The molecular phenotype of circulating tumor cells is associated with clinical outcome of patients with breast cancer. The aim of this study was to enhance the prognostic accuracy of the circulating tumor cell phenotype in metastatic breast cancer by incorporating miRNA into a combined prediction model.
Keywords: Breast cancer, Circulating tumor cells, MicroRNA, Prognosis, Metastasis
Abstract
Background.
The molecular phenotype of circulating tumor cells (CTCs) was associated with clinical outcome of patients with breast cancer. CTCs isolated from patients with metastatic breast cancer (MBC) display a unique microRNA (miRNA) expression profile. The aim of this study was to enhance the prognostic accuracy of the CTC phenotype in patients with MBC, by incorporating miRNA into a combined prediction model.
Subjects, Materials, and Methods.
CTCs were detected by CellSearch and enriched by magnetic cell sorting. miRNA deep sequencing and quantitative polymerase chain reaction were used to screen and verify potentially CTC‐specific miRNA candidates. Patients with MBC were enrolled from two independent cohorts, and overall survival (OS) and chemotherapy response were analyzed.
Results.
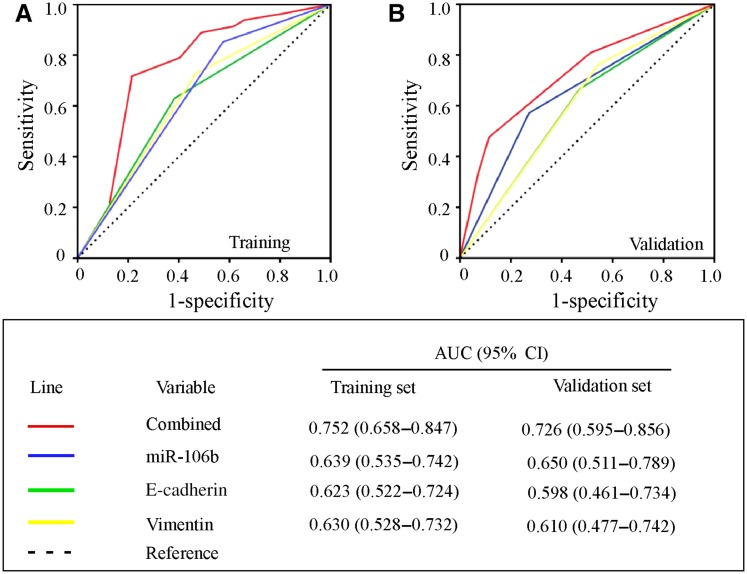
We screened and identified that miR‐106b was an upregulated molecule in patients with MBC with CTC ≥5/7.5 mL (n = 16) compared with patients with CTC = 0/7.5 mL (n = 16) and healthy donors (n = 8). The expression of CTC‐specific miR‐106b correlated with vimentin and E‐cadherin in CTC and acted as an independent factor for predicting OS (hazard ratio 2.157, 95% confidence interval [CI] 1.098–4.239, p = .026). Although CTC‐specific miR‐106b, E‐cadherin, and vimentin showed a prognostic potential independently, the prognostic performance for OS based on the combination of three markers was significantly enhanced in Cohort 1 (area under the curve [AUC] 0.752, 95% CI 0.658–0.847, n = 128) and further validated in Cohort 2 (AUC 0.726, 95% CI 0.595–0.856, n = 91). Besides, a combined model incorporating miR‐106b was associated with therapy response.
Conclusion.
The phenotypic assemblies of CTC incorporating miR‐106b show enhanced prognostic accuracy of overall survival in patients with MBC.
Implications for Practice.
In order to enhance the prognostic accuracy of the circulating tumor cell (CTC) phenotype in patients with metastatic breast cancer (MBC), this study screened and identified a CTC‐specific microRNA (miRNA), miR‐106b, as an upregulated molecule based on the comparison of miRNA profile between CTCs, primary tumors, and healthy blood donors. By incorporating miR‐106b into a combined prediction model, the prognostic accuracy of the CTC phenotype for patients with MBC was greatly improved in both the training and validation cohorts. This work provides clinical evidence supporting the prognostic potential of CTC‐specific miRNA for patients with MBC. These results indicate that developing CTC‐specific miRNAs as new biomarkers will help to further optimize personalized therapy.
关键词。乳腺癌 • 循环肿瘤细胞 • 微小 RNA • 预后 • 转移
摘要
背景。循环肿瘤细胞 (CTC) 的分子表型与乳腺癌患者的临床结果相关。从转移性乳腺癌 (MBC) 患者中分离出的 CTC 显示出独特的微小 RNA(miRNA)表达谱。本研究的目的是通过将miRNA 添加进组合预测模型,来提高 MBC 患者 CTC 表型的预后准确性。
受试者、材料和方法。采用细胞搜索法检测 CTC,采用磁细胞分选法富集 CTC。采用miRNA 深度测序技术和定量聚合酶链反应筛选并验证可能的 CTC 特异性miRNA 候选物。从两个独立队列中纳入 MBC 患者,并分析总生存率 (OS) 和化疗反应。
结果。我们筛选并确定,与 CTC = 0/7.5 mL (n = 16) 的患者和健康供体(n = 8)相比,在 CTC ≥ 5/7.5 mL (n = 16)的 MBC 患者中,miR‐106b 是一种上调分子。CTC 特异性 miR‐106b 的表达与 CTC 中波形蛋白和 E‐钙粘蛋白相关,并作为预测 OS 的独立因素 [风险比 2.157,95% 置信区间 (CI)1.098–4.239,p = 0.026]。尽管 CTC 特异性 miR‐106b、E‐钙粘蛋白和波形蛋白各自显示了预后潜力,但基于三个标志物组合的 OS 预后表现在队列 1 [曲线下面积 (AUC)0.752,95% CI 0.658‐0.847,n = 128]中显著增强,并在队列 2(AUC 0.726,95% CI 0.595‐0.856, n = 91)中得到进一步验证。此外,包含 miR‐106b 的组合模型与治疗反应相关。
结论。包含 miR‐106b 的 CTC 表型组合显示,MBC 患者总生存率的预后准确性增强。
实践意义:为了提高转移性乳腺癌(MBC)患者循环肿瘤细胞(CTC)表型的预后准确性,本研究在比较 CTC、原发性肿瘤和健康供体之间的微小RNA (miRNA)谱的基础上,筛选并确定了一种 CTC 特异性miRNA,即 miR‐106b,作为上调分子。通过将 miR‐106b 添加进组合预测模型,在训练队列和验证队列中,MBC 患者 CTC 表型的预后准确性都有了很大提高。本研究提供了临床证据,证明 CTC 特异性miRNA 对 MBC 患者的预后潜力。这些结果表明,开发 CTC 特异性miRNA 作为新的生物标志物将有助于进一步优化个体化治疗。
Introduction
Metastasis is associated with the presence of peripheral blood circulating tumor cells (CTCs), which are suggested to be potential seeds for hematogenous cancer metastasis [1], [2]. Several studies have demonstrated that the presence of CTCs before the initiation and after the completion of adjuvant chemotherapy is associated with poor clinical outcome [3], [4]. In metastatic breast cancer (MBC), the assessment of CTCs before and shortly after the initiation of chemotherapy may predict progression‐free survival (PFS) and overall survival (OS) [5], [6]. Although the presence of chromosomal alterations confirmed the malignant nature of CTCs [7], [8], only some are capable of promoting metastasis [9]. Regarding their individual gene expression, characterizing the molecular phenotypes of the CTCs, rather than their blood concentration and/or number, is essential to further understand their metastatic potential as well as identify novel markers related to patients’ prognosis.
