ABSTRACT
Paracrine and endocrine roles have increasingly been ascribed to extracellular vesicles (EVs) generated by multicellular organisms. Central to the biogenesis, content, and function of EVs are their delimiting lipid bilayer membranes. To evaluate research progress on membranes and EVs, the International Society for Extracellular Vesicles (ISEV) conducted a workshop in March 2018 in Baltimore, Maryland, USA, bringing together key opinion leaders and hands-on researchers who were selected on the basis of submitted applications. The workshop was accompanied by two scientific surveys and covered four broad topics: EV biogenesis and release; EV uptake and fusion; technologies and strategies used to study EV membranes; and EV transfer and functional assays. In this ISEV position paper, we synthesize the results of the workshop and the related surveys to outline important outstanding questions about EV membranes and describe areas of consensus. The workshop discussions and survey responses reveal that while much progress has been made in the field, there are still several concepts that divide opinion. Good consensus exists in some areas, including particular aspects of EV biogenesis, uptake and downstream signalling. Areas with little to no consensus include EV storage and stability, as well as whether and how EVs fuse with target cells. Further research is needed in these key areas, as a better understanding of membrane biology will contribute substantially towards advancing the field of extracellular vesicles.
KEYWORDS: Extracellular vesicles, Exosomes, membranes, ISEV workshop, position paper, biogenesis, uptake, fusion, technology
Introduction
Extracellular vesicles (EVs) are delimited by double-leaflet lipid membranes and released by cells of most, if not all, organisms. EVs remove waste components from the cell, can be used to share nutrition, and serve as cell-to-cell communicators by carrying and transferring bioactive enzymes and molecules and molecular information [1,2]. These mechanisms are important for normal homeostasis and regulatory functions, including development. EVs also play roles in disease processes, responding to and mediating inflammation and contributing to the development and progression of diseases [3–6]. These characteristics of EVs are also promising for theranostic applications [7–10], and for disease detection [11–18].
The generic term “EV” covers a broad range of vesicles [19], but it is not yet certain whether phenotypic heterogeneity is mirrored by functional heterogeneity. Two potential classes of EVs are determined by two major cellular sites of EV biogenesis [20,21]: the plasma membrane (PM) and the endosomal system. Although “exosome” was initially used for vesicles shed from the PM [22], the term was later adopted to specifically refer to intraluminal vesicles (ILVs) formed in the multivesicular body (MVB) that were released from the cell by fusion of the MVB with the PM [23–25]. In contrast, the terms “microvesicle,” “ectosome,” and “microparticle” are used to describe vesicles shed directly from the PM [26]. In addition to their biogenesis, EV subclasses may also be defined by size, shape, density, surface molecules, internal cargo, membrane components, cell type of origin, or function.
Membranes are central to the identity and function of EVs. Specific lipids and membrane proteins in EVs may be used to reveal the cell type or the subcellular site of origin [27–29]. Bioactive lipids and integral or otherwise membrane-associated proteins may directly engage signalling pathways of cells and influence target cell-specificity of some EV populations [8]. Membrane-associated proteins also appear to be involved in EV uptake into cellular compartments, such as the endosomal/lysosomal system [30,31]. The EV membrane protects internal contents, mainly derived from the parent cell cytosol that may be transferred to recipient cells if EV-cell fusion occurs. From an experimental perspective, to show that EVs are present in a preparation, one must demonstrate the presence of an intact lipid bilayer that encloses cytosolic material [32,33] and maintains its integrity.
In order to reconcile and stimulate discussion on membranes and EVs, the International Society for Extracellular Vesicles (ISEV) conducted two targeted surveys and convened a workshop in March 2018 (Baltimore, MD) to collect and synthesize the input of EV and membrane biology scientists. The goal of this process was to gather expert opinions and define questions for future research. Four specific topics of interest, corresponding to four sessions and roundtable discussions at the Workshop, were; 1) the roles of membranes in EV biogenesis and release; 2) membranes and EV uptake and fusion; 3) technologies and strategies used to study EVs and membranes; and 4) EV functional transfer and functional assays (Text box 1). An overarching goal was to learn how better to utilize existing technologies for the study of EV membranes, and to understand what novel techniques might be required. In addition to 55 junior and senior membrane biologists and engineers who contributed to the Workshop on-site, other ISEV scientists were in close communication with the organizers to ensure a balanced, interdisciplinary approach. This position paper was drafted to summarize current perceptions and opportunities as well as important questions in the study of EVs and their membranes. Areas of relative consensus are identified within the field, and areas where there is broader disagreement have also been highlighted. A number of specific recommendations are made for topics that require further study or technological development, which should help to focus and drive the field forward.
Text Box 1.
Roundtable topics, moderators, and descriptions.
|
Roundtable #1: Biogenesis and Release Moderators: Matias Ostrowski, Hang Yin, Roosmarijn Vandenbroucke |
| EV subpopulations are commonly defined by site or mechanism of biogenesis. Roundtable 1 aimed to identify outstanding questions about how various molecules and pathways influence formation and release of EVs. |
| Roundtable #2: Uptake and Fusion |
| Moderators: Pieter Vader, Jeanne Sisk, David Carter |
| After release, EVs may exert effects through autocrine, paracrine, or endocrine processes, all of which require interaction of EV membranes with target cells. This roundtable focused on current knowledge of EV-cell interactions, including uptake and fusion, and experimental approaches needed to dissect mechanisms. |
| Roundtable #3: Technologies and Strategies |
| Moderators: Marca Wauben, Paolo Bergese |
| Since unique technologies and approaches may be needed to study EVs and their membranes, this roundtable discussion sought to identify technologies, experimental methods, and models that have not yet been well applied to EV studies or should be further developed to enable more sophisticated analysis of EVs and membranes. |
| *Roundtable #4: Transfer and Functional Assays |
| Moderators: Jan Lötvall, Daniel Anthony |
| How EVs transfer cargo to recipient cells and how to assess effects of transfer were the considerations for this roundtable discussion. Also discussed were best practices for conducting and reporting EV studies (especially visualization), use of in vitro generated EVs for in vivo uptake studies, and the future of EV-based therapeutics. |
| *Because of substantial content overlap of Roundtable 4 with Roundtables 1–3, information from this roundtable has been integrated into other sections below. |
Membranes and EVs workshop pre- and post-surveys
An important part of the Workshop was gathering the opinion of experts who participated or were involved in the organization. Prior to the Workshop, a seven-question survey was circulated to planners and registrants to obtain opinions about the state of the field and identify outstanding questions (Table 1).
Table 1.
Workshop pre-survey questions.
| Pre-Workshop Survey Questions |
|---|
| What, in your opinion, are the top publications (up to 3) in the last five years that have addressed important questions of EV/membrane biology? |
| What are the most pressing current questions in EV biogenesis? (Up to 4) |
| What are the most pressing questions surrounding EV uptake and/or fusion? (Up to 4) |
| What important questions remain about EV component loading (natural or artificial) – this includes lipids, proteins, internal cargo? (Up to 4) |
| What are the most important unanswered questions about EV function as related to target cell interaction/uptake/fusion? (Up to 4) |
| To help answer outstanding EV/membrane questions, are there any technologies, methods, or models that have not yet been developed or fully applied? If so, what are they? Or what are the capabilities you would want? (Open response) |
| Do you have any position or opinion related to this workshop that you suspect some of your colleagues would disagree with? If so, what? (Up to 3) |
| Other comments or suggestions? (Open response) |
Following the Workshop, a 42-question survey was released to assess opinions post-Workshop using questions in a Likert-scale format: participants were asked to read a statement and rate on a scale of 0–10 whether they disagreed (0–4), agreed (6–10), or did not feel there was enough evidence to support or refute the statement (5). Participants were asked to refrain from entering responses for questions if they felt they lacked sufficient knowledge to answer. Three tables were constructed listing the survey questions pertaining to each overarching discussion topic; EV biogenesis, uptake, and technologies for studying EVs (Table 2–4).
Table 2.
Survey questions regarding EV biogenesis.
| EV Biogenesis Survey Questions | ||
|---|---|---|
| Figure 1 | Budding into the multivesicular body (MVB) as intraluminal vesicles (ILVs) and budding from the plasma membrane are the two major EV biogenesis pathways, with at least partially independent molecular machinery of biogenesis. | |
| It is possible that what we refer to as MVBs are actually physical extensions of the plasma membrane and not late endosomes. | ||
| It is currently possible to distinguish, using protein, lipid, or other markers, an “exosome” (former ILV in the MVB) from a “microvesicle” (from the plasma membrane) after the respective vesicle has left the cell. | ||
| Size can be used to separate EVs by biogenesis pathway. | ||
| EVs from the endosomal system are smaller, on average, than EVs that bud from the plasma membrane. | ||
| We know the basic size distribution of EVs from biofluids and cell culture. | ||
| Figure 2 | Excluding apoptotic bodies and other “macrovesicles”, the average diameter of most EV populations is: | Significantly smaller than 100 nm |
| Roughly in the 100–150 nm range | ||
| Significantly larger than 150 nm | ||
| Figure 3 | Asymmetric distribution of lipids (inner, outer leaflet) is the same in EVs as in the cell membrane of origin and remains stable over time. | |
| The inner and outer sides of the EV membrane are revealed by inner and outer domains of proteins in the expected orientation relative to the cell. That is, the cellular membrane topology is maintained by EVs. | ||
| Figure 4 | The weight of the evidence supports preferential packaging of certain miRNAs or other RNA cargo into specific subsets of EVs. | |
| The RNA Cargo of larger EVs correlates with cellular expression, but that of small EVs does not. | ||
| Figure 5 | It is possible to create cells or organisms that do not produce EVs. | |
| EV biogenesis is essential for life, as evidenced by lethality of TSG101 knockouts and knockouts of multiple biogenesis-linked proteins. | ||
| Figure 6 | Lipid-raft domains (endosome-like domains, rich in cholesterol, etc.) play a role in EV biogenesis; without them, many EVs would not form. | |
| nSMase2 is not involved in biogenesis of all EV subtypes in all cells, hence discrepant results of nSMase2 blocking. | ||
| Energetic requirements of EV biogenesis are largely unknown. | ||
Table 4.
Survey questions regarding current technologies for studying EVs.
| EV-Technology Survey Questions | ||
|---|---|---|
| Figure 12 | Different measurement technologies are biased to certain EV size ranges. | |
| Optical scattering methods of EV measurement such as nanoparticle tracking are not specific to EVs. | ||
| Lipid dyes form artefactual particles on their own and with non-EV materials; results of lipid dye experiments are unreliable unless one can effectively separate EVs and artefacts by flotation gradients. | ||
| Figure 13 | With which statement do you agree more? | High-resolution single EV analysis by flow cytometry is now possible for labs with access to a standard flow cytometer. |
| It remains necessary to have specialized equipment, reagents, and expertise to perform single EV flow analysis for EVs below about 500 nm in diameter. | ||
| Figure 14 | Fluorescence triggering in EV flow cytometry allows better resolution than scatter. | |
| Better generic dyes of EVs are needed for flow cytometry and other investigations. | ||
| Development of reagents such as single chain antibodies, aptamers, and less bulky fluorophores is needed to improve sensitivity of EV flow. | ||
| Figure 15 | It is currently possible to make artificial EVs that faithfully mimic genuine EVs | |
| It is currently possible to affect EV distribution to tissues by manipulating EV surface features. | ||
| New animal models and more relevant in vitro systems are needed to address questions about production and function of subsets of EVs. | ||
Shown in Table 2 are 16 questions focusing on the fundamentals of EV biogenesis, the ways in which EV sub-populations are identified, the influences of membrane composition on EV biogenesis, and EV cargo packaging mechanisms. Table 3 outlines 16 questions used to gauge participants’ views on EV uptake, fusion, and stability. Ten questions pertaining to the necessity of novel assay development and the future of EV engineering are shown in Table 4. A summary of the responses, along with specific recommendations that emerged from the Workshop survey and discussions, is presented in Table 5. The table indicates areas of consensus, broad agreement, non-consensus, and recommendations for future EV research.
Table 3.
Survey questions regarding EV uptake.
| EV Uptake Survey Questions | |||
|---|---|---|---|
| Figure 7 | Most cell types, sooner or later, internalize at least a proportion of stained EVs, seemingly regardless of the cells of origin. | ||
| EV-cell fusion is most likely to occur through endosomal uptake and acidification. | |||
| Proteins on the EV surface are required for most fusion events between EVs and cellular membranes. | |||
| EV-cell fusion events are actually quite rare in vivo, and may involve minority subpopulations of EVs and specific uptake pathways. | |||
| Current in vitro studies of uptake (anything involving 2D tissue culture plastic substrates) are not worthwhile as unrepresentative of in vivo biology. | |||
| Figure 8 | Rank the following from 1 (most likely) to 5 (least likely). Interacting with target cells, EVs exert effects by: |
Signalling through proteins displayed on the target cell surface or in the endosomal lumen | |
| Transferring functional proteins | |||
| Transferring functional lipids | |||
| Transferring functional RNA molecules | |||
| Serving as a form of nutrition/molecular recycling for the recipient cell | |||
| Figure 9 | Current technologies are adequate to measure both functional and physical stability of EVs. | ||
| Physical stability of EVs (defined here as the tendency to maintain vesicular form) is related to size. | |||
| Figure 10 | Regarding freeze-thaw of EVs: | All EVs are generally resistant to freeze-thaw damage | |
| Small EVs are generally resistant to freeze-thaw damage | |||
| EVs are damaged both physically and functionally | |||
| EVs are damaged functionally, but may show the same physical characteristics | |||
| We still don’t know enough to answer this question | |||
| Figure 11 | In vitro EV transfer experiments are highly time-dependent, and the relevance to timing/EV stability in vivo is often unclear. | ||
| Dose-response studies are essential in establishing any effect of EVs. | |||
| Most EVs in vivo are bioactive. | |||
| EVs in circulation (blood) are less likely to be bioactive and are cleared rapidly. | |||
| EVs are most likely to have a signalling function in tissue, i.e. locally. | |||
| Tumour-bearing mice accumulate more EVs in cancer tissue mostly because of vascular leakiness. | |||
| The apparently low rates of EV:cell fusion indicated by systems such as the Cre/lox stoplight system may reflect sensitivity or idiosyncrasies of the assay and not imply that fusion is really so rare. | |||
Table 5.
