Abstract
Stimuli-responsive compounds that provide on-site, controlled antimicrobial activity promise an effective approach to prevent infections, reducing the need for systemic antibiotics. We present one novel pH-sensitive quaternary pyridinium salts (QPS) whose antibacterial activity is boosted by low pH and controlled by adjusting the pH between 4 and 8. Particularly, this compound selectively inhibits growth of acid-producing bacteria within a multispecies community. The successful antibacterial action of this QPS maintains the environmental pH above 5.5, a threshold pH, below which demineralization/erosion takes place. The design, synthesis, and characterization of this QPS and its short-chain analog are discussed. In addition, their pH-sensitive physicochemical properties in aqueous and organic solutions are evaluated by UV-Vis spectroscopy, dynamic light scattering, and nuclear magnetic resonance spectroscopy. Furthermore, the mechanism of action reveals a switchable assembly that is triggered by acid/base interaction and formed by tightly stacked π-conjugated systems and base moieties. Finally, a model is proposed to recognize the correlated but different mechanisms of pH-sensitivity and acid-induced, pH-controlled antibacterial efficacy. We anticipate that successful application of these QPSs and their derivatives will provide protections against infection and erosion through targeted treatments to acid-producing bacteria and modulation of environmental pH.
Keywords: Target treatment, acid-producing bacteria, erosion, quaternary ammonium salt, π-conjugated systems
Graphical Abstract

BRIEFS. Target treatment of acid-producing bacteria is achieved by a pH-sensitive compound that selectively inhibits growth of acid-producing bacteria within a multispecies community, which consequentially modulates the environmental pH.
1. Introduction
Advanced materials that provide on-site, targeted microbial treatment represent new strategies to prevent infection and circumvent drug accumulation in the environment.1–6 The natural analogs of these materials are short cationic amphiphilic peptides, such as cathelicidins, in mammals’ innate host defense systems.6 These peptides serve as both antibiotics and immune modulators.7 Specifically, a precision-guided antimicrobial peptide has been successfully engineered to selectively suppress specific pathogenic bacterium8 and achieve targeted modulating of human microbial ecology.9 However, in many clinical and environmental settings, it is not one particular bacterium, but rather multiple species with similar physiological prosperities, such as acid-production in the pathogenesis of dental caries.10 In addition, the high cost of such manufacturing peptides and their complex mechanism of action demand new approaches in material development.
Smart materials with on-demand antimicrobial efficacy are especially desirable, as they generate stimuli-responsive antibacterial activity on-site. Such materials offer a new strategy to treat infection locally with less antimicrobial agent, thus improving antimicrobial stewardship and potentially reducing antimicrobial resistance.11–15 For instance, optically-controlled antibacterial activity successfully enhanced the efficacy of drugs through light irradiation.11–13 In another work, the antibacterial activity of a supramolecular complex was reversibly switched on and off through assembly and disassembly of a cationic poly(phenylene vinylene) derivative.14–15 However, in all these pioneering works, an external trigger, either light or chemicals, was required, and only nonspecific broad-spectrum antimicrobial activities were delivered.
Herein, we introduce a new quaternary pyridinium salt (QPS) that exhibits pH-controlled antibacterial activity that selectively inhibits the growth of acid-producing bacteria. This molecule, (E)-1-hexadecyl-4-((4-(methacryloyloxy)phenyl)diazenyl)-pyridinium bromide (named Azo-QPS-C16), is capable of adjusting its antibacterial performance within a physiological pH range from 4 to 8. The antibacterial efficacy of Azo-QPS-C16 may also be triggered locally by acidic metabolic products of nearby bacteria, resulting in subsequent killing of these bacteria, circumventing the need for external means of intervention. For example, Streptococcus mutans (S. mutans) and other acidogenic Streptococcus spp. within dental biofilms consumes sugars and generates acids, presenting a significant threat to dental tissue health and biomedical devices including orthopedic implants and dental restorations.16–18 Accordingly, Azo-QPS-C16 attacks acid-producing bacteria in response to their own metabolic activity. Moreover, a model is proposed based on the pH-sensitive physiochemical properties of this molecule and its short-chain analog to understand its acid-induced, pH-controlled antibacterial efficacy. We anticipate that successful application of Azo-QPS-C16 and its derivatives will provide an economically suitable and controllable solution to specifically treat acid-producing bacteria and equip devices with pH-sensitive protection against infection and erosion, consequently avoid the chronic accumulation of active antimicrobials in environment.
2. Results and Discussion
2.1. Synthesis and experimental design.
The Azo-QPS-C16 and its short chain analog (Azo-QPS-C2) were synthesized in three steps starting from 4-aminopyridine and phenol (Scheme 1). First, a 4-(4-hydroxyphenylazo) pyridine core was obtained through an azo coupling reaction. Second, the core was functionalized by methacryloyl chloride to equip a polymerizable head group19 onto the phenyl ring. This head group may be copolymerized with other C=C functional groups, thus enabling the final molecule to be anchored onto polymers and filler particles. Third, a sixteen-carbon tail was attached to the pyridine ring through refluxing with 1-bromohexadecane. The Azo-QPS-C16 was obtained as a red powder in 49 % overall yield. When ethyl bromide was used in the third step, Azo-QPS-C2 was prepared. These compounds were characterized via 1H NMR (proton nuclear magnetic resonance spectroscopy, Figure. S1a) and 13C NMR (carbon nuclear magnetic resonance spectroscopy, Figure S1b) spectroscopies, and high-resolution electrospray ionization (ESI) mass spectrometry. Only the trans- isomers of these QPSs were identified through NMR, i.e., no cis-trans photo-isomerization was detected, possibly due to their ultrafast isomerization rate. 20
Scheme 1.
