Abstract
Polyunsaturated fatty acids (PUFAs) play a role in a wide variety of physiological processes. They are produced by a series of desaturation and elongation reactions. -6-desaturase is a membrane-bound enzyme that catalyzes the conversion of -linolenic acid (C18:3-3) and linoleic acid (C18:2-6) to stearidonic acid (18:4-3) and -linolenic acid (18:3-6). It is encoded by the FADS2 gene located on bovine chromosome 29. The aim of this study was to identify a single nucleotide polymorphism in the FADS2 gene and to determine possible associations with milk fatty acid composition in two breeds of dairy cattle, i.e., Jersey and Polish Holstein-Friesian. Direct DNA sequencing revealed the presence of an A-to-G substitution in intron 3 of the FADS2 gene (rs209202414). Both populations were genotyped with an appropriate PCR-RFLP assay. The following genotype distributions were observed: for Jerseys, AA 0.24, AG 0.63, and GG 0.13; for Polish Holstein-Friesians, AA 0.17, AG 0.40, and GG 0.43. In Jerseys, statistically significant relationships were found between the FASD2 genotypes and the following milk fatty acids: lauric (), behenic (), lignoceric (), oleic (), eicosatrienoic (), and docosadienoic (). In Polish Holstein-Friesian cows, significant associations were observed for erucic () and docosahexaenoic () acids. The study indicated the A-to-G substitution (rs209202414) in the bovine FADS2 gene as a potential genetic marker for fatty acid composition in cattle milk.
1. Introduction
The feeding of dairy cows is the main factor impacting milk fat composition. Pasture intake reduces the concentration of saturated fatty acids (SFAs) in the milk of grazing cows (Couvreur et al., 2007; Frigo et al., 2015; Hanuš et al., 2016; Ponnampalamet al., 2018). Furthermore, genetic factors influence fatty acid (FA) variability. The FA profile in milk changes during lactation, emphasizing the relationship between the physiological status of cow and milk composition (Bastin et al., 2011). The effects on milk FA composition are also breed-dependent. The greatest breed differences are observed between Holstein and Jersey milk (with the higher concentrations of SFAs in Jerseys) (Arnould and Soyeurt, 2009). Some authors have reported that milk fat composition is modulated by the polymorphisms in genes involved in milk fat synthesis processes, like DGAT1 and SCD1 (Carvajal et al., 2016, Tzompa-Sosa et al., 2016).
Dietary long-chain polyunsaturated fatty acids (PUFAs) increased intestinal FADS2 mRNA abundance but had modest effects on its level in the liver of suckling pigs (Jacobi et al., 2011). PUFAs regulate fatty acid desaturase (FADS1, FADS2) activity in the liver and adipocyte tissue (Nakamura and Nara, 2004; Ralston et al., 2015). Hatanaka et al. (2016) reported that long-chain polyunsaturated fatty acid (LC-PUFA, > C20) intake is crucial for the growth of -6-desaturase knockout (D6D-KO) mice. The FADS2 indel polymorphism in the European grayling was found to be associated with muscle FA composition (Renaville et al., 2013). Matsumoto et al. (2014) found that the SNP (g. 823 > ) in the FADS2 promoter had a significant effect on several beef quality traits, including beef marbling score, whereas Takahashi et al. (2016) reported a highly significant association between the rs211580559 SNP in exon 7 of the FADS2 gene and intramuscular C18:2(-6) composition. In the transcriptomic study, Wang et al. (2017) pointed to FADS2 as a strong candidate gene that may be associated with intramuscular fat deposition. Recently, Gol et al. (2018) reported that the polymorphism in the porcine FADS2 gene is linked to arachidonic acid metabolism.
Fatty acid desaturase-2 (FADS2) is a component of the lipid metabolic pathway and converts essential FA into LC-PUFA by the introduction of a double bond between carbon atoms at positions 6 and 7 of FA (14). FADS2 is a rate-limiting enzyme involved in the conversion of linoleic acid (LA; 18:2-6) into -linolenic acid (GLA; 18:3:-6) and that of -linolenic acid (ALA; 18:3-3) into stearidonic acid (SDA; 18:4-3).
Some genome-wide association studies showed that the FADS locus is one of the strongest genetic predictors of plasma phospholipid PUFA (Lemaitre et al., 2011; Tanaka et al., 2009). Ibeagha-Awemu et al. (2014) demonstrated positive associations between three SNP within the FADS2 gene and the milk PUFA in Canadian Holstein cows. Therefore, the main aim of this study was to analyze the associations between the FADS2 gene polymorphism and milk fat composition in two breeds of dairy cattle (Polish Holstein-Friesian and Jersey).
2. Materials and methods
2.1. Animals
The study involved 150 Holstein-Friesian cows housed in a conventional free-stall barn in West Pomeranian Province, Poland, and 104 Jersey cows kept in a tie-stall barn in Greater Poland Province. Only healthy animals from 2–5 years old were included. The nutrition and management of cows were quite similar. Feeding was based on a total mixed ration (TMR), mainly composed of maize silage, grass haylage, maize cereals, oat cereals, soybean meals, and mineral–vitamin mixtures. No ethical consent was required for the present study since the milk samples were collected during milking and the blood samples during routine veterinary visits.
2.2. SNP identification and genotyping
Genomic DNA was isolated from whole peripheral blood using the salting-out method (MasterPure™ DNA Purification Kit for Blood, Epicentre, Madison, Wisconsin, USA). Exons 1, 3, and 12 of the bovine FADS2 gene were amplified using the primers given in Table 1. The reference sequence of the FADS2 gene located on chromosome 29 (GenBank Acc. No. NC_037356.1) was used.
Table 1.