Summary of topics on which there is largely agreement, relative consensus, or clear lack of consensus; a set of specific recommendations are included.
| Consensus | Most agree | No consensus | Recommendations | |
|---|---|---|---|---|
| Biogenesis, size and content | Fusion of MVBs and budding from the PM are the main routes of EV biogenesis | It is not possible to distinguish microvesicles from exosomes using size, proteins, lipids or other markers, though most agree that MVs are on average larger | Most EV populations are <100 or 100-150nm | Researchers should consider the entire trafficking landscape of intracellular vesicular organelles which can directly, or indirectly affect EV biogenesis and secretion |
| EV biogenesis is essential for life | MVBs may actually be physical extensions of the plasma membrane and not late endosomes | Substantial additional work is required to identify specific markers of EV subtypes released across or within cell types | ||
| It is not possible to generate cells or animals that don’t produce any vesicles | RNA cargo of larger EVs shows better correlation with cellular expression compared to that of smaller EVs | Improved separation and characterization technologies are required for unbiased and accurate counting and sizing of EVs | ||
| The energetics of EV biogenesis are unknown | Asymmetry of lipids (in inner/outer leaflet) and proteins (orientation of proteins) is maintained relative to the cell | Reproducible in vitro assay systems which closely mimic the physiological context are needed to study EV cargo loading | ||
| Lipid rafts are important in EV biogenesis, and nSMase2 is not involved in the biogenesis of ALL EV subtypes | Unbiased genetic screens and small molecule modulator screens may be needed to resolve unappreciated and combinatorial contributions to EV biogenesis | |||
| There is some specific loading of cargo into specific subsets of EVs | The roles of various sphingomyelinases, ceramides, and lipid rafts in EV biogenesis requires further investigation | |||
| Transfer, uptake and biodistribution |
The apparent low rates of EV:cell fusion indicated by systems such as the Cre/lox stoplight system reflect the sensitivity and idiosyncrasies of the assay and do not imply that fusion is really so rare | Fusion is most likely to occur through endosomal uptake and acidification | EV:cell fusion events are rare, involving a minority subpopulation of EVs and specific uptake pathway | Further experiments are required to decipher the rules of cellular targeting and uptake by EVs |
| EV transfer experiments are time-dependent | Most cells eventually internalize at least some stained EVs | Greater understanding of how EVs induce their most important effects (direct interactions, transfer of different cargo etc) is needed | ||
| EVs accumulate in cancer tissue in xenografts because of vascular leakiness | EVs in vivo are bioactive; there is less consensus on whether EVs in circulation are bioactive, with most believing that EVs are most likely to have signalling functions locally within tissues | Serial or differential dosing may be necessary for in vivo studies aimed at understanding the function or biodistribution of EVs | ||
| Proteins on the EV are required for fusion | Improved methodology, including imaging and staining, is required for the study of EV biodistribution | |||
| The most significant interaction of EVs with cells is via signalling that occurs through proteins displayed on the target cell surface or in the endosomal lumen | There is a need for advanced animal models to study the physiological importance of EV-mediated cargo transfer between cells and tissue | |||
| It is possible to affect EV distribution to tissues by manipulating EV surface features | The field needs to establish guidelines for defining and/or concluding which EV subpopulations and associated cargo are involved in homeostatic maintenance and pathological responses | |||
| Even studies in 2D culture systems are worthwhile as a representation of at least some aspects of in vivo uptake | ||||
| Methodology | Dose-response studies are essential for establishing functions for EVs | Lipid dyes can form artefactual particles making results of experiments less reliable unless one can effectively separate these by flotation | It is possible to make EVs that faithfully mimic genuine EVs | Dose-dependency is a key factor to include in experimental design |
| Fluorescence triggering in EV flow cytometry allows better resolution than scatter | Free dye controls are needed for experiments using stained EVs | |||
| High-resolution single EV analysis by flow cytometry requires specialized equipment, reagents and expertise | Better generic EV dyes and tags are needed for flow cytometry and other studies | |||
| Optical scattering methods of EV measurement are not specific to EVs | There is a need for reagents such as single chain antibodies, aptamers and less bulky fluorophores to improve the sensitivity of EV detection in flow cytometry | |||
| Different measurement technologies are biased to certain EV size ranges | New animals and more relevant in vitro systems are needed to address questions about production and function of subsets of EVs | |||
| Improved and novel methodology is needed for single EV analysis | ||||
| Improved imaging techniques are needed for EV analysis at different levels of resolution | ||||
| Storage and stability | Current technologies are not adequate to measure both functional and physical stability of EVs | Stability of EVs is related to their size | More work is needed to understand the effects of storage on the stability and function of EVs | |
| Freeze-thawing affects the structure and/or functionality of EVs |
EV biogenesis and release
The Pre-Workshop survey returned many questions about biogenesis. Cell biology approaches have identified members of the endosomal sorting complex required for transport (ESCRT) pathway that contribute to EV biogenesis, but what are the requirements for these factors at different subcellular sites? Might molecular redundancies make it necessary to knock out or modulate multiple components of the biogenesis machinery to fully understand how EVs are formed? What about ESCRT-independent pathways? Do different biogenesis pathways give rise to vesicles with distinct functions, or conversely, does shared machinery at different locations (PM vs endosome) give rise to vesicles with comparable function? Another set of questions involves the molecular composition of released EVs. Do EVs released by a single cell have a diversity of form and function? What molecular associations are driven by proximity and random incorporation versus active loading, even energy-dependence? Do reports of specific incorporation of cargo molecules rely too much on non-physiologic experimental manipulations, or do they reflect physiologic realities? Some questions even addressed what might be called “limited” release, when some vesicles remain at or near the surface of the parent cell, held in place by tethering or adhesion proteins [34,35]. These questions portray a field that has clearly taken large strides forward, but in which ample opportunities remain for additional discovery.
Based on the survey, six overarching questions were formulated to guide the roundtable discussion during the workshop. Consensus positions and additional research needs are presented in Sections 3.1–3.6.
How are EVs formed, and what are their defining characteristics?
A long-held assumption is that EVs form chiefly at two subcellular sites: the PM (microvesicles, ectosomes, microparticles), and the endosomal system/MVB (i.e. exosomes). Almost all survey respondents agreed to some extent with this concept (94%, Figure 1). ISEV standardization efforts [32,33] suggest that multiple membrane-associated proteins should be measured to demonstrate the presence of the lipid bilayer and thus EVs. Beyond this necessary, yet general EV characterization, questions remain about the existence of biogenesis-specific membrane markers, and whether they can be generalized to EVs from many cell types. Several workshop participants suggested TyA, C1q, Arrestin domain-containing protein 1 (ARRDC1), and CD73 as putative markers of PM-derived EVs, while several tetraspanins, including CD61, CD63, CD81, ESCRT proteins, such as TSG101, and Alix, as well as syntenin, flotillin, and heat shock proteins were proposed as specific markers of endosome-derived EVs. However, the subcellular distribution of tetraspanin markers can be cell type-specific and many endosomal proteins traffic through the plasma membrane on the way to the endosome. Furthermore, ESCRT proteins have been shown to be released in PM-derived EVs, as well as MVB-derived EVs, questioning their specificity as markers. One useful suggestion for studying EV-specific protein markers was combining techniques, e.g. western blot, electron microscopy (EM), live imaging combining pH sensors with EV markers such as tetraspanins (e.g. pHLuorin-TSPAN systems [36]), and proteomics. It is recommended that substantial additional work will be undertaken to identify specific markers of EV subtypes released across cell types or even within specific cell types.
Figure 1.
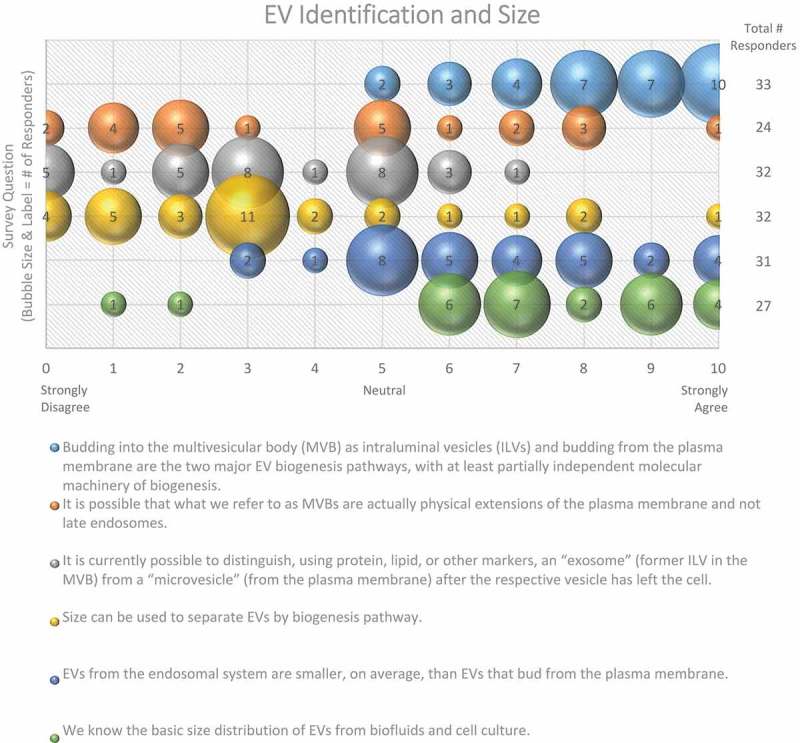
EV Identification and size. Six questions regarding EV identification and sizing were administered in the post-workshop survey. For each question, participants’ answers are depicted horizontally on a Likert-scale from 0 to 10, with bubble size reflecting of the number of responders at each point on the scale. Most responders believe that there are multiple distinct pathways for vesicle biogenesis that result in heterogeneity in terms of size. Identifying vesicles from these pathways based on size, protein or lipid markers remains difficult.
Size is also commonly used to distinguish EV subpopulations, following the assumption that exosomes are smaller than ectosomes; however, even if this assumption is valid for the average EV, small EVs (e.g. <100 nm in diameter) can be shed from the PM, and larger ILVs have been observed in MVBs, which could give rise to larger exosomes. Some groups have observed cell-type-specific differences in the size of ILVs in MVBs, further complicating the relationship between EV size and biogenesis. The majority (78%) of post-workshop survey respondents agree that size alone cannot be used to definitively categorize EV subpopulations (Figure 1). To further complicate matters, the field has not yet reached a consensus regarding the overall size distribution of EVs, as about half (48%) of the survey respondents believe that the average diameter is between 100 and 150 nm, while the other half (45%) believe it is less than 100 nm (Figure 2). This difference of opinion could arise from separation and characterization technologies that do not adequately recover or detect very small or very large EVs, or the employment of different techniques across laboratories. Recent technological advancements, such as the use of asymmetric flow field-flow fractionation for EV characterization, have revealed that some cell types release two distinct subpopulations of small EVs (sEVs; 60–80 nm and 90–120 nm), as well as a third population of small (~35 nm), non-membranous nanoparticles, referred to as “exomeres” [37].
Figure 2.
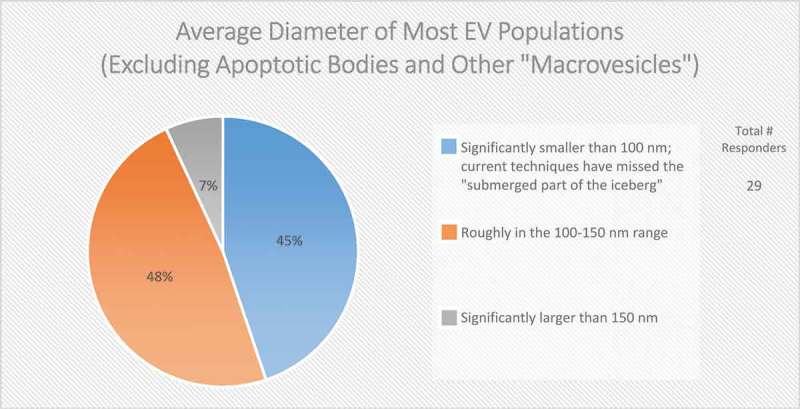
The average diameter of EVs (Excluding apoptotic bodies and other “macrovesicles”). In the post-workshop survey, participants were asked to choose from the three listed options. Responders believe that most EV populations are less than 150 nm in size. Those vesicles less than 100 nm in size are difficult to detect using techniques based on light scattering.
The results of the survey confirm that to aid in our understanding of EV biogenesis, we must address several considerations. How are EVs different from their parent cell in terms of membrane architecture and cargo? Can we identify key regulators or signalling pathways necessary for biogenesis through the use of genetic manipulations? What are the physiological consequences of inhibiting EV biogenesis? What is the relative contribution (quantitatively) of PM vs MVB pathways for EV biogenesis? Researchers should consider the entire trafficking landscape of intracellular vesicular organelles which can directly, or indirectly affect EV biogenesis and secretion.
Does the topology of EV membrane lipids and proteins reflect that of the cell?
Phospholipids are distributed asymmetrically across cellular lipid bilayers, and intracellular compartments can differ in their lipid composition (e.g. differences in PM and MVB membranes). This distribution determines, among other things, curvature, and the fluidic and electrostatic properties of lipid membranes [38]. Phosphatidylserines (PS), phosphatidylethanolamines (PE), and phosphatidylinositols (PI) are ubiquitous phospholipids located predominantly in the inner leaflet (cytosolic side) of the PM, due to the action of phospholipid flippases. Interestingly, PS and PE have been reported in the outer leaflet of EV membranes [35,39], which may be a by-product of their biogenesis and may be important for their function. PS exposure is known to serve as an “eat me” signal for engulfment and may, therefore, influence EV uptake. Workshop participants had contrasting opinions about whether EV phospholipid distribution mirrors that of the cell, and whether lipid distribution remains stable over time (Figure 3).
Figure 3.
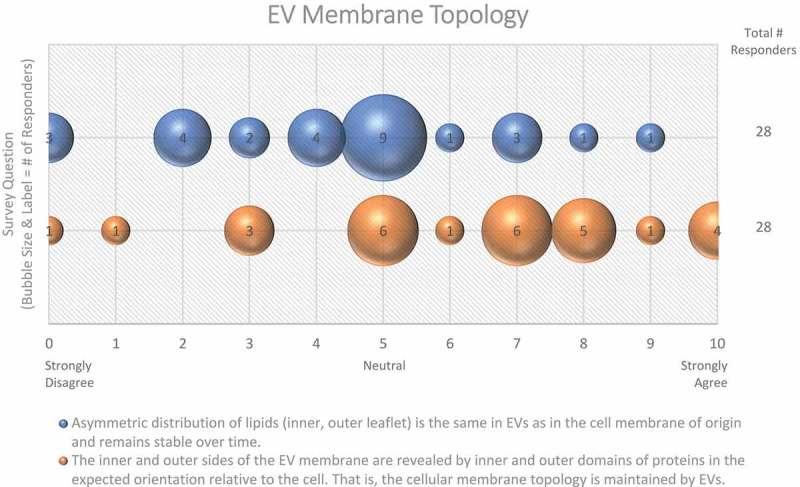
EV membrane topology. Two questions regarding EV membrane topology were administered in the post-workshop survey. For each question, participants’ answers are depicted horizontally on a Likert-scale from 0 to 10, with bubble size reflecting of the number of responders at each point on the scale. Responders are uncertain as to whether the lipid distribution of EV membranes is the same as the original cell membrane.
The majority of workshop participants (61%) suggested that the membrane topology of the cell is maintained in EVs (Figure 3). However, there is evidence that some EV membrane proteins have an “inside-out” topology [40]. Protease digestion assays, membrane permeabilization, and antibodies targeting outer or inner epitopes of specific membrane-associated EV proteins may be useful to investigate the topology and subcellular origin of specific EV proteins, and reveal whether this is a result of EV biogenesis or an artefact of EV purification methods.
How do membranes influence EV cargo loading or sorting?
Despite considerable interest, specific EV cargo sorting mechanisms are still unclear, although several have been proposed [41,42]. Ubiquitin-dependent ESCRT sorting mechanisms [43] and tetraspanin-enriched microdomains (TEMs) have been proposed to sort proteins into EVs [44]. Sorting of RNAs has also been postulated (reviewed in [45]). While the majority (61%) of Workshop survey respondents believe that the packaging of certain RNAs into EV subsets occurs (Figure 4), this is still an area of intense ongoing research. Conceptually, the larger the EV, the more likely it is to incorporate a given cytoplasmic entity, whereas sEV contents are more likely to be restricted to molecules in close proximity to membranes. This concept is supported by some data, such as the finding that large EVs (lEVs) and their parent cells have highly correlated RNA expression profiles, while RNA expression of sEVs differs significantly from that of the source cell [46]. Likewise, larger cargo, such as full-length mRNAs with associated proteins, may not easily fit into sEVs without efficient packaging mechanisms. On the other hand, enveloped viruses can fit nucleic acids of up to 30 kb into vesicles of ~80 nm in diameter. Thus, size constraints apply, but perhaps the organization of the packaged materials is even more relevant.
Figure 4.
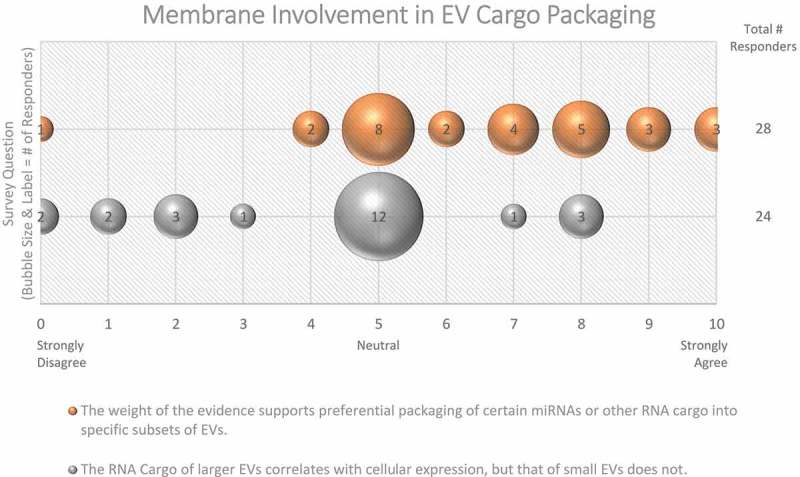
Membrane Involvement in EV cargo packaging. Two questions regarding the involvement of membranes in EV cargo packaging were administered in the Post-Workshop survey. For each question, participants’ answers are depicted horizontally on a Likert-scale from 0 to 10, with bubble size reflecting of the number of responders at each point on the scale. Responders are not sure whether miRNA or RNA cargo is specific to certain subtypes of EVs.
Related to understanding how protein–RNA interactions may allow for specific cargo loading into EVs, standing questions in the field revolve around the involvement of ribonucleoproteins (RNPs) like Argonaute2 (Ago2) [47], ELAV-like protein 1/human antigen R (HuR) [45,48], and hnRNPA2B1 [49], and the sequence-specificity of effects. If RNA loading is sequence-specific, it may also be dependent on cell type and altered during stress conditions. Assessing physiological relevance is also important. For example, introducing high concentrations of highly purified proteins or synthetic RNAs into cell lysates may result in associations that are physiologically irrelevant, and/or induce off-target effects. Finally, perhaps not all EV “cargo” is contained inside the EV. RNA and DNA alike have been reported in association with the outside of the membrane as well; however, this may be an artefact of the isolation procedure [45]. The importance of developing reproducible in vitro assay systems to study EV cargo loading which closely mimics the physiological context cannot be overemphasized. Successful adaptation of such working models is recommended as it will help answer many questions related to the targeting and packaging of EV cargo molecules.
