Abstract
The Epidermal Growth Factor Receptor (EGFR) and the Transforming Growth Factor-beta (TGF-β) are key regulators of hepatocarcinogenesis. Targeting EGFR was proposed as a promising therapy; however, poor success was obtained in human hepatocellular carcinoma (HCC) clinical trials. Here, we describe how EGFR is frequently downregulated in HCC patients while TGF-β is upregulated. Using 2D/3D cellular models, we show that after EGFR loss, TGF-β is more efficient in its pro-migratory and invasive effects, inducing epithelial to amoeboid transition. EGFR knock-down promotes loss of cell-cell and cell-to-matrix adhesion, favouring TGF-β-induced actomyosin contractility and acquisition of an amoeboid migratory phenotype. Moreover, TGF-β upregulates RHOC and CDC42 after EGFR silencing, promoting Myosin II in amoeboid cells. Importantly, low EGFR combined with high TGFB1 or RHOC/CDC42 levels confer poor patient prognosis. In conclusion, this work reveals a new tumour suppressor function for EGFR counteracting TGF-β-mediated epithelial to amoeboid transitions in HCC, supporting a rational for targeting the TGF-β pathway in patients with low EGFR expression. Our work also highlights the relevance of epithelial to amoeboid transition in human tumours and the need to better target this process in the clinic.
Keywords: Liver, Migration, Adhesion, Matrix, Myosin II
Abbreviations: EGFR, Epidermal Growth Factor Receptor; TGF-β, Transforming Growth Factor-beta; pMLC2, phospho-Myosin Light Chain II; EMT, Epithelial-to-mesenchymal transition
Highlights
-
•
EGFR expression is low and heterogeneous in a great percentage of HCC patients.
-
•
EGFR loss in HCC cells facilitates TGF-β pro-migratory and invasive functions.
-
•
EGFR silenced HCC cells respond to TGF-β inducing epithelial-amoeboid transition.
-
•
TGF-β upregulates RHOC and CDC42 and actomyosin contractility in EGFR silenced cells.
-
•
Low EGFR combined with high TGFB1 or RHOC/CDC42 levels confer poor HCC prognosis.
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most common liver tumours and its prevalence is rapidly arising, presenting a high frequency of relapse and metastasis [1]. Transforming Growth Factor-beta (TGF-β) is a liver tumour growth suppressor, inhibiting proliferation and inducing cell death [2,3]. However, paradoxically, TGF-β also modulates processes such as invasion, immune regulation and remodelling of the microenvironment, which cancer cells may exploit to their advantage [4].
Epidermal Growth Factor Receptor (EGFR) belongs to a tyrosine kinase receptor family with essential roles in cell proliferation, survival, differentiation, adhesion and migration. Alterations in its activity have been implicated in the development and growth of many tumours [5]. We have previously described a cross-talk between TGF-β and EGFR pathways that allows HCC cells to escape from TGF-β-induced suppressor actions [6]. TGF-β plays a dual role inducing both pro- and anti-apoptotic signals, the latter being mediated by transactivation of the EGFR pathway, through upregulation of EGFR ligands and activation of the metalloprotease TACE/ADAM17 that mediates ligand shedding [7]. Liver tumour cells that overcome pro-apoptotic effects of this cytokine undergo TGF-β-induced epithelial to mesenchymal transition (EMT) acquiring cancer stem cell properties, which contribute to tumour cell migration and invasion, as well as drug resistance [[8], [9], [10]].
Pro-migratory actions of TGF-β in HCC cells are mainly attributed to its capacity to induce EMT [11,12]. However, it is well recognized that tumour cells can use several modes of migration. Cancer cells can disseminate as individual cells or expand in solid strands, sheets or clusters using collective strategies [13]. Individual cell migration modes comprise a range of behaviours and - at the end of the spectra - elongated-mesenchymal and rounded-amoeboid modes of cell migration have been clearly identified [14]. Elongated-mesenchymal migration relies on stronger adhesion levels that are dependent on Rac-driven actin polymerization [15]. On the other hand, formin dependent actin dynamics [16], Rho-ROCK [17] and Cdc42-PAK2 [18] control Myosin II to generate actomyosin contractility needed for rounded-amoeboid migration [19]. Thus, high levels of Myosin II and lower levels of adhesion are characteristic of amoeboid types of migration, where the functional protrusions are blebs [20]. Almost all the studies on HCC cell migration are mainly based on acquisition of EMT features [21]. Once cells have lost their epithelial behaviour, little is known about the contribution of different modes of migration to HCC dissemination.
In spite of the potential pro-tumorigenic role of the EGFR pathway in HCC [22,23], when moving to human clinical trials, EGFR monoclonal antibodies, such as cetuximab, and EGFR tyrosine kinase inhibitors, such as gefitinib or erlotinib, showed none or only a modest activity in advanced HCC patients [[24], [25], [26]]. The aim of this work was to understand how EGFR is regulated in HCC and if it has a possible contribution to TGF-β driven pro-migratory and invasive effects.
2. Materials and methods
2.1. Cell lines
Hep3B and PLC/PRF/5 cell lines were obtained from European Collection of Cell Cultures (ECACC, Porton Down, Salisbury, Wiltshire, UK). Hep3B were grown in MEM medium (supplemented with non-essential amino acids), and PLC/PRF/5 in DMEM, both media supplemented with 10% FBS, in a humidified atmosphere at 37 °C, 5% CO2.
2.2. Knockdown assays
Hep3B and PLC/PRF/5 cells were stable silenced for EGFR using different plasmids, either alone or in combination, selected from Mission SH, Sigma-Aldrich (Madrid, Spain) as well as a control unspecific shRNA (sh-).
2.3. Analysis of gene expression
RNeasy Mini Kit (Ref. 74104, Qiagen, Valencia, CA, USA) was used for total RNA isolation. Reverse transcription (RT) was carried out with random primers using High Capacity RNA to cDNA Master Mix Kit (Ref. 4387406, Applied Biosystems, Foster City, CA, USA). Expression levels were determined in duplicates in a LightCycler® 480 Real Time PCR System, using the LightCycler® 480 SYBR Green I Master Mix (Ref. 04887352001, Roche Diagnostics GmbH, Mannheim, Germany).
2.4. Immunohistochemistry and histology analyses
Immunohistochemical analyses were performed using standard procedures [27]. Antibodies used and conditions are summarized in Supplementary Table III.
2.5. Statistical analyses
Statistical analyses were performed as an estimation of the associated probability to a Student's t-test (95% confidence interval) or as Two-way ANOVA method, depending on the involved conditions. Experiments were carried out at least 3 independent times with 2–3 technical replicates. Data were represented as mean ± standard error of the mean (SEM). Differences between groups were compared using Student's t-test (when comparing two groups) or Two-way ANOVA with Tukey's multiple comparison post-hoc test (differences between groups considering two independent variables). For survival analysis, expression data from the cohort of HCC patients and The Cancer Genome Atlas (TCGA) database were categorized using the median. Kaplan–Meier method using the log-rank test was used to estimate survival curves. For data from human samples, statistical significance was determined by Linear correlation analysis. In all cases, statistical calculation was developed using GraphPad Prism software (GraphPad for Science Inc., San Diego, CA, USA), except survival analysis which were performed using SPSS (IBM, North Harbour, Portsmouth, UK). Differences were considered statistically significant at p < 0.05 (* or #), p < 0.01 (** or ##) and p < 0.001 (*** or ###).
For further procedures and details see Supplementary Materials and Methods.
