Abstract
Background:
The role of hepatitis B surface antigen (HBsAg) levels in predicting the risk of developing hepatocellular carcinoma (HCC) has remained unclear. The aim of this study was to obtain the most up-to-date estimated measure of the association between HBsAg levels and the development of HCC in patients.
Methods:
We performed a systematic review by searching for relevant studies on PubMed, Scopus, ProQuest and the Cochrane Central Register of Controlled Trials from January 2002 to November 2017. We presented the effects of HBsAg levels at each cut-off value as the odds ratios (ORs) at 95% confidence interval (CI). We also investigated HCC and its potential risk factors including HBeAg, and HBV DNA. We registered our protocol with the International Prospective Register of Systematic Reviews (PROSPERO) with the registration number CRD42018081138.
Results:
We selected 10 studies representing 12 541 cases. At the 100 IU/ml cut-off, the OR for HCC at the high HBsAg level versus the low level was 4.99 (95% CI, 3.01–8.29) with high inconsistency (I2=79%). At the 1,000 IU/ml threshold, the pooled OR for HCC at the high HBsAg versus the low level was 2.46 (95% CI, 2.15–2.83) with low variance. We also found correlations between the risk of HCC and male gender (OR=2.12), hepatitis B e-antigen positivity (OR=2.99), or hepatitis B (HBV) viral load ≥ 2,000 IU/ml (OR=4.37).
Conclusion:
Our study revealed that HBsAg levels ≥ 100 IU/ml, and notably >1,000 IU/ml, are associated with an increased risk of HCC development.
Key Words: Hepatitis B surface antigens, hepatocellular carcinoma, chronic hepatitis B infections, meta-analysis
Introduction
In the 2015 World Health Organization report, the percentage of people living with chronic hepatitis B infections was 3.5% (257 million persons) of the global population; it is estimated that fewer than 1.7 million (8%) of the infected patients received anti-viral treatment (WHO, 2017). Of the adult patients living with chronic hepatitis B (CHB) infections, 20%–30% have been estimated to develop cirrhosis or hepatocellular carcinoma (HCC) in the future, and on average, there were 340,000 annual deaths from hepatitis B-induced HCC between 1990 and 2010 (Lozano et al., 2012; WHO, 2019). A variety of genetic factors and viral markers have been used to screen for HBV-related HCC.
The presence of hepatitis B surface antigen (HBsAg) is the primary indicator for confirming HBV infection. The presence of HBsAg has shown a strong association with that of the covalently closed circular deoxyribonucleic acid (cccDNA) that functions as a model for transcription of all viral ribonucleic acids (RNAs) and greatly correlates with the viral load (Werle-Lapostolle et al., 2004). The formation of cccDNA is the key mechanism of HBV tumourigenesis, but measuring cccDNA levels remains challenging. As a result, HBsAg quantification has become a commonly used proxy (Werle-Lapostolle et al., 2004; Guo and Guo, 2015; Valaydon and Locarnini, 2017). In a systematic review involving 6,028 patients with CHB published in 2015, 335 of them contracted HCC during the 14.7-year follow-up, and the risk of HCC in patients with high serum HBsAg levels (≥1,000 IU/ml) was greater than that among patients with low HBsAg levels (Qu et al., 2015).
Although new clinical research has focussed on the effect of the quantitation of HBsAg on the prediction of HCC development in different populations, stages, and cut-off values, no results have been applied to establish routine practices yet (Lee et al., 2015; Chien et al., 2016; Liu et al., 2016; Yang et al., 2016; Kumar et al., 2017). We have perceived an increase in the number of publications of HBsAg on HCC, and thus decided to analyse their data to try to obtain a better-estimated measure of the association between HBsAg levels and HCC. We also investigated different cut-off values of the HBsAg level used as a biomarker for HCC development in patients living with chronic hepatitis B infections.
Materials and Methods
We performed a systematic review and meta-analysis based on our priori protocol registered at PROSPERO (registration number CRD42018081138). The protocol was approved of exemption by the Institutional Review Board of the Faculty of Tropical Medicine, Mahidol University, Thailand (MUTM-EXMPT 2017-006).
Search strategy and eligibility criteria
We searched for published articles from January 2002 to November 2017 on PubMed, Scopus, Cochrane Central Register of Controlled Trials (CENTRAL) and ProQuest. Our search terms included chronic hepatitis B infection, HBsAg and HCC. We supplemented our search by reviewing reference lists for all appropriate studies (Table 1).
Table 1.
