increase vitamin B6 content in rice prompted us to examine its impact on growth and yield of rice. In the first instance, we characterized selected 35S lines grown hydroponically in a climate chamber (28°C under a 16/8 h light/dark regime) for 15 days. Under these conditions, there were only limited (if any) impacts on the selected lines with respect to lengths and fresh weight of leaves and roots (Figure 3a,b). We also assessed mature plants of the 35S lines grown under greenhouse conditions for effects on plant height, number of tillers and leaf dry weight, as well as the number of days from seed germination to panicle initiation (Table 1a). Evaluation of these parameters revealed a minor (8–13%) but statistically significant decrease in plant height of four of the 35S lines (35S‐12a, 35S‐31, 35S‐32, and 35S‐35) compared to the empty vector control plants. In contrast, the tiller number per plant, leaf dry weight, and time to panicle initiation were not altered (Table 1a). We also analyzed seed production parameters, specifically seed yield (based on weight), seed setting rate, number of panicles per plant and number of seeds per plant (Table 1b). There was no statistically significant difference in the number of panicles per plant and seed setting rate. The number of seeds per plant was decreased modestly in two lines (35S‐31 and 35S‐32) compared to the empty vector control line. However, a statistically significant decrease in seed yield (17–42%) was observed in all transgenic lines compared to empty vector control plants, with the exception of line 35S‐12b. The measured difference was mainly due to the reduced mass of 100 seeds for all yield‐affected lines.
Figure 3.
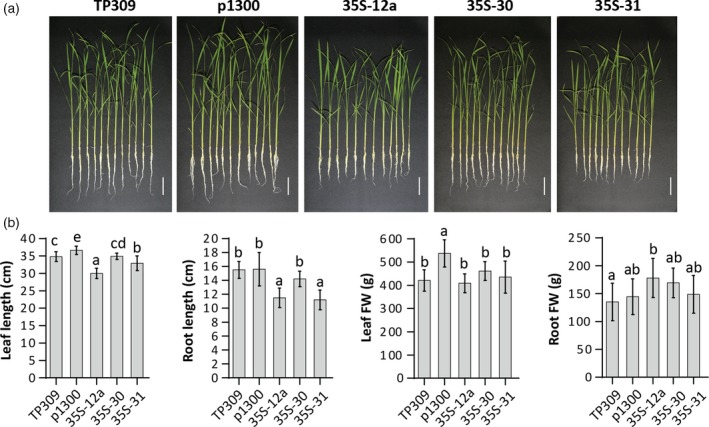
Phenotypic characterization of vitamin B6 enhanced rice lines.
Plantlets in the T3 generation of the 35S lines indicated were assessed for a phenotype compared to wild‐type (TP309) and the empty vector transgenic control (p1300) after growth for 15 days under hydroponic conditions.
(a) Pictures are of plantlets at 15 days of growth under hydroponic conditions. The scale bar represents 5 cm.
(b) Leaf length, root length, leaf fresh weight (FW) and root fresh weight of the rice lines. Average ± SD of 12 biological replicates. Tukey's multiple comparison test (P < 0.05).
Table 1.
Phenotypic characterization of selected transgenic rice lines in the T2 generation. (a) Evaluation of selected 35S lines at maturity grown under greenhouse conditions (plant height, number of tillers per plant and leaf dry weight) and determination of the number of days from seed germination to the panicle initiation, compared to wild‐type (TP309) and the empty vector transgenic line (p1300). (b) Evaluation of seed production at maturity
| a | Plant phenotype at maturity | Days to reach panicle initiation | ||
|---|---|---|---|---|
| Plant height (cm) | Number of tillers/plant | Leaf dry weight (g) | ||
| TP309 | 94.2 [±3.8] | 4.7 [±0.9] | 18.1 [±3.4] | 115.3 [±4.8] |
| p1300 | 93.9 [±4.7] | 5.0 [±1.0] | 17.4 [±2.1] | 111.1 [±6.0] |
| 35S‐12a | 87.2 [±2.1]** | 5.3 [±1.0] | 18.8 [±4.3] | 108.6 [±4.6] |
| 35S‐12b | 91.1 [±5.6] | 5.0 [±0.7] | 15.9 [±3.1] | 112.1 [±4.4] |
| 35S‐30 | 92.4 [±4.1] | 4.8 [±0.8] | 18.0 [±3.9] | 110.8 [±4.1] |
| 35S‐31 | 81.6 [±2.4]** | 5.0 [±1.1] | 15.2 [±2.6] | 108.8 [±3.4] |
| 35S‐32 | 82.7 [±4.2]** | 5.3 [±1.5] | 14.3 [±5.7] | 115.3 [±7.0] |
| 35S‐35 | 86.3 [±6.2]* | 5.8 [±0.7] | 16.3 [±3.7] | 111.9 [±4.1] |
| b | Plant seed production | ||||
|---|---|---|---|---|---|
| Yield (g) | Seed setting rate (%) | Mass of 100 seeds (g) | Number of seeds/plant | Number of panicles/plant | |
| TP309 | 7.4 [±1.8] | 83.6 [±2.4]* | 2.4 [±0.2] | 332.8 [±64.2]* | 4.6 [±0.7] |
| p1300 | 6.6 [±1.4] | 75.4 [±9.0] | 2.6 [±0.1] | 261.0 [±53.0] | 4.3 [±0.9] |
| 35S‐12a | 5.4 [±0.9]* | 77.4 [±3.8] | 2.1 [±0.0]** | 256.4 [±39.1] | 4.9 [±0.6] |
| 35S‐12b | 5.5 [±1.1] | 66.7 [±8.6] | 2.6 [±0.1] | 216.8 [±49.1] | 5.0 [±0.7] |
| 35S‐30 | 5.0 [±1.1]* | 78.1 [±6.5] | 2.0 [±0.1]** | 250.7 [±55.3] | 4.4 [±0.9] |
| 35S‐31 | 4.3 [±0.6]** | 85.5 [±4.7]** | 2.1 [±0.1]** | 204.1 [±26.0]* | 4.2 [±0.4] |
| 35S‐32 | 3.8 [±1.6]** | 82.2 [±7.2] | 2.1 [±0.0]** | 182.3 [±73.1]* | 4.3 [±1.2] |
| 35S‐35 | 5.5 [±1.0]* | 79.9 [±7.7] | 2.1 [±0.1]** | 259.2 [±46.2] | 5.0 [±1.0] |
Average ± SD of nine biological replicates. Student's t‐test (p1300 versus transgenic line), *P < 0.05, **P < 0.01.
Increased vitamin B6 content does not improve stress performance in rice
Given the considerable enhancement in vitamin B6 content of the generated transgenic rice plants, we next considered if stress tolerance was improved in these lines. To test for abiotic stress tolerance, we subjected 10‐day‐old hydroponically grown rice plantlets to salt stress for 15 days. While application of salt (150 mm sodium chloride) caused a decrease in plant growth in comparison with non‐stressed plantlets, there was no statistically significant phenotypic difference between controls (wild‐type TP309 and the empty vector control line p1300) and the selected transgenic lines accumulating higher levels of vitamin B6 (Figure 4a,b). To test for biotic stress tolerance, wild‐type TP309 and transgenic lines p1300 and 35S‐12a (that accumulates high levels of vitamin B6 in leaves) grown under greenhouse conditions were inoculated with two Xanthomonas oryzae pv. oryzae strains, PX071 (Leach et al., 1992) and BAI3 (Gonzalez et al., 2007a), which both cause rice leaf blight. The evaluation of the lesion length 15 days post‐inoculation showed that all wild‐type and the transgenic line 35S‐12a did not display a statistically significant difference in disease‐related symptoms compared to controls (Figure 4c).
Figure 4.
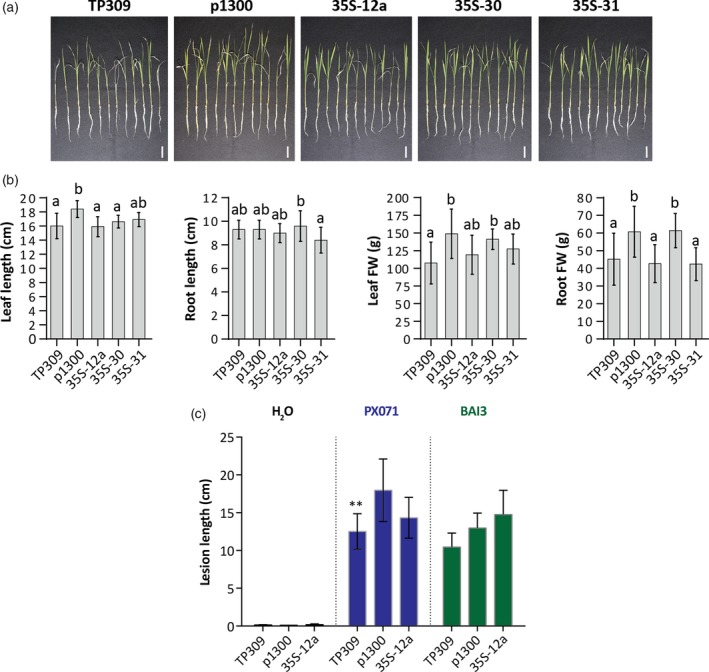
Phenotypic characterization of vitamin B6 enhanced rice lines under stress conditions.
(a) Pictures of plantlets as indicated from the T3 generation grown in hydroponic culture 15 days after application of salt stress (150 mm NaCl). The scale bar represents 3 cm.