Epithelial‐mesenchymal transition (EMT) is considered to be the crucial event in the metastatic process of cancer, involving aberrant expression of the EMT‐related molecules and the acquisition of a migratory mesenchymal phenotype [10]. The morphological and phenotypical changes of EMT undergone by cancer cells also exist in CTCs of breast cancer [11]. CTCs that undergo EMT lose cell‐cell contacts and polarity, downregulate epithelial‐associated genes, and acquire mesenchymal gene expression [12], [13], [14]. E‐cadherin and vimentin act as the core protein of the epithelial adherence junction [15], [16]. Several studies have demonstrated that vimentin is expressed in CTCs of patients with breast cancer [17], [18]. CTCs displaying upregulated vimentin were associated with clinical response to therapy and disease progression [19], [20], suggesting that the molecular phenotype of CTCs including EMT was associated with the clinical outcome of patients with breast cancer.
It has been reported that CTCs isolated from a cohort of patients with MBC by the CellSearch Profile Kit display a unique miRNA expression profile [21], indicating that an extensive miRNA characterization of CTCs will hold great promise and improve the currently available prognostic models on the basis of primary tissue. However, little is known regarding the correlation between the noncoding molecules in CTCs and EMT‐related characteristics as well as their potential clinical relevance.
In the current study, in order to identify new miRNAs for CTC phenotype classifications and enhance the prognostic value of the CTC phenotype for patients with MBC, we screened the CTC‐specific miRNAs based on the comparison of miRNA deep sequencing between CTCs and primary tumors and verified miR‐106b as a key molecule with the highest upregulation in CTCs. Furthermore, we quantified the expression level of miR‐106b in CTCs of patients with MBC and investigated the prognostic role of the CTC phenotype incorporating CTC‐specific miR‐106b for patients with MBC in two independent cohorts.
Subjects, Materials, and Methods
Patients
This study enrolled patients initially diagnosed with MBC between March 2012 and December 2015 in the Sun Yat‐sen Memorial Hospital (SYSMH; Guangzhou, China) and the Sun Yat‐sen University Cancer Center (SYSUCC; Guangzhou, China). All patients were required to have clinical and radiologic evidence of MBC with either measurable or evaluable disease. Metastatic diseases were measured or evaluated using clinical and radiological methods in accordance with the RECIST version 1.1 criteria. All patients had Eastern Cooperative Oncology Group performance status 0–1.
Prior adjuvant chemotherapy and/or hormone treatment were allowed. The estrogen receptor (ER) and the progesterone receptor (PR) status of the primary tumors were determined by immunohistochemistry. Human epidermal growth receptor 2 (HER2) expression was evaluated using the HercepTest (Dako, Denmark) and assessed according to the DAKO‐score; samples with score 2+ were further analyzed by fluorescent in situ hybridization.
Before treatment, all patients had a complete clinical and laboratory evaluation, chest and abdominal computed tomography scans, and whole‐body bone scan. Reassessment of the disease status was also performed every 8 weeks. Response to treatment was assessed using the RECIST 1.1 criteria.
Blood Sample Collection
Prior to the administration of systemic therapy, 7.5 mL of blood was drawn from patients in CellSave tubes (Veridex LLC) for CTC enumeration by CellSearch. CTC enumeration and characterization were confirmed by independent reviewers.
Fifty milliliters of fresh blood was collected for sorting and enriching CTCs using magnetic activated cell sorting (MACS; Miltenyi Biotec, Bergisch Gladbach, Germany) if CTC count ≥5/7.5 mL after primary enumeration. To avoid contamination with epithelial cells from the skin, all blood samples were obtained at the middle of vein puncture after the first 1 mL of blood was discarded.
Enumeration of CTCs Using the CellSearch System
Enumeration of CTCs was performed at the central laboratory of our institution using the U.S. Food and Drug Administration‐approved CellSearch system by using CELLSEARCH Circulating Tumor Cell Kit. The process has been described in the manufacturer's instructions. Unfavorable CTC enumeration was defined as ≥5 CTCs in 7.5 ml of peripheral blood.
CTCs Sorting and Enrichment
CTCs were isolated from 50 mL EDTA blood by MACS. Briefly, samples were layered over Ficoll‐Paque (1.077, density) and centrifuged at 400g for 30 minutes at room temperature; peripheral blood mononuclear cells (PBMCs) were present at the interphase. PBMCs were resuspended at 5 × 107 cells in 300 mL solution containing 100 mL FcR Blocking Reagent (130‐059‐901, Miltenyi Biotec), 100 mL CD45 Microbeads (130‐045‐801, Miltenyi Biotec), and 100 mL CD15 Microbeads (130‐091‐058, Miltenyi Biotec). After depletion of CD45+ and CD15+ cells by magnetic separation with autoMACS Pro Separator, 100 mL CD326 (also known as Epithelial Cell Adhesion Molecule, EpCAM) Microbeads (130‐095‐500, Miltenyi Biotec) per 5 × 107 cells was added for 30‐minute incubation at 4°C. The magnetically charged CD326+ and CD326‐ cell fractions were eluted as the EpCAM+ and EpCAM− CTCs. The purity of epithelial cells was determined by immunofluorescent staining with an anticytokeratin antibody (10 μg/mL, ab41825; Abcam, UK), which was more than 95%. For a cell to be identified as a CTC, it had to meet two criteria: (a) positive staining for a tumor‐specific marker by immunocytochemistry (cytokeratin) and (b) positive scoring upon review by the cytopathologist.
Screening and Quantification of CTC‐Specific miRNAs
Total RNA was extracted from primary tumors and CTCs enriched by EpCAM‐based CellSearch from 50 mL of blood of six patients with MBC. MiRNA Deep Sequencing on Illumina HiSeq 2500 sequencing platform with 10 M reads (Illumina, San Diego, CA; Guangzhou RiboBio Company, Guangzhou, China) was used to screen CTC‐specific miRNA candidates from these samples. Real‐time polymerase chain reaction (PCR) was used to quantify CTC‐specific miRNAs expression levels. Complementary DNA (cDNA) was generated using the miScript Reverse Transcription (RT) Kit (Qiagen GmbH, Hilden, Germany). Briefly, real‐time PCR was performed using the miScript SYBR Green PCR Kit (Qiagen) on an Mx3005P QPCR System (Stratagene, La Jolla, CA). The specificity of this RT‐PCR technique was confirmed by dissociation curve analysis using the Mx3005P QPCR System (Stratagene) according to the manufacturer's instruction. Bulge‐Loop miRNA primers were offered by RiboBio Company, Guangzhou, China. Transcripts of U6 small RNA were quantified for normalization of the levels of miRNAs [22], [23].
ΔCt values were used to normalize and calculate the concentration of miRNAs. The experiment was repeated three times and the data were analyzed blind. The 2−ΔΔCt value was the difference in ΔCt between patients and controls, and the normalized miRNAs expression levels were calculated with the formula 2−ΔΔCt.
