Abstract
Background
Circular RNAs (circRNAs), a subclass of non-coding RNAs, play essential roles in tumorigenesis and aggressiveness. Our previous study has identified that circAGO2 drives gastric cancer progression through activating human antigen R (HuR), a protein stabilizing AU-rich element-containing mRNAs. However, the functions and underlying mechanisms of circRNAs derived from HuR in gastric cancer progression remain elusive.
Methods
CircRNAs derived from HuR were detected by real-time quantitative RT-PCR and validated by Sanger sequencing. Biotin-labeled RNA pull-down, mass spectrometry, RNA immunoprecipitation, RNA electrophoretic mobility shift, and in vitro binding assays were applied to identify proteins interacting with circRNA. Gene expression regulation was observed by chromatin immunoprecipitation, dual-luciferase assay, real-time quantitative RT-PCR, and western blot assays. Gain- and loss-of-function studies were performed to observe the impacts of circRNA and its protein partner on the growth, invasion, and metastasis of gastric cancer cells in vitro and in vivo.
Results
Circ-HuR (hsa_circ_0049027) was predominantly detected in the nucleus, and was down-regulated in gastric cancer tissues and cell lines. Ectopic expression of circ-HuR suppressed the growth, invasion, and metastasis of gastric cancer cells in vitro and in vivo. Mechanistically, circ-HuR interacted with CCHC-type zinc finger nucleic acid binding protein (CNBP), and subsequently restrained its binding to HuR promoter, resulting in down-regulation of HuR and repression of tumor progression.
Conclusions
Circ-HuR serves as a tumor suppressor to inhibit CNBP-facilitated HuR expression and gastric cancer progression, indicating a potential therapeutic target for gastric cancer.
Keywords: Circular RNAs, Human antigen R, Gastric cancer, CCHC-type zinc finger nucleic acid binding protein
Background
Gastric cancer is one of the leading causes of cancer-related death, mainly due to high rate of recurrence and distant metastasis in advanced cases [1]. Thus, it is urgent to elucidate the mechanisms underlying the progression of gastric cancer. Human antigen R (HuR) is a member of the embryonic lethal abnormal visual protein (ELAV) family that is involved in nervous system development, cellular proliferation, and migration [2]. As a RNA binding protein (RBP), HuR increases mRNA stability of target genes through binding to poly-U elements or AU-rich elements (AREs) in 3′-untranslated region (3′-UTR) [3], and plays an important role in tumor development, recurrence, invasion, and metastasis [4]. Studies have shown that HuR is highly expressed in breast cancer, colorectal cancer, gastric cancer, and prostate cancer, and is closely related to clinicopathological features, lymph node metastasis, low survival rate, and poor prognosis of cancer patients [5–9]. However, the mechanisms regulating HuR expression during gastric cancer progression still remain to be elucidated.
Circular RNAs (circRNAs), a group of transcripts characterized by closed continuous loops stably existing in tissues and cells, are essential regulators of gene expression, while dysregulated circRNAs have been identified in almost all types of cancers [10]. Previous studies show that circRNAs play multiple important roles in cellular physiology via acting as microRNA (miRNA) sponges, RBP-binding molecules, transcriptional regulators, or templates for protein translation. The inspiring miRNA sponging activity has been proven in CDR1as and ciRS-7 [11, 12], and ciRS-7 contains 70 target sites of miRNA-7 to act as a competing endogenous RNA. However, the majority of circRNAs harbor few binding sites for a single miRNA, and high-throughput sequencing analysis reveals that miRNA sponging mechanism cannot be widely applied across the properties of circRNAs [13]. Recent studies have shown the emerging roles of circRNAs in control of gene expression via physical interaction with proteins. For example, circ-Amotl1 derived from angiomotin-like 1 (AMOTL1) promotes tumorigenesis by binding to and retaining c-Myc within the nuclei in breast cancer [14]. Circ-CTNNB1 generated by β-catenin (CTNNB1) activates Wnt signaling pathway by interacting with DEAD-box polypeptide 3 [15]. In addition, circ-EIF3J generated by eukaryotic translation elongation factor 3 J (EIF3J) interacts with U1 snRNP and RNA polymerase II to promote parental gene transcription [16]. In our previous work, we have demonstrated that HuR is highly expressed in gastric cancer tissues and cells, while circRNA derived from Argonaute 2 (circAGO2) binds and promotes the recruitment of HuR protein to the 3′-UTR of target genes, and facilitates the proliferation, invasion, and metastasis of gastric cancer cells, suggesting that circRNA can regulate HuR activity in gastric cancer [17]. Meanwhile, the roles of circRNAs in regulating HuR expression in gastric cancer still remain largely elusive.
In this study, we identify a circRNA consisting of exons 3, 4, and 5 of HuR (circ-HuR) as a novel tumor suppressor in gastric cancer. We discover that circ-HuR is significantly down-regulated in gastric cancer, and effectively inhibits the growth, invasion, and metastasis of gastric cancer cells. Mechanistically, circ-HuR directly interacted with CCHC-type zinc finger nucleic acid binding protein (CNBP), and acted as an inhibitor to restrain the binding of CNBP to HuR promoter, resulting in repression of HuR expression and tumor progression, which indicates the essential roles of circ-HuR and CNBP in gastric cancer progression.
Materials and methods
Patient tissues and cell culture
Tumor and adjacent normal (peritumor) tissues of 81 gastric cancer cases were obtained at surgery in Union Hospital of Tongji Medical College, with demographic and clinicopathological details indicated in Additional file 1: Table S1. The Institutional Review Board of Tongji Medical College approved human tissue study (approval number: 2011-S085). All procedures were carried out in accordance with guidelines set forth by Declaration of Helsinki. Written informed consent was obtained from all patients. Fresh tumor tissues were validated by pathological diagnosis, frozen in liquid nitrogen, and stored at − 80 °C. Human embryonic kidney HEK293T (CRL-11268) cells and gastric cancer cell lines AGS (CRL-1739), MKN-45 (JCRB0254), MKN-74 (JCRB0255) and NCI-N87 (CRL-5822) were obtained from American Type Culture Collection (Rockville, MD) and Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan), authenticated by short tandem repeat (STR) profiling, and applied for study within 6 months following resuscitation of frozen aliquots. Cell lines were cultured in RPMI 1640 medium (Gibco, Carlsbad, CA, USA) supplied with 10% fetal bovine serum (Gibco) at 37 °C in a humidified atmosphere of 5% CO2.
RT-PCR and real-time quantitative RT-PCR (qRT-PCR)
Total RNA was isolated according to the instructions of RNeasy Mini Kit (QIAGEN, Stockach, Germany). For circRNA detection, treatment with RNase R (3 U/mg, Epicenter, Madison, WI, USA) was undertaken at 37 °C for 15 min, and cDNA was synthesized by using reverse transcription kit (Takara, Dalian, China). Genomic DNA (gDNA) was isolated with DNA Mini Kit (QIAGEN). Quantification of mRNA, circular RNA and gDNA was performed by using a SYBR Green PCR Kit (Takara), primers (Additional file 1: Table S2) and Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). The levels of circRNA and mRNA were normalized to those of β-actin and determined by 2-△△Ct method.
Western blot assay
Cellular proteins were extracted with RIPA lysis buffer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Western blot assay was performed as previously described [18, 19], with antibodies specific for HuR (ab200342), cyclin D2 (CCND2, ab207604), CTNNB1 (ab32572), CNBP (ab48027), enolase 1 (ENO1, ab155102), nucleosome assembly protein 1 like 1 (NAP1L1, ab33076), matrix metalloproteinases 14 (MMP-14, ab51074), c-Myc (ab32072), Flag (ab45766), β-actin (ab125402, Abcam lnc., Cambridge, MA, USA), heparin binding growth factor (HDGF, A10435), non-POU domain containing octamer binding (NONO, A5282), splicing factor proline and glutamine rich (SFPQ, A0958, ABclonal, Wuhan, China), glutathione S-transferase (GST, sc-33,614), histone H3 (sc-24,516), or glyceraldehyde 3-phosphate dehydrogenase (GAPDH, sc-47,724, Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Plasmid construction and stable transfection
Linear circ-HuR and hsa_circ_23897 were synthesized by TSINGKE (Wuhan, China) and inserted into pLCDH-ciR (Geenseed Biotech Co., Guangzhou, China). Mutation of back-splicing elements of circ-HuR or hsa_circ_23897 vector was prepared with GeneTailor™ Site-Directed Mutagenesis System (Invitrogen, Carlsbad, CA, USA) and primers (Additional file 1: Table S3). Human CNBP cDNA (540 bp) was subcloned into CV186 (Genechem Co., Ltd., Shanghai, China), while its truncations were obtained by PCR amplification (Additional file 1: Table S3) and inserted into pCMV-3Tag-1A or pGEX-6P-1 (Addgene, Cambridge, MA, USA), respectively. Two independent single guide RNAs (sgRNAs) targeting downstream region of CNBP transcription start site (Additional file 1: Table S4) were inserted into dCas9-BFP-KRAB (Addgene). Oligonucleotides specific for short hairpin RNAs (shRNAs) against circ-HuR (Additional file 1: Table S4) were inserted into GV298 (Genechem Co., Ltd). Lentiviral vectors were co-transfected with packaging plasmids psPAX2 and pMD2G into HEK293T cells. Infectious lentiviruses were harvested at 36 and 60 h after transfection, followed with concentration by ultracentrifugation (2 h at 120,000 g). Stable cell lines were obtained by selection with puromycin.