(b) Leaf length, root length, leaf fresh weight (FW) and root fresh weight of 35S lines in (a) compared to wild‐type (TP309) and the empty vector transgenic control (p1300). Average ± SD of 12 biological replicates. Tukey's multiple comparison test (P < 0.05).
(c) Lesion length in rice leaves inoculated with Xanthomonas oryzae. Lesion length in leaves of 35S‐12a, T3 generation, 15 days after inoculation with PX071 (Leach et al., 1992) and BAI3 (Gonzalez et al., 2007a) Xanthomonas oryzae pv. oryzae strains compared to wild‐type (TP309) and the empty vector transgenic control (p1300). Inoculation with water was performed as a negative control. Average ± SD of four replicates for water and 8–12 replicates for bacterial inoculation (PX071‐TP309 (n =8), PX071‐p1300 (n = 7), PX071‐35S‐12a (n = 8), BAI3‐TP309 (n = 12), BAI3‐p1300 (n = 9), BAI3‐35S‐12a (n = 8)). Student's t‐test (p1300 versus transgenic line), **P < 0.01.
Vitamer profiling of rice lines with enhanced vitamin B6 content
As outlined above, vitamin B6 is a family of compounds (some of which may have as yet undefined non‐coenzyme roles); we selected two 35S lines (35S‐12a, 35S‐31) for detailed B6 vitamer profiling, using a previously established HPLC method (Szydlowski et al., 2013; Li et al., 2015). The distribution of B6 vitamers was analyzed by HPLC in leaves, unpolished and polished seeds. In addition to peaks corresponding to the six standards used (PMP, PM, PNP, PN, PLP, PL), up to six peaks (numbered 1−6) with retention times different from the known standards could be observed in the plant extracts (Figures 5a and S3). As a large proportion of vitamin B6 can be stored in plants as β‐glucosides (Gregory, 1998), we treated rice leaf extracts with β‐glucosidase to determine if any of the unknown peaks could be assigned to glucosylated derivatives. All observed unknown peaks responded to the treatment (with the exception of peak 5, which was not detected in these leaf extracts), either decreasing (peaks 1/2 and 3) or increasing (peaks 4 and 6) in response to the treatment (Figure S3a). Peaks 1/2 and 3 were separately collected from untreated leaf extracts and treated with β‐glucosidase. Within the assigned 1/2 peak, peak 1 decreased in abundance upon treatment with β‐glucosidase (Figure S3b) but no new peak appeared and therefore could not be assigned to any known B6 vitamer with this method. Peak 2 did not respond to the β‐glucosidase treatment (Figure S3b). The disappearance of peak 3 with a concomitant increase in PN, allowed us to clearly assign this peak to glucosylated PN (PN‐Glu) (Figure S3b). We therefore assigned peak 3 as PN‐Glu equivalents and used it to calculate the abundance of this vitamer. A comparison of the chromatograms of the controls (wild‐type TP309 and the empty vector control line p1300) and transgenic rice plants (35S‐12a and 35S‐31 lines) show clear differences in vitamer distribution in leaves and in unpolished seeds (Figure 5a). Vitamer quantification of these 35S transgenic lines revealed that the main contributors to vitamin B6 accumulation in leaves and unpolished seeds were PN‐Glu (up to 57.8‐fold increase in leaves and 2.9‐ to 4.3‐fold increase in unpolished seeds) and the unphosphorylated forms (6.6‐ to 9.2‐fold increase in leaves and up to 2.0‐fold increase in unpolished seeds with a major contribution of PM and PN) (Figure 5b; Table 2). No increase was observed in phosphorylated forms except in leaves of the 35S‐12a line, which showed a 1.7‐fold increase, primarily due to increased PLP levels (Table 2a). The transgenic lines did not display significantly higher accumulation of vitamin B6 in polished seeds as compared to the wild‐type control (Figure 5b; Table 2c), corroborating the results observed for the yeast bioassay (Figure 2). The major contributors to vitamin B6 content in polished seeds were PN‐Glu and unphosphorylated vitamers (Table 2c). With the HPLC technique, total vitamin B6 can be calculated as the sum of all quantified vitamers, including PN‐Glu as assigned in this study (Figure S3). Compared to the empty vector control line, the transgenic 35S lines showed up to a 28.3‐fold increase in leaves, up to a 3.1‐fold increase in unpolished seeds and up to a 2.2‐fold increase in polished seeds (Figure 5b; Table 2). For polished seeds, it should be noted that the best performing line (35S‐12a) did not display a statistically significant increase compared to the wild‐type control TP309. Both TP309 and p1300 controls had similar vitamin B6 levels in leaves, but displayed a minor yet statistically significant difference in both unpolished and polished seeds (Figure 5b; Table 2). Moreover, the comparison of vitamin B6 levels in unpolished seeds (1.02 to 3.15 ng mg fresh weight (FW)−1) and polished seeds (0.13 to 0.29 ng mg FW−1) (Figure 5b; Table 2b,c) suggested that vitamin B6 mainly accumulates in the embryo and/or aleurone layer, corroborating the analysis of total vitamin B6 content using the yeast bioassay (Figure 2). These results are in agreement with previous studies reporting that other essential micronutrients are stored largely in the husk, aleurone and embryo of rice seeds (Kennedy et al., 2003; Lucca et al., 2006; Bhullar and Gruissem, 2013; Dong et al., 2016).
Figure 5.
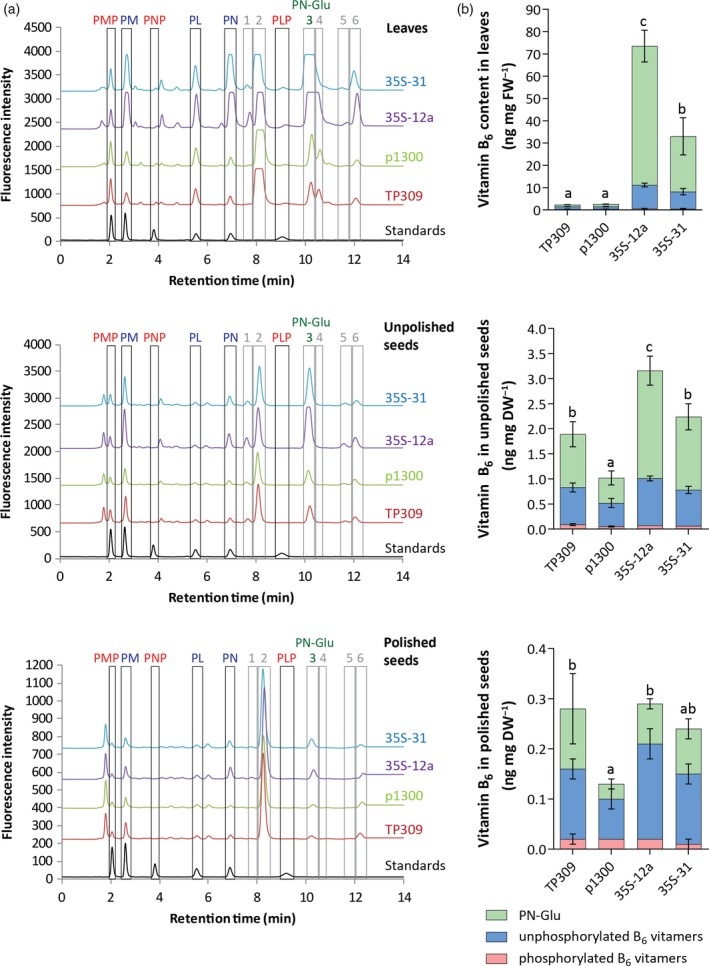
Profiling of B6 vitamers in rice lines enhanced in vitamin B6 content in the T2 generation.
(a) HPLC chromatograms of leaf, unpolished seed and polished seed extracts of T2 generation plants grown in the greenhouse. Selected 35S lines are compared with the wild‐type (TP309) and empty vector control (p1300). To facilitate the visualization, the profile of TP309 was offset by 700, p1300 by 1500, 35S‐12a by 2300 and 35S‐31 by 3100, relative to the baseline for the leaf extracts; the profile of TP309 was offset by 600, p1300 by 1300, 35S‐12a by 2000 and 35S‐31 by 2800, relative to the baseline for the unpolished seed extracts; the profile of TP309 was offset by 160, p1300 by 340, 35S‐12a by 500 and 35S‐31 by 670, relative to the baseline for the polished seed extracts. Profiles for the leaf samples 35S‐12a and 35S‐31 appear saturated for PN and PN‐Glu peaks but vitamin B6 quantification was performed on diluted samples having unsaturated signals. The numbers annotate peaks that do not correspond to the standards used. Peak 3 could be assigned as glucosylated pyridoxine (PN‐Glu) based on its correlation with a corresponding increase in PN content after treatment with β‐glucosidase (see Supplementary Figure S3).
(b) Vitamin B6 content (PN‐Glu (green); unphosphorylated B6 vitamers (blue); phosphorylated B6 vitamers (pink)) in rice leaves, unpolished seeds and polished seeds according to the HPLC analysis. Average ± SD of four biological replicates. Tukey's multiple comparison test (P < 0.05) for total vitamin B6 content.
Table 2.
