Abstract
Background
Prostate-specific membrane antigen (PSMA; folate hydrolase) prostate cancer (PC) expression has theranostic utility.
Objective
To elucidate PC PSMA expression and associate this with defective DNA damage repair (DDR).
Design, setting, and participants
Membranous PSMA (mPSMA) expression was scored immunohistochemically from metastatic castration-resistant PC (mCRPC) and matching, same-patient, diagnostic biopsies, and correlated with next-generation sequencing (NGS) and clinical outcome data.
Outcome measurements and statistical analysis
Expression of mPSMA was quantitated by modified H-score. Patient DNA was tested by NGS. Gene expression and activity scores were determined from mCRPC transcriptomes. Statistical correlations utilised Wilcoxon signed rank tests, survival was estimated by Kaplan-Meier test, and sample heterogeneity was quantified by Shannon's diversity index.
Results and limitations
Expression of mPSMA at diagnosis was associated with higher Gleason grade (p = 0.04) and worse overall survival (p = 0.006). Overall, mPSMA expression levels increased at mCRPC (median H-score [interquartile range]: castration-sensitive prostate cancer [CSPC] 17.5 [0.0–60.0] vs mCRPC 55.0 [2.8–117.5]). Surprisingly, 42% (n = 16) of CSPC and 27% (n = 16) of mCRPC tissues sampled had no detectable mPSMA (H-score <10). Marked intratumour heterogeneity of mPSMA expression, with foci containing no detectable PSMA, was observed in all mPSMA expressing CSPC (100%) and 37 (84%) mCRPC biopsies. Heterogeneous intrapatient mPSMA expression between metastases was also observed, with the lowest expression in liver metastases. Tumours with DDR had higher mPSMA expression (p = 0.016; 87.5 [25.0–247.5] vs 20 [0.3–98.8]; difference in medians 60 [5.0–95.0]); validation cohort studies confirmed higher mPSMA expression in patients with deleterious aberrations in BRCA2 (p < 0.001; median H-score: 300 [165–300]; difference in medians 195.0 [100.0–270.0]) and ATM (p = 0.005; 212.5 [136.3–300]; difference in medians 140.0 [55.0–200]) than in molecularly unselected mCRPC biopsies (55.0 [2.75–117.5]). Validation studies using mCRPC transcriptomes corroborated these findings, also indicating that SOX2 high tumours have low PSMA expression.
Conclusions
Membranous PSMA expression is upregulated in some but not all PCs, with mPSMA expression demonstrating marked inter- and intrapatient heterogeneity. DDR aberrations are associated with higher mPSMA expression and merit further evaluation as predictive biomarkers of response for PSMA-targeted therapies in larger, prospective cohorts.
Patient summary
Through analysis of prostate cancer samples, we report that the presence of prostate-specific membrane antigen (PSMA) is extremely variable both within one patient and between different patients. This may limit the usefulness of PSMA scans and PSMA-targeted therapies. We show for the first time that prostate cancers with defective DNA repair produce more PSMA and so may respond better to PSMA-targeting treatments.
Keywords: Prostate-specific membrane antigen, Prostate cancer, Castration-resistant prostate cancer, Defective DNA repair, Theranostics, Tumour heterogeneity, BRCA2, Treatment resistance
Take Home Message
Membranous prostate-specific membrane antigen (mPSMA) expression is upregulated in many, but not all, prostate cancers. Importantly, prostate-specific membrane antigen (PSMA) expression demonstrates marked intra- and interpatient heterogeneity, limiting the clinical utility of PSMA theranostics. Defective DNA repair gene aberrations are associated with significantly higher mPSMA expression levels in metastatic castration-resistant prostate cancer and may serve as predictive biomarkers for PSMA-targeted therapies.
1. Introduction
Prostate-specific membrane antigen (PSMA) is a type II transmembrane protein with enzymatic function as a folate hydrolase-carboxypeptidase [1]. PSMA is predominantly localised to five sites: the prostate, the kidney, salivary glands, nervous system glia, and the small bowel jejunal brush border. In astrocytes and Schwann cells, PSMA has been shown to modulate signal transduction pathways by hydrolysing the neurotransmitter N-acetyl aspartylglutamate (NAAG). At the jejunal brush border, PSMA assists in folate absorption by removing the C-terminal glutamates from dietary folates. The role of PSMA in the prostate is less clear, although it has been implicated in folate and glutamate cellular uptake, which are key to DNA synthesis and repair, amino acid and polyamine generation, and PI3K-Akt signalling [1], [2], [3].
Nevertheless, PSMA is widely recognised to be overexpressed in prostate cancer (PC) [4], [5], emerging as a promising target for theranostics. However, concerns remain regarding this strategy. For example, although multiple positron-emission tomography/computed tomography (PET/CT) studies indicate the superiority of PSMA-PET over choline PET [6], the reported sensitivity of these diagnostic techniques encompasses a broad range [7]. Similarly, while radiolabelled anti-PSMA therapies such as 177Lu-PSMA-617 appear well tolerated and result in prostate-specific antigen (PSA) level decreases of ≥50% in 30–60% of metastatic castration-resistant prostate cancer (mCRPC) patients [8], [9], nonresponder rates of approximately 30% have been reported [9], [10], [11], with antitumour activity being higher when PSMA-PET based patient selection is pursued.