RNA fluorescence in situ hybridization (RNA-FISH)
Digoxin (DIG)-labeled antisense or sense probe for circ-HuR junction sequence (Additional file 1: Table S4) was synthesized. The probes for GAPDH and U1 were generated by in vitro transcription of PCR products (Additional file 1: Table S2) using DIG Labeling Kit (MyLab Corporation, Beijing, China). Hybridization was undertaken using Fluorescent In Situ Hybridization kit (RiboBio, Guangzhou, China) following the manufacturer’s instructions, while the nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The images were analyzed via a Nikon A1Si Laser Scanning Confocal Microscope (Nikon Instruments Inc., Japan).
Dual-luciferase reporter assay
Human HuR promoter (1341 bp) or c-Myc promoter (1363 bp) was amplified from gDNA using primer sets (Additional file 1: Table S3) and subcloned into pGL3-Basic (Promega, Madison, WI, USA). Human MMP-14 promoter reporter was previously described [20]. Mutation of CNBP binding site was established using GeneTailor™ Site-Directed Mutagenesis System (Invitrogen) and primers (Additional file 1: Table S3). Human CNBP activity luciferase reporter was established by inserting oligonucleotides containing four canonical CNBP binding sites (Additional file 1: Table S3) into pGL3-Basic (Promega). Dual-luciferase assay was performed according to the manufacturer’s instructions (Promega).
Biotin-labeled RNA pull-down and mass spectrometry
Biotin-labeled oligonucleotide probes (Additional file 1: Table S4) targeting junction sites of circRNAs were synthesized (Invitrogen). Using Biotin RNA Labeling Mix kit (Roche, Indianapolis, IN, USA) and T7 RNA polymerase, the biotin-labeled RNA probes for circRNAs were in vitro transcribed as previously described [15, 17]. RNA pull-down assay was performed at room temperature, and retrieved proteins were detected by mass spectrometry analysis at Wuhan Institute of Biotechnology (Wuhan, China).
Fluorescence immunocytochemical staining
Cells were plated on coverslip and incubated with antibody specific for CNBP (ab48027, Abcam lnc.) at 4 °C overnight, and incubated with Alexa Fluor 594 goat anti-rabbit IgG and DAPI. The images were photographed under a Nikon A1Si Laser Scanning Confocal Microscope (Nikon Instruments Inc., Japan).
Chromatin immunoprecipitation (ChIP)
ChIP assay was undertaken in accordance with the manual of EZ-ChIP kit (Upstate Biotechnology, Temacula, CA, USA), with antibodies specific for CNBP (ab48027, Abcam lnc.). Primer sets were listed in Additional file 1: Table S2.
Cross-linking RNA immunoprecipitation (RIP) assay
Cells were cross-linked at 254 nm by ultraviolet light, and RIP assay was performed according to the instructions of Magna RIPTM Kit (Millipore, Bedford, MA, USA), with antibodies specific for CNBP (ab48027), ENO1 (ab155102), NAP1L1 (ab33076), Flag (ab45766, Abcam lnc.), HDGF (A10435), NONO (A5282), SFPQ (A0958, ABclonal). Co-precipitated RNAs were detected by RT-PCR or real-time qRT-PCR with specific primers (Additional file 1: Table S2).
In vitro binding assay
Four truncations of CNBP were amplified with primers (Additional file 1: Table S3), and subcloned into pCMV-3Tag-1A or pGEX-6P-1 (Addgene). GST-tagged CNBP protein was produced from E. coli as previously described [18, 21]. The Flag-CNBP, GST-CNBP, and circ-HuR complexes were pulled down using GST or Flag beads (Sigma, St. Louis, MO, USA). Circ-HuR was measured by RT-PCR with divergent primers (Additional file 1: Table S2), whereas protein was detected by SDS-PAGE and western blot.
RNA electrophoretic mobility shift assay (EMSA)
Biotin-labeled circular probe of circ-HuR was prepared as described above. RNA EMSA was conducted according to the instructions of LightShift Chemiluminescent RNA EMSA Kit (Thermo Fisher Scientific, Inc.)
In vitro cell viability, growth, and invasion assays
The in vitro viability, growth, and invasive capabilities of cancer cells were detected by MTT colorimetric, soft agar, and matrigel invasion assays as described previously [15, 21].
Xenografts in nude mice
All animal experiments were carried out in accordance with NIH Guidelines for the Care and Use of Laboratory Animals, and approved by the Animal Care Committee of Tongji Medical College (approval number: Y20080290). In vivo tumor growth and experimental metastasis studies were performed with blindly randomized four-week-old male BALB/c nude mice (n = 5 per group). For tumor growth studies, AGS cells (1 × 106) stably transfected with vectors were injected into the upper back of nude mice (n = 5 per group). For metastasis studies, AGS cells (0.4 × 106) stably transfected with vectors were injected from tail vein of nude mice (n = 5 per group). The tumor volume and survival time of each mouse were monitored and recorded, while xenografts were imaged using In-Vivo Xtreme II small animal imaging system (Bruker Corporation, Billerica, MA) and detected by hematoxylin and eosin (H&E) and immunohistochemical staining.
Immunohistochemistry
Immunohistochemical staining and quantitative evaluation was performed as previously described [15, 21], with antibodies specific for Ki-67 (sc-23,900, Santa Cruz Biotechnology; 1:100 dilution) or CD31 (ab28364, Abcam Inc.; 1:100 dilution). The degree of positivity was measured according to percentage of positive cancer cells.
Statistical analysis
All data were shown as mean ± standard error of the mean (SEM) processed by GraphPad Prism 5.0 (La Jolla, USA). Student’s t-test, analysis of variance (ANOVA), and chi-square analysis were used to evaluate the difference. Pearson’s correlation coefficient assay was used to analyze expression correlation. Log-rank test and Cox regression models were used to assess survival difference and hazard ratio. All statistical tests were two-sided and considered statistically significant when P values less than 0.05.