3. Results
3.1. Low levels of EGFR concomitant with high activation of the TGF-β pathway is observed in HCC patients
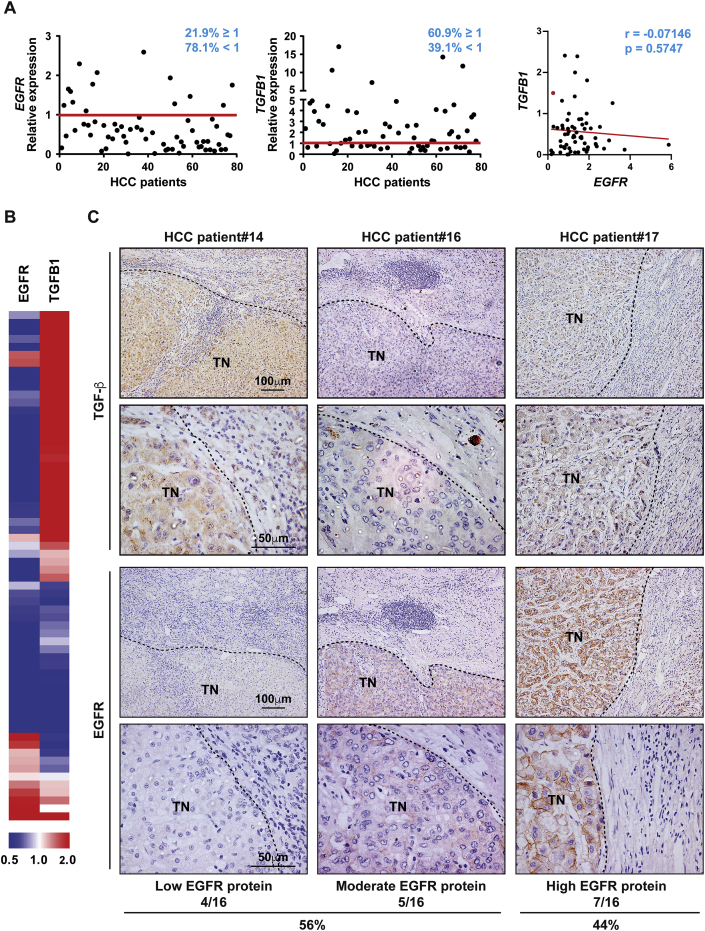
We first explored how expression levels of EGFR and TGF-β were distributed in tumour and non-tumour tissues from a cohort of 64 HCC patients (Supplementary Table IV). In spite of the heterogeneity among HCC tumours, most tumour tissues from HCC patients expressed low levels of EGFR while expressing high levels of TGFB1 (Fig. 1A and B). We observed that 78% of patients presented low EGFR mRNA levels (Fig. 1A). When correlation between EGFR and TGFB1 expression was analysed, we found a tendency (although not statistically significant) to lower levels of EGFR in patients with high TGFB1 expression. Nevertheless, we cannot exclude that the changes in the expression of these genes could be independent events. We extended the analysis to a higher number of HCC patients using Mas Liver database (n = 115) from Oncomine (https://www.oncomine.org/). This analysis revealed decreased EGFR expression in HCC compared to normal liver or cirrhotic liver. On the other hand, high TGFB1 expression was significantly increased from a cirrhotic stage compared to normal liver (Supplementary Fig. S1). Furthermore, using the Human Protein Atlas and TCGA (The Cancer Genome Atlas) dataset for pan-cancer RNA expression of EGFR, we observed that liver cancer is amongst the cancers harbouring lower EGFR levels (Supplementary Fig. S2). Immunohistochemical analysis in our patient cohort revealed that most HCC tissues presented high TGF-β protein levels in tumour cells, as well as in the surrounding tissue (Fig. 1C), concomitant with pSMAD2 staining (Supplementary Fig. S3), confirming activation of the pathway. Several patterns of EGFR protein expression were found in tissues from HCC patients with 56% of them presenting either low or moderate EGFR protein expression (Fig. 1C). These data show that low EGFR concomitant with high TGF-β expression is a common event in HCC patients.
Fig. 1.
Tissues from HCC patients express low levels of EGFR concomitant with high levels of TGF-β. A)EGFR and TGFB1 expression analysed by qRT-PCR in a cohort of 64 HCC patients, where relative expression of each HCC tumour tissue versus its respective surrounding tissue was calculated and represented as % (cut-off ≥ 1) (left and middle). Linear correlation analysis between EGFR and TGFB1 tumour expression in the same cohort of HCC patients. Each dot represents relative expression of each HCC tumour tissue (right). B) Representation of EGFR and TGFB1 expression on a heatmap of the same cohort of HCC patients. C) Immunohistochemistry of TGF-β and EGFR in tissues from the same cohort of HCC patients. Representative 10× and 40× images are shown. Abbreviation: TN, tumour nodule.
3.2. EGFR silencing induces cellular changes in HCC that are amplified in the presence of TGF-β
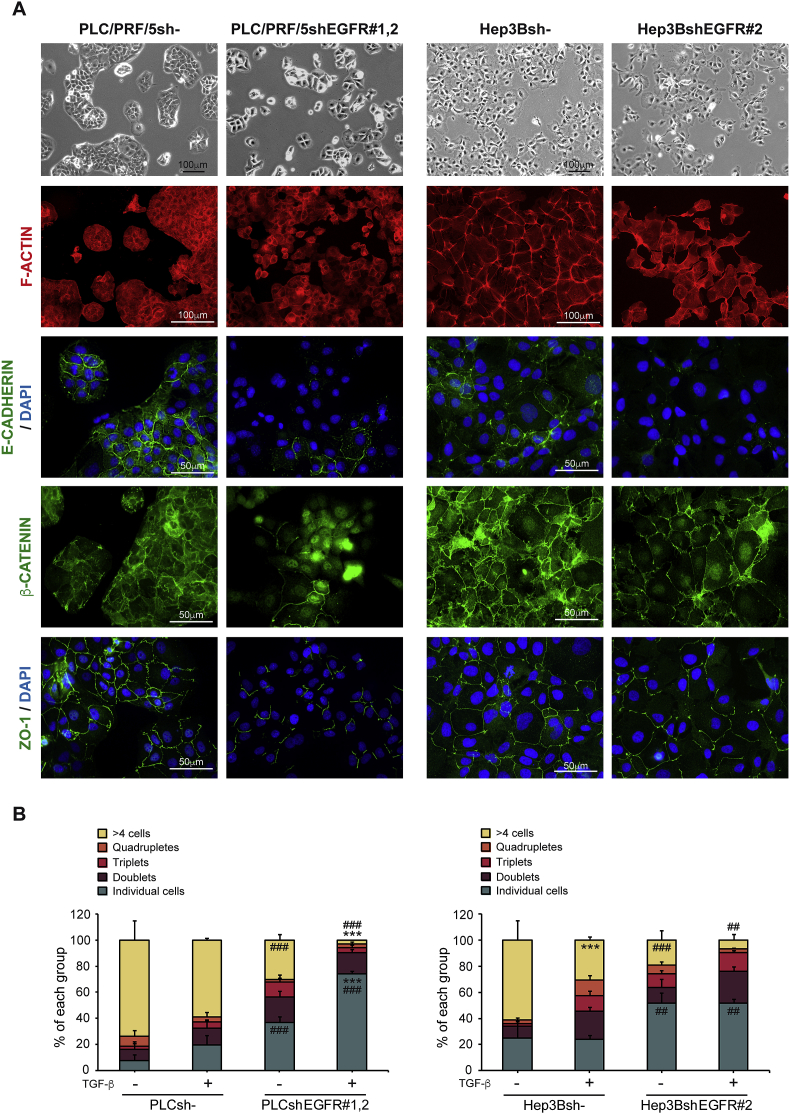
To further understand how EGFR could regulate cellular responses to TGF-β, two HCC cell lines were used (Supplementary Table V). PLC/PRF/5 cell line was chosen as a model of epithelial phenotype and well-defined parenchymal areas (known as clusters), while Hep3B cell line shows a mixed epithelial and mesenchymal behaviour, and a more relaxed parenchymal structure (Supplementary Fig. S4). shRNA against EGFR resulted in decreased response to EGF treatment (Supplementary Fig. S5) and profound changes in the parenchymal structures (Fig. 2A and Supplementary Fig. S6A). This was paralleled by dissolution of cell-cell contacts, measured by loss of junctional E-CADHERIN, β-CATENIN nuclear translocation and a decreased ZO-1 at tight junctions (Fig. 2A and Supplementary Fig. S6A). Furthermore, silencing of EGFR reduced number of cells in clusters, increasing the number of single cells, a phenomenon that was potentiated upon TGF-β treatment (Fig. 2B and Supplementary Fig. S6B). These data suggest that EGFR plays a role in the organization of parenchymal structures including how E-CADHERIN is recruited to cell-cell junctions.