Stepwise synthesis of Azo-QPS-C16
The quaternary pyridinium salt with long alkyl chain (QPS-C16) was chosen as our active antibacterial agent. This amphiphilic structure’s biocidal performance comes from its capability to freely associate with cell membranes, forming mixed-micelle aggregates with hydrophobic membrane components that solubilize membrane and lyse the cell.21 Furthermore, the phenyl-azo-pyridinium core forms pH-sensitive assembly that regulates the biocidal activity of Azo-QPS-C16 according to the environmental pH. Moreover, adding acid/base also adjusts the assembly status. Specifically, as illustrated in Figure 1a, the biocidal activity is turned on in an acidic environment or by adding acid (locally) through disassembly of the agglomerates. Addition of acid leads to disassembly of the bulky agglomerates, and releases Azo-QPS-C16 molecules, consequently increases the number of active antimicrobial sites, thus enhance antibacterial efficacy (antibacterial on). This process is reversed (antibacterial off) in a neutral or mild basic environment.
Figure 1.
pH-sensitive, reversible spectroscopic properties and assembly behavior of Azo-QPS-C16. (a) A cartoon illustrates the assembly and disassembly between Azo-QPS-C16 and its nanosized aggregates, mediated by pH switch or addition of base/acid, for reversible control of antibacterial activity. (b) UV-vis spectra of Azo-QPS-C16 in DMSO solution with presence of trimethylamine (TEA). (c) Changes in UV-vis absorbance peak at 554 nm with stepwise addition of TEA and TFA; inset shows corresponding photo image of the solution. (d) Particle size distribution of solution samples showed in (c), with respective step numbers indicated.
2.2. Spectroscopic properties and assembly behaviors
The Azo-QPS-C16 exhibits pH-sensitive and acid/base-induced physicochemical properties in aqueous solutions and organic solutions, respectively. In aqueous solutions, Azo-QPS-C16 (0.01 mmol/L, 5.7 μg/mL) appear clear orange and purple in acidic and basic conditions, respectively, consistent with the red-shift observed in UV-vis spectra (Figure 1b). Intensification of a peak centered at 554 nm and simultaneous decrease of a peak at 347 nm are proportional to a change in pH from 4.1 to 7.9 (Figure S2a). In organic solutions, adding base leads to similar red-shift in absorbance peaks. In DMSO solutions (0.05 mmol/L, 28.6 μg/mL), the decrease of peak intensity at 347 nm is proportional to an increase in molar ratio of triethylamine (TEA) to QPS (Figure S2b). This change in absorption spectra is reversible. As shown in Figure 1c, reversible switching of colors in DMSO was demonstrated over three cycles through successive additions of 5 molar equivalents TEA and then trifluoroacetic acid (TFA): addition of base immediately increase the absorbance peak at 554 nm, while TFA treatment quickly regenerates the original peak intensity.
Concurrently as color changed, a switchable assembly behavior of Azo-QPS-C16 was observed via dynamic light scattering (DLS). Particles with an average hydrodynamic diameter of 51 ± 19 nm were detected when TEA was added, occurring simultaneously with the appearance of the absorbance peak at 554 nm. No particles were found after treated with TFA. As TEA/TFA cycled, particles of the same size distribution formed and disassembled (Figure 1d). Atomic force microscopy (AFM, Figure S3) and transmission electron microscopy (TEM, Figure S4) indicate that particles between 30 nm and 100 nm diameter form upon addition of TEA to Azo-QPS-C16, agreeing well with the DLS data.
The short chain Azo-QPS-C2 showed similar switchable assembly in terms of appearance and particle size distribution (Figure S5). Its UV-Vis spectrum has two peaks at 350 nm and 540 nm, comparing to 347 nm and 554 nm in the spectra of Azo-QPS-C16. DLS determined that these two compounds had the same hydrodynamic diameter and particle size distribution when base was added. Such a good match indicates that the chain length of the tails has ignorable impact on the assembly of these compounds. This switchable aggregation and spectroscopic property is believed to be related to a head-to-tail π-π stacking which will be explained with our model later in this manuscript.
2.3. Acid-enhanced antibacterial efficacy and selective inhibition of acid-producing bacteria
The agglomeration of Azo-QPS-C16 is associated with the acid-induced, pH-sensitive antimicrobial activity, as the molecule can regulate the number of active antibacterial sites through reversible assembly, in response to pH changes or addition of acid. Our molecules are effective on both Gram-negative (Escherichia coli) and Gram-positive (Streptococcus mutans) bacteria. The ability of bacteria to proliferate after exposure to Azo-QPS-C16 in different pH buffers (pH 4.1, 5.8 and 7.9) was characterized by adding growth medium to the pretreated bacterial cells and measuring the optical density (OD) at 600 nm every 15 min. Figure 2a and 2b show the growth curve of E. coli after treated in pH 4.1 and pH 7.9 buffers, respectively. Exposure to 2.5 µg/mL of Azo-QPS-C16 at pH 4.1 fully inhibited E. coli growth throughout the whole experimental period (19 h). However, exposure at pH 7.9 required a much higher concentration (40 µg/mL) to inhibit cell growth. Obvious growth of E. coli was observed for all buffer control measurements including different pH values with and without dimethyl sulfoxide (DMSO). The pH dependence of bactericidal activity of Azo-QPS-C16 in terms of minimum bactericidal concentration (MBC) was evaluated by inoculating cultures onto a Lysogeny Broth (LB) agar plate after treating the cells with a two-fold dilution series of Azo-QPS-C16 for 30 min. As pH increased, the MBC increased consecutively (Figure 2c). A 16-fold difference in MBC was observed between pH 4.1 and pH 7.9.