Results
Circ-HuR is down-regulated and decreases HuR expression in gastric cancer
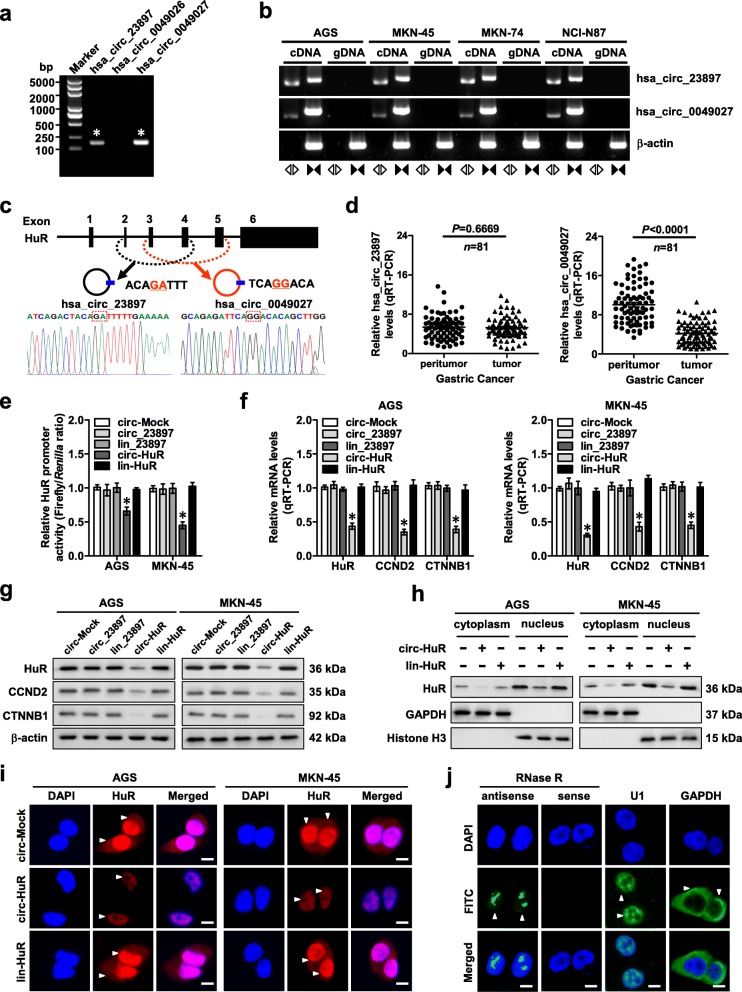
Analysis of circRNA sequencing databases circBase (http://www.circbase.org/) and circRNADb (http://reprod.njmu.edu.cn/circrnadb) revealed 3 potential circRNAs derived from HuR. Among them, hsa_circ_23897 and hsa_circ_0049027 were validated by PCR amplification using divergent primers from cDNA, but not from genomic DNA, of gastric cancer cell lines (Fig. 1a, b). Hsa_circ_23897 was consisted of exons 2, 3, and 4 (446 nt), while hsa_circ_0049027 was composed of exons 3, 4, and 5 of HuR (484 nt), and both of them were validated by Sanger sequencing (Fig. 1c). Endogenous hsa_circ_23897 and hsa_circ_0049027 were resistant to RNase R digestion, while linear mRNA of HuR in AGS and MKN-45 cells was significantly reduced by RNase R treatment (Additional file 1: Figure S1a). In gastric tissues and cell lines, hsa_circ_0049027 was remarkably down-regulated, while hsa_circ_23897 levels were not significantly altered (Fig. 1d and Additional file 1: Figure S1b). To investigate the functions of these circRNAs, hsa_circ_23897 (circ_23897), hsa_circ_0049027 (termed as circ-HuR), and their corresponding linear transcript (lin_23897 and lin-HuR) constructs were stably transfected into AGS and MKN-45 cells (Additional file 1: Figure S1c). Ectopic expression of circ-HuR, but not of circ_23897, lin_23897, or lin-HuR, attenuated the promoter activity of HuR (Fig. 1e) and decreased the transcript and protein levels of HuR and its downstream genes CCND2 [22] and CTNNB1 [23] in AGS and MKN-45 cells (Fig. 1f, g). Meanwhile, stable transfection of two shRNAs targeting the junction of circ-HuR resulted in its down-regulation (Additional file 1: Figure S1d) and up-regulation of HuR and its downstream genes (CCND2 and CTNNB1) in gastric cancer cells (Additional file 1: Figure S1e, f). Accordingly, ectopic expression of circ-HuR, rather than lin-HuR, attenuated the HuR expression in both nucleus and cytoplasm of gastric cancer cells (Fig. 1h, i). RNA-FISH assay indicated the endogenous nuclear enrichment of circ-HuR in AGS cells (Fig. 1j). Additionally, subcellular fractionation assay showed the nuclear localization of exogenous circ-HuR in AGS and MKN-45 cells stably transfected with circ-HuR (Additional file 1: Figure S1g). As a RBP involved in post-transcriptional control of gene expression, the mRNA-stabilizing function of HuR is linked to its localization in the cytoplasm [24, 25]. Neither ectopic expression nor knockdown of circ-HuR affected the half-life of HuR mRNA (Additional file 1: Figure S2a), whereas over-expression of circ-HuR decreased the half-life and transcript levels of CCND2 and CTNNB1 in a HuR-dependent manner (Additional file 1: Figure S2b, c). These results indicated that circ-HuR was down-regulated and suppressed the expression of HuR in gastric cancer.
Fig. 1.
Circ-HuR is down-regulated and decreases HuR expression in gastric cancer. a RT-PCR assay with divergent primers indicating the detection of three circRNAs derived from HuR in AGS cells. b PCR assay with divergent and convergent primers showing the amplification of circRNAs from cDNA or genomic DNA (gDNA) of gastric cancer cell lines, while β-actin was used as a negative control. c Schematic illustration indicating the generation of hsa_circ_23897 and hsa_circ_0049027 from its host gene, and validation by Sanger sequencing. d Real-time qRT-PCR assay showing the relative levels (normalized to β-actin) of hsa_circ_23897 or hsa_circ_0049027 in the peritumor and tumor tissues of gastric cancer (n = 81). e Dual-luciferase assay indicating the promoter activity of HuR in AGS and MKN-45 cells stably transfected with empty vector (circ-Mock), hsa_circ-23897 (circ_23897), linear circ-23897 (lin_23897), circ-HuR, or linear circ-HuR (lin-HuR). f and g Real-time qRT-PCR (f, normalized to β-actin, n = 5) and western blot (g) assays revealing the transcript and protein levels of HuR and its downstream target genes in AGS and MKN-45 cells stably transfected with circ-Mock, circ-23897, lin_23897, circ-HuR, or lin-HuR. h and i Western blot (h) and immunofluorescence (i) assays showing the cytoplasmic and nuclear accumulation of HuR in AGS and MKN-45 cells stably transfected with circ-Mock, circ-HuR, or lin-HuR. Nuclei were stained by DAPI (blue). Scale bar, 10 μm. j RNA-FISH assay indicating the nuclear localization of circ-HuR in AGS cells using an antisense probe (green), while sense probe was used as a negative control. U1 and GAPDH were applied as positive controls. Scale bar, 10 μm. Student’s t-test and ANOVA analyzed the difference in (d-f). *P < 0.01 vs. circ-Mock. Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (a-c) and (e-j)
Over-expression of circ-HuR suppresses the growth and aggressiveness of gastric cancer
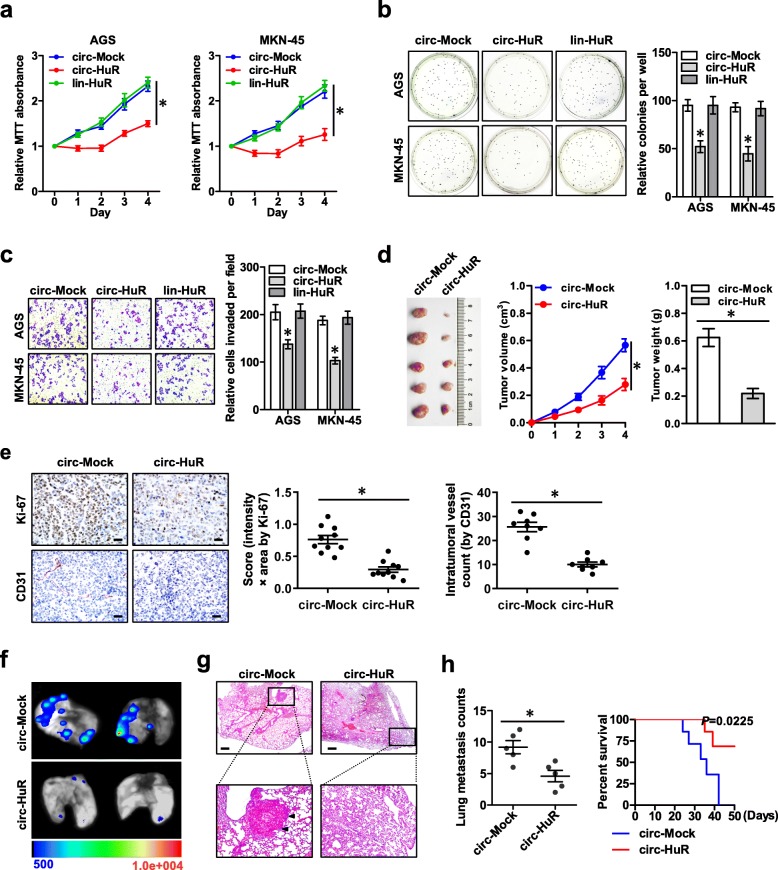
To explore the functions of circ-HuR in gastric cancer, its impacts on tumorigenesis and aggressiveness were investigated in AGS and MKN-45 cells with stable transfection of circ-HuR or its linear transcript (lin-HuR). In MTT colorimetric assay, stable ectopic expression of circ-HuR, but not of lin-HuR, decreased the viability of gastric cancer cells, than those stably transfected with empty vector (circ-Mock) (Fig. 2a). In soft agar and matrigel invasion assays, stable over-expression of circ-HuR reduced the growth and invasion capability of AGS and MKN-45 cells, respectively (Fig. 2b-c), while ectopic expression of lin-HuR did not affect these biological features of cancer cells (Fig. 2b-c). Consistently, stably transfection of circ-HuR resulted in a significant decrease in the growth and weight of xenograft tumors formed by subcutaneous injection of AGS cells into athymic nude mice (Fig. 2d). Immunohistochemical staining showed lower Ki-67 proliferation index and less CD31-positive microvessels of xenograft tumors formed by AGS cells stably transfected with circ-HuR (Fig. 2e). Importantly, athymic nude mice treated with tail vein injection of AGS cells stably transfected with circ-HuR displayed less lung metastatic colonies and greater survival probability (Fig. 2f-h). These results suggested that over-expression of circ-HuR suppressed the growth and aggressiveness of gastric cancer in vitro and in vivo.