Quantification of EMT‐Related Molecules in CTCs
RNA was extracted from CTCs using the Recover All Total Nucleic Acid Isolation Kit (Ambion, Life Technologies, Carlsbad, CA) as described. Total RNA from each sample was quantified by NanoDrop ND‐1000 (ThermoFisher). cDNA was synthesized from total RNA using the ImProm‐II Reverse Transcriptase system (Promega) according to the manufacturer's protocol. The mRNA levels were measured by quantitative real‐time PCR, which was performed in an ABI 7500 Real‐time PCR system using SYBR Green PCR Master Mix (Applied Biosystems). Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) were used as standards for the absolute quantification of each cDNA. The amount of target gene expression could be calculated from the standard curve, which plotted the cycle threshold (Ct) value and the corresponding value of the amount of input DNA. The quantitative PCR primers used are listed in supplemental online Table 1.
Statistical Analysis
Statistical analyses were performed using SPSS version 17.0 (SPSS, Chicago, IL). Student's t test and one‐way analysis of variance analyses were performed to analyze the differences between the groups. A chi‐square test was applied to analyze the relationship between miR‐106b expression and EMT‐related molecules of CTC. OS was defined as the time elapsed between inclusion and death from any cause. Survival curves were plotted according to the Kaplan‐Meier method. Statistical significance between survival curves was assessed using the log‐rank test. Multivariate analysis was done by the Cox proportional hazards model with prognostic factors. The Cox regression model was performed for the combined three prognostic factors. The receiver operating characteristic (ROC) curves were employed to test the sensitivity and specificity of variables in predicting OS. For the other analyses, a p value of <.05 was considered to be statistically significant.
Results
Patient Characteristics
From March 2012 to December 2015, a total of 128 patients with MBC in SYSMH (Cohort 1) and 91 patients with MBC in SYSUCC (Cohort 2) were enrolled in this study. Cohort 1 was used as a training set in our study, whereas Cohort 2 was used as a validation set. A flow chart outlining the selection of patients from SYSMH (Cohort 1) is shown in supplemental online Figure 1. The patients from Cohort 2 were selected and investigated in a similar manner as those in Cohort 1. All enrolled patients had five or more CTCs in 7.5 mL of blood enumerated at baseline section by CellSearch. Patient characteristics of Cohort 1 and Cohort 2 at baseline are summarized in Table 1 and supplemental online Table 2. The median age (range) at initial diagnosis of these patients with breast cancer was 51 (23–69) years, and the median age at study entry was 56 (29–72) years. Notably, the majority of patients had ER‐positive (ER+; 96/128, 75%), PR‐positive (87/128, 67.9%), and HER2‐negative (101/128, 78.9%) primary tumors. In most patients, the tumor had disseminated into more than one organ (87/128, 68%), and approximately half of the patients had both bone and visceral metastases (70/128, 54.7%). All patients received front‐line chemotherapy. Among them, 16 (12.5%) patients received fluorouracil, epirubicin, and cyclophosphamide/epirubicin and cyclophosphamide regimen, 42 (32.8%) patients received epirubicin and taxane chemotherapy regimen, and 53 (41.4 %) patients received taxane chemotherapy regimen. Moreover, there were 17 (13.3 %) patients who received chemotherapy regimens without epirubicin and taxane. In addition, 24 of 27 (88.9 %) patients with HER2‐positive tumors received anti‐HER2 therapy (trastuzumab or lapatinib or both). All ER+ patients received endocrine therapy. Follow‐up data were available for 128 patients with a median follow‐up of 13.1 months (range, 2.5–29.7 months) for OS.
Table 1. Characteristics of patients in Cohort 1 and Cohort 2.
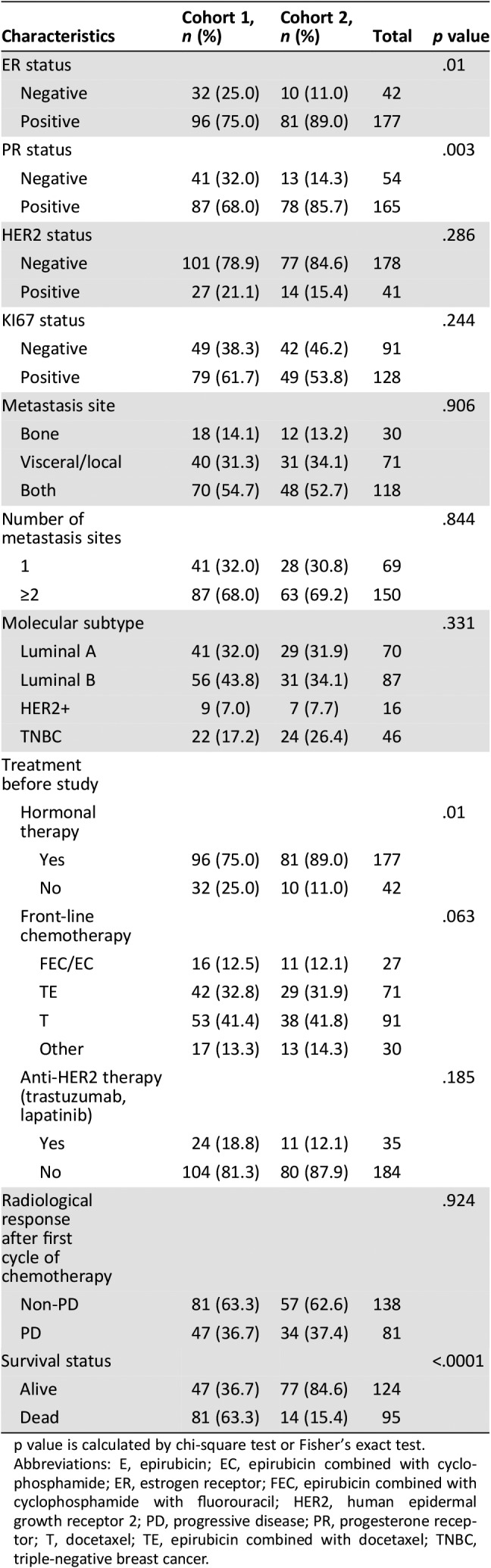
p value is calculated by chi‐square test or Fisher's exact test.
Abbreviations: E, epirubicin; EC, epirubicin combined with cyclophosphamide; ER, estrogen receptor; FEC, epirubicin combined with cyclophosphamide with fluorouracil; HER2, human epidermal growth receptor 2; PD, progressive disease; PR, progesterone receptor; T, docetaxel; TE, epirubicin combined with docetaxel; TNBC, triple‐negative breast cancer.