One potential explanation for these discrepancies is heterogeneity of PSMA expression [12]. However, the extent of, implications of, and mechanisms underlying this phenomenon remain poorly understood due, in part, to previous studies investigating PSMA expression commonly using antibodies, such as the 7E11 anti-PSMA monoclonal antibody, that recognise the intracellular portion of PSMA [4], [5], [13]. As such, these antibodies are susceptible to overestimating PSMA expression, given that they recognise cytoplasmic PSMA, which is not clinically actionable, as evidenced by early clinical trials of radioimmunoconjugates using these antibodies that showed poor antitumour activity [14], [15]. Subsequently, new antibodies that recognise the extracellular portion of PSMA have been developed [16], allowing more accurate assessment of PSMA expression. However, despite this, more recent publications have been limited by a lack of differentiation between cytoplasmic PSMA expression and more clinically relevant membranous PSMA (mPSMA) [17], [18]. We therefore elected to evaluate, by immunohistochemistry (IHC), the variability of mPSMA protein expression in mCRPC biopsies and, when available, the matching, same-patient, diagnostic, castration-sensitive prostate cancer (CSPC) biopsies, to determine the extent of PSMA heterogeneity and how this may impact the clinical utility of PSMA-theranostic strategies. Furthermore, given that intratumour heterogeneity can be significantly increased by genomic instability [19], we have also pursued next-generation sequencing (NGS) studies to determine whether deleterious DNA damage repair (DDR) aberrations, which are a well-described contributor to genomic instability [20], [21], are associated with PSMA protein expression and whether DDR biomarkers can have utility for guiding PSMA-directed therapies.
2. Patients and methods
2.1. Patients and tissue samples
All patients had mCRPC treated at the Institute of Cancer Research and Royal Marsden Hospital (ICR/RMH), provided written informed consent, and enrolled into institutional protocols approved by the RMH ethics review committee (reference no. 04/Q0801/60). Patient clinical data were retrospectively collected from the Royal Marsden NHS Foundation Trust electronic patient record system.
2.1.1. ICR/RMH test cohort
Sixty patients were identified as having sufficient formalin-fixed, paraffin-embedded (FFPE) mCRPC biopsies available for assessment, of whom 38 also had matched, same-patient, diagnostic CSPC tissue samples. All tissue blocks were freshly sectioned and considered for IHC analyses only if adequate material was present (≥50 tumour cells). All CSPC biopsies demonstrated adenocarcinoma.
2.1.2. ICR/RMH validation cohort
An independent cohort with known DDR aberrations (BRCA2 homozygous deletion, n = 10; ATM loss by IHC, n = 10; CDK12 mutation, n = 6; mismatch repair, n = 9), as determined by NGS, and with sufficient FFPE mCRPC tissue for IHC was compiled to validate the findings of the ICR/RMH test cohort.
2.1.3. International Stand Up To Cancer/Prostate Cancer Foundation validation cohort
A total of 163 mCRPC transcriptomes generated by the International Stand Up To Cancer/Prostate Cancer Foundation (SU2C/PCF) Prostate Cancer Dream Team were downloaded and reanalysed [22].
2.2. Antibody validation
Antibody specificity was determined by Western blot comparing detection of PSMA protein expression in LnCaP whole cell lysates cultured with either nontargeting control siRNA or ON-TARGETplus pooled PSMA siRNA (Dharmacon; GE Healthcare: Chicago, Illinois, United States).
2.3. Immunohistochemisty
PSMA IHC was performed using a mouse anti-PSMA antibody (M3620; monoclonal [clone 3E6]; DAKO; Agilent Technologies: Santa Clara, California, United States). Antigen retrieval was achieved by microwaving slides in antigen retrieval buffer (Tris/EDTA buffer pH 8.1 diluted 1:10) for 18 min at 800 W prior to incubation with anti-PSMA antibody (1:100 dilution) for 1 h at room temperature. The reaction was visualised using the EnVision system (K4061; DAKO). Antibody specificity was confirmed in LnCaP (positive control) and PC3 (negative control) xenograft models (Fig. 1A). Membranous PSMA quantification for each sample was determined by a pathologist blinded to clinical and molecular data using modified H-scores ([{% of weak staining} × 1] + [{% of moderate staining} × 2] + [{% of strong staining} × 3]), to determine the overall percentage of mPSMA positivity across the entire stained tumour sample, yielding a range from 0 to 300 [23].
Fig. 1.
Membranous PSMA (mPSMA) protein expression at the time of prostate cancer (PC) diagnosis is heterogeneous and is associated with a worse overall survival. (A) Immunohistochemical (IHC) antibody validation performed using PC3 cell mouse xenograft, which does not express PSMA protein, and PSMA-expressing LnCaP cell mouse xenograft, demonstrating positive brown membranous and cytoplasmic staining. (B) Antibody signal specificity confirmed by detection of a single band in LnCaP whole-cell lysates by Western blot, with downregulation following treatment with pooled PSMA siRNA compared with nontargeting control siRNA. (C) Graphical representation of mPSMA protein expression in tissue samples obtained from patients with castration-sensitive prostate cancer (CSPC) obtained at diagnosis of PC. Expression of mPSMA protein quantified by H-score and presented in order of increasing H-score. Degree of heterogeneity in mPSMA protein expression observed within samples measured by Shannon's diversity index (SDI) and depicted as heat map ranging from low heterogeneity (white) to high heterogeneity (orange). Of the 38 CSPC tissue samples analysed (median H-score [interquartile range]: 17.5 [0.0–60.0]), 16 (42%; 95% CI [28–58%]) were found to be negative for mPSMA protein expression (H-score <10). Among the 26 remaining samples, as mPSMA protein expression increased, so too did the degree of intrasample heterogeneity, with all CSPC biopsies that expressed mPSMA (PSMA 1+, PSMA 2+, or PSMA 3+) also exhibiting areas of tumour with no detectable PSMA expression (PSMA 0). (D–G) Examples of microscopic appearance of PSMA IHC. All scale bars are set to 100 μm. (D) Prostatectomy demonstrating prostatic adenocarcinoma with no detectable PSMA expression. (E) Prostate biopsy containing prostate cancer cells with strong brown positive membranous and cytoplasmic staining for PSMA. (F) Prostatectomy and (G) prostate biopsy showing heterogeneity in PSMA expression with areas of strong PSMA expression (PSMA 2+ and PSMA 3+) interspersed amongst tumour cells with no detectable PSMA protein expression (PSMA 0). (H) High PSMA protein expression (H-score > median) at the time of prostate cancer diagnosis associated with significantly shorter median OS. Bx = biopsy; CI = confidence interval; HR = hazard ratio; OS = overall survival; PSMA = prostate-specific membrane antigen.