Search Terms
| Concepts | Search terms | Search details |
|---|---|---|
| Population | 1.Chronic hepatitis B (MeSH) | "hepatitis b, chronic"[MeSH Terms] |
| 2. Chronic hepatitis B infection (KW) | "Chronic HBV infection"[All Fields] | |
| Combine 1, 2 with “OR” | "hepatitis b, chronic"[MeSH Terms] OR "Chronic HBV infection"[All Fields] | |
| Index test | 3. Hepatitis B surface antigen (MeSH) | "hepatitis b surface antigens"[MeSH Terms] |
| 4. Quantification of Hepatitis B surface antigen (KW) | Quantification[All Fields] AND ("hepatitis b surface antigens"[MeSH Terms] OR "hepatitis b surface antigens"[All Fields] OR "hepatitis b surface antigen"[All Fields]) | |
| 5. HBsAg (KW) | "HBsAg"[All Fields] | |
| 6. Quantitative serum HBsAg level (KW) | Quantitative[All Fields] AND ("serum"[MeSH Terms] OR "serum"[All Fields]) AND ("hepatitis b surface antigens"[MeSH Terms] OR "hepatitis b surface antigens"[All Fields] OR "hbsag"[All Fields]) AND level[All Fields] | |
| 7. Quantitative Hepatitis B surface antigen (KW) | "Quantitative Hepatitis B surface antigen"[All Fields] | |
| 8. HBsAg quantification (KW) | "HBsAg quantification"[All Fields] | |
| 9. qHBsAg (KW) | "qHBsAg"[All Fields] | |
| Combine 3, 4, 5, 6, 7, 8, 9 with “OR” | "qHBsAg"[All Fields] OR ((((("hepatitis b surface antigens"[MeSH Terms] OR (Quantification[All Fields] AND ("hepatitis b surface antigens"[MeSH Terms] OR "hepatitis b surface antigens"[All Fields] OR "hepatitis b surface antigen"[All Fields]))) OR "HBsAg"[All Fields]) OR (Quantitative[All Fields] AND ("serum"[MeSH Terms] OR "serum"[All Fields]) AND ("hepatitis b surface antigens"[MeSH Terms] OR "hepatitis b surface antigens"[All Fields] OR "hbsag"[All Fields]) AND level[All Fields])) OR "Quantitative Hepatitis B surface antigen"[All Fields]) OR "HBsAg quantification"[All Fields]) | |
| Outcome | 10. Liver neoplasm (MeSH) | "carcinoma, hepatocellular"[MeSH Terms] |
| 11. Liver neoplasm (MeSH) | "liver neoplasms"[MeSH Terms] | |
| 12. Liver cancer (KW) | "Liver cancer"[All Fields] | |
| 13. Hepatoma (KW) | "Hepatoma"[All Fields] | |
| 14. Hepatic cancer (KW) | "Hepatic cancer"[All Fields] | |
| 15. Malignant hepatic tumor (KW) | "Malignant hepatic tumor"[All Fields] | |
| 16. Hepatic neoplasm (KW) | "hepatic neoplasm"[All Fields] | |
| 17. HBV-related hepatocarcinogenesis (KW) | "HBV-related hepatocarcinogenesis"[All Fields] | |
| 18. Liver carcinogenesis (KW) | "Liver carcinogenesis"[All Fields] | |
| Combine 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 with “OR” | ((((((("carcinoma, hepatocellular"[MeSH Terms] OR "liver neoplasms"[MeSH Terms]) OR "Liver cancer"[All Fields]) OR "Hepatoma"[All Fields]) OR "Hepatic cancer"[All Fields]) OR "Malignant hepatic tumor"[All Fields]) OR "hepatic neoplasm"[All Fields]) OR "HBV-related hepatocarcinogenesis"[All Fields]) OR "Liver carcinogenesis"[All Fields] | |
| Final Search | Combine “population”, “index test”, “outcome” with “AND” Filters: studies published in the last ten years, written in English, and human studies only |
(("qHBsAg"[All Fields] OR ((((("hepatitis b surface antigens"[MeSH Terms] OR (Quantification[All Fields] AND ("hepatitis b surface antigens"[MeSH Terms] OR "hepatitis b surface antigens"[All Fields] OR "hepatitis b surface antigen"[All Fields]))) OR "HBsAg"[All Fields]) OR (Quantitative[All Fields] AND ("serum"[MeSH Terms] OR "serum"[All Fields]) AND ("hepatitis b surface antigens"[MeSH Terms] OR "hepatitis b surface antigens"[All Fields] OR "hbsag"[All Fields]) AND level[All Fields])) OR "Quantitative Hepatitis B surface antigen"[All Fields]) OR "HBsAg quantification"[All Fields])) AND (((((((("carcinoma, hepatocellular"[MeSH Terms] OR "liver neoplasms"[MeSH Terms]) OR "Liver cancer"[All Fields]) OR "Hepatoma"[All Fields]) OR "Hepatic cancer"[All Fields]) OR "Malignant hepatic tumor"[All Fields]) OR "hepatic neoplasm"[All Fields]) OR "HBV-related hepatocarcinogenesis"[All Fields]) OR "Liver carcinogenesis"[All Fields])) AND ("hepatitis b, chronic"[MeSH Terms] OR "Chronic HBV infection"[All Fields]) AND (("2002/01/01"[PDAT] : "2017/12/31"[PDAT]) AND "humans"[MeSH Terms] AND English[lang]) |
Our eligibility criteria for studies included: (a) various designs (cohort, case-control and cross-sectional studies or randomised trials); (b) participants presenting chronic hepatitis B infections and no limitations regarding age, country, or clinical setting; (c) available values for quantitative HBsAg tests at baseline; (d) newly developed HCC confirmed by standard criteria; (e) English language and (f) possibility of contacting authors for missing data.