Fig. 2.
Over-expression of circ-HuR suppresses the growth and aggressiveness of gastric cancer. a MTT colorimetric assay showing the viability of AGS and MKN-45 cells stably transfected with empty vector (circ-Mock), circ-HuR, or lin-HuR. b and c Soft agar (b) and matrigel invasion (c) assays indicating the in vitro growth and invasion of AGS and MKN-45 cells stably transfected with circ-Mock, circ-HuR, or lin-HuR. d Representative (left panel), in vivo growth curve (middle panel), and weight at the end points (right panel) of xenograft tumors formed by subcutaneous injection of AGS cells stably transfected with circ-Mock or circ-HuR into the dorsal flanks of nude mice (n = 5 for each group). e Representative images (left panel) and quantification (right panel) of immunohistochemical staining showing the expression of Ki-67 and CD31 within xenograft tumors formed by hypodermic injection of AGS cells stably transfected with circ-Mock or circ-HuR (n = 5 for each group). Scale bars: 50 μm. f-h Representative images (f), H&E staining (g, arrowheads), and quantification (h, left panel) of lung metastatic colonization and Kaplan-Meier curves (h, right panel) of nude mice treated with tail vein injection of AGS cells stably transfected with mock or circ-HuR (n = 5 for each group). Scale bar: 100 μm. ANOVA and Student’s t-test analyzed the difference in a-e and h. Log-rank test for survival comparison in (h). *P < 0.01 vs. circ-Mock. Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (a-c)
Circ-HuR interacts with CNBP protein in gastric cancer cells
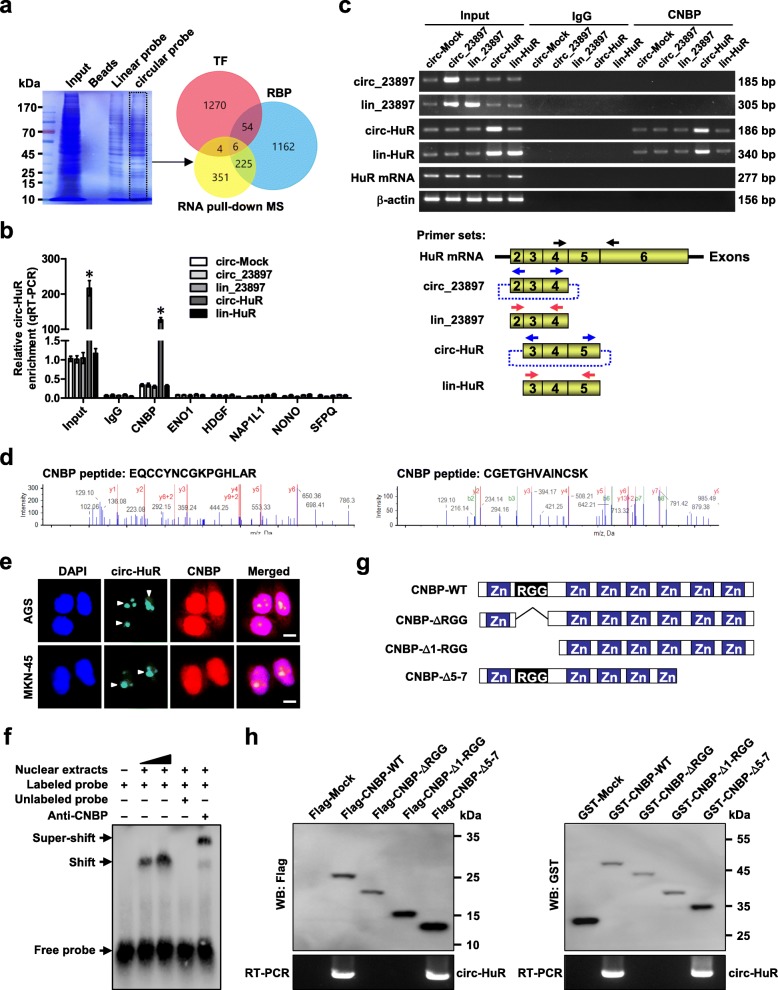
To identify potential protein partner of circ-HuR, we performed proteomic analysis of circ-HuR-associated protein complex in AGS cells by biotin-labeled circular or linear RNA pull-down assay. The circular probe was gained by ligation of linear circ-HuR transcript in vitro and RNase R digestion (Additional file 1: Figure S3a). Mass spectrometry (MS) assay revealed 586 differential proteins between circular and linear circ-HuR pull-down groups, and overlapping analysis with established RBPs (http://www.ablife.cc) and transcription factors (TFs, http://www.genomatix.de) indicated six potential circ-HuR-interacting partners (Fig. 3a and Additional file 2: Table S5). By using probes generated by ligation of linear transcript or synthesized as oligonucleotides targeting junction site, further validating RNA pull-down assay indicated the interaction of circ-HuR with CNBP, rather than ENO1, HDGF, NAP1L1, NONO, or SFPQ, which was increased by over-expression of circ-HuR (Additional file 1: Figure S3b, c). Meanwhile, there was no interaction of CNBP with lin-HuR or circ_23897 in AGS cells (Additional file 1: Figure S3b, c). RIP assay also indicated the binding of circ-HuR to CNBP, rather than ENO1, HDGF, NAP1L1, NONO, or SFPQ (Fig. 3b). Moreover, transfection of circ-HuR, but not of lin-HuR, circ_23897, or lin_23897, led to its increased enrichment in RNA co-precipitated by CNBP antibody in AGS cells (Fig. 3b, c and Additional file 1: Figure S3d). The identified peptides of CNBP from MS assay were shown in Fig. 3d. Dual RNA-FISH and immunofluorescence assay confirmed the co-localization of circ-HuR and CNBP in AGS and MKN-45 cells (Fig. 3e). RNA EMSA using cyclized probes showed that circ-HuR interacted with endogenous CNBP in the nuclear extracts of AGS cells (Fig. 3f). In vitro binding assay indicated that Arg-Gly-Gly (RGG) box domain (22–42 amino acids), but not other domains, of Flag-tagged or GST-tagged CNBP protein was crucial for its interaction with circ-HuR (Fig. 3g, h). Through bioinformatics analysis using UCSC Genome Browser (http://genome.ucsc.edu/), we found the CNBP binding motif (5′-AATGAGA-3′) [26] within HuR promoter, implicating the roles of CNBP in regulating HuR expression. These results suggested that circ-HuR interacted with CNBP, a protein with features of both RBP and TF, in gastric cancer cells.
Fig. 3.