2.4. Determination of deleterious DNA damage repair aberrations
Targeted NGS was available for 56 patients within the ICR/RMH test cohort and performed using DNA extracted from tumour biopsy and germline DNA samples according to a previously published protocol [24], [25]. Libraries were constructed from 40 ng of DNA using a customised GeneRead DNAseq Mix-n-Match v2 panel (Qiagen: Hilden, Germany) and sequenced on the MiSeq Sequencer (Illumina: San Diego, California, United States) at a mean depth of 874×. Sequencing results were used to classify patients within the study cohort as being either positive or negative for deleterious genomic aberrations in DNA-repair genes, as listed in Supplementary Table 1 [24].
2.5. Gene expression and activity scores
Paired-end transcriptome sequencing reads were aligned to the human reference genome (GRCh37/hg19) using Tophat2 (v2.0.7). Gene expression, Fragments Per Kilobase of transcript per Million mapped reads (FPKM), was calculated using Cufflinks [26]. Double-strand break repair score was calculated through cumulative measurement of 19 genes in the homologous recombination repair pathway from the Molecular Signatures Database (M11429).
2.6. Statistical analysis
H-scores were reported as median values and interquartile ranges. Comparisons of mPSMA expression between CSPC and mCRPC tissue samples, and correlations with NGS data were determined using the Wilcoxon matched-pair signed rank test. Overall survival (OS) was defined as time from diagnostic biopsy to the date of death. Median OS was estimated using the Kaplan-Meier method, with hazard ratios determined by Cox regression. Sample heterogeneity was quantified using Shannon's diversity index (SDI) [27]. All analyses were conducted using Stata v13.1; graphs were generated using GraphPad Prism v7.
3. Results
3.1. Antibody validation
To emulate PSMA-targeted diagnostics and theranostics currently under clinical evaluation, anti-PSMA subclone 3E6, which targets an extracellular epitope of PSMA, was chosen over the 7E11 subclone, which recognises the intracellular portion of PSMA. Antibody specificity was validated by Western blot demonstrating a reduction in detectable PSMA protein expression following treatment with PSMA-specific siRNA compared with nontargeting control siRNA (Fig. 1B and Supplementary Fig. 2).
3.2. Expression of mPSMA protein at diagnosis of PC is heterogeneous and associated with a worse overall survival
Expression levels of mPSMA protein were evaluated in 38 CSPC PC biopsies obtained at diagnosis (median H-score [interquartile range]: 17.5 [0.0–60.0]). Interestingly, 16 (42%; 95% confidence interval [CI; 28–58%]) patient samples had no detectable expression of mPSMA (H-score <10). Furthermore, amongst the remaining biopsies that expressed PSMA, there was not only apparent interpatient heterogeneity, but also marked intrapatient heterogeneity in expression, with all evaluated tissue samples exhibiting areas without detectable PSMA (Fig. 1C). To quantify this intratumour heterogeneity, each tumour biopsy was assigned an SDI score; this revealed that not only did all biopsies with detectable mPSMA exhibit heterogeneous expression, but also that the degree of intrapatient heterogeneity increased in parallel with mPSMA H-score. To determine the clinical significance of mPSMA at PC diagnosis, the association of mPSMA expression with clinical characteristics and OS was determined. Higher levels of mPSMA expression were associated with a higher Gleason grade (p = 0.04; Supplementary Table 2) and a worse overall survival (hazard ratio [95% CI]: H-score <17.5 = 1.00 vs H-score ≥17.5 = 2.97 [1.38–6.43]; p = 0.006) when dichotomised by the median H-score (Fig. 1H). Taken together, these data indicate that in addition to a large proportion of patients exhibiting no detectable PSMA expression in their tumour biopsies at diagnosis of PC, when mPSMA is present, its expression is inherently heterogeneous. Furthermore, although the patient cohort presented here is relatively limited in size, making inferences on the impact of PSMA expression on survival more challenging, our results suggest that mPSMA expression at PC diagnosis is associated with a poor prognosis and a more aggressive histological phenotype, meriting further evaluation in larger prospective datasets.
3.3. Expression of mPSMA protein is upregulated in mCRPC, but demonstrates marked intra- and interpatient heterogeneity
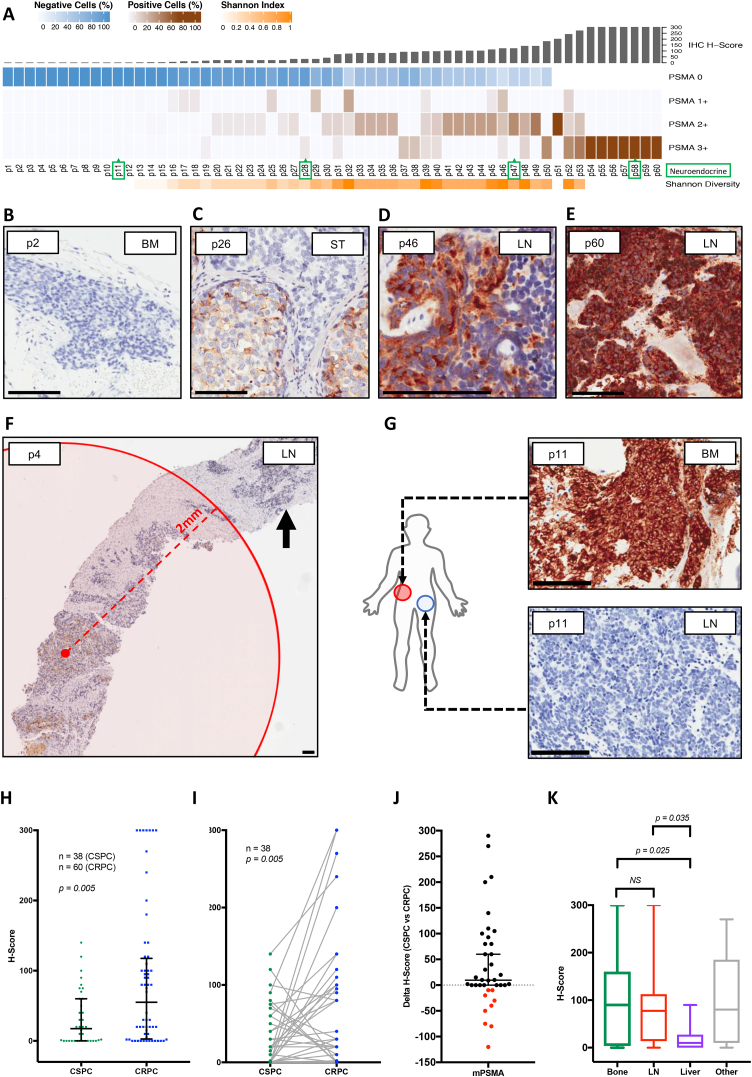
Median mCRPC mPSMA H-score was 55.0 ([2.8–117.5]; n = 60). Importantly, 16 (27% [17–39%]) mCRPC biopsies had no detectable expression of mPSMA, with only one patient (patient 11) having evidence of neuroendocrine differentiation. Interestingly, a comparison with matched, same-patient CSPC tissue samples revealed that half of these samples with no mPSMA protein expression were also negative for mPSMA expression at PC diagnosis. In keeping with our findings in CSPC biopsies, 37 of the 44 (84% [71–92%]) mCRPC biopsies expressing mPSMA demonstrated marked heterogeneity in expression (Fig. 2A), with SDI scores similarly increasing in line with the H-score. While the degree of heterogeneity in mPSMA expression varied between patient samples, common patterns of mPSMA expression were identified. These included the following: (1) PSMA-positive and PSMA-negative cells interspersed within a single area of prostate adenocarcinoma (Fig. 2C); (2) clusters of adenocarcinoma cells with strong