Fig. 2.
Effect of EGFR silencing on cell-cell contacts in HCC cells. Unsilenced and EGFR silenced PLC/PRF/5 and Hep3B cells were cultured on plastic. A) Phase contrast microscopy photographs, and immunostaining of F-ACTIN (red), E-CADHERIN, β-CATENIN and ZO-1 (green), and DAPI (blue: nuclei) in both HCC cell lines. Representative 20× and 40× images of one of the clones of each cell line are shown. B) Quantification of the number of cells per group of PLC/PRF/5 and Hep3B cells treated or not with TGF-β (2 ng/mL) during 72 h. Data are mean ± SEM of 3 independent experiments, and ≥5 fields per condition were quantified. Two-way ANOVA was used: ***p < 0.001 compared to control untreated cells in each cell line (unsilenced or silenced); ##p < 0.01 and ###p < 0.001 compared to unsilenced cells in each condition (untreated or treated).
3.3. EGFR silencing promotes a decrease in cell-to-matrix adhesion and increased TGF-β-induced migration
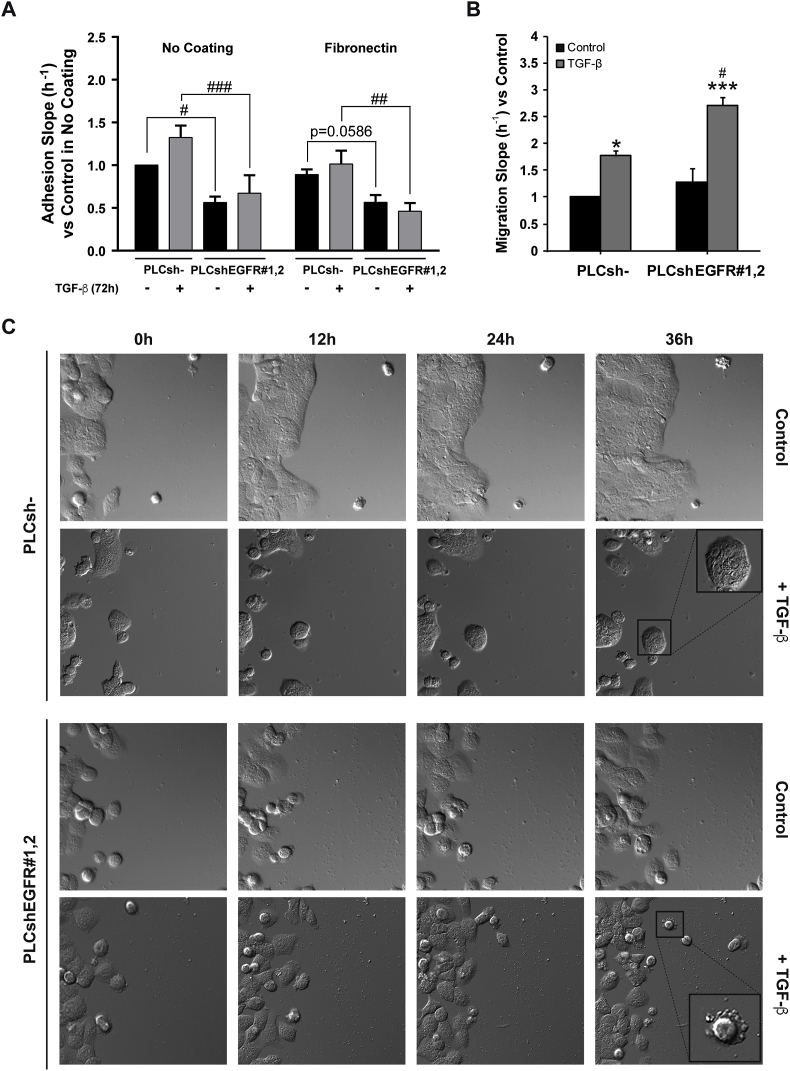
We next explored how EGFR silencing affected cell-to-matrix adhesion. Using xCELLigence System, we observed that EGFR silencing induced a significant decrease in the adhesive capacity of HCC cells plated either in no-coated or in fibronectin-coated plates (Fig. 3A). PLC/PRF/5 cells treated with TGF-β displayed changes in focal adhesion complex distribution, as measured by VINCULIN immunofluorescence (Supplementary Fig. S7). Indeed, cells separating from the cluster structure reorganized focal adhesions that were now localised at the cell edge (Supplementary Fig. S7). Interestingly, EGFR silencing altered vinculin staining and upon EGFR silencing, TGF-β did not promote change in the distribution of focal adhesions (Supplementary Fig. S7). Similar results were observed in Hep3B cells (Supplementary Fig. S8). These observations were confirmed using fibronectin-coated plates (Supplementary Fig. S9). Overall, these results suggest that EGFR silencing reduces cell-to-matrix adhesion and modifies TGF-β regulation of focal adhesions.
Fig. 3.
Effect of EGFR silencing on cell-to-matrix adhesion and migration capacity in HCC cells, untreated or treated with TGF-β. A) Unsilenced and EGFR silenced PLC/PRF/5 cells were treated with TGF-β (2 ng/mL) during 68 h, then trypsinized and plated in the xCELLigence system for a real-time adhesion assay. Adhesion was then assessed during 4 h, and it is expressed as relative to PLC/PRF/5 unsilenced and untreated cells in no coating and fibronectin coating conditions. B) Unsilenced and EGFR silenced PLC/PRF/5 cells were treated with TGF-β (2 ng/mL) during 64 h, then trypsinized and plated in the xCELLigence system for a real-time migration assay. Migration was then assessed during 8 h, and it is expressed as relative to PLC/PRF/5 unsilenced and untreated cells. Data are mean ± SEM of 3 independent experiments performed in biological duplicates in A and in biological quadruplicates in B. Two-way ANOVA was used: *p < 0.05 and ***p < 0.001 compared to control untreated cells in each cell line (unsilenced or silenced); #p < 0.05, ##p < 0.01 and ###p < 0.001 compared to unsilenced cells in each condition (untreated or treated). C) EGFR unsilenced and silenced PLC/PRF/5 cells were treated with TGF-β (2 ng/mL) and migration was assessed for 60 h in a 2D matrix. Representative 20× images of the first 36 h obtained from time-lapse migrating video microscopy analysis are shown.
We next tested if the effects in adhesion could impact the migratory capacity of these cells. Real-time migration assay showed that TGF-β treatment increased the migration of PLC/PRF/5 cells (Fig. 3B). However, after EGFR silencing, TGF-β conferred cells a significantly higher migratory capacity (Fig. 3B). Time-lapse video microscopy confirmed these results (Fig. 3C and Supplementary Videos 1–8). Importantly, we observed that increased migration was accompanied by blebbing activity in the migratory cells, especially in PLC/PRF/5 cells.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.canlet.2019.08.011.
The following are the supplementary data related to this article:
Therefore, these data suggest that EGFR attenuation enhances TGF-β-induced effects in HCC cell migration, promoting amoeboid cell migration.
3.4. Effect of EGFR silencing on the epithelial to mesenchymal transition genes in HCC cells
EMT genes have been linked to migration and invasion induced by TGF-β in HCC [11,12]. We next analysed EMT-gene expression changes. Consistent with our previous data [10], PLC/PRF/5 cells did not undergo a full EMT in response to TGF-β (Supplementary Fig. S10). Despite increased expression of VIM and CDH2, they retained CDH1 levels. After TGF-β treatment of PLC/PRF/5 cells, no differences in EMT-transcription factors were detected except for ZEB2, which could be driving the pro-migratory program (Supplementary Fig. S10). Interestingly, EGFR silencing, regardless of TGF-β treatment, dramatically decreased CDH1 mRNA levels, consistent with our previous observations (Fig. 2), but also decreased CDH2 levels (Supplementary Fig. S10). Thus, silencing EGFR allows TGF-β to act under conditions where cell-cell adhesions are reduced. On the other hand, Hep3B cells undergo a full EMT after TGF-β treatment and EGFR silencing did not interfere with the full EMT program (Supplementary Fig. S10).