Figure 2.
Acid-enhanced antibacterial behaviors of Azo-QPS-C16 compound. E. coli growth curves after treatment with different concentrations of Azo-QPS-C16 for 45 min at (a) pH 4.1 and (b) pH 7.9. Each data point represents the average of 3 values, and each error bar represents one standard deviation and serves as the estimate of measurement uncertainty. (c) MBC values of Azo-QPS-C16 against E. coli and S. mutans at different pH values. (d) Biomass measurement of bacteria treated with combination of sucrose and Azo-QPS-C16, data collected from both supernatant (blue column) and biofilm (red column); pH values were obtained during OD measurement. (e) DGGE fingerprints from extracted community DNA.
We observed similar pH-sensitive antibacterial activity of Azo-QPS-C16 towards the Gram-positive, lactic acid producing, cariogenic bacterium S. mutans. According to MBC assessments (Figure 2c), Azo-QPS-C16 is 8-fold more effective against S. mutans in acidic than in mildly basic conditions. In contrast, Azo-QPS-C2 did not inhibit the growth of either E. coli or S. mutans in any of the pH conditions and concentrations (up to 1000 µg/mL) evaluated. This difference confirms that the length of the chain attached to nitrogen of the pyridinium salt plays a vital role in antibacterial activities of the QPS.
The selective killing/inhibition of acid-producing bacteria by Azo-QPS-C16 was examined using an in vitro multispecies biofilm model that simulates oral microbial community.22 Biofilms were established as described in Experimental Section. The effects of Azo-QPS-C16 on microbial community profile of biofilm as well as unattached planktonic cells in the presence and absence of sucrose—the cariogenic dietary carbohydrate, was monitored. Planktonic and biofilm cells were quantified using optical density at 600 nm to reflect the biomass; while the microbial profiles were analyzed by polymerase chain reaction denaturing gradient gel electrophoresis (PCR-DGGE). Data showed that, in the absence of sucrose, the pH of overnight culturing medium remained above 7.5, and the addition of Azo-QPS-C16 alone resulted in only moderate reduction in biomass of both biofilm and unattached planktonic cells compared to negative control, with an increase in the relative abundance of bacterial species such as Enterobacter spp. and Klebsiella spp and decrease in that of Streptococcus spp, including Streptococcus agalactiae. The addition of sucrose alone in the culturing medium achieved the highest biomass in both planktonic portion and biofilm among all the tested conditions (Figure 2d). However, the increase in biomass was not uniform. Certain oral species, such as S. agalactiae were enriched; while other bacterial species including Enterobacter spp. and Klebsiella spp. (Figure 2e, sample# 2 and 6) was reduced in relative abundance. More importantly, the inclusion of sucrose resulted in a drastic pH reduction (pH 4.5 compared to pH 8.0 in negative control) of the culturing medium (Figure 2d), which is in agreement with the fact that many oral bacteria, particularly cariogenic streptococci are able to ferment sucrose and produce acid as byproducts. The culturing condition with a reduced pH favors the growth of acidogenic and aciduric species, such as streptococcus spp., while inhibits the growth of less aciduric bacteria, such as Enterobacteria spp. and Klepsiella spp..23 Interestingly, while maintaining similar level of biomass in planktonic portion, the presence of Azo-QPS-C16 abolished the sucrose-induced increase in biofilm biomass. PCR-DGGE analysis revealed that the presence of Azo-QPS-C16 almost eliminated the species, including S. agalactiae, which were enriched in sucrose only group, while increased relative abundance of Enterobacter spp. and Klebsiella spp. Most intriguingly, the presence of Azo-QPS-C16 maintained the pH of the culturing medium above 5.5, a critical pH value below which demineralization of enamel and dentin takes place.10 The higher pH value even in the presence of sucrose could likely be explained by the more targeted killing of sucrose-fermenting, acid-producing bacteria, such as Streptococcus spp. by Azo-QPS-C16 and the subsequent outgrowth of non-acid producers, including Enterobacter spp. and Klebsiella spp. Meanwhile, many acid-producing Streptococcus spp are also good biofilm-formers due to their ability to metabolize carbohydrates and generate exopolysaccharides which contributes to their enhanced biofilm formation ability. The killing/inhibition of these species could result in reduced biomass. These intriguing results are supportive of Azo-QPS-C16’s capability to selectively attack acid-producing bacteria, thus modulate the environment pH and prevent demineralization and erosion of tooth.