PSMA expression intertwined within regions of PSMA-null tumour cells (Fig. 2D); and (3) discrete nodular PSMA expression, with regions of PSMA-negative tumour cells segregated away from regions of PSMA expressing cells, a number of which were >2 mm apart, which is the maximal tissue penetrance of the beta particle emitter 177Lu (Fig. 2F). Furthermore, heterogeneous mPSMA expression between metastases from the same patient was also observed (Fig. 2G). To determine whether mPSMA expression levels were dependent on the type of tissue biopsied, mPSMA expression was correlated with mCRPC biopsy site. No difference was observed between samples obtained from bone (n = 25) and lymphoid tissue (n = 22); however, interestingly, a significant reduction in mPSMA expression was observed in liver metastases (n = 8), with none of these biopsies demonstrating overt neuroendocrine differentiation. Taken together, these data indicate that while mPSMA increases in mCRPC, a significant proportion of patients exhibit no detectable PSMA. When present, however, mPSMA expression in mCRPC exhibits marked intra- and interpatient heterogeneity, resulting in clinically significant distances between areas of PSMA-expressing and PSMA-null cells, potentially impacting the clinical utility of PSMA theranostics. Furthermore, while absence of mPSMA expression at diagnosis was associated with a lack of mPSMA mCRPC expression, there was poor concordance between mPSMA expression at diagnosis of PC and at castration-resistant disease.
Fig. 2.
Expression of mPSMA protein is upregulated in mCRPC, but demonstrates marked intra- and interpatient heterogeneity. (A) Graphic representation of mPSMA protein expression in mCRPC tissue samples. Expression of mPSMA protein quantified by H-score and presented in order of increasing H-score. Intrasample heterogeneity of mPSMA protein expression quantified by SDI and depicted as a heat map ranging from low heterogeneity (white) to high heterogeneity (orange). Tumours with histopathological evidence of neuroendocrine differentiation, confirmed by detectable synaptophysin and/or chromogranin A by IHC, are highlighted by green boxes around patient identifiers. Overall mPSMA expression levels were higher in mCRPC (55.0 [2.8–117.5]; n = 60); however, as seen in CSPC tissue biopsies, 27% (n = 16; 95% CI [17–39%]) of mCRPC patient biopsies had no detectable expression of mPSMA (H-score <10). Interestingly, eight (50% [28–72%]) of these patients were also negative for mPSMA expression at the diagnosis of PC. Expression of mPSMA in mCRPC also demonstrated marked intra- and intertumour heterogeneity, with 84% ([71–92%]; n = 37) of mCRPC biopsies that expressed mPSMA (n = 44; PSMA 1+, PSMA 2+,or PSMA 3+) also exhibiting areas of tumour with no detectable PSMA expression (PSMA 0), and only 10 (23% [13–37%) mPSMA-positive mCRPC samples not containing regions of tumour cells with no mPSMA protein expression (PSMA 0). (B–E) Examples of microscopic appearance of PSMA IHC. All scale bars are set to 100 μm. (B) Bone marrow trephine demonstrating prostatic adenocarcinoma with no detectable PSMA expression. (C) Soft tissue metastasis showing adenocarcinoma cells with moderate (PSMA 2+) PSMA expression (brown) interspersed and juxtaposed alongside tumour cells with no detectable PSMA expression (PSMA 0). (D) Lymph node biopsy exhibiting strongly positive PSMA expressing cells (PSMA 2+ and PSMA 3+) intertwined within a region of PSMA-negative (PSMA 0) tumour cells. (E) Lymph node biopsy with strong positive membranous and cytoplasmic staining for PSMA (PSMA 3+) with no PSMA negative cells. (F) Lymph node biopsy highlighting the clinical significance of PSMA intratumour heterogeneity. Discrete nodule of tumour cells with no PSMA protein expression (arrow) segregated away from the region of PSMA-expressing cells (brown) and located beyond the maximal tissue penetrance of 177Lu (red circle). (G) Bone and lymph node mCRPC biopsies from the same patient demonstrating significant heterogeneity between metastases. (H) Expression levels of mPSMA significantly higher (p = 0.005) in mCRPC biopsies (55.0 [2.8–117.5]; n = 60) compared with CSPC biopsies (17.5 [0.0–60.0]; n = 38). (I) Comparison with matched, same-patient CSPC tissue samples (n = 38), however, revealed poor concordance in mPSMA expression levels between CSPC and mCRPC samples. (J) Expression of mPSMA increases in most patients on progression to mCRPC; however, nine patients had no change in expression levels (change in H-score <5), while nine (24% [13–39%]) had less mPSMA protein expression at mCRPC. Interestingly, eight (89% [57–99%]) of the patients with no change in mPSMA expression were negative for mPSMA expression at the diagnosis of PC (H-score <10), and remained so on progression to mCRPC. (K) Comparison of mPSMA expression levels relative to the type of tissue biopsied. No difference was observed between samples obtained from bone (n = 25) and lymphoid tissue (n = 22); however, significantly lower mPSMA expression was seen in liver metastases (n = 8), none of which demonstrating overt evidence of neuroendocrine differentiation. BM = bone marrow; CI = confidence interval; CSPC = castration-sensitive prostate cancer; HR = hazard ratio; LN = lymph node; mCRPC = metastatic castration-resistant PC; mPSMA = membranous PSMA; NS = not significant; PC = prostate cancer; PSMA = prostate-specific membrane antigen; SDI = Shannon's diversity index; ST = soft tissue.
