Abstract
Lesson Learned.
Circulating tumor cells, microRNA markers, or other biomarkers merit examination as part of correlative scientific analyses in prospective clinical trials.
Background.
Platinum chemotherapy resistance occurs in approximately 25% of patients with ovarian carcinoma; however, no biomarkers of ovarian carcinoma chemoresistance have been validated. We performed a prospective trial designed to identify tumor‐based predictive biomarkers of platinum resistance.
Methods.
Tumor specimens were collected from 29 women with newly diagnosed histopathologically proven primary ovarian carcinoma. Of these, 23 women had specimens accessible for assessment and outcome data available regarding chemosensitive versus chemoresistance status via review of the medical record. Tumor slices were stained with antibodies against two microRNAs (miRNAs 29b and 199a) differentially expressed in chemoresistant ovarian cancer cell lines. Additionally, blood samples obtained at the time of diagnosis were analyzed for the presence of circulating tumor cells (CTCs).
Results.
The average age of the patients was 64 years, and 82.6% had high‐grade epithelial carcinomas. The baseline median CA‐125 was 464 (range 32–2,782). No statistically significant differences were observed in miR29b or 199a expression in platinum‐resistant/refractory versus platinum‐sensitive tumors. Furthermore, the presence of CTCs was not found to be statistically significantly predictive of eventual platinum resistance.
Conclusion.
Our analysis showed no differences in miR29b and 199a expression, and differences in baseline CTCs in women with newly diagnosed ovarian tumors were not statistically significant.
Abstract
经验总结
• 作为前瞻性临床试验相关科学分析的一部分,值得检查循环肿瘤细胞、微小 RNA 标志物或其他生物标志物。
摘要
背景。约 25% 的卵巢癌患者出现铂类化疗耐药性;但是,卵巢癌耐药性的生物标志物尚未得到验证。我们进行了一项前瞻性试验,旨在确定基于肿瘤的铂类耐药性预测生物标志物。
方法。从 29 名新诊断的经组织病理证实的原发性卵巢癌患者中收集肿瘤标本。其中,23 名女性通过查阅病历获得了关于化疗敏感性和耐药性状态评估和结果数据的标本。肿瘤切片用两种微小 RNA 抗体(miRNA 29b 和 199a)染色,在耐药卵巢癌细胞系中差异表达。此外,对诊断时采集的血样进行循环肿瘤细胞(CTC)存在性分析。
结果。患者平均年龄为 64 岁,82.6% 的患者患有高级别上皮癌。基线中位 CA‐125 为 464(范围32–2 782)。在铂类耐药/难治性与铂类敏感性肿瘤中,miR29b 或 199a 的表达无统计学上的显著差异。此外,CTC 的存在并不能在统计学上显著预测最终的铂类耐药性。
结论。我们的分析显示,miR29b 和 199a 的表达无差异,新诊断卵巢肿瘤女性患者的基线 CTC 差异在统计学上不显著。
Discussion
Platinum chemotherapy resistance is a clinical designation, assessed clinically based on recurrence or progression of malignant disease within 6 months after cessation of platinum treatment. This designation is not based on identification of any testable validated biomarkers. We hypothesized that there exist histopathologic features and/or molecular expression profiles of malignant ovarian tumors that are predictive of platinum chemotherapy resistance.
We have previously reported that CTCs were more prevalent in women with metastases to the ovary than in primary ovarian carcinomas [1]. We speculated that CTCs may also predict chemoresistance in newly diagnosed primary ovarian malignancies. Women with newly diagnosed complex pelvic masses evaluated at the University of Minnesota Gynecologic Oncology Clinic from April 1, 2014, to June 30, 2016, were recruited for this prospective clinical study (Table 1). Written informed consent was obtained from all participants. In this study, we detected differences in CTCs between platinum‐resistant and platinum‐sensitive disease; however, these differences were not statistically significant. Limitations of this study include the small sample size and low yield of CTCs using the CellSearch method [1]. Newer technologies aimed at improving sensitivity of CTC detection, with addition of further capabilities of pharmacogenomic profiling of CTCs, will better enhance abilities to harness these blood‐based biomarkers to determine their predictive value in identifying chemoresistance.