HPLC analysis of B6 vitamers in leaves, unpolished seeds and polished seeds of wild‐type and transgenic rice. B6 vitamer distribution in leaves (a), dry unpolished seeds (b) and dry polished seeds (c) of the control and transgenic rice lines (T2 generation). Total unphosphorylated B6 vitamers corresponds to the PM, PN and PL contents for each replicate; total phosphorylated B6 vitamers correspond to the PMP, PNP and PLP contents for each replicate; total vitamin B6 corresponds to the unphosphorylated vitamers, the phosphorylated vitamers and the PN‐Glu contents for each replicate
| Vitamers (ng mg FW−1) | Unphosphorylated | Phosphorylated | Glucosylated | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PM | PN | PL | Total | Fold change | PMP | PNP | PLP | Total | Fold change | PN‐Glu | Fold change | Total vit. B6 | Fold change | |
| a. Leaves | ||||||||||||||
| TP309 | 0.2a [±0.02] | 0.32a [±0.06] | 0.32a [±0.02] | 1.13a [±0.04] | (×1.0) | 0.13a [±0.02] | nd | 0.22a [±0.05] | 0.35a [±0.06] | (×1.0) | 0.85a [±0.05] | (×0.8) | 2.33a [±0.05] | (×0.9) |
| p1300 | 0.20a [±0.01] | 0.31a [±0.04] | 0.64a [±0.05] | 1.15a [±0.08] | 0.13a [±0.02] | nd | 0.24a [±0.05] | 0.37a [±0.05] | 1.08a [±0.06] | 2.60a [±0.17] | ||||
| 35S‐12a | 2.09c [±0.16] | 7.21c [±0.81] | 1.27c [±0.01] | 10.57c [±0.85] | (×9.2) | 0.11a [±0.01] | nd | 0.50b [±0.09] | 0.61b [±0.08] | (×1.7) | 62.37c [±7.13] | (×57.8) | 73.56c [±7.47] | (×28.3) |
| 35S‐31 | 1.49b [±0.44] | 5.13b [±0.89] | 1.05b [±0.18] | 7.67b [±1.41] | (×6.6) | 0.11a [±0.02] | trace | 0.35a,b [±0.12] | 0.51a,b [±0.15] | (×1.4) | 24.86b [±8.36] | (×23.0) | 33.04b [±9.76] | (×12.7) |
| b. Unpolished seeds | ||||||||||||||
| TP309 | 0.34b [±0.03] | 0.27a,b [±0.08] | 0.13b [±0.01] | 0.74b,c [±0.09] | (×1.6) | 0.07c [±0.01] | trace | 0.01b [±0.00] | 0.09b [±0.02] | (×1.9) | 1.06a,b [±0.25] | (×2.1) | 1.90b [±0.35] | (×1.9) |
| p1300 | 0.21a [±0.02] | 0.16a [±0.05] | 0.09a [±0.01] | 0.47a [±0.09] | 0.04a [±0.01] | nd | 0.01b [±0.00] | 0.05a [±0.01] | 0.50a [±0.14] | 1.02a [±0.20] | ||||
| 35S‐12a | 0.45c [±0.02] | 0.40c [±0.03] | 0.09a [±0.00] | 0.94c [±0.05] | (×2.0) | 0.06b [±0.00] | trace | 0.01b [±0.00] | 0.07a,b [±0.00] | (×1.4) | 2.15d [±0.29] | (×4.3) | 3.15d [±0.30] | (×3.1) |
| 35S‐31 | 0.33b [±0.04] | 0.29b,c [±0.03] | 0.09a [±0.01] | 0.72b [±0.07] | (×1.5) | 0.05b [±0.00] | nd | 0.01a [±0.00] | 0.06a [±0.00] | (×1.2) | 1.46b,c [±0.26] | (×2.9) | 2.23b,c [±0.32] | (×2.2) |
| c. Polished seeds | ||||||||||||||
| TP309 | 0.06c [±0.01] | 0.05b [±0.01] | 0.04a [±0.00] | 0.14b [±0.02] | (×1.7) | 0.01b [±0.00] | nd | 0.01a [±0.00] | 0.02a [±0.01] | (×1.0) | 0.12b [±0.07] | (×3.6) | 0.28b [±0.10] | (×2.1) |
| p1300 | 0.03a [±0.01] | 0.02a [±0.00] | 0.03a [±0.01] | 0.08a [±0.02] | 0.01a [±0.00] | nd | 0.01a [±0.00] | 0.02a [±0.00] | 0.03a [±0.01] | 0.13a [±0.02] | ||||
| 35S‐12a | 0.05b,c [±0.01] | 0.11d [±0.01] | 0.04a [±0.01] | 0.19c [±0.03] | (×2.3) | 0.01a,b [±0.00] | nd | 0.01a [±0.00] | 0.02a [±0.00] | (×1.1) | 0.08a,b [±0.01] | (×2.5) | 0.29b [±0.02] | (×2.2) |
| 35S‐31 | 0.04a,b [±0.01] | 0.06b,c [±0.01] | 0.04a [±0.01] | 0.14b [±0.02] | (×1.6) | 0.01a [±0.00] | nd | 0.00a [±0.01] | 0.01a [±0.01] | (×0.7) | 0.09a,b [±0.02] | (×3.6) | 0.23a,b [±0.03] | (×1.8) |
Average ± SD of four biological replicates. SD indicated in brackets. Tukey's multiple comparison test (P < 0.05). nd, not detected. trace: B6 vitamer content of at least one replicate could not be detected. Vitamin B6 content in wild‐type TP309 and in transgenic rice lines was compared with the empty vector control p1300 (fold change in brackets).
Low transgene expression in seeds accounts for the limited increase in vitamin B6 content
Given the disparity between the ability to enhance vitamin B6 contents in rice leaves but not to the same extent in polished seeds in this study, we decided to further probe rice vitamin B6 biosynthesis. Recent studies have predicted three catalytic PDX1 loci (Os7g01020, Os10g01080, and Os11g48080) in the rice genome (Moccand et al., 2014; Dell'Aglio et al., 2017). As they shared the highest similarity to Arabidopsis PDX1.3, they were annotated OsPDX1.3a‐c, respectively (Dell'Aglio et al., 2017). In the present study, we annotated the single homolog of PDX2 in rice (Os2g03740), named hereafter OsPDX2 (Figure 1). We analyzed the expression of OsPDX1.3a, OsPDX1.3b, OsPDX1.3c and OsPDX2 in leaves, unpolished and polished seeds of wild‐type TP309, as well as selected 35S transgenic lines and the empty vector control line. Interestingly, while OsPDX1.3a appears to be expressed ubiquitously in the tissues examined, OsPDX1.3b is substantially more abundant in leaves compared with seeds, whereas OsPDX1.3c is most abundant in seeds (Figure 6). These observations might suggest a specialization of OsPDX1.3b and OsPDX1.3c expression patterns in leaves and seeds, respectively. The expression of OsPDX1.3b was in agreement with publicly available expression data in Genevestigator (Hruz et al., 2008) (Figure S4) but the high expression of OsPDX1.3c in seeds was not noted in this resource. OsPDX2 appears to be expressed at similar levels in all three tissue samples (Figure 6). In general, expression levels of the endogenous genes were unaltered in either leaves, or unpolished or polished seeds in the transgenic lines (Figure 6).
Figure 6.

Real‐time quantitative PCR analysis of rice PDX gene expression in leaves, unpolished seeds, and polished seeds.
Expression levels of OsPDX1.3a (a), OsPDX1.3b (b), OsPDX1.3c (c) and OsPDX2 (d) in selected 35S rice lines (T2 generation) compared with the wild‐type (TP309) and empty vector control line (p1300). Average ± SD of three biological replicates. Tukey's multiple comparison test (P < 0.05) for each sample type (i.e., leaves, unpolished seeds, polished seeds). Statistics are not shown as no significant difference was found.
As the endogenous rice PDX transcripts appeared to be unaltered, we were prompted to investigate the Arabidopsis transgene expression at both the transcript and protein level in an effort to explain the differential accumulation of vitamin B6. In the 35S lines, while substantial transcript expression of Arabidopsis PDX1.1 and PDX2 was observed in all tissues examined, expression was higher (up to 10‐fold) in leaves compared with either unpolished or polished seeds (Figure 7a). We also examined protein levels using antibodies specific to Arabidopsis PDX1.1 and PDX2, respectively (Tambasco‐Studart et al., 2007; Raschke et al., 2011). Both Arabidopsis PDX1.1 and PDX2 could be clearly detected in leaves of the 35S lines examined, immunostaining of which was not observed in the empty vector control line (Figure 7b). Interestingly, however, the Arabidopsis PDX1.1 protein could not be detected in unpolished or polished seeds of the transgenic lines (Figure 7b). While this may be accounted for by the lower transcript levels in these tissues, the inability to immunolabel the protein despite the high detection level in the leaves suggests that another mechanism contributes to the low level of protein in the seed tissue. The accumulation of PDX2 could not be deciphered due to cross‐reacting protein in these tissues (Figure 7b).
Figure 7.
