Abstract
Background and objectives:
Hepatocellular carcinoma (HCC) is a potential health problem in Egypt because of the high prevalence of HCV infection. Using alpha-fetoprotein for diagnosis is unsatisfactory especially in early stages. Many studies showed that microRNAs (miRNA) expression may be associated with the development and progression of various types of cancer including HCC and it may serve as biomarkers for diagnosis. This study examined two miRNAs which are miRNA-122 and miRNA-224 if it could serve as biomarkers for diagnosis of HCC.
Methods:
This study included 20 patients with HCV-induced HCC and 20 patients with HCV-induced liver cirrhosis for comparison. As well as 20 healthy volunteers as controls. All participants were subjected to history taking, clinical examination, and determination of serum alpha-fetoprotein. Quantification of plasma miRNA-122 and miRNA-224 was done by real-time quantitative PCR.
Results:
Our results showed that levels of miRNA-122 were significantly lower in HCC group compared to the cirrhosis group and controls, while levels of miRNA-224 were significantly higher. The levels of both miRNAs have a correlation with tumor size. Moreover, the diagnostic accuracy of miRNA-122 (sensitivity 95%, specificity 81%, p-value <0.001) and of miRNA-224 (sensitivity 85%, specificity 79%, p-value <0.001) in discriminating patients with HCC from patients with liver cirrhosis were higher than that of alpha-fetoprotein (sensitivity 70%, specificity 70%, p-value <0.05). In addition, combining any one of these miRNAs with alpha-fetoprotein will increase the diagnostic accuracy compared to using each marker alone.
Conclusion:
miRNA-122 and miRNA-224 could serve as biomarkers for the diagnosis of HCC.
Key Words: Hepatocellular carcinoma, miRNA-122, miRNA-224
Introduction
Hepatocellular carcinoma (HCC) is a common primary liver cancer that has a very bad prognosis. Its mortality rate is increasing despite the global efforts for early detection and improving the treatment options (Okajima et al., 2016; Fu et al., 2018).
Most of HCC cases are caused by liver cirrhosis because of hepatitis B (HBV) or C (HCV) virus infections. Other factors that increase the risk for HCC include chronic alcohol consumption, non-alcoholic liver disease, exposure to aflatoxins and certain inherited liver diseases as hemochromatosis (Mizuguchi et al., 2016).
Currently, HCC diagnosis depends primarily on finding a hepatic focal lesion on imaging techniques, however it is not always suitable for the early detection of small lesions (Lin et al., 2016). In addition to serum alpha-fetoprotein (AFP) which was accepted to be a tumor marker for HCC, early detection of small tumors will be missed in a number of patients as alpha-fetoprotein has limited sensitivity and specificity in the early stages of HCC (Terentiev and Moldogazieva, 2013).
Recognizing the molecular targets of HCC may help in increasing the survival rate of patients suffering from such grave illness (Okajima et al., 2016). Recently, many researches elucidated that some molecules and signaling pathways have a crucial role in the pathogenesis of HCC like PI3K/Akt/mTOR, Hedgehog, Ras/Raf/MAPK, Wnt-β-catenin, HGF/c-Met, and EGFR. Furthermore, genomic instability and alterations are considered to have a role in HCC (Urabe et al., 1996; Wilkens et al., 2004).
MicroRNAs (miRNAs) which are small non-coding RNAs molecules that are involved in gene expression post-transcriptionally. It regulates cell cycle and apoptosis and its expression has a role in the pathogenesis of cancer and cancer progression. It has been observed that dysregulation of miRNAs expression occur in specific liver diseases, like hepatitis, cirrhosis and liver cancers (Lee et al., 1993). Specific miRNAs were found to have negative regulatory effects on gene expression and these may act as an oncogenes or tumor suppressors (Pineau et al., 2010; Yu et al., 2011).
MIR-122 is abundant in the liver tissue compared to other tissues as detected by RNA-sequencing. Its hepatocyte-specific expression early in liver injury in addition to its direct interaction with HCV RNA has made it an attractive biomarker in liver diseases (Thakral and Ghoshal, 2015).
Also, MiR-224 has a role in the development and progression of HCC in light of the way that it tethers to 3′UTR regions of the genes that are engaged in the process of cell apoptosis, proliferation, and invasion (Ma et al., 2012; Zhang et al., 2013; Li et al., 2014; Hung et al., 2018).
Our study aim was to investigate the value of serum Mir-122 and MiR-224 in the diagnosis of hepatocellular carcinoma (HCC).
Materials and Methods
Study population
A total number of 60 individuals (34 males and 26 females) were enrolled in our cross-sectional study that was done from March 2018 to December 2018.
The patients have been selected from inpatients and outpatient clinics of the National Liver Institute, Shebin Elkom, Menoufia, Egypt. The selected subjects were divided into 3 groups:
(Group I), which comprises 20 patient with HCV-induced HCC, (Group II) comprising 20 patient with liver cirrhosis due to chronic hepatitis C, and (Group III) comprising 20 healthy viral infection-free volunteers of matched age and sex as a control group.