Study selection process
A researcher (TTV) screened titles and abstracts. Selected full-text articles were analysed to determine their eligibility criteria fit. Then, we searched for missing information from study authors to handle eligibility criteria-related problems. We documented all the reasons for excluding articles.
Data collection process and quality assessment
Two independent reviewers (KP and TTV) extracted data and filled out the data extraction forms. Any controversies between the two reviewers were resolved via discussion, or by getting a third opinion (from CK). The review authors independently evaluated the methodological quality of the studies by using the Quality Assessment of Diagnostic Accuracy Studies version-2 tool (QUADAS-2) (Whiting et al., 2011).
Data analysis
We analysed the data for the two cut-off points of HBsAg levels: 100 and 1,000 IU/ml. The HBsAg level effect is presented as ORs with 95% confidence interval (CI). We performed an inverse variance meta-analysis to combine effects across studies. We considered an overall OR as statistically significant using a z-test when the P-value < 0.05. We assessed heterogeneity using Cochrane’s Q test with a significance level p < 0.05 and with an I2 test. The I2 statistic had three ranges of I2 values: low (25%), moderate (50%) and high (75%). We applied fixed effects meta-analysis when there was no evidence of heterogeneity across study effects; otherwise, we applied random effects meta-analysis. In addition, we explored the sources of heterogeneity through subgroup meta-analyses when relevant. Egger’s test was used to assess publication bias at a significant threshold of 0.05. We used the Review Manager 5.3 to perform meta-analyses and R version 3.4.2 to draw funnel plots and perform Egger’s tests.
Results
Study selection
We ran our search using the four databases PubMed, Scopus, CENTRAL and ProQuest. We found 594 articles after eliminating duplicates. After the first round of scanning articles by assessing the titles and abstracts, we included 33 in the second round. We contacted the authors from four studies with initial insufficient data for our analysis and obtained the requested data from 3 studies that were included in the analysis. Of the 33 studies, 10 were relevant to our systematic review and meta-analysis (Figure 1) (Tseng et al., 2012; Lee et al., 2013; Kawanaka et al., 2014; Ahn et al., 2015; Lee et al., 2015; Chien et al., 2016; Liu et al., 2016; Liu et al., 2017; Tseng et al., 2017; Yang et al., 2017).
Figure 1.
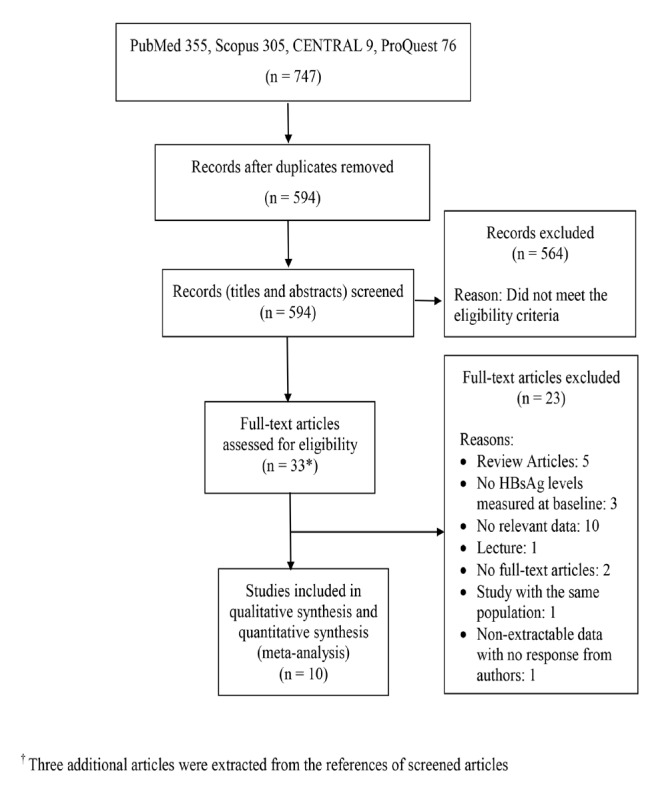
Flow Dagram of Study Inclusion
Characteristics of 10 included studies
Characteristics of each primary study are shown in Table 2. The total number of patients enrolled in the 10 studies was 12 541, with sample sizes ranging from 167 to 3,340. All of the studies were conducted in Asia (70% in Taiwan). None of the participants from 10 studies presented HIV co-infections. Half of the studies were performed in hospitals and half in communities. Only one study, had participants receiving nucleotide analogue treatments before the start of the study (Kawanaka et al., 2014).
