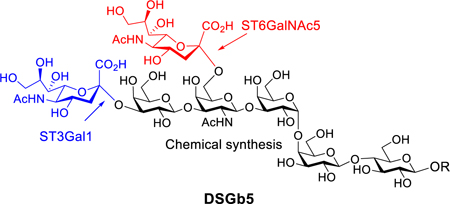
Abstract
Disialosyl globopentaosylceramide (DSGb5) is often expressed by renal cell carcinomas. To investigate properties of DSGb5, we have prepared its oligosaccharide moiety by chemically synthesizing Gb5 which was enzymatically sialylated using the mammalian sialyltransferases ST3Gal1 and ST6GalNAc5. Glycan microarray binding studies indicate that Siglec-7 does not recognize DSGb5, and preferentially binds Neu5Acα(2,8)Neu5Ac containing glycans.
Graphical Abstract
The oligosaccharide of the tumor-associated antigen DSGb5 was synthesized in a chemoenzymatic manner, by exploiting the regioselective mammalian enzymes ST3Gal1 and ST6GalNAc5. Unexpectedly, the resulting compound did not show binding to Siglec-7 on a microarray.
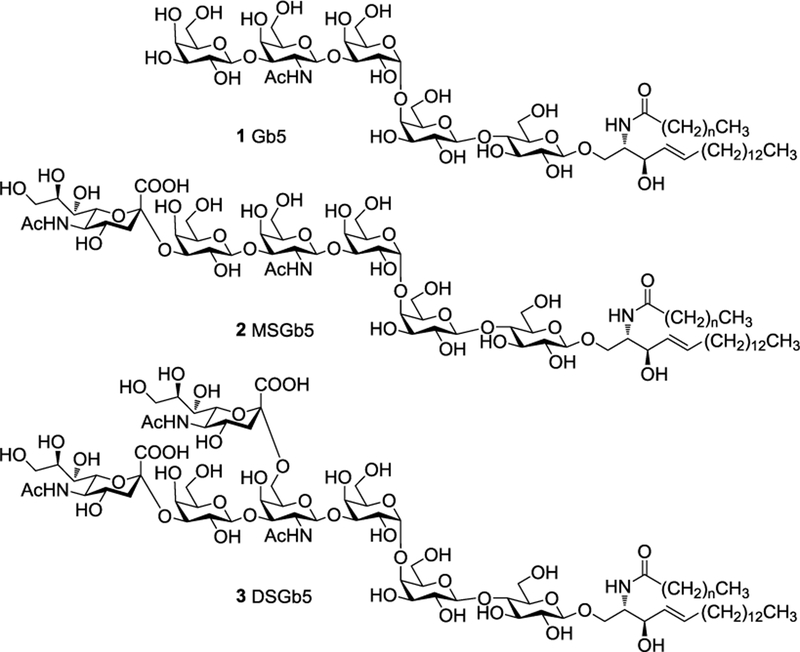
Glycosphingolipids (GSPs) are a diverse group of biomolecules that are composed of ceramide modified by a glycan. They decorate the cell surface of all vertebrate cells and play important roles in a variety of cellular processes such as cell signalling, trafficking, adhesion, proliferation, and immune modulation.1 GSPs are also critically involved in embryogenesis and are expressed in stage dependent manner. For example, globopentaosyl ceramide (1, Gb5Cer or SSEA-3, Fig. 1) and monosialyl Gb5Cer (2, MSGb5Cer or SSEA-4), which belong to the globo-series of GSPs, are well established markers of pluripotent stem cells and their expression is often used for embryonic stem cell (ES) characterization. Recently, a number of other GSPs, including Gb4, Lc4, fucosyl Lc4Cer, Globo H, and disialyl Gb5 (3, DSGb5), were identified in undifferentiated human ES and iPS cells.2 It was found that during ES differentiation, the biosynthesis of globo- and lacto-series GSPs declines and switches to the formation of ganglioside type GSPs.3
Figure 1.
Chemical structures of Gb5 (1), MSGb5 (2) and DSGb5 (3) where the fatty acid chain length can vary between n = 14 – 22.