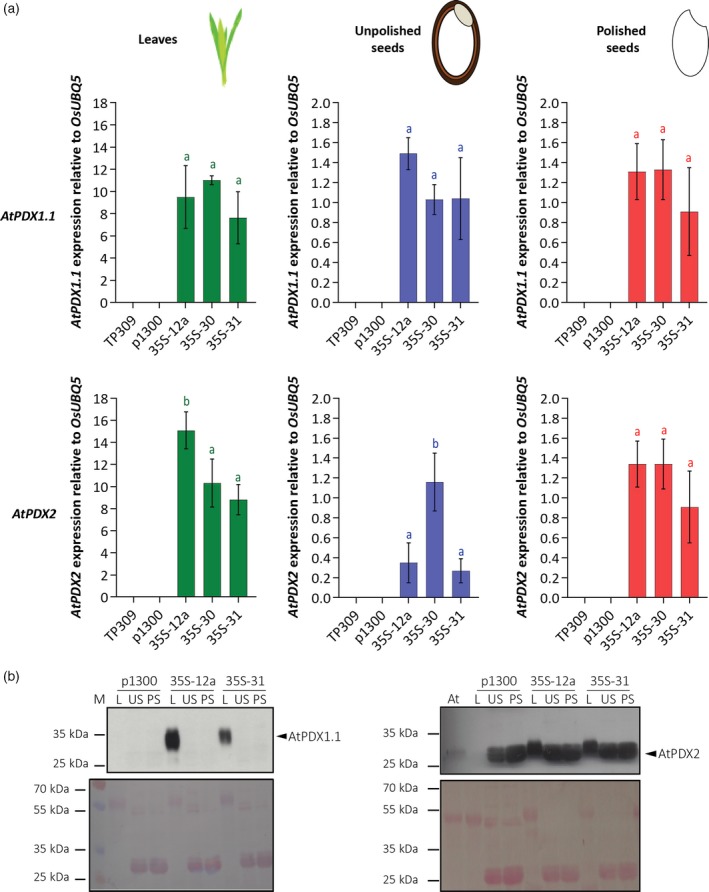
PDX transgene expression and protein accumulation in leaves, unpolished seeds, and polished seeds.
(a) AtPDX1.1 and AtPDX2 transcript expression levels in 35S transgenic rice lines (T2 generation) compared with wild‐type (TP309) and the empty vector control (p1300). Average ± SD of three biological replicates. Tukey's multiple comparison test (P < 0.05) for each tissue.
(b) Western blot analysis of AtPDX1.1 and AtPDX2 protein abundance (upper panel) and Ponceau staining of the nitrocellulose membrane (lower panel) in control and transgenic rice lines. Here, 50 μg of total rice proteins and 30 μg of total Arabidopsis leaf proteins were probed with peptide antibodies specific to AtPDX1.1 (Raschke et al., 2011) or AtPDX2 (Tambasco‐Studart et al., 2007). M, protein molecular‐weight marker; L, leaves; US, unpolished seeds; PS, polished seeds; At, Arabidopsis thaliana.
Discussion
In the present study, we report vitamin B6 pathway engineering in a monocot species and its impact on plant growth and development. Considerable enhancement of vitamin B6 levels could be achieved by expressing the Arabidopsis PDX1.1 and PDX2 genes in rice. This enhancement was most pronounced in rice leaves but was more limited in seeds. This may be explained by the higher transcript levels of the AtPDX genes observed in leaves rather than seeds. Indeed, the AtPDX1.1 and AtPDX2 proteins could be detected in leaves but not in seeds. Although, the CaMV35S promoter is considered to be less active in seed tissue compared with leaf tissue, there was a significant increase of AtPDX transcripts in both unpolished and polished seeds from the selected transgenic rice lines and to similar levels. Yet, an increase in vitamin B6 was only observed in transgenic unpolished seeds and no significant enrichment could be detected in the rice endosperm. Previous studies show that transgenes driven by the 35S promoter translate into detectable levels of the corresponding proteins in rice seeds (Furtado et al., 2008; Long et al., 2013). However, our immunochemical analyses failed to detect the Arabidopsis PDX proteins in either polished or unpolished seeds. This would seem to suggest that the accumulation of the protein is the limiting parameter for vitamin B6 production in this organ and may be most pronounced in the endosperm tissue. This observation warrants further investigation and could be suggestive of the fact that these proteins are tightly controlled (or repressed) in this tissue compared to leaves. In line with this hypothesis, we generated a small pool of rice lines expressing AtPDX1.1 and AtPDX2 under the control of the endosperm‐specific globulin (Glob) promoter (included in Figure S1). We could observe transgene expression in these Glob lines in seed tissue (Figure S5a, b), and notably at similar levels to those observed in the 35S lines (compare with Figure 7a). By contrast, expression of the AtPDX transgenes was not detected in leaf tissue of the rice Glob lines, as expected (Figure S5a, b). However, as for the 35S lines, the AtPDX proteins could not be detected in seed tissue of the Glob lines (Figure S5c, d). Moreover, while we did observe an increase in vitamin B6 content in the seeds of the Glob lines generated (albeit a small pool) (Figure S5e, f), the increase was similar to that observed in the seeds of the 35S lines (Figure 2e, f). Also similar to the 35S lines, the levels of vitamin B6 in polished seeds of the Glob lines was not above the wild‐type level (Figure S5e, f). As expected there was no increase in vitamin B6 content in the leaves or roots of the Glob lines compared with the control lines (Figure S5g, h). Although a more rigorous study with the use of endosperm‐specific promoters is required in the future, our analysis here suggests that enhancing seed vitamin B6 content could be constrained by the ability to express the PDX1 proteins and may indicate an endogenous regulatory mechanism of these proteins in rice seeds. Of course, a potentially limited availability of precursors in endosperm tissue might also represent another bottleneck for upregulated vitamin B6 biosynthesis therein.
Increasing vitamin B6 levels had very little impact on rice growth and development. Nonetheless, a significant yield penalty was observed by assessing the mass of 100 seeds in selected transgenic rice lines. Expression of PDX transgenes in other plant species, including Arabidopsis, cassava and potato (Raschke et al., 2011; Li et al., 2015; Bagri et al., 2018) was not reported to be associated with yield penalty. However, previous work in Arabidopsis showed a positive correlation between vitamin B6 content and seed size, yet a yield penalty in terms of number of seeds was observed (Raschke et al., 2011). Recent work in maize indicated that vitamin B6 is essential for embryo development but has a limited role in endosperm development (Yang et al., 2017), therefore the mechanism behind the observations in rice may not be restricted to monocots and may be species‐dependent. The vitamin B6 biosynthesis de novo pathway in rice has so far only been partially described. Based on homology with genes previously discovered in Arabidopsis (Tambasco‐Studart et al., 2007; Raschke et al., 2011), there are three homologs for PDX1 in rice (PDX1.3a‐c) (Dell'Aglio et al., 2017), and we identified a single homolog for PDX2. The expression profiles of OsPDX1.3b and OsPDX1.3c in leaves and in seeds, respectively, suggest organ‐specific expression for these two homologs. The expression level of OsPDX1.3b in photosynthetic tissue (i.e., leaf) was considerably higher than the other two PDX1.3 genes. It is of note in the present study, that while we demonstrate that AtPDX1.1 and AtPDX2 from Arabidopsis are functional in rice based on the significant increase in total vitamin B6 content in leaves, they have little impact on the endogenous PDX gene expression levels in rice.
The salt stress assays carried out here suggest that increased vitamin B6 content in unpolished seeds and leaves of transgenic rice lines does not impact their performance under these conditions. Similarly, disease symptom evaluation upon inoculation with the bacterial leaf blight pathogen Xanthomonas oryzae pv. oryzae did not reveal significant differences between control lines and a transgenic line accumulating high levels of vitamin B6. This contrasts with the previously reported positive contribution of vitamin B6 to plant responses to environmental stress in various other species, which were notably members of the eudicot group (Arabidopsis, tomato) (Raschke et al., 2011; Zhang et al., 2014, 2015). However, it should be noted that the evidence for vitamin B6 contribution to plant performance under biotic and abiotic stresses mostly comes from mutant lines or virus induced gene‐silenced lines impaired in vitamin B6 biosynthesis (Titiz et al., 2006; Zhang et al., 2014, 2015). It remains to be determined whether the positive contribution of vitamin B6 to stress responses in planta is due to a direct effect based on its reported antioxidant properties and/or an indirect effect based on its role as a coenzyme, possibly for an enzyme contributing to antioxidant capacities (Fitzpatrick, 2011). Whether the non‐responsiveness to environmental stress observed here is species or group (eudicots versus monocots)–dependent would require analysis in other monocot and eudicot plant species with enhanced vitamin B6 content.
Interestingly, the B6 vitamer profiles of rice wild‐type TP309 leaves determined in this study differ from those measured in cassava wild‐type cv.60444 leaves (Li et al., 2015). In particular, the contribution of PN‐Glu to total vitamin B6 is lower for rice leaves (36%) as compared to cassava leaves (57%). Noticeably, PN‐Glu represents a major proportion of vitamin B6 pools in rice unpolished (56%) and polished (43%) seeds. The profiles of B6 vitamers in transgenic cassava (Li et al., 2015) and rice (this study) upon ectopic expression of AtPDX1.1 and AtPDX2 transgenes indicate that the increase in vitamin B6 biosynthesis translates into similar accumulation of B6 vitamers, that is, PN‐Glu also represents a major accumulating form of vitamin B6 (Table 2). The function of PN‐Glu in planta is unknown, but it is suggested to be an unreactive/inert form of vitamin B6 (Gregory, 1998) and its preferential accumulation in these transgenic rice and cassava lines suggests that it could participate in the preservation of vitamin B6 homeostasis. Indeed, balancing of B6 vitamers has recently been demonstrated to be essential for normal plant development (Colinas et al., 2016). Due to the reduced bioavailability of glucosylated vitamers in humans (approximately 50%, Gregory, 2012), the identification of genetic factors and conditions controlling the interconversion of glucosylated vitamers to the non‐glucosylated equivalents will be of high importance for future strategies targeted toward B6 vitamin biofortification of crops. In general, there have been contrasting results for metabolic engineering of B vitamins in rice endosperm. Transgenic rice expressing folate (vitamin B9) biosynthetic genes from Arabidopsis (AtGTPCHI and AtADCS) under the control of Glob and glutelin B 1 promoters, respectively, displayed a substantial increase of folate content in rice endosperm (Storozhenko et al., 2007), which was further accentuated in a later study by the expression of a synthetic folate binding protein to enhance stability (Blancquaert et al., 2015). In contrast, constitutive overexpression of endogenous vitamin B1 biosynthesis genes (OsTHIC and OsTHI1) in rice led to significant increases in leaves and to a more limited extent in unpolished seed, but most of the gain was lost during polishing (Dong et al., 2016). While identifying bottlenecks in vitamin B6 biosynthesis in the rice endosperm should be prioritized, future strategies to enrich vitamin B6 content in rice endosperm might also benefit from the identification and characterization of vitamin B6 transporters, as well as other factors contributing to vitamin accumulation and stabilization, such as vitamin B6 binding proteins.