Primer sequences used for the amplifications of the bovine FADS2 exons (1, 3, and 12).
| Region | Primer sequences (5–3) | Product | Annealing |
|---|---|---|---|
| length | temp. (C) | ||
| Exon 1 | F1: GGAGGAGAAGACAAAAGCCGA | 437 | 60 |
| |
R1:TGAGCGCCGTAGACACTTTT |
|
|
| Exon 3 | F3: TCCCAGATCACCGAGGACTT | 292 | 60 |
| |
R3: TTCAGAGCGTTGGCACCTAG |
|
|
| Exon 12 | F12: CGGGCAACTGGTCCCTTTAT | 389 | 60 |
| R12: GTCCCATGACCAAGTGCCTC |
PCR amplifications were performed in a total volume of 15 L containing 50 ng of genomic DNA, 1.5 mM , 0.2 mM of each dNTP, 15 pmol of each primer, and 0.3 U of Taq polymerase (Eur, Poland). The following thermal profile was applied: 5 min at 94 C, 32 cycles of 30 s at 94 C, 30 s at the annealing temperature, and 30 s at 72 C; and a final extension of 5 min at 72 C. The PCR products were separated in agarose gel (1 %, 30 min, 120 V) and then extracted using the GEL/PCR Purification GPB Mini Kit (GenoPlast Biochemicals, Poland). Finally, the samples were sent for sequencing to an external laboratory (Genomed, Poland). A PCR-RFLP assay has been developed for the genotyping of an A-to-G substitution (rs209202414) in intron 3 of the FADS2 gene. The PCR conditions were the same as those described above (Primers F3 and R3, Table 1). A total of 10 L of the PCR product was digested with 2 U of TseFI restriction enzyme (SibEnzyme Ltd, Russia). Subsequently, the restriction fragments were separated in a 3 % agarose gel (60 min, 120 V) stained with ethidium bromide.
2.3. Milk samples and fatty acid composition
Milk samples for the determination of fatty acid composition were collected from cows after the 90th day of lactation to avoid the period of negative energy balance and to maximize the period of de novo milk fat synthesis in the mammary gland. The samples were transported to the laboratory and kept frozen until further processing. Total lipids were extracted from each sample using a chloroform–methanol solution according to Folch et al. (1957). FAs were transformed into fatty acid methyl esters (FAMEs) with the basic method using boron trifluoride according to the Polish standards (PN-EN ISO 12966-2: 2011). The FAME composition was analyzed by gas chromatography with mass spectrometer (Clarus 600 GC/MS system, PerkinElmer, USA) equipped with an Elite-5MS capillary column (length: 60 m; inner diameter: 0.25 mm; film thickness: 0.25 m). Helium with a constant flow of 1 mL min was used as the carrier gas. The sample volume was 1 mL (split ratio, ). The injector temperature was 290 C. The column started at a temperature of 110 C and was ramped up to 180 C at a rate of 5 C per minute, then 15 min at 180 C, followed by the gradient of 5 C per minute up to 290 C and then 5 min at this temperature. The temperature of transfer line was 290 C. For mass spectrometry, the selected-ion recording technique was used with the ionization energy of 70 eV and an ion source temperature of 200 C. The individual FAMEs were identified by the comparison of their retention times with that of the standard compound (Supelco™ 37 Component FAME Mix, Sigma-Aldrich, Germany). A total of 37 fatty acids were investigated in milk samples. However, only fatty acids with an even number of carbon atoms were considered in the association analyses, since only these are synthesized de novo, elongated, and desaturated in the mammary gland. The peaks were analyzed with TurboMass software (PerkinElmer Inc., Waltham, MA, USA).
2.4. Statistical analysis
Statistical analyses were performed using the appropriate R packages (R Core Team, 2015). An additive relationship matrix was constructed based on a three-generation pedigree using the kinship2 R package (Therneau et al., 2014). The following linear model (Eq. 1) was constructed and estimated using the lmekin function of the coxme R package (Therneau, 2015):
| 1 |
where is the phenotypic value of each trait, is the overall mean, is the fixed effect corresponding to the genotype of polymorphisms, LS is the fixed effect of lactation number and lactation season, is the regression coefficient for cow age (), is the regression coefficient for days in milk (DIM), is the random polygenic effect for all known pedigree relationships, and is the random residual.
3. Results
3.1. SNP identification and genotyping
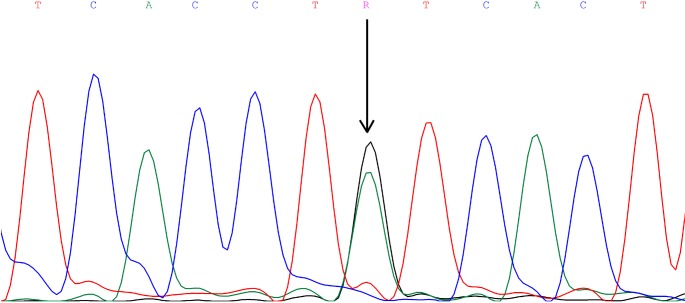
DNA fragments overlapping exons 1, 3, and 12 with the parts of adjacent introns of the FADS2 gene were sequenced. These analyses revealed the presence of an to substitution at position 23 of intron 3 (Fig. 1; GenBank rs209202414). The PCR products amplified with the F3 and R3 primers (Table 1) were digested with TseFI restriction enzyme. After electrophoresis, the following genotypes were observed: GG (205, 72, 15 bp), AG (205, 87, 72, 15 bp), and AA (205, 87 bp). The 15 bp fragments were not detectable (Fig. 2). The following genotype distributions were observed: for Jerseys, AA 0.24, AG 0.63, and GG 0.13; for Polish Holstein-Friesians, AA 0.17, AG 0.40, and GG 0.43. According to the chi-squared test, these distributions differed significantly (; ). In the Jersey group, the major allele was , while in the Polish Holstein-Friesian group, the allele was prevalent (Table 2).
Figure 1.
The rs209202414 A-to-G polymorphism in intron 3 of the bovine FADS2 gene revealed by DNA sequencing.
Figure 2.
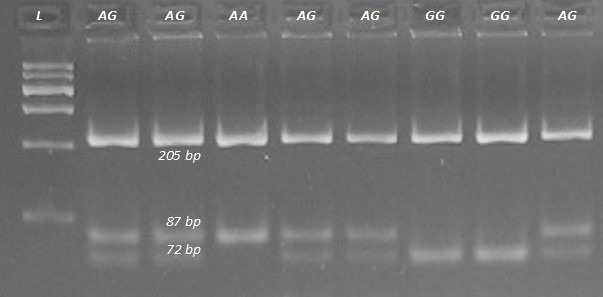
Genotyping the rs209202414 A-to-G polymorphism in the bovine FADS2 gene. Digestion with TseFI restriction enzyme revealed the AA, AG, and GG genotypes. is a 600 bp DNA ladder.
Table 2.