3.4. Expression of mPSMA protein in mCRPC is associated with deleterious DNA repair aberrations
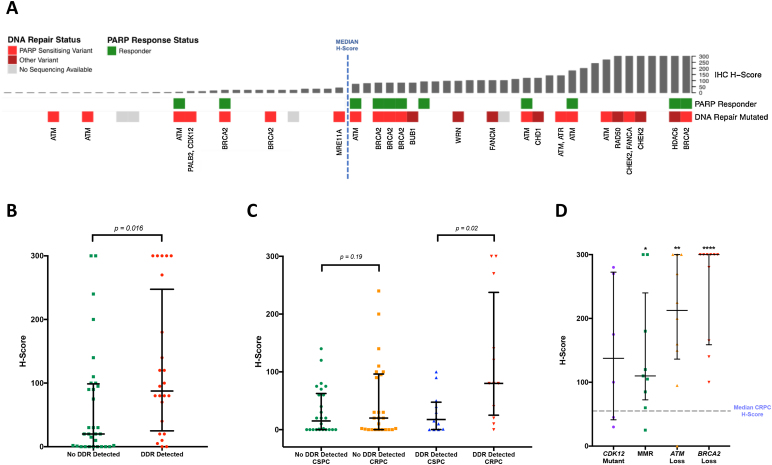
In light of the observed marked heterogeneity of mPSMA expression, and given that intratumour heterogeneity can be significantly increased by genomic instability [19], targeted NGS data were analysed to identify potential associations between mPSMA expression and deleterious DDR aberrations (Fig. 3A). Interestingly, mCRPC patients with deleterious DDR aberrations were found to have significantly higher (p = 0.016) mPSMA expression than those without these aberrations (DDR 87.5 [25.0–247.5] vs no DDR 20 [0.3–98.8]; difference in medians 60 [5.0–95.0]; Fig. 3B). Furthermore, when comparing the change in mPSMA protein expression between matched, same-patient CSPC and mCRPC tissue samples, we observed that while mPSMA expression increased significantly in mCRPC patients with detectable DDR aberrations (p = 0.02), this was not the case for patients without detectable DDR gene mutations (p = 0.19) (Coef = 35; 95% CI: 2–68; p = 0.04; Fig. 3C). Interestingly, in keeping with these findings, although not statistically significant, amongst the 37 patients within our patient cohort who received treatment with a PARP inhibitor, nine of the 11 (82%) patients who responded to therapy had a mPSMA H-score above the median (55.0 [2.8–117.5]). To validate the association between mPSMA expression and deleterious DDR aberrations, mCRPC mPSMA expression levels were evaluated in an independent PC cohort with known DDR aberrations. Tumours with deleterious DDR aberrations were found to have significantly higher mPSMA expression levels than unselected mCRPC biopsies (BRCA2 300 [165–300], difference in medians 195.0 [100.0–270.0], p ≤ 0.001; ATM 212.5 [136.3–300], difference in medians 140.0 [55.0–200.0], p = 0.005; CDK12 137.5 [41.3–272.5], difference in medians 45 [−20.0 to 175.0], p = 0.1; MMR 110.0 [72.5–240.0], difference in medians 60 [5.0–110.0], p = 0.03; Fig. 3D). Taken together, these data suggest that deleterious DDR aberrations are associated with significantly higher mPSMA expression levels in mCRPC.
Fig. 3.
Expression of mPSMA protein in mCRPC is significantly higher in patients with deleterious DNA repair genomic aberrations. (A) Bar graph representing mPSMA protein expression in mCRPC patient biopsies (n = 60) presented in order of increasing H-score. Median H-score (55.0 [2.8–117.5]) demarcated by blue dashed line. Deleterious DNA damage repair (DDR) aberrations, as detected by next-generation sequencing (NGS) of patient tissue samples, are indicated by red boxes with affected gene labelled below; PARP inhibitor sensitising aberrations are depicted as dark red boxes and genomic variants of currently unknown significance as light red boxes. Green boxes depict patients with documented response to PARP inhibitor therapy. Grey boxes depict patients for whom mCRPC NGS data were not available (n = 4). (B) Patients with deleterious DDR aberrations in genes involved in the DNA repair pathway have significantly higher (p = 0.016; Wilcoxon rank-sum analysis) levels of mPSMA expression than those without these (DDR 87.5 [25.0–247.5] vs no DDR 20 [0.3–98.8]; difference in medians 60 [5.0–95.0]). (C) Expression of mPSMA increases significantly (p = 0.02) in mCRPC, compared with CSPC, in patients with detected deleterious DDR aberrations (median CSPC H-score = 17.5 [0.3–47.5] vs median mCRPC H-score = 80 [25–237.5]), unlike patients without DDR aberrations (median CSPC H-score = 15 [0–62.5] vs median mCRPC H-score = 20 [0–96.3]; p = 0.19) (Coef = 35; 95% CI: 2–68; p = 0.04). (D) Association between mPSMA expression and deleterious DDR aberrations was validated in an independent cohort with known DDR aberrations. Tumours with DDR aberrations had significantly higher mPSMA expression (BRCA2 300 [165–300], difference in medians 195.0 [100.0–270.0], p = < 0.001; ATM 212.5 [136.3–300], difference in medians 140.0 [55.0–200.0], p = 0.005; CDK12 137.5 [41.3–272.5], difference in medians 45 [−20.0 to 175.0], p = 0.1; MMR 110.0 [72.5–240.0], difference in medians 60 [5.0–110.0], p = 0.032) than unselected mCRPC biopsies, as shown by the dashed line (55.0 [2.75–117.5]). All p values were calculated using Mann-Whitney test. * p < 0.05, ** p < 0.01, *** p < 0.001. CI = confidence interval; CSPC = castration-sensitive prostate cancer; mCRPC = metastatic castration-resistant prostate cancer; mPSMA = membranous prostate-specific membrane antigen.