Exclusion criteria
Patients with a history of another malignancy, patients with liver diseases due to other causes than viral hepatitis C and previous treatment for HCV or HCC.
All individuals were subjected to detailed history taking, physical examination, radiological investigations (abdominal ultrasound and CT), and laboratory investigations including complete blood count, kidney, and liver function tests (albumin, total bilirubin, direct bilirubin ALT and AST), HBV and HCV serology. In addition, HCV Reverse-Transcriptase Polymerase Chain Reaction (RT-PCR) for patients in group I and II was done.
Liver cirrhosis was diagnosed based on history, findings on clinical examination, laboratory results, and imaging including abdominal ultrasonography and CT. Child-Pugh score was used to assess the severity of the liver disease. HCC was diagnosed clinically based on dynamic imaging (abdominal ultrasound and triphasic CT), and/or serum alpha-fetoprotein with underlying chronic liver disease. Tumor staging was done using Barcelona clinic liver cancer (BCLC) staging (Llovet et al., 1999).
Methods
Blood sample
Seven ml of venous blood were withdrawn by venipuncture; 5 ml was transferred into a plain tube, left to clot, centrifuged for 10 min at 4,000 R.P.M. The serum obtained was stored at −80 ºC until analysis of serum AFP level, liver function tests, liver enzymes, and hepatitis viral markers. The remaining 2 ml of blood were collected into EDTA containing tube for HCV RT-PCR, RNA extraction and estimation of Gene expression of MicroRNA122 and 224.
Hepatitis C Virus Antibody (ANTI-HCV) was determined by electrochemiluminescence immunoassay “ECLA”, using Cobas immunoassay analyzer (Feinstone et al., 1975).
Determination of Hepatitis B Surface Antigen (HBsAg) in serum by using (Sorin Biomedica Co. kits, Italy) (Boniolo et al., 1982).
Serum aminotransferases (ALT and AST) were determined by kinetic UV optimized method IFCC (ELTEC Kit, England) (Bergmeyer et al., 1986).
Serum total bilirubin determination was done using DIAMOND diagnostics Kit, Germany (Doumas et al., 1973).
Serum albumin was determined by the quantitative method of enhanced specificity of bromocresol green colorimetric by Diamond Diagnostics kit, Germany (Pinnell and Northam, 1978).
Prothrombin time is determined by STA-Stago Compact CT autoanalyzer (Colman et al., 1994).
RT-PCR for HCV; nucleic acid extraction was done by QIAGEN viral RNA Mini Extraction Kit.
Serum AFP was done by enzyme-linked immunosorbent assay (ELISA) using IMMULITE 1000 system by a kit supplied by Siemens Medical Solutions Diagnostics, USA (Gitlin, 1975).
Assay of microRNA 122 and 224 gene expression
MicroRNA was first effectively isolated from plasma by using kits supplied by Qiagen™ RNA extraction kit MiRNeasy kit, according to manufacturer instructions.
Once microRNA has been purified it needs to be quantified by using NanoDrop® N50 nanophotometer Implen GmbH and Implen, Inc. Schatzbogen 52 81829 München, Germany to determine both RNA concentration and purity. The extracted microRNA product was stored at –80°C for reverse transcription step. The extracted microRNA was reverse transcribed (RT) to synthesize single-stranded cDNA using the Qiagen®miScript II RT Kit. RT master mix was prepared by adding the following components to a nuclease-free microcentrifuge tube: 4 μl 5×miScript HiSpec Buffer, 2μl 10×miScript Nuclease Mix, 2 μl RNase-free water, 2 μl miScript Reverse Transcriptase Mix, then a 10 μl Template RNA was added to each a 20-µl reaction tube containing reverse-transcription master mix. Reverse transcription was carried out at 37°C for 60 minutes and 95°C for 5 minutes on Applied Biosystems 2720 thermal cycler (Bioline, Singapore, USA) cDNA product was diluted to 5 ng/ul for determining transcript levels by real-time quantitative PCR.
Diluted cDNA generated by the miScript II RT Kit was used as the template for real-time PCR with the miScript SYBR Green PCR Kit produced by Qiagen. A reaction mix was prepared as follow: 12.5 μl 2x QuantiTect SYBR Green PCR Master Mix, 2.5 μl 10x miScript Universal Primer based on mRNA sequences obtained from the miRBase database for (miRNA-122 and miRNA-224), 2.5 μl Template cDNA and 3.5 μl RNase-free water. Program the real-time cycler Applied Biosystems®7500 thermal cycler (Applied Biosystems, Foster City, CA, USA) according to the following conditions:95°C for 15 min (initial denaturation step), the denaturation at 94°C for 15 s, annealing for 30 s at 55°C, extension for 30 s at 70°C, for 40 cycles. Amplification of U6 small RNA was performed with each experimental sample as an endogenous control.
Relative quantification (RQ) expression levels were calculated using the comparative 2−ΔΔCt method with Applied Biosystems 7500 software version 2.0.1. Where the amount of the target miRNA-122 and miRNA-224 were normalized to an endogenous reference U6 small RNA and relative to control.