Table 2.
Characteristics of 10 Selected Studies
| Study (author, year) | Liu | Chien | Lee | Lee | Tseng | Yang | Ahn | Kawanawa | Tseng | Liu |
|---|---|---|---|---|---|---|---|---|---|---|
| 2016 | 2016 | 2015 | 2013 | 2012 | 2017 | 2015 | 2014 | 2017 | 2017 | |
| Country | Taiwan | Taiwan | Taiwan | Taiwan | Taiwan | China | Korea | Japan | Taiwan | Taiwan |
| Setting of data collection | Community | Community | Hospital | Community | Hospital | Community | Hospital | Hospital | Hospital | Community |
| Study design | Retrospective cohort | Prospective cohort | Cross-sectional | Prospective cohort | Retrospective cohort | Case-control | Retrospective cohort | Prospective cohort | Retrospective cohort | Case-control |
| Sample size (n) | 1529 | 301 | 412 | 3340 | 2688 | 432 | 527 | 167 | 2075 | 1070 |
| Age (years) | Mean age: 46.6 | < 40: 22 (7.3%) ≥ 40: 279 (92.7%) |
Mean ± SD: 68.6 ± 6.9 | < 40: 1147 ≥ 40: 2193 |
< 40: 1407 ≥ 40: 1281 |
Case group: 56.37 ± 9.24 Control group: 58.12 ± 9.18 |
Mean ± SD: 48.8 ± 9.9 | Mean ± SD: 49.2 ± 11.1 |
< 40: 1076 ≥ 40: 999 |
Mean age: 58.7 |
| Gender | ||||||||||
| Male/Female | 993/536 | 240/61 | 262/150 | 2030/1310 | 1634/1054 | 317/115 | 311/216 | 112/55 | 1222/853 | 794/276 |
| HBV genotype (n) | B: 775 B + C/C: 754 |
B: 128 C: 125 B + C: 2 |
NA | C: 718 B or B + C: 1480 |
B: 1308 C: 312 |
NA | NA | A: 3 B: 4 C: 144 Undetermined: 16 |
B: 1612 C: 371 Undetermined: 92 |
NA |
| Stage of CHB (n) | Inactive CHB: 77 Immune reactivation: 497 Unknown: 752 |
Unknown: 301 | Unknown: 412 | Unknown: 3340 | Inactive CHB: 2688 | Unknown: 432 | Unknown: 527 | Unknown: 167 | Inactive CHB: 2075 | Unknown: 1070 |
| ALT mean/median, SD/ranges (U/L) | NA | NA | 105.4 ± 252.7 | NA | NA | NA | 37 (24–66) | NA | NA | NA |
| Qualitative HBeAg Positive/ negative (n/n) |
0/1529 | 83/215 | 15/307 | 504/2836 | 523/2165 | 93/339 | 0/527 | 81/86 | 372/1703 | NR/NR |
| HBV DNA (IU/ml) | ||||||||||
| < 2,000 (n) | 980 | 97 | 72 | 1899 | 1068 | 220 | 0 | NA | 876 | 389 |
| ≥ 2,000 (n) | 549 | 204 | 144 | 1441 | 1097 | 212 | 527 | NA | 1703 | 681 |
| HIV coinfection (n) | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| HCV coinfection, Yes/No (n/n) | 0/1529 | NA/NA | 0/412 | 0/3340 | 0/2688 | NA/NA | NA/NA | NA/NA | 0/2075 | NA/NA |
| Treatment received/ Medications |
No/NA | No/NA | No/NA | No/NA | No/NA | Unknown/NR | No/NA | Yes/ Nucleotide analogues |
No/NA | No/NA |
| Duration of follow-up (years) | 12 | 12 | Mean ± SD: 7.1 ± 6.3 |
12 | Mean ± SD: 14.7 ± 4.3 |
† Case group: 4.42 † Control group: 9.53 |
Median (ranges): 3.6 (0.5–5.8) |
Median (ranges): 5.8 (2–13.1) |
Mean ± SD: 16.02 ± 4.75 |
NR |
| HBsAg cut-off value (IU/ml) |
1000 | 100 1000 |
100 | 100 1000 |
100 1000 |
100 1000 |
1000 | 2000 | 1000 | 100 1000 |
†, Median age in the case and the control group; NA, not applicable; NR, not reported; SD, standard deviation; n, number; HBV, hepatitis B virus; CHB, chronic hepatitis B; ALT, alanine aminotransferase; HBeAg, hepatitis B e-antigen; HBV DNA, hepatitis B virus deoxyribonucleic acid; HIV, human immunodeficiency virus; HCV, hepatitis C virus; HBsAg, hepatitis B surface antigen
Methodology assessments
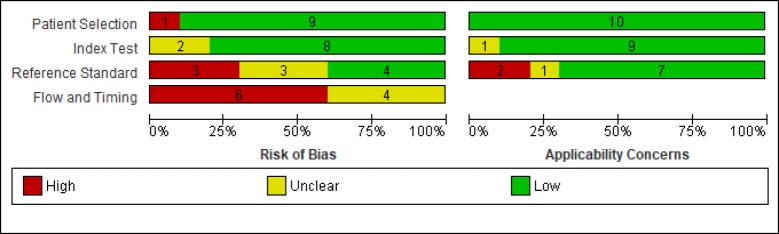
The risk of bias and applicability concerns of the 10 selected studies is displayed in Figure 2. Most studies presented a low risk of bias respecting patient selection and HBsAg level testing. Thirty percent of studies were at a high risk of bias in terms of HCC diagnosis. The risk of bias in most studies (60%) had to do with flow and timing. All of the studies had low applicability risks, except for the standard reference test, i.e. HCC diagnosis was done for participants with suspected HCC via different screening methods, such as serum alpha-fetoprotein (AFP) and abdominal ultrasound.