In the present study we demonstrated the potential of the PDX transgene approach to considerably enhance the vitamin B6 content in rice and with no impact on endogenous PDX gene expression or overall growth and development, with the exception of a modest impact on seed yield. This study in a monocot species has provided some interesting insights into potential regulatory pathways of vitamin B6 sequestration in plants and opened up areas for future fundamental investigation as well as applied studies that will enable significant vitamin B6 enrichment in rice endosperm.
Experimental procedures
Binary vector construction
The constitutive expression vector containing p35S::AtPDX1.1‐p35S::AtPDX2 (Arabidopsis thaliana PDX1.1, At2g38230 and PDX2, At5g60540) previously described (Li et al., 2015) was used in this study. The endosperm‐specific expression vector containing pGlob::AtPDX1.1‐pGlob::AtPDX2 was generated by substituting the cauliflower mosaic virus (CaMV) 35S promoters in p35S::AtPDX1.1‐p35S::AtPDX2 with a 1.0 kb Globulin (Glob) promoter sequence from rice (le Qu and Takaiwa, 2004). Both expression vectors were constructed in the pCAMBIA1300 backbone. A schematic representation of the T‐DNA region is provided in Figure 2.
Rice transformation and molecular characterization of transgenic lines
Binary vectors were introduced into Agrobacterium tumefaciens strain EHA105 (Hood et al., 1993) and rice transformation, selection on hygromycin and regeneration were conducted according to a previously established protocol (Nishimura et al., 2006). Isolation of genomic DNA from rice leaves was performed based on a previously established protocol (Sheu et al., 1996). Frozen ground tissue was suspended in 900 μL of urea extraction buffer (without sarkosine) and extracted with 1 volume of phenol:chloroform:isoamylalcohol (25:24:1, pH 7.5–8.0) at room temperature for 15 min. The recovered aqueous phase was mixed with 0.1 volume of 3 m sodium acetate (pH 5.2), 1 μL RNase A (20 mg mL−1) and 1 volume of isopropanol. The extract was incubated for 20 min at −80°C and centrifuged at 16 100 g for 3 min at room temperature. The DNA pellet was washed with 70% followed by 100% ethanol, vacuum dried and re‐suspended in sterile Milli‐Q water. The identification of transformants containing both AtPDX1.1 and AtPDX2 transgenes was done by PCR. Primers are detailed in Table S1. Southern blot analyses were conducted for the confirmation of T‐DNA insertion and determination of transgene copy number, using a DIG‐dUTP‐labeled hptII probe, synthesized by PCR (PCR DIG probe synthesis kit; Roche Diagnostics AG, Risch‐Rotkreuz, Switzerland) (Table S1), following a previously described procedure (Vanderschuren et al., 2009).
Plant material
Dried seeds of Oryza sativa ssp. japonica cv. Taipei 309 were de‐husked and surface‐sterilized in 70% ethanol for 30 sec, then in a 1.5% sodium hypochlorite solution including 0.01% Tween 20 for 30 min under vigorous shaking. Surface‐sterilized seeds were rinsed five times with sterile water before being placed in sterile plastic jars containing Murashige and Skoog (MS) medium (1× MS salts including vitamins (Duchefa, Haarlem, Netherlands), 3% (w/v) sucrose, 0.3% (w/v) gelrite; pH 5.8) containing 50 mg L−1 of hygromycin (Carl Roth, Karlsruhe, Germany) for selection of transgenic lines. Seeds were stratified in the dark at 28°C for 48 h before being transferred to a climate chamber (28°C under a 16/8 h light/dark regime) for 12 days. Plantlets were subsequently transferred to soil and grown under greenhouse conditions (12 h light at 30°C and 70% humidity, 12 h dark at 20°C and 60% humidity). Either T2 or T3 generation plants were used for analyses, as indicated. Nine replicates per line were distributed in three pots of three replicates, organized in three blocks in the greenhouse. The three or four replicates analyzed came from the three blocks and the different tissues were sampled as follows: leaves, three leaves from three different tillers of 45‐day‐old plants (vegetative stage) were pooled and frozen in liquid nitrogen; roots, during harvest, root samples were collected, rinsed with tap water and frozen in liquid nitrogen; fresh mature seeds, 1–2 weeks before harvesting, mature seeds were sampled. Some seeds were de‐husked only (referred to as fresh unpolished seeds) and frozen in liquid nitrogen. Other seeds were de‐husked and the aleurone layer was peeled off using forceps and a razor blade (Kuwano et al., 2011). The inner starchy endosperm (referred to as fresh polished seeds) and the embryo were collected separately and frozen in liquid nitrogen. Dry mature seed samples were collected from harvesting fully ripened panicles and drying for 5 days at 37°C. The dried seeds were either de‐husked only (referred to as dry unpolished seeds) and stored at −80°C or de‐husked and polished for 2 min (seed polisher PEARLEST, Kett) to obtain the starchy endosperm (referred to as dry polished seeds) and stored at −80°C.
Stress treatments
Salt stress
For the salt stress treatment, de‐husked seeds were germinated in water for 4 days, including 48 h in the dark to synchronize germination. Rice seedlings were then hydroponically grown in Yoshida's solution (0.7 mm K2SO4, 2 mm Ca(NO3)2, 0.1 mm KH2PO4, 0.5 mm MgSO4, 0.1 mm KCl, 10 μm H3BO3, 0.5 μm MnSO4, 0.2 μm CuSO4, 0.01 μm (NH4)Mo7O24, 0.5 μm ZnSO4, 100 μm Fe‐EDTA) (Kobayashi et al., 2005) in a climate chamber (28°C under a 16/8 h light/dark regime) for 6 days. Four concentrations of NaCl (50, 100, 150, 200 mm) were initially evaluated. The concentration of 150 mm NaCl was used in this study because of the intermediate visible effects on rice plantlet morphology. Salt stress treatment was applied for 15 days using Yoshida's solution supplemented with 150 mm NaCl, with changes of the solution every 2 days. Plantlets were grown simultaneously in Yoshida's solution without salt as controls.
Leaf blight assay
Xanthomonas oryzae pv. oryzae pathogenicity assays were performed under greenhouse conditions (12 h light at 28°C and 80% humidity, 12 h dark at 25°C and 70% humidity). TP309 wild‐type and transgenic line 35S‐12a were grown for 4–5 weeks and inoculated with two different Xanthomonas oryzae pv. oryzae strains: PX071 (Leach et al., 1992) and BAI3 (Gonzalez et al., 2007a). Leaf clip inoculation was performed as previously described (Kauffman et al., 1973) with bacterial suspensions grown up to a cell density value of OD600 nm = 0.2. Lesion length was measured 15 days post‐inoculation.
Vitamin B6 quantification
Yeast bioassay
Yeast bioassays for vitamin B6 content were performed according to a method previously established with the Saccharomyces carlsbergensis American Type Culture Collection 9080 strain (Tambasco‐Studart et al., 2005). The amount of plant material required for vitamin B6 extraction varied between organs: leaves (50 mg), roots (100 mg), dry seeds (50 mg), fresh seeds (20 mg), embryo (20 mg). Frozen ground tissues were re‐suspended in 20 mm sulfuric acid (ratio: 100 mg tissue/1 mL extraction buffer), incubated for 30 min in the dark at room temperature and the extract was sterilized for 1 h at 100°C. After extraction, the solution was adjusted to pH 5.7 using 3 m sodium acetate and centrifuged. The supernatant was then treated with acid phosphatase (0.2 U/10 μL in 50 μL plant extract) (Sigma‐Aldrich, St. Louis, MI, USA) and β‐glucosidase (0.2 U/10 μL in 50 μL plant extract) (Sigma‐Aldrich, St. Louis, MI, USA) for 12–15 h at 37°C to convert phosphorylated and glucosylated forms of vitamin B6 to free forms. Total vitamin B6 content was calculated using the linear range of a dose−response curve constructed with known amounts of commercial pyridoxine hydrochloride (Sigma‐Aldrich, St. Louis, MI, USA).
High‐performance liquid chromatography measurement
Quantification of B6 vitamers was performed essentially according to a previously established HPLC method with minor modifications (Szydlowski et al., 2013). B6 vitamers were extracted from frozen ground tissues using 50 mm ammonium acetate pH 4.0 (ratio: 100 mg tissue/200 μL ammonium acetate). The extract was vortexed for 10 min, centrifuged at 16 100 g for 15 min at room temperature and the supernatant incubated for 3 min at 99°C. The extract was again centrifuged at 16 100 g for 15 min at room temperature and the supernatant was used for analysis. The extract was analyzed on an Agilent Technologies 1200 HPLC instrument to determine B6 vitamer profiles, using a Sunfire C18 column (Waters, Milford, MA, USA), 4.6 × 150 mm, 3.5 μm particle diameter. The chromatography and detection method used was previously described (Szydlowski et al., 2013). The amount of glucosylated PN vitamer (PN‐Glu) was extrapolated from the increase in PN content observed after treatment of an equal amount of the original extract with β‐glucosidase (Sigma‐Aldrich, catalog number 49 290, 10 μL of 15 mg mL−1 stock dissolved in 50 mm ammonium acetate pH 4.0) for 2–4 h at 37°C and then boiled as above, prior to injection. Vitamer contents were calculated using the linear range of standard curves constructed with known amounts of PN, PM, PL, PNP, PMP and PLP (Colinas et al., 2016). Measurements for vitamers showing saturated signals on the largest injection volume were re‐injected with a smaller volume and/or diluted prior to analysis.