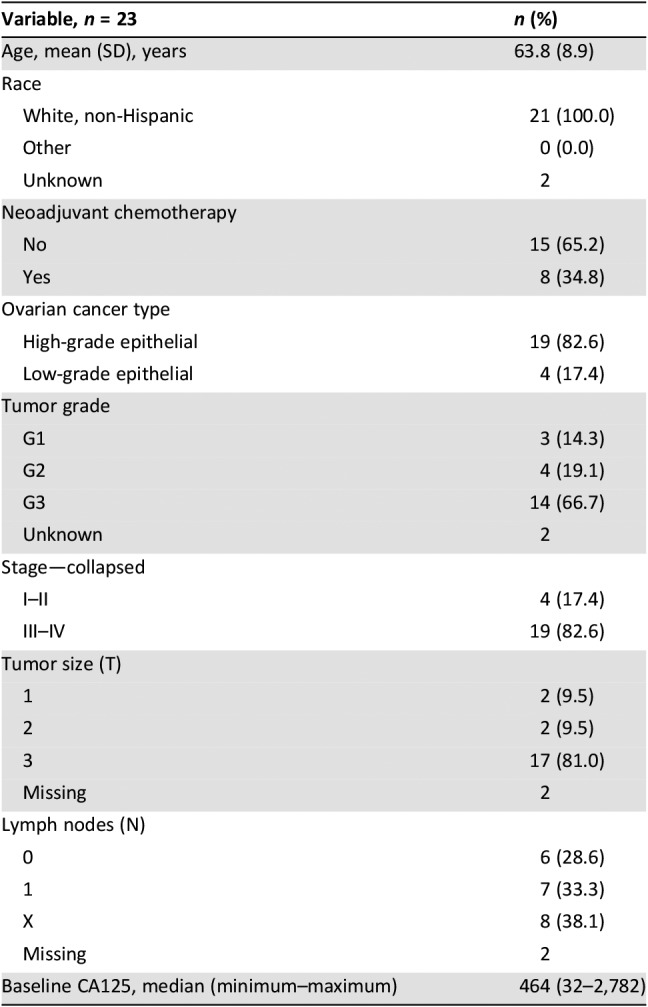
Table 1. Patient demographic and clinical characteristics.
We further hypothesized that tumoral expression of specific miRNAs from tissue at the time of surgical resection can be used as predictive biomarkers of chemoresistance to platinum‐based chemotherapy. MiRNAs are small noncoding forms of RNA that are increasingly being investigated as signals that stimulate increased cellular proliferation and growth in many forms of cancer, including ovarian adenocarcinoma [2]. Profiling studies have identified several candidate miRNAs, such as the miRNA‐200 and miRNA let‐7 families, that could be potentially associated with chemotherapy resistance. We detected no differences in miR29b or miRNA 199a expression in tumor tissue.
Trial Information
- Disease
Ovarian cancer
- Stage of Disease/Treatment
Primary
- Prior Therapy
None
- Type of Study
Phase I/II, nontherapeutic trial—biomarker only
- Primary Endpoint
Correlative endpoint
- Secondary Endpoint
Chemoresistance
- Investigator's Analysis
Correlative endpoints not met but clinical activity observed
Patient Characteristics
- Number of Patients, Male
0
- Number of Patients, Female
23
- Age
Median: 64 years
- Number of Prior Systemic Therapies
Median: 0
- Performance Status: ECOG
Unknown
- Cancer Types or Histologic Subtypes
Carcinoma, 23
Primary Assessment Method
- Title
New assessment
- Number of Patients Screened
29
- Number of Patients Enrolled
23
- Number of Patients Evaluable for Toxicity
0
- Number of Patients Evaluated for Efficacy
23
- Evaluation Method
Tumor marker
- Response Assessment CR
n = 0
- Response Assessment PR
n = 0
- Response Assessment SD
n = 0
- Response Assessment PD
n = 0
- Response Assessment OTHER
n = 23
- (Median) Duration Assessments Duration of Treatment
6 months
Assessment, Analysis, and Discussion
- Completion
Study completed
- Investigator's Assessment
Correlative endpoints not met but clinical activity observed
Materials and Methods
This study was approved by the institutional review board at the University of Minnesota (UMN; Study 1312M46201). For assessment of circulating tumor cells (CTCs), whole blood samples were obtained at the time of diagnosis or initial clinic evaluation as we recently reported [1]. To perform CTC enumeration, 7.5 cc of whole blood was collected in a CellSearch collection tube prior to surgery. More specifically, for patients who had surgery performed as their initial form of treatment (i.e., no neoadjuvant therapy), blood samples were collected on average 10.3 days prior to surgery (median = 8 days; range 0–31 days). For patients who received neoadjuvant chemotherapy treatment prior to surgery, blood was collected on average 9.6 days prior to start of the chemotherapy course (median = 8 days; range 2–17 days). Carcinoma cells were positively selected using magnetic beads conjugated to an anti‐epithelial cell adhesion molecule (EpCAM) antibody. CTC enumeration was performed using photomicroscopic image analysis after staining cells with DAPI, anti‐CD45, and an anticytokeratin cocktail consisting of CK8, CK18, and CK19. CTCs were identified as positive if they were EpCAM positive, CK positive, DAPI positive, and CD45 negative and had the morphology of a single intact carcinoma cell (i.e., no cell clusters identified).