Each run was completed using melting curve analysis to confirm the specificity of the amplification and absence of primer dimers.
Statistical Analysis
Analysis of data was done by the use of IBM SPSS statistics software version 20 (SPSS Inc., Chicago, IL). Mean and standard deviation (SD) were used for the presentation of numerical data while frequency and percentage were used for the presentation of qualitative data. Examining the relation between qualitative variables was done using Chi-square test.
Mann–Whitney test was used to compare between two groups for non-normally distributed quantitative data while Kruskal–Wallis test [non-parametric ANOVA (analysis of variance)] was used for comparison between three groups. Based on Kruskal–Wallis distribution, post-hoc was used for pair-wise comparison. Examining the correlation between miRNAs and the clinicopathological features of HCC was done using the Spearman correlation test.
Receiver operating characteristic (ROC) curve analysis was done for evaluating the diagnostic value of serum miRNAs in HCC beside prediction of the best possible cutoff values. Logistic regressions were used to evaluate the ability of combined tests to discriminate HCC patients from cirrhotic and controls.
Results
Patient’s demographic and biochemical data: Analysis of the clinical and laboratory data of the three different groups are presented in Table 1. For statistical analysis, there was no statistically significant difference between the studied groups as regards the age and gender. Males were predominant in the HCC and cirrhosis group.
Table 1.
Demographic and Laboratory Parameters of the Different Studied Groups
| Variables | Group I | Group II (Cirrhosis) | Group III (Control) | (P-value) |
|---|---|---|---|---|
| mean ± SE | (HCC) (n = 20) | (n = 20) | (n = 20) | |
| Gender | ||||
| Male n (%) | 16 (80) | 13 (65) | 10 (50) | |
| Female n (%) | 4 (20) | 7 (35) | 10 (50) | 0.094 |
| Age (years) | 56.95± 1.73 | 56.50± 2.1 | 52.35± 1.4 | 0.137 |
| AST (U/L) | 78.25± 15.14 a | 77.45± 10.5 a | 31± 1.6 | 0.003 |
| ALT (U/L) | 86.07 ± 17.6 a | 80.25± 12.9 a | 22.73± 1.5 | 0.001 |
| ALB (g/dl) | 3.40± 0.1a | 3.77± 0.1a | 4.25± 0.1 | <0.001 |
| T- Bil (mg/dl) | 1.42± 0.2 a | 1±0.1 a | 0.86± 0.05 | 0.03 |
| INR | 1.15± 0.08 a | 1.14± 0.03 a | 0.94± 0.02 | 0.01 |
| PLT (×103/μL) | 132± 13.7 a | 152.70± 20.96 a | 299.35± 19.7 | <0.001 |
| AFP (ng/ml) | 463.3± 193.28 a,b | 25.3±5.9 a,b | 9.92± 0.73 | 0.008 |
| MiRNA-122 (RQ) | 0.103± 0.06 a,b | 2.98± 3.32 a | 1.96± 0.43 | 0.001 |
| MiRNA-224 (RQ) | 5.4± 1.3 a,b | 1.89± 0.6 a | 1.02± 0.3 | 0.001 |
a, Significantly differs from control group; b, Significantly differs from cirrhotic group; SE, standard error; AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALB, Albumin; T. Bil, total bilirubin; INR, international normalized ratio; AFP, alpha-fetoprotein; miRNA, micro RNA; RQ, Relative quantity.
Analysis of the biochemical results showed that, there was statistically significant difference between HCC and cirrhosis group when compared to the control group as regard AST, ALT, serum albumin, total bilirubin, INR, platelet count and alpha-fetoprotein level (P < 0.05).
Expression of miRNA-122 in serum
The results showed that there is a statistically significant difference between the studied groups as regard serum miRNA-122 expression. When comparing the HCC group with the cirrhosis and control group, there is a significant decrease in miRNA-122 expression. While miRNA-122 expression in the cirrhosis group is significantly increased when compared to the control group.
Expression of miRNA-224 in serum
The results showed that there was a statistically significant difference between the studied groups as regard serum miRNA-224 expression. Expression of miRNA-224 was significantly higher in the HCC group when compared to the cirrhosis and control group. Compared to the control group, the cirrhosis group showed significantly higher expression of miRNA-224. So overall, the expression of miRNA-224 among the different group showed a stepwise increase (HCC > cirrhosis > control).
Tumor characteristics in the HCC group: It is shown in Table 2. Most of the patients were classified as grade A according to Child-Pugh classifications (80%). According to BCLC staging, 10 patients were in stage A (50%), 3 patients in stage B (15%) and 7 patients were in stage C (35%).
Table 2.