Circ-HuR interacts with CNBP protein in gastric cancer. a Coomassie bright blue staining (left panel), mass spectrometry (MS) assay, and overlapping analysis (Venn diagram, right panel) with established RBP and TF databases revealing the proteins pulled down by biotin-labeled linear or circular forms of circ-HuR from the lysates of AGS cells. b RIP and real-time qRT-PCR assays showing the relative interaction between circ-HuR and six proteins in AGS cells stably transfected with empty vector (circ-Mock), hsa_circ-23897 (circ_23897), linear circ-23897 (lin_23897), circ-HuR, or linear circ-HuR (lin-HuR), with normalization to input of cells transfected with circ-Mock. c RIP assay with primer sets (lower panel) indicating the interaction between circ-HuR and CNBP in AGS cells stably transfected with circ-Mock, circ_23897, lin_23897, circ-HuR, or lin-HuR (upper panel). d MS assay depicting the identified CNBP peptides pulled down by circ-HuR. e Dual RNA-FISH and immunofluorescence staining assay indicating the co-localization of circ-HuR (green) and CNBP (red) in AGS and MKN-45 cells, with nuclei staining with DAPI (blue). Scale bar, 5 μm. f RNA EMSA determining the interaction between endogenous CNBP protein and biotin-labeled circular probe of circ-HuR (arrowheads), with CNBP antibody incubation or competition using an excess of unlabeled circular probe of circ-HuR. g Schematic diagram revealing the domains of CNBP truncations. h In vitro binding assay showing the enriched circ-HuR levels detected by RT-PCR (lower panel) after incubation with full-length or truncations of Flag-tagged or GST-tagged recombinant CNBP protein validated by western blot (upper panel). ANOVA analyzed the difference in (b). *P < 0.01 vs. circ-Mock. Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (b, c, e, f, and h)
CNBP promotes HuR expression, growth, and aggressiveness of gastric cancer
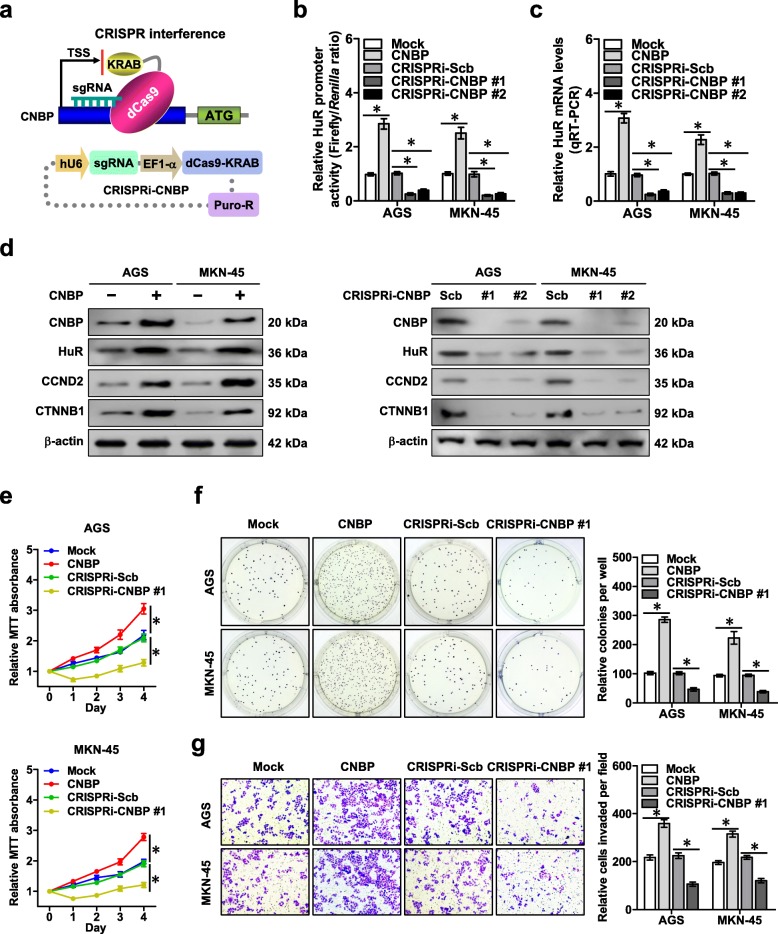
To investigate the roles of CNBP in gastric cancer progression, dCas9-based Clustered regularly interspaced short palindromic repeats (CRISPR) interference (CRISPRi) [27–30] was applied for CNBP knockdown (Fig. 4a). Stable transfection of CNBP and two independent gRNAs against CNBP (CRISPRi-CNBP) into AGS and MKN-45 cells resulted in increased and reduced levels of CNBP, respectively (Additional file 1: Figure S4a). Notably, neither ectopic expression nor knockdown of circ-HuR affected the CNBP transcript and protein levels in AGS and MKN-45 cells (Additional file 1: Figure S4b, c). ChIP and quantitative PCR (qPCR) indicated endogenous binding of CNBP to HuR promoter, which was increased and decreased by over-expression or knockdown of CNBP, respectively (Additional file 1: Figure S4d). Stable ectopic expression or knockdown of CNBP promoted and attenuated the promoter activity of HuR in AGS and MKN-45 cells, respectively (Fig. 4b). Furthermore, the transcript and protein levels of HuR and its downstream genes CCND2 and CTNNB1 were increased and decreased in AGS and MKN-45 cells with stable over-expression or knockdown of CNBP, respectively (Fig. 4c, d and Additional file 1: Figure S4e). In MTT colorimetric, soft agar, and matrigel invasion assays, stable over-expression or knockdown of CNBP facilitated and reduced the viability, growth and invasiveness of AGS and MKN-45 cells, respectively (Fig. 4e–g). Mining of a well-defined gastric cancer dataset derived from Kaplan-Meier Plotter (http://kmplot.com/analysis) revealed that higher CNBP (P = 1.9 × 10− 3 and P = 5.4 × 10− 4) or HuR (P = 8.5 × 10− 5 and P = 4.0 × 10− 5) expression was associated with lower overall survival (OS) and first progression (FP) survival probability of patients (Additional file 1: Figure S4f). These results reveled that CNBP promoted the HuR expression and tumor progression in gastric cancer.
Fig. 4.
CNBP promotes HuR expression, growth, and aggressiveness of gastric cancer. a Schematic illustration of dCas9-based CRISPRi for CNBP and small guide RNA (sgRNA) targeting region. b Dual-luciferase assay revealing the promoter activity of HuR in AGS and MKN-45 cells stably transfected with empty vector (mock), CNBP, CRISPRi-Scb, CRISPRi-CNBP #1, or CRISPRi-CNBP #2. (c) and (d) Real-time qRT-PCR (c, normalized to β-actin, n = 5) and western blot (d) assays showing the transcript and protein levels of HuR in AGS and MKN-45 cells stably transfected with mock, CNBP, CRISPRi-Scb, CRISPRi-CNBP #1, or CRISPRi-CNBP #2. e MTT colorimetric assay indicating the viability of AGS and MKN-45 cells stably transfected with mock, CNBP, CRISPRi-Scb, or CRISPRi-CNBP #1. f and g Soft agar (f) and matrigel invasion (g) assays showing the in vitro growth and invasion of AGS and MKN-45 cells stably transfected with mock, CNBP, CRISPRi-Scb, or CRISPRi-CNBP #1. ANOVA analyzed the difference in (b, c and e-g). *P < 0.01 vs. mock or CRISPRi-Scb. Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (b-g)
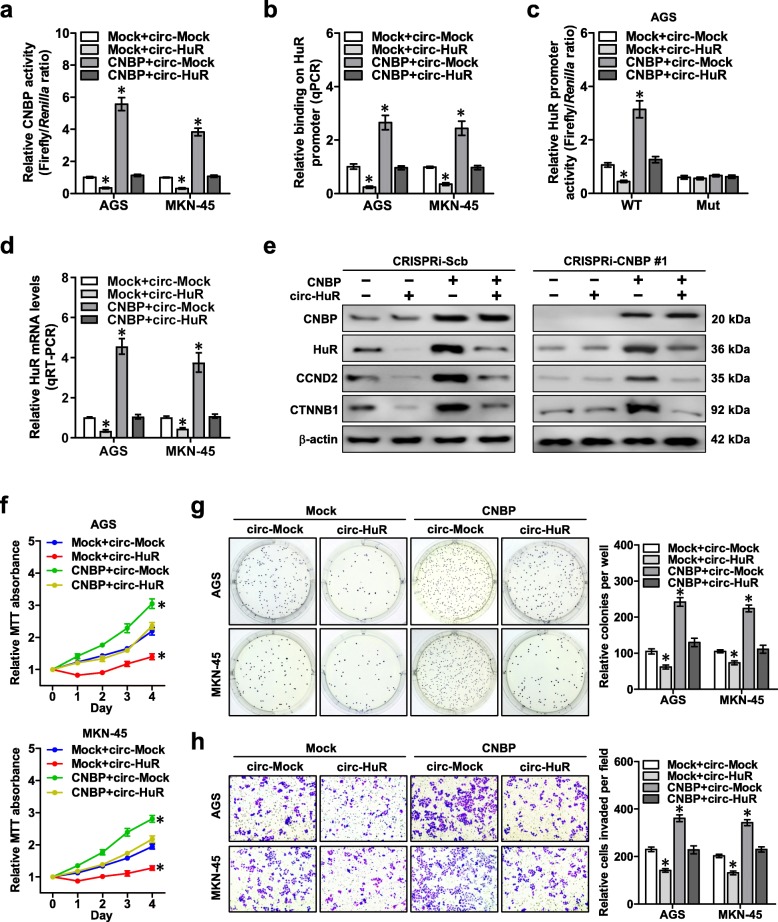
Circ-HuR suppresses HuR expression, growth, and invasion of gastric cancer cells via repressing CNBP transactivation
We further investigated the interplay effects between circ-HuR and CNBP in regulating HuR expression and gastric cancer progression. In a cohort of 81 gastric cancer cases, lower circ-HuR expression (P = 3.0 × 10− 3) and higher expression of CNBP (P = 7.0 × 10− 4) or HuR (P = 3.5 × 10− 2) was associated with lower survival probability of patients (Additional file 1: Figure S4 g). In addition, the circ-HuR (R = -0.602, P < 1.0 × 10− 4) or CNBP (R = 0.683, P < 1.0 × 10− 4) levels were negatively and positively correlated with those of HuR in these gastric cancer tissues, respectively (Additional file 1: Figure S4h). Cox regression analysis revealed that distant metastasis [hazard ratio (HR) = 2.809, P = 0.002], tumor-node-metastasis (TNM) stage (HR = 2.519, P = 0.015), circ-HuR levels (HR = 0.616, P = 0.012), and CNBP expression (HR = 2.643, P = 0.003) were prognostic factors for gastric cancer patients (Additional file 1: Table S1).