Which proteins and lipids control EV biogenesis and secretion?
Genetic or epigenetic manipulation has implicated several families of proteins in EV biogenesis and release, such as Rab GTPases, ARRDC1, and ESCRT complexes [50,51]. The Rab family of small GTPases plays a critical role in intracellular trafficking, and several Rabs, including Rab27a, Rab27b, Rab35, and Rab11, have been implicated in EV release [52–58]. In the case of exosomes, it should be considered that their release machinery will include both molecules affecting the formation of ILVs, the transport of MVBs to the PM, and the fusion of MVBs with the PM. In the case of microvesicles, the machinery includes proteins involved in trafficking to the PM, as well as factors at or in the PM [59]. ARRDC1 mediates the release of a subpopulation of PM-derived EVs known as ARMMS (ARRDC1-mediated microvesicles) [60] and possibly endosome-derived EVs [61]. Gut explants from ARRDC1 and ARRDC4 knockout animals showed markedly reduced EV release [62], while in vitro overexpression of ARRDC1 significantly increased EV secretion [63].
To what extent can knockout or knockdown of EV biogenesis proteins abrogate EV release, and what consequences does this have for the cell or organism? The majority (59%) of Workshop participants do not believe it is possible to create an organism or cell that does not produce any EVs (Figure 5). Knockout of TSG101 appears to be lethal in some models [64,65], as the protein is essential for many important cellular functions, such as endosomal receptor sorting independent of EV secretion [66]. Not all knockouts of EV proteins are lethal or produce overt phenotypes; however, the lack of overt phenotypic changes in a mouse does not necessarily equate with a lack of important function. For example, in vivo knockout of ARRDC1 reduces EV plasma concentrations by ~50% in mice and confers no behavioural differences in normal settings; however, a phenotype may emerge after induction of non-physiological conditions. Overall, the data support the existence of independent and redundant biogenesis pathways with multiple components. Unbiased genetic screens and small molecule modulator screens may be needed to resolve unappreciated contributors to EV biogenesis. Similarly, manipulating multiple factors may be necessary to understand some mechanisms involved in EV biogenesis.
Figure 5.
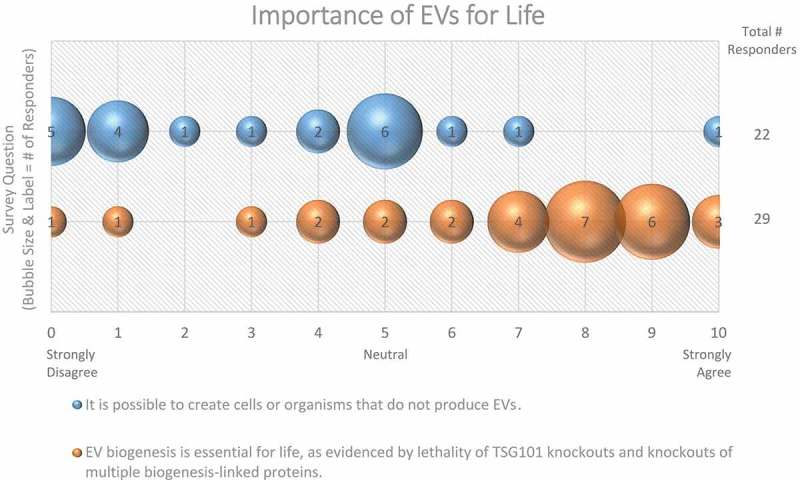
Importance of EVs for Life. Two questions regarding the importance of EVs for life were administered in the post-workshop survey. For each question, participants’ answers are depicted horizontally on a likert-scale from 0 to 10, with bubble size reflecting of the number of responders at each point on the scale. The majority of responders believe that EV production is necessary for cell and organism survival.
Aside from membrane proteins, lipids are also believed to influence EV formation. PE is thought to play a critical role in regulating membrane fusion and curvature, and therefore may be involved in EV biogenesis or function [35,59]. PI has also been implicated in EV release through one of its by-products. Hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) yields inositol trisphosphate (IP3) and diacylglycerol (DAG); DAG contributes to MVB formation, and to the secretion of EVs [67,68].
Sphingomyelin is a sphingolipid normally found in the outer leaflet of membranes (extracellular or luminal side). Enzymes such as neutral sphingomyelinase (nSMase) and acid sphingomyelinase (aSMase) convert sphingomyelin into phosphocholine and ceramide, which alters membrane fluidity and promotes microdomain formation. Interestingly, 62% of survey respondents believe that lipid rafts/microdomains contribute to the formation of vesicles [69,70] (Figure 6). nSMase inhibitors, such as GW4869, have been shown to significantly reduce small EV release from some [71], but not all systems [72], and even results in a compensatory increase in large EVs in some systems [73]. Conversely, overexpressing nSMase2 increases ILV formation, which is thought to occur via an ESCRT-independent biogenesis pathway [71]. More than half (59%) of ISEV workshop participants doubted nSMase2 involvement in the biogenesis of all EV subtypes (Figure 6), but there is also evidence that aSMases are involved in EV release [74]. Thus, the roles of various sphingomyelinases, ceramide, and lipid rafts in EV biogenesis require further investigation.
Figure 6.
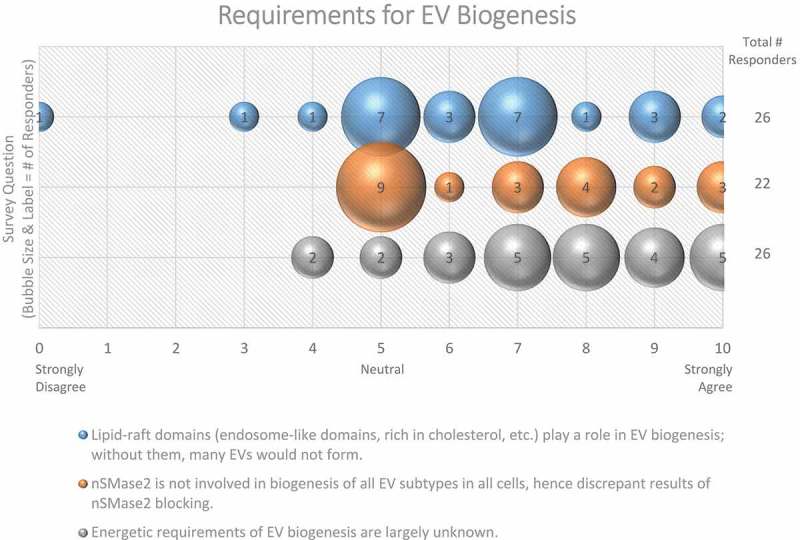
Requirements for EV Biogenesis. Three questions regarding requirements for EV biogenesis were administered in the post-workshop survey. For each question, participants’ answers are depicted horizontally on a likert-scale from 0 to 10, with bubble size reflecting of the number of responders at each point on the scale. Responders believe that the roles of lipid-raft domains, nSMase2, and the energetic requirements of EV biogenesis need to be further explored.
How are external signalling agents and membrane components involved in EV release?
Beyond genetic manipulation, biogenesis pathways can be altered by external signals. For example, serotonin stimulates EV release from microglia [75], and histamine induces EV release from cervical carcinoma cells. Inflammatory signals generally alter EV release [76]. For example, dendritic cells exposed to lipopolysaccharide (LPS) produce significantly more, and more immunogenic EVs compared with non-exposed controls [77,78]. Exposure to dsDNA has also been shown to induce inflammation and EV release [79], and IgE-mediated mast cell activation leads to the rapid release of a distinct EV subset [27]. Despite low levels of constitutive EV release by B cells, upon stimulation via CD40 and IL4 receptor, EVs displaying MHCI, MHCII, and surface antibodies were released, likely participating in immune responses [80]. Similarly, stimulation of toll-like receptors (TLRs) induces the release of exosomes with pro-inflammatory activity [81]. Thus, EV biogenesis is also influenced by external factors.
What are the energetic requirements of EV biogenesis?
The field has yet to determine the energetic requirements for biogenesis and release of EVs. Is metabolic adaptation required to provide energy for EV secretion and/or precursors for the biosynthesis of the lipids, proteins, etc. that will be incorporated into EVs? Indeed, there are very few studies that show the relationship between cellular metabolism and EV release. One recent study suggests that EV secretion from tumour cells heavily relies on aerobic glycolysis [82]. Others have shown that activation of hypoxia-inducible factor-1α (HIF-1α) can induce transcription of both Rab22a [83] and Rab27a [84], and indirectly leads to increased EV secretion. Thus, metabolic regulators play an important role in EV production, and further research is needed to directly link metabolism and EV production.
Contact, uptake, and fusion
In the Pre-Workshop survey, planners and registrants were asked about outstanding questions regarding EV uptake and/or fusion. From the responses, one can clearly appreciate that this topic is greatly understudied, as the field has yet to find answers to some basic questions. For example, it remains largely unclear how EVs interact with cells, and what dictates the next step (signalling, uptake or fusion) of the bound EVs. The molecular drivers (e.g. proteins, lipids, sugars, nucleic acids) of EV-cell interactions are largely unknown, including how these vary among cell types, with cell state, or among EV subtypes. What molecules or conditions drive EV endocytosis, and how do they affect EVs’ intracellular destination? Do EVs deliver their cargo into the cytosol after membrane fusion in a similar manner to viruses and if so, does this occur at the plasma membrane or an endosomal membrane? How often do fusion events occur and what determines this? Is the field focusing too much on fusion at the expense of lysosomal degradation or signalling, giving the perhaps false impression that fusion is the main fate of EVs and that cargo delivery to the cytosol is their most important role?
Based on this list of questions from the survey responders, we formulated four overarching questions that formed the basis of the roundtable discussion during the Workshop, and of the Post-Workshop survey. Consensus on these topics is presented in sections 4.1–4.4.
The most important unanswered questions about EV function focused mainly on EV tissue distribution and their ability to cross anatomic barriers, and on their true importance for life under physiological and pathological conditions. These questions were discussed during the roundtable on transfer and functional assays. Consensus on these topics based on this discussion and the Post-Workshop survey is presented in sections 4.5 and 4.6, and recommendations stemming from the survey and Workshop discussions are summarized in Table 5.
What are the general mechanisms of cell-EV uptake?
EVs might exert effects on cells by contact, uptake, fusion, degradation, or a combination of these modalities. As an example of each, “contact” would include interactions between EV surface proteins and cell surface proteins; “uptake” would be internalization of the EV into the endosomal system (for example); “fusion” would be fusion of EV and cellular membranes, with EV contents being released into the cell cytoplasm. The latter might occur at the plasma membrane or only after “uptake.” While there are many reports demonstrating EV contact, endocytosis, and endolysosomal degradation, direct evidence for EV fusion with cellular membranes is scarce. General mechanisms for cell-EV uptake remain unclear, with many questions arising:
Is cellular uptake of EVs mediated by specific processing mechanisms? Is the primary purpose of EV uptake cellular communication and signalling, or is uptake just a byproduct of clearance or homeostatic mechanisms? Does uptake also confer function? Are there differential uptake pathways for different subpopulations of EVs (small vs large EVs)? Where do EVs go once they are endocytosed by individual cells; do they fuse with endo/lysosomal membranes to reach the cytoplasm, go to the ER [85], end up being degraded in lysosomes, or do something completely different? How is EV cargo unloaded into a cell? How does EV-based transfer of genetic material and cellular signals differ from that mediated by tunnelling nanotubes?
Three quarters of survey responders agreed that most cell types eventually internalize at least a proportion of EVs, seemingly regardless of the cell of origin (Figure 7). Despite the possibility of non-specific uptake, three-quarters of responders agreed that proteins on the EV surface are required for most fusion events between EVs and cellular membranes (Figure 7). Cells could also recognize lipids on the surface of the EVs, allowing for non-specific uptake or fusion events to occur. However, specificity likely determines many of the interactions between EVs and their target cells, so it is plausible that many of these interactions are mediated by receptor-ligand binding.
Figure 7.
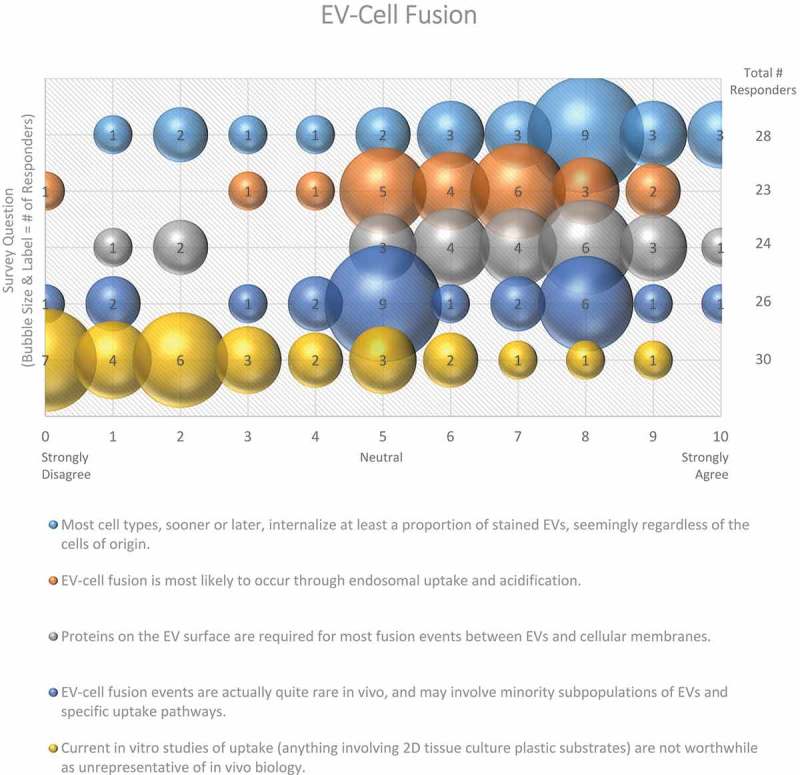
EV-cell fusion. Five questions regarding EV-cell fusion were administered in the post-workshop survey. For each question, participants’ answers are depicted horizontally on a Likert-scale from 0 to 10, with bubble size reflecting of the number of responders at each point on the scale. Responders agree that recipient cells internalize EVs from different cell types through endosomal uptake and acidification, and that proteins on the EV surface are responsible for fusion events. Survey participants are not sure how frequent EV-cell fusion events are in vivo.
How specific are EV-cell interactions, and how can we experimentally investigate them?
The EV-cell interaction is likely a handshake: factors on the surface of both membranes contribute. Molecules exposed on the EV surface, including fibronectin, PS, and integrins, interact with heparan sulphate proteoglycans [86], T-cell immunoglobulin- and mucin-domain-containing molecules [87], and cell-associated extracellular matrix [8] present on recipient cells. Chemokines and their receptors may act as bridges linking the two membranes together, as they are expressed on both EVs and cells [88,89]. Although follicular dendritic cells do not express MHC-II, their surface can become fully decorated with vesicles that do (likely derived from B-cells) [90]. Additionally, the charge or reactivity of EVs, glycolipids on the EV surface, and even membrane curvature can all influence how EVs interact with certain recipient cells.
Survey participants were asked to rank several potential mechanisms of EV-cell interaction through which EVs exert their effects from the most (1) to least (5) likely (Figure 8). Signalling through membrane proteins was ranked as the most likely mechanism. The next two most likely mechanisms were considered to be the delivery of functional proteins and RNAs, respectively. Transfer of functional lipids and nutrition (molecular recycling) were the next most likely.
Figure 8.
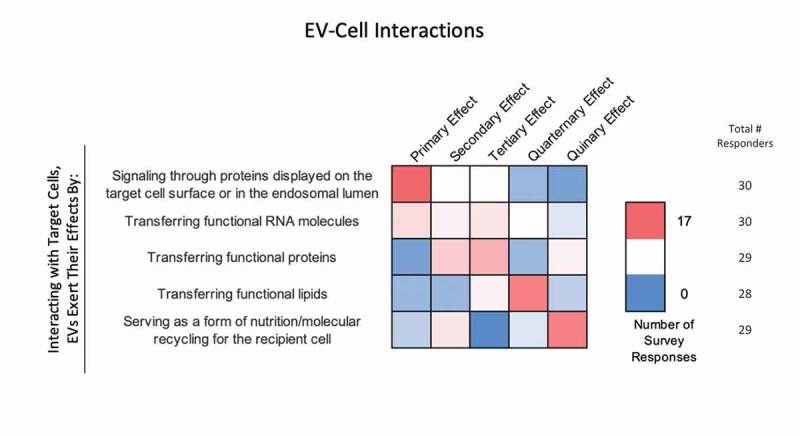
EV-cell interactions. In the post-workshop survey, participants were asked to rank order the most to least likely ways in which EVs interact with target cells. Answers are depicted in a heat map, with pink shades indicating a higher number of responders, and blue indicating a lower number of responders. Responders believe that EVs primarily interact with target cells by signalling through proteins displayed on the target-cell surface or endosomal lumen. Transferring functional RNA, proteins and lipids is seen as a secondary effect. Most believe that EVs are indirectly a form of nutrition or molecular recycling for recipient cells.