Figure 2.
Risk of Bias and Applicability Concerns Graph of 10 Included Studies
Figure 3 illustrates the risks of bias and applicability across individual studies. The methodological qualities of the two studies by Chien et al published in 2016 and by Lee et al published in 2015 were the highest (Lee et al., 2015; Chien et al., 2016). We found one study had risks of reference standard, bias, flow and timing and applicability of findings (Yang et al., 2017).
Figure 3.

Risk of Bias and Applicability Concerns Summary of 10 Included Studies
Association between the baseline level of HBsAg (at 100 IU/ml) and the risk of HCC in patients with chronic hepatitis B infection
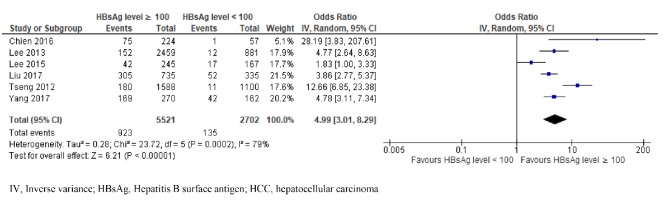
There were six studies investigating the correlation between the baseline HBsAg level and the HCC risk in patients with chronic HBV infection using the HBsAg threshold of 100 IU/ml (Tseng et al., 2012; Lee et al., 2013; Lee et al., 2015; Chien et al., 2016; Liu et al., 2017; Yang et al., 2017). Among 8,223 participants, 1,058 presented HCC during the follow-ups (923 participants with high HBsAg and 135 with low HBsAg levels). We detected a high heterogeneity of ORs among these studies (χ2 = 23.72, p = 0.0002, I2 = 79%) (Figure 4). Under a random effects model, the patients with high HBsAg level had a 4.99 times higher chance of developing HCC than those with a low HBsAg level (OR = 4.99; 95% CI, 3.01–8.29; p < 0.00001).
Figure 4.
Comparison of High and Low HBsAg Level (at 100 IU/ml Cut-Off) and Risk of HCC in Patients with Chronic HBV Infection
Factors that could potentially cause heterogeneity of ORs across studies included gender, advanced age, HCV coinfection, HBV genotype C and data collection setting. Using the data available, we further explored the sources of heterogeneity by performing a subgroup analysis at the HBsAg threshold of 100 IU/ml: the association between the baseline HBsAg level and the risk of HCC development in studies conducted in the hospitals or in the communities. We found no difference between the hospital setting and the community setting subgroups (χ2 = 0, p = 0.95).
Association between baseline level of HBsAg (at 1000 IU/ml) and risk of HCC
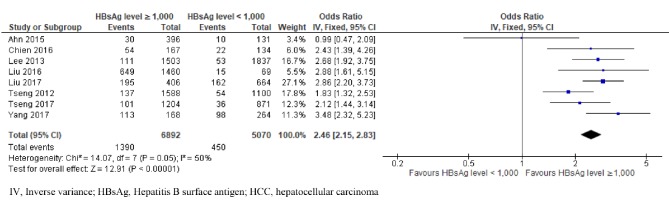
We analysed data from eight studies reporting on the HBsAg level at 1,000 on the risk of HCC in patients with chronic HBV infection (Tseng et al., 2012; Lee et al., 2013; Ahn et al., 2015; Chien et al., 2016; Liu et al., 2016; Liu et al., 2017; Tseng et al., 2017; Yang et al., 2017). A total of 1,840 patients presented HCC, and among them, 75.5% (1,390) had high HBsAg levels and 24.5% (450) had low HBsAg levels. We observed low heterogeneity (χ2 = 14.07, p = 0.05, I2 = 50%). Using a fixed effect model, participants with high HBsAg levels had a 2.46-fold rise in the risk of HCC development compared with those who had low levels (OR = 2.46; 95% CI, 2.15–2.83; P < 0.00001) (Figure 5).
Figure 5.