A number of GSPs that are expressed during early embryogenesis reoccur during oncogenesis. For example, Gb5 and MSGb5 are highly expressed in breast cancer,4 testicular germ cell tumors,5 and aggressive human renal cell carcinomas.6 Furthermore, GSLs such as Globo-H are overexpressed by many epithelial cell cancers and occur on cancer stem cells. The overexpression of these glycolipids appears to promote tumorigenicity,7 immune suppression,8 and enhances cancer cell motility and invasiveness.9 The chemical synthesis of the oligosaccharide moieties of these glycolipids has received considerable attention,10 which has made it possible to examine biological properties of individual GSPs and opened the way to develop immune-therapeutic strategies such as experimental cancer vaccines.11
DSGb5 (Fig 1), which is also expressed during early embryogenesis, is often observed in renal cell carcinomas.12 Cell culture experiments indicate that DSGb5 promotes cell migration.13 It has therefore been postulated that DSGb5 may play a role in metastasis and potentially can serve as a marker for patients at risk for metastasis.14 There is also data to suggest that DSGb5 is a ligand for sialic acid-binding Ig-like lectin-7 (Siglec-7), which is expressed on natural killer (NK) cells thereby inhibiting NK-cell cytotoxicity.15
The biological properties ascribed to DSGb5 are mainly based on association studies and knockdown or overexpression of glycosyl transferases involved in its biosynthesis, and hence there is lack of direct biochemical evidence that this GSP mediates biological processes such as immune suppression by engaging with Siglec-7. In part, this is due to the fact that DSGb5 cannot easily be isolated from natural sources.
Here, we report a strategy for the preparation of the oligosaccharide moiety of DSGb5 by chemically synthesizing the pentasaccharide Gb5, which was modified by the mammalian glycosyl transferases ST3Gal1 and ST6GalNAc5. This compound and a number of glycans derived from other globosides and gangliosides were printed as a glycan microarray and their interaction with Siglec-7 was investigated.
A challenging aspect of the preparation of DSGb5 is the regio- and stereoselective introduction of the α2,3- and α2,6-linked sialosides.16 Recently, a number of microbial sialyl transferases have been described that make it possible to prepare gangliosides from the ganglio-, lacto-, and globo-series having α2,8-Neu5Ac-α2,3-Neu5Ac and/or α2,3-sialosides at the terminal galactose.17 These enzymes (e.g. PmST1 and CstII) can readily be expressed in E. coli making it straightforward to install these sialosides. To date, no microbial sialyl transferase has been identified that can selectively install a α2,6-linked sialoside at GalNAc of a Gal-β1,3-GalNAc epitope. Recently, considerable progress has been made in the expression of human sialyl transferases,18 and therefore we were compelled to investigate whether DSGb5 can be prepared by human ST3Gal1 and ST6GalNAc6, which are enzymes that can install an α2,3- and α2,6- sialoside at the terminal Gal and internal GalNAc, respectively.19