These data indicate that EMT genes are induced to different extents after TGF-β stimulation but a full-EMT gene induction is not strictly needed for TGF-β to exert pro-migratory effects in HCC.
3.5. After EGFR loss, TGF-β activates Myosin II driven amoeboid invasion
We have previously reported that PLC/PRF/5 cells are able to undergo epithelial to amoeboid transition [28], concomitant with loss in cell-to-matrix adhesion, leading to amoeboid efficient invasion [29]. We could already observe amoeboid-like migration in 2D-substrates (Fig. 3C). Since amoeboid behaviour is favoured in pliable environments, we next used complex collagen I matrices to recapitulate the liver cancer microenvironment. Similar to 2D culture, the number of cells per group decreased in HCC cells cultured on collagen I upon TGF-β treatment. EGFR silencing promoted parenchymal disruption, amplifying TGF-β effects (Supplementary Figs. 11A and B). Importantly, in these matrices the parenchymal structures were much smaller when compared to the experiments in 2D (Fig. 2B), indicating that a collagen matrix induces loss of cell-cell contacts and gain of cell-to-matrix contacts, promoting individual migratory phenotypes.
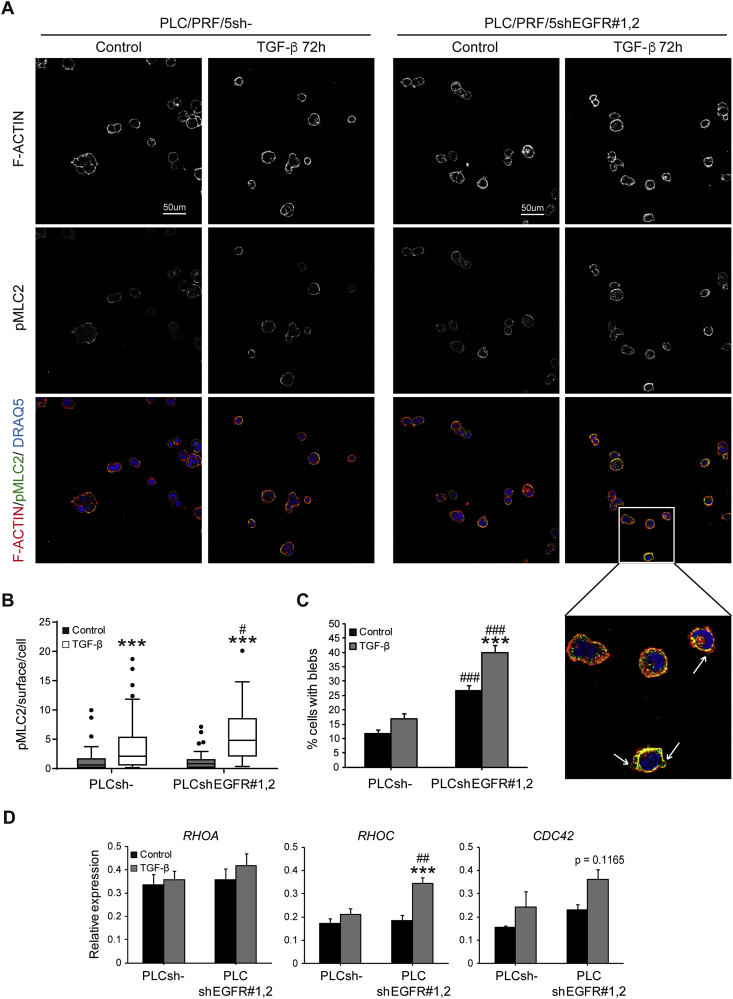
Amoeboid cell migration is characterized by an actin rich cortex, high levels of Myosin II in the cortex and blebs as functional protrusions [13]. TGF-β treatment in PLC/PRF/5 led to significantly higher Myosin II levels although had modest effects in bleb promotion (Fig. 4A–C). Importantly, EGFR silencing, combined with TGF-β treatment, induced most efficiently high Myosin II in the cell cortex, cell blebbing and amoeboid behaviour (Fig. 4A–C). Time-lapse video microscopy confirmed these observations (Supplementary videos 9–12).
Fig. 4.
Effect of EGFR silencing on actomyosin contractility in HCC cells after TGF-β treatment when cultured on top of a matrix of collagen I. Unsilenced and EGFR silenced PLC/PRF/5 cells were cultured on plastic and treated with TGF-β (2 ng/mL) during 48 h. Cells were then trypsinized and cultured on top of a bovine collagen I matrix for 24 h more, up to a total of 72 h with TGF-β treatment. A) Representative 40× confocal images of one stack of immunostaining of pMLC2 (green), F-ACTIN (red), and DRAQ5 (blue: nuclei) are shown. B) Quantification of pMLC2 immunostaining intensity per surface and cell. C) Quantification of the percentage of cells with blebs. D) qRT-PCR analysis of RHOA (RHOA), RHOC (RHOC) and CDC42 (CDC42) mRNA levels in unsilenced and EGFR silenced PLC/PRF/5 cells after 72 h of TGF-β treatment (2 ng/mL) when cells were cultured on plastic. Data in B and C are mean ± SEM of 3 independent experiments performed in duplicates and ≥5 fields per condition were quantified. Data in D are mean ± SEM of ≥3 independent experiments. Two-way ANOVA was used: ***p < 0.001 compared to control untreated cells in each cell line (unsilenced or silenced); #p < 0.05, ##p < 0.01 and ###p < 0.001 compared to unsilenced cells in each condition (untreated or treated).
Supplementary video related to this article can be found at https://doi.org/10.1016/j.canlet.2019.08.011.
The following are the supplementary data related to this article:
Rho GTPases are important for sustaining bleb based migration and Myosin II activation [15,18]. Interestingly, after EGFR silencing, PLC/PRF/5 cells treated with TGF-β significantly upregulated RHOC and a tendency was also observed in CDC42, both regulators of amoeboid migration (Fig. 4D). Similar results were obtained in Hep3B cells, where EGFR silencing allowed TGF-β to significantly increase Myosin II activity and the percentage of cells with blebs (Supplementary Figs. S12A–C). Notably, Hep3B cells treated with TGF-β upregulated RHOC and CDC42 regardless of EGFR status (Supplementary Fig. S12D).
In summary, attenuation of EGFR facilitates TGF-β-induced Myosin II activation, in part via regulation of Rho GTPases expression levels.