Genotypic frequencies of the rs209202414 SNP in the bovine FADS2 gene.
| Genotype |
Allele |
|||||
|---|---|---|---|---|---|---|
| Breed | AA | AG | GG | |||
| Jersey | 104 | 0.24 | 0.63 | 0.13 | 0.55 | 0.45 |
| Holstein-Friesian | 150 | 0.17 | 0.40 | 0.43 | 0.37 | 0.63 |
3.2. The association of genotype with fatty acid composition in the milk of Polish Holstein-Friesian and Jersey cattle
Fatty acid composition in the milk fat of Jersey and Polish Holstein-Friesian cows is given in Tables 3 and 4. The association analysis indicated significant differences in some FA content between cows carrying different FADS2 genotypes. In Jersey cattle, significant associations were recorded between the FADS2 (rs209202414) polymorphism and the following milk FA: lauric (), behenic (), lignoceric (9), oleic (), eicosatrienoic (), and docosadienoic (). In Polish Holstein-Friesian cows, significant associations were observed for erucic () and docosahexaenoic () acids.
Table 3.
The association of FADS2 polymorphism with the fatty acid composition (%) in the milk of Jersey cows.
| Trait | Total ( 104) | Genotype |
||||
|---|---|---|---|---|---|---|
| AA | AG | GG | ||||
| ( 25) | ( 65) | ( 14) | ||||
| MY | Milk yield (kg) | 21.381 3.945 | 20.048 3.354 | 21.698 3.763 | 22.286 5.295 | n.s. |
| FY | Fat yield (kg) | 1.065 0.213 | 1.028 0.186 | 1.071 0.215 | 1.108 0.251 | n.s. |
| FP | Fat (%) | 5.018 0.706 | 5.182 0.791 | 4.953 0.676 | 5.024 0.689 | n.s. |
| C6:0 | Caproic | 2.705 0.388 | 2.666 0.326 | 2.699 0.405 | 2.8 0.423 | n.s. |
| C8:0 | Caprylic | 1.692 0.285 | 1.645 0.2 | 1.695 0.306 | 1.763 0.317 | n.s. |
| C10:0 | Capric | 3.467 0.447 | 3.397 0.412 | 3.487 0.479 | 3.495 0.363 | n.s. |
| C12:0 | Lauric | 4.001 0.55 | 3.914 0.514 | 4.026 0.588 | 4.044 0.43 | 0.0486 |
| C14:0 | Myristic | 12.459 1.172 | 12.356 1.132 | 12.514 1.24 | 12.384 0.947 | n.s. |
| C16:0 | Palmitic | 37.721 2.864 | 37.369 2.926 | 37.979 2.955 | 37.148 2.297 | n.s. |
| C18:0 | Stearic | 12.6 1.645 | 12.556 1.498 | 12.625 1.813 | 12.562 1.043 | n.s. |
| C20:0 | Arachidic | 0.107 0.02 | 0.108 0.013 | 0.107 0.021 | 0.109 0.026 | n.s. |
| C22:0 | Behenic | 0.028 0.007 | 0.027 0.006 | 0.028 0.007 | 0.03 0.009 | 0.0199 |
| C24:0 | Lignoceric | 0.021 0.006 | 0.02 0.004 | 0.021 0.006 | 0.023 0.007 | 0.0209 |
| C14:1 | Myristoleic | 1.333 0.367 | 1.422 0.399 | 1.305 0.346 | 1.306 0.404 | n.s. |
| C16:1 | Palmitoleic | 1.589 0.374 | 1.634 0.386 | 1.565 0.345 | 1.623 0.488 | n.s. |
| C18:1-9c | Oleic | 16.487 3.085 | 17.105 3.39 | 16.152 3.125 | 16.938 2.147 | 0.0386 |
| C18:1-9t | Elaidic | 1.036 0.189 | 0.993 0.185 | 1.049 0.199 | 1.053 0.143 | n.s. |
| C18:2-6c | Linoleic | 1.997 0.342 | 1.992 0.326 | 1.995 0.357 | 2.013 0.321 | n.s. |
| C18:3-3 | -Linolenic | 0.115 0.052 | 0.112 0.046 | 0.118 0.055 | 0.106 0.045 | n.s. |
| C18:3-6 | -Linolenic | 0.013 0.004 | 0.012 0.004 | 0.013 0.005 | 0.013 0.004 | n.s. |
| C20:1 | Eicosenoic | 0.006 0.002 | 0.007 0.002 | 0.006 0.002 | 0.006 0.002 | n.s. |
| C20:2 | Eicosadienoic | 0.006 0.002 | 0.007 0.001 | 0.006 0.002 | 0.006 0.002 | n.s. |
| C20:3-3 | Eicosatrienoic | 0.08 0.03 | 0.076 0.025 | 0.082 0.032 | 0.082 0.03 | 0.0113 |
| C20:3-6 | Eicosatrienoic | 0.01 0.004 | 0.01 0.003 | 0.01 0.004 | 0.011 0.005 | n.s. |
| C20:4-6 | Arachidonic | 0.125 0.036 | 0.125 0.031 | 0.124 0.037 | 0.131 0.043 | n.s. |
| C20:5-3 | Eicosapentaenoic | 0.023 0.007 | 0.022 0.005 | 0.024 0.007 | 0.025 0.008 | n.s. |
| C22:1-9 | Erucic | 0.025 0.007 | 0.024 0.005 | 0.025 0.007 | 0.027 0.008 | n.s. |
| C22:2 | Docosadienoic | 0.002 0.001 | 0.002 0.001 | 0.002 0.001 | 0.003 0.001 | 0.0181 |
| C22:6-3 | Docosahexaenoic | 0.002 0.001 | 0.002 0.001 | 0.002 0.001 | 0.002 0.001 | n.s. |
| C24:1 | Nervonic | 0.004 0.001 | 0.004 0.001 | 0.004 0.001 | 0.004 0.001 | n.s. |
Different superscripts within rows indicate statistically significant differences (). n.s. – non-significant.
Table 4.