Comparison of High and Low HBsAg Level (at 1,000 IU/ml Cut-Off) and Risk of HCC in Patients with Chronic HBV Infection
Association between gender and risk of HCC in patients with chronic HBV infection
We were able to combine data from seven studies to obtain a summary OR (Tseng et al., 2012; Lee et al., 2013; Kawanaka et al., 2014; Chien et al., 2016; Liu et al., 2017; Tseng et al., 2017; Yang et al., 2017). Our results showed that men had a higher risk of developing HCC than women (OR, 2.12; 95% CI, 1.52–2.95; p < 0.00001), with moderate heterogeneity (χ2 = 20.48, p = 0.002, I2 = 71%).
Association between hepatitis B e-antigen (HBeAg) status and HCC risk
We found accessible data for six studies (Tseng et al., 2012; Lee et al., 2013; Kawanaka et al., 2014; Chien et al., 2016; Tseng et al., 2017; Yang et al., 2017). Pooled data on the risk of developing HCC among participants with HBeAg positivity compared with the risk in those with HBeAg negativity identified HBeAg positivity as a risk factor for HCC development (OR = 2.99; 95% CI, 1.3–6.89; p = 0.01), with significant heterogeneity (χ2 = 99.76, p < 0.00001, I2 = 95%).
Association between the HBV DNA level and risk of HCC
We analysed data from seven studies reporting on the difference in HBV DNA levels and the risk of HBV-related HCC (Tseng et al., 2012; Lee et al., 2013; Chien et al., 2016; Liu et al., 2016; Liu et al., 2017; Tseng et al., 2017; Yang et al., 2017). Our results showed that participants with HBV DNA levels ≥ 2000 IU/ml had a 4.37 times higher risk of developing HCC than those with lower HBV DNA level (OR, 4.37; 95% CI, 2.96–6.46; p < 0.00001), with high heterogeneity (χ2 = 33.32, p < 0.00001, I2 = 82%).
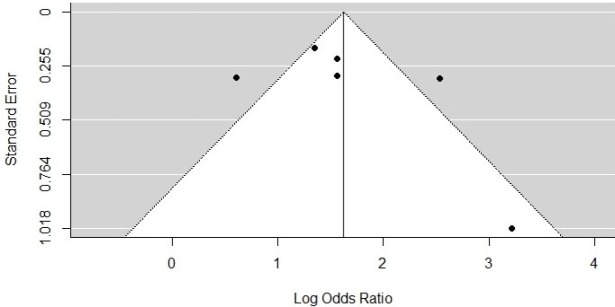
Publication bias
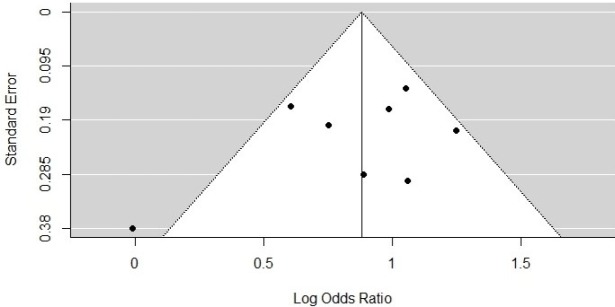
We visualised publication bias using a funnel plot with six studies included under the cut-off of 100 IU/ml (Figure 6) and eight studies under the cut-off of 1,000 IU/ml (Figure 7). Using Egger’s test, we saw no evidence of publication bias (p = 0.4423 for the HBsAg cut-off of 100 IU/ml; p = 0.3647 for the HBsAg cut-off of 1,000 IU/ml).
Figure 6.
Funnel Plot for the Incidence of HCC at High HBsAg Level versus Low HBsAg Level (100 IU/ml)
Figure 7.
Funnel Plot for the Incidence of HCC of High HBsAg Level (1,000 IU/ml) versus Low HBsAg Level
Discussion
We searched four databases to identify articles published during the past 15 years (January 2002 to November 2017) that fit our eligibility criteria. We selected 10 studies for our review. Most of the populations included in the study were classified as inactive HBV infections and had not received antiviral therapy. Qu et al., (2015) have published a meta-analysis on the effect of serum HBsAg levels for predicting clinical outcomes of patients with chronic HBV infection, but only two studies in their analysis had the baseline HBsAg levels, and they only evaluated the cut-off at 1,000 IU/ml. In our systematic review, we updated and assessed the association with HCC development across the two different available cut-offs (100 and 1,000 IU/ml).
We designed a well-prepared analysis in which we extracted data from the original studies with different HBsAg cut-off points when available; we also combined ORs across studies at each cut-off. Moreover, learning from the previous meta-analysis, we increased the number of research databases (PubMed, Scopus, CENTRAL and ProQuest) to avoid heterogeneity and publication bias.