Characteristics of the Tumor in HCC Group (n = 20)
| n (%) | |
|---|---|
| Number of the focal lesions | |
| Single | 10 (50%) |
| Multiple (2-3) | 10 (50%) |
| Location of the focal lesions | |
| Rt. Lobe | 10 (50%) |
| Lt. Lobe | 4 (20%) |
| Both | 6 (30%) |
| Size of the tumor by CT (cm) | |
| ≤ 5 cm | 11 (55%) |
| > 5 cm | 9 (45%) |
| Ascites | |
| No | 17 (85%) |
| Yes (mild) | 3 (15%) |
| Child-Pugh score | |
| Stage A | 16 (80%) |
| Stage B | 4 (20%) |
| BCLC | |
| Stage A | 10 (50%) |
| Stage B | 3 (15%) |
| Stage C | 7 (35%) |
Rt., right; Lt., left; CT, computed tomography; Barcelona Clinic Liver Cancer.
Correlations between the studied miRNAs and clinical parameters in the HCC group: As shown in Table 3, there was no statistically significant correlation between miRNA-122 or miRNA-224 expressions and the laboratory results (AST, ALT, serum albumin, total bilirubin, INR, platelet count). However, regarding their correlation with the tumor characteristics, miRNA-122 has a significant inverse correlation while miRNA-224 has a significant direct correlation with the tumor size.
Table 3.
Correlations between the Studied miRNAs and the Clinical Parameters in the HCC Group
| miRNA-122 | miRNA-224 | |||
|---|---|---|---|---|
| r | P-value | r | P-value | |
| ALT | -0.067 | 0.778 | 0.171 | 0.47 |
| AST | 0.006 | 0.98 | 0.112 | 0.637 |
| ALB | -0.002 | 0.993 | 0.24 | 0.308 |
| T. Bil | -0.114 | 0.631 | 0.312 | 0.18 |
| INR | -0.164 | 0.49 | -0.041 | 0.863 |
| Platelets | -0.233 | 0.322 | 0.088 | 0.713 |
| Focal lesion size | -0.534 | 0.015* | 0.622 | 0.032* |
| Focal lesion number | 0.037 | 0.876 | -0.353 | 0.126 |
| AFP | -0.099 | 0.454 | 0.134 | 0.308 |
| CTP classification | -0.202 | 0.211 | 0.107 | 0.511 |
| BCLC | 0.267 | 0.255 | -0.049 | 0.838 |
| miRNA-122 | - | - | 0.001 | 0.996 |
| miRNA-224 | 0.001 | 0.996 | - | - |
*Significant correlation
ROC analysis of the studied miRNAs:
As shown in Table 4, at the cutoff point of 1.19 (RQ), miRNA-122 can discriminate patients with HCC from patients with liver cirrhosis with a sensitivity of 95%, a specificity of 81%, positive predictive value (76.9%) and negative predictive value (95%). While at a cutoff point of 0.99 (RQ), miRNA-224 can discriminate HCC patients from cirrhotic patients with a sensitivity of 85%, a specificity of 79%, positive predictive value (78%) and negative predictive value (80%). While AFP showed lower sensitivity and specificity (70% and 70% respectively).
Table 4.
Diagnostic Performance of Alpha-Fetoprotein, miRNA-122, and miRNA-224 for Discriminating HCC from Cirrhosis and Contro
| AFP | miRNA-122 | miRNA-224 | AFP | miRNA-122 | miRNA-224 | AFP+ miRNA-122 | AFP+ miRNA-224 | |
|---|---|---|---|---|---|---|---|---|
| AUC | 0.71 | 0.98 | 0.8 | 0.68 | 0.93 | 0.77 | 0.96 | 0.83 |
| Cut-off value | 11 | 0.99 | 1.06 | 11.5 | 1.19 | 0.99 | - | - |
| sensitivity | 70 | 90 | 85 | 70 | 95 | 85 | 98 | 94 |
| specificity | 65 | 70 | 85 | 70 | 81 | 79 | 87 | 79 |
| PPV | 66.7 | 76.9 | 85 | 70 | 76.9 | 78 | 89 | 82.5 |
| NPV | 68.4 | 98 | 85 | 70 | 95 | 80 | 97 | 93 |
| P-value | 0.02 | <0.001 | <0.001 | 0.013 | <0.001 | 0.004 | <0.001 | 0.002 |
AUC, area under the curve; PPV, positive predictive value; NPV, negative predictive value.