The association of FADS2 polymorphism with the fatty acid composition (%) in the milk of Polish Holstein-Friesian cows.
| Trait | Total ( 150) | Genotype |
||||
|---|---|---|---|---|---|---|
| AA | AG | GG | ||||
| ( 25) | ( 60) | ( 65) | ||||
| MY | Milk yield (kg) | 30.842 8.402 | 31.54 7.276 | 30.78 8.004 | 30.631 9.229 | n.s. |
| FY | Fat yield (kg) | 1.267 0.357 | 1.25 0.259 | 1.288 0.362 | 1.254 0.387 | n.s. |
| FP | Fat (%) | 4.147 0.627 | 4.036 0.695 | 4.207 0.55 | 4.134 0.668 | n.s. |
| C6:0 | Caproic | 2.25 0.437 | 2.314 0.377 | 2.199 0.416 | 2.272 0.476 | n.s. |
| C8:0 | Caprylic | 1.309 0.245 | 1.349 0.244 | 1.307 0.242 | 1.296 0.251 | n.s. |
| C10:0 | Capric | 3.053 0.56 | 3.076 0.488 | 3.091 0.595 | 3.01 0.556 | n.s. |
| C12:0 | Lauric | 3.694 0.643 | 3.717 0.597 | 3.731 0.706 | 3.652 0.605 | n.s. |
| C14:0 | Myristic | 12.33 1.444 | 12.418 1.246 | 12.229 1.571 | 12.391 1.407 | n.s. |
| C16:0 | Palmitic | 41.182 4.881 | 41.62 4.92 | 41.386 5.032 | 40.826 4.775 | n.s. |
| C18:0 | Stearic | 9.209 2.518 | 8.714 1.878 | 9.414 2.83 | 9.21 2.432 | n.s. |
| C20:0 | Arachidic | 0.054 0.019 | 0.05 0.011 | 0.055 0.02 | 0.054 0.02 | n.s. |
| C22:0 | Behenic | 0.017 0.007 | 0.016 0.005 | 0.016 0.007 | 0.019 0.009 | n.s. |
| C24:0 | Lignoceric | 0.016 0.01 | 0.014 0.005 | 0.014 0.006 | 0.018 0.013 | n.s. |
| C14:1 | Myristoleic | 1.361 0.49 | 1.394 0.45 | 1.325 0.476 | 1.381 0.522 | n.s. |
| C16:1 | Palmitoleic | 2.175 0.702 | 2.028 0.531 | 2.17 0.783 | 2.236 0.68 | n.s. |
| C18:1-9c | Oleic | 16.523 3.259 | 16.558 3.435 | 16.258 3.472 | 16.753 3.013 | n.s. |
| C18:1-9t | Elaidic | 0.994 0.297 | 1.015 0.319 | 0.993 0.263 | 0.987 0.322 | n.s. |
| C18:2-6c | Linoleic | 2.822 0.659 | 2.754 0.603 | 2.823 0.564 | 2.848 0.761 | n.s. |
| C18:3-3 | -Linolenic | 0.282 0.094 | 0.262 0.079 | 0.285 0.09 | 0.288 0.102 | n.s. |
| C18:3-6 | -Linolenic | 0.012 0.004 | 0.012 0.004 | 0.012 0.004 | 0.012 0.003 | n.s. |
| C20:1 | Eicosenoic | 0.004 0.002 | 0.004 0.002 | 0.004 0.002 | 0.004 0.002 | n.s. |
| C20:2 | Eicosadienoic | 0.004 0.002 | 0.004 0.002 | 0.004 0.002 | 0.004 0.002 | n.s. |
| C20:3-3 | Eicosatrienoic | 0.066 0.026 | 0.063 0.02 | 0.068 0.029 | 0.065 0.026 | n.s. |
| C20:3-6 | Eicosatrienoic | 0.007 0.003 | 0.007 0.003 | 0.007 0.003 | 0.008 0.003 | n.s. |
| C20:4-6 | Arachidonic | 0.139 0.043 | 0.14 0.027 | 0.141 0.05 | 0.137 0.043 | n.s. |
| C20:5-3 | Eicosapentaenoic | 0.02 0.006 | 0.02 0.005 | 0.02 0.006 | 0.02 0.006 | n.s. |
| C22:1-9 | Erucic | 0.022 0.016 | 0.02 0.015 | 0.019 0.014 | 0.025 0.018 | 0.046 |
| C22:2 | Docosadienoic | 0.004 0.003 | 0.003 0.002 | 0.003 0.002 | 0.004 0.003 | n.s. |
| C22:6-3 | Docosahexaenoic | 0.001 0.002 | 0.001 0.001 | 0.003 0.001 | 0.002 0.003 | 0.0469 |
| C24:1 | Nervonic | 0.004 0.004 | 0.003 0.001 | 0.004 0.001 | 0.005 0.005 | n.s. |
Different superscripts within rows indicate statistically significant differences (). n.s. – non-significant.
4. Discussion
The most important non-genetic factor significantly affecting fat content, and milk fatty acid profile in particular, is nutrition. It is associated with the fact that the fats contained in the feed are in part the source of fatty acids in the milk of ruminants, which are also the product of the reactions occurring in the rumen and lactocytes (Palmquist, 2006). The fatty acid composition in the milk samples of cows in the present study corresponded with the results obtained for dairy breeds. The most represented group of FAs in milk was SFA, followed by monounsaturated fatty acid (MUFA) and PUFA, which is consistent with the previous results (Carvajal et al., 2016; Hanuš et al., 2016; Vranković et al., 2017). Vranković et al. (2017) showed a similar FA composition in the milk of Holstein cows (C10:0 3.053, C12:0 3.694, C14:0 12.33 vs. C10:0 3.00, C12:0 3.70, C14:0 12.03) at the 150th day of lactation.
Ibeagha-Awemu et al. (2014) found significant associations of several polymorphisms in the FADS cluster with oleic acid, AA, dihomo-γ-linolenic acid (DGLA), SFA, and MUFA indices, but not with C20:5-3, C20:5-6, or C22:6-3 in the milk of Canadian Holstein cows. The authors suggested a possible involvement of these SNPs in FA synthesis and indicated them as potential genetic markers in the breeding programs increasing the content of milk FAs that are valuable for human health.