2.4. The mechanism of switchable pH-sensitive assembly.
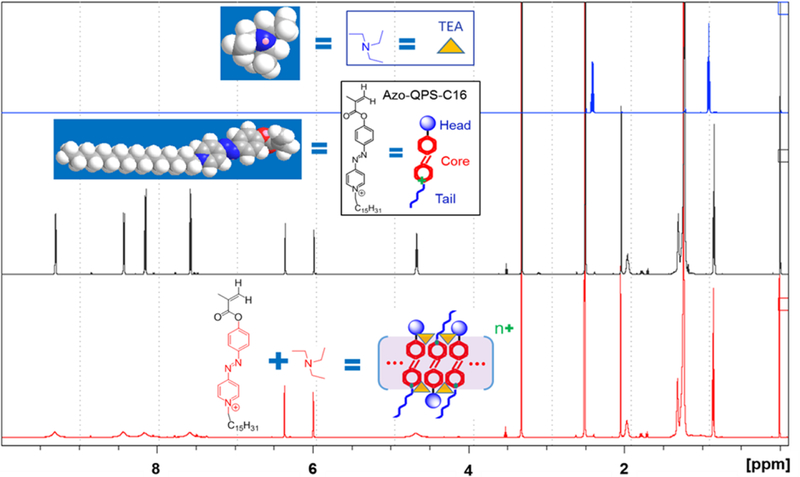
The chemistry revealed by NMR spectra further confirms the key role of the phenyl-azo-pyridinium core in the assembly of Azo-QPS-C16/2. In addition, NMR spectra suggest that there are interactions between these compounds and the base. Figure 3 shows the spectra of TEA, Azo-QPS-C16, and their equimolar mixture (based on the sample preparation). The spectrum of the mixture differs in a number of ways from that of the components. First, the peaks of protons on the aromatic rings and the first carbon of the QPS’s tail yield very broad resonance signals; second, the protons on TEA are unobservable; third, no peak shift or change in integration are identified in the peaks of the other protons; and finally, no new peaks appear. Peak broadening and unobservable protons are likely due to assembly, which has been reported in supramolecular gels formed by low molecular weight species 24 and agrees well with the particle formation determined by DLS. The assembly significantly increases the correlation time, consequently, leads to a very short transversal relaxation time, and very broad or unobservable signals. Moreover, the lack of TEA protons suggests that it participates in the assembly. TEA is a base and may interact with the Azo-QPS-C16 which is weakly acidic, pKa = 5.33. The product of this interaction may be a chemical complex formed by the base and the Azo-QPS-C16. Based on the above NMR results combined with the base-induced red-shift in UV-vis spectra and assembly determined by DLS, we believe that the interaction of TEA with Azo-QPS-C16 triggers assembly of tightly stacked π-conjugated cores, formed by two or more Azo-QPS-C16 molecules and the base. A model is suggested in Figure 3, which uses a sandwich stacking conformation where the cores of the trans-isomer of Azo-QPS-C16 molecules are aligned in parallel. Further, the Azo-QPS-C16 molecules are packed with an alternating head-to-tail arrangement that minimizes the potential repulsion among the QPS moieties due to the positive charge of the pyridinium salt. This stacking model also maximizes the participation of TEA as its interaction with the Azo-QPS-C16 is most likely taking place close to the QPS moieties. The base triggered broadening resonance signals of the protons on the first carbon of the pyridinium tail strongly suggest such a possibility. As a result of charge repulsion and potential steric hindrance, head-to-tail arrangement is more favored than head-to-head or tail-to-tail arrangements and is likely to create the most impenetrable packing of the phenyl-azo-pyridinium core. Furthermore, TEA and its interaction with Azo-QPS-C16 molecules may also serve as a shutter to the access of the already tightly stacked core, thus isolating the core from the rest of the Azo-QPS-C16 molecules. Consequently, distinct observability of resonance signals is detected by NMR spectrometer. Finally, the interaction and the condensed stacking redistribute the positive charge of the QPS moiety within the assembly. The charge of the assembly (n+) is related to the number (n) of the participating Azo-QPS-C16 molecules.
Figure 3.
NMR spectra of TEA (blue), Azo-QPS-C16 (black), and the equimolar mixture of these two (red) in anhydrous deuterated DMSO. The intensity of the spectra was adjusted for the demonstration purposes. The insets are 3D space-filling models, chemical structures and cartoons of the corresponding chemicals. The red-colored portion of the chemical structures in the bottom layer highlights the protons unobservable to NMR due to the assembly which results in very broad resonance signals.
The NMR spectra of Azo-QPS-C2 illuminate the same phenomena of base-induced assembly (Figure S6) and further validate our observations in DLS. In addition, the intermediate compound, Azo-QPS, does not form such base-induced assembly. The color of Azo-QPS solution did not change upon addition of base, and no broad resonance signals nor unobservable protons were observed in the NMR spectra (Figure S7). Such distinct differences identify the roles of the pyridinium chain for the pH sensing and assembly. Specifically, it transforms a basic pyridine derivative into an pyridinium salt which is liable to acid-base interaction. Such interaction is exchangeable between organic solutions and aqueous solutions through adjusting the chemistry of the base. As an example, when moisture-containing deuterated DMSO was used for the NMR evaluation of TEA/Azo-QPS-C16 mixtures instead of anhydrous DMSO, the protons of TEA appeared, while the protons of Azo-QPS-C16 behaved the same as they did in anhydrous solvents. In this case, the OH− ions replaced TEA in the assembly. Accordingly, in aqueous solutions such as those used in the bacterial studies, OH− ions may participate in the triggering and assembly.