For assessment of microRNA (miRNA) expression, an miRNA array of chemosensitive (A2780) and chemoresistant (C200) ovarian cell lines was performed using the Nanostring protocol to identify the top 100 miRNAs that were differentially expressed between the two cell lines (Digital Genomics). The top five miRNAs for each cell line were internally validated using quantitative reverse transcriptase polymerase chain reaction. Two candidates were identified, miRNA 199a and 29b, that were significantly different between the two cell lines (miRNA‐29b higher in C200/platinum‐resistant cells; miRNA‐199a higher in A2780/platinum‐sensitive cells). We used standard immunohistochemistry and in situ hybridization techniques to determine correlation of expression of miRNA 199a and 29 with platinum resistance in tumors resected from women with newly diagnosed primary ovarian carcinomas. Slides from formalin‐fixed paraffin‐embedded (FFPE) biopsy or surgical samples of primary ovarian tumors were obtained from the University of Minnesota Tissue Procurement Facility. miRNAs 199a and 29b were visualized using Exiqon's miRCURY LNA microRNA in situ hybridization Optimization Kit (FFPE; kit1 90001). All probes were supplied and/or ordered through Exiqon (Qiagen, Seoul, South Korea). LNA U6 snRNA probe, 5' DIG‐labeled as positive control at 1 nM. LNA scrambled microRNA probe, double‐DIG‐labeled as negative control at 40 nM. Double‐DIG‐labeled miRCURY LNA Detection probes for miR‐199a and miR‐29b were optimized at 40 nM.
Routine staining with hematoxylin and eosin was also performed for tissue identification. Slides were visualized using light microscope (5× objective lens) and scored as either positive or nondetectable by two pathologists (M.K. and M.A.L.).
Patient demographic and clinical data were extracted from the electronic medical record, including age at diagnosis, race, pelvic mass origin and histology, tumor grade and stage, tumor size, and lymph node status. Serum levels of CA‐125 were assessed per standard of care. Information was entered into the password‐protected REDCap database available through the UMN Clinical and Translational Science Institute [3].
Statistical Analysis
Demographic and clinical characteristics were summarized using descriptive statistics. The associations between presence of CTCs (0, >0), miRNA 199b and miRNA29b, and chemoresistance were compared using Fisher's exact tests. Data were analyzed using SAS version 9.4 (SAS Institute, Cary, NC), and p values <.05 were considered statistically significant.
Results and Discussion
Ovarian cancer is an insidious disease; thus, the majority of cases are advanced (stage III/IV) at the time of diagnosis [4]. After diagnosis and initial treatment, ovarian cancer declares itself as a refractory and morbid malignancy. Approximately 15,000 women in the U.S. die each year from complications of this disease. However, current approaches to the treatment of ovarian cancer have failed to significantly improve clinical outcomes and overall survival, despite recent advances in developing targeted therapies for the treatment of similarly aggressive cancers, including lung, melanoma, and renal cell carcinomas. Currently, platinum‐based chemotherapy drugs are the backbone of treatment of advanced and recurrent ovarian cancer, but a major clinical problem is development of tumor resistance to platinum therapy. A total of 70%–90% of patients experience recurrence of this disease [5]; approximately 30% are classified as having a platinum‐resistant subtype [6]. To date, there are no clear biologic explanations or biomarkers of platinum resistance. Identifying and validating biomarkers that can predict response to platinum chemotherapy will help risk‐stratify patients at the time of diagnosis into categories that will help direct their plan of treatment. If a biomarker or panel of biomarkers can predict platinum resistance, then the next step would be to perform a prospective clinical trial in which patients with upregulated resistance biomarkers are treated with an alternative approach, such as another drug with or without platinum, to determine how the biomarkers can best be used in clinical management.