Oleic acid is a health-beneficial product of the delta-9 desaturation of stearic acid, catalyzed by SCD. Burdge and Wootton (2002) demonstrated that docosahexaenoic acid (DHA) is produced internally through a series of desaturation and elongation reactions from the dietary precursor, -linolenic acid. The positive effect of DHA on health has been extensively reviewed (Calder and Yaqoob, 2009; Ponnampalam et al., 2018). However, no associations were found between the FADS2 polymorphism and the C18:2-6c, C18:3-3, and D6D indices. This may be a result of dietary PUFA precursor (LA and ALA) susceptibility to biohydrogenation in the rumen (Chikunya et al., 2004). Appropriate supplementation of dairy cow diets may change the proportion between milk SFA and MUFA/PUFA concentrations. In the study by Kliem et al. (2019) on the use of whey protein and rapeseed oil gel as feed supplements in Holsteins, an incremental inclusion of whey protein gel caused a linear increase in MUFA and PUFA and the same decrease in SFA. Bougouin et al. (2019), investigating an effect of starch-rich or lipid-supplemented diets in lactating Holstein cows, found a higher milk SFA concentration and lower MUFA and trans-10 C18:1 concentrations in the animals fed diets containing the Ca salts of palm oil and starch from maize grain and wheat in comparison with those comprising extruded rapeseeds and sunflower seeds, whereas the levels of trans-11 C18:1 were unchanged. Finally, Santillo et al. (2016) observed an increased level of SFA, MUFA (mainly due to the contribution of C18:1 cis-9), and PUFA in Italian Simmental cows supplemented with dietary whole flaxseed.
The PUFA level in an organism is related to many positive health outcomes and plays a crucial role in its function. Some of these effects are determined by the LC-PUFA (Tosi et al., 2014). Animals are unable to synthesize essential fatty acids (EFAs), but they can convert them (from the diet) to more unsaturated FA with a longer carbon chain (Nakamura and Nara, 2004). The desaturation and elongation processes of omega-3 acids are carried out by desaturases and elongases leading to the formation of LC-PUFA (Cormier et al., 2014).
In humans, Al-Hilal et al. (2013) reported that the FADS polymorphisms are very important regulators of LC-PUFA synthesis and explained the variance of several fatty acids. Similar results were published by Boschetti et al. (2015), who demonstrated relationships between genotype and desaturating ability and, consequently, a significant impact on the PUFA content in poultry meat. Fast-growing chickens showed lower expression of hepatic FADS1 and FADS2 and thus a significantly lower content of, for example, 18:2(-6) and 20:4(-6) FA () in breast meat. Other factors can also modulate FADS2 activity. Cho et al. (1999) showed that dietary PUFA can abolish the level of hepatic FADS2 mRNA in human. Takeuchi et al. (2010) reported that a high level of dietary PUFA can suppress the transcription of SREBP-1c, a major transcription factor involved in the upregulation of FADS2 expression. Diet components may affect SREBP1 expression or activity. In the study by Li et al. (2018) on fatty acid composition in the muscles of Yanbian Yellow steers, the expression of SREBP1 increased with age in the animals fed a corn-based finishing diet with an increasing proportion of corn in the ration (every 4 months). Han et al. (2012), investigating the expression of lipogenic genes in lactating Holsteins, found that the expression of SREBP1 in the mammary gland was downregulated in the animals fed the Ca salts of conjugated linoleic acid (CLA), whereas Harvatine and Bauman (2006) reported that treatments causing milk fat depression (in the form of a low forage, high oil diet, and the trans-10, cis-12 CLA infusions) decreased the expression of SREBP1 in the bovine mammary gland.
In beef cattle, Matsumoto et al. (2014) found a significant effect of the SNP (g.-823 > ) in the promoter region of the FADS2 gene on carcass traits and fatty acid composition. In Japanese Black cattle, the percentage of C14:0 in the GG animals was higher than that of the GA ones. Subcutaneous fat thickness of the GG individuals was thinner than that of the GA ones, which led to higher yield estimates for the former. The beef marbling score of Holstein animals carrying the GG genotype was significantly higher than that of the GA individuals. An analogous relationship (although non-significant) was observed in Japanese Black cattle. Finally, the percentage of C16:0 was higher for the GG genotype compared with the GA genotype, and the percentage of MUFA was higher in the GA animals than that in the GG animals with a higher percentage of SFA in the latter. A later study on Japanese Black steers (Takahashi et al., 2016) showed that a highly significant association existed between the rs211580559 SNP ( > in exon 7) and intramuscular C18:2(-6) composition (with the CC individuals having significantly higher C18:2(-6) composition than the CT ones), whereas no significant relationships between this SNP and other investigated fatty acids (C14:0, C14:1, C16:0, C16:1, C18:0 and C18:1) were found. Beak et al. (2019) analyzed an SNP (rs109772589) in the FADS2 gene for its possible association with the fatty acid profile in Hanwoo beef cattle. However, all genotyped animals had the same GA genotype. Therefore, no further analysis was performed.
The differences in the milk FA profile between Jersey and Polish Holstein-Friesian cows may be determined by inter-breed variations in milk FA composition, which has been previously reported (Palladino et al., 2010). A nutrigenomic study showed that cows fed ALA- or LA-rich diets had increased PUFA and decreased SFA levels in milk compared with a control diet, which resulted from a diet-specific differential regulation of genes involved in FA metabolism in the mammary gland. The authors postulated that a lower level of SFA was due to the suppression of genes involved in FA metabolism and synthesis, and a higher level of PUFA was a consequence of the increased availability and incorporation of substrates used for milk PUFA synthesis (Ibeagha-Awemu et al., 2016). Different genetic variants may affect the level of FA or indices. In the study by Ding et al. (2016) on the role of selected SNP in the FADS gene cluster (FADS1, FADS2, and FADS3) on the PUFA concentration in the breast milk of Chinese women, the rs1535 SNP and two-locus haplotypes in the FADS2 gene as well as a two-locus haplotype in the FADS1 gene were associated with the GLA and AA concentrations with the minor allele carriers having lower concentrations of these acids. On the other hand, the three-locus haplotype in the FADS2 gene significantly affected concentrations of GLA but not AA. The cited authors also showed that the individuals homozygous for an SNP in the FADS3 gene had lower concentrations of ALA and LA in their breast milk. Mychaleckyj et al. (2018), investigating breast milk fatty acid composition in Bangladeshi mothers, showed that AA is the primary FA in breast milk influenced by genetic variation at the FADS1/2/3 locus and that the most significant genetic association at this locus was with the fraction of AA at the rs174556 SNP. Finally, Kgwatalala et al. (2009) reported that one of the analyzed regulatory variants in the SCD1 gene was associated with higher C10 and C12 desaturase indices and higher contents of C10:1 and C12:1 in the milk of Holstein cows.