The above results demonstrate that Azo-QPS-C16 is multifunctional and pH-sensitive. In solution, the pH-sensitivity is triggered by acid-base interaction and leads to different assembly stages of tightly stacked π-conjugated phenyl-azo-pyridinium core and base. The participation of basic moieties may be OH− ions in aqueous solutions or moisture-containing solvents. The pH sensitivity is closely interrelated to the chemistry of the phenyl-azo-pyridinium core. The chain length of the QPS tail has minimal impact on the pH-sensitivity in solutions. However, the long carbon chain is vital to the acid-enhanced antibacterial efficacy, which is determined by a combination of multiple factors including the amphiphilic properties, cations, charge density and counter ions.21 The distinct impact of the chain length to pH-sensitive assembly and antibacterial efficacy offer a tool to design and prepare pH-sensitive materials with a broad range of antibacterial efficacy.
3. Conclusions
Above all, the discovery in this study provides valuable tools in material design to prepare multifunctional stimuli-responsive materials. Successful application of Azo-QPS-C16 and its derivatives promise a smart and self-regulating defense systems for medical devices. Eventually, an optimal combination of pH-sensitivity and acid-induced antibacterial efficacy may be obtained to provide targeted antibacterial treatment for infection and erosion, particularly in fighting dental caries, one of the most prevalent and costly chronic infectious diseases.
4. Experimental Section
General information for synthesis and characterization.
Commercially available materials purchased from Alfa Aesar (Tewksbury, MA, USA), Sigma-Aldrich (Saint Louis, MO, USA) and TCI America (Portland, OR, USA) were used as received. Proton and carbon nuclear magnetic resonance (1H and 13C NMR) spectra were recorded on a Bruker instrument (600 MHz, Billerica, MA, USA) using 5 mm tubes (Figure S1). Chemical shifts were recorded in parts per million (ppm, δ) relative to tetramethylsilane (δ = 0.00), dimethylsulfoxide (δ = 2.50) or chloroform (δ = 7.26). 1H NMR splitting patterns are designated as singlet (s), doublet (d), triplet (t), quartet (q), dd (doublet of doublets), and m (multiplets). High-resolution mass spectra (MS) were recorded on a JEOL AccuTOF (Peabody, MA, USA) for ESI-TOF-MS analysis using methanol as the solvent (Needle voltage = 2100 V, Temperature = 250 C degree, Postive ion mode, and CsI as the internal standard). UV-vis spectra were recorded on a Thermo Spectronic Genesys 5 UV-vis spectrophotometer (Thermo Scientific, Waltham, MA USA) using quartz cuvettes with 1 cm path length at 298 K after baseline correction. Column chromatography was performed on silica gel (VWR, 230–400 mesh). Analytical thin-layer chromatography (TLC) was carried out on an EMD Millipore (Billerica, MA, USA) 60 F254 pre-coated silica gel plate (0.2 mm thickness). Visualization was performed using UV radiation (254 nm).
Preparation of 4-((4-methacryloyloxy)phenylazo)pyridine (Azo-QPS).
The compound was synthesized following previous reported procedures with minor modifications.19 5 g (53.2 mmol) of phenol and 4 g (58.0 mmol) of sodium nitrite were dissolved in 20 mL of 10 % (by mass) sodium hydroxide aqueous solution, and the mixture was stirred in an ice-bath at (0 to 4) °C. The mixture was added dropwise to a pre-cooled solution made from 6 g (63.8 mmol) of 4-aminopyridine in hydrogen chloride aqueous solution. The reaction was stirred in the ice-bath for 30 minutes and then stirred at room temperature overnight. The pH of the reaction was adjusted to 6 to 7 with 10 % (by mass) sodium hydroxide, and the precipitate was collected by filtration and dried in air. The azo product was used in the next step without further purification. 4 g (20.1 mmol) of the azo and 1.25 equivalent of trimethylamine was dissolved in tetrahydrofuran (THF). One equivalent of methacryloyl chloride was added to the reaction dropwise. The reaction was stirred at room temperature for 2 h. 4-((4-methacryloyloxy)phenylazo)pyridine was purified by column chromatography with an 87 % yield.
Preparation of (E)-1-hexadecyl-4-((4-(methacryloyloxy)phenyl)diazenyl)-pyridinium bromide (Azo-QPS-C16).
In a round bottom flask, Azo-QPS was refluxed with 1.5 equivalent of 1-Bromohexadecane in acetonitrile for 5 days. The dark red product was further purified by recrystallization with ether and acetone, affording 78 % yield of Azo-QPS-C16 as a dark red powder. The overall yield of these three steps was 49 %. 1H NMR (600 MHz, CDCl3) δ 9.64 (d, J = 6.0 Hz, 2 H), 8.30 (d, J = 6.0 Hz, 2 H), 8.10 (d, J = 6.0, 11.0 Hz, 2 H), 7.40 (d, J = 16.0, 2 H), 6.42 (s, 1 H), 5.86 (s, 1 H), 5.11 (t, J = 7.4 Hz, 2 H), 2.08 (m, 5 H), 1.33 (m, 26 H), 0.88 (m, 3 H) ppm; 13C NMR (600 MHz, CDCl3) δ 165.01, 160.42, 156.39, 149.86, 147.21, 135.34, 128.44, 126.27, 123.07, 120.50, 62.03, 32.11, 31.93, 31.17, 29.70, 29.69, 29.66, 29.64, 29.60, 29.51, 29.37, 29.10, 28.77, 26.15, 22.70, 18.32, 14.12 ppm. Hi-Resolution MS (ESI): m/z calculated. for C31H46N3O2+, 492.3585; found [M]+: C31H46N3O2+, 492.3599.