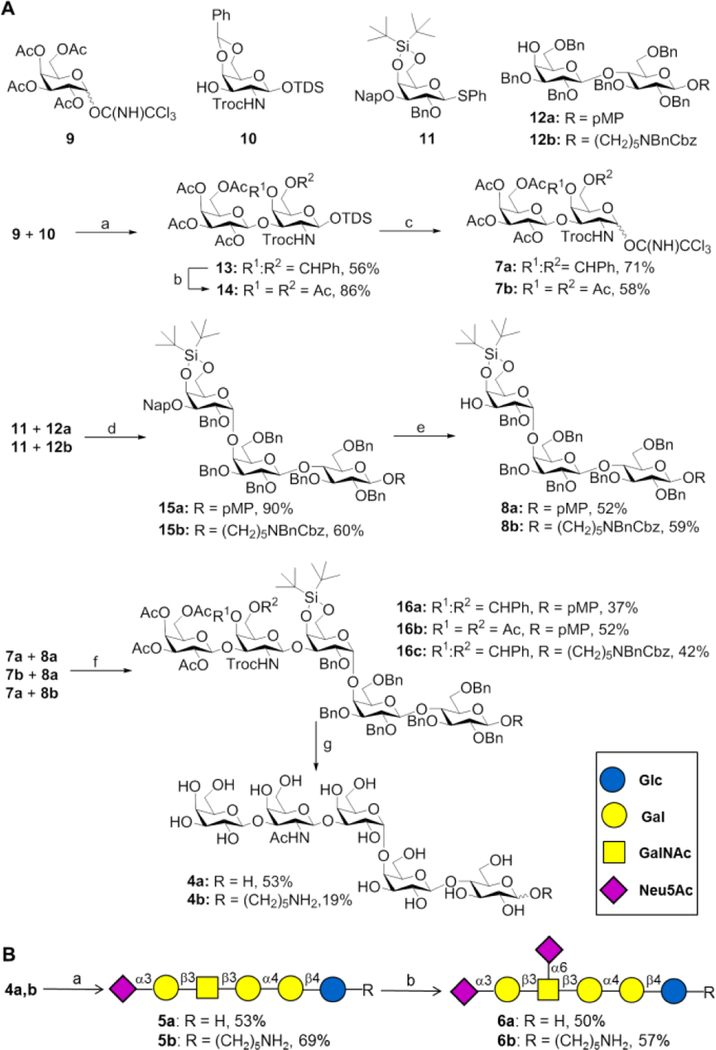
It was envisaged that the oligosaccharide moiety of Gb5 (4a) could be assembled by block coupling of disaccharide 7a with trisaccharide 8a, which in turn were expected to be available from building blocks 9, 10, 11 and 12 (Scheme 1). We anticipated that a chemical strategy to prepare Gb5 would be more attractive than reported chemoenzymatic20 or enzymatic approaches using microbial enzymes because these give low conversions, requiring large quantities of enzyme and can be promiscuous to give unwanted side products such as Gb5 modified by additional β1,3-Gal that is difficult to remove.17b, 21 Thus, a TMSOTf catalyzed glycosylation of trichloroacetimidate 9 with galactosamine acceptor 10 gave disaccharide 13 in a yield of 56% as only the β-anomer. The anomeric TDS protecting group of 13 could easily be cleaved by treatment with HF-pyridine in pyridine to give a lactol, which was converted into trichloroacetimidate 7a by reaction with trichloroacetonitrile in the presence of Cs2CO3. The use of DBU as the base resulted in substantially lower yield due to hydrolysis of the base sensitive Troc protecting group.
Scheme 1.
Chemoenzymatic synthesis of DSGb5. (A) Chemical synthesis of Gb5. Reagents and conditions: (a) TMSOTf, CH2Cl2, 4 Å MS, −35 °C; (b) i. 80% AcOH, 80 °C, ii. Ac2O, pyridine, DMAP; (c) i. HF·pyridine, pyridine, ii. Cs2CO3, Cl3CCN, CH2Cl2, 0 °C; (d) NIS, TfOH, CH2Cl2, 4 Å MS, −30 °C; (e) DDQ, CH2Cl2/PBS buffer; (f) TMSOTf, CH2Cl2, 4 Å MS, −30 °C; (g) i. HF·pyridine, ii. NaOH, THF, 80 °C, iii. Ac2O, pyridine, iv. CAN, CH3CN/H2O, 0 °C (for 16a and 16b), v. NaOMe, MeOH, vi. Pd(OH)2/C, H2, MeOH/H2O/AcOH. (B) Enzymatic extension to produce DSGb5. Reagents and conditions: (a) ST3Gal1, CMP-Neu5Ac, CIAP, MgCl2 (20 mM), sodium cacodylate buffer (50 mM, pH 7.5), 37 °C; (b) ST6GalNAc5, CMP-Neu5Ac, CIAP, MgCl2 (20 mM), sodium cacodylate buffer (50 mM, pH 7.5), 37 °C.
Trisaccharide acceptor 8a was prepared by a glycosylation of thioglycosyl donor 11 with lactosyl acceptor 12a using NIS/TMSOTf as the promoter system. The glycosylation proceeded with absolute α-anomeric selectivity due to the presence of the bulky 4,6-di-O-tert-butyl-silane protecting group that sterically blocks the β-face of the acceptor.22 The trisaccharide was isolated in a yield of 90% after purification by silica column chromatography. Treatment of 15a with DDQ in a mixture of DCM and PBS (24/1 v/v) gave, after purification by silica column chromatography, acceptor 8a in a yield of 52%.