In recent years, there have been several genome-wide association studies on milk fat traits (Grisart et al., 2002; Daetwyler et al., 2008; Moioli et al., 2007). The majority of associated SNPs were located in intergenic and intronic regions (Ibeagha-Awemu et al., 2016). Intronic SNPs may affect highly conserved elements and cis-acting RNA, which can impact RNA splicing and the rate of mRNA transcription (Millar et al., 2010; Hong et al., 2018).
5. Conclusions
This study showed a significant association between the FADS2 polymorphism and milk fatty acid composition in Jersey and Polish Holstein-Friesian cattle. The differences between breeds may result from the inter-individual variation in milk FA metabolism. The study indicated the A-to-G substitution (rs209202414) in the bovine FADS2 gene as a potential genetic marker for fatty acid composition in cattle milk.
Data availability
The data used in the present study are confidential and therefore not publicly available.
Author contributions
WSP and ML contributed to the conception and design of the study, acquisition of data, and drafting the manuscript. DZ and AD contributed to the analysis of data, interpretation of results, and drafting the manuscript. ZS, YHY, and YHC contributed to the conception and design of the study, drafting the manuscript, and critical revision of the manuscript. All the authors approved the final version of the manuscript to be published.
Financial support
This research has been supported by the National Science Centre of Poland (grant no. UMO-2013/11/N/NZ9/04631).
Review statement
This paper was edited by Steffen Maak and reviewed by Andrés Carvajal and one anonymous referee.
References
- Al-Hilal M, Alsaleh A, Maniou Z, Lewis FJ, Hall WL, Sanders TAB, O'Dell SD, MARINA study team Genetic variation at the FADS1-FADS2 gene locus influences delta-5 desaturase activity and LC-PUFA proportions after fish oil supplement . J Lipid Res. 2013;54:542–551. doi: 10.1194/jlr.P032276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnould VM, Soyeurt H. Genetic variability of milk fatty acids. J Appl Genet. 2009;50:29–39. doi: 10.1007/BF03195649. [DOI] [PubMed] [Google Scholar]
- Bastin C, Gengler N, Soyeurt H. Phenotypic and genetic variability of production traits and milk fatty acid contents across days in milk for Walloon Holstein first-parity cows. J Dairy Sci. 2011;94:4152–4163. doi: 10.3168/jds.2010-4108. [DOI] [PubMed] [Google Scholar]
- Beak SH, Lee Y, Lee EB, Kim KH, Kim JG, Bok JD, Kang SK. Study on the fatty acid profile of phospholipid and neutral lipid in Hanwoo beef and their relationship to genetic variation. J Anim Sci Technol. 2019;61:69–76. doi: 10.5187/jast.2019.61.2.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschetti E, Bordoni A, Meluzzi A, Castellini C, Dal Bosco A, Sirri F. Fatty acid composition of chicken breast meat is dependent on genotype-related variation of FADS1 and FADS2 gene expression and desaturating activity . Animal. 2016;10:700–708. doi: 10.1017/S1751731115002712. [DOI] [PubMed] [Google Scholar]
- Bougouin A, Martin C, Doreau M, Ferlay A. Effects of starch-rich or lipid-supplemented diets that induce milk fat depression on rumen biohydrogenation of fatty acids and methanogenesis in lactating dairy cows. Animal. 2019;13:1421–1431. doi: 10.1017/S1751731118003154. [DOI] [PubMed] [Google Scholar]
- Burdge GC, Wootton SA. Conversion of -linolenic acid to eicosapentaenoic, docosapentaenoic and docosahexaenoic acids in young women . Br J Nutr. 2002;88:411–420. doi: 10.1079/BJN2002689. [DOI] [PubMed] [Google Scholar]
- Calder PC, Yaqoob P. Omega-3 polyunsaturated fatty acids and human health outcomes. Biofactors. 2009;35:266–272. doi: 10.1002/biof.42. [DOI] [PubMed] [Google Scholar]
- Carvajal AM, Huircan P, Dezamour JM, Subiabre I, Kerr B, Morales R, Ungerfeld EM. Milk fatty acid profile is modulated by DGAT1 and SCD1 genotypes in dairy cattle on pasture and strategic supplementation . Genet Mol Res. 2016;15:1–12. doi: 10.4238/gmr.15027057. [DOI] [PubMed] [Google Scholar]
- Chikunya S, Demirel G, Enser M, Wood JD, Wilkinson RG, Sinclair LA. Biohydrogenation of dietary -3 PUFA and stability of ingested vitamin E in the rumen, and their effects on microbial activity in sheep . Br J Nutr. 2004;91:539–550. doi: 10.1079/BJN20031078. [DOI] [PubMed] [Google Scholar]
- Cho HP, Nakamura M, Clarke SD. Cloning, expression, and fatty acid regulation of the human delta-5 desaturase. J Biol Chem. 1999;274:37335–37339. doi: 10.1074/jbc.274.52.37335. [DOI] [PubMed] [Google Scholar]
- Cormier H, Rudkowska I, Lemieux S, Couture P, Julien P, Vohl MC. Effects of FADS and ELOVL polymorphisms on indexes of desaturase and elongase activities: results from a pre-post fish oil supplementation. Genes Nutr. 2014;9:437. doi: 10.1007/s12263-014-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur S, Hurtaud C, Marnet PG, Faverdin P, Peyraud JL. Composition of milk fat from cows selected for milk fat globule size and offered either fresh pasture or a corn silage-based diet. J Dairy Sci. 2007;90:392–403. doi: 10.3168/jds.S0022-0302(07)72640-1. [DOI] [PubMed] [Google Scholar]
- Daetwyler HD, Schenkel FS, Sargolzaei M, Robinson JA. A genome scan to detect quantitative trait loci for economically important traits in Holstein cattle using two methods and a dense single nucleotide polymorphism map. J Dairy Sci. 2008;91:3225–3236. doi: 10.3168/jds.2007-0333. [DOI] [PubMed] [Google Scholar]