Preparation of (E)-1-ethyl-4-((4-(methacryloyloxy)phenyl)diazenyl)-pyridinium bromide (Azo-QPS-C2).
In a round bottom flask, Azo-QPS was refluxed with 1.5 equivalent of 1-Bromoethane in acetonitrile for 5 days. The dark red product was further purified by recrystallization with ether and acetone, affording 20 % yield of Azo-QPS-C2 as a dark red powder. 1H NMR (600 MHz, CDCl3) δ 9.77 (d, J = 6.0 Hz, 2 H), 8.33 (d, J = 6.0 Hz, 2 H), 8.11 (d, J = 6.0, 11.0 Hz, 2 H), 7.42 (d, J = 16.0, 2 H), 6.43 (s, 1 H), 5.87 (s, 1 H), 5.22 (t, J = 7.4 Hz, 2 H), 2.12 (s, 3H), 1.80 (t, J = 6.0 Hz, 3 H) ppm; 13C NMR (600 MHz, CDCl3) δ 165.00, 160.47, 156.35, 149.84, 147.05, 135.33, 128.41, 126.26, 123.06, 120.62, 57.26, 18.32, 17.34 ppm.
Bacterial strains and growth conditions.
Streptococcus mutans (S. mutans, UA159) and Escherichia coli (E. coli, K12) were purchased from ATCC (American Type Culture Collection, Manassas, VA, USA). Todd Hewitt Broth (THB), and Lysogeny Broth (LB) powder and agar were purchased from BD (Becton and Dickinson Company, Franklin Lakes, NJ, USA). Planktonic cultures were inoculated from 25 % (by volume) glycerol stocks stored at −80 °C. E. coli cultures were grown in LB at 37 °C in a shaker-incubator. S. mutans cultures were grown in THB at 37 °C with 5 % (by volume) CO2 overnight.
Bacterial growth curves.
Overnight cultures of E. coli and S. mutans were diluted directly into buffers with different pH values (100 mmol/L, pH 4.1 sodium acetate buffer, pH 5.8 sodium phosphate buffer, and pH 7.9 sodium phosphate buffer) to an optical density at 600 nm (OD600) of approximately 0.001.
In a 96-well plate, 2 µL of Azo-QPS-C16 at twofold dilutions in dimethyl sulfoxide (DMSO) were added to 98 µL of bacteria suspension per well for a final concentration ranging from 1.25 µg/mL to 40 µg/mL. After 45 min of treatment at room temperature, 10 µL from each well was transferred to a new 96-well plate already containing 100 µL growth media per well to measure the growth curves. E. coli were cultured at 37 °C, and the OD600 of each well was measured by a Tecan Spark microplate reader (Männedorf, Switzerland) every 15 min for up to 19 h, with 30 s shaking before each measurement. S. mutans were grown at 37 °C with 5 % CO2, and the OD600 of each well was measured periodically using a Molecular Devices (Sunnyvale, CA) SpectraMax M5 microplate reader. All experiments were conducted in triplicate, and repeated at least three times on different days.
MBC determination.
MBC values were determined by colony formation on agar plates. Bacteria were grown and incubated for 45 min with Azo-QPS-C16 as described above, and then 5 µL from each well were spotted onto an agar plate and incubated for 48 h to allow colony formation. The presence or absence of bacterial growth was determined by naked eye. All experiments were conducted in triplicate and repeated at least three times on different days.
Growth of saliva-derived multispecies Biofilms.
Saliva-derived biofilms were established using a previously published protocol. 22 Briefly, saliva samples were collected and pooled from 5 healthy volunteers (approved under UCLA IRB 13–001075) and used to inoculate SHI medium25 containing the following 4 different combination of supplements: 1) Azo-QPS-C16 (40 µg/ml); 2) sucrose (60 mM); 3) Azo-QPS-C16 (40 µg/ml) and sucrose (60 mM); and 4) negative control. Then, 1 mL was added to each well of a 24-well plate that had been pre-coated with sterilized, cell-free saliva. Plates were incubated at 37°C for 16 h in microaerobic conditions (2 % O2, 5 % CO2, balanced with nitrogen) to allow biofilm formation. After incubation, planktonic portion was collected, optical density was measured at 600 nm and cells were pelleted for DNA isolation. Meanwhile, 1 ml of medium was added to the biofilm portion, biofilm cells were dispersed by scraping and vortex to break the cell aggregates. Optical density was measured at 600 nm and cells were collected for DNA isolation.
PCR-DGGE analysis.
Total genomic DNA of bacterial samples was isolated using the MasterPure™ DNA purification kit (Epicentre). DNA quality and quantity were determined by a Nanodrop 2000 Spectrophotometer. Amplification of bacterial 16S rRNA genes by PCR was carried out as described previously.26 Briefly, the universal primer set, Bac1 (5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCGACTACGTGCCAGCAGCC-3′) and Bac2 (5′-GGACTACCAGGGTATCTAATCC-3′), were used to amply an approximately 300-bp internal fragment of the 16S rRNA gene. Each 50-μl PCR reaction contained 100 ng of purified genomic DNA, 40 pmol of each primer, 200 μM of each dNTP, 4.0 mM MgCl2, 5 μl of 10X PCR buffer, and 2.5 U of Taq DNA polymerase (Invitrogen). Cycling conditions were 94 °C for 3 min, followed by 30 cycles of 94 °C for 1 min, 56 °C for 1 min and 72 °C for 30 s, with a final extension period of 5 min at 72 °C. The resulting PCR products were evaluated by electrophoresis in 1.0 % agarose gels.