Our results showed that at the HBsAg cut-off of 100 IU/ml, the odds ratio of developing HCC among patients with chronic HBV infection with an HBsAg level ≥ 100 is 4.99 times that compared with patients with an HBsAg level < 100, but the heterogeneity is high. In a study by Lee and colleagues, the percentage of elderly participants developing HCC during the follow-up was lower in those with an HBsAg level < 100 IU/ml at baseline (OR = 1.83; 95% CI, 1.00–3.33), which results in a 63% reduction in the chance of developing HCC in comparison to our results (OR = 4.99; 95% CI, 3.01–8.29) (Lee et al., 2015). However, in our analysis of this cut-off at 100 IU/ml, we found high heterogeneity. To evaluate this finding, we compared one group with studies performed in the hospital to the other group with studies in the community, but we found no difference. We believe the advanced age in the patients in Lee’s study, may explain the difference in the chance of developing HCC, but data on age were unavailable to compute this subgroup.
We also found that at the cut-off of 1,000 IU/ml, the patients with an HBsAg ≥ 1,000 IU/ml have a 2.46-fold increased OR of developing HCC (95% CI, 2.15–2.83), which is slightly higher than the published OR of 2.21 (95% CI, 1.52–3.22) (Qu et al., 2015). However, our more recent data were obtained from eight studies with a total of 11,962 samples, which is considerably higher than the number in the review by Qu et al; therefore our study should provide a more precise overall OR for this threshold. In addition, we did not found evidence of publication bias.
Variables such as male gender, HBeAg positivity and high hepatitis B viral load were significantly associated with HCC development, with moderate-to-high heterogeneity. We also observed one study using an HBsAg cut-off of 2,000 IU/ml, and while we did not include that data in our meta-analysis, the study showed that levels of HBsAg ≥2,000 IU/mL were not associated with HCC development (p = 0.64) after receiving long-term anti-viral therapy (Kawanaka et al., 2014). Our results on the well-established factors, namely, male gender, HBeAg positivity and a high HBVDNA level (≥ 2,000 IU/ml) are consistent with earlier findings (Yang et al., 2002; Chen et al., 2006; Poh et al., 2015; Zheng et al., 2017). However, these results should be regarded with some degree of scepticism due to the moderate-to-high variations among studies.
We acknowledge some limitations to our meta-analysis. First, we lacked data on some confounding factors such as the HBV genotypes (genotypes B and C account for most patients in Asia), the HBV stages, age, duration of follow-up, qualitative HBeAg and fibrosis stages, because most included publications in our review studied patients with an unknown stage of liver cirrhosis attributed to a high rate of HCC (Fernandez-Rodriguez and Gutierrez-Garcia, 2014). This made it difficult to explore the heterogeneity of aetiologies. Second, a verification bias was introduced in these chosen studies as participants underwent a gold standard for HCC diagnosis (e.g. histology or two typical imaging findings) once HCC manifestations had been identified during the clinical follow-up. We could omit some HCC cases, bringing about a low incidence of diagnosed HCC outcomes.
In conclusion, without publication bias evidences, the meta-analyses at both HBsAg cut-offs 100 IU/ml and 1,000 IU/ml are associated with an increased risk of HCC development. In addition, risk factors such as male gender, HBeAg positivity and high HBV DNA level have been correlated with a greater risk of HCC development. Therefore, our results may help physicians customize a better HCC surveillance system among patients with chronic HBV infection. Finally, our meta-analysis suggests a need for studies on the value of the HBsAg levels in subgroup populations, especially in those with advanced age, different stages of chronic HBV infection and different HBV genotypes.
Funding Statement
This work was supported by the Faculty of Tropical Medicine, Mahidol University, Thailand Research Fund [MRG6080025]. The funders had no role in the study design, data collection, analysis, decision to publish or preparation of the manuscript.
Acknowledgments
We would like to express our gratitude to the staff members of the Office of Research Services (Faculty of Tropical Medicine, Mahidol University, Thailand), to Professor Jia-Horng Kao (Division of Hepatology/Gastroenterology, National Taiwan University Hospital), to Assistant Professor Dong Hyun Sinn (Department of Medicine, Samsung Medical Centre), and, to Dr. Rajneesh Kumar (Department of Gastroenterology and Hepatology, Singapore General Hospital), who provided valuable data for the project.