3.6. TGF-β and EGFR expression and distribution in tumours in vivo
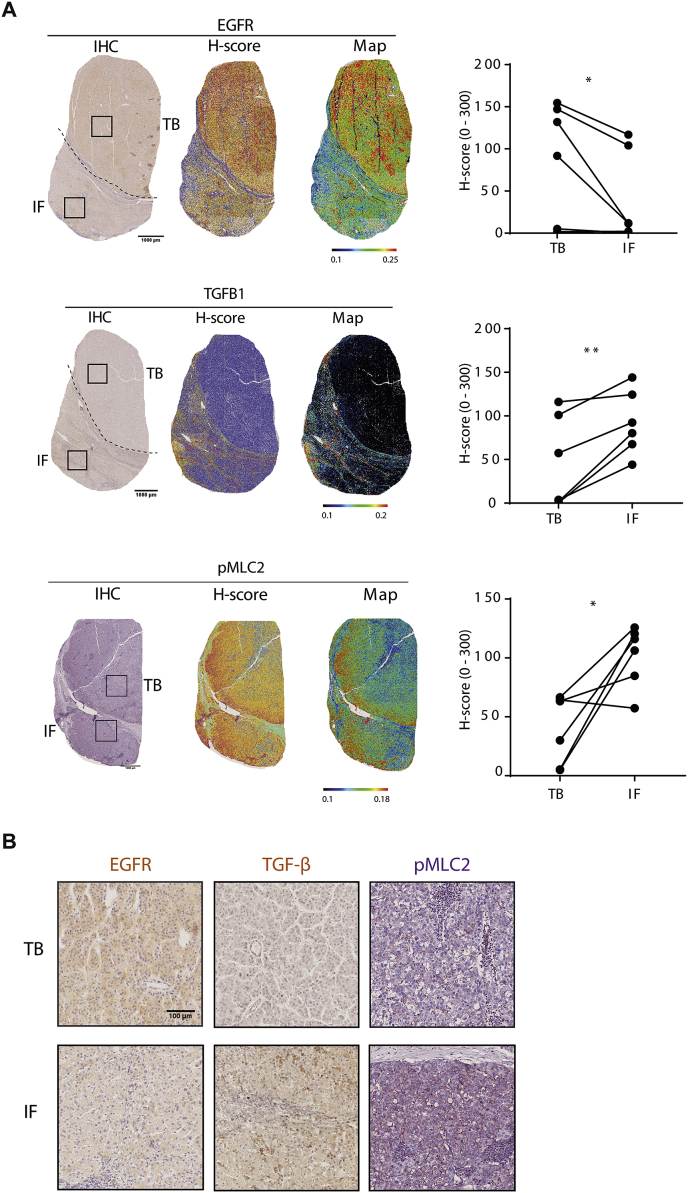
To validate our observations in the human clinical setting, we used digital pathology to assess EGFR, TGF-β expression levels and Myosin II activity in different regions of HCC tumours. We divided tumours in areas defined as tumour body (TB) and invasive front (IF), following similar criteria as in melanoma patients [30]. H-Scores showed how EGFR levels were downregulated in the invasive fronts of tumours while TGF-β and pMLC2 levels were clearly upregulated (Fig. 5). Interestingly, within a single tumour, EGFR and TGF-β levels may change across the full tumour and may present inverse positivity. This data highlights regional and heterogeneous expression levels of EGFR and TGF-β in HCC tumours and suggests that the invasive fronts are areas enriched in cells with high levels of TGF-β with reduced EGFR expression.
Fig. 5.
Immunohistochemistry (IHC) analysis and quantification for EGFR, TGF-β and pMLC2 markers in whole section samples. A) Each marker (EGFR, TGF-β or pMLC2) is represented using the IHC staining (left), its H-score (middle) representation and rainbow map (right) using digital pathology approach. Scale bar: 1000 μm. B) Representative images for EGFR, TGF-β or pMLC2 corresponding to the TB and IF areas highlighted in A. Scale bar: 100 μm. Graphs type: “before and after” using lines and dots to represent TB and IF matched for each patient. Student's Paired t-test was used: *p < 0.05, **p < 0.01. Abbreviations: TB, tumour body; IF, invasive front.
3.7. HCC patients with low EGFR/High TGFB1 expressions revealed poor prognosis
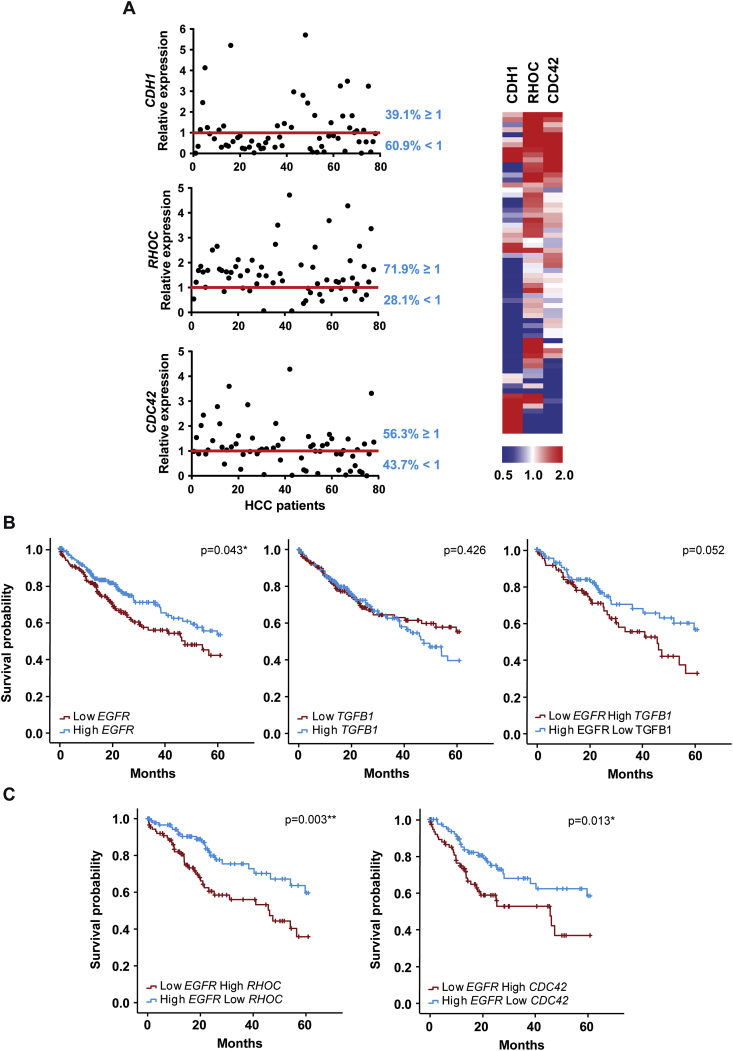
With the aim of exploring the relevance of decreased expression of EGFR in the molecular characteristics of the tumours, we performed transcriptomic analysis of our 64 patients' cohort. Data revealed that most tumour tissues from HCC patients presented low expression levels of CDH1 and very high levels of RHOC, while a moderate increase was found for CDC42 (Fig. 6A). Expression of EGFR inversely correlated with CDH1 (although not statistically significant, due to the number of patients analysed), but no correlation was found with RHOC or CDC42, whose changes in expression must depend on the combination with other factors (such as TGFB1) (Supplementary Fig. S13). Furthermore, using TCGA data we observed that patients with low expression of EGFR had significantly worse prognosis compared with patients that presented high expression of the receptor (n = 327 patients). However, patients expressing high levels of TGFB1 did not show significant worse prognosis 5 years after being diagnosed (Fig. 6B), as had been previously suggested [31]. Although the number of patients was not sufficient for a strong statistical analysis, a similar pattern was observed when this analysis was made in the 64 patients’ cohort from our hospital (Supplementary Fig. S14). Interestingly, the stratification of patients considering both EGFR and TGFB1 expressions indicated that low levels of EGFR and high levels TGFB1 conferred the worst prognosis (Fig. 6B and Supplementary Fig. S14). Moreover, patients with low EGFR and high RHOC or CDC42 levels had also significantly worse prognosis, as compared with patients with high EGFR and low Rho GTPases expressions (Fig. 6C and Supplementary Fig. S14A). Finally, due to the different histological expression pattern of EGFR in IF/TB (Fig. 5), we performed the survival analysis to compare the patients with high/low EGFR expression level in IF/TB. Results strongly suggested worse prognosis in patients with low IF/TB EGFR expression (Supplementary Fig. S14B). In tumour grade 3/4, a high percentage of tumours with high expression of TGFB1 or RHOC expressed low levels of EGFR (Supplementary Table VI). These findings reinforce the idea of loss in EGFR expression as a potential switch in the progression of HCC to a higher tumour grade and, thus, a worst prognosis for HCC patients.
Fig. 6.