In dual-luciferase assay with a reporter containing four canonical CNBP binding sites, ectopic expression or knockdown of CNBP facilitated and attenuated the transactivation of CNBP in AGS and MKN-45 cells, respectively (Additional file 1: Figure S5a), while stable over-expression or knockdown of circ-HuR suppressed and increased the CNBP transactivation in these cells (Additional file 1: Figure S5b). Notably, ectopic expression of circ-HuR attenuated the increase of CNBP transactivation, CNBP enrichment, promoter activity and expression levels of HuR induced by over-expression of CNBP in AGS and MKN-45 cells (Fig. 5a-e and Additional file 1: Figure S5c). However, neither CNBP nor circ-HuR affected the activity of HuR promoter containing mutant CNBP binding site (Fig. 5c and Additional file 1: Figure S5c). Ectopic expression of circ-HuR did not affect the HuR levels in AGS cells stably transfected with CRISPRi-CNBP #1 (Fig. 5e). To further investigate the effects of circ-HuR on CNBP transactivation, two established CNBP target genes MMP-14 [31] and c-Myc [32] were chosen for studies. As shown in Additional file 1: Figure S5d-g, stable ectopic expression of CNBP facilitated the CNBP enrichment, promoter activity, transcripts and protein levels of MMP-14 and c-Myc in AGS and MKN-45 cells, which were prevented by over-expression of circ-HuR. Consistently, stable knockdown of circ-HuR increased the CNBP enrichment, promoter activity, transcripts and protein expression levels of HuR, MMP-14 and c-Myc in these gastric cancer cells, which were rescued by CRISPRi-mediated knockdown of CNBP (Fig. S5 h-k). Importantly, stable circ-HuR over-expression attenuated the increase in viability, growth, and invasiveness of AGS and MKN-45 cells induced by ectopic expression of CNBP (Fig. 5f–h). These results showed that circ-HuR suppressed HuR expression, growth, and invasion of gastric cancer cells via repressing CNBP transactivation.
Fig. 5.
Circ-HuR suppresses HuR expression, growth, and invasion of gastric cancer cells via repressing CNBP transactivation. a Dual-luciferase assay revealing the transactivation of CNBP in AGS and MKN-45 cells stably transfected with empty vector (mock) or CNBP, and those co-transfected with circ-Mock or circ-HuR. b ChIP and qPCR assays showing the changes in binding of CNBP to HuR promoter in AGS and MKN-45 cells stably transfected with mock or CNBP, and those co-transfected with circ-Mock or circ-HuR. c and d Dual-luciferase (c) and real-time qRT-PCR (d) assays indicating the activity of HuR promoter with wild type (WT) or mutant (Mut) CNBP binding site and transcript levels (normalized to β-actin, n = 4) of HuR in AGS and MKN-45 cells stably transfected with mock or CNBP, and those co-transfected with circ-Mock or circ-HuR. e Western blot assay showing the expression of CNBP, HuR, CCND2, and CTNNB1 in AGS cells stably transfected with CRISPRi-Scb or CRISPRi-CNBP #1, and those co-transfected with mock, CNBP, circ-Mock, or circ-HuR. f MTT colorimetric assay indicating the viability of AGS and MKN-45 cells stably transfected with mock or CNBP, and those co-transfected with circ-Mock or circ-HuR. g and h Soft agar (g) and matrigel invasion (h) assays showing in vitro growth and invasion of AGS and MKN-45 cells stably transfected with mock or CNBP, and those co-transfected with circ-Mock or circ-HuR. ANOVA analyzed the difference in (a-d and f-h). *P < 0.01 vs. mock+ circ-Mock. Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (a-h)
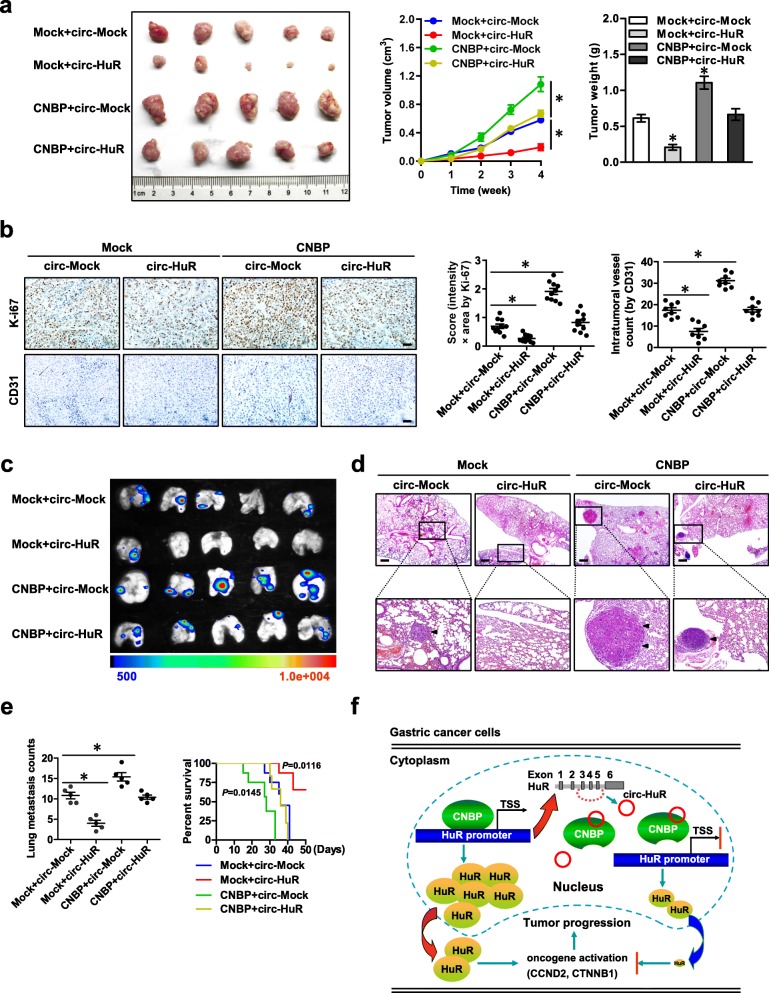
Circ-HuR suppresses gastric cancer progression by inhibiting CNBP transactivation in vivo
To further confirm the in vitro findings, we observed the biological roles of circ-HuR and CNBP in vivo. Consistently, stable transfection of CNBP resulted in an obvious increase in the growth and weight of subcutaneous xenograft tumors formed by AGS cells in nude mice, which were attenuated by ectopic expression of circ-HuR (Fig. 6a). Immunohistochemical staining revealed the increase of Ki-67 proliferation index and CD31-positive microvessels in xenograft tumors formed by AGS cells stably over-expressing CNBP, which were prevented by ectopic expression of circ-HuR (Fig. 6b). Importantly, athymic nude mice treated with tail vein injection of AGS cells stably transfected with CNBP displayed more lung metastatic colonies and poorer overall survival probability, while stable over-expression of circ-HuR attenuated these effects (Fig. 6c-e). Collectively, these results indicated that circ-HuR suppressed gastric cancer progression by inhibiting CNBP transactivation in vivo.
Fig. 6.
Circ-HuR suppresses gastric cancer progression by inhibiting CNBP transactivation in vivo. a Representative (left panel), in vivo growth curve (middle panel), and weight at the end points (right panel) of xenograft tumors formed by subcutaneous injection of AGS cells stably transfected with empty vector (mock) or CNBP, and those co-transfected with circ-Mock or circ-HuR into the dorsal flanks of nude mice (n = 5 for each group). b Representative images (left panel) and quantification (right panel) of immunohistochemical staining showing the expression of Ki-67 and CD31 within xenograft tumors formed by hypodermic injection of AGS cells stably transfected with mock or CNBP, and those co-transfected with circ-Mock or circ-HuR (n = 5 for each group). Scale bars: 50 μm. c-e Representative images (c), H&E staining (d, arrowheads), and quantification (e, left panel) of lung metastatic colonization and Kaplan-Meier curves (e, right panel) of nude mice treated with tail vein injection of AGS cells stably transfected with mock or CNBP, and those co-transfected with circ-Mock or circ-HuR (n = 5 for each group). Scale bar: 100 μm. f The mechanisms underlying circ-HuR-suppressed tumor progression: as a nuclear circRNA, circ-HuR interacts with CNBP to inhibit its binding to HuR promoter, resulting in down-regulation of HuR and suppression of gastric cancer progression. ANOVA analyzed the difference in (a, b, e). Log-rank test for survival comparison in (e). *P < 0.01 vs. mock+ circ-Mock. Data are shown as mean ± SEM (error bars)
Discussion
With the development of genomic analysis platform, a large number of abundant and conserved circRNAs have been identified in human tissues and cells [33–35]. Recent studies suggest that circRNAs are able to function as tumor drivers or tumor suppressors in multiply ways [10, 14, 16, 36–39]. Besides action mode of binding miRNA, circRNAs participate in parental gene expression at transcriptional level [16, 40]. In this study, we discover that circ-HuR (hsa_circ_0049027), a novel nuclear circRNA, is down-regulated in gastric cancer. Over-expression of circ-HuR suppresses the growth and aggressiveness of gastric cancer in vitro and in vivo. Mechanistically, circ-HuR inhibits the enrichment of CNBP on the promoter of HuR, resulting in reduction of HuR expression (Fig. 6f). These findings highlight a novel tumor suppressive circRNA in regulating tumor growth and aggressiveness, presenting a promising therapeutic target for gastric cancer.