Glycosylation of disaccharide 7a and trisaccharide 8a in the presence of TMSOTf in DCM at −30 °C resulted in the formation of pentasaccharide 16a in a yield of 59% as a separable mixture of α/β anomers (β/α= 1.7). Optimization of the reaction conditions revealed that the overall yield of the glycosylation could be increased by lowering the reaction temperature, however, this did not affect the poor anomeric selectivity (SI, Table S1). Fortunately, the use of glycosyl donor 7b, having acetyl esters instead of a benzylidene acetal at the 4,6-diol of GalNTroc,23 gave in a TMSOTf mediated glycosylation with acceptor 8a at −50 °C, pentasaccharide 16b as only the β-anomer in an isolated yield of 52%.
Pentasaccharide 16a was deprotected by the six-step procedure to give Gb5 (4a). Thus, the silyl protecting group was cleaved by treatment with HF·pyridine which was followed by hydrolysis of the acetyl esters and Troc protecting group with aqueous NaOH in THF with heating (80 oC). The resulting compound was acetylated with acetic anhydride in pyridine and then the anomeric methoxyphenyl (MP) protecting group was oxidatively removed by cerium ammonium nitrate (CAN) in a mixture of acetonitrile and H2O. Finally, deacetylation under Zemplén conditions (cat. NaOMe in MeOH) followed by hydrogenation over Pd(OH)2/C in a mixture of MeOH/H2O/HOAc afforded Gb5 (4a) in an overall yield of 53% after purification by Bio-Gel P-2 size exclusion chromatography followed by semi-preparative HPLC using a HILIC column (XBridge® Amide 5 μm, 4.6 mm x 250 mm, Waters).
Next, attention was focused on the enzymatic sialylation of 4a to give the oligosaccharide moiety of DSGb5. Thus, 4a was treated with ST3Gal1 in the presence of CMP-Neu5Ac (1.5 eq.) in sodium cacodylate buffer (pH = 7.5, 50 mM) containing MgCl2 (20 mM) at 37 °C. The reaction was performed in the presence of CIAP to hydrolyse CMP which may cause product inhibition.24 Analysis of the reaction mixture by TLC and MALDI-TOF MS indicated that after an incubation time of 4 days, all starting material had been converted into product. Interestingly, Gb5 proved to be a rather poor substrate for the microbial α2,3-sialyl transferase (PmST1) and even after prolonged incubation, only partial conversion was observed (∼30%). Next, compound 5a was treated with recombinant ST6GalNAc6 and surprisingly, no product formation was detected. Gratifyingly, the use of ST6GalNAc5 could readily add the second sialoside to provide DSGb5 (6a). We found this enzyme requires an α2,3-linked sialoside for activity and the use of Gal(β1,3)GalNAc did not give product whereas Neu5Ac(α2,3)Gal(β1,3)GalNAc was readily modified by ST6GalNAc5. These results indicate that the biosynthesis of DSGb5 involves an orchestrated attachment of the sialosides in which the 2,3-linked Neu5Ac is first installed, followed by the introduction of the 2,6-sialoside. MSGb5 (5a) and DSGb5 (6a) were purified by Bio-Gel P-2 size exclusion column chromatography followed HPLC using a HILIC column and the resulting compounds were fully characterized by high resolution mass spectrometry and multi-dimensional NMR. A ROESY experiment showed close proximity of H-4 and H-6 of GalNAc with H-3ax of branching Neu5Ac confirming proper connectivity of the α2,6-sialoside of DSGb5.
We also prepared the oligosaccharides of Gb5, MSGb5 and DSGb5 modified by an amino pentyl linker (4b, 5b and 6b, respectively) in a similar fashion by using acceptor 12b instead of 12a. These compounds were printed as a microarray to explore ligand requirements of Siglec-7.
