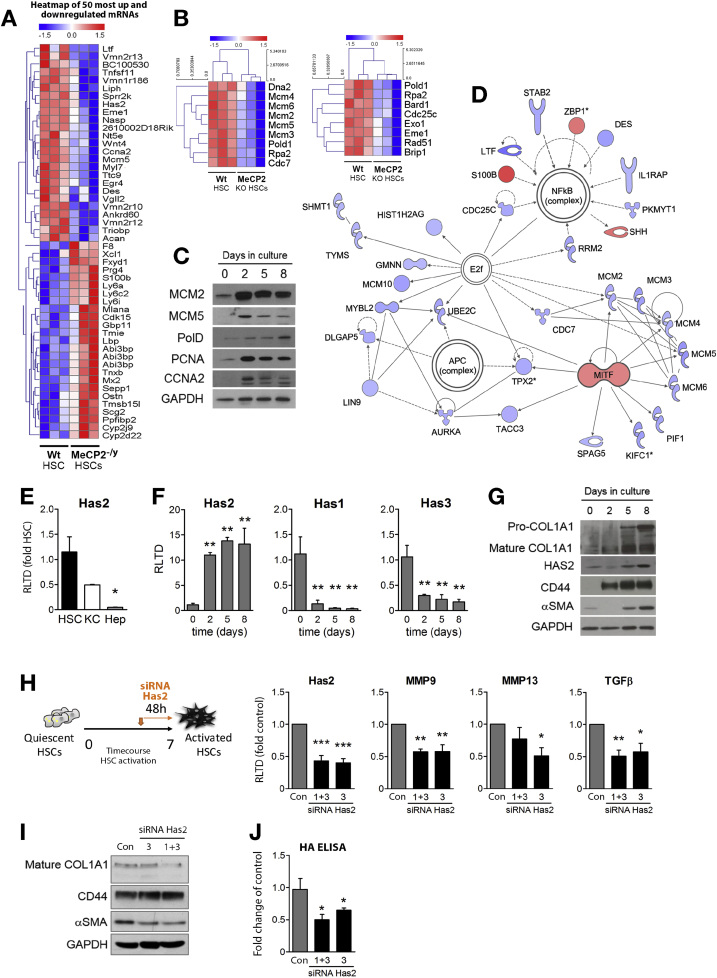
Figure 2.
Mecp2 regulates DNA replication and repair systems; Has2 is involved in HSC transdifferentiation. (A) Heatmap displaying the results of mRNA microarray performed on activated HSCs isolated from 3 control and 3 Mecp2–/y mice. The top 50 most up-regulated and down-regulated genes are shown. Blue denotes down-regulation; red, up-regulation; white, unchanged. (B) Heatmap displaying the most down-regulated genes with a fundamental role in DNA repair responses (C) Western blot for MCM2, MCM5, POLD1, PCNA, CCNA2, and GAPDH in WT HSCs after 0, 2, 5, and 8 days in culture. (D) IPA was used to form a network of focus genes that are downstream targets of differentially expressed mRNAs. Blue nodes signify that a gene was down-regulated in Mecp2–/y myofibroblasts; red signifies up-regulation. Symbol shapes signify the nature of the encoded protein, and unshaded symbols signify genes relevant to the pathway but not differentially expressed in our data set. (E) mRNA level of Has2 from HSCs, Kupffer cells (KC), and hepatocytes (Hep). (F) mRNA levels of Has2, Has1, and Has3 in HSCs after 0, 2, 5, and 8 days in culture. (G) Western blot for COL1A1, HAS2, the receptor CD44, α-SMA, and GAPDH in HSCs after 0, 2, 5, and 8 days in culture. (H) Schematic showing the silencing experiment: HSCs were cultured for 5 days, then transfected with a cocktail of Has2 targeting siRNA (1 + 3), single Has2 targeting siRNA (3) or a negative control (Con) and harvested 48 hours later on day 7. mRNA levels of Has2, MMP9, MMP13, and transforming growth factor β in control and Has2 siRNA–transfected HSCs. Error bars represent mean ± SEM. Statistical significance was determined by ANOVA; *P < .05, **P < .01, ***P < .001. (I) Western blot for Col1A1, CD44, α-SMA, and GAPDH and (J) HA enzyme-linked immunosorbent assay in HSCs transfected with Has2 siRNA (as in H).