References
- Ahn JM, Sinn DH, Gwak GY, et al. Prediction of clinical outcomes in Hepatitis B E antigen negative chronic Hepatitis B patients with elevated Hepatitis B virus DNA levels. PLoS One. 2015;10:e0144777. doi: 10.1371/journal.pone.0144777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Yang HI, Su J, et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- Chien J, Liu J, Lee MH, et al. Risk and predictors of hepatocellular carcinoma for chronic hepatitis B patients with newly developed cirrhosis. J Gastroenterol Hepatol. 2016;31:1971–7. doi: 10.1111/jgh.13422. [DOI] [PubMed] [Google Scholar]
- Fernandez-Rodriguez CM, Gutierrez-Garcia ML. Prevention of hepatocellular carcinoma in patients with chronic hepatitis B. World J Gastrointest Pharmacol Ther. 2014;5:175–82. doi: 10.4292/wjgpt.v5.i3.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JT, Guo H. Metabolism and function of hepatitis B virus cccDNA: Implications for the development of cccDNA-targeting antiviral therapeutics. Antiviral Res. 2015;122:91–100. doi: 10.1016/j.antiviral.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawanaka M, Nishino K, Nakamura J, et al. Quantitative levels of Hepatitis B virus DNA and surface antigen and the risk of Hepatocellular Carcinoma in patients with Hepatitis B receiving long-term nucleos(t)ide analogue therapy. Liver Cancer. 2014;3:41–52. doi: 10.1159/000343857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Testoni B, Fresquet J, et al. Development of hepatocellular carcinoma in chronic hepatitis B patients with advanced fibrosis is independent of viral genotype. J Med Virol. 2017;89:845–8. doi: 10.1002/jmv.24707. [DOI] [PubMed] [Google Scholar]
- Lee MH, Yang HI, Liu J, et al. Prediction models of long-term cirrhosis and hepatocellular carcinoma risk in chronic hepatitis B patients: risk scores integrating host and virus profiles. Hepatology. 2013;58:546–54. doi: 10.1002/hep.26385. [DOI] [PubMed] [Google Scholar]
- Lee P-L, Chen J-J, Tung H-D, et al. Serum hepatitis B surface antigen level might predict cirrhosis and hepatocellular carcinoma in older patients with chronic hepatitis B. Adv Dig Med. 2015;2:102–7. [Google Scholar]
- Liu J, Hu HH, Lee MH, et al. Serum levels of M2BPGi as short-term predictors of Hepatocellular Carcinoma in untreated Chronic Hepatitis B patients. Sci Rep. 2017;7:14352. doi: 10.1038/s41598-017-14747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yang HI, Lee MH, et al. Serum levels of Hepatitis B surface antigen and DNA can predict inactive carriers with low risk of disease progression. Hepatology. 2016;64:381–9. doi: 10.1002/hep.28552. [DOI] [PubMed] [Google Scholar]
- Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh Z, Goh BB, Chang PE, et al. Rates of cirrhosis and hepatocellular carcinoma in chronic hepatitis B and the role of surveillance: a 10-year follow-up of 673 patients. Eur J Gastroenterol Hepatol. 2015;27:638–43. doi: 10.1097/MEG.0000000000000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L-S, Liu J-X, Zhang H-F, et al. Effect of serum hepatitis B surface antigen levels on predicting the clinical outcomes of chronic hepatitis B infection: A meta-analysis. Hepatol Res. 2015;45:1004–13. doi: 10.1111/hepr.12444. [DOI] [PubMed] [Google Scholar]
- Tseng T-C, Liu C-J, Su T-H, et al. Fibrosis-4 index helps identify HBV carriers with the lowest risk of Hepatocellular Carcinoma. Am J Gastroenterol. 2017;112:1564. doi: 10.1038/ajg.2017.254. [DOI] [PubMed] [Google Scholar]
- Tseng TC, Liu CJ, Yang HC, et al. High levels of hepatitis B surface antigen increase risk of hepatocellular carcinoma in patients with low HBV load. Gastroenterology. 2012;142:1140–9. doi: 10.1053/j.gastro.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Valaydon ZS, Locarnini SA. The virological aspects of hepatitis B. Best Pract Res Clin Gastroenterol. 2017;31:257–64. doi: 10.1016/j.bpg.2017.04.013. [DOI] [PubMed] [Google Scholar]
- Werle-Lapostolle B, Bowden S, Locarnini S, et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology. 2004;126:1750–8. doi: 10.1053/j.gastro.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Whiting PF, Rutjes AS, Westwood ME, et al. Quadas-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- WHO. Global hepatitis report. WHO. 2017. 978-92-4-156545-5. [Google Scholar]
- WHO. Hepatitis B. 2019. [Accessed 15 July 2019]. http://www.who.int/mediacentre/factsheets/fs204/en/
- Yang HI, Lu SN, Liaw YF, et al. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med. 2002;347:168–74. doi: 10.1056/NEJMoa013215. [DOI] [PubMed] [Google Scholar]
- Yang HI, Tseng TC, Liu J, et al. Incorporating serum level of Hepatitis B surface antigen or omitting level of Hepatitis B virus DNA does not affect calculation of risk for Hepatocellular Carcinoma in patients without cirrhosis. Clin Gastroenterol Hepatol. 2016;14:461–8. doi: 10.1016/j.cgh.2015.10.033. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gao J, Tan YT, et al. Individual and combined effects of hepatitis B surface antigen level and viral load on liver cancer risk. J Gastroenterol Hepatol. 2017;33:1131–7. doi: 10.1111/jgh.14032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Zhu YJ, Wang HY, et al. Gender disparity in hepatocellular carcinoma (HCC): multiple underlying mechanisms. Sci China Life Sci. 2017;60:575–84. doi: 10.1007/s11427-016-9043-9. [DOI] [PubMed] [Google Scholar]