Gene expression quantification using real‐time quantitative PCR (RT‐qPCR)
RNA extraction
Total RNA from leaves was extracted according to a previously established method with some modifications (Chang et al., 1993). Frozen ground tissue was mixed with 1 mL of extraction buffer (2% cetyl trimethylammonium bromide, 2% polyvinylpyrrolidone K‐30, 100 mm Tris HCl, 25 mm ethylenediaminetetraacetic acid, 2 m NaCl, 0.5 g L−1 spermidine) and 2% β‐mercaptoethanol, incubated at 50°C with shaking at 400–500 rpm for 15 min and extracted twice with 1 volume of chloroform:isolamylalcohol (24:1, pH 7.5–8.0). Nucleic acids from the recovered aqueous phase were precipitated with 1 mL of ice cold absolute ethanol for 30 min at −80°C and centrifuged at 16 100 g for 30 min at 4°C. The pellet was washed with 80% ethanol and re‐suspended in diethylpyrocarbonate (DEPC)‐treated water. RNA was precipitated in 2 m lithium chloride overnight at −20°C and centrifuged at 16 100 g for 30 min at 4°C. The RNA pellet was washed with 80 and 100% ethanol, vacuum dried and re‐suspended in DEPC‐treated water.
RNA extraction from fresh rice seeds was performed using an established protocol (Singh et al., 2003).
RT‐qPCR analysis
cDNA was synthesized using random hexamer oligonucleotide primers with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific AG, Basel, Switzerland) according to the manufacturer's instructions, with 2 μg total RNA for leaves and 3 to 5 μg total RNA for fresh seeds. RT‐qPCR reactions were performed using the LightCycler 480 II (Roche Diagnostics AG, Risch‐Rotkreuz, Switzerland) system and Fast SYBR® Green Master Mix (Applied Biosystems, altham, MA, USA, Thermo Fisher Scientific, Basel, Switzerland). The reaction mixture contained 1 μL of DEPC‐treated water, 1 μm of each primer, 5 μL SYBR master mix and 2 μL cDNA diluted 10‐fold for leaves and undiluted for seeds. PCR conditions were as follows: initial denaturation for 2 min at 95°C followed by 40 cycles of denaturation for 10 sec at 95°C, annealing for 20 sec at 60°C and extension for 30 sec at 72°C. Relative target gene expression levels were normalized to the rice reference gene UBQ5 (Jain et al., 2006) and calculated using the delta delta CT method (Livak and Schmittgen, 2001). Sequences of primers used for RT‐qPCR analysis are detailed in Table S2.
In silico identification of the rice PDX2 gene
The rice ortholog of Arabidopsis thaliana AtPDX2 (At5g60540) was identified by BLASTing the Arabidopsis protein sequences from the TAIR10 database (Lamesch et al., 2012) against the available translated Oryza sativa v7_JGI genome in Phytozome (Ouyang et al., 2007).
Immunochemical analyses
Protein extraction was performed according to a previously established protocol with some modifications (Svozil et al., 2014). Total proteins were extracted from frozen ground tissues in 1 mL sodium dodecyl sulfate (SDS) buffer (4% SDS, 40 mm Tris‐base, 5 mm MgCl2, and 2 × protease inhibitor mix (Roche, Basel, Switzerland)) for 20 min at room temperature. Cell debris was centrifuged for 15 min at 16 100 g and the supernatant transferred to a fresh tube. Protein concentration was determined using the Pierce™ BCA Protein Assay Kit (Thermo Fischer Scientific). Immunodetection was based on a previously established protocol for Arabidopsis (Titiz et al., 2006), including some modifications. Proteins were separated on 12% SDS‐PAGE gels and subsequently transferred onto nitrocellulose membranes. Membranes were blocked in Tris‐buffer saline solution containing 0.05% Tween‐20 and 20% milk powder overnight at 4°C. AtPDX1.1 protein was detected with specific antibodies raised against the peptide GEGAMTETKQKSP (Raschke et al., 2011) and AtPDX2 protein was detected with antibodies raised against the recombinant Arabidopsis protein (Tambasco‐Studart et al., 2007). Blots were incubated with primary antibodies (1:1000 dilutions) for 1 h and then probed with a Goat Anti‐Rabbit IgG (H+L)‐HRP conjugated secondary antibody (Bio‐Rad, Hercules, CA, USA) (dilution 1:5000) for 1 h, before chemiluminescent detection.
Statistical analyses
Detailed description of statistical analysis is provided in Data S1.
Data statement
Supporting data can be accessed as Supplementary Information on The Plant Journal website.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Figure S1. Molecular characterization of the generated dual‐expressing AtPDX1.1 and AtPDX2 transgenic rice lines in the T0 generation.
Figure S2. Analysis of total vitamin B6 contents of transgenic rice lines in the T1 generation.
Figure S3. Assignment of a glucosylated B6 vitamer in rice leaf extracts.
Figure S4. Rice PDX gene expression patterns across different tissues.
Figure S5. PDX transgene expression, protein accumulation and vitamin B6 content in the rice Glob lines.
Table S1. Primers used for the molecular characterization of generated transgenic rice lines.
Table S2. Primers used for real‐time quantitative PCR analysis.
Data S1. Statistical analysis.
Acknowledgements
Financial support is gratefully acknowledged from the Swiss National Science Foundation (grants 31003A‐141117/1 and 31003A‐162555 to TBF; grant 31003A‐140911 to WG, HV, and TBF), the VELUX Foundation, the University of Geneva and the ETH Zurich. The authors thank Irene Zurkirchen (ETH Zurich) for support in the greenhouse.
Contributor Information
Teresa B. Fitzpatrick, Email: theresa.fitzpatrick@unige.ch.
Hervé Vanderschuren, Email: herve.vanderschuren@uliege.be.
References
- Bagri, D.S. , Upadhyaya, D.C. and Kumar, A. (2018) Overexpression of PDX‐II gene in potato (Solanum tuberosum L.) leads to the enhanced accumulation of vitamin B6 in tuber tissues and tolerance to abiotic stresses. Plant Sci. 272, 267–275. [DOI] [PubMed] [Google Scholar]
- Bhullar, N.K. and Gruissem, W. (2013) Nutritional enhancement of rice for human health: the contribution of biotechnology. Biotechnol. Adv. 31, 50–57. [DOI] [PubMed] [Google Scholar]
- Bilski, P. , Li, M.Y. , Ehrenshaft, M. , Daub, M.E. and Chignell, C.F. (2000) Vitamin B6 (pyridoxine) and its derivatives are efficient singlet oxygen quenchers and potential fungal antioxidants. Photochem. Photobiol. 71, 129–134. [DOI] [PubMed] [Google Scholar]
- Blancquaert, D. , Van Daele, J. , Strobbe, S. , Kiekens, F. , Storozhenko, S. , De Steur, H. , Gellynck, X. , Lambert, W. , Stove, C. and Van Der Straeten, D. (2015) Improving folate (vitamin B9) stability in biofortified rice through metabolic engineering. Nat. Biotechnol. 33, 1076–1078. [DOI] [PubMed] [Google Scholar]
- Burns, K.E. , Xiang, Y. , Kinsland, C.L. , McLafferty, F.W. and Begley, T.P. (2005) Reconstitution and biochemical characterization of a new pyridoxal‐5′‐phosphate biosynthetic pathway. J. Am. Chem. Soc. 127, 3682–3683. [DOI] [PubMed] [Google Scholar]
- Chang, S. , Puryear, J. and Cairney, J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116. [Google Scholar]
- Chen, H. and Xiong, L. (2005) Pyridoxine is required for post‐embryonic root development and tolerance to osmotic and oxidative stresses. Plant J. 44, 396–408. [DOI] [PubMed] [Google Scholar]
- Chen, H. and Xiong, L. (2009) Enhancement of vitamin B6 levels in seeds through metabolic engineering. Plant Biotechnol. J. 7, 673–681. [DOI] [PubMed] [Google Scholar]
- Colinas, M. and Fitzpatrick, T.B. (2015) Natures balancing act: examining biosynthesis de novo, recycling and processing damaged vitamin B metabolites. Curr. Opin. Plant Biol. 25, 98–106. [DOI] [PubMed] [Google Scholar]
- Colinas, M. and Fitzpatrick, T.B. (2016) Interaction between vitamin B6 metabolism, nitrogen metabolism and autoimmunity. Plant Signal. Behav. 11, e1161876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colinas, M. , Eisenhut, M. , Tohge, T. , Pesquera, M. , Fernie, A.R. , Weber, A.P. and Fitzpatrick, T.B. (2016) Balancing of B6 vitamers is essential for plant development and metabolism in Arabidopsis. Plant Cell, 28, 439–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon, A. , Miersch, O. , Felix, G. , Camp, R.G. and Apel, K. (2005) Concurrent activation of cell death‐regulating signaling pathways by singlet oxygen in Arabidopsis thaliana . Plant J. 41, 68–80. [DOI] [PubMed] [Google Scholar]