Selection of Potentially CTC‐Specific miRNA Candidates
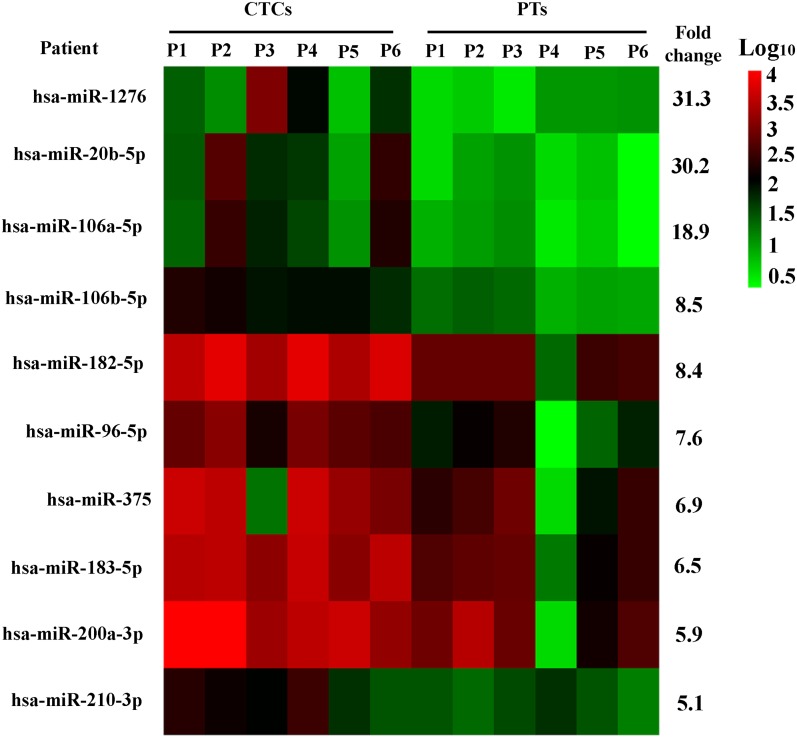
To found out CTC‐specific miRNAs, we first used an MiRNA Deep Sequencing to screen the miRNA candidates in primary tumors and CTCs enriched by EpCAM‐based CellSearch from 50 mL of blood of six patients with MBC (supplemental online Table 3) among 2,588 miRNAs in the MiRNA Sequencing (Fig. 1) based on the following standards: fold changes were more than 5 folds compared with the primary tumor in all six patients with MBC, basal expression level of normalized intensity were more than 100, miRNA candidates were reported to play functional roles in promoting breast cancer metastasis. Finally, 10 miRNAs were picked out as upregulated candidates (supplemental online Table 4), and these 10 upregulated miRNAs could be sensitivity nonhomogeneously amplified with a minimum number of miRNA in sensitive multiplex stem‐loop cDNA approach with the TaqMan‐based linear preamplification method. We further compared these 10 miRNA transcript levels in cells enriched by MACS (CMs) of blood from healthy blood donors (HBDs; n = 8) and CTC samples from patients with MBC (n = 32) by RT‐PCR (supplemental online Table 3). We observed that only miR‐106b was significantly upregulated in CTCs ≥5 samples (n = 16) compared with CTCs = 0 samples (n = 16) and HBD‐CSs (supplemental online Table 5). This suggests that miR‐106b might be a potentially CTC‐specific miRNA, and we selected it for our further investigation.
Figure 1.
MiRNA Deep Sequencing reveals differentially expressed microRNAs between primary tumors and CTCs from six patients with metastatic breast cancer.
Abbreviations: CTCs, circulating tumor cells; P, patient; PTs, primary tumors.
Association of miR‐106b and EMT‐Related Molecules in CTC
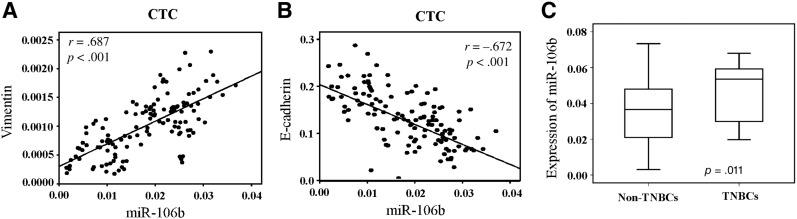
It has been reported that miR‐106b regulates the EMT of cancer cells and promotes tumor progression [24]. It is also noted that, upon EMT inducement, cancer cells exhibit morphological changes and a couple of EMT‐related molecules including E‐cadherin, vimentin, ck19, Snail, and N‐cadherin. Therefore, we further quantified the expression level of miR‐106b and the above EMT‐related molecules in CTCs using quantitative RT‐PCR, and analyzed their correlations. As a result, we found that the expression of CTC‐specific miR‐106b was positively correlated with vimentin (r = .687, p < .001; Fig. 2A) and negatively correlated with the expression level of E‐cadherin (r = −.672, p < .001; Fig. 2B) but had no significant correlations with other molecules (supplemental online Fig. 2). In subtype analysis, we found that the expression level of CTC‐specific miR‐106b was significantly higher in patients with triple‐negative breast cancer (TNBC) compared with non‐TNBC subtypes (p = .011; Fig. 2C). These data suggest that miR‐106b is a specific biomarker for EMT‐related phenotype of CTC.
Figure 2.
MiR‐106b is a specific biomarker for CTC phenotypes. Correlation of the expression of miR‐106b with vimentin (A, n = 128) and E‐cadherin (B, n = 128). (C): Quantification of miR‐106b by real‐time quantitative polymerase chain reaction in TNBCs (n = 16) and non‐TNBCs (n = 112).
Abbreviations: CTC, circulating tumor cell; TNBCs, triple‐negative breast cancers.
Association of CTC‐Specific miR‐106b, E‐cadherin, and Vimentin with Clinical Outcome
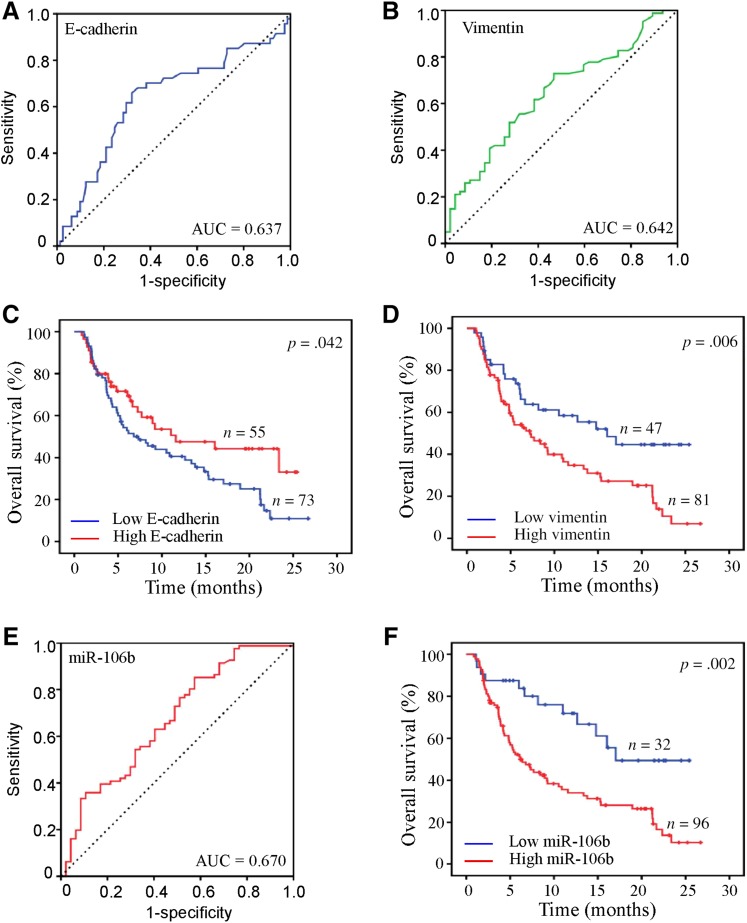
It is well known that the number and phenotypes of CTC correlates with prognosis of patients with MBC [17], [25]. Therefore, we further investigated the clinical relevance of the above CTC‐specific molecules with the prognosis of patients with MBC. First, we individually assessed the prognostic performance of CTC‐specific miR‐106b, E‐cadherin, and vimentin by plotting the ROC curve in 128 patients. Consistent with other studies [26], [27], in our study, CTC‐specific E‐cadherin and vimentin showed a prognostic value for patients with MBC, respectively (E‐cadherin, AUC = 0.637, Fig. 3A; vimentin, AUC = 0.642, Fig. 3B). The lower‐expression group of E‐cadherin (hazard ratio [HR] 1.624, 95% confidence interval [CI] 1.017–2.594, p = .042; Fig. 3C) and higher‐expression group of vimentin (HR 1.993, 95% CI 1.218–3.259, p = .006; Fig. 3D) showed poorer OS in Kaplan‐Meier curve analysis. Interestingly, higher prognostic performance of CTC‐specific miR‐106b for patients with MBC was observed in ROC analysis (AUC = 0.670, 95% CI 0.572–0.769; Fig. 3E). In order to further analyze the association between CTC‐specific miR‐106b and prognosis, an optimal cutoff value (0.0105) was determined by ROC analysis and Youden index calculated by sensitivity and specificity, which was widely used for determination of optimal cutoff value in diagnostic test and prognosis prediction by other studies [28]. Based on this cutoff value, 128 patients with MBC of Cohort 1 and 91 of Cohort 2 were divided into miR‐106 high‐expression and low‐expression subgroups. As shown by Kaplan‐Meier curve analysis, the miR‐106 high‐expression group was correlated with a poorer OS (HR 2.707, 95% CI 1.463–5.008, p = .002; Fig. 3F), compared with the low‐expression group.