Certain conditions may need to be met before EV uptake can occur. For example, dendritic cell-derived EVs can only bind activated T-cells that express high-affinity LFA1 [91]. Cellular uptake of EVs may also be regulated by competitive mechanisms. For example, when working with unfractionated peripheral blood mononuclear cells (PBMC), MSC-EVs are mainly taken up by monocytes and only scarcely by T and NK cells; yet adding EVs to singularly cultured T or NK cells increases uptake significantly [92]. Furthermore, small and large EVs may have different membrane compositions that could give rise to divergent uptake mechanisms.
EV uptake in vivo also shows selectivity. Red blood cells are the most abundant cells in the body and produce a high number of EVs that contain unique miRNAs [93,94]. These EVs are only found in blood/serum and are usually not observed in the surrounding tissue. EVs present in the circulation and interstitial fluid of zebrafish are taken up only by endothelial cells and macrophages, but not muscle cells (despite being bathed in EVs) [95]. These observations support the notion that there are likely differential uptake mechanisms for EVs, depending on cell type and/or EV subpopulation.
Specific proteins or lipids that may prevent uptake in one cell type versus another by inhibiting docking, fusion, or internalization of EVs have not been identified. An interesting consideration is how the extracellular matrix (ECM) may influence EV processing or uptake in neighbouring cells. Unlike cell culture models, the in vivo ECM may contain proteins and proteases that contribute to or prevent EV uptake. As such, experiments conducted in vitro likely do not recapitulate in vivo uptake mechanisms accurately.
How do variables such as dose, time, pH, and temperature affect EV uptake?
“One dose does not fit all”. EV uptake by cells should be determined through specific, well-designed dose-dependent experiments. Comparison of dose-dependent uptake profiles of human serum small EVs by murine and human cell lines showed that EVs from the same source could be internalized by one, several or all of the cell lines depending on the dose [96]. Therefore, considering a fixed EV dose for different target cell lines may entail misleading results.
With respect to time, EV uptake has been reported in as little as 15 min [87], which is consistent with endocytosis rates, while lysosomal degradation of EVs has been reported within hours [85,97]. If EV cargo is delivered to cells so rapidly, a rethink of days-long experiments after a single addition of EVs may, depending on the functional readout, be in order.
Considering pH, ILVs are formed in the acidic lumen of the MVB, so exosomal proteins should be somewhat pH resistant. Nevertheless, pH can alter EV interactions with cells. For example, some viral membrane fusion proteins are inactive at pH 7, but undergo conformational changes at pH 5, leading to membrane fusion [98]. An EV that has entered the endosomal pathway will also undergo progressive acidification. As a fusion of EVs and cellular membranes would lead to the delivery of the EV content into the cell cytoplasm [9], understanding how pH influences the cell–EV interactions is an interesting avenue of research that requires further work.
Regarding temperature, it was previously shown that EVs are taken up by cells at 37°C, but EVs can bind to the cell surface when endocytosis is blocked at 4°C [99,100]. Experimental manipulation of temperature might provide valuable mechanistic clues into EV uptake.
Membrane fusion and stability
In the context of EV uptake and function, it is important to define EV structural and functional stability. Structural stability is the tendency of an EV to remain intact over time and under different conditions. Functional stability is the retention of a particular effect. On questions of EV stability, over half (67%) of survey respondents agreed that current technologies for studying EVs are not adequate to measure both functional and physical stability of EVs (Figure 9). When asked how freeze-thaw cycles may affect EV stability, 50% felt that more must be learned about potentially detrimental effects (Figure 10).
Figure 9.
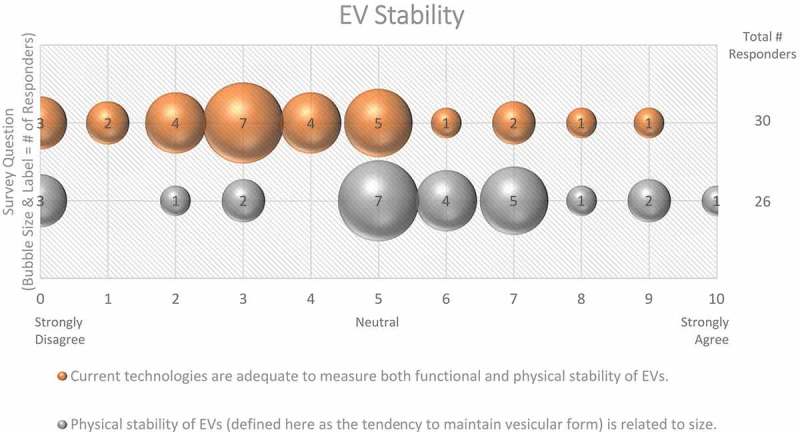
EV stability. Two questions regarding EV stability were administered in the post-workshop survey. For each question, participants’ answers are depicted horizontally on a Likert-scale from 0 to 10, with bubble size reflecting of the number of responders at each point on the scale. While EVs are physically stable, most survey participants believe that current technologies need to be improved to simultaneously measure the functional and physical stability of EVs.
Figure 10.
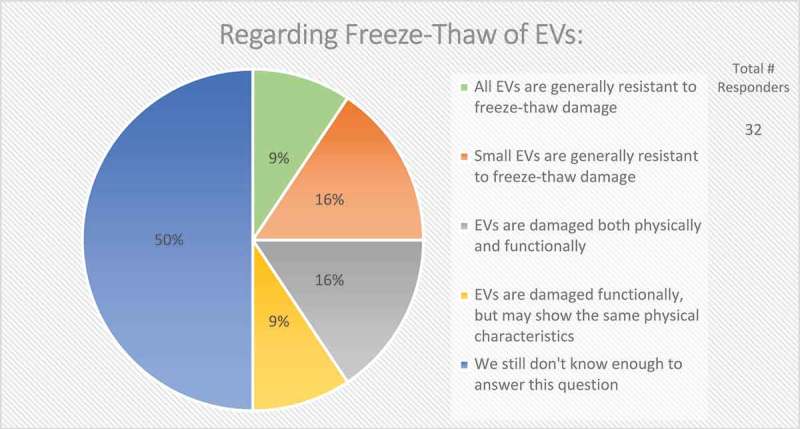
Storage of EVs. In the post-workshop survey, participants were asked to choose from five options whether or not they believe freeze-thawing causes damage to EVs. Responders agree that we do not know enough about how freeze-thawing affects EV stability, uptake, and functionality.
What about the relationship between fusogenicity and stability? Is an EV that fuses with a cellular membrane inherently “unstable”? Here, the molecular composition is likely to define membrane stability and tendency to fuse. Although membranes are coated with sugars and proteins, it is the lipid composition that most heavily dictates thermodynamic favourability for fusion. In principle, all vesicle-cell membrane fusions are thermodynamically favourable. The barriers to fusion are however kinetic, which may be lowered in relation to lipid compositions. For example, the insertion of cholesterol influences curvature and fusion properties of small vesicles [101–103]. Cone-shaped phospholipids induce high curvature, while those of cylindrical shape are found in more planar membranes. Fusion is thermodynamically favourable for more curved vesicles, as it brings the vesicles to a lower energy state. Small vesicles have a higher curvature and tighter membranes, lending to a very high-energy state and a possible predisposition for fusion. Supporting this likelihood, a portion of synthetic small unilamellar vesicles fuses with each other given enough time [104,105]. Contrastingly, the recovery of EVs as small as 30–50 nm in diameter from biological and culture fluids suggests a certain stability of even small EVs or subsets thereof. EVs may be more likely to fuse after isolation than when in a complex mixture such as plasma. However, current techniques cannot easily distinguish between a large EV and a similarly-sized EV that formed by the fusion of several smaller vesicles. When asked about the relationship between EV size and stability, 50% of respondents believed that size has a large impact on physical stability (Figure 9). EV-cell fusion is thermodynamically favourable but high kinetic barriers prevent spontaneous membrane coalescence. Lowering kinetic barriers can alter lipid membranes, including increased bilayer curvature, and the presence of protein-based fusion catalysts.
The functional capacity of an EV is likely dependent on membrane protein topology. Overall, membrane protein composition appears to be heavily influenced by EV size, the cell type of origin and cellular activation state. One technique to study EV membrane composition is free radical incorporation or electron-dense lipid labelling. Incorporating free radicals in membrane samples can provide useful topological information on both the membrane and/or associated/integral proteins based on the preferential localization of the radical used [106,107]. On the other hand, using lipids containing radicals at different positions in the tail could help provide information about the transmembrane region itself [108]. Some new techniques to make pseudo-membranes containing differently shaped phospholipids can allow for further study of how composition can influence membrane curvature and fusion by using nuclear magnetic resonance (NMR) [109].
Many questions remain to be answered about EV functional stability: Is the physical structure or the encapsulated content most important? If the structure of an EV has been compromised, does that necessarily affect its carrier/delivery function? Also, does the presence or absence of cargo reciprocally influence EV stability?
Finally, it is worth noting that our knowledge on EV membrane interactions with inorganic nanostructures and surfaces is poor and fragmented, albeit of key importance for future EV processing and engineering technologies (e.g. colloidal gold nanoplasmonic assays [110], microfluidics/lab-on-chip applications [111], EV supported lipid bilayers [112]).
In vivo administration of EVs: how membrane components and associates affect distribution
One of the most alluring aspects of EVs is the potential for the delivery of therapeutic drugs/molecules, which requires an understanding of the biodistribution of EVs introduced to healthy and diseased organisms. When introducing EVs to in vivo models, many factors need to be considered, including dose, route of administration, source of the administered EVs, and techniques for assessing biodistribution (i.e. labelling).
An overwhelming majority (87%) of workshop survey respondents suggested that EV transfer experiments are highly time dependent, and that the in vivo relevance is often unclear (Figure 11). Since the local physiological concentrations of EVs are often unknown and may change depending on disease state, dose–response studies to optimize EV concentrations for the creation of uptake profiles is critical; 90% of participants agreed (see also MISEV2018 [33]) on the importance of dose-response to establish any effect of EVs for both in vitro and in vivo studies (Figure 11). The half-life of exogenously administered EVs in circulation has been reported to be relatively short (in the range of tens of minutes), so serial or differential dosing may be necessary and will likely require optimization. By increasing the half-life of EVs, for example, through PEGylation [113] or by incorporation of a “don’t eat me” signal such as CD47 [114], higher plasma and/or tissue levels may be reached.
Figure 11.
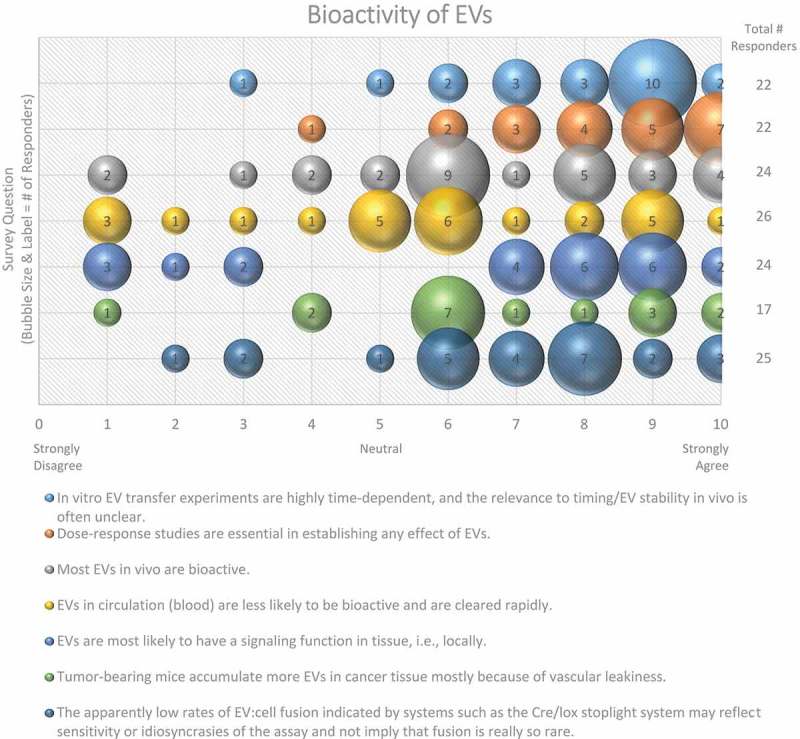
Bioactivity of EVs. Seven questions regarding the bioactivity of EVs were administered in the post-workshop survey. For each question, participants’ answers are depicted horizontally on a Likert-scale from 0 to 10, with bubble size reflecting of the number of responders at each point on the scale. Responders believe that the use of EVs for in vitro transfer experiments is time-dependent, that dose–response studies are important, and that EVs have a greater functional impact in the local tissue environment. Survey participants are undecided on how to determine and identify bioactive EVs. The survey reveals the need for improved technology for the study of EV-cell fusion.
Administration route (e.g. intraperitoneal (i.p.), tail vein, intramuscular, or interventricular injection or intranasal inhalation) can also influence EV biodistribution, and thereby the concentration that reaches the desired target. Subcutaneous injections lead to accumulation of EVs in the lymphatic system [115,116], while those injected directly into the tail vein are cleared by the liver and spleen [117,118]. These data are consistent with previous work in the field of synthetic nanomedicine, which indicate that the vast majority of injected nanoparticles aggregate in liver and spleen, with only very subtle accumulation in specific tissues [119,120].
The concept of “targeting” is perhaps somewhat misleading for EVs under normal conditions, necessitating careful descriptions [121]. Unlike cells, EVs cannot actively seek targets via signal gradients, reports of motility notwithstanding [122]. Instead, passive accumulation, which is mainly driven by EVs’ physiochemical properties, seems the likely dominant distribution mechanism, with “targeting” determined by different affinities for cells with which chance interaction occurs.
Nevertheless, in inflammatory conditions including certain cancers, injected nanoparticles may accumulate in inflamed tissue due to vascular leakiness [114]. For example, in vivo targeting of EVs to the heart is difficult because of the intact endothelial barrier; yet, if the tissue is infarcted, the chance of infiltration will increase due to vascular leakiness [123]. Similarly, EVs are more likely to enter the brain when it is inflamed [124], and EV accumulation within kidneys is significantly increased in animal models of acute kidney injury [125]. Nevertheless, only about 44% of Workshop survey respondents felt that EVs accumulate in cancerous tissue (Figure 11).
To study EV biodistribution, EV labelling or some other method is needed for tracking and visualization. Theoretically, a membrane labelling dye may largely disappear upon fusion, making it challenging to study functional EV biodistribution. An alternative approach might be to trace EV cargo, e.g. through measuring delivery of Cre recombinase mRNA or protein via EVs, using animals harbouring reporter systems such as a Cre-sensitive colour-switch reporter [126]. A clear recommendation stemming from the Workshop is the need for an improved methodology to study EV biodistribution.
What is the physiological role of EVs in homeostasis and disease?
While the field has not identified which is the most significant EV-related disease, cancer is the most heavily studied, and EVs have been strongly implicated in metastasis and the formation of tumour microenvironments. Other intensively studied pathologies include neurodegenerative disorders, such as Alzheimer’s disease, Parkinson’s disease, and prion diseases [127,128], as well as diabetes and acute phase responses. Understanding the basic physiological functions of EVs will allow for further exploration of their roles in homeostatic maintenance or disease dissemination. Another recommendation from this Workshop is the need for advanced animal models to study the physiological importance of EV-mediated cargo transfer between cells and tissue; these are required to conclude how EV-mediated communication impacts disease development. This is especially true when exploring the potential secondary effects of EVs. For example, EVs released from damaged lung can induce pulmonary hypertension when injected into healthy mice. However, these effects appear to be modulated by EV-induced alterations in bone marrow cells, which then promote pulmonary hypertension [129,130]. Furthermore, the field needs to establish guidelines for defining and/or concluding which EV subpopulations and associated cargo are involved in homoeostatic maintenance and pathological responses. One approach is to screen EV populations before and after (patho)physiological stimulations/treatment regimens, with the subsequent utilization of techniques like RNA sequencing, proteomics, and lipidomics. Implementation of multi-omics and longitudinal cohorts can aid in enhancing the overall understanding of EV biology and composition and can serve as quality controls for EV subsets of interest.