Analysis of EGFR, TGFB1 and related genes expression in tissues from HCC patients shows prognosis significance. A) E-CADHERIN (CDH1), RHOC (RHOC) and CDC42 (CDC42) expression analysed by qRT-PCR where relative expression of each HCC tumour tissue versus its respective surrounding tissue was calculated and represented as % (cut-off ≥ 1). Representation of EGFR and TGFB1 expression on a heatmap of the same cohort of HCC patients is on the right. Kaplan-Meier estimation of 5 years survival in The Cancer Genome Atlas (TCGA) of HCC patients according to EGFR expression, TGFB1 expression and the combination of both (B), as well as according to EGFR expression combined with RHOC and CDC42 expression (C) (N = 327 HCC patients). Kaplan–Meier method using the log-rank test was used: *p < 0.05 and **p < 0.01.
Altogether, our results propose an unexpected role for EGFR loss that can contribute to HCC progression especially when TGF-β/RhoC/Cdc42 levels are increased.
4. Discussion
Hepatocellular carcinoma treatment is challenging, as the mechanisms underlying tumour progression are still largely unknown. Overexpression of EGFR occurring in some human cancers has been reported in the literature, correlating with more aggressive tumours, metastasis and poor patient survival [32]. However, in HCC, clinical studies with EGFR inhibitors have so far shown only modest results [22,23]. Indeed, further studies are needed to improve the molecular understanding of the EGFR-induced signalling pathways that may control HCC development and progression. Results shown here try to shed light on it, showing that EGFR expression is low and heterogeneous in a great percentage of HCC patients and suggesting that EGFR loss facilitates some of TGF-β pro-invasive and metastatic functions.
To spread within tissues, tumour cells use migration mechanisms that are similar to those that occur in normal cells during physiological processes. Multiple environmental factors, such as chemokines, cytokines like TGF-β, and growth factors like EGF, can induce and regulate tumour-cell motility, thereby contributing to invasion. However, depending on the cell type and tissue environment, cells can migrate individually, when cell-cell junctions are absent, or collectively as strands, sheets or clusters, when cell-cell adhesions are retained [13]. Although TGF-β is well known to promote EMT and contribute to metastasis in this way [11,12,33], our results demonstrate that HCC cells also respond to TGF-β inducing an epithelial to amoeboid transition after silencing EGFR. In melanoma, a non-epithelial tumour, TGF-β induces amoeboid migration [30]. Melanocytes derive from neural crest - a multipotent cell population arising from ectoderm - that undergoes EMT during development. Interestingly, here we observe that melanoma is also a tumour with nearly absent EGFR expression, which could indicate similar regulatory mechanisms in these two tumour types that have very different developmental origin.
Here we show that EGFR is implicated in the maintenance of cell-cell contacts and parenchymal structures between HCC cells. Indeed, a disruption of cell-cell adhesion is observed after EGFR silencing. After that, TGF-β can better access individual cells and increase their migratory potential. There is evidence in the literature supporting a role for EGFR in cell-cell junction regulation. As such, EGFR co-precipitated with E-cadherin in keratinocytes when cell-cell junctions were formed and - in the absence of EGFR - these cell-cell contacts are abolished [34]. Moreover, E-cadherin is responsible of integrating mechano-transduction and EGFR signalling in epithelial barriers, controlling adhesion and cortical contractility through the localization and activation status of EGFR [35]. Results presented here reveal that when reducing EGFR levels, TGF-β treatment was more efficient in increasing Myosin II activity and membrane blebbing, characteristics of amoeboid-type of migration. In this scenario, Myosin II is no longer restricted to cell-cell junctions that have been lost, but rather has re-localised to the cell cortex. Interestingly, we observed amoeboid migration even in a 2D environment by time-lapse microscopy, highlighting the strong effects induced by TGF-β treatment upon EGFR silencing in the more parenchymal cells. It is known that rounded-amoeboid type of movement is driven by high Myosin II levels regulated by Rho signalling (RhoA and RhoC) [20] or Cdc42 [18]. In fact, we recently described that HCC cells undergo an epithelial to amoeboid transition when silencing NOX4, a NADPH oxidase involved in many processes such as adhesion and proliferation [28]. RHOC and CDC42 levels were increased in NOX4 silenced cells, similar to what we observe after EGFR silencing and TGF-β-treatment. Interestingly, RhoC has also been described as a key gene upregulated in metastatic melanoma [36], pointing at further homologies between these two tumour types. RhoC is a key regulator of Myosin II dependent actomyosin contractility, suggesting that its high expression levels is key for regulating Myosin II in HCC. Furthermore, high Myosin II levels coupled to lower adhesion are key for fast amoeboid migration [37]. These features are recapitulated in the work we present herein.
The role of ErbB family in inducing a mesenchymal migratory phenotype might be tumour and context dependent. It was proposed that EGFR favours EMT [[38], [39], [40]], but some reports indicate that targeting EGFR in carcinomas may promote an infiltrative invasion front composed of mesenchymal-like cells [41]. Supporting our observations here, EGF contributes to the final acquisition of an epithelial, mature, phenotype in foetal hepatocytes in culture [42]. In the same line of evidence, due to their effects on cell-cell adhesion, EGFR mediates collective migration in the border cells during Drosophila oogenesis to read guidance cues secreted by the oocyte [43].
Our observations in HCC patients show that low EGFR and high TGFB1 expression have prognostic value. This could indicate that in such patients TGF-β has acquired pro-tumorigenic signalling. Importantly, we report that a relevant percentage of HCC patients expressed higher levels of RHOC and CDC42 within the tumour when compared with non-tumour areas. More importantly, low expression of EGFR combined with high expression of either RHOC or CDC42 is associated with worse prognosis in HCC patients. In accordance, we have previously shown that RHOC and CDC42 are significantly increased in HCC metastasis compared to primary lesions [28]. Interestingly, our results also indicate that low IF/TB EGFR expression at the histological level also suggests worse prognosis.
EGFR plays a hepatoprotective role in chronic liver diseases [22,44,45]. Here we propose that EGFR could also play a tumour suppressor role in advanced stages of HCC. In line with this evidence, deletion of EGFR in hepatocytes led to increased hepatocarcinogenesis [44]. Lower expression of EGFR in the tumor cells would increase the accessibility of liver macrophages to EGFR ligands, which further contributes to increased TGF-β production and liver tumor progression. Interestingly, another member of the receptor family: ERBB4, was also proposed to suppress development of HCC and has prognostic value in patients [46]. Further work will be necessary to better understand how EGFR expression is down-regulated in liver tumor cells. Analysis of response elements present in the EGFR promoter (from Gene Cards Human Gene Data Base) indicated potential binding of Hepatocyte Nuclear Factor 4-alpha (HNF4A), which plays essential roles during liver development. Its expression is maximal in differentiated hepatocytes, but decays in HCC [47]. It is tempting to speculate that transcription factors, such as HNF4A, involved in the differentiation of hepatocytes, could potentially control the expression of EGFR that regulates hepatocyte final differentiation [42,48].
Our work also highlights the relevance of the epithelial to amoeboid transition in human tumours and the need to better target this process in the clinic. Up to date, there are limited reports showing how HCC undergoes epithelial to amoeboid or mesenchymal to amoeboid transitions, due to lack of physiologically relevant experimental settings. Of note, one study showed that depletion of HAb18G/CD147 induced amoeboid migration in HCC [49]. Interestingly, anti-CD147 therapy in tumours resulted in EGFR decreased protein levels [50]. Overall, data here emphasize the importance of patient stratification for HCC treatment. We propose that TGFB1, RHOC or CDC42 expression levels should be assessed before using anti-EGFR therapy, as it can be acting as a tumour suppressor, especially if it is concomitant with a high TGFB1 expression. In this line, a large subset of HCC patients with low EGFR and high TGFB1 levels would benefit from targeting either the TGF-β or the Rho-ROCK-Myosin II pathway.
Authorship agreement and conflict of interest disclosure
All co-authors agree to the submission to Cancer Letters and agree with the content and presentation of the paper. Declarations of interest: none.