- Ding Z, Liu GL, Li X, Chen XY, Wu YX, Cui CC, Zhang X, Yang G, Xie L. Association of polyunsaturated fatty acids in breast milk with fatty acid desaturase gene polymorphisms among Chinese lactating mothers. Prostaglandins Leukot Essent Fatty Acids. 2016;109:66–71. doi: 10.1016/j.plefa.2016.03.009. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane-Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Frigo E, Samorè AB, Reghenzani L, Bergomi N, Strillacci MG, Schiavini F, Prinsen RT, Cozzi MC, Serra M, Rossoni A, Bagnato A. Variation of milk components in the Italian Brown cattle. J Dairy Res. 2015;82:485–490. doi: 10.1017/S0022029915000540. [DOI] [PubMed] [Google Scholar]
- Gol S, Pena RN, Rothschild MF, Tor M, Estany J. A polymorphism in the fatty acid desaturase-2 gene is associated with the arachidonic acid metabolism in pigs. Sci Rep. 2018;8:14336. doi: 10.1038/s41598-018-32710-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisart B, Coppieters W, Farnir F, Karim L, Ford C, Berzi P, Cambisano N, Mni M, Reid S, Simon P, Spelman R, Georges M, Snell R. Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition . Genome Res. 2002;12:222–231. doi: 10.1101/gr.224202. [DOI] [PubMed] [Google Scholar]
- Han LQ, Pang K, Li HJ, Zhu SB, Wang LF, Wang YB, Yang GQ, Yang GY. Conjugated linoleic acid-induced milk fat reduction associated with depressed expression of lipogenic genes in lactating Holstein mammary glands. Genet Mol Res. 2012;11:4754–4764. doi: 10.4238/2012.September.17.2. [DOI] [PubMed] [Google Scholar]
- Hanuš O, Krížová L, Samková E, Špicka J, Kucera J, Klimešová M, Roubal P, Jedelská R. The effect of cattle breed, season and type of diet on the fatty acid profile of raw milk. Arch Anim Breed. 2016;59:373–380. doi: 10.5194/aab-59-373-2016. [DOI] [Google Scholar]
- Harvatine KJ, Bauman DE. SREBP1 and thyroid hormone responsive spot 14 (S14) are involved in the regulation of bovine mammary lipid synthesis during diet-induced milk fat depression and treatment with CLA. J Nutr. 2006;136:2468–2474. doi: 10.1093/jn/136.10.2468. [DOI] [PubMed] [Google Scholar]
- Hatanaka E, Harauma A, Yasuda H, Watanabe J, Nakamura MT, Salem Jr N, Moriguchi T. Essentiality of arachidonic acid intake in murine early development. Prostaglandins Leukot Essent Fatty Acids. 2016;108:51–57. doi: 10.1016/j.plefa.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Hong MJ, Yoo SS, Choi JE, Kang HG, Do SK, Lee JH, Lee WK, Lee J, Lee SY, Cha SI, Kim CH, Lee EB, Cho S, Jheon S, Park JY. Functional intronic variant of SLC5A10 affects DRG2 expression and survival outcomes of early-stage non-small-cell lung cancer. Cancer Sci. 2018;109:3908–3909. doi: 10.1111/cas.13814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibeagha-Awemu EM, Akwanji KA, Beaudoin F, Zhao X. Associations between variants of FADS genes and omega-3 and omega-6 milk fatty acids of Canadian Holstein cows . BMC Genet. 2014;15:25. doi: 10.1186/1471-2156-15-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibeagha-Awemu EM, Li R, Ammah AA, Dudemaine PL, Bissonnette N, Benchaar C, Zhao X. Transcriptome adaptation of the bovine mammary gland to diets rich in unsaturated fatty acids shows greater impact of linseed oil over safflower oil on gene expression and metabolic pathways. BMC Genomics. 2016;17:104. doi: 10.1186/s12864-016-2423-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi SK, Lin X, Corl BA, Hess HA, Harrell RJ, Odle J. Dietary arachidonate differentially alters desaturase-elongase pathway flux and gene expression in liver and intestine of suckling pigs. J Nutr. 2011;141:548–553. doi: 10.3945/jn.110.127118. [DOI] [PubMed] [Google Scholar]
- Kgwatalala PM, Ibeagha-Awemu EM, Hayes JF, Zhao X. Stearoyl-CoA desaturase 1 3'UTR SNPs and their influence on milk fatty acid composition of Canadian Holstein cows. J Anim Breed Genet. 2009;126:394–403. doi: 10.1111/j.1439-0388.2008.00796.x. [DOI] [PubMed] [Google Scholar]
- Kliem KE, Humphries DJ, Grandison AS, Morgan R, Livingstone KM, Givens DI, Reynolds CK. Effect of a whey protein and rapeseed oil gel feed supplement on milk fatty acid composition of Holstein cows. J Dairy Sci. 2019;102:288–300. doi: 10.3168/jds.2018-15247. [DOI] [PubMed] [Google Scholar]
- Lemaitre RN, Tanaka T, Tang W, Manichaikul A, Foy M, Kabagambe EK, Nettleton JA, King IB, Weng LC, Bhattacharya S, Bandinelli S, Bis JC, Rich SS, Jacobs Jr DR, Cherubini A, McKnight B, Liang S, Gu X, Rice K, Laurie CC, Lumley T, Browning BL, Psaty BM, Chen YD, Friedlander Y, Djousse L, Wu JH, Siscovick DS, Uitterlinden AG, Arnett DK, Ferrucci L, Fornage M, Tsai MY, Mozaffarian D, Steffen LM. Genetic loci associated with plasma phospholipid -3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium . PLoS Genet. 2011;7:e1002193. doi: 10.1371/journal.pgen.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Yan CG, Gao QS, Yan Y, Choi SH, Smith SB. Adipogenic/lipogenic gene expression and fatty acid composition in chuck, loin, and round muscles in response to grain feeding of Yanbian Yellow cattle. J Anim Sci. 2018;96:2698–2709. doi: 10.1093/jas/sky161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H, Nogi T, Tabuchi I, Oyama K, Mannen H, Sasazaki S. The SNPs in the promoter regions of the bovine FADS2 and FABP4 genes are associated with beef quality traits. Livest Sci. 2014;163:34–40. doi: 10.1016/j.livsci.2014.02.016. [DOI] [Google Scholar]