The Siglecs are a family of transmembrane cell surface receptors expressed on hemopoietic cells that can bind specific sialic acid containing glycoconjugates.25 Such binding events results in inhibitory signals that dampen innate and adaptive immune responses. Siglec‐7 is predominantly expressed on natural killer (NK) cells, and its engagement with specific sialoglycans on target cells results in inhibition of NK cellular toxicity. Over-expression of Siglec-7 ligands on cancer cells is a proposed mechanism of immune escape, and reversal of such interactions may lead to a new class of checkpoint inhibitors.26 In vitro binding studies have indicated that Siglec-7 has a preference for glycoconjugates bearing a Neu5Acα(2,8)Neu5Ac motif such as present in b-series of gangliosides including GD3, GD2, GT1b and GQ1b.27 It has been shown that the expressing of GD3 on a target cells leads to suppression of NK mediated cytolytic activity in a Siglec-7 dependent manner.28 There are also indications that glycans bearing an internal branching α(2,6)-linked sialic acid at GalNAc or GlcNAc, such as present in LSTb, disiayl Lewisa and DSGb5, can also be recognized by Siglec-7.15a, 27, 29
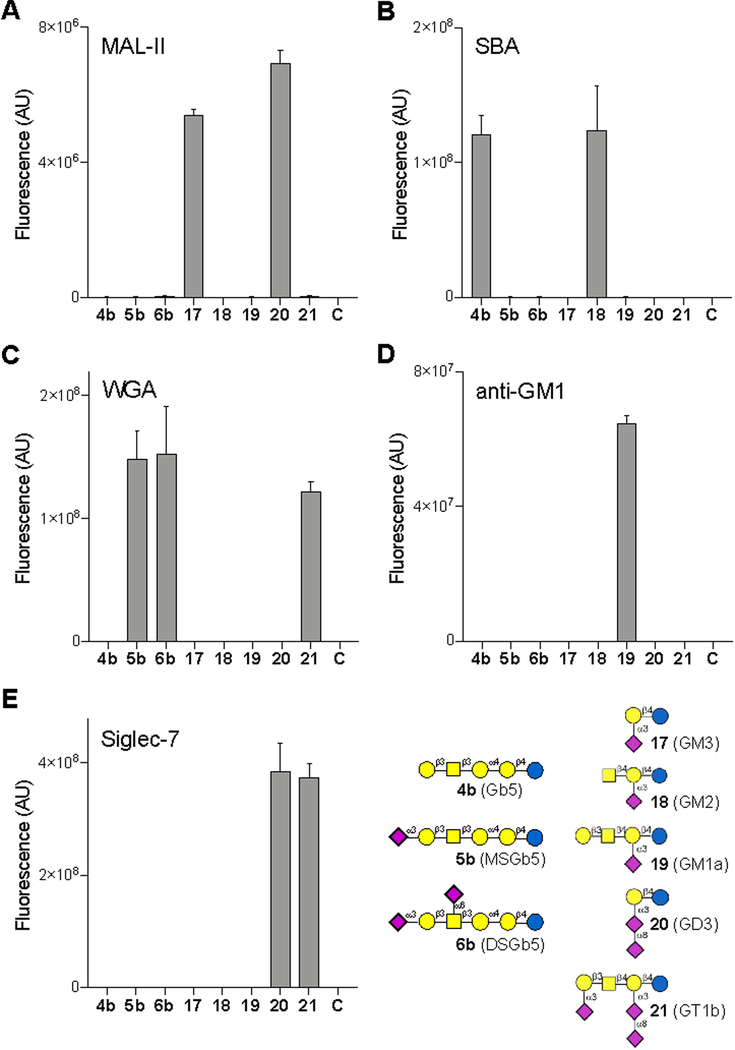
A glycan microarray was created by piezoelectric non-contact printing of compounds 4b, 5b and 6b and GM3 (17), GM2 (18), GM1a (19), GD3 (20) and GT1b (21)17b, 30 on N-hydroxysuccinimide (NHS)-activated glass slides as replicates of 6. To validate proper printing, the microarray was examined for binding of the biotinylated lectins Maackia amurensis leukagglutinin (MAL-II), soybean agglutinin (SBA) and wheat germ agglutinin (WGA). These lectins were preincubated with Streptavidin-AlexaFluor635 and binding of immobilized oligosaccharides was established by measuring fluorescence intensity using a microarray scanner. As anticipated, MAL-II, which is known to recognize Neu5Ac(α2,3)Gal(β1,4)GlcNAc/Glc, did bind GM3 (17) and GM1 (20) having such an oligosaccharide fragment (Fig. 2). SBA recognized compounds 4b and 18, which contain a terminal Gal and GalNAc residue, respectively that are known to be ligands for this lectin. WGA, which binds the GlcNAc moiety but also some forms of sialic acid, bound compounds 5b, 6b and 21, indicating it has a preference for Neu5Ac(α2,3)Gal(β1,3)GalNAc containing oligosaccharides. Next, the array was incubated with biotin-conjugated ganglioside GM1 polyclonal antibody, and as anticipated only binding to GM1 was detected. Interestingly, a similar binding experiment with recombinant human Siglec-7 comp showed binding to GD3 (20) and GT1b (21), however no recognition of DSGb5 (6b) was observed. These results indicate that Siglec-7 has a strong preference for α2,8-Neu5Ac-α2,3-Neu5Ac containing oligosaccharides, and has low or no affinity for DSGb5, which has a branching α2,6- and a terminal α2,3-sialoside.31 The previously proposed interaction of DSGb5 with Siglec-7 was based on the observation that cells that express DSGb5 bind to Siglec-7 transfected COS‐7 cells. Furthermore, knockdown of ST6GalNAc6 of cancer renal cancer cells resulted in a reduced expression of DSGb5 and a substantial lower binding of a Siglec-7-Fc fusion protein.15 ST6GalNAc6 has been implicated in the biosynthesis of various other gangliosides including GM1b, GT1b and GD1a, and it is likely that knockdown of ST6GalNAc6 results in a lower expression of these gangliosides, which may affect Siglec-7 binding.32 Furthermore, this transferase is also involved in the biosynthesis of disiayl Lewisa, which is also a proposed ligand for Siglec-7.33
Figure 2.