Technologies and strategies for studying EVs: considerations and limitations
The results of the Pre-Workshop survey confirm that there remain many uncertainties and controversies in the EV field. For example, do the results of EV-uptake experiments using cells cultured in 2D really reflect what occurs in vivo? Does fluorescent labelling of purified EVs lead to artefacts caused by aggregations of the dye? How does the curvature of the vesicle and the surface it binds to affect the fusion or function of the EV? Do current methodologies for isolation or enrichment alter the function of EVs and must we “live with” the current limitations in separation technology? What is the role of heterogeneity in EV function and what new tools are needed to assess EV heterogeneity? Some reasons why these questions have not yet been resolved include the relative youth of the EV field, a lack of robust tools and technologies for studying EVs, as well as the lack of proper reporting and consistent terminology in a largely heterogeneous approach to analysing EVs across the community.
Many of the most frequently cited “influential studies” highlighted in the pre-workshop survey related to characterizing EV subpopulations [131] and developing novel technologies for studying EVs with single-particle sensitivity [132–135]. The efforts of the Society to summarize and update the field with current knowledge were also appreciated by the citation of previous ISEV position papers [33,45,136]. Participants also indicated the importance of standardization and reporting initiatives such as EV-TRACK [137].
The need for new technologies and methodologies was also evident from the verbatim feedback in the survey. There was a particular focus on the need for new or improved imaging techniques. Several applications of electron microscopy (EM) were suggested, including 3D scanning electron microscopy (3D-SEM) combined with labelling techniques such as APEX (a monomeric peroxidase that withstands EM sample preparation) and correlative light and electron microscopy (CLEM). Many EVs are below the diffraction limit of light, so super-resolution microscopy methods need to be developed further to visualize EVs with better resolution. Tracking of single EVs is challenging due to their small size and paucity of molecular content; imaging individual EVs could be enabled by the use of brighter tags such as tandem fluorescent proteins or SunTag [138]. Improved techniques for quantifying the occurrence and effects of EV transfer, including in the in vivo setting, were raised as an important requirement in the field. Furthermore, there are other biophysical properties of EVs that should also be investigated, including stiffness, adhesiveness, aggregation and morphology. Techniques such as atomic force microscopy (AFM), scanning probe microscopy, cryoEM and neutron-scattering techniques could be used to measure these properties. A need for improved isolation and labelling of EVs was also reported in the survey. Taken together, these comments highlight the need for improved technologies in the EV field. This was also evident from the range of talks at the Workshop and the round-table discussions. The current “limitations” should be viewed as opportunities: new approaches combined with a greater understanding of membrane biology will allow us to gain new insight into EV biology and begin to resolve some of the uncertainty in the field.
Experimental issues with purification and characterization of EVs
EVs are challenging to study due to their small size (mostly <1 µm diameter), heterogeneity and lack of discriminative markers [139]. Although differential ultracentrifugation is the historical workhorse for EV isolation [140], it may introduce artefacts such as disruption of membrane topology, EV aggregation [141] and decoration of EVs with other components present in the sample. Theoretically, filtration by gravity flow should be gentler, but pushing samples forcefully though a small-pored filter may, besides trapping EVs in the filter, also damage or break EV membranes [136]. Altering properties of EVs during separation may also affect functionality. Other important questions to consider include: do our current separation techniques over-purify or select for more “stable” EVs? Are we removing important signalling molecules or cofactors from the supernatant when we remove EVs from their biological medium? How does the starting sample volume and type (i.e. the biological matrix in which the EVs are present) influence EV stability? Is one isolation technique better for certain biological sample types than another? Many of these questions still need to be resolved by basic analytical experimentation.
Likewise, the best practices for EV characterization, counting and sizing are still not universally agreed upon. When working with samples derived from mixed populations of cells, such as a biofluid or supernatant from mixed cultures, the limited knowledge of cell-specific EV markers makes identifying the cell of origin challenging. Ongoing multiomics-based research approaches should assist in the identification of tissue/cell-specific markers. There are also a range of methods for counting and sizing EVs, including nanoparticle tracking analysis (NTA), tunable resistive pulse sensing (TRPS), Raman spectroscopy, flow cytometry, single-particle interferometric reflectance imaging sensing (SP-IRIS) and electron microscopy. These vary in their accuracy, resolution, strengths and weaknesses [133,135]. Unsurprisingly, the vast majority (90%) of survey responders agree that different measurement technologies are biased towards certain EV size ranges (Figure 12).
Figure 12.
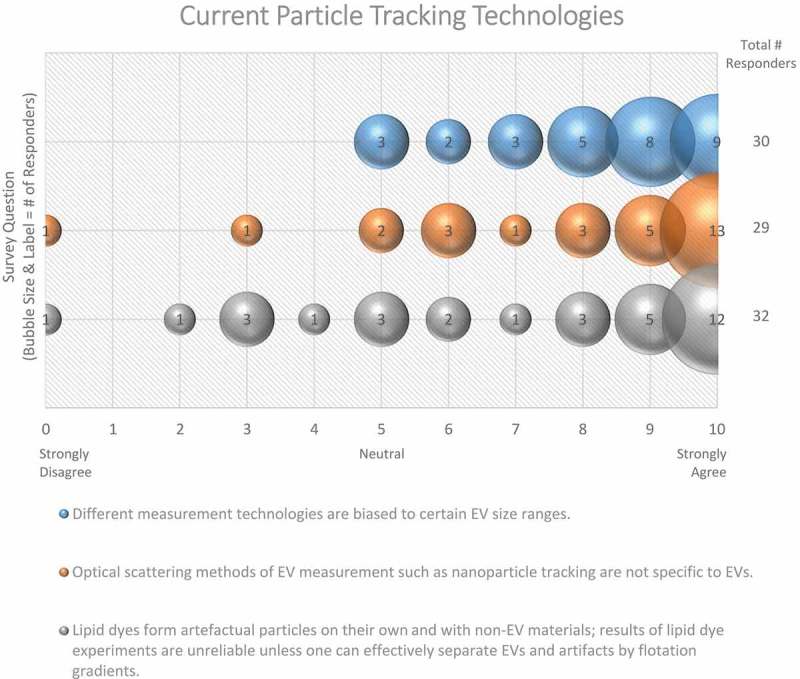
Current particle tracking technologies. Three questions regarding current particle tracking technologies were administered in the post-workshop survey. For each question, participants’ answers are depicted horizontally on a Likert-scale from 0 to 10, with bubble size reflecting of the number of responders at each point on the scale. Survey participants require improved, non-biased technologies for determining EV size. The use of lipid dye can cause experimental artefacts.
Exploring current EV fluorescent labelling techniques
Many experiments incorporate the use of fluorescent labels to visualize EV production, uptake, and biodistribution. Although there is currently no efficient pan-EV marker, many uptake studies utilize lipophilic dyes such as PKH and Bodipy. The mechanism of action of PKH is intercalating, with saturated acyl-chain lipids inserting into lipid-raft structures. While PKH does not form highly ordered micelles, it can form micelle-like structures if it is not below the critical micelle concentration [142]. Above this concentration, the properties can change and give rise to artefacts and background [142,143]. Bodipy is another popular dye which some groups prefer over PKH as there is reduced background and limited overlap with signals from EVs. Alternatives to lipophilic dyes include CFSE and maleimide-thiol coupling of dyes to EVs [144]. Interestingly, some labelling protocols recommend BSA to quench dye labelling of EVs. However, BSA may contain bovine EVs. Protocols for best EV labelling practices are needed to discern between labelled EVs, aggregates of dye, and label incorporated by contaminants such as those in BSA. Free dye controls are important, to show clearance of unbound dye, and that all fluorescent entities observed are EV-associated. Flotation gradients may assist with separation of labelled EVs from dye aggregates, but other contaminants may remain [143].
The majority (72%) of workshop participants concur that lipid dye experiments are unreliable unless the proper controls are used to discern between labelled EVs and aggregated dye (Figure 12). An endogenous method to label EVs by using an amphiphilic Near Infrared (NIR)-fluorescent probe, in which cells carry out the labelling prior to secretion of the EVs, has been recently proposed [145]. It does not require the use of immuno-labels, reagents for conjugation reactions or chromatographic purifications.
Another strategy for the labelling of vesicles is via fluorescent tagging of EV proteins. For example, tagging of tetraspanin proteins such as CD63 or CD81 with fluorescent proteins such as GFP or mCherry allows EVs to be visualized and tracked. Innovative approaches in which pH-dependent tags are employed, have been used to visualize EVs following fusion of the MVB with the PM [146]. This approach depends on the acidic environment of the multivesicular body and a neutral environment outside the cell. Alternatively, tagging abundant fluorescent reporters with a degradation motif (degron) is a way to specifically label EVs and remove the reporter from the source cell, allowing the observation of autocrine interactions in vivo [35,59,147]. Issues that need to be considered for any protein-tagging approach include the half-life of the fluorescent tag, the brightness of the tag, the limits of resolution of the microscopy technique used, and the possibility that tagged EVs (or indeed dye-labelled EVs) may have altered cargo or function. Another issue is that the visualization of EVs that lack the fluorescently tagged protein would be “invisible” and would thus be missing from any analysis. In this respect, general EV membrane labelling using, for example, fluorescent proteins fused to farnesylation or palmitoylation signals may be preferred [148,149]. It is recommended that further work is undertaken to optimize and establish the best methodology for the fluorescent tagging or labelling of EVs.
Single-vesicle analysis techniques – where are we and what do we need?
The survey revealed that single-vesicle analysis techniques are one of the most frequently suggested technological advancements that are needed for the field, with high-resolution single-vesicle flow cytometry being particularly in demand, offering high-throughput analysis with a commonly available tool. Of note, 70–75% of EV researchers use flow cytometry (FC) for targeted phenotyping [150]. Almost every survey respondent (97%) valued the development of single-vesicle flow analysis for EVs below ~500 nm in diameter (Figure 13). A reliable and accurate EV-nanoflow technique with the use of specialized cytometers with higher scatter sensitivity to measure small particles would allow for profound advances in understanding both membrane composition and internal cargo through state-of-the-art sorting and subsequent analysis. Because the size of the majority of EVs is below the wavelength of visible light, the majority (68%) of survey respondents believe that fluorescent triggering in EV-flow cytometry allows for better resolution than scatter (Figure 14); however, since no EV-specific generic labelling strategy is available, other non-EV components (e.g. lipoprotein particles), cell debris and/or EV membrane fragments may also be detected. Furthermore, the small size of EVs renders them particularly prone to swarm effects often leading to erroneous interpretation of data, as many instruments are still not sensitive enough to detect such small particles [151,152]. Of significance, there still needs to be established the ideal fluorescent dye for fluorescent triggering which is capable of staining all EVs in a preparation.
Figure 13.
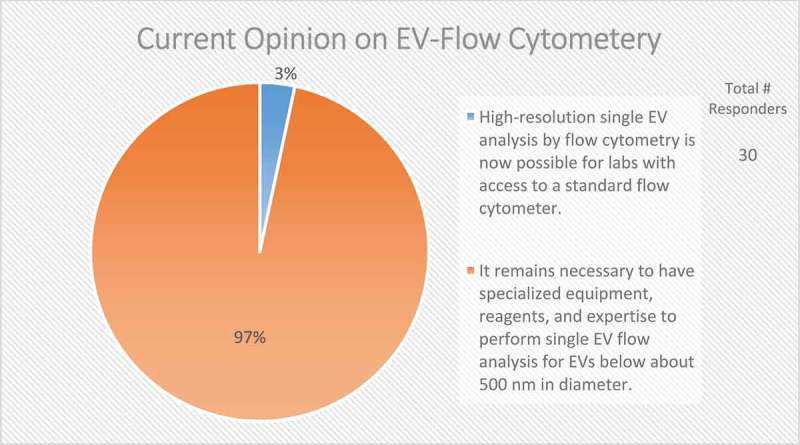
Current opinion on EV-flow cytometry. In the post-workshop survey, participants were asked to choose between two options regarding the current status of applying flow cytometry to the study of EVs. Almost all responders to this question call for specialized equipment, reagents and expertise to characterize single EVs through flow cytometry.
Figure 14.
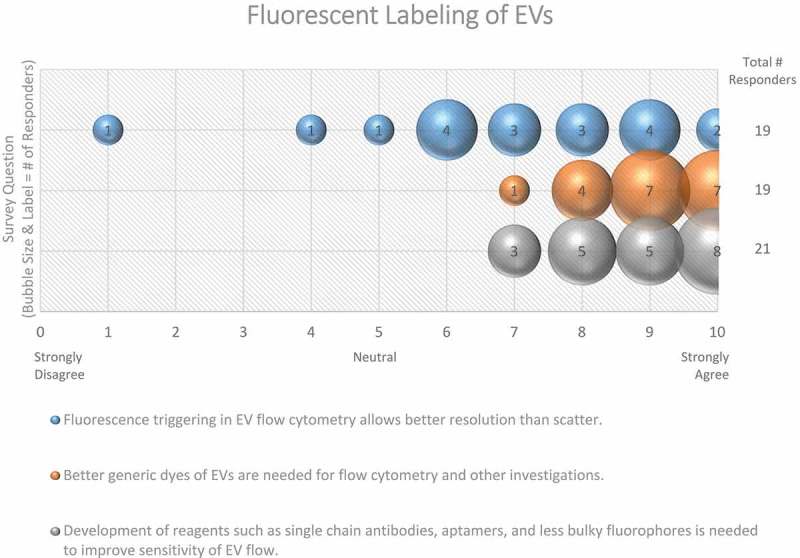
Fluorescent labelling of EVs. Three questions regarding fluorescent labelling of EVs were administered in the post-workshop survey. For each question, participants’ answers are depicted horizontally on a Likert-scale from 0 to 10, with bubble size reflecting of the number of responders at each point on the scale. Survey question participants acknowledge that better dyes and reagents are needed to study EVs using flow cytometry. Still, using fluorescent flow cytometry to study EVs provides better resolution than scatter.
Depending on EV size, it is estimated that about 1–100 copies of a particular protein could be present on the EV surface [153]. This low number of proteins present on the EV surface may hamper the immune-detection by fluorescent antibodies of EVs or EV subsets. The development of brighter fluorophores, alternatives for fluorescent-antibodies, as well as the ability to increase the signal-to-noise ratio by optimizing flow cytometers and settings for EV analysis will lead to improved single-EV detection. Instead of labelling proteins on the EV surface, highly specific fluorescent lipid dyes may hold potential for fluorescent-based EV analysis. In that respect, proteins that bind different lipids can be useful to distinguish subpopulations of EVs and could be applied to the single-vesicle analysis, including flow cytometry [154]. Myristoylated alanine-rich C-kinase substrate (MARCKS) peptides [155,156] could also be of potential use to label vesicles based on membrane curvature. Ultimately, the combination of generic lipid dyes or lipid detection together with specific labelling of EVs with fluorescent antibodies or other reagents to label EV surface proteins offers new possibilities to define and eventually sort specific EV subpopulations. However, with multi-fluorescent approaches precautions should be taken for quenching effects due to the small surface area of the EV.
In general, every survey respondent (100%) agreed that the development of reagents, such as single-chain antibodies, aptamers, and less bulky fluorophores, is needed to improve the sensitivity of EV-flow cytometric-based analysis (Figure 14).
Currently, high-resolution flow-cytometric single-EV analysis utilizes slow flow rates and although sorting of single EVs can be performed [151], long sort times and diluted samples result in a limited amount of sorted EVs, which restricts the possibilities for downstream analysis. Additionally, the purity of the sorted EV populations should be analysed and demonstrated by other methods and potential effects of the sorting process and subsequent sample processing on EV integrity should be evaluated.
Altogether, there is a growing awareness of the physical limitations of commonly used flow cytometers developed for cell analysis which are using different optics and laser configurations tuned for cellular analysis. However, the field is moving towards better instrumentation for flow cytometric analysis of EVs, either by optimizing the existing flow cytometers designed for cell analysis or by designing novel instruments built for small particle analysis. As a reaction to the demand of more specialized flow cytometers, manufacturers incorporate certain instrument designs to increase the capability of sorting and phenotyping EVs <500nm. Some instruments are capable of detecting EVs down to 100 nm. Examples are imaging flow cytometry, spectral flow cytometry, high-sensitivity flow cytometry, and nanoscale flow cytometry [157].
Besides the development of more advanced flow cytometry for EV research, better transparency about reporting instrument configuration is needed. This will allow the interpretation of flow cytometry data from one instrument to another. Applying strict and well-defined controls in each reaction will also increase the reproducibility and rigour of EV-flow cytometry experiments. To advance the EV-flow cytometry field a three-society workgroup was established in 2015; the ISEV-ISAC-ISTH EV Flow cytometry workgroup (http://www.evflowcytometry.org/). They are establishing a framework for the minimal requirements for accurately reporting EV measurement by flow cytometry.