In this study, 23 patients with newly diagnosed ovarian cancer were enrolled with appropriate samples for the analysis. The mean age of participants was 63.8 ± 8.9 years, and all were white, non‐Hispanic (Table 1). All women underwent surgical staging, and 34.8% received neoadjuvant chemotherapy prior to surgery. Most (82.6%) had advanced‐stage (III/IV) disease and high‐grade tumors (66.7%). The median baseline CA‐125 was 464 units/mL (range 32–2,782 units/mL).
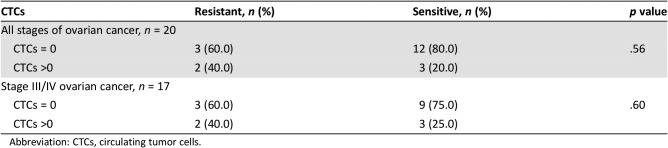
We compared CTC values in the chemoresistant and chemosensitive patient populations to determine whether detection of CTCs prior to treatment could serve as a potential predictive biomarker of platinum resistance (Table 2). We found that five patients had detectable CTCs (>0), with two of five (40%) having chemoresistant disease. Of patients with no detectable CTCs, 80% (3/15) had chemosensitive disease (p = .56). No statistically significant association was observed between CTC detection and chemoresistant status in patients diagnosed with primary ovarian malignancy at any stage. Although 17 of 20 patients with detectable CTCs had stage III or IV form of the disease, subanalysis of these patients did not show a statistically significant predictive value of CTC detection for chemoresistance. As we have discussed in our prior publication on CTCs in ovarian cancer, the system used for CTC identification and collection is very limited because of the low sensitivity of this test [1]. Newer technologies have emerged that provide a higher rate of detection; thus, it is conceivable that future studies may find correlations to chemoresistance with the more sensitive detection methods.
Table 2. CTCs at baseline among study participants.
Abbreviation: CTCs, circulating tumor cells.
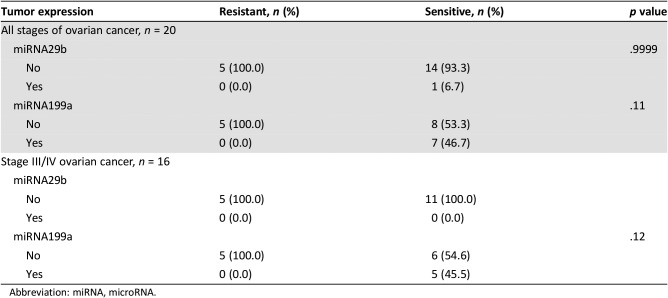
In addition to CTCs, we evaluated whether miRNAs predict chemoresistance in our cohort of patients with ovarian cancer. MiRNAs are small (18–24 nucleotides) noncoding RNA molecules that inhibit gene expression by suppressing the translation process [7]. Many studies have been performed evaluating miRNAs in ovarian cancer. MiRNA signatures of cisplatin‐sensitive and ‐resistant cell lines have identified the let‐7 family as well as other miRNAs that are variably expressed in ovarian cancer [8]. Twist1, which has been associated with stemness in multiple cancer cell lines, was shown to regulate the expression of the miR199a2/214 pathway [9]. Multiple studies have documented the role of miRNA in ovarian cancer tumorigenesis [10]. Dysregulation of miRNA expression, with both upregulation or downregulation, can have a role in the dissemination and progression of cancers, and manipulation of this milieu can offer possible therapeutic targets [11]. Our screen of miRNA differentially expressed between platinum‐resistant and platinum‐sensitive ovarian cancer cell lines identified two candidates: miRNA 199a and 29b. Recent studies have shown that several miRNAs regulate hypoxia‐inducible factor one‐alpha (HIF1α) in multiple different cancer subtypes [12]. Specifically, miRNA 199a has been involved in the regulation of HIF1α, and in our microarray analysis, it was found to be upregulated in ovarian cancer cell lines that are chemosensitive. In our validation analysis, we found a variable change in miR199 between the chemosensitive and chemoresistant cell lines in regard to expression levels in both hypoxia and FBxW7. However, the candidate miRNAs differentially expressed in our cell lines (miRNA 199a and miRNA 29b) were not statistically significantly associated with chemoresistant status in our patient cohort (p = .11 and p = .9999, respectively; Table 3). A limitation of our miRNA screen is that assessment of differential miRNA expression was performed using microarray technology that has in recent years been surpassed by newer methods with higher sensitivities and specificities. Thus, more sensitive assays in current use may yield more refined results that could lead to validation in human studies.