3.5. Metastatic CRPC transcriptome analyses: PSMA, double-strand break repair, BRCA2, and epithelial-mesenchymal transition
To further explore the association between PSMA expression and DNA repair, analysis of RNA-sequencing data obtained from 163 mCRPC transcriptomes was performed (SU2C/PCF validation cohort). This demonstrated an inverse correlation between PSMA mRNA expression, and both BRCA2 mRNA expression (p < 0.001; Fig. 4A) and double-strand break repair (p < 0.001; Fig. 4C), which, taken together, suggest that high PSMA expression in mCRPC is associated with BRCA2 loss and a reduction in homology-dependent DNA repair. Furthermore, since RB1 can be codeleted with BRCA2, with emerging endocrine resistance and luminal to basal transition with associated hyperexpression of SOX2, we correlated SOX2 expression with PSMA expression [28], [29]. PSMA expression correlated negatively and robustly with SOX2 (p < 0.001; Fig. 4D), SNAI1 (p < 0.001; Fig. 4E), SNAI2 (p = 0.039; Fig. 4F), NCAM1 (p < 0.001; Fig. 4G), and EON2 (p < 0.001; Fig. 4H) expression levels, suggesting downregulation of PSMA expression with luminal to basal transition and the neuroendocrine-like phenotype.
Fig. 4.
PSMA mRNA expression is inversely correlated with BRCA2 mRNA expression and double-strand break repair. Analysis of RNA-sequencing data obtained from 163 mCRPC transcriptomes demonstrating an inverse correlation between PSMA mRNA expression, and (A) BRCA2 mRNA expression (p < 0.001) and (B) ATM mRNA expression (p = 0.059), associating PSMA expression with BRCA2 loss. (C) An inverse correlation was observed between PSMA mRNA expression and an mRNA signature of double-strand break repair activity (p < 0.001) calculated through cumulative measurement of 19 genes involved in homologous recombination DNA repair, as determined from the Molecular Signatures Database (M11429), indicating that as PSMA mRNA expression increases, DDR activity decreases. (D) PSMA expression was also found to be inversely correlated with SOX2 mRNA expression (p < 0.001), as well as with (E) SNAI1 (p < 0.001), (F) SNAI2 (p = 0.039), (G) NCAM1 (p < 0.001), and (H) ENO2 (p < 0.001) expression levels, suggesting a downregulation of PSMA expression with luminal to basal transition and the neuroendocrine-like phenotype. DDR = defective DNA repair; FPKM = Fragments Per Kilobase of transcript per Million mapped reads; mCRPC = metastatic castration-resistant prostate cancer; PSMA = prostate-specific membrane antigen.
4. Discussion
The protein PSMA, which is arguably critical to folate and glutamate PC cell uptake necessary for maintaining genomic stability, has become a key target for PC theranostics [8], [9], [10], [11]. Early-phase trials of PSMA-targeting theranostics, including immunoconjugates, radioimmunoconjugates carrying alpha- and beta-emitting particles, and bispecific antibodies targeting CD3 and PSMA, have reported antitumour activity against mCRPC. These trials have often recruited patients with positive PSMA-ligand uptake on PET [10], [11]; however, this patient selection is not always being pursued so as to minimise cost. De novo and acquired resistance to these agents has also been reported, and is likely to be at least in part related to the absence or loss of PSMA expression, although the mechanisms for these have not been fully elucidated to date. A better understanding of inter- and intratumour heterogeneity in PSMA expression is now key to successful development of these agents. We now present the first robust evidence that PC PSMA expression is strongly associated with DNA repair defective tumours, perhaps explaining why therapeutics that generate double-strand DNA breaks, such as radioimmunoconjugates, have had antitumour activity against PC. We propose that future trials with these agents should consider targeting DDR PC. These data are supported by a recent case report of an exceptional response to 177Lu in a patient harbouring BRCA2 loss [30].
While our analyses presented here focus on mPSMA, given that we believe that this represents a more clinically relevant measure of PSMA expression in PC, we acknowledge that differentiation between membranous and cytoplasmic PSMA expression, particularly in cases where cytoplasmic PSMA expression is high, may be a limitation of our work. Despite this, our data herein indicate that the inherent intratumour heterogeneity of PSMA expression in PC represents a significant potential contributor to resistance to these therapies. Furthermore, while our conclusions are limited by the retrospective nature of our study and the relatively small size of our patient cohorts, we envision that trials that do not mandate evidence of PSMA expression prior to enrolment will inevitably observe lower rates of response. Moreover, our data indicating that liver metastases in mCRPC have very low PSMA expression also raise concern that patients suffering from these may not benefit from these therapies, with these tumours perhaps being more likely to have an emerging basal phenotype [31], [32], [33]. Further studies are now needed to determine whether this will be a commoner site of resistance to PSMA-targeting therapies.