Microarray results of the synthetic compounds with (A) MAL-II, (B) SBA, (C) WGA, (D) anti-GM1 and (E) Siglec-7. C indicates buffer control. Bars represent the mean ± SD.
Although no binding was observed on the glycan microarray, it cannot be excluded that the ceramide moiety of DSGb5 can modulate Siglec-7 binding by organizing it into microdomains for low-affinity high-avidity multivalent interactions.34 Therefore, our future studies will focus on the preparation of DSGb5 having a ceramide moiety for binding and cellular activation studies. Such a compound will also be valuable to explore other cellular roles of DSGb5 such as promoting tumor cell migration.13 Furthermore, a very recent study, employing a library of isogenic HEK293 cells with combinatorically engineered glycosylation capacities, indicated that Siglec-7 recognizes core 2 O-glycans having 2,3- and 2,6-linked sialosides which will also be an important target for future synthesis.35 It is clear that the ability of glycans having an internal α2,6-sialoside at GlcNAc and GalNAc to mediate cellular activation in Siglec-7 dependent manner needs further investigation.
Conclusions
In conclusion, DSGb5 was synthesized by a chemoenzymatic approach in which the oligosaccharide moiety of Gb5 was assembled chemically by a block coupling approach followed by enzymatic sialylation using the mammalian sialyltransferases, ST3Gal1 and ST6GalNAc5 to install an α2,3- and α2,6-linked sialoside, respectively. Glycan microarray binding studies indicate that the oligosaccharide moiety of DSGb5 is not recognized by Siglec-7, and it is likely this ganglioside promotes tumorigenesis through other mechanisms such as increasing cell migration and invasion.
Supplementary Material
Acknowledgements
This research was supported by Utrecht University. Siglec-7 was a kind gift from Dr. R.L. Schnaar.
Footnotes
Electronic Supplementary Information (ESI) available: Materials and methods, analytical data and copies of NMR spectra. See DOI: 10.1039/x0xx00000x
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.(a) Schnaar RL, Gerardy-Schahn R and Hildebrandt H, Physiol. Rev, 2014, 94, 461; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Furukawa K, Ohmi Y, Tajima O, Ohkawa Y, Kondo Y, Shuting J, Hashimoto N and Furukawa K, in Gangliosides in Health and Disease eds. Schnaar RL and Lopez PHH, Elsevier B.V., 2018, vol. 156, pp. 265. [DOI] [PubMed] [Google Scholar]
- 2.Ho MY, Yu AL and Yu J, Glycoconj. J, 2017, 34, 765. [DOI] [PubMed] [Google Scholar]
- 3.Liang YJ, Kuo HH, Lin CH, Chen YY, Yang BC, Cheng YY, Yu AL, Khoo KH and Yu J, Proc. Natl. Acad. Sci. U. S. A, 2010, 107, 22564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Chang WW, Lee CH, Lee P, Lin J, Hsu CW, Hung JT, Lin JJ, Yu JC, Shao LE, Yu J, Wong CH and Yu AL, Proc. Natl. Acad. Sci. U. S. A, 2008, 105, 11667; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hung TC, Lin CW, Hsu TL, Wu CY and Wong CH, J. Am. Chem. Soc, 2013, 135, 5934; [DOI] [PubMed] [Google Scholar]; (c) Sivasubramaniyan