Figure 3.
Association of circulating tumor cell (CTC)‐specific molecules with clinical outcome. Receiver operating characteristic curves evaluated the prognostic accuracy for E‐cadherin (A), vimentin (B), and miR‐106b (E) in CTCs. Kaplan‐Meier survival curves for E‐cadherin (C), vimentin (D), and miR‐106b (F) in CTCs.
Abbreviation: AUC, area under the curve.
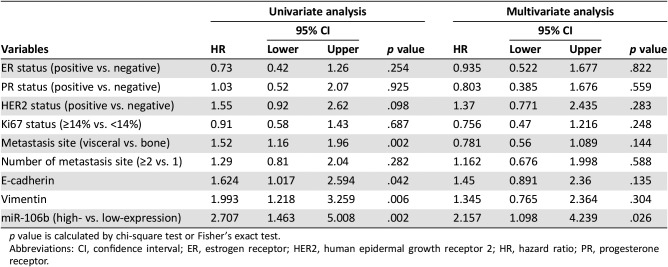
In addition, univariate analysis found that patients with visceral metastasis (HR 1.52, 95% CI 1.16–1.96, p = .002), high expression of miR‐106 and vimentin, and low expression of E‐cadherin were more likely to have poor OS. However, in multivariate analysis, only CTC‐specific miR‐106b was an independent factor for predicting OS (HR 2.157, 95% CI 1.098–4.239, p = .026; Table 2).
Table 2. Univariate and multivariate analysis for overall survival.
p value is calculated by chi‐square test or Fisher's exact test.
Abbreviations: CI, confidence interval; ER, estrogen receptor; HER2, human epidermal growth receptor 2; HR, hazard ratio; PR, progesterone receptor.
Assessment and Validation of the Prognostic Accuracy of the Three‐Molecule Combination Including CTC‐Specific miR‐106b, E‐cadherin, and Vimentin
Because of the limited prognostic accuracy of CTC‐specific E‐cadherin and vimentin and relative higher prognostic performance of CTC‐specific miR‐106b for patients with MBC, we further incorporated miR‐106b into a Cox regression model to compare the prognostic accuracy between the multimolecule assemblies and single biomarker. As a result, we found that the new model combining the expression of three molecules was significantly more accurate than any other single biomarker evaluated in our study for predicting overall survival (AUC 0.752, 95% CI 0.658–0.847; Fig. 4A).
Figure 4.
Comparison of the prognostic accuracy of single and combined factors. (A): Training set. (B): Validation set.
Abbreviations: AUC, area under the curve; CI, confidence interval.
This result was further validated in Cohort 2. In Cohort 2, the clinicopathological characterization of 91 selected patients with MBC was similar to that of patients from SYSMH (Cohort 1), including 81 (89%) ER‐positive patients and 14 (15.4%) HER2‐positive patients (supplemental online Table 2). Notably, most patients were Luminal subtype (60 of 91, 65.9%) and triple negative (24 of 91, 26.4%), and only 7 patients were HER2+ subtype. In addition, we found that the distribution of miR‐106b expression in Cohort 2 was similar to Cohort 1 (supplemental online Fig. 3). Therefore, we used the same cutoff value to divide Cohort 2 into miR‐106b high‐ and low‐expression subgroups. We verified that the new model combining the expression of three molecules also showed a significantly prognostic value for predicting overall survival in the validation cohort (AUC 0.726, 95% CI 0.595–0.856; Fig. 4B).
In subtype analysis, we made ROC analysis to test the prognostic accuracy in all 219 patients (Cohort 1 and Cohort 2) and found that the prognostic accuracy of the three‐molecule combination including CTC‐specific miR‐106b, E‐cadherin, and vimentin was still significant in luminal subtypes (AUC = 0.693, 95% CI 0.609–0.776, n = 157; supplemental online Fig. 4A) and the TNBC subtype (AUC = 0.710, 95% CI 0.522–0.898, n = 48; supplemental online Fig. 4B).