Polyacrylamide gels at an 8 % concentration were prepared with a denaturing urea/formamide gradient between 40 % (containing 2.8 M urea and 16 % (v/v) formamide) and 70 % (containing 4.9 M urea and 28 % (v/v) formamide). Approximately 300 ng of the PCR product were applied per lane. The gels were submerged in 1 × TAE (Tris-Acetate-EDTA) buffer (40 mM Tris base, 40 mM glacial acid acetic, 1 mM EDTA) and the PCR products were separated by electrophoresis for 17 h at 58 °C using a fixed voltage of 60 V in the Bio-Rad DCode System (Bio-Rad laboratories, Inc. Hercules, CA, USA). After electrophoresis, the gels were rinsed and stained for 15 min in 1 × TAE buffer containing 0.5 μg/ml ethidium bromide, followed by 10 min of de-staining in 1 × TAE buffer. DGGE profile images were digitally recorded using the Molecular Imager Gel Documentation system (BioRad). Diversity Database Software (BioRad) was used to assess the change in the relative intensity of bands corresponding to bacterial species of interest.
Identification of bacterial species in DGGE gel.
Bands of interest were excised from the DGGE gels and transferred to a 1.5 ml microfuge tube containing 10 μl of sterile ddH2O. Tubes were incubated at 4 °C overnight before the recovered DNA samples were reamplified with the universal primer set (Bac1 and Bac2). The PCR products were purified using the QIAquick PCR purification kit (Qiagen) and sequenced at the UCLA Sequencing and Genotyping Core Facility. The obtained partial 16S rRNA gene sequences (about 300bp) were used to BLAST search against the HOMD (http://www.homd.org) and NCBI (www.ncbi.nlm.nih.gov) databases. Sequences with 98–100 % identity to those deposited in the public domain databases were considered to be positive identification of taxa.
DLS studies.
Dynamic light scattering (DLS) was performed using a Nicomp 380 ZLS DLS system (Particle Sizing Systems Inc., Santa Barbara, CA). Samples were contained in disposable glass culture tubes. Prior to sample loading, the tubes were blown with nitrogen gas and filled with 0.4 mL of the dye sample. The tubes were sealed using parafilm and then centrifuged at 14 000 rpm (19 000 g) for 5 min to separate larger particles and dust. Samples were placed in the instrument and allowed to attain thermal equilibrium. Autocorrelation function was acquired for 3 min at 173° scattering angle. A Laplace inversion based Nicomp routine was used to find the distribution of decay rates. The decay rates are linked to the effective-sphere hydrodynamic diameters by the appropriate equations.27 The size distribution is reported as a discrete set of diameter bins and corresponding intensity-weighted fractions as probabilities assigned to diameter bins.
Supplementary Material
ACKNOWLEDGMENT
This work was funded by the National Institute of Dental and Craniofacial Research (U01DE023752 and DE020102). Financial support was also provided through the American Dental Association (ADA) and ADA Foundation. The authors thank Drs. Drago Skrtic, Nicole Ritzert, Ryan Nieuwendaal, Lisa Fredin, Kathleen Schwarz, Thomas Allison, and Thomas Moffat for their discussion and technical recommendations and the Center for Nanoscale Science and Technology (CNST) at NIST for technical support.
Funding Sources
National Institute of Dental and Craniofacial Research (U01DE023752 and DE020102). Financial support was also provided through the American Dental Association (ADA) and ADA Foundation.
Footnotes
Supporting Information. Additional figures including NMR, AFM, TEM and UV-vis spectra are available free of charge.