K, Harichandan A, Schilbach K, Mack AF, Bedke J, Stenzl A, Kanz L, Niederfellner G and Buhring HJ, Glycobiology, 2015, 25, 902; [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Cheung SK, Chuang PK, Huang HW, Hwang-Verslues WW, Cho CH, Yang WB, Shen CN, Hsiao M, Hsu TL, Chang CF and Wong CH, Proc. Natl. Acad. Sci. U. S. A, 2016, 113, 960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olie RA, Fenderson B, Daley K, Oosterhuis JW, Murphy J and Looijenga LH, Br. J. Cancer, 1996, 74, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ando R, Tokuda N, Yamamoto T, Ikeda K, Hashimoto N, Taguchi R, Fan X, Furukawa K, Niimura Y, Suzuki A, Goto M and Furukawa K, Glycoconj. J, 2016, 33, 169. [DOI] [PubMed] [Google Scholar]
- 7.Hakomori S and Zhang Y, Chem. Biol, 1997, 4, 97. [DOI] [PubMed] [Google Scholar]
- 8.(a) Tsai YC, Huang J, Cheng R,JY, Lin JJ, Hung JT, Wu YY, Yeh KT and Yu AL, J. Cancer Sc. Ther, 2013, 5, 264; [Google Scholar]; (b) Wondimu A, Liu Y, Su Y, Bobb D, Ma JS, Chakrabarti L, Radoja S and Ladisch S, Cancer Res, 2014, 74, 5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ono M and Hakomori S, Glycoconj. J, 2004, 20, 71. [DOI] [PubMed] [Google Scholar]
- 10.(a) Lassaletta JM and Schmidt RR, Tetrahedron Lett, 1995, 36, 4209; [Google Scholar]; (b) Ishida H, Miyawaki R, Kiso M and Hasegawa A, J. Carbohydr. Chem, 1996, 15, 163; [DOI] [PubMed] [Google Scholar]; (c) Hsu CH, Chu KC, Lin YS, Han JL, Peng YS, Ren CT, Wu CY and Wong CH, Chem. Eur. J, 2010, 16, 1754. [DOI] [PubMed] [Google Scholar]
- 11.Danishefsky SJ, Shue YK, Chang MN and Wong CH, Acc. Chem. Res, 2015, 48, 643. [DOI] [PubMed] [Google Scholar]
- 12.Saito S, Levery SB, Salyan ME, Goldberg RI and Hakomori S, J. Biol. Chem, 1994, 269, 5644. [PubMed] [Google Scholar]
- 13.Kawasaki Y, Ito A, Kakoi N, Shimada S, Itoh J, Mitsuzuka K and Arai Y, Tohoku J. Exp. Med, 2015, 236, 1. [DOI] [PubMed] [Google Scholar]
- 14.Itoh J, Ito A, Shimada S, Kawasaki Y, Kakoi N, Saito H, Mitsuzuka K, Watanabe M, Satoh M, Saito S and Arai Y, Glycoconj. J, 2017, 34, 267. [DOI] [PubMed] [Google Scholar]
- 15.(a) Ito A, Handa K, Withers DA, Satoh M and Hakomori S, FEBS Lett, 2001, 498, 116; [DOI] [PubMed] [Google Scholar]; (b) Kawasaki Y, Ito A, Withers DA, Taima T, Kakoi N, Saito S and Arai Y, Glycobiology, 2010, 20, 1373. [DOI] [PubMed] [Google Scholar]
- 16.Chen X and Varki A, ACS Chem. Biol, 2010, 5, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Yu H, Huang S, Chokhawala H, Sun M, Zheng H and Chen X, Angew. Chem. Int. Ed, 2006, 45, 3938; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yu H, Li Y, Zeng J, Thon V, Nguyen DM, Ly T, Kuang HY, Ngo A and Chen X, J. Org. Chem, 2016, 81, 10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moremen KW, Ramiah A, Stuart M, Steel J, Meng L, Forouhar F, Moniz HA, Gahlay G, Gao Z, Chapla D, Wang S, Yang JY, Prabhakar PK, Johnson R, Rosa MD, Geisler C, Nairn AV, Seetharaman J, Wu SC, Tong L, Gilbert HJ, LaBaer J and Jarvis DL, Nat. Chem. Biol, 2018, 14, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Saito