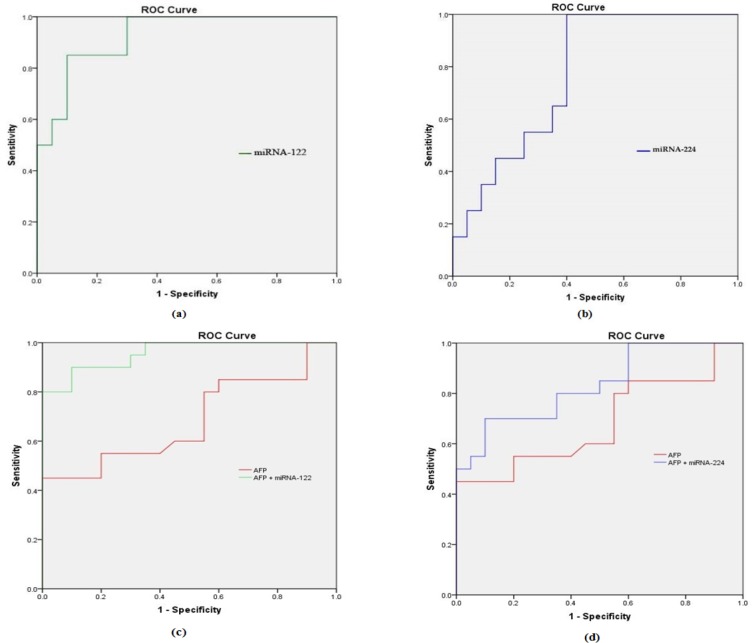
The diagnostic performance of both miRNA-122 and miRNA-224 are higher than AFP in discriminating HCC patients from both cirrhotic patients and controls. Moreover combining each of the studied miRNAs with AFP will increase the diagnostic accuracy as shown in Figure 1.
Figure 1.
Receiver Operating Characteristic Curve (ROC) Analysis of (a) miRNA-122, (b) miRNA-224, (c) AFP and AFP + miRNA-122 and (d) AFP and AFP + miRNA-224 for discriminating HCC group from cirrhotic group
Discussion
Hepatocellular carcinoma (HCC) is a complex disease with multiple variables that vary from country to another (Marrero et al., 2010). It is considered one of the potential health burdens in Egypt due to the high prevalence of hepatitis C viral infection (Elghazaly et al., 2018).
Complications of liver biopsy in addition to the unsatisfactory accuracy of alpha-fetoprotein make the need for a new sensitive and specific biomarker for early detection of HCC mandatory (Mamdouh et al,. 2017; Russo et al., 2018).
MicroRNAs are small non-coding RNA molecules that regulate the expression of genes at the post-transcriptional level. It has an important role in cancer development and progression as it acts as oncogenes or tumor suppressors (Nagy et al., 2018). Moreover, as it modulates the HCC functions, it may have a therapeutic potential as targeted molecular therapy (Xu et al., 2018).
Our study’s aim is to assess the role of two plasma miRNAs, which are miRNA-122 and miRNA-224 as diagnostic markers in HCV-induced HCC.
In the present study, analysis of the clinical data of the three different groups revealed that AST, ALT, serum bilirubin and INR were significantly elevated, while serum albumin and platelet count are decreased in the HCC and cirrhosis groups compared to controls. These results are expected and simply explained by the damage of the adjacent hepatocytes resulting from tumor growth and interference with their blood supply or continues hepatocyte necrosis due to active cirrhosis or chronic active hepatitis (El–Serag and Rudolph, 2007; Okonkwo et al., 2011; Domingo et al., 2013; Baghdady et al., 2014). In addition to decreased thrombopoietin, which is a glycoprotein synthesized primarily in the liver and regulates the production of peripheral platelets (Liu et al., 2018).
Also, there was a statistically significant difference between HCC and cirrhotic patients compared to controls regarding serum AFP level. Many studies observed that Alpha-fetoprotein is overexpressed in HCC (Qu et al., 2011; Köberle et al., 2013; El-Abd et al., 2015; Naz et al., 2018) and Chu et al., (2001) found that the severity of cirrhosis was significant predictors of elevated serum AFP and that higher serum AFP levels were significantly correlated with advanced cirrhosis in patients with chronic HCV.
In our study, plasma miRNA-122 was significantly decreased in HCC patients compared to cirrhotic patients and controls. This supports the concept of its antitumor function (Bandiera et al., 2015). Similarly, many studies reported the same (Xu et al., 2011; Qi et al., 2013; Amr et al., 2017; Weis et al., 2019). In contrary, few studies showed that miRNA-122 level was higher in HCC patients when compared to patients without HCC. This could be explained by that these studies included patients with HBV infection. They postulated that miRNA-122 expression is downregulated in HCC while its blood levels are increased due to its release from tumor tissue (Qi et al., 2011; Hung et al., 2016).
Koberle et al., (2013) reported no significant differences in miRNA-122 levels in HCC patients compared to patients without HCC. This is could be explained by that, his study included patients with multiple disease etiologies rather than HCV. Similar to that, El-Abd et al., (2015) found no statistical difference in miRNA-122 levels between HCC patients and those without HCC. However, there was a lower level in HCC patients, but it didn’t reach statistical significance.
In the current study, miRNA-122 expression was significantly higher in cirrhotic patients compared to controls. Some studies found the same observation and it could be explained by miRNA-122 leakage from apoptotic cells into the blood (Laterza et al., 2009; Xu et al., 2011; Amr et al., 2017).