REFERENCES
- 1.Levin-Reisman I; Ronin I; Gefen O; Braniss I; Shoresh N; Balaban NQ, Antibiotic tolerance facilitates the evolution of resistance. Science 2017, 355 (6327), 826–830. [DOI] [PubMed] [Google Scholar]
- 2.Harms A; Maisonneuve E; Gerdes K, Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354 (6318), aaf4268:1–11. [DOI] [PubMed] [Google Scholar]
- 3.Hurdle JG; O’Neill AJ; Chopra I; Lee RE, Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol 2011, 9 (1), 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clatworthy AE; Pierson E; Hung DT, Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol 2007, 3 (9), 541–548. [DOI] [PubMed] [Google Scholar]
- 5.Buffie CG; Bucci V; Stein RR; McKenney PT; Ling LL; Gobourne A; No D; Liu H; Kinnebrew M; Viale A; Littmann E; van den Brink MRM; Jenq RR; Taur Y; Sander C; Cross JR; Toussaint NC; Xavier JB; Pamer EG, Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015, 517 (7533), 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hancock REW; Sahl HG, Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol 2006, 24 (12), 1551–1557. [DOI] [PubMed] [Google Scholar]
- 7.Nizet V; Ohtake T; Lauth X; Trowbridge J; Rudisill J; Dorschner RA; Pestonjamasp V; Piraino J; Huttner K; Gallo RL, Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 2001, 414 (6862), 454–457. [DOI] [PubMed] [Google Scholar]
- 8.Li L; He J; Eckert R; Yarbrough D; Lux R; Anderson M; Shi WY, Design and Characterization of an Acid-Activated Antimicrobial Peptide. Chem. Biol. Drug Des. 2010, 75 (1), 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo LH; McLean JS; Yang Y; Eckert R; Kaplan CW; Kyme P; Sheikh O; Varnum B; Lux R; Shi WY; He XS, Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. Proceedings of the National Academy of Sciences of the United States of America 2015, 112 (24), 7569–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinberg I, A mixed-bacteriae ecological approach to understanding the role of the oral bacteria in dental caries causation: An alternative to Streptococcus mutans and the specific-plaque hypothesis. Crit. Rev. Oral Biol. Med. 2002, 13 (2), 108–125. [DOI] [PubMed] [Google Scholar]
- 11.Velema WA; van der Berg JP; Szymanski W; Driessen AJM; Feringa B, Orthogonal Control of Antibacterial Activity with Light. ACS Chem. Biol 2014, 9 (9), 1969–1974. [DOI] [PubMed] [Google Scholar]
- 12.Velema WA; van der Berg JP; Hansen MJ; Szymanski W; Driessen AJM; Feringa BL, Optical control of antibacterial activity. Nat. Chem 2013, 5 (11), 924–928. [DOI] [PubMed] [Google Scholar]
- 13.Velema WA; Szymanski W; Feringa BL, Photopharmacology: Beyond Proof of Principle. Journal of the American Chemical Society 2014, 136 (6), 2178–2191. [DOI] [PubMed] [Google Scholar]
- 14.Bai HT; Yuan HX; Nie CY; Wang B; Lv FT; Liu LB; Wang S, A Supramolecular Antibiotic Switch for Antibacterial Regulation. Angewandte Chemie-International Edition 2015, 54 (45), 13208–13213. [DOI] [PubMed] [Google Scholar]
- 15.Qian WJ; Texter J; Yan F, Frontiers in poly(ionic liquid)s: syntheses and applications. Chem. Soc. Rev 2017, 46 (4), 1124–1159. [DOI] [PubMed] [Google Scholar]
- 16.Tanner ACR; Mathney JMJ; Kent RL; Chalmers NI; Hughes CV; Loo CY; Pradhan N; Kanasi E; Hwang J; Dahlan MA; Papadopolou E; Dewhirst FE, Cultivable Anaerobic Microbiota of Severe Early Childhood Caries. J. Clin. Microbiol 2011, 49 (4), 1464–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ajdic D; McShan WM; McLaughlin RE; Savic G; Chang J; Carson MB; Primeaux C; Tian RY; Kenton S; Jia HG; Lin SP; Qian YD; Li SL; Zhu H; Najar F; Lai HS; White J; Roe BA; Ferretti JJ, Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proceedings of the National Academy of Sciences of the United States of America 2002, 99 (22), 14434–14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merkel KD; Erdmann JM; McHugh KP; Abu-Amer Y; Ross FP; Teitelbaum SL, Tumor necrosis factor-alpha mediates orthopedic implant osteolysis. Am. J. Pathol 1999, 154 (1), 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang LJ; Chen SJ; Guo XZ; Zhang Y; Zhang HQ, Azobenzene-containing molecularly imprinted polymer microspheres with photo- and thermoresponsive template binding properties in pure aqueous media by atom transfer radical polymerization. Langmuir 2012, 28 (25), 9767–9777. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Amoros J; Massed WA; Nonell S; Velasco D, Fast Isomerizing Methyl Iodide Azopyridinium Salts for Molecular Switches. Org Lett 2010, 12 (15), 3514–3517. [DOI] [PubMed] [Google Scholar]
- 21.Chen CZS; Cooper SL, Interactions between dendrimer biocides and bacterial membranes. Biomaterials 2002, 23 (16), 3359–3368. [DOI] [PubMed] [Google Scholar]
- 22.Edlund A; Yang Y; Hall AP; Guo LH; Lux R; He XS; Nelson KE; Nealson KH; Yooseph S; Shi WY; McLean JS, An in vitro biofilm model system maintaining a highly reproducible species and metabolic diversity approaching that of the human oral microbiome. Microbiome 2013, 1 (25), 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo LH; McLean JS; Lux R; He XS; Shi WY, The well-coordinated linkage between acidogenicity and aciduricity via insoluble glucans on the surface of Streptococcus mutans. Scientific Reports 2015, 5 (18015), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Escuder B; Llusar M; Miravet JF, Insight on the NMR study of supramolecular gels and its application to monitor molecular recognition on self-assembled fibers. J. Org. Chem 2006, 71 (20), 7747–7752. [DOI] [PubMed] [Google Scholar]
- 25.Tian Y; He X; Torralba M; Yooseph S; Nelson KE; Lux R; McLean JS; Yu G; Shi W, Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Mol. Oral Microbiol 2010, 25 (5), 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He XS; Tian Y; Guo LH; Ano T; Lux R; Zusman DR; Shi WY, In Vitro Communities Derived from Oral and Gut Microbial Floras Inhibit the Growth of Bacteria of Foreign Origins. Microb. Ecol 2010, 60 (3), 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koppel DE, Analysis of macromolecular polydispersity in intensity correlation spectroscopy - method of cumulants. J. Chem. Phys 1972, 57 (11), 4814–4820. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.