S, Aoki H, Ito A, Ueno S, Wada T, Mitsuzuka K, Satoh M, Arai Y and Miyagi T, J. Biol. Chem, 2003, 278, 26474; [DOI] [PubMed] [Google Scholar]; (b) Senda M, Ito A, Tsuchida A, Hagiwara T, Kaneda T, Nakamura Y, Kasama K, Kiso M, Yoshikawa K, Katagiri Y, Ono Y, Ogiso M, Urano T, Furukawa K, Oshima S and Furukawa K, Biochem. J, 2007, 402, 459; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Gupta R, Matta KL and Neelamegham S, Biochem. Biophys. Res. Commun, 2016, 469, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S-P, Hsiao W-C, Yu C-C, Chien W-T, Lin H-J, Huang L-D, Lin C-H, Wu W-L, Wu S-H and Lin C-C, Adv. Synth. Catal, 2014, 356, 3199. [Google Scholar]
- 21.(a) Su DM, Eguchi H, Yi W, Li L, Wang PG and Xia C, Org. Lett, 2008, 10, 1009; [DOI] [PubMed] [Google Scholar]; (b) Tsai TI, Lee HY, Chang SH, Wang CH, Tu YC, Lin YC, Hwang DR, Wu CY and Wong CH, J. Am. Chem. Soc, 2013, 135, 14831. [DOI] [PubMed] [Google Scholar]
- 22.Kumagai D, Miyazaki M and Nishimura S, Tetrahedron Lett, 2001, 42, 1953. [Google Scholar]
- 23.Wang Z, Gilbert M, Eguchi H, Yu H, Cheng J, Muthana S, Zhou L, Wang PG, Chen X and Huang X, Adv. Synth. Catal, 2008, 350, 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unverzagt C, Kunz H and Paulson JC, J. Am. Chem. Soc, 1990, 112, 9308. [Google Scholar]
- 25.Crocker PR, Paulson JC and Varki A, Nat. Rev. Immunol, 2007, 7, 255. [DOI] [PubMed] [Google Scholar]
- 26.Hudak JE, Canham SM and Bertozzi CR, Nat. Chem. Biol, 2014, 10, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaji T, Teranishi T, Alphey MS, Crocker PR and Hashimoto Y, J. Biol. Chem, 2002, 277, 6324. [DOI] [PubMed] [Google Scholar]
- 28.Nicoll G, Avril T, Lock K, Furukawa K, Bovin N and Crocker PR, Eur. J. Immunol, 2003, 33, 1642. [DOI] [PubMed] [Google Scholar]
- 29.Blixt O, Collins BE, van den Nieuwenhof IM, Crocker PR and Paulson JC, J. Biol. Chem, 2003, 278, 31007. [DOI] [PubMed] [Google Scholar]
- 30.Li T, Liu L, Wei N, Yang JY, Chapla DG, Moremen KW and Boons GJ, Nat. Chem, 2019, 11, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A similar finding was reported during the preparation of this manuscript: Li PJ, Huang SY, Chiang PY, Fan CY, Guo LJ, Wu DY, Angata T and Lin CC, Angew. Chem. Int. Ed, 2019, In press. [Google Scholar]
- 32.Takashima S and Tsuji S, Trends Glycosc. Glycotechnol, 2011, 23, 178. [Google Scholar]
- 33.Miyazaki K, Ohmori K, Izawa M, Koike T, Kumamoto K, Furukawa K, Ando T, Kiso M, Yamaji T, Hashimoto Y, Suzuki A, Yoshida A, Takeuchi M and Kannagi R, Cancer Res, 2004, 64, 4498. [DOI] [PubMed] [Google Scholar]
- 34.Simons K and Toomre D, Nat. Rev. Mol. Cell Biol, 2000, 1, 31. [DOI] [PubMed] [Google Scholar]
- 35.Narimatsu Y, Joshi HJ, Nason R, Van Coillie J, Karlsson R, Sun L, Ye Z, Chen YH, Schjoldager KT, Steentoft C, Furukawa S, Bensing BA, Sullam PM, Thompson AJ, Paulson JC, Bull C, Adema GJ, Mandel U, Hansen L, Bennett EP, Varki A, Vakhrushev SY, Yang Z and Clausen H, Mol. Cell, 2019, 75, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.