Association of the Three‐Molecule Combination and Clinical Response
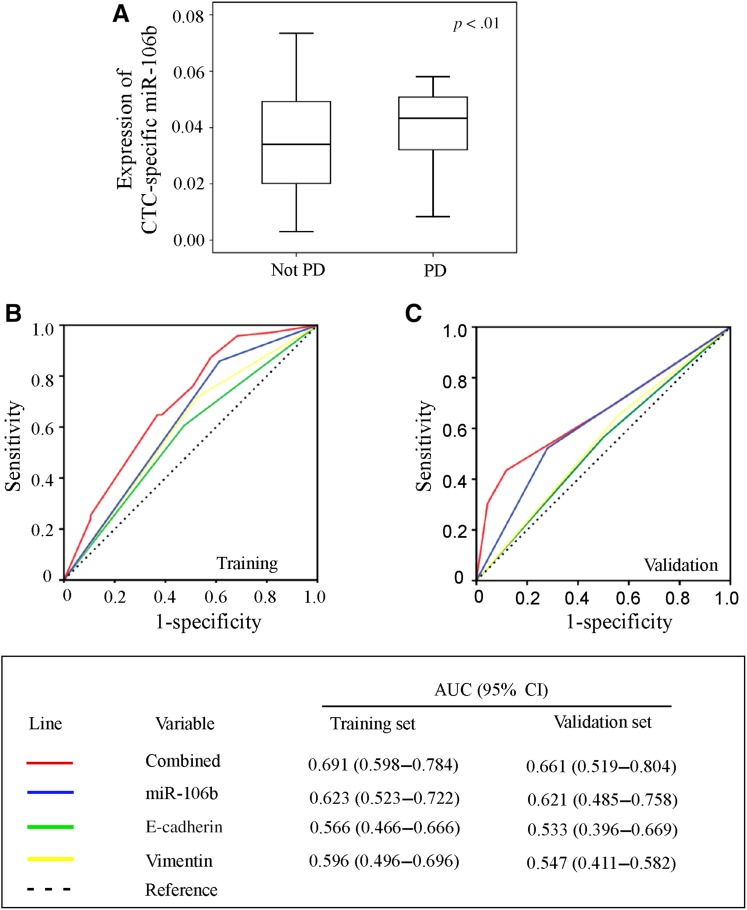
The correlation between expression of CTC‐specific miR‐106b and clinical response was further analyzed. Patients with progression at the radiological examination following two cycles of first‐line chemotherapy showed higher expression of CTC‐specific miR‐106b (Fig. 5A, p < .01). Further analysis showed that the combination of CTC‐specific miR‐106b containing E‐cadherin and vimentin could significantly predict the clinical response to therapy in patients with MBC (AUC 0.691, 95% CI 0.598–0.784; Fig. 5B). This result was further validated in Cohort 2 with 91 patients with MBC (AUC 0.661, 95% CI 0.519–0.804; Fig. 5C).
Figure 5.
(A): Correlations between expression of CTC‐specific miR‐106b and clinical response. Comparison of the prognostic accuracy of single and combined factors in the training set (B) and validation set (C).
Abbreviations: AUC, area under the curve; CI, confidence interval; CTC, circulating tumor cell; PD, progressive disease.
Discussion
In this study, we found that miR‐106b was significantly upregulated in CTCs compared with primary tumor and CMs of blood from HBDs. The expression level of CTC‐specific miR‐106b was correlated with EMT‐related phenotype of CTC. In addition, high expression of CTC‐specific miR‐106b was correlated with a poor OS and acted as an independent factor for predicting OS. More importantly, by incorporating miRNA expressions into a combined prediction model, the prognostic accuracy of CTC phenotype in patients with MBC was significantly enhanced.
Quantification of CTCs in breast [29], colorectal [30], and pancreatic cancer [31] has been shown to correlate with survival. However, less than 0.01% of CTCs seem to survive in the target organ for colonization and subsequent growth. An intriguing area of active research is whether molecular characterization of CTC before clinical manifestation could better predict metastatic risk and facilitate individualized therapeutic strategies to inhibit target organ colonization and metastatic growth. Of note, the activation of an EMT is favorable for the CTC population [32]. It has been reported that the presence of mesenchymal markers such as vimentin on CTCs more accurately predicted worse prognosis than the expression of cytokeratin [20], [33].
In our study, we found that CTC‐specific miR106b was positively related to the expression level of vimentin in CTCs. It has been reported that overexpression of miR‐106b was observed in a variety of human tumors, which plays an oncogenic role in tumor progression and prognosis, including colorectal cancer [34], [35], gastric cancer [36], [37], hepatocellular carcinoma [38], [39], glioma tumors [40], renal cell carcinoma [41], [42], and head and neck squamous cell carcinomas [43]. MiR‐106b plays a functional role in promoting proliferation, migration, and invasion of cancer cells both in vitro and in vivo [44], [45], [46]. Clinical data show that high expression level of miR‐106b is associated with metastasis and poor prognosis [34], [47]. Consistent with these findings, we found that patients with MBC with high CTC‐specific miR‐106b expression had shorter progression‐free survival, suggesting that this phenotypic attribute becomes salient when considering each of the steps in the cascade of events required for metastatic success.
Our clinical data showed that in both the training and validation set, the combination of CTC‐specific miR‐106b, E‐cadherin, and vimentin was significantly more accurate than any single molecule assessed in our study for predicting overall survival and clinical response to therapy. Several lines of explanation may be responsible for this enhanced prognostic value of this molecule combination. First, molecular phenotypes of CTCs were associated with various behavior of cancer cells including anoikis resistance metastasis and therapeutic resistance. It has been reported that miR‐106b promotes anoikis resistance [24] and EMT processes in breast cancer cells by targeting RB [48] and Snail gene [49] . Acquisition of a more malignant phenotype for CTC might easily explain the resistance to elimination strategies such as chemotherapy, local antigrowth signaling, and immune attack by the host. Second, according to our results, CTC‐specific miR‐106b was expressed at a higher level in mesenchymal‐like CTCs that highly expressed vimentin, suggesting that there is a coordination between miR‐106b and EMT‐related phenotypes of CTCs. Third, mechanically, our previous study demonstrated that miR‐106b determines the effect of transforming growth factor β, a key EMT inducer, on the tumor behavior of breast cancer cells by targeting RB [48]. Several studies have also demonstrated that a variety of genes were identified as the target of miR‐106b including caspase‐7 [47], PTEN [50], and Smad7 [17], [20], [51], which contribute to miR‐106b‐mediated EMT of cancer cells. In this scenario, CTC‐specific miR‐106b plays a critical role in regulating the EMT of CTCs, indicating that CTCs with higher expression of vimentin and miR‐106b show a higher motility in the bloodstream and more easily interact with the intravascular microenvironment, extravagate through the microvasculature, and interact with the metastatic microenvironment of target organs. Consistent with this notion, the clinical relevance was further demonstrated by our clinical data that CTC‐specific miR‐106b was an independent factor for predicting overall survival and therapy response of patients with MBC. These are responsible for incorporating miR‐106b into the molecular phenotype of CTC and could further enhance the prognostic accuracy of CTC.
Increasing evidence shows that the phenotype of CTCs was related to disease prognosis and holds great promise in guiding treatment decision making in patients with breast cancer. Therefore, identifying new molecules to refine the molecular phenotype of CTC will help the clinicians to better formulate appropriate therapy strategies for individual patients. The EMT‐related biomarkers including vimentin and E‐cadherin have been reported to detect the phenotype of CTC [17], [20], [52], and CTCs displaying upregulated vimentin were associated with clinical response to therapy and disease progression [17], [20]. With the development of sequencing technology and in‐depth understanding of the mechanism for cancer progression, it is necessary to take into account more gene information and then characterize the detailed molecular phenotype of CTC to improve the prognostic performance of established CTC‐related clinical models. In our study, we screened and validated that miR‐106b was an upregulated biomarker in patients with MBC. The expression of CTC‐specific miR‐106b was correlated with vimentin and E‐cadherin in CTC and acted as an independent factor for predicting OS in patients with MBC. The prognostic performance for OS based on the combination of three markers (miR‐106b, E‐cadherin, and vimentin) was significantly enhanced and can be further validated in an external validation cohort. Our results suggest that incorporating miRNA into molecular phenotypes of circulating tumor cells enhances the prognostic accuracy. In addition, this new model is feasible, efficient, and cost‐effective for clinical application.