Alternative approaches to single-EV analysis are also being developed, including 3D-SEM, CLEM, AFM and Raman spectroscopy. However, these methods are less commonly available and do not offer high throughput analysis. AFM has been used to quantify the physical characteristics of EVs, such as stiffness, as well as for the visualization of EV budding. “Label free” methods are attractive, as labelling by itself can alter the EVs. The use of optical tweezers for studying and manipulating single, large EVs is also gaining traction [158]. Capillary electrophoresis techniques may also be implemented to allow for separation of EVs of different sizes or composition. Cryo-EM is another good tool for visualizing EVs since EM with immunogold labelling is the only technique that combines morphological information at high resolution and the specificity of labelling. A recommendation arising from this Workshop is for the field to improve and develop single-vesicle analysis techniques that will allow researchers to ask new, important questions related to all aspects of EV biology.
What can the EV field learn from engineers and virologists?
To make genuine leaps forward it is recommended that the EV field must adopt a more multidisciplinary approach, seeking expertise from specialists in other disciplines, including engineering, physics, imaging and chemistry. A biomimetic approach to studying EVs involves producing synthetic analogues. Survey participants had mixed views on current capabilities to engineer truly artificial EVs, with about one-third believing it is presently possible, and almost half (42%) thinking it is not (Figure 15), though this could be partly due to different interpretations of the term “artificial EVs”. Indeed, engineering EVs from biological sources is done routinely [159]. For example, EVs could be loaded with therapeutic cargo, like siRNAs or other drugs; successful delivery of this cargo can be used for therapeutic purposes or, as described above, to analyse EV uptake or distribution. One key point of discussion was the difference between intracellular and extracellular loading of engineered EV cargo. In other words, should artificial cargo be added by manipulating donor cells which then load the cargo for us, or should we isolate EVs and artificially insert (or add to the surface) cargo via experimental perturbations such as electroporation or pH-dependent opening? The choice of how to artificially load cargo in engineered EVs could affect the utilization of the cargo in recipient cells. Another way to engineer EVs is to alter the surface composition to potentially re-direct the bio-targeting of vesicles [160]. Whether a top-down approach (starting with native EVs and modifying them to see how this affects function) or a bottom-up approach (generating artificial EVs from purified components) is taken, a better understanding of membrane biology will enhance our ability to engineer, understand and use EVs.
Figure 15.
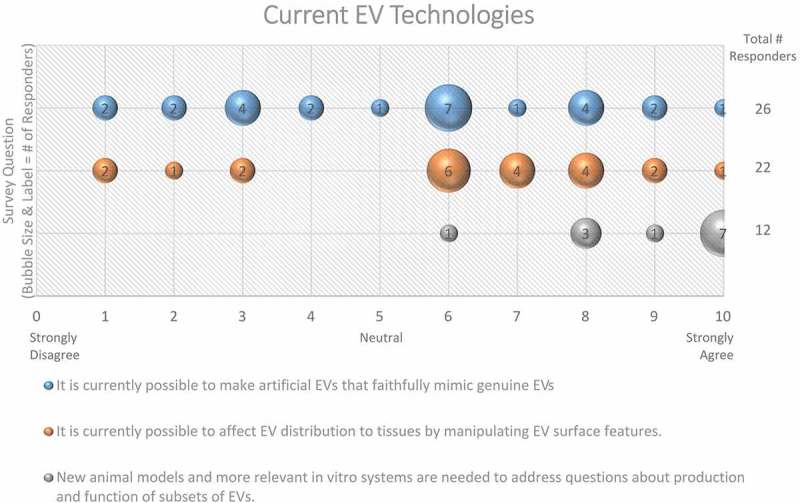
Current EV Technologies. Three questions regarding current EV technologies were administered in the post-workshop survey. For each question, participants’ answers are depicted horizontally on a Likert-scale from 0 to 10, with bubble size reflecting of the number of responders at each point on the scale. Responders agree that new animal and in vitro models are needed to address questions concerning EV production and function. Survey participants are not sure whether artificial EVs can mimic genuine EVs, or that manipulation of EV surface features will affect biodistribution.
Another area discussed in the Workshop was that the EV field can learn a great deal from the virology field. Enveloped virus morphogenesis, secretion, and entry into target cells are well understood and are likely to reflect, at least partly, EV biogenesis, target-cell contact, endocytosis, and cargo delivery processes. Enveloped viruses encode proteins that drive particle (vesicle) assembly in producer cells, and particle (vesicle) disassembly in target cells. There may be valuable dividends to searches for EV-associated proteins that are functionally similar to those operating in enveloped virus infections. The virology field could lend a great deal of information as to how to artificially design EVs to target specific cell types and develop assays to indicate delivery of RNA or protein into the cytosol. The same mechanisms utilized for viral RNA packaging may also participate in EV RNA packaging. Fostering collaborations with virologists should be encouraged as it will likely aid in understanding EV release and uptake pathways.
What new techniques or experimental approaches do we need to study EVs?
To fully understand EV biology, physiology, and pathology, the field needs more than just better labelling techniques and the ability to perform single-EV analysis; the recommendation from the Workshop is that a raft of new technological approaches is required. Unsurprisingly, almost all survey participants (97%) agree that new animal models and more relevant in vitro systems are needed to address questions about production and function of subsets of EVs (Figure 15). Animal models may be a viable option for understanding the production of EV subsets (MVB biogenesis, transport to PM, fusion with the PM, and budding from the PM). Genetic manipulations to track the transfer and uptake of EVs within an animal [126] may address these questions. The use of zebrafish [95], Drosophila melanogaster [161–163], or C. elegans [35,59,164] as genetic model organisms to study EV biogenesis may initially be more useful than rodent models, and are gaining traction within the field [165].
When using in vitro experimental methods, the environment in which EVs are generated, i.e. single-cell readouts, 2D cell culture, or organoids, is important. Will the use of organoids provide a more realistic indication of what is occurring with EV production and cargo transport than other culture methods? Understanding how EVs interact within a three-dimensional structure may be more helpful and translatable than traditional culture methods and is a goal for the future.
Reporting in EV studies
There are many factors that need to be addressed and reported when performing EV work, and many of these are outlined in both the 2014 and 2018 MISEV reports [32,33]. Transparency is needed to help the field reconcile conflicting data from different laboratories. For example, some data suggest that the passage number of cells can strongly impact EVs in terms of number, cargo, and markers (e.g. CD63) [166], making it difficult to draw conclusions as to whether or not observed effects were due to experimental manipulations. If a certain cell type in question does require supplementation with FBS, how are EVs depleted from serum? Even a 24-h ultracentrifugation does not remove all of the bovine RNA present in FBS, potentially leading to artefactual results [167]. If a cell type can survive in the absence of FBS, how does serum-starvation change cellular RNA profiles or influence assay readouts? Additionally, by culturing cells in serum-free conditions, clonal populations are being selected for survival, which could heavily influence experimental outcomes of downstream assays. This is also highly relevant to experiments performed using clonal cell lines generated by CRISPR/Cas9-mediated genome engineering. It is also important to understand the normal physiology of the cells being utilized for experiments prior to perturbing them. Careful experimental design is required when attempting to recapitulate a disease within a dish. Additionally, more primary cell work is required, as cell lines can behave in a dissimilar manner [168]. Overall, critical evaluation of how model systems reflect actual biology is essential. To identify large-scale, significant results, experimental consistency is key. We must strive to encourage and ensure high replicability and reproducibility within the field, which depends on critical evaluation of the consistency across experiments and detailed reporting.
The future of EV therapeutics
A major question in EV therapeutics is under what conditions the FDA, EMA and other regulatory agencies will approve therapeutic use of EVs. The clinical requirements for EV therapeutics and assays must be achieved before approval can be granted [169]. Cell-based therapies have been approved, so there is hope for EVs in the future. Future implementation of these therapies will focus heavily on the safety to bio-therapeutic ratio. There is a strong feeling in the EV community that regulatory agencies will eventually support the use of EV therapeutics. How such agencies would categorize EVs also needs to be considered; i.e. will EVs be considered cell-like agents, or pharmaceutical agents? While the origin of cell culture-derived EVs is clear, the mechanism by which they convey function in vivo remains unknown. Another consideration is that we still do not isolate a pure homogenous population of vesicles; EV heterogeneity is an important problem/opportunity for the field [170,171]. A challenge for the therapeutic use of EVs is, therefore, the ability to purify the vesicles effectively enough. Thus, further work on the fundamental cell biology of EVs is essential to realize their therapeutic potential [172].
Conclusion
EV research is rapidly advancing, but many unanswered questions remain regarding basic EV biology and mechanisms of action. The primary goal of this workshop was to gauge the field’s understanding of the role of membranes in EV biogenesis, uptake/fusion, and the types of technologies needed to study these events. Through focused discussions and surveys administered to Workshop participants, considerable gaps in knowledge were identified and outline substantial opportunities for future research.
The field generally agrees that there are two main EV biogenesis pathways, which give rise to distinct EV subpopulations (exosomes and MVs). Despite this, many participants pointed out that we do not currently possess the technology to discern these vesicles from one another once they have entered the extracellular space. Gaining a better understanding of the fundamental properties of EV biogenesis, including the role of varying molecular components and changes in membrane topology during EV formation will undoubtedly aid in developing techniques to differentiate EVs derived from the endocytic pathway versus those shed from the plasma membrane. Other major considerations that have yet to be addressed are the energetic requirements needed for EV biogenesis, how various stimuli alter the activation of these mechanisms, and whether these mechanisms are conserved across cell types and species.
There remain many unanswered questions regarding the basic mechanisms by which EV-cell interactions occur; however, it is likely that these communications necessitate surface molecules present on both EVs and cells. Whether a predominant mechanism (i.e. signalling at the cell surface, endocytosis, or membrane fusion) presides and the level of specificity by which these interactions occur has yet to be determined. The impact of other factors that may influence EV-cell interactions including experimental timing, pH, temperature, membrane composition, EV concentration and stability, and the presence/absence of extracellular matrix all requires further investigation. The cellular consequences of these interactions are heavily understudied; basic knowledge as to how and where EV cargo is unloaded into cells, and the role of EVs in homeostasis has not yet been elucidated. As EVs are being considered as potential therapeutics, in vivo administration and biodistribution studies are now widely conducted. When designing these essential experiments, special considerations for the route of administration, how to best assess/measure biodistribution and careful data interpretation are required.
Development of new techniques for isolating and studying EVs is crucial for advancing the field. Indeed, differential ultracentrifugation, the current gold standard for EV isolation, may unintentionally introduce artefacts and change the inherent properties of the isolated particles. Similarly, the quantification and characterization of isolated vesicles using popular methodologies like nanoparticle tracking analysis may also generate biased or inaccurate data. Despite the great potential of fluorescent labels and dyes for studying EVs, there are numerous shortcomings associated with current products, and implementation of proper controls is often overlooked in the experimental design. Single-vesicle analysis tools are some of the most sought-after technological advancements in the field and are indeed on the horizon. Regardless of the currently available technologies, one of the most important things scientists can do for the study of EVs is to provide transparent reporting in their publications to allow for data replication and proper interpretation. It is hoped that by providing this summary of where consensus is present (or lacking) along with the recommendations for future work that this ISEV position paper will provide a supporting framework for the field to move forward.
Funding Statement
KWW and AER are supported in part by US NIH R01 DA047807; KWW and YH were supported in part by US NIH R01 DA040385 and R56 AG057430, AS is supported by the National Science Foundation Graduate Research Fellowship under Grant No. 1746891; PB is supported by the National Interuniversity Consortium of Materials Science and Technology INSTM and the Center for Colloid and Surface Science (CSGI) through evFOUNDRY (Horizon 2020- Future and emerging technologies (H2020-FETOPEN), ID: 801367); UE is supported by NIH 1K23HL126101-01A1; DWF and NNH are supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging; SMJ is supported by NIH (HL141611, GM130923); NSF (1750542); SK is supported by NIH R01 MH113645; GL is supported by Fondation ARC (PJA20171206453), Cancéropôle Île-de-France (Emergence 2018) and French National Research Agency (ANR-10-IDEX-0001-02 PSL*, ANR-11-LABX-0043 and ANR-18-CE15-0008); AML is supported by The South-Eastern Norwegian Regional Health Authority, The Research Council of Norway, The Norwegian Cancer Society; QL is supported in part by the US NIH R01HL139496, R01ES029097, R01ES030302 and P30ES000002. VM is supported in part by grants from Maryland Stem Cell Research Fund (2016‐MSCRFE‐2739) and National Institutes of Health (R56 AG‐057430); CM received support through NIH T32 OD011089. SS is supported by NIH/NHLBI: R01 HL124187, R01 HL140469, R01HL145644; NYSTEM: C32562GG; AHA:17GRNT33460554; Foundation Leducq: 17CVD04; GvN and FV are supported by Fondation pour la Recherche Medicale (contract AJE20160635884), to GvN, the Fondation ARC pour la Recherche sur le Cancer fellowship (PJA 20161204808) to FJV; AVV is supported by BIO2015-72124-EXP. RTI2018-095214-B-I00 MICINN Ministerio de Ciencia Innovación y Universidades. INNVAL17/22 University Hospital Marqués de Valdecilla-IDIVAL.; AMW is supported by Deutsche Forschungsgemeinschaft (DFG) grant WE5719/2-1; HY is supported by the National Key R&D Program of China Grand No. 2017YFA0505200, the National Natural Science Foundation of China Grand No.20151300651 and No.20181371453, Beijing Outstanding Young Scientist program Grand No. BJJWZYJH01201910003013; DRFC is supported by BBSRC (BB/P006205/1), Cancer Research UK (A28052).
Acknowledgments
The authors would like to thank J. Woodland Pomeroy (Johns Hopkins University) for facilitating all aspects of the planning and execution of the workshop. The Workshop Organizing Committee consisted of Kenneth W. Witwer (Chair; Johns Hopkins University), Norman J. Haughey (Johns Hopkins University), Steven M. Jay (University of Maryland), Alicia Llorente (Oslo University Hospital), Vasiliki Machairaki (Johns Hopkins University), Nicole Noren Hooten (National Institute on Aging, National Institutes of Health), Jeanne Sisk (Johns Hopkins University), Marca Wauben (Universiteit Utrecht), and Hang Yin (Tsinghua University). Roundtable moderators were Daniel Anthony (University of Oxford), Paolo Bergese (University of Brescia), David Raul Francisco Carter (Oxford Brookes University), Jan Lötvall (University of Gothenburg), Matias Ostrowski (University of Buenos Aires), Jeanne Sisk (Johns Hopkins University), Pieter Vader (University Medical Center Utrecht), Roosmarijn E Vandenbroucke (VIB-UGent Center for Inflammation Research), Marca Wauben (Utrecht University), and Hang Yin (University of Colorado Boulder). Oral session chairs included Amrita Datta Chaudhuri (Johns Hopkins University), Yiyao Huang (Johns Hopkins University), Steven M. Jay (University of Maryland), Vasiliki Machairaki (Johns Hopkins University), Nicole Noren Hooten (National Institute on Aging, National Institutes of Health), Ashley E. Russell (Johns Hopkins University), and Zhiwei Wu (Nanjing University).
Disclosure statement
Wauben (MHMW) had a collaborative research agreement with BD Biosciences Europe, Erembodegem, Belgium, to optimize analysis of EVs with the BD Influx.