- Dell'Aglio, E. , Boycheva, S. and Fitzpatrick, T.B. (2017) The pseudoenzyme PDX1.2 sustains vitamin B6 biosynthesis as a function of heat stress. Plant Physiol. 174, 2098–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow, S.A. , Walls, A.A. and Daub, M.E. (2005) Regulation of biosynthetic genes and antioxidant properties of vitamin B6 vitamers during plant defense responses. Physiol. Mol. Plant Pathol. 66, 244–255. [Google Scholar]
- Denslow, S.A. , Rueschhoff, E.E. and Daub, M.E. (2007) Regulation of the Arabidopsis thaliana vitamin B6 biosynthesis genes by abiotic stress. Plant Physiol. Biochem. 45, 152–161. [DOI] [PubMed] [Google Scholar]
- Di Salvo, M. , Safo, M. and Contestabile, R. (2012) Biomedical aspects of pyridoxal 5′‐phosphate availability. Front. Biosci. (Elite Ed.) 4, 897–913. [DOI] [PubMed] [Google Scholar]
- Dong, W. , Thomas, N. , Ronald, P.C. and Goyer, A. (2016) Overexpression of thiamin biosynthesis genes in rice increases leaf and unpolished grain thiamin content but not resistance to Xanthomonas oryzae pv. oryzae . Front. Plant Sci. 7, 616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick, T.B. (2011) Vitamin B6 in plants: more than meets the eye. Adv. Bot. Res. 59, 1–38. [Google Scholar]
- Fitzpatrick, T.B. , Amrhein, N. , Kappes, B. , Macheroux, P. , Tews, I. and Raschle, T. (2007) Two independent routes of de novo vitamin B6 biosynthesis: not that different after all. Biochem. J. 407, 1–13. [DOI] [PubMed] [Google Scholar]
- Fudge, J. , Mangel, N. , Gruissem, W. , Vanderschuren, H. and Fitzpatrick, T.B. (2017) Rationalising vitamin B6 biofortification in crop plants. Curr. Opin. Biotechnol. 44, 130–137. [DOI] [PubMed] [Google Scholar]
- Furtado, A. , Henry, R.J. and Takaiwa, F. (2008) Comparison of promoters in transgenic rice. Plant Biotechnol. J. 6, 679–693. [DOI] [PubMed] [Google Scholar]
- Gonzalez, C. , Szurek, B. , Manceau, C. , Mathieu, T. , Sere, Y. and Verdier, V. (2007a) Molecular and pathotypic characterization of new Xanthomonas oryzae strains from West Africa. Mol. Plant Microbe Interact. 20, 534–546. [DOI] [PubMed] [Google Scholar]
- Gonzalez, E. , Danehower, D. and Daub, M.E. (2007b) Vitamer levels, stress response, enzyme activity, and gene regulation of Arabidopsis lines mutant in the pyridoxine/pyridoxamine 5′‐phosphate oxidase (PDX3) and the pyridoxal kinase (SOS4) genes involved in the vitamin B6 salvage pathway. Plant Physiol. 145, 985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Grebmer, K. , Saltzman, A. , Birol, E. , Wiesmann, D. , Prasai, N. , Yin, S. , Yohannes, Y. , Menon, P. , Thompson, J. and Sonntag, A. (2014) 2014 Global hunger index: The challenge of hidden hunger. Welthungerhilfe, International Food Policy Research Institute, and Concern Worldwide. Bonn, Washington, DC, and Dublin.
- Gregory, J.F. III (1998) Nutritional properties and significance of vitamin glycosides. Annu. Rev. Nutr. 18, 277–296. [DOI] [PubMed] [Google Scholar]
- Gregory, J.F. III (2012) Accounting for differences in the bioactivity and bioavailability of vitamers. Food Nutr. Res. 56, 5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, J.F. III and Ink, S.L. (1987) Identification and quantification of pyridoxine beta‐glucoside as a major form of vitamin B6 in plant‐derived foods. J. Agric. Food Chem. 35, 76–82. [Google Scholar]
- Hanson, A.D. , Beaudoin, G.A. , McCarty, D.R. and Gregory, J.F. (2016) Does abiotic stress cause functional B vitamin deficiency in plants? Plant Physiol. 172, 2082–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux, M. , Ksas, B. , Szewczyk, A. , Rumeau, D. , Franck, F. , Caffarri, S. and Triantaphylides, C. (2009) Vitamin B6 deficient plants display increased sensitivity to high light and photo‐oxidative stress. BMC Plant Biol. 9, 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann, H. and Mooney, S. (2010) Vitamin B6: a molecule for human health? Molecules, 15, 442–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero, S. and Daub, M.E. (2007) Genetic manipulation of vitamin B6 biosynthesis in tobacco and fungi uncovers limitations to up‐regulation of the pathway. Plant Sci. 172, 609–620. [Google Scholar]
- Herrero, S. , Gonzalez, E. , Gillikin, J.W. , Velez, H. and Daub, M.E. (2011) Identification and characterization of a pyridoxal reductase involved in the vitamin B6 salvage pathway in Arabidopsis. Plant Mol. Biol. 76, 157–169. [DOI] [PubMed] [Google Scholar]
- Hood, E. , Gelvin, S. , Melchers, L. and Hoekema, A. (1993) New Agrobacterium helper plasmids for gene‐transfer to plants. Transgenic Res. 2, 208–218. [Google Scholar]
- Hruz, T. , Laule, O. , Szabo, G. , Wessendorp, F. , Bleuler, S. , Oertle, L. , Widmayer, P. , Gruissem, W. and Zimmermann, P. (2008) Genevestigator v3: a reference expression database for the meta‐analysis of transcriptomes. Adv. Bioinformatics, 2008, 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, S. , Zhang, J. , Wang, L. and Huang, L. (2013) Effect of abiotic stress on the abundance of different vitamin B6 vitamers in tobacco plants. Plant Physiol. Biochem. 66, 63–67. [DOI] [PubMed] [Google Scholar]
- Jain, M. , Nijhawan, A. , Tyagi, A.K. and Khurana, J.P. (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real‐time PCR. Biochem. Biophys. Res. Commun. 345, 646–651. [DOI] [PubMed] [Google Scholar]
- Kauffman, H.E. , Reddy, A.P.K. , Hsieh, S.P.Y. and Merca, S.D. (1973) An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae . Plant Dis. Rep. 57, 537–541. [Google Scholar]
- Kennedy, G. , Burlingame, B. and Nguyen, V.N. (2003) Nutritional contribution of rice and impact of biotechnology and biodiversity in rice‐consuming countries. Proceedings of the 20th Session of the International Rice Commission: FAO corporate document repository. Bangkok.
- Kobayashi, T. , Suzuki, M. , Inoue, H. , Itai, R.N. , Takahashi, M. , Nakanishi, H. , Mori, S. and Nishizawa, N.K. (2005) Expression of iron‐acquisition‐related genes in iron‐deficient rice is co‐ordinately induced by partially conserved iron‐deficiency‐responsive elements. J. Exp. Bot. 56, 1305–1316. [DOI] [PubMed] [Google Scholar]
- Kuwano, M. , Masumura, T. and Yoshida, K.T. (2011) A novel endosperm transfer cell‐containing region‐specific gene and its promoter in rice. Plant Mol. Biol. 76, 47–56. [DOI] [PubMed] [Google Scholar]
- Lamesch, P. , Berardini, T.Z. , Li, D. et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, J.E. , Rhoads, M.L. , Vera Cruz, C.M. , White, F.F. , Mew, T.W. and Leung, H. (1992) Assessment of genetic diversity and population structure of Xanthomonas oryzae pv. oryzae with a repetitive DNA element. Appl. Environ. Microbiol. 58, 2188–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K.T. , Moulin, M. , Mangel, N. , Albersen, M. , Verhoeven‐Duif, N.M. , Ma, Q. , Zhang, P. , Fitzpatrick, T.B. , Gruissem, W. and Vanderschuren, H. (2015) Increased bioavailable vitamin B6 in field‐grown transgenic cassava for dietary sufficiency. Nat. Biotechnol. 33, 1029–1032. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Long, X. , Liu, Q. , Chan, M. , Wang, Q. and Sun, S.S. (2013) Metabolic engineering and profiling of rice with increased lysine. Plant Biotechnol. J. 11, 490–501. [DOI] [PubMed] [Google Scholar]
- Lucca, P. , Poletti, S. and Sautter, C. (2006) Genetic engineering approaches to enrich rice with iron and vitamin A. Physiol. Plant. 126, 291–303. [Google Scholar]
- Lum, H.K. , Kwok, F. and Lo, S.C. (2002) Cloning and characterization of Arabidopsis thaliana pyridoxal kinase. Planta, 215, 870–879. [DOI] [PubMed] [Google Scholar]
- Meng, F. , Wei, Y. and Yang, X. (2005) Iron content and bioavailability in rice. J. Trace Elem. Med. Biol. 18, 333–338. [DOI] [PubMed] [Google Scholar]
- Moccand, C. , Kaufmann, M. and Fitzpatrick, T.B. (2011) It takes two to tango: defining an essential second active site in pyridoxal 5′‐phosphate synthase. PLoS One, 6, e16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moccand, C. , Boycheva, S. , Surriabre, P. , Tambasco‐Studart, M. , Raschke, M. , Kaufmann, M. and Fitzpatrick, T.B. (2014) The pseudoenzyme PDX1.2 boosts vitamin B6 biosynthesis under heat and oxidative stress in Arabidopsis. J. Biol. Chem. 289, 8203–8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, A. , Aichi, I. and Matsuoka, M. (2006) A protocol for Agrobacterium‐mediated transformation in rice. Nat. Protoc. 1, 2796–2802. [DOI] [PubMed] [Google Scholar]