In our study, there was still some weakness that deserves to be acknowledged. First, the molecular mechanisms by which miR‐106b and EMT‐related molecules in CTCs regulate tumor progression were not presented in this study. However, several reasons are responsible for this limitation. As far as we know, the isolation and enrichment as well as ex vivo culture of viable CTCs are still technically challenging, and the frequency of CTC in the peripheral blood of patients with solid tumors varies among individual patients with different prior systemic therapies. In recent years, although a few papers reported that the genome alternation of CTC lines and suspended cancer cells may be related to the drug sensitivity in both in vitro and xenografts models in breast cancer [53], the evidence is indirect and not strong enough because of the above limitations. Second, the downstream targeted molecules of miR‐106b related to EMT should be further tested in our future studies.
Conclusion
Our work provides clinical evidence supporting the prognostic potential of CTC‐specific miRNA for patients with MBC. These results indicate that developing CTC‐specific miRNAs as new biomarkers will help to further optimize personalized therapy.
See http://www.TheOncologist.com for supplemental material available online.
Acknowledgments
This work is supported by the National Key R&D Program of China (2017YFC1309103 and 2017YFC1309104); the Natural Science Foundation of China (81672594,81772836 and 81872139); National Science Foundation of Guangdong Province (2017A030313554); Sun Yat‐sen Memorial Hospital Cultivation Project for Clinical Research (SYS‐C‐201805); and Clinical Innovation Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (2018GZR0201004). This work was permitted by the Sun Yat‐sen University Ethic Committee of Sun Yat‐sen Memorial Hospital (201108). All patients signed an informed consent form before being enrolled in this study.
Contributed equally.
Author Contributions
Conception/design: Weige Tan, Chang Gong
Provision of study materials or patients: Weige Tan, Xinhua Xie.
Collection and/or assembly of data: Weige Tan, Gehao Liang, Luyuan Tan, Zihao Liu, Yun Ling, Wenjing Zhong, Zhenluan Tian, Wanyi Lin
Data analysis and interpretation: Weige Tan, Gehao Liang, Xinhua Xie
Manuscript writing: Weige Tan, Gehao Liang, Xinhua Xie, Wenguo Jiang, Andrew J. Sanders, Chang Gong
Financial approval of manuscript: Weige Tan, Gehao Liang, Xinhua Xie, Wenguo Jiang, Luyuan Tan, Andrew J. Sanders, Zihao Liu, Yun Ling, Wenjing Zhong, Zhenluan Tian, Wanyi Lin, Chang Gong
Disclosures
The authors indicated no financial relationships.
References
- 1.Maheswaran S, Haber DA. Circulating tumor cells: a window into cancer biology and metastasis. Curr Opin Genet Dev 2010;20:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim MY, Oskarsson T, Acharyya S et al. Tumor self‐seeding by circulating cancer cells. Cell 2009;139:1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xenidis N, Perraki M, Kafousi M et al. Predictive and prognostic value of peripheral blood cytokeratin‐19 mRNA‐positive cells detected by real‐time polymerase chain reaction in node‐negative breast cancer patients. J Clin Oncol 2006;24:3756–3762. [DOI] [PubMed] [Google Scholar]
- 4.Stathopoulou A, Vlachonikolis I, Mavroudis D et al. Molecular detection of cytokeratin‐19‐positive cells in the peripheral blood of patients with operable breast cancer: Evaluation of their prognostic significance. J Clin Oncol 2002;20:3404–3412. [DOI] [PubMed] [Google Scholar]
- 5.Cristofanilli M, Hayes DF, Budd GT et al. Circulating tumor cells: A novel prognostic factor for newly diagnosed metastatic breast cancer. J Clin Oncol 2005;23:1420–1430. [DOI] [PubMed] [Google Scholar]
- 6.Cristofanilli M, Budd GT, Ellis MJ et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781–791. [DOI] [PubMed] [Google Scholar]
- 7.Fehm T, Sagalowsky A, Clifford E et al. Cytogenetic evidence that circulating epithelial cells in patients with carcinoma are malignant. Clin Cancer Res 2002;8:2073–2084. [PubMed] [Google Scholar]
- 8.Bozionellou V, Mavroudis D, Perraki M et al. Trastuzumab administration can effectively target chemotherapy‐resistant cytokeratin‐19 messenger RNA‐positive tumor cells in the peripheral blood and bone marrow of patients with breast cancer. Clin Cancer Res 2004;10:8185–8194. [DOI] [PubMed] [Google Scholar]
- 9.Klein CA, Blankenstein TJ, Schmidt‐Kittler O et al. Genetic heterogeneity of single disseminated tumour cells in minimal residual cancer. Lancet 2002;360:683–689. [DOI] [PubMed] [Google Scholar]
- 10.Thiery JP. Epithelial‐mesenchymal transitions in tumour progression. Nat Rev Cancer 2002;2:442–454. [DOI] [PubMed] [Google Scholar]
- 11.Yu M, Bardia A, Wittner BS et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013;339:580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakajima S, Doi R, Toyoda E et al. N‐cadherin expression and epithelial‐mesenchymal transition in pancreatic carcinoma. Clin Cancer Res 2004;10:4125–4133. [DOI] [PubMed] [Google Scholar]
- 13.Cano A, Perez‐Moreno MA, Rodrigo I et al. The transcription factor snail controls epithelial‐mesenchymal transitions by repressing E‐cadherin expression. Nat Cell Biol 2000;2:76–83. [DOI] [PubMed] [Google Scholar]
- 14.Behrens J. Cadherins and catenins: Role in signal transduction and tumor progression. Cancer Metastasis Rev 1999;18:15–30. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Yan Q, Long X et al. Vimentin affects the mobility and invasiveness of prostate cancer cells. Cell Biochem Funct 2008;26:571–577. [DOI] [PubMed] [Google Scholar]
- 16.McInroy L, Maatta A. Down‐regulation of vimentin expression inhibits carcinoma cell migration and adhesion. Biochem Biophys Res Commun 2007;360:109–114. [DOI] [PubMed] [Google Scholar]
- 17.Satelli A, Brownlee Z, Mitra A et al. Circulating tumor cell enumeration with a combination of epithelial cell adhesion molecule‐ and cell‐surface vimentin‐based methods for monitoring breast cancer therapeutic response. Clin Chem 2015;61:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamilton G, Rath B. Mesenchymal‐epithelial transition and circulating tumor cells in small cell lung cancer. Adv Exp Med Biol 2017;994:229–245. [DOI] [PubMed] [Google Scholar]
- 19.Satelli A, Batth IS, Brownlee Z et al. Potential role of nuclear PD‐L1 expression in cell‐surface vimentin positive circulating tumor cells as a prognostic marker in cancer patients. Sci Rep 2016;6:28910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satelli A, Batth I, Brownlee Z et al. EMT circulating tumor cells detected by cell‐surface vimentin are associated with prostate cancer progression. Oncotarget 2017;8:49329–49337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sieuwerts AM, Mostert B, Bolt‐de Vries J et al. mRNA and microRNA expression profiles in circulating tumor cells and primary tumors of metastatic breast cancer patients. Clin Cancer Res 2011;17:3600–3618. [DOI] [PubMed] [Google Scholar]