References
- [1].van Niel G, D’Angelo G, Raposo G.. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–33. Available from: http://www.nature.com/doifinder/10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- [2].Mathieu M, Martin-Jaular L, Lavieu G, et al. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21(1):9–17. [DOI] [PubMed] [Google Scholar]
- [3].Lee Y, El AS, Wood MJA. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. 2012;21(1):R125–34. Available from: https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/hmg/21/R1/10.1093/hmg/dds317/2/dds317.pdf?Expires=1501692232&Signature=CufUkuolAsCDJuyvphs7BZ0FxPUq9VkTxeWcc59XjAneU~1Yk8pPjJoM04o5v0LPzkI~aiNaPBpEq2c-4UXe-BGIZ9Oj6hEpr~ijjHOBHdONL83LUtgh [DOI] [PubMed] [Google Scholar]
- [4].Povero D, Eguchi A, Li H, et al. Circulating extracellular vesicles with specific proteome and liver MicroRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS One. 2014;9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Diderot- UD. Extracellular vesicles in coronary. Nat Publ Gr. 2017;14(5):259–272. [DOI] [PubMed] [Google Scholar]
- [6].Buzas EI, György B, Nagy G, et al. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol. 2014;10(6):356–364. Available from: http://www.nature.com/articles/nrrheum.2014.19 [DOI] [PubMed] [Google Scholar]
- [7].Alvarez-Erviti L, Seow Y, Yin H, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. 2011;29(4):3–4. Available from: https://www.nature.com/nbt/journal/v29/n4/pdf/nbt.1807.pdf [DOI] [PubMed] [Google Scholar]
- [8].Hoshino A, Costa-Silva B, Shen T-L, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. Available from: http://www.nature.com/doifinder/10.1038/nature15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Parolini I, Federici C, Raggi C, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284(49):34211–34222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tan A, Rajadas J, Seifalian AM. Exosomes as nano-theranostic delivery platforms for gene therapy. Adv Drug Deliv Rev. 2013;65(3):357–367. [DOI] [PubMed] [Google Scholar]
- [11].Rabinowits G, Gerçel-Taylor C, JM D, et al. Exosomal microRNA: A diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–46. [DOI] [PubMed] [Google Scholar]
- [12].Mizutani K, Terazawa R, Kameyama K, et al. Isolation of prostate cancer-related exosomes. Anticancer Res. 2014;34(7):3419–3423. [PubMed] [Google Scholar]
- [13].Lance RS, Drake RR, Troyer DA. Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Expert Rev Anticancer Ther. 2011;11(9):1341–1343. [DOI] [PubMed] [Google Scholar]
- [14].Tokuhara T, Hattori N, Ishida H, et al. Clinical significance of CD151 gene expression in non-small cell lung cancer. Clin Cancer Res. 2001;7(12):4109–4114. [PubMed] [Google Scholar]
- [15].Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22635005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Logozzi M, De MA, Lugini L, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4(4). Available from: https://www.ncbi.nlm.nih.gov/pubmed/19381331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li Y, Zhang Y, Qiu F, et al. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis. 2011;32(15):1976–1983. [DOI] [PubMed] [Google Scholar]
- [19].György B, Szabó TG, Pásztói M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68(16):2667–2688. Available from: http://link.springer.com/10.1007/s00018-011-0689-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Heijnen HF, Schiel AE, Fijnheer R, et al. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and-granules. Blood. 1999;94(11):3791–3799. Available from: www.bloodjournal.org [PubMed] [Google Scholar]
- [21].Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23420871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Trams EG, Lauter CJ, Salem N, et al. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta. 1981;645(1):63–70. [DOI] [PubMed] [Google Scholar]
- [23].Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97(2):329–339. Available from: http://www.ncbi.nlm.nih.gov/pubmed/6309857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Johnstone RM, Adam M, Hammond JR, et al. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262(19):9412–9420. [PubMed] [Google Scholar]
- [25].Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364–372. [DOI] [PubMed] [Google Scholar]
- [27].Groot Kormelink T, Arkesteijn GJA, van de Lest CHA, et al. Mast cell degranulation is accompanied by the release of a selective subset of extracellular vesicles that contain mast cell–specific proteases. J Immunol. 2016;197(8):3382–3392. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27619994 [DOI] [PubMed] [Google Scholar]
- [28].Skotland T, Hessvik NP, Sandvig K, et al. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J Lipid Res. 2018;60(1):9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Osteikoetxea X, Balogh A, Szabó-Taylor K, et al. Improved characterization of EV preparations based on protein to lipid ratio and lipid properties. PLoS One. 2015;10(3):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mulcahy LA, Pink RC, Carter DRF. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ofir-Birin Y, Abou Karam P, Rudik A, et al. Monitoring extracellular vesicle cargo active uptake by imaging flow cytometry. Front Immunol. 2018;9:1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lötvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the international society for extracellular vesicles. J Extracell Vesicles. 2014;3(1):26913 Available from: https://www.tandfonline.com/doi/full/10.3402/jev.v3.26913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750 Available from: https://www.tandfonline.com/doi/full/10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Edgar JR, Manna PT, Nishimura S, et al. Tetherin is an exosomal tether. Elife. 2016;5 Available from https://elifesciences.org/articles/17180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wehman AM, Poggioli C, Schweinsberg P, et al. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. Elegans Embryos Curr Biol. 2011;21(23):1951–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bebelman M, van Niel G, Pegtel M, et al. Real-time imaging assay of multivesicular body-PM fusion to quantify exosome release from single cells. Protoc Exch. 2018; Available from: http://www.nature.com/protocolexchange/protocols/6837 [DOI] [PubMed] [Google Scholar]
- [37].Zhang H, Freitas D, Kim HS, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018;20(3):332–343. Available from: http://www.nature.com/articles/s41556-018-0040-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Vance JE, Steenbergen R. Metabolism and functions of phosphatidylserine. Prog Lipid Res. 2005;44(4):207–234. Available from: https://www.sciencedirect.com/science/article/pii/S0163782705000214?via%3Dihub [DOI] [PubMed] [Google Scholar]
- [39].Lea J, Sharma R, Yang F, et al. Detection of phosphatidylserine-positive exosomes as a diagnostic marker for ovarian malignancies: a proof of concept study. Oncotarget. 2017;8(9):14395–14407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cvjetkovic A, Jang SC, Konečná B, et al. Detailed analysis of protein topology of extracellular vesicles-evidence of unconventional membrane protein orientation. Sci Rep. 2016;6:36338 Available from https://www.nature.com/articles/srep36338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol. 2016;36(3):301–312. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27053351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Janas T, Janas MM, Sapoń K, et al. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015;589(13):1391–1398. Available from: https://www.sciencedirect.com/science/article/pii/S001457931500294X [DOI] [PubMed] [Google Scholar]
- [43].Gibbings DJ, Ciaudo C, Erhardt M, et al. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11(9):1143–1149. Available from: http://www.nature.com/doifinder/10.1038/ncb1929 [DOI] [PubMed] [Google Scholar]
- [44].Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I, et al. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem. 2013;288(17):11649–11661. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23463506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mateescu B, Kowal EJK, van Balkom BWM, et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J Extracell Vesicles. 2017;6(1):1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wei Z, Batagov AO, Schinelli S, et al. Coding and noncoding landscape of extracellular RNA released by human glioma stem cells. Nat Commun. 2017;8(1):1145 Available from: http://www.nature.com/articles/s41467-017-01196-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].McKenzie AJ, Hoshino D, Hong NH, et al. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 2016;15(5):978–987. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27117408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mukherjee K, Ghoshal B, Ghosh S, et al. Reversible HuR‐microRNA binding controls extracellular export of miR‐122 and augments stress response. EMBO Rep. 2016;17(8):1184–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4(1):2980 Available from: http://www.nature.com/articles/ncomms3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Colombo M, Moita C, van Niel G, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126(Pt 24):5553–5565. [DOI] [PubMed] [Google Scholar]
- [51].Lu A, Wawro P, DW M, et al. Genome-wide interrogation of extracellular vesicle biology using barcoded miRNAs. Elife. 2018;7 Available from https://elifesciences.org/articles/41460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ostrowski M, Carmo NB, Krumeich S, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1):19–30. Available from: http://www.nature.com/doifinder/10.1038/ncb2000 [DOI] [PubMed] [Google Scholar]
- [53].Sinha S, Hoshino D, Hong NH, et al. Cortactin promotes exosome secretion by controlling branched actin dynamics. J Cell Biol. 2016;214(2):197–213. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27402952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hoshino D, Kirkbride KC, Costello K, et al. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013;5(5):1159–1168. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24290760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Savina A, Furlán M, Vidal M, et al. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278(22):20083–20090. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12639953 [DOI] [PubMed] [Google Scholar]
- [56].Savina A, Fader CM, Damiani MT, et al. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic. 2005;6(2): 131–143. Available from. [DOI] [PubMed] [Google Scholar]
- [57].Hsu C, Morohashi Y, Yoshimura S, et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A–C. J Cell Biol. 2010;189(2):223–232. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20404108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yeung V, Webber JP, Dunlop EA, et al. Rab35-dependent extracellular nanovesicles are required for induction of tumour supporting stroma. Nanoscale. 2018;10(18):8547–8559. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29693684 [DOI] [PubMed] [Google Scholar]
- [59].Beer KB, Rivas-Castillo J, Kuhn K, et al. Extracellular vesicle budding is inhibited by redundant regulators of TAT-5 flippase localization and phospholipid asymmetry. Proc Natl Acad Sci U S A. 2018;115(6):E1127–36. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29367422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Nabhan JF, Hu R, Oh RS, et al. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. PNAS. 2012;109(11):4146–4151. Available from: http://www.pnas.org/content/pnas/109/11/4146.full.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Anand S, Foot N, Ang C-S, et al. Arrestin-domain containing protein 1 (Arrdc1) regulates the protein cargo and release of extracellular vesicles. Proteomics. 2018;18(17):1800266 Available from: http://www.ncbi.nlm.nih.gov/pubmed/30035390 [DOI] [PubMed] [Google Scholar]
- [62].Mackenzie K, Foot NJ, Anand S, et al. Regulation of the divalent metal ion transporter via membrane budding. Cell Discov. 2016;2(1):16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang Q, Lu Q. Plasma membrane-derived extracellular microvesicles mediate non-canonical intercellular NOTCH signaling. Nat Commun. 2017;8(1):709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Krempler A, Henry MD, Triplett AA, et al. Targeted deletion of the Tsg101 gene results in cell cycle arrest at G1/S and p53-independent cell death. J Biol Chem. 2002;277(45):43216–43223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Morris CR, Stanton MJ, Manthey KC, et al. A knockout of the Tsg101 gene leads to decreased expression of ErbB receptor tyrosine kinases and induction of autophagy prior to cell death. Basu S, editor. PLoS One. 2012;7(3):e34308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Lu Q, Hope LW, Brasch M, et al. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc Natl Acad Sci. 2003;100(13):7626–7631. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12802020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Alonso R, Rodríguez MC, Pindado J, et al. Diacylglycerol kinase alpha regulates the secretion of lethal exosomes bearing Fas ligand during activation-induced cell death of T lymphocytes. J Biol Chem. 2005;280(31):28439–28450. Available from: http://www.jbc.org/content/280/31/28439.full.pdf [DOI] [PubMed] [Google Scholar]
- [68].Alonso R, Rodriguez M, Marsh M, et al. Diacylglycerol kinase alpha regulates the formation and polarisation of mature multivesicular bodies involved in the secretion of Fas ligand-containing exosomes in T lymphocytes. Cell Death Differ. 2011;18(10):1161–1173. Available from: https://www.nature.com/articles/cdd2010184.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].de Gassart A, Geminard C, Fevrier B, et al. Lipid raft-associated protein sorting in exosomes. Blood. 2003;102(13):4336–4344. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12881314 [DOI] [PubMed] [Google Scholar]
- [70].Valapala M, Vishwanatha JK. Lipid raft endocytosis and exosomal transport facilitate extracellular trafficking of annexin A2. J Biol Chem. 2011;286(35):30911–30925. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21737841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Trajkovic K, Hsu C, Chiantia S, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 80 2008;319(5867):1244–1247. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18309083 [DOI] [PubMed] [Google Scholar]
- [72].Phuyal S, Hessvik NP, Skotland T, et al. Regulation of exosome release by glycosphingolipids and flotillins. Febs J. 2014;281(9):2214–2227. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24605801 [DOI] [PubMed] [Google Scholar]
- [73].Menck K, Sönmezer C, Worst TS, et al. Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. J Extracell Vesicles. 2017;6(1):1378056 Available from: http://www.ncbi.nlm.nih.gov/pubmed/29184623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Bianco F, Perrotta C, Novellino L, et al. Acid sphingomyelinase activity triggers microparticle release from glial cells. Embo J. 2009;28(8):1043–1054. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19300439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Glebov K, Löchner M, Jabs R, et al. Serotonin stimulates secretion of exosomes from microglia cells. Glia. 2015;63(4):626–634. Available from. [DOI] [PubMed] [Google Scholar]
- [76].Thery C, Ostrowski M, Segura E, et al. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9(8):581–593. [DOI] [PubMed] [Google Scholar]
- [77].Obregon C, Rothen-Rutishauser B, Gitahi SK, et al. Exovesicles from human activated dendritic cells fuse with resting dendritic cells, allowing them to present alloantigens. Am J Pathol. 2006;169(6):2127–2136. Available from: https://www.sciencedirect.com/science/article/pii/S0002944010626727?via%3Dihub#fig1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Segura E, Nicco C, Lombard B, et al. ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood. 2005;106(1):216–223. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15790784 [DOI] [PubMed] [Google Scholar]
- [79].Iliev D, Strandskog G, Nepal A, et al. Stimulation of exosome release by extracellular DNA is conserved across multiple cell types. Febs J. 2018;285(16):3114–3133. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/febs.14601 [DOI] [PubMed] [Google Scholar]
- [80].Saunderson SC, Schuberth PC, Dunn AC, et al. Induction of exosome release in primary B cells stimulated via CD40 and the IL-4 receptor. J Immunol. 2008;180(12):8146–8152. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18523279 [DOI] [PubMed] [Google Scholar]
- [81].Srinivasan S, Su M, Ravishankar S, et al. TLR-exosomes exhibit distinct kinetics and effector function. Sci Rep. 2017;7(1):41623 Available from: http://www.nature.com/articles/srep41623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wei Y, Wang D, Jin F, et al. Pyruvate kinase type M2 promotes tumour cell exosome release via phosphorylating synaptosome-associated protein 23. Nat Commun. 2017;8:14041 Available from http://www.nature.com/doifinder/10.1038/ncomms14041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Wang T, Gilkes DM, Takano N, et al. Hypoxia-inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc Natl Acad Sci. 2014;111(31):E3234–42. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24938788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Zhang F, Li R, Yang Y, et al. Specific decrease in B-cell-derived extracellular vesicles enhances post-chemotherapeutic CD8+ T cell responses. Immunity. 2019;50(3):738–750. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1074761319300330 [DOI] [PubMed] [Google Scholar]
- [85].Heusermann W, Hean J, Trojer D, et al. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J Cell Biol. 2016;213(2):173–184. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27114500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Purushothaman A, Bandari SK, Liu J, et al. Fibronectin on the surface of myeloma cell-derived exosomes mediates exosome-cell interactions. J Biol Chem. 2016;291(4):1652–1663. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26601950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Feng D, Zhao W-L, Ye -Y-Y, et al. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11(5):675–687. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20136776 [DOI] [PubMed] [Google Scholar]
- [88].Berenguer J, Lagerweij T, Zhao XW, et al. Glycosylated extracellular vesicles released by glioblastoma cells are decorated by CCL18 allowing for cellular uptake via chemokine receptor CCR8. J Extracell Vesicles. 2018;7(1):1446660 Available from: https://www.tandfonline.com/doi/full/10.1080/20013078.2018.1446660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Mack M, Kleinschmidt A, Bruhl H, et al. Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat Med. 2000;6(7):769–775. [DOI] [PubMed] [Google Scholar]
- [90].Denzer K, van Eijk M, Kleijmeer MJ, et al. Follicular dendritic cells carry MHC class II-expressing microvesicles at their surface. J Immunol. 2000;165(3):1259–1265. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10903724 [DOI] [PubMed] [Google Scholar]
- [91].Nolte-’t Hoen ENM, Buschow SI, Anderton SM, et al. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood. 2009;113(9):1977–1981. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19064723 [DOI] [PubMed] [Google Scholar]
- [92].Di Trapani M, Bassi G, Midolo M, et al. Differential and transferable modulatory effects of mesenchymal stromal cell-derived extracellular vesicles on T, B and NK cell functions. Sci Rep. 2016;6(1):24120 Available from: http://www.ncbi.nlm.nih.gov/pubmed/27071676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Kirschner MB, Kao SC, Edelman JJ, et al. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6(9):e24145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Pritchard CC, Kroh E, Wood B, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev Res. 2012;5(3):492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Verweij FJ, Revenu C, Arras G, et al. Live tracking of inter-organ communication by endogenous exosomes in vivo. Dev Cell. 2019;48(4):573–589.e4. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30745143 [DOI] [PubMed] [Google Scholar]