Our results demonstrated that there were no significant correlations between miRNA-122 expression and clinical parameters in the HCC group (AST, ALT, serum albumin, total bilirubin, INR, platelet count, focal lesion number, AFP, Child-Pugh classification and BCLC), while there was an inverse correlation with tumor size. El Garem et al., (2014) found no correlation between miRNA-122 and both the liver synthetic functions and AFP in the HCC patients however, he found a correlation between miRNA-122 and higher ALT and AST levels in the chronic hepatitis groups. This augments the theory of initial increase in miRNA-122 levels due to inflammation and necrosis of the hepatocytes, then it will be followed by a drop with the development of fibrosis. While, Amr et al., (2017) found a significant inverse correlation between miRNA-122 and AST, ALT, AFP Level and tumor size in the HCC group.
In the present work, miR-224 was significantly overexpressed in the plasma of HCC patients compared to cirrhotic patients and healthy controls. This agrees with those reported by many studies (Lin et al., 2016; Amr et al., 2017; Mamdouh et al., 2017). miRNA-224 promotes the development and progression of HCC through suppressing apoptosis via targeting apoptosis inhibitor 5 (API-5) or acting as an oncogene which activates the AKT signaling pathway, thus promoting tumor cell proliferation and invasion (Ladeiro et al., 2008; Wang et al., 2008; Ma et al., 2012).
In our study, there were no significant correlations between miRNA-244 expression and the clinical parameters in the HCC group (AST, ALT, serum albumin, total bilirubin, INR, platelet count, focal lesion number, AFP, Child-Pugh classification and BCLC), while there was a direct correlation with tumor size. A study conducted by Lin et al., (2016) showed that there was no correlation between miRNA-224 and the clinicopathological features in HCC patients except the BCLC stage. Amr et al., (2017) found a significant direct correlation between miRNA-224 and AST, ALT, AFP and tumor size and a significant inverse correlation with serum albumin. While Okajima et al., (2016) found a direct correlation between plasma miRNA-224 and tumor size.
It is well established that Egypt has a high prevalence rate of hepatitis C viral infection and this usually leads to liver cirrhosis and HCC (Elghazaly et al., 2018). So, we performed ROC analysis for the evaluation of the diagnostic accuracy of miRNA-122 and miRNA-224 in discriminating HCV-induced HCC patients from patients with liver cirrhosis and controls.
ROC analysis of miRNA-122 in discriminating HCC from cirrhotic patients showed an AUC of 0.93 with a sensitivity of 95%, a specificity of 81%, PPV of 76.9, NPV of 95, P-value (<0.001) at a cutoff of 1.19 (RQ). While miRNA-224 showed an AUC of 0.77 with a sensitivity of 85%, a specificity of 71%, PPV of 78, NPV of 80, P-value (0.004) at a cutoff of 0.99 (RQ). However, AFP showed AUC of 0.68, sensitivity of 70 %, specificity of 70 %, PPV of 70, NPV of 70, P-value (<0.05) at cutoff 11.5 ng/mL.
So, both miRNAs have better diagnostic performance than AFP in discriminating patients with HCC from patients with liver cirrhosis. Moreover, combining each of these miRNAs with AFP will increase the diagnostic accuracy compared to using each marker alone.
These findings are similar to previous studies which found that miRNA-122 and miRNA-224 have better diagnostic performance than AFP in diagnosing HCC (Wang and Lee, 2011; Xu et al., 2011; Muawia et al., 2015; Amr et al., 2017).
In summary, our study showed that expression of miRNA-122 was decreased, while the expression of miRNA-224 was increased in plasma of HCC patients compared to patients with liver cirrhosis and controls and both of them have a correlation with the tumor size. In addition, they have better diagnostic accuracy in discriminating patients with HCC from those without. So, it could be promising biomarkers for the diagnosis of HCC. However, more studies with large numbers of candidates with different etiologies of HCC are needed before clinical practice.
Our study has some limitations, such as a relatively small number of patients, and uni-central study. So, a larger number multi-centric study is recommended in the future work.
In conclusion, serum miRNA-122 and miRNA-224 could be used as biomarkers for diagnosis of HCV-induced HCC with better accuracy than AFP.
Acknowledgments
The authors gratefully acknowledge the contribution of the patients and volunteers in this study.
Funding
The authors received no specific funding for this work.Research involving Human Participants The National Liver Institute research ethics committee approved this study and it has been performed according to Declaration of Helsinki ethical standards.
Compliance with Ethical Standards
The National Liver Institute research ethics committee approved this study and it has been performed according to Declaration of Helsinki ethical standards. All participants included in the study provided informed consent before enrollment.