- 22.Stathopoulou A, Gizi A, Perraki M et al. Real‐time quantification of CK‐19 mRNA‐positive cells in peripheral blood of breast cancer patients using the lightcycler system. Clin Cancer Res 2003;9:5145–5151. [PubMed] [Google Scholar]
- 23.Chen ZH, Zhang GL, Li HR et al. A panel of five circulating microRNAs as potential biomarkers for prostate cancer. Prostate 2012;72:1443–1452. [DOI] [PubMed] [Google Scholar]
- 24.Piao J, You K, Guo Y et al. Substrate stiffness affects epithelial‐mesenchymal transition of cervical cancer cells through miR‐106b and its target protein DAB2. Int J Oncol 2017;50:2033–2042. [DOI] [PubMed] [Google Scholar]
- 25.Schramm A, Friedl TW, Schochter F et al. Therapeutic intervention based on circulating tumor cell phenotype in metastatic breast cancer: Concept of the DETECT study program. Arch Gynecol Obstet 2016;293:271–281. [DOI] [PubMed] [Google Scholar]
- 26.Yamashita N, Tokunaga E, Iimori M et al. Epithelial paradox: Clinical significance of coexpression of E‐cadherin and vimentin with regard to invasion and metastasis of breast cancer. Clin Breast Cancer 2018;18:e1003–e1009. [DOI] [PubMed] [Google Scholar]
- 27.Liu T, Zhang X, Shang M et al. Dysregulated expression of Slug, vimentin, and E‐cadherin correlates with poor clinical outcome in patients with basal‐like breast cancer. J Surg Oncol 2013;107:188–194. [DOI] [PubMed] [Google Scholar]
- 28.Perkins NJ, Schisterman EF. The Youden Index and the optimal cut‐point corrected for measurement error. Biom J 2005;47:428–441. [DOI] [PubMed] [Google Scholar]
- 29.Budd GT, Cristofanilli M, Ellis MJ et al. Circulating tumor cells versus imaging—Predicting overall survival in metastatic breast cancer. Clin Cancer Res 2006;12:6403–6409. [DOI] [PubMed] [Google Scholar]
- 30.Cohen SJ, Punt CJ, Iannotti N et al. Relationship of circulating tumor cells to tumor response, progression‐free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:3213–3221. [DOI] [PubMed] [Google Scholar]
- 31.de Bono JS, Scher HI, Montgomery RB et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration‐resistant prostate cancer. Clin Cancer Res 2008;14:6302–6309. [DOI] [PubMed] [Google Scholar]
- 32.Charpentier M, Martin S. Interplay of stem cell characteristics, EMT, and microtentacles in circulating breast tumor cells. Cancers (Basel) 2013;5:1545–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mego M, Gao H, Lee BN et al. Prognostic value of EMT‐circulating tumor cells in metastatic breast cancer patients undergoing high‐dose chemotherapy with autologous hematopoietic stem cell transplantation. J Cancer 2012;3:369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang GJ, Li JS, Zhou H et al. MicroRNA‐106b promotes colorectal cancer cell migration and invasion by directly targeting DLC1. J Exp Clin Cancer Res 2015. Jul 30;34:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishida N, Nagahara M, Sato T et al. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin Cancer Res 2012;18:3054–3070. [DOI] [PubMed] [Google Scholar]
- 36.LArki P, Ahadi A, Zare A et al. Up‐regulation of miR‐21, miR‐25, miR‐93, and miR‐106b in gastric cancer. Iran Biomed J 2018;22:367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng Q, Jin C, Chen W et al. Downregulation of serum miR‐17 and miR‐106b levels in gastric cancer and benign gastric diseases. Chin J Cancer Res 2014;26:711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen CS, Su ZR, Lee YP et al. miR‐106b promotes cancer progression in hepatitis B virus‐associated hepatocellular carcinoma. World J Gastroenterol 2016;22:5183–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi F, Huang M, Pan Y et al. A genetic variant in the promoter region of miR‐106b‐25 cluster predict clinical outcome of HBV‐related hepatocellular carcinoma in Chinese. PLoS One 2014;9:e85394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang A, Hao J, Wang K et al. Down‐regulation of miR‐106b suppresses the growth of human glioma cells. J Neurooncol 2013;112:179–189. [DOI] [PubMed] [Google Scholar]
- 41.Sun K, Jia Z, Duan R et al. Long non‐coding RNA XIST regulates miR‐106b‐5p/P21 axis to suppress tumor progression in renal cell carcinoma. Biochem Biophys Res Commun 2019;510:416–420. [DOI] [PubMed] [Google Scholar]
- 42.Xiang W, He J, Huang C et al. miR‐106b‐5p targets tumor suppressor gene SETD2 to inactive its function in clear cell renal cell carcinoma. Oncotarget 2015;6:4066–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao CY, Guo ZJ, Cao L et al. Expression of mir‐106b in esophageal squamous cell carcinoma [in Chinese]. Nan Fang Yi Ke Da Xue Xue Bao 2016;36:1667–1671. [PubMed] [Google Scholar]
- 44.Zhang GJ, Li JS, Zhou H et al. MicroRNA‐106b promotes colorectal cancer cell migration and invasion by directly targeting DLC1. J Exp Clin Cancer Res 2015;34:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang X, Zhao Q, Geng T et al. MiR‐106b regulates the apoptosis and tumorigenesis of hepatocellular carcinoma via targeting Zinc finger and BTB domain‐containing protein 7A (Zbtb7a). J Biochem Mol Toxicol 2018;32:e22169. [DOI] [PubMed] [Google Scholar]
- 46.Lu J, Wei JH, Feng ZH et al. miR‐106b‐5p promotes renal cell carcinoma aggressiveness and stem‐cell‐like phenotype by activating Wnt/beta‐catenin signalling. Oncotarget 2017;8:21461–21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hudson RS, Yi M, Esposito D et al. MicroRNA‐106b‐25 cluster expression is associated with early disease recurrence and targets caspase‐7 and focal adhesion in human prostate cancer. Oncogene 2013;32:4139–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang P, Chen A, He W et al. BMP‐2 induces EMT and breast cancer stemness through Rb and CD44. Cell Death Discov 2017;3:17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu Y, Liu Y, Li WH et al. P2Y2 receptor promotes the migration and invasion of breast cancer cells via EMT‐related genes Snail and E‐cadherin. Oncol Rep 2018;39:138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Chen D, Liang S et al. miR‐106b promotes cell invasion and metastasis via PTEN mediated EMT in ESCC. Oncol Lett 2018;15:4619–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dai F, Liu T, Zheng S et al. MiR‐106b promotes migration and invasion through enhancing EMT via downregulation of Smad 7 in Kazakh's esophageal squamous cell carcinoma. Tumour Biol 2016;37:14595–14604. [DOI] [PubMed] [Google Scholar]
- 52.Hong Y, Zhang Q. Phenotype of circulating tumor cell: Face‐off between epithelial and mesenchymal masks. Tumour Biol 2016;37:5663–5674. [DOI] [PubMed] [Google Scholar]
- 53.Khosravi F, Trainor PJ, Lambert C et al. Static micro‐array isolation, dynamic time series classification, capture and enumeration of spiked breast cancer cells in blood: The nanotube‐CTC chip. Nanotechnology 2016;27:44LT03. [DOI] [PMC free article] [PubMed] [Google Scholar]