CNBP is a highly conserved zinc finger protein consisting of seven tandem repeats of CCHC zinc finger and an arginine/glycine-rich region, and contributes to embryonic development, organogenesis, and tumorigenesis [26, 31, 41, 42]. As a transcription factor, CNBP binds to the promoters of target genes such as macrophage colony-stimulating factor (CSF1) [43], MMP-14 [31], and c-Myc [32]. CNBP expression is elevated in human tumor tissues, and is associated with proliferation, invasion, and migration of tumor cells in vitro and in vivo [31, 44]. In the current study, mining of public gastric cancer datasets reveals that higher expression of CNBP is associated with poor outcome of patients. Gain- and loss-of-function studies show that CNBP facilitates HuR expression, growth, and aggressiveness of cancer cells, suggesting the oncogenic roles of CNBP in gastric cancer progression. Additionally, our results indicate that RGG box domain is essential for the transcriptional activity of CNBP. We identify that circ-HuR is able to suppress the transcriptional activity of CNBP, and functions as an endogenous inhibitor for repressing the binding of CNBP to HuR promoter, resulting in down-regulation of HuR and its target genes involved in cancer progression.
HuR protein is able to regulate the expression of labile mRNAs containing AU-rich elements through increasing their half-life [45]. As a RBP shuttling between the nucleus and cytoplasm, HuR exerts a fundamental role in tumor progression, and its cytoplasmic presence is intimately linked to mRNA stabilizing function [5, 46]. Many HuR target genes are necessary for cell growth and proliferation, such as cyclin A, cyclin B1, cyclin D2 [22, 47], hypoxia-inducible factor 1α (HIF-1α), vascular endothelial growth factor (VEGF), cyclooxygenase-2 (COX-2), and β-catenin [8, 23, 48]. Thus, it has been a promising tumor therapeutic approach via modulating the expression or activity of HuR. For example, treatment with MS-444, an inhibitor interfering the RNA binding and trafficking of HuR, results in loss of viability and induction of apoptosis in malignant glioma cells [49]. Additionally, CMLD-2, a disruptor of interaction between HuR and mRNA targets, exerts antitumor effects in thyroid cancer cells by decreasing cell viability and increasing apoptosis [50], highlighting HuR as a promising therapeutic target for cancers. In this study, our evidence shows that circ-HuR is able to inhibit HuR expression and suppress the growth and aggressiveness of gastric cancer in vitro and in vivo, suggesting a potential therapeutic approach for cancers.
Conclusions
This study indicates that a novel nuclear circRNA, termed as circ-HuR, is down-expressed in gastric cancer tissues and cells. Circ-HuR acts as an inhibitor of CNBP transactivation by directly interacting with its RGG domain. Interestingly, CNBP promotes the expression of HuR at transcription level via binding to HuR promoter in gastric cancer cells. Meanwhile, circ-HuR restrains the transcription of HuR by inhibiting CNBP transactivation, and suppresses the growth and aggressiveness of gastric cancer in vitro and in vivo. This study extends our knowledge about the regulation of HuR expression and gastric cancer progression by circRNA, and provides a potential target for treatment of gastric cancer.
Supplementary information
Additional file 1: Figure S1. Expression of HuR-derived circRNAs and their effects on HuR expression in gastric cancer cells. Figure S2. Effects of circ-HuR on expression of HuR and downstream genes in gastric cancer cells. Figure S3. Interaction between circ-HuR and CNBP protein in gastric cancer cells. Figure S4. Roles of circ-HuR and CNBP in regulating transcription of HuR and downstream genes. Figure S5. circ-HuR regulates CNBP transactivation and target gene expression in gastric cancer cells. Table S1. Univariate and multivariate analysis of prognostic factors in gastric cancer patients. Table S2. Primer sets used for RT-PCR, qPCR, RIP, and ChIP. Table S3. Primer sets used for constructs. Table S4. Oligonucleotide sets used for CRISPR-dCas9, short hairpin RNAs, or probe.
Additional file 2: Table S5. Over-lapping analysis of circ-HuR-binding protein with RNA binding protein and transcription factor.
Acknowledgements
We appreciate Dr. Jouko Lohi for providing MMP-14 promoter vector.
Abbreviations
- ATG
Translational start codon
- bHLH
Basic helix-loop-helix
- CCND2
Cyclin D2
- circRNAs
Circular RNAs
- c-Myc
MYC proto-oncogene
- CNBP
CCHC-type zinc finger nucleic acid binding protein
- CRISPR
Clustered regularly interspaced short palindromic repeats
- CTNNB1
Catenin beta 1
- dCas9
Dead mutant of Cas9 endonuclease
- EF1-α
Elongation factor-1α promoter
- ENO1
Enolase 1
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- HDGF
Heparin binding growth factor
- hU6
human U6 promoter
- HuR
Human antigen R
- MMP-14
Matrix metallopeptidase 14
- NAP1L1
Nucleosome assembly protein 1 like 1
- NONO
Non-POU domain containing octamer binding
- Puro-R
Puromycin resistance
- qRT-PCR
Quantitative real-time PCR
- RIP
RNA immunoprecipitation
- RNA-FISH
RNA fluorescence in situ hybridization
- RT-PCR
Reverse transcription PCR
- SFPQ
Splicing factor proline and glutamine rich
- sgRNA
Small guide RNA
- shRNA
Short hairpin RNA
- sh-Scb
Scramble shRNA
- TSS
Transcriptional start site
Authors’ contributions
FY and AH conceived and performed most of the experiments; DL and JW accomplished some of in vitro experiments; YG, YL, HL, YC, and XW accomplished in vivo studies; FY and AH undertook the mining of publicly available datasets; KH critically reviewed the manuscript; QT and LZ wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was granted by the National Natural Science Foundation of China (81472363, 81572423, 81672500, 81773094, 81772967, 81874085, 81874066, 81802925, 81903011, 81903008), and Fundamental Research Funds for the Central Universities (2019kfyRCPY032).
Availability of data and materials
The data supporting the conclusions of this article are presented within the article and its Additional files.
Ethics approval and consent to participate
All animal experiments were approved by the Animal Care Committee of Tongji Medical College. The Institutional Review Board of Tongji Medical College approved the human tissue study. All procedures were carried out in accordance with guidelines set forth by Declaration of Helsinki.
Consent for publication
Written informed consent was obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Feng Yang, Anpei Hu and Dan Li contributed equally to this work.
Contributor Information
Feng Yang, Email: fengyangunion@163.com.
Anpei Hu, Email: anpei_hu@163.com.
Dan Li, Email: d.li26@hotmail.com.
Jianqun Wang, Email: jianqun_wang@126.com.
Yanhua Guo, Email: yanhuaguo99@hotmail.com.
Yang Liu, Email: yang_liu2000@126.com.
Hongjun Li, Email: hongjun_liwh@163.com.
Yajun Chen, Email: yajun_chentj@126.com.
Xiaojing Wang, Email: xiaojingwangunion@163.com.
Kai Huang, Email: kaihuangunion@gmail.com.
Liduan Zheng, Phone: 86-27-85726129, Email: ld_zheng@hotmail.com.
Qiangsong Tong, Phone: 86-27-85350762, Email: qs_tong@hotmail.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12943-019-1094-z.