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Amr KS, Elmawgoud Atia HA, Elazeem Elbnhawy RA, et al. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis. 2017;4:215–21. doi: 10.1016/j.gendis.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghdady I, Fouad F, Sayed M, et al. Serum markers for the early detection of hepatocellular carcinoma in patients with chronic viral hepatitis C infection. Menoufia Med J. 2014;27:544. [Google Scholar]
- Bandiera S, Pfeffer S, Baumert TF, et al. miR-122–a key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448–57. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Bergmeyer HU, Horder M, Rej R. IFCC methods for alanine aminotransferase. J Clin Chem Clin Biochem. 1986;24:481–95. [PubMed] [Google Scholar]
- Boniolo A, Davis M, Matheza R. The use of ELISA in screening of HBsAg. J Immunol. 1982;49:1–5. [Google Scholar]
- Chu CW, Hwang SJ, Luo JC, et al. Clinical, virologic, and pathologic significance of elevated serum alpha-fetoprotein levels in patients with chronic hepatitis C. J Clin Gastroenterol. 2001;32:240–4. doi: 10.1097/00004836-200103000-00014. [DOI] [PubMed] [Google Scholar]
- Colman RW, Hirsh J, Marder VJ, Salzman EW. Hemostasis and thrombosis: basic principles and clinical practice. Philadelphia: J.B. Lippincott; 1994. pp. 559–5. [Google Scholar]
- Domingo EO, Lingao AL, Lao JY, et al. The significance of common laboratory tests in hepatocellular carcinoma. Philippi J Intern Med. 2013;51:1–3. [Google Scholar]
- Doumas BT, Perry BW, Sasse EA, et al. Standardization in bilirubin assays: evaluation of selected methods and stability of bilirubin solutions. Clin Chem. 1973;19:984–93. [PubMed] [Google Scholar]
- El-Abd NE, Fawzy NA, El-Sheikh SM, et al. Circulating miRNA-122, miRNA-199a, and miRNA-16 as Biomarkers for early detection of hepatocellular carcinoma in Egyptian patients with chronic Hepatitis C virus infection. Mol Diagn Ther. 2015;19:213–20. doi: 10.1007/s40291-015-0148-1. [DOI] [PubMed] [Google Scholar]
- El-Garem H, Ammer A, Shehab H, et al. Circulating microRNA, miR-122 and miR-221 signature in Egyptian patients with chronic hepatitis C related hepatocellular carcinoma. World J Hepatol. 2014;6:818. doi: 10.4254/wjh.v6.i11.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El–Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–76. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- Elghazaly H, Gaballah A, Eldin NB. Clinic-pathological pattern of hepatocellular carcinoma (HCC) in Egypt. Ann Oncol. 2018;29:mdy151. [Google Scholar]
- Feinstone SM, Kapikian AZ, Purcell RH, et al. Transfusion-associated hepatitis not due to viral hepatitis type A or B. N Engl J Med. 1975;292:767–70. doi: 10.1056/NEJM197504102921502. [DOI] [PubMed] [Google Scholar]
- Fu X-T, Shi Y-H, Zhou J, et al. MicroRNA-30a suppresses autophagy-mediated anoikis resistance and metastasis in hepatocellular carcinoma. Cancer lett. 2018;412:108–17. doi: 10.1016/j.canlet.2017.10.012. [DOI] [PubMed] [Google Scholar]
- Gitlin D. Normal biology of α-fetoprotein. Ann N Y Acad Sci. 1975;259:7–16. doi: 10.1111/j.1749-6632.1975.tb25397.x. [DOI] [PubMed] [Google Scholar]
- Hung CH, Hu TH, Lu SN, et al. Circulating micro RNAs as biomarkers for diagnosis of early hepatocellular carcinoma associated with hepatitis B virus. Int J Cancer. 2016;138:714–20. doi: 10.1002/ijc.29802. [DOI] [PubMed] [Google Scholar]
- Hung JH, Li CH, Yeh CH, et al. MicroRNA-224 down-regulates Glycine N-methyltransferase gene expression in Hepatocellular Carcinoma. Sci Rep. 2018;8:12284. doi: 10.1038/s41598-018-30682-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köberle V, Kronenberger B, Pleli T, et al. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur J Cancer. 2013;49:3442–9. doi: 10.1016/j.ejca.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008a;47:1955–63. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- Laterza OF, Lim L, Garrett-Engele PW, et al. Plasma MicroRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55:1977–83. doi: 10.1373/clinchem.2009.131797. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Li Q, Ding C, Chen C, et al. miR-224 promotion of cell migration and invasion by targeting Homeobox D 10 gene in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2014;29:835–42. doi: 10.1111/jgh.12429. [DOI] [PubMed] [Google Scholar]
- Lin L, Lu B, Yu J, et al. Serum miR-224 as a biomarker for detection of hepatocellular carcinoma at early stage. Clin Res Hepatol Gastroenterol. 2016;40:397–404. doi: 10.1016/j.clinre.2015.11.005. [DOI] [PubMed] [Google Scholar]
- Liu F, Zhou H, Cao L, et al. Risk of reduced platelet counts in patients with nonalcoholic fatty liver disease (NAFLD): a prospective cohort study. Lipids Health Dis. 2018;17:221. doi: 10.1186/s12944-018-0865-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Seminars in liver disease by Thieme Medical Publishers; 1999. pp. 329–38. [DOI] [PubMed] [Google Scholar]