Funding
This work was supported by 1) the Ministry of Science, Innovation and Universities, Spain: SAF2015-64149-R and RTI2018-094079-B-100 to I.F. (cofounded by FEDER funds/Development Fund-a way to build Europe); 2) National Biomedical Research Institute on Liver and Gastrointestinal Diseases: CIBEREHD-CB17/04/00017 to I.F. (funded by Instituto de Salud Carlos III, Spain); 3) People Programme (Marie Curie Actions) of the FP7-2013, under REA grant agreement #PITN-GA-2012-316549 (IT-LIVER) to I.F. and A.M.; 4) Cancer Research UK (CRUK): C33043/A12065 and C33043/A24478 and Barts Charity to V.S-M, O.M. and E.C-M.; 5) Royal Society RG110591 to V.S-M.; 6) Fundación Ramón Areces to E.C-M.
CRediT authorship contribution statement
Judit López-Luque: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Visualization, Project administration. Esther Bertran: Conceptualization, Methodology, Supervision. Eva Crosas-Molist: Software, Formal analysis, Writing - review & editing. Oscar Maiques: Methodology, Software. Andrea Malfettone: Methodology. Laia Caja: Methodology, Writing - review & editing. Teresa Serrano: Resources, Supervision. Emilio Ramos: Resources, Supervision. Victoria Sanz-Moreno: Writing - review & editing, Supervision, Conceptualization, Funding acquisition. Isabel Fabregat: Conceptualization, Validation, Formal analysis, Writing - original draft, Writing - review & editing, Supervision, Project administration, Funding acquisition, Visualization.
Acknowledgements
We thank CERCA Programme/Generalitat de Catalunya for institutional support. We acknowledge technical support of Dr. Benjamín Torrejón (Scientific and Technical Services, University of Barcelona, CCiTUB) and Dr. Carme Casal (Optical Microscopy Facility, Center of Regenerative Medicine in Barcelona, CMRB).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.canlet.2019.08.011.
Contributor Information
Judit López-Luque, Email: jlopezl@idibell.cat.
Esther Bertran, Email: ebertran@idibell.cat.
Eva Crosas-Molist, Email: e.crosas-molist@qmul.ac.uk.
Oscar Maiques, Email: o.m.carlos@qmul.ac.uk.
Andrea Malfettone, Email: a.malfettone@gmail.com.
Laia Caja, Email: laia.caja@imbim.uu.se.
Teresa Serrano, Email: teresasp2016@icloud.com.
Emilio Ramos, Email: eramos@bellvitgehospital.cat.
Victoria Sanz-Moreno, Email: v.sanz-moreno@qmul.ac.uk.
Isabel Fabregat, Email: ifabregat@idibell.cat.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Llovet J.M., Zucman-Rossi J., Pikarsky E., Sangro B., Schwartz M., Sherman M. Hepatocellular carcinoma. Nat Rev Dis Primer. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 2.Carr B.I., Hayashi I., Branum E.L., Moses H.L. Inhibition of DNA synthesis in rat hepatocytes by platelet-derived type beta transforming growth factor. Cancer Res. 1986;46(5):2330–2334. [PubMed] [Google Scholar]
- 3.Sánchez A., Alvarez A.M., Benito M., Fabregat I. Apoptosis induced by transforming growth factor-beta in fetal hepatocyte primary cultures: involvement of reactive oxygen intermediates. J. Biol. Chem. 1996;271(13):7416–7422. doi: 10.1074/jbc.271.13.7416. [DOI] [PubMed] [Google Scholar]
- 4.Moustakas A., Heldin C.-H. Induction of epithelial-mesenchymal transition by transforming growth factor β. Semin. Cancer Biol. 2012;22(5–6):446–454. doi: 10.1016/j.semcancer.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Scaltriti M., Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin. Cancer Res. 2006;12(18):5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 6.Caja L., Sancho P., Bertran E., Fabregat I. Dissecting the effect of targeting the epidermal growth factor receptor on TGF-β-induced-apoptosis in human hepatocellular carcinoma cells. J. Hepatol. 2011;55(2):351–358. doi: 10.1016/j.jhep.2010.10.041. [DOI] [PubMed] [Google Scholar]
- 7.Murillo M.M., del Castillo G., Sánchez A., Fernández M., Fabregat I. Involvement of EGF receptor and c-Src in the survival signals induced by TGF-beta1 in hepatocytes. Oncogene. 2005;24(28):4580–4587. doi: 10.1038/sj.onc.1208664. [DOI] [PubMed] [Google Scholar]
- 8.Caja L., Bertran E., Campbell J., Fausto N., Fabregat I. The transforming growth factor-beta (TGF-β) mediates acquisition of a mesenchymal stem cell-like phenotype in human liver cells. J. Cell. Physiol. 2011;226(5):1214–1223. doi: 10.1002/jcp.22439. [DOI] [PubMed] [Google Scholar]
- 9.Fernando J., Malfettone A., Cepeda E.B., Vilarrasa-Blasi R., Bertran E., Raimondi G. A mesenchymal-like phenotype and expression of CD44 predict lack of apoptotic response to sorafenib in liver tumor cells. Int. J. Cancer. 2015;136(4):E161–E172. doi: 10.1002/ijc.29097. [DOI] [PubMed] [Google Scholar]
- 10.Malfettone A., Soukupova J., Bertran E., Crosas-Molist E., Lastra R., Fernando J. Transforming growth factor-β-induced plasticity causes a migratory stemness phenotype in hepatocellular carcinoma. Cancer Lett. 2017;392:39–50. doi: 10.1016/j.canlet.2017.01.037. [DOI] [PubMed] [Google Scholar]
- 11.Franco D.L., Mainez J., Vega S., Sancho P., Murillo M.M., de Frutos C.A. Snail1 suppresses TGF-beta-induced apoptosis and is sufficient to trigger EMT in hepatocytes. J. Cell Sci. 2010;123(Pt 20):3467–3477. doi: 10.1242/jcs.068692. [DOI] [PubMed] [Google Scholar]
- 12.Bertran E., Crosas-Molist E., Sancho P., Caja L., Lopez-Luque J., Navarro E. Overactivation of the TGF-β pathway confers a mesenchymal-like phenotype and CXCR4-dependent migratory properties to liver tumor cells. Hepatology. 2013;58(6):2032–2044. doi: 10.1002/hep.26597. [DOI] [PubMed] [Google Scholar]
- 13.Friedl P., Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer. 2003;3(5):362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 14.Pandya P., Orgaz J.L., Sanz-Moreno V. Modes of invasion during tumour dissemination. Mol Oncol. 2017;11(1):5–27. doi: 10.1002/1878-0261.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanz-Moreno V., Gadea G., Ahn J., Paterson H., Marra P., Pinner S. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135(3):510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 16.Hu S., Dasbiswas K., Guo Z., Tee Y.-H., Thiagarajan V., Hersen P. Long-range self-organization of cytoskeletal myosin II filament stacks. Nat. Cell Biol. 2017;19(2):133–141. doi: 10.1038/ncb3466. [DOI] [PubMed] [Google Scholar]
- 17.Sahai E., Marshall C.J. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 2003;5(8):711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 18.Gadea G., Sanz-Moreno V., Self A., Godi A., Marshall C.J. DOCK10-mediated Cdc42 activation is necessary for amoeboid invasion of melanoma cells. Curr. Biol. 2008;18(19):1456–1465. doi: 10.1016/j.cub.2008.08.053. [DOI] [PubMed] [Google Scholar]
- 19.Ito M., Nakano T., Erdodi F., Hartshorne D.J. Myosin phosphatase: structure, regulation and function. Mol. Cell. Biochem. 2004;259(1–2):197–209. doi: 10.1023/b:mcbi.0000021373.14288.00. [DOI] [PubMed] [Google Scholar]
- 20.Sanz-Moreno V., Marshall C.J. The plasticity of cytoskeletal dynamics underlying neoplastic cell migration. Curr. Opin. Cell Biol. 2010;22(5):690–696. doi: 10.1016/j.ceb.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 21.Giannelli G., Koudelkova P., Dituri F., Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J. Hepatol. 2016;65(4):798–808. doi: 10.1016/j.jhep.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Berasain C., Avila M.A. The EGFR signalling system in the liver: from hepatoprotection to hepatocarcinogenesis. J. Gastroenterol. 2014;49(1):9–23. doi: 10.1007/s00535-013-0907-x. [DOI] [PubMed] [Google Scholar]