- Millar DS, Horan M, Chuzhanova NA, Cooper DN. Characterisation of a functional intronic polymorphism in the human growth hormone (GH1) gene. Hum Genomics. 2010;4:289–301. doi: 10.1186/1479-7364-4-5-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moioli B, Contarini G, Avalli A, Catillo G, Orrù L, De Matteis G, Masoero G, Napolitano F. Short communication: Effect of stearoyl-coenzyme A desaturase polymorphism on fatty acid composition of milk. J Dairy Sci. 2007;90:3553–3558. doi: 10.3168/jds.2006-855. [DOI] [PubMed] [Google Scholar]
- Mychaleckyj JC, Nayak U, Colgate ER, Zhang D, Carstensen T, Ahmed S, Ahmed T, Mentzer AJ, Alam M, Kirkpatrick BD, Haque R, Faruque ASG, Petri Jr. WA. Multiplex genomewide association analysis of breast milk fatty acid composition extends the phenotypic association and potential selection of FADS1 variants to arachidonic acid, a critical infant micronutrient . J Med Genet. 2018;55:459–468. doi: 10.1136/jmedgenet-2017-105134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura MT, Nara TY. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu Rev Nutr. 2004;24:345–376. doi: 10.1146/annurev.nutr.24.121803.063211. [DOI] [PubMed] [Google Scholar]
- Palladino RA, Buckley F, Prendiville R, Murphy JJ, Callan J, Kenny DA. A comparison between Holstein-Friesian and Jersey dairy cows and their F(1) hybrid on milk fatty acid composition under grazing conditions. J Dairy Sci. 2010;93:2176–2184. doi: 10.3168/jds.2009-2453. [DOI] [PubMed] [Google Scholar]
- Palmquist DL. Advanced Dairy Chemistry, Vol. 2, Lipids. 3rd Edn. Boston, MA: Springer US; 2006. Milk Fat: Origin of fatty acids and influence of nutritional factors thereon; pp. 43–92. [Google Scholar]
- , Ponnampalam EN, Hopkins DL, Jacobs JL. Increasing omega-3 levels in meat from ruminants under pasture-based systems. Rev Sci Tech. 2018;37:57–70. doi: 10.20506/rst.37.1.2740. [DOI] [PubMed] [Google Scholar]
- Ralston JC, Matravadia S, Gaudio N, Holloway GP, Mutch DM. Polyunsaturated fatty acid regulation of adipocyte FADS1 and FADS2 expression and function. Obesity (Silver Spring) 2015;23:725–728. doi: 10.1002/oby.21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing, Version 3.2.3. Vienna, Austria: Foundation for Statistical Computing; 2015. [Google Scholar]
- Renaville B, Tulli F, Bruno M, Tibaldi E, Messina M. Fatty acid desaturase 2 (FADS2) insertion/deletion polymorphism impact on muscle fatty acid profile in European grayling (Thymallusthymallus) . Br J Nutr. 2013;110:1559–1564. doi: 10.1017/S0007114513001049. [DOI] [PubMed] [Google Scholar]
- Santillo A, Caroprese M, Marino R, d'Angelo F, Sevi A, Albenzio M. Fatty acid profile of milk and Cacioricotta cheese from Italian Simmental cows as affected by dietary flaxseed supplementation. J Dairy Sci. 2016;99:2545–2551. doi: 10.3168/jds.2015-10419. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Hayashi M, Ushizawa K, Nishino K, Haga Y, Saito T, Fujimori Y, Iwama N, Takeda H, Komatsu M, Malau-Aduli AEO. Association of bovine fatty acid desaturase 2 gene single-nucleotide polymorphisms with intramuscular fatty acid composition in Japanese Black steers. Open J Anim Sci. 2016;6:105–115. doi: 10.4236/ojas.2016.62013. [DOI] [Google Scholar]
- Takeuchi Y, Yahagi N, Izumida Y, Nishi M, Kubota M, Teraoka Y, Yamamoto T, Matsuzaka T, Nakagawa Y, Sekiya M, Iizuka Y, Ohashi K, Osuga J, Gotoda T, Ishibashi S, Itaka K, Nagai R, Yamada N, Kadowaki T, Shimano H. Polyunsaturated fatty acids selectively suppress sterol regulatory element-binding protein-1 through proteolytic processing and autoloop regulatory circuit. J Biol Chem. 2010;285:11681–11691. doi: 10.1074/jbc.M109.096107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Shen J, Abecasis GR, Kisialiou A, Ordovas JM, Guralnik JM, Singleton A, Bandinelli S, Cherubini A, Arnett D, Tsai MY, Ferrucci L. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009;5:e1000338. doi: 10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau T, Atkinson E, Sinnwell J, Schaid D, Mcdonnell S. [last access: 17 July 2019];Kinship2: Pedigree functions. R package version 1.6. . 2014 available at: http://CRAN.R-project.org/package=kinship2 .
- Therneau TM. [last access: 17 July 2019];Coxme: mixed effects cox models, R package version 2.2-5 . 2015 available at: http://CRAN.R-project.org/package=coxme .
- Tosi F, Sartori F, Guarini P, Olivieri O, Martinelli N. Delta-5 and delta-6 desaturases: crucial enzymes in polyunsaturated fatty acid-related pathways with pleiotropic influences in health and disease. Adv Exp Med Biol. 2014;824:61–81. doi: 10.1007/978-3-319-07320-0_7. [DOI] [PubMed] [Google Scholar]
- Tzompa-Sosa DA, van Valenberg HJF, van Aken GA, Bovenhuis H. Milk fat triacylglycerols and their relations with milk fatty acid composition, DGAT1 K232A polymorphism, and milk production traits. J Dairy Sci. 2016;99:3624–3631. doi: 10.3168/jds.2015-10592. [DOI] [PubMed] [Google Scholar]
- Vranković L, Aladrović J, Octenjak D, Bijelić D, Cvetnić L, Stojević Z. Milk fatty acid composition as an indicator of energy status in Holstein dairy cows. Arch Anim Breed. 2017;60:205–212. doi: 10.5194/aab-60-205-2017. [DOI] [Google Scholar]
- Wang X, Zhang Y, Zhang X, Wang D, Jin G, Li B, Xu F, Cheng J, Zhang F, Wu S, Rui S, He J, Zhang R, Liu W. The comprehensive liver transcriptome of two cattle breeds with different intramuscular fat content. Biochem Bioph Res Co. 2017;490:1018–1025. doi: 10.1016/j.bbrc.2017.06.157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used in the present study are confidential and therefore not publicly available.