References
- 1.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 2.Zucal C, D'Agostino V, Loffredo R, Mantelli B, NatthakanThongon LP, Latorre E, Provenzani A. Targeting the multifaceted HuR protein, benefits and caveats. Curr Drug Targets. 2015;16:499–515. doi: 10.2174/1389450116666150223163632. [DOI] [PubMed] [Google Scholar]
- 3.Doller A, el-S A, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol Cell Biol. 2008;28:2608–2625. doi: 10.1128/MCB.01530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vazquez-Chantada M, Fernandez-Ramos D, Embade N, Martinez-Lopez N, Varela-Rey M, Woodhoo A, Luka Z, Wagner C, Anglim PP, Finnell RH, et al. HuR/methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation, and carcinogenesis. Gastroenterology. 2010;138:1943–1953. doi: 10.1053/j.gastro.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotta-Loizou I, Giaginis C, Theocharis S. Clinical significance of HuR expression in human malignancy. Med Oncol. 2014;31:161. doi: 10.1007/s12032-014-0161-y. [DOI] [PubMed] [Google Scholar]
- 6.Heinonen M, Bono P, Narko K, Chang SH, Lundin J, Joensuu H, Furneaux H, Hla T, Haglund C, Ristimaki A. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer Res. 2005;65:2157–2161. doi: 10.1158/0008-5472.CAN-04-3765. [DOI] [PubMed] [Google Scholar]
- 7.Denkert C, Koch I, von Keyserlingk N, Noske A, Niesporek S, Dietel M, Weichert W. Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Mod Pathol. 2006;19:1261–1269. doi: 10.1038/modpathol.3800645. [DOI] [PubMed] [Google Scholar]
- 8.Mrena J, Wiksten JP, Thiel A, Kokkola A, Pohjola L, Lundin J, Nordling S, Ristimaki A, Haglund C. Cyclooxygenase-2 is an independent prognostic factor in gastric cancer and its expression is regulated by the messenger RNA stability factor HuR. Clin Cancer Res. 2005;11:7362–7368. doi: 10.1158/1078-0432.CCR-05-0764. [DOI] [PubMed] [Google Scholar]
- 9.Niesporek S, Kristiansen G, Thoma A, Weichert W, Noske A, Buckendahl AC, Jung K, Stephan C, Dietel M, Denkert C. Expression of the ELAV-like protein HuR in human prostate carcinoma is an indicator of disease relapse and linked to COX-2 expression. Int J Oncol. 2008;32:341–347. [PubMed] [Google Scholar]
- 10.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–881. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 12.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 13.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, Ma J, Li X, Zeng Y, Yang Z, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F, Fang E, Mei H, Chen Y, Li H, Li D, Song H, Wang J, Hong M, Xiao W, et al. Cis-acting circ-CTNNB1 promotes β-catenin signaling and cancer progression via DDX3-mediated transactivation of YY1. Cancer Res. 2019;79:557–571. doi: 10.1158/0008-5472.CAN-18-1559. [DOI] [PubMed] [Google Scholar]
- 16.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, Li D, Song H, Wang J, Hong M, et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–1364. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li D, Wang X, Mei H, Fang E, Ye L, Song H, Yang F, Li H, Huang K, Zheng L, et al. Long noncoding RNA pancEts-1 promotes neuroblastoma progression through hnRNPK-mediated β-catenin stabilization. Cancer Res. 2018;78:1169–1183. doi: 10.1158/0008-5472.CAN-17-2295. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Song H, Mei H, Fang E, Wang X, Yang F, Li H, Chen Y, Huang K, Zheng L, et al. Armadillo repeat containing 12 promotes neuroblastoma progression through interaction with retinoblastoma binding protein 4. Nat Commun. 2018;9:2829. doi: 10.1038/s41467-018-05286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang X, Mei H, Qu H, Zhao X, Li D, Song H, Jiao W, Pu J, Huang K, Zheng L, et al. miRNA-584-5p exerts tumor suppressive functions in human neuroblastoma through repressing transcription of matrix metalloproteinase 14. Biochim Biophys Acta. 2015;1852:1743–1754. doi: 10.1016/j.bbadis.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Zhao X, Li D, Pu J, Mei H, Yang D, Xiang X, Qu H, Huang K, Zheng L, Tong Q. CTCF cooperates with noncoding RNA MYCNOS to promote neuroblastoma progression through facilitating MYCN expression. Oncogene. 2016;35:3565–3576. doi: 10.1038/onc.2015.422. [DOI] [PubMed] [Google Scholar]
- 22.Sun Q, Tripathi V, Yoon JH, Singh DK, Hao Q, Min KW, Davila S, Zealy RW, Li XL, Polycarpou-Schwarz M, et al. MIR100 host gene-encoded lncRNAs regulate cell cycle by modulating the interaction between HuR and its target mRNAs. Nucleic Acids Res. 2018;46:10405–10416. doi: 10.1093/nar/gky696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 24.Atasoy U, Watson J, Patel D, Keene JD. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- 25.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee E, Lee TA, Kim JH, Park A, Ra EA, Kang S, Choi HJ, Choi JL, Huh HD, Lee JE, et al. CNBP acts as a key transcriptional regulator of sustained expression of interleukin-6. Nucleic Acids Res. 2017;45:3280–3296. doi: 10.1093/nar/gkx071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Y, Shen W, Zhang J, Yang B, Liu YN, Qi H, Yu X, Lu SY, Chen Y, Xu YZ, et al. CRISPR interference-based specific and efficient gene inactivation in the brain. Nat Neurosci. 2018;21:447–454. doi: 10.1038/s41593-018-0077-5. [DOI] [PubMed] [Google Scholar]
- 31.Lee E, Lee TA, Yoo HJ, Lee S, Park B. CNBP controls tumor cell biology by regulating tumor-promoting gene expression. Mol Carcinog. 2019;58:1492–1501. doi: 10.1002/mc.23030. [DOI] [PubMed] [Google Scholar]
- 32.Michelotti EF, Tomonaga T, Krutzsch H, Levens D. Cellular nucleic acid binding protein regulates the CT element of the human c-myc protooncogene. J Biol Chem. 1995;270:9494–9499. doi: 10.1074/jbc.270.16.9494. [DOI] [PubMed] [Google Scholar]
- 33.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang G, Li S, Yang N, Zou Y, Zheng D, Xiao T. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8–18. doi: 10.1016/j.canlet.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15:995–1005. doi: 10.1080/15476286.2018.1486659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, et al. Novel role of FBXW7 circular RNA in repressing Glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, Zhang H, Yang P, Cui X, Ren Y, et al. A noncoding regulatory RNAs network driven by circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. 2019. 10.1002/hep.30795. [DOI] [PubMed]
- 40.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 41.Calcaterra NB, Armas P, Weiner AM, Borgognone M. CNBP: a multifunctional nucleic acid chaperone involved in cell death and proliferation control. IUBMB Life. 2010;62:707–714. doi: 10.1002/iub.379. [DOI] [PubMed] [Google Scholar]
- 42.Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, Naegeli HU, Ottl J, Oberer L, Guenat C, Moss S, et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat Chem Biol. 2007;3:508–515. doi: 10.1038/nchembio.2007.14. [DOI] [PubMed] [Google Scholar]
- 43.Konicek BW, Xia X, Rajavashisth T, Harrington MA. Regulation of mouse colony-stimulating factor-1 gene promoter activity by AP1 and cellular nucleic acid-binding protein. DNA Cell Biol. 1998;17:799–809. doi: 10.1089/dna.1998.17.799. [DOI] [PubMed] [Google Scholar]
- 44.D'Amico D, Antonucci L, Di Magno L, Coni S, Sdruscia G, Macone A, Miele E, Infante P, Di Marcotullio L, De Smaele E, et al. Non-canonical hedgehog/AMPK-mediated control of polyamine metabolism supports neuronal and medulloblastoma cell growth. Dev Cell. 2015;35:21–35. doi: 10.1016/j.devcel.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levine TD, Gao F, King PH, Andrews LG, Keene JD. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/MCB.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan H, Strickland A, Madhu V, Johnson ZI, Chand SN, Brody JR, Fertala A, Zheng Z, Shapiro IM, Risbud MV. RNA binding protein HuR regulates extracellular matrix gene expression and pH homeostasis independent of controlling HIF-1α signaling in nucleus pulposus cells. Matrix Biol. 2019;77:23–40. doi: 10.1016/j.matbio.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J, Hjelmeland AB. Anti-cancer effects of the HuR inhibitor, MS-444, in malignant glioma cells. Cancer Biol Ther. 2019;20:979–988. doi: 10.1080/15384047.2019.1591673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allegri L, Baldan F, Roy S, Aube J, Russo D, Filetti S, Damante G. The HuR CMLD-2 inhibitor exhibits antitumor effects via MAD2 downregulation in thyroid cancer cells. Sci Rep. 2019;9:7374. doi: 10.1038/s41598-019-43894-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Expression of HuR-derived circRNAs and their effects on HuR expression in gastric cancer cells. Figure S2. Effects of circ-HuR on expression of HuR and downstream genes in gastric cancer cells. Figure S3. Interaction between circ-HuR and CNBP protein in gastric cancer cells. Figure S4. Roles of circ-HuR and CNBP in regulating transcription of HuR and downstream genes. Figure S5. circ-HuR regulates CNBP transactivation and target gene expression in gastric cancer cells. Table S1. Univariate and multivariate analysis of prognostic factors in gastric cancer patients. Table S2. Primer sets used for RT-PCR, qPCR, RIP, and ChIP. Table S3. Primer sets used for constructs. Table S4. Oligonucleotide sets used for CRISPR-dCas9, short hairpin RNAs, or probe.
Additional file 2: Table S5. Over-lapping analysis of circ-HuR-binding protein with RNA binding protein and transcription factor.
Data Availability Statement
The data supporting the conclusions of this article are presented within the article and its Additional files.