The association between PSMA and deleterious DDR aberrations also raises many interesting questions for future study. Importantly, we do not propose that deleterious DDR aberrations alone cause increased expression of PSMA. Rather, we hypothesise that the biological consequences for tumour cells of having deleterious aberrations in DDR genes potentially promote the overexpression of PSMA. Deleterious DDR aberrations are associated with replication stress, placing increased demand for metabolic precursors such as folate and glutamate, which are crucial to DNA synthesis and repair. As such, given the enzymatic capability of PSMA to yield glutamate and folate monoglutamate from polyglutamated folates [2], and its reported role as a folate transporter [1], one possible explanation to account for this association is that PSMA overexpression in cells with deleterious DDR aberrations represents an adaptive cellular response. In this setting, PSMA overexpression may be being driven by the increased requirement of these cells for cellular metabolites such as folate and glutamate. Alternatively, PSMA has also been reported to promote PI3K-Akt signalling [3], [34]. This has been demonstrated to suppress DNA repair by homologous recombination, which is defective in cells with deleterious DDR aberrations such as BRCA2 loss, and also to enhance nonhomologous end joining [35]. This again hints towards PSMA serving as a survival mechanism for PC cells with DDR. Most likely, however, the mechanisms underlying the expression of PSMA are multifactorial, involving a complex interaction between numerous cellular processes. In spite of these uncertainties, it is clear that a deeper understanding of the biology of PSMA in PC is now needed, firstly, to account for this association, but also to elucidate pharmacological strategies that can manipulate PSMA expression, for example, perhaps through generating PC genomic instability or replication stress. Such strategies may inform on the development of optimal drug combinations for PSMA-targeting therapeutics.
5. Conclusions
Expression of mPSMA is upregulated in many but not all PCs, and exhibits marked intra- and interpatient heterogeneity. DDR gene aberrations are associated with significantly higher mPSMA expression levels and merit further evaluation as predictive biomarkers of response for PSMA-targeted therapies in larger, prospective clinical trials.
Author contributions: Johann S. de Bono had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Paschalis, de Bono.
Acquisition of data: Paschalis, Riisnaes, Rodrigues, Gurel, Bertan, Ferreira, Seed, Yuan, Welti, Neeb, Sumanasuriya, Rescigno, Bianchini, Tunariu, Carreira, Sharp.
Analysis and interpretation of data: Paschalis, Rodrigues, Gurel, Seed, Yuan, Carreira, Sharp, de Bono.
Drafting of the manuscript: Paschalis, Sharp, de Bono.
Critical revision of the manuscript for important intellectual content: Paschalis, Sheehan, Riisnaes, Rodrigues, Ferreira, Lambros, Sumanasuriya, Rescigno, Bianchini, Tunariu, Carreira, Sharp, Oyen, de Bono.
Statistical analysis: Paschalis, Dolling.
Obtaining funding: de Bono.
Administrative, technical, or material support: None.
Supervision: de Bono.
Other: None.
Financial disclosures: Johann S. de Bono certifies that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Funding/Support and role of the sponsor: This study was supported by Movember Foundation, Prostate Cancer UK (PCUK), Cancer Research UK (CRUK), and Prostate Cancer Foundation (PCF).
Associate Editor: Giacomo Novara
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.eururo.2019.06.030.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Yao V., Berkman C.E., Choi J.K., O'Keefe D.S., Bacich D.J. Expression of prostate-specific membrane antigen (PSMA), increases cell folate uptake and proliferation and suggests a novel role for PSMA in the uptake of the non-polyglutamated folate, folic acid. Prostate. 2010;70:305–316. doi: 10.1002/pros.21065. [DOI] [PubMed] [Google Scholar]
- 2.Tisman G. Modulation of one-carbon metabolism by B vitamins: implications for transformation and progression of prostate cancer. In: Spiess P.E., editor. Prostate cancer—from bench to bedside. IntechOpen; 2011. [Google Scholar]
- 3.Caromile L.A., Dorthe K., Rahman M.M. PSMA redirects cell survival signaling from the MAPK to the PI3K-AKT pathways to promote the progression of prostate cancer. Sci Signal. 2017;10(470) doi: 10.1126/scisignal.aag3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silver D.A., Pellicer I., Fair W.R., Heston W.D., Cordon-Cardo C. Prostate-specific membrane antigen expression in normal and malignant human tissues. Clin Cancer Res. 1997;3:81–85. [PubMed] [Google Scholar]
- 5.Bostwick D.G., Pacelli A., Blute M., Roche P., Murphy G.P. Prostate specific membrane antigen expression in prostatic intraepithelial neoplasia and adenocarcinoma: a study of 184 cases. Cancer. 1998;82:2256–2261. doi: 10.1002/(sici)1097-0142(19980601)82:11<2256::aid-cncr22>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 6.Afshar-Oromieh A., Zechmann C.M., Malcher A. Comparison of PET imaging with a (68)Ga-labelled PSMA ligand and (18)F-choline-based PET/CT for the diagnosis of recurrent prostate cancer. Eur J Nucl Med Mol Imaging. 2014;41:11–20. doi: 10.1007/s00259-013-2525-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perera M., Papa N., Christidis D. Sensitivity, specificity, and predictors of positive (68)Ga-prostate-specific membrane antigen positron emission tomography in advanced prostate cancer: a systematic review and meta-analysis. Eur Urol. 2016;70:926–937. doi: 10.1016/j.eururo.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 8.Eibername> M., Fendler W.P., Rowe S.P. Prostate-specific membrane antigen ligands for imaging and therapy. J Nucl Med. 2017;58(Suppl. 2) doi: 10.2967/jnumed.116.186767. 67s–76s. [DOI] [PubMed] [Google Scholar]