- [96].Busatto S, Giacomini A, Montis C, et al. Uptake profiles of human serum exosomes by murine and human tumor cells through combined use of colloidal nanoplasmonics and flow cytofluorimetric analysis. Anal Chem. 2018;90(13):7855–7861. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29870225 [DOI] [PubMed] [Google Scholar]
- [97].Morelli AE, Larregina AT, Shufesky WJ, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104(10):3257–3266. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15284116 [DOI] [PubMed] [Google Scholar]
- [98].Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15(7):690–698. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18596815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Costa Verdera H, Gitz-Francois JJ, Schiffelers RM, et al. Cellular uptake of extracellular vesicles is mediated by clathrin-independent endocytosis and macropinocytosis. J Control Release. 2017;266:100–108. Available from http://www.ncbi.nlm.nih.gov/pubmed/28919558 [DOI] [PubMed] [Google Scholar]
- [100].Svensson KJ, Christianson HC, Wittrup A, et al. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem. 2013;288(24):17713–17724. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23653359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhang J, Xue R, Ong W-Y, et al. Roles of cholesterol in vesicle fusion and motion. Biophys J. 2009;97(5):1371–1380. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19720025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Najafinobar N, Mellander LJ, Kurczy ME, et al. Cholesterol alters the dynamics of release in protein independent cell models for exocytosis. Sci Rep. 2016;6(1):33702 Available from: http://www.ncbi.nlm.nih.gov/pubmed/27650365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Grouleff J, Irudayam SJ, Skeby KK, et al. The influence of cholesterol on membrane protein structure, function, and dynamics studied by molecular dynamics simulations. Biochim Biophys Acta - Biomembr. 2015;1848(9):1783–1795. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25839353 [DOI] [PubMed] [Google Scholar]
- [104].Lee JY, Schick M. Calculation of free energy barriers to the fusion of small vesicles. Biophys J. 2008;94(5):1699–1706. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18024495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Lentz BR, Carpenter TJ, Alford DR. Spontaneous fusion of phosphatidylcholine small unilamellar vesicles in the fluid phase. Biochemistry. 1987;26(17):5389–5397. Available from: http://pubs.acs.org/doi/abs/10.1021/bi00391a026 [DOI] [PubMed] [Google Scholar]
- [106].Tesch DM, Nevzorov AA. Sensitivity enhancement and contrasting information provided by free radicals in oriented-sample NMR of bicelle-reconstituted membrane proteins. J Magn Reson. 2014;239:9–15. Available from http://www.ncbi.nlm.nih.gov/pubmed/24355622 [DOI] [PubMed] [Google Scholar]
- [107].Al-Abdul-Wahid MS, Neale C, Pomes R, et al. A solution NMR approach to the measurement of amphiphile immersion depth and orientation in membrane model systems. J Am Chem Soc. 2009;131:6452–6459. Available from http://www.ncbi.nlm.nih.gov/pubmed/19415935 [DOI] [PubMed] [Google Scholar]
- [108].Chu S, Maltsev S, Emwas AH, et al. Solid-state NMR paramagnetic relaxation enhancement immersion depth studies in phospholipid bilayers. J Magn Reson. 2010;207:89–94. Available from http://www.ncbi.nlm.nih.gov/pubmed/20851650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Park SH, Berkamp S, Cook GA, et al. Nanodiscs versus macrodiscs for NMR of membrane proteins. Biochemistry. 2011;50:8983–8985. Available from http://www.ncbi.nlm.nih.gov/pubmed/21936505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Maiolo D, Paolini L, Di Noto G, et al. Colorimetric nanoplasmonic assay to determine purity and titrate extracellular vesicles. Anal Chem. 2015;87(8):4168–4176. Available from: http://pubs.acs.org/doi/10.1021/ac504861d [DOI] [PubMed] [Google Scholar]
- [111].Contreras-Naranjo JC, Wu H-J, Ugaz VM. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab Chip. 2017;17(21):3558–3577. Available from http://xlink.rsc.org/?DOI=C7LC00592J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Montis C, Busatto S, Valle F, et al. Biogenic supported lipid bilayers from nanosized extracellular vesicles. Adv Biosyst. 2018;2(4):1700200 Available from: http://doi.wiley.com/10.1002/adbi.201700200 [Google Scholar]
- [113].Kooijmans SAA, Fliervoet LAL, van der Meel R, et al. PEGylated and targeted extracellular vesicles display enhanced cell specificity and circulation time. J Control Release. 2016;224:77–85. [DOI] [PubMed] [Google Scholar]
- [114].Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Royo F, Cossio U, Ruiz de Angulo A, et al. Modification of the glycosylation of extracellular vesicles alters their biodistribution in mice. Nanoscale. 2019;11:1531–1537. Available from http://www.ncbi.nlm.nih.gov/pubmed/30623961 [DOI] [PubMed] [Google Scholar]
- [116].Hu L, Wickline SA, Hood JL. Magnetic resonance imaging of melanoma exosomes in lymph nodes. Magn Reson Med. 2015;74(1):266–271. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25052384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Lai CP, Mardini O, Ericsson M, et al. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano. 2014;8:483–494. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24383518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Wiklander OP, Nordin JZ, O’Loughlin A, et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles. 2015;4:26316 Available from http://www.ncbi.nlm.nih.gov/pubmed/25899407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Lundy DJ, Chen K-H, Toh EK-W, et al. Distribution of systemically administered nanoparticles reveals a size-dependent effect immediately following cardiac ischaemia-reperfusion injury. Sci Rep. 2016;6(1):25613 Available from: http://www.ncbi.nlm.nih.gov/pubmed/27161857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].van der Meel R, Fens MHAM, Vader P, et al. Extracellular vesicles as drug delivery systems: lessons from the liposome field. J Control Release. 2014;195:72–85. Available from http://www.ncbi.nlm.nih.gov/pubmed/25094032 [DOI] [PubMed] [Google Scholar]
- [121].Reineke J. Terminology matters: there is no targeting, but retention. J Control Release. 2018;273:180–183. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29360476 [DOI] [PubMed] [Google Scholar]
- [122].Cvjetkovic A, Crescitelli R, Lässer C, et al. Extracellular vesicles in motion. Matters. 2017;3(6):e201704000003 Available from: https://sciencematters.io/articles/10.19185/matters.201704000003 [Google Scholar]
- [123].Allijn IE, Czarny BMS, Wang X, et al. Liposome encapsulated berberine treatment attenuates cardiac dysfunction after myocardial infarction. J Control Release. 2017;247:127–133. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28065862 [DOI] [PubMed] [Google Scholar]
- [124].Balusu S, Van Wonterghem E, De Rycke R, et al. Identification of a novel mechanism of blood–brain communication during peripheral inflammation via choroid plexus‐derived extracellular vesicles. EMBO Mol Med. 2016;8(10):1162–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Grange C, Tapparo M, Bruno S, et al. Biodistribution of mesenchymal stem cell-derived extracellular vesicles in a model of acute kidney injury monitored by optical imaging. Int J Mol Med. 2014;33(5):1055–1063. Available from: https://www.spandidos-publications.com/10.3892/ijmm.2014.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Zomer A, Steenbeek SC, Maynard C, et al. Studying extracellular vesicle transfer by a Cre-loxP method. Nat Protoc. 2015;11(1):87–101. Available from: http://www.nature.com/doifinder/10.1038/nprot.2015.138 [DOI] [PubMed] [Google Scholar]
- [127].Quek C, Hill AF. The role of extracellular vesicles in neurodegenerative diseases. Biochem Biophys Res Commun. 2017;483(4):1178–1186. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27659705 [DOI] [PubMed] [Google Scholar]
- [128].You Y, Ikezu T. Emerging roles of extracellular vesicles in neurodegenerative disorders. Neurobiol Dis. 2019;130:104512 Available from http://www.ncbi.nlm.nih.gov/pubmed/31229685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Aliotta JM, Sanchez-Guijo FM, Dooner GJ, et al. Alteration of marrow cell gene expression, protein production, and engraftment into lung by lung-derived microvesicles: a novel mechanism for phenotype modulation. Stem Cells. 2007;25(9):2245–2256. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17556595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Aliotta JM, Pereira M, Wen S, et al. Exosomes induce and reverse monocrotaline-induced pulmonary hypertension in mice. Cardiovasc Res. 2016;110(3):319–330. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26980205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci. 2016;113(8):E968–77. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26858453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Chen C, Zong S, Wang Z, et al. Imaging and intracellular tracking of cancer-derived exosomes using single-molecule localization-based super-resolution microscope. ACS Appl Mater Interfaces. 2016;8(39):25825–25833. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27617891 [DOI] [PubMed] [Google Scholar]
- [133].van der Pol E, Coumans FAW, Grootemaat AE, et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014;12(7):1182–1192. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24818656 [DOI] [PubMed] [Google Scholar]
- [134].Ter-Ovanesyan D, Kowal EJK, Regev A, et al. Imaging of isolated extracellular vesicles using fluorescence microscopy. Methods Mol Biol (Clifton, NJ). 2017;233–241. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28828661 [DOI] [PubMed] [Google Scholar]
- [135].Maas SLN, de Vrij J, van der Vlist EJ, et al. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. J Control Release. 2015;200:87–96. Available from http://www.ncbi.nlm.nih.gov/pubmed/25555362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Witwer KW, Buzás EI, Bemis LT, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:20360 Available from http://www.ncbi.nlm.nih.gov/pubmed/24009894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Van Deun J, Mestdagh P, Agostinis P, et al. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods. 2017;14(3):228–232. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28245209 [DOI] [PubMed] [Google Scholar]
- [138].Tanenbaum ME, Gilbert LA, Qi LS, et al. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159(3):635–646. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25307933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Ramirez MI, Amorim MG, Gadelha C, et al. Technical challenges of working with extracellular vesicles. Nanoscale. 2018;10(3):881–906. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29265147 [DOI] [PubMed] [Google Scholar]
- [140].Théry C, Amigorena S, Raposo G, et al. Isolation and characterization of exosomes from cell culture supernatants and biological fluids In: Current protocols in cell biology. Hoboken, NJ, USA: John Wiley & Sons, Inc; 2006. p. Unit 3.22 Available from: http://www.ncbi.nlm.nih.gov/pubmed/18228490 [DOI] [PubMed] [Google Scholar]
- [141].Linares R, Tan S, Gounou C, et al. High-speed centrifugation induces aggregation of extracellular vesicles. J Extracell Vesicles. 2015;4(1):29509 Available from: http://www.ncbi.nlm.nih.gov/pubmed/26700615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Pužar Dominkuš P, Stenovec M, Sitar S, et al. PKH26 labeling of extracellular vesicles: characterization and cellular internalization of contaminating PKH26 nanoparticles. Biochim Biophys Acta - Biomembr. 2018;1860(6):1350–1361. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29551275 [DOI] [PubMed] [Google Scholar]
- [143].van der Vlist EJ, Nolte-’t Hoen ENM, Stoorvogel W, et al. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat Protoc. 2012;7(7):1311–1326. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22722367 [DOI] [PubMed] [Google Scholar]
- [144].Roberts-Dalton HD, Cocks A, Falcon-Perez JM, et al. Fluorescence labelling of extracellular vesicles using a novel thiol-based strategy for quantitative analysis of cellular delivery and intracellular traffic. Nanoscale. 2017;9(36):13693–13706. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28880029 [DOI] [PubMed] [Google Scholar]
- [145].Monopoli MP, Zendrini A, Wu D, et al. Endogenous exosome labelling with an amphiphilic NIR-fluorescent probe. Chem Commun. 2018;54(52):7219–7222. Available from http://xlink.rsc.org/?DOI=C8CC02135J. [DOI] [PubMed] [Google Scholar]
- [146].Verweij FJ, Bebelman MP, Jimenez CR, et al. Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J Cell Biol. 2018;217(3):1129–1142. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29339438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Beer KB, Fazeli G, Judasova K, et al. Degron-tagged reporters probe membrane topology and enable the specific labelling of membrane-wrapped structures. Nat Commun. 2019;10(1):3490 Available from: http://www.nature.com/articles/s41467-019-11442-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Lai CP, Kim EY, Badr CE, et al. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat Commun. 2015;6(1):7029 Available from: http://www.nature.com/articles/ncomms8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Gabrielli M, Battista N, Riganti L, et al. Active endocannabinoids are secreted on extracellular membrane vesicles. EMBO Rep. 2015;16(2):213–220. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25568329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Gardiner C, Di VD, Sahoo S, et al. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles. 2016;5(1):32945 Available from: http://www.ncbi.nlm.nih.gov/pubmed/27802845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Kormelink TG, Arkesteijn GJA, Nauwelaers FA, et al. Prerequisites for the analysis and sorting of extracellular vesicle subpopulations by high-resolution flow cytometry. Cytom Part A. 2016;89(2):135–147. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25688721 [DOI] [PubMed] [Google Scholar]
- [152].Libregts SFWM, Arkesteijn GJA, Németh A, et al. Flow cytometric analysis of extracellular vesicle subsets in plasma: impact of swarm by particles of non-interest. Available from J Thromb Haemost. 2018;167:1423–1436. [DOI] [PubMed] [Google Scholar]
- [153].Nolan JP, Jones JC. Detection of platelet vesicles by flow cytometry. Platelets. 2017;28(3):256–262. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28277059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Lai RC, Tan SS, Yeo RWY, et al. MSC secretes at least 3 EV types each with a unique permutation of membrane lipid, protein and RNA. J Extracell Vesicles. 2016;5(1):29828 Available from: http://www.ncbi.nlm.nih.gov/pubmed/26928672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Morton LA, Tamura R, de Jesus AJ, et al. Biophysical investigations with MARCKS-ED: dissecting the molecular mechanism of its curvature sensing behaviors. Biochim Biophys Acta - Biomembr. 2014;1838(12):3137–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Morton LA, Yang H, Saludes JP, et al. MARCKS-ED peptide as a curvature and lipid sensor. ACS Chem Biol. 2013;8(1):218–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Gomes J, Lucien F, Cooper T, et al. Analytical considerations in nanoscale flow cytometry of extracellular vesicles to achieve data linearity. Thromb Haemost. 2018;118(09):1612–1624. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30112749 [DOI] [PubMed] [Google Scholar]
- [158].Prada I, Amin L, Furlan R, et al. A new approach to follow a single extracellular vesicle–cell interaction using optical tweezers. Biotechniques. 2016;60(1):35–41. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26757810 [DOI] [PubMed] [Google Scholar]
- [159].Murphy DE, de Jong OG, Brouwer M, et al. Extracellular vesicle-based therapeutics: natural versus engineered targeting and trafficking. Exp Mol Med. 2019;51(3):32 Available from: http://www.nature.com/articles/s12276-019-0223-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Kooijmans SAA, Gitz-Francois JJJM, Schiffelers RM, et al. Recombinant phosphatidylserine-binding nanobodies for targeting of extracellular vesicles to tumor cells: a plug-and-play approach. Nanoscale. 2018;10(5):2413–2426. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29334397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Gross JC, Chaudhary V, Bartscherer K, et al. Active Wnt proteins are secreted on exosomes. Nat Cell Biol. 2012;14(10):1036–1045. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22983114 [DOI] [PubMed] [Google Scholar]
- [162].Korkut C, Li Y, Koles K, et al. Regulation of postsynaptic retrograde signaling by presynaptic exosome release. Neuron. 2013;77(6):1039–1046. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23522040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Ashley J, Cordy B, Lucia D, et al. Retrovirus-like gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell. 2018;172(1–2):262–274.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Hyenne V, Apaydin A, Rodriguez D, et al. RAL-1 controls multivesicular body biogenesis and exosome secretion. J Cell Biol. 2015;211(1):27–37. Available from: http://www.jcb.org/lookup/doi/10.1083/jcb.201504136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Beer KB, Wehman AM. Mechanisms and functions of extracellular vesicle release in vivo—what we can learn from flies and worms. Cell Adhes Migrat. 2017; 11 (2):135––150.. doi: 10.1080/19336918.2016.1236899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Patel DB, Gray KM, Santharam Y, et al. Impact of cell culture parameters on production and vascularization bioactivity of mesenchymal stem cell-derived extracellular vesicles. Bioeng Transl Med. 2017;2(2):170–179. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28932818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Wei Z, Batagov AO, Carter DRF, et al. Fetal bovine serum RNA interferes with the cell culture-derived extracellular RNA. Sci Rep. 2016;6(1):31175 Available from: http://www.ncbi.nlm.nih.gov/pubmed/27503761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Liu Y, Mi Y, Mueller T, et al. Multi-omic measurements of heterogeneity in HeLa cells across laboratories. Nat Biotechnol. 2019;37(3):314–322. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30778230 [DOI] [PubMed] [Google Scholar]
- [169].Ayers L, Pink R, Carter DRF, et al. Clinical requirements for extracellular vesicle assays. J Extracell Vesicles. 2019;8(1):1593755 Available from: http://www.ncbi.nlm.nih.gov/pubmed/30949310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Tkach M, Kowal J, Théry C. Why the need and how to approach the functional diversity of extracellular vesicles. Philos Trans R Soc B Biol Sci. 2018;373(1737):20160479 Available from: http://rstb.royalsocietypublishing.org/lookup/doi/10.1098/rstb.2016.0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Zabeo D, Cvjetkovic A, Lässer C, et al. Exosomes purified from a single cell type have diverse morphology. J Extracell Vesicles. 2017;6(1):1329476 Available from: http://www.ncbi.nlm.nih.gov/pubmed/28717422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Lener T, Gimona M, Aigner L, et al. Applying extracellular vesicles based therapeutics in clinical trials – an ISEV position paper. J Extracell Vesicles. 2015;4(1):30087 Available from: http://www.ncbi.nlm.nih.gov/pubmed/26725829 [DOI] [PMC free article] [PubMed] [Google Scholar]