- Ma D, Tao X, Gao F, et al. miR-224 functions as an onco-miRNA in hepatocellular carcinoma cells by activating AKT signaling. Oncol Lett. 2012;4:483–8. doi: 10.3892/ol.2012.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamdouh S, Khorshed F, Aboushousha T, et al. Evaluation of Mir-224, Mir-215 and Mir-143 as serum biomarkers for HCV associated hepatocellular carcinoma. Asian Pac J Cancer Prev. 2017;18:3167–71. doi: 10.22034/APJCP.2017.18.11.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrero JA, Kudo M, Bronowicki JP. The challenge of prognosis and staging for hepatocellular carcinoma. Oncologist. 2010;15:23–33. doi: 10.1634/theoncologist.2010-S4-23. [DOI] [PubMed] [Google Scholar]
- Mizuguchi Y, Takizawa T, Yoshida H, et al. Dysregulated miRNA in progression of hepatocellular carcinoma: A systematic review. Hepatol Res. 2016;46:391–406. doi: 10.1111/hepr.12606. [DOI] [PubMed] [Google Scholar]
- Muawia S, El-Said H, Kamel T. Correlation of miR-122 with Bcl-w is a paradigm for the role of micro RNAs in the liver injury development. Int J Biol Sci Appl. 2015;2:86–96. [Google Scholar]
- Nagy A, Lanczky A, Menyhart O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz Z, Usman S, Saleem K, et al. Alpha-fetoprotein: A fabulous biomarker in hepatocellular, gastric and rectal cancer diagnosis. Biomed Res. 2018;29:2478–83. [Google Scholar]
- Okajima W, Komatsu S, Ichikawa D, et al. Circulating microRNA profiles in plasma: identification of miR-224 as a novel diagnostic biomarker in hepatocellular carcinoma independent of hepatic function. Oncotarget. 2016;7:53820–36. doi: 10.18632/oncotarget.10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonkwo UC, Nwosu MN, Ukah C, et al. The clinical and pathological features of hepatocellular carcinoma in Nnewi, Nigeria. Niger J Med. 2011;20:366–71. [PubMed] [Google Scholar]
- Pineau P, Volinia S, McJunkin K, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–9. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinnell AE, Northam BE. New automated dye-binding method for serum albumin determination with bromcresol purple. Clin Chem. 1978;24:80–6. [PubMed] [Google Scholar]
- Qi P, Cheng S-q, Wang H, et al. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One. 2011;6:e28486. doi: 10.1371/journal.pone.0028486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Wang J, Katayama H, et al. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma. 2013;60:135. doi: 10.4149/neo_2013_018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu KZ, Zhang K, Li H, et al. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol. 2011;45:355–60. doi: 10.1097/MCG.0b013e3181f18ac2. [DOI] [PubMed] [Google Scholar]
- Russo FP, Imondi A, Lynch EN, et al. When and how should we perform a biopsy for HCC in patients with liver cirrhosis in 2018? A review. Dig Liver Dis. 2018;50:640–6. doi: 10.1016/j.dld.2018.03.014. [DOI] [PubMed] [Google Scholar]
- Terentiev AA, Moldogazieva NT. Alpha-fetoprotein: a renaissance. Tumor Biol. 2013;34:2075–91. doi: 10.1007/s13277-013-0904-y. [DOI] [PubMed] [Google Scholar]
- Thakral S, Ghoshal K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr Gene Ther. 2015;15:142–50. doi: 10.2174/1566523214666141224095610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urabe Y, Nouso K, Higashi T, et al. Telomere length in human liver diseases. Liver. 1996;16:293–7. doi: 10.1111/j.1600-0676.1996.tb00748.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lee AT, Ma JZ, et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J Biol Chem. 2008;283:13205–15. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lee CG. Role of miR-224 in hepatocellular carcinoma: a tool for possible therapeutic intervention? Epigenomics. 2011;3:235–43. doi: 10.2217/epi.11.5. [DOI] [PubMed] [Google Scholar]
- Weis A, Marquart L, Calvopina DA, et al. Serum MicroRNAs as biomarkers in Hepatitis C: Preliminary evidence of a MicroRNA panel for the diagnosis of Hepatocellular Carcinoma. Int J Mol Sci. 2019;20:864. doi: 10.3390/ijms20040864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkens L, Flemming P, Gebel M, et al. Induction of aneuploidy by increasing chromosomal instability during dedifferentiation of hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2004;101:1309–14. doi: 10.1073/pnas.0305817101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wu C, Che X, et al. Circulating MicroRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50:136–42. doi: 10.1002/mc.20712. [DOI] [PubMed] [Google Scholar]
- Xu X, Tao Y, Shan L, et al. The role of MicroRNAs in Hepatocellular carcinoma. J Cancer. 2018;9:3557–69. doi: 10.7150/jca.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D-C, Li Q-G, Ding X-W, et al. Circulating microRNAs: potential biomarkers for cancer. Int J Mol Sci. 2011;12:2055–63. doi: 10.3390/ijms12032055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Takahashi S, Tasaka A, et al. Involvement of microRNA-224 in cell proliferation, migration, invasion, and anti-apoptosis in hepatocellular carcinoma. J Gastroenterol Hepatol. 2013;28:565–75. doi: 10.1111/j.1440-1746.2012.07271.x. [DOI] [PubMed] [Google Scholar]