- 9.Rahbar K., Schmidt M., Heinzel A. Response and tolerability of a single dose of 177Lu-PSMA-617 in patients with metastatic castration-resistant prostate cancer: a multicenter retrospective analysis. J Nucl Med. 2016;57:1334–1338. doi: 10.2967/jnumed.116.173757. [DOI] [PubMed] [Google Scholar]
- 10.Ahmadzadehfar H., Wegen S., Yordanova A. Overall survival and response pattern of castration-resistant metastatic prostate cancer to multiple cycles of radioligand therapy using [(177)Lu]Lu-PSMA-617. Eur J Nucl Med Mol Imaging. 2017;44:1448–1454. doi: 10.1007/s00259-017-3716-2. [DOI] [PubMed] [Google Scholar]
- 11.Rahbar K., Ahmadzadehfar H., Kratochwil C. German multicenter study investigating 177Lu-PSMA-617 radioligand therapy in advanced prostate cancer patients. J Nucl Med. 2017;58:85–90. doi: 10.2967/jnumed.116.183194. [DOI] [PubMed] [Google Scholar]
- 12.Mannweiler S., Amersdorfer P., Trajanoski S. Heterogeneity of prostate-specific membrane antigen (PSMA) expression in prostate carcinoma with distant metastasis. Pathol Oncol Res. 2009;15:167–172. doi: 10.1007/s12253-008-9104-2. [DOI] [PubMed] [Google Scholar]
- 13.Ross J.S., Sheehan C.E., Fisher H.A. Correlation of primary tumor prostate-specific membrane antigen expression with disease recurrence in prostate cancer. Clin Cancer Res. 2003;9:6357–6362. [PubMed] [Google Scholar]
- 14.Deb N., Goris M., Trisler K. Treatment of hormone-refractory prostate cancer with 90Y-CYT-356 monoclonal antibody. Clin Cancer Res. 1996;2:1289–1297. [PubMed] [Google Scholar]
- 15.Kahn D., Austin J.C., Maguire R.T. A phase II study of [90Y] yttrium-capromab pendetide in the treatment of men with prostate cancer recurrence following radical prostatectomy. Cancer Biother Radiopharm. 1999;14:99–111. doi: 10.1089/cbr.1999.14.99. [DOI] [PubMed] [Google Scholar]
- 16.Murphy G.P., Greene T.G., Tino W.T., Boynton A.L., Holmes E.H. Isolation and characterization of monoclonal antibodies specific for the extracellular domain of prostate specific membrane antigen. J Urol. 1998;160(6 Pt 2):2396–2401. doi: 10.1097/00005392-199812020-00006. [DOI] [PubMed] [Google Scholar]
- 17.Perner S., Hofer M.D., Kim R. Prostate-specific membrane antigen expression as a predictor of prostate cancer progression. Hum Pathol. 2007;38:696–701. doi: 10.1016/j.humpath.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Minner S., Wittmer C., Graefen M. High level PSMA expression is associated with early PSA recurrence in surgically treated prostate cancer. Prostate. 2011;71:281–288. doi: 10.1002/pros.21241. [DOI] [PubMed] [Google Scholar]
- 19.Burrell R.A., McGranahan N., Bartek, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 20.Knijnenburg T.A., Wang L., Zimmermann M.T. Genomic and molecular landscape of DNA damage repair deficiency across the Cancer Genome Atlas. Cell Rep. 2018;23 doi: 10.1016/j.celrep.2018.03.076. 239–254.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgoulis A., Vorgias C.E., Chrousos G.P., Rogakou E.P. Genome Instability and γH2AX. Int J Mol Sci. 2017;18:1979. doi: 10.3390/ijms18091979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson D., Van Allen E.M., Wu Y.M. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Detre S., Saclani Jotti G., Dowsett M. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mateo J., Carreira S., Sandhu S. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodall J., Mateo J., Yuan W. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 2017;7:1006–1017. doi: 10.1158/2159-8290.CD-17-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C., Roberts A., Goff L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maley C.C., Aktipis A., Graham T.A. Classifying the evolutionary and ecological features of neoplasms. Nat Rev Cancer. 2017;17:605–619. doi: 10.1038/nrc.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seed G., Yuan W., Mateo J. Gene copy number estimation from targeted next-generation sequencing of prostate cancer biopsies: analytic validation and clinical qualification. Clin Cancer Res. 2017;23:6070–6077. doi: 10.1158/1078-0432.CCR-17-0972. [DOI] [PubMed] [Google Scholar]
- 29.Mu P., Zhang Z., Benelli M. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science. 2017;355:84–88. doi: 10.1126/science.aah4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crumbaker M, L. Emmett, L.G. Horvath, and A.M. Joshua, Exceptional response to 177lutetium prostate-specific membrane antigen in prostate cancer harboring DNA repair defects. JCO Precis Oncol. In press. 10.1200/PO.18.00237. [DOI] [PubMed]
- 31.Pouessel D., Gallet B., Bibeau F. Liver metastases in prostate carcinoma: clinical characteristics and outcome. BJU Int. 2007;99:807–811. doi: 10.1111/j.1464-410X.2006.06663.x. [DOI] [PubMed] [Google Scholar]
- 32.Beltran H., Tagawa S.T., Park K. Challenges in recognizing treatment-related neuroendocrine prostate cancer. J Clin Oncol. 2012;30:e386–e389. doi: 10.1200/JCO.2011.41.5166. [DOI] [PubMed] [Google Scholar]
- 33.Shou J., Zhang Q., Wang S., Zhang D. The prognosis of different distant metastases pattern in prostate cancer: a population based retrospective study. Prostate. 2018;78:491–497. doi: 10.1002/pros.23492. [DOI] [PubMed] [Google Scholar]
- 34.Kaittanis C., Andreou C., Hieronymus H. Prostate-specific membrane antigen cleavage of vitamin B9 stimulates oncogenic signaling through metabotropic glutamate receptors. J Exp Med. 2018;215:159–175. doi: 10.1084/jem.20171052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, K.M. Turner, W.K. Alfred Yung, K. Chen, and W. Zhang Q., Turner K.M., Alfred Yung W.K., Chen K., Zhang W. Role of AKT signaling in DNA repair and clinical response to cancer therapy. Neuro Oncol. 2014;16:1313–1323. doi: 10.1093/neuonc/nou058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.