- 23.Komposch K., Sibilia M. EGFR signaling in liver diseases. Int. J. Mol. Sci. 2015;17(1) doi: 10.3390/ijms17010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu A.X., Stuart K., Blaszkowsky L.S., Muzikansky A., Reitberg D.P., Clark J.W. Phase 2 study of cetuximab in patients with advanced hepatocellular carcinoma. Cancer. 2007;110(3):581–589. doi: 10.1002/cncr.22829. [DOI] [PubMed] [Google Scholar]
- 25.O'Dwyer P.J., Giantonio B.J., Levy D.E., Kauh J.S., Fitzgerald D.B., Benson A.B. Gefitinib in advanced unresectable hepatocellular carcinoma: results from the Eastern cooperative oncology group's study E1203. J. Clin. Oncol. 2006;24(18_suppl) 4143–4143. [Google Scholar]
- 26.Philip P.A., Mahoney M.R., Allmer C., Thomas J., Pitot H.C., Kim G. Phase II study of Erlotinib (OSI-774) in patients with advanced hepatocellular cancer. J. Clin. Oncol. 2005;23(27):6657–6663. doi: 10.1200/JCO.2005.14.696. [DOI] [PubMed] [Google Scholar]
- 27.López-Luque J., Caballero-Díaz D., Martinez-Palacián A., Roncero C., Moreno-Càceres J., García-Bravo M. Dissecting the role of epidermal growth factor receptor catalytic activity during liver regeneration and hepatocarcinogenesis. Hepatology. 2016;63(2):604–619. doi: 10.1002/hep.28134. [DOI] [PubMed] [Google Scholar]
- 28.Crosas-Molist E., Bertran E., Rodriguez-Hernandez I., Herraiz C., Cantelli G., Fabra À. The NADPH oxidase NOX4 represses epithelial to amoeboid transition and efficient tumour dissemination. Oncogene. 2017;36(21):3002–3014. doi: 10.1038/onc.2016.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y.-J., Le Berre M., Lautenschlaeger F., Maiuri P., Callan-Jones A., Heuzé M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 2015;160(4):659–672. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Cantelli G., Orgaz J.L., Rodriguez-Hernandez I., Karagiannis P., Maiques O., Matias-Guiu X. TGF-β-Induced transcription sustains amoeboid melanoma migration and dissemination. Curr. Biol. 2015;25(22):2899–2914. doi: 10.1016/j.cub.2015.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coulouarn C., Factor V.M., Thorgeirsson S.S. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47(6):2059–2067. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hynes N.E., MacDonald G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009;21(2):177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 33.Fabregat I., Caballero-Díaz D. Transforming growth factor-β-induced cell plasticity in liver fibrosis and hepatocarcinogenesis. Front Oncol. 2018;8:357. doi: 10.3389/fonc.2018.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erasmus J.C., Welsh N.J., Braga V.M.M. Cooperation of distinct Rac-dependent pathways to stabilise E-cadherin adhesion. Cell. Signal. 2015;27(9):1905–1913. doi: 10.1016/j.cellsig.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rübsam M., Mertz A.F., Kubo A., Marg S., Jüngst C., Goranci-Buzhala G. E-cadherin integrates mechanotransduction and EGFR signaling to control junctional tissue polarization and tight junction positioning. Nat. Commun. 2017;8(1):1250. doi: 10.1038/s41467-017-01170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark E.A., Golub T.R., Lander E.S., Hynes R.O. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406(6795):532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 37.Friedl P., Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147(5):992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 38.Lorch J.H., Klessner J., Park J.K., Getsios S., Wu Y.L., Stack M.S. Epidermal growth factor receptor inhibition promotes desmosome assembly and strengthens intercellular adhesion in squamous cell carcinoma cells. J. Biol. Chem. 2004;279(35):37191–37200. doi: 10.1074/jbc.M405123200. [DOI] [PubMed] [Google Scholar]
- 39.Yue P., Zhang X., Paladino D., Sengupta B., Ahmad S., Holloway R.W. Hyperactive EGF receptor, Jaks and Stat3 signaling promote enhanced colony-forming ability, motility and migration of cisplatin-resistant ovarian cancer cells. Oncogene. 2012;31(18):2309–2322. doi: 10.1038/onc.2011.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S.V., Subramaniam D., Cyriac G.C., Abdul-Khalek F.J., Giaccone G. Emerging protein kinase inhibitors for non-small cell lung cancer. Expert Opin. Emerg. Drugs. 2014;19(1):51–65. doi: 10.1517/14728214.2014.873403. [DOI] [PubMed] [Google Scholar]
- 41.Basu D., Bewley A.F., Sperry S.M., Montone K.T., Gimotty P.A., Rasanen K. EGFR inhibition promotes an aggressive invasion pattern mediated by mesenchymal-like tumor cells within squamous cell carcinomas. Mol. Cancer Ther. 2013;12(10):2176–2186. doi: 10.1158/1535-7163.MCT-12-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sánchez A., Pagan R., Alvarez A.M., Roncero C., Vilaró S., Benito M. Transforming growth factor-beta (TGF-beta) and EGF promote cord-like structures that indicate terminal differentiation of fetal hepatocytes in primary culture. Exp. Cell Res. 1998;242(1):27–37. doi: 10.1006/excr.1998.4088. [DOI] [PubMed] [Google Scholar]
- 43.Inaki M., Vishnu S., Cliffe A., Rørth P. Effective guidance of collective migration based on differences in cell states. Proc. Natl. Acad. Sci. U. S. A. 2012;109(6):2027–2032. doi: 10.1073/pnas.1115260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanaya H., Natarajan A., Komposch K., Li L., Amberg N., Chen L. EGFR has a tumour-promoting role in liver macrophages during hepatocellular carcinoma formation. Nat. Cell Biol. 2014;16(10):972–977. doi: 10.1038/ncb3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santamaría E., Rodríguez-Ortigosa C.M., Uriarte I., Latasa M.U., Urtasun R., Alvarez-Sola G. The epidermal growth factor receptor ligand amphiregulin protects from cholestatic liver injury and regulates bile acids synthesis. Hepatology. 2019;69(4):1632–1647. doi: 10.1002/hep.30348. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y., Song L., Ni H., Sun L., Jiao W., Chen L. ERBB4 acts as a suppressor in the development of hepatocellular carcinoma. Carcinogenesis. 2017;38(4):465–473. doi: 10.1093/carcin/bgx017. [DOI] [PubMed] [Google Scholar]
- 47.Ishiyama T., Kano J., Minami Y., Iijima T., Morishita Y., Noguchi M. Expression of HNFs and C/EBP alpha is correlated with immunocytochemical differentiation of cell lines derived from human hepatocellular carcinomas, hepatoblastomas and immortalized hepatocytes. Cancer Sci. 2003;94(9):757–763. doi: 10.1111/j.1349-7006.2003.tb01515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gruppuso P.A., Curran T.R., Mead J.E., Fausto N., Oh W. Fetal growth factors as determinants of intrauterine hepatic growth. Diabetes. 1991;40(Suppl 2):51–55. doi: 10.2337/diab.40.2.s51. [DOI] [PubMed] [Google Scholar]
- 49.Zhao P., Zhang W., Wang S.-J., Yu X.-L., Tang J., Huang W. HAb18G/CD147 promotes cell motility by regulating annexin II-activated RhoA and Rac1 signaling pathways in hepatocellular carcinoma cells. Hepatology. 2011;54(6):2012–2024. doi: 10.1002/hep.24592. [DOI] [PubMed] [Google Scholar]
- 50.Frederick J.W., Sweeny L., Hartman Y., Zhou T., Rosenthal E.L. Epidermal growth factor receptor inhibition by anti-CD147 therapy in cutaneous squamous cell carcinoma. Head Neck. 2016;38(2):247–252. doi: 10.1002/hed.23885. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.