- Ollilainen, V.M. (1999) HPLC analysis of vitamin B6 in foods. Agri. Food Sci. 8, 519–618. [Google Scholar]
- Ouyang, S. , Zhu, W. , Hamilton, J. et al. (2007) The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 35, D883–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani, R. and Peracchi, A. (2003) A genomic overview of pyridoxal‐phosphate‐dependent enzymes. EMBO Rep. 4, 850–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Qu, Q. and Takaiwa, F. (2004) Evaluation of tissue specificity and expression strength of rice seed component gene promoters in transgenic rice. Plant Biotechnol. J. 2, 113–125. [DOI] [PubMed] [Google Scholar]
- Raschke, M. , Boycheva, S. , Crevecoeur, M. , Nunes‐Nesi, A. , Witt, S. , Fernie, A.R. , Amrhein, N. and Fitzpatrick, T.B. (2011) Enhanced levels of vitamin B6 increase aerial organ size and positively affect stress tolerance in Arabidopsis. Plant J. 66, 414–432. [DOI] [PubMed] [Google Scholar]
- Raschle, T. , Amrhein, N. and Fitzpatrick, T.B. (2005) On the two components of pyridoxal 5′‐phosphate synthase from Bacillus subtilis . J. Biol. Chem. 280, 32291–32300. [DOI] [PubMed] [Google Scholar]
- Raschle, T. , Arigoni, D. , Brunisholz, R. , Rechsteiner, H. , Amrhein, N. and Fitzpatrick, T.B. (2007) Reaction mechanism of pyridoxal 5′‐phosphate synthase. Detection of an enzyme‐bound chromophoric intermediate. J. Biol. Chem. 282, 6098–6105. [DOI] [PubMed] [Google Scholar]
- Raschle, T. , Speziga, D. , Kress, W. , Moccand, C. , Gehrig, P. , Amrhein, N. , Weber‐Ban, E. and Fitzpatrick, T.B. (2009) Intersubunit cross‐talk in pyridoxal 5′‐phosphate synthase, coordinated by the C terminus of the synthase subunit. J. Biol. Chem. 284, 7706–7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristila, M. , Strid, H. , Eriksson, L.A. , Strid, A. and Savenstrand, H. (2011) The role of the pyridoxine (vitamin B6) biosynthesis enzyme PDX1 in ultraviolet‐B radiation responses in plants. Plant Physiol. Biochem. 49, 284–292. [DOI] [PubMed] [Google Scholar]
- Ruiz, A. , Garcia‐Villoria, J. , Ormazabal, A. , Zschocke, J. , Fiol, M. , Navarro‐Sastre, A. , Artuch, R. , Vilaseca, M.A. and Ribes, A. (2008) A new fatal case of pyridox(am)ine 5′‐phosphate oxidase (PNPO) deficiency. Mol. Genet. Metab. 93, 216–218. [DOI] [PubMed] [Google Scholar]
- Sang, Y. , Barbosa, J.M. , Wu, H. , Locy, R.D. and Singh, N.K. (2007) Identification of a pyridoxine (pyridoxamine) 5′‐phosphate oxidase from Arabidopsis thaliana . FEBS Lett. 581, 344–348. [DOI] [PubMed] [Google Scholar]
- Sang, Y. , Locy, R.D. , Goertzen, L.R. , Rashotte, A.M. , Si, Y. , Kang, K. and Singh, N.K. (2011) Expression, in vivo localization and phylogenetic analysis of a pyridoxine 5′‐phosphate oxidase in Arabidopsis thaliana . Plant Physiol. Biochem. 49, 88–95. [DOI] [PubMed] [Google Scholar]
- Savenstrand, H. , Brosche, M. and Strid, A. (2004) Ultraviolet‐B signalling: Arabidopsis brassinosteroid mutants are defective in UV‐B regulated defence gene expression. Plant Physiol. Biochem. 42, 687–694. [DOI] [PubMed] [Google Scholar]
- Seck, P.A. , Diagne, A. , Mohanty, S. and Wopereis, M.C.S. (2012) Crops that feed the world 7: Rice. Food Security, 4, 7–24. [Google Scholar]
- Sheu, J.J. , Yu, T.S. , Tong, W.F. and Yu, S.M. (1996) Carbohydrate starvation stimulates differential expression of rice alpha‐amylase genes that is modulated through complicated transcriptional and posttranscriptional processes. J. Biol. Chem. 271, 26998–27004. [DOI] [PubMed] [Google Scholar]
- Shi, H. and Zhu, J.K. (2002) SOS4, a pyridoxal kinase gene, is required for root hair development in Arabidopsis. Plant Physiol. 129, 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Xiong, L. , Stevenson, B. , Lu, T. and Zhu, J.K. (2002) The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance. Plant Cell, 14, 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, G. , Kumar, S. and Singh, P. (2003) A quick method to isolate RNA from wheat and other carbohydrate‐rich seeds. Plant Mol. Biol. Rep. 21, 93–93. [Google Scholar]
- Singh, U. , Praharaj, C.S. , Singh, S.S. and Singh, N.P. (2016) Biofortification of Food Crops. New Delhi, Heidelberg, New York, Dordrecht, London: Springer India. [Google Scholar]
- Storozhenko, S. , De Brouwer, V. , Volckaert, M. , Navarrete, O. , Blancquaert, D. , Zhang, G.F. , Lambert, W. and Van Der Straeten, D. (2007) Folate fortification of rice by metabolic engineering. Nat. Biotechnol. 25, 1277–1279. [DOI] [PubMed] [Google Scholar]
- Strobbe, S. and Van Der Straeten, D. (2018) Toward Eradication of B‐Vitamin Deficiencies: Considerations for Crop Biofortification. Front. Plant Sci. 9, 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svozil, J. , Hirsch‐Hoffmann, M. , Dudler, R. , Gruissem, W. and Baerenfaller, K. (2014) Protein abundance changes and ubiquitylation targets identified after inhibition of the proteasome with syringolin A. Mol. Cell Proteomics, 13, 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szydlowski, N. , Bürkle, L. , Pourcel, L. , Moulin, M. , Stolz, J. and Fitzpatrick, T.B. (2013) Recycling of pyridoxine (vitamin B6) by PUP1 in Arabidopsis. Plant J. 75, 40–52. [DOI] [PubMed] [Google Scholar]
- Tambasco‐Studart, M. , Titiz, O. , Raschle, T. , Forster, G. , Amrhein, N. and Fitzpatrick, T.B. (2005) Vitamin B6 biosynthesis in higher plants. Proc. Natl Acad. Sci. USA, 102, 13687–13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambasco‐Studart, M. , Tews, I. , Amrhein, N. and Fitzpatrick, T.B. (2007) Functional analysis of PDX2 from Arabidopsis, a glutaminase involved in vitamin B6 biosynthesis. Plant Physiol. 144, 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, B. and Amoroso, L. (2011) Combating Micronutrient Deficiencies: Food‐based Approaches. Rome: CAB International and FAO. [Google Scholar]
- Titiz, O. , Tambasco‐Studart, M. , Warzych, E. , Apel, K. , Amrhein, N. , Laloi, C. and Fitzpatrick, T.B. (2006) PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J. 48, 933–946. [DOI] [PubMed] [Google Scholar]
- Vanderschuren, H. , Alder, A. , Zhang, P. and Gruissem, W. (2009) Dose‐dependent RNAi‐mediated geminivirus resistance in the tropical root crop cassava. Plant Mol. Biol. 70, 265–272. [DOI] [PubMed] [Google Scholar]
- Vanderschuren, H. , Boycheva, S. , Li, K.T. , Szydlowski, N. , Gruissem, W. and Fitzpatrick, T.B. (2013) Strategies for vitamin B6 biofortification of plants: a dual role as a micronutrient and a stress protectant. Front. Plant Sci. 4, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.Z. , Ding, S. , Wang, Y. , Li, C.L. , Shen, Y. , Meeley, R. , McCarty, D.R. and Tan, B.C. (2017) Small kernel2 encodes a glutaminase in vitamin B6 biosynthesis essential for maize seed development. Plant Physiol. 174, 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Liu, B. , Li, X. , Ouyang, Z. , Huang, L. , Hong, Y. , Zhang, H. , Li, D. and Song, F. (2014) The de novo biosynthesis of vitamin B6 is required for disease resistance against Botrytis cinerea in tomato. Mol. Plant Microbe Interact. 27, 688–699. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Jin, X. , Ouyang, Z. , Li, X. , Liu, B. , Huang, L. , Hong, Y. , Zhang, H. , Song, F. and Li, D. (2015) Vitamin B6 contributes to disease resistance against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea in Arabidopsis thaliana . J. Plant Physiol. 175, 21–25. [DOI] [PubMed] [Google Scholar]
- Zhao, C. , Piao, S. , Wang, X. et al. (2016) Plausible rice yield losses under future climate warming. Nat. Plants, 3, 16202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Molecular characterization of the generated dual‐expressing AtPDX1.1 and AtPDX2 transgenic rice lines in the T0 generation.
Figure S2. Analysis of total vitamin B6 contents of transgenic rice lines in the T1 generation.
Figure S3. Assignment of a glucosylated B6 vitamer in rice leaf extracts.
Figure S4. Rice PDX gene expression patterns across different tissues.
Figure S5. PDX transgene expression, protein accumulation and vitamin B6 content in the rice Glob lines.
Table S1. Primers used for the molecular characterization of generated transgenic rice lines.
Table S2. Primers used for real‐time quantitative PCR analysis.
Data S1. Statistical analysis.