Table 3. miRNA expression among study participants.
Abbreviation: miRNA, microRNA.
In conclusion, there is a strong need for a clinically validated biomarker, or set of biomarkers, that could identify treatment‐refractory ovarian cancers. Although unsuccessful in the present study, CTCs, miRNA markers, or other biomarkers merit examination as part of correlative science analyses in prospective clinical trials.
Tables
Acknowledgments
We thank Michael Franklin, M.S., for editorial review of the manuscript.
Footnotes
UMN Study Identifier: 1312M46201
Sponsor: Clinical and Translational Science Institute, University of Minnesota
Principal Investigator: Emil Lou
IRB Approved: Yes
Disclosures
Emil Lou: Novocure, LLC (C/A), Boston Scientific (H), Novocure, LLC; Intima Capital (RF), Minnetronix; Nomocan (Other: uncompensated expenses); Michael A. Linden: Cell Signaling Technology, Royalties for licensed technology (IP); Jazz Pharmaceuticals (C/A), France Foundation, Cellgene (H); Melissa A. Geller: Tesaro, Inc., Gynecologic Oncology Group, NRG Oncology Foundation, GOG Foundation, Genentech, Inc., Morphotek, Inc., American Cancer Society, Inc., Fate Therapeutics, Inc. (RF). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Lou E, Vogel RI, Teoh D et al. Assessment of circulating tumor cells as a predictive biomarker of histology in women with suspected ovarian cancer. Lab Med 2018;49:134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhlmann JD, Rasch J, Wimberger P et al. MicroRNA and the pathogenesis of ovarian cancer‐‐A new horizon for molecular diagnostics and treatment? Clin Chem Lab Med 2012;50:601–615. [DOI] [PubMed] [Google Scholar]
- 3.Harris PA, Taylor R, Thielke R et al. Research electronic data capture (REDCap)‐‐A metadata‐driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tapia G, Diaz‐Padilla I. Molecular mechanisms of platinum resistance in ovarian cancer. In: Diaz‐Padilla I, ed. Ovarian Cancer: A Clinical and Translational Update. London, UK: IntechOpen, 2013:205–223. [Google Scholar]
- 5.National Cancer Institute . SEER Cancer Statistics Review, 1975–2005. Available at http://seer.cancer.gov/csr/1975_2005/. Accessed April 26, 2019.
- 6.Markman M, Bookman MA. Second‐line treatment of ovarian cancer. The Oncologist 2000;5:26–35. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C. MicroRNomics: A newly emerging approach for disease biology. Physiol Genomics 2008;33:139–147. [DOI] [PubMed] [Google Scholar]
- 8.Kumar S, Kumar A, Shah PP et al. MicroRNA signature of cis‐platin resistant vs. cis‐platin sensitive ovarian cancer cell lines. J Ovarian Res 2011;4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin G, Chen R, Alvero AB et al. Twisting stemness, inflammation and proliferation of epithelial ovarian cancer cells through miR199a2/214. Oncogene 2010;29:3545–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Leva G, Croce CM. The role of microRNAs in the tumorigenesis of ovarian cancer. Front Oncol 2013;3:153. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009;10:704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen G, Li X, Jia YF et al. Hypoxia‐regulated microRNAs in human cancer. Acta Pharmacol Sin 2013;34:336–341. [DOI] [PMC free article] [PubMed] [Google Scholar]