Abstract
Friedreich’s ataxia is caused by large homozygous, intronic expansions of GAA repeats in the frataxin (FXN) gene, resulting in severe downregulation of its expression. Pathogenic repeats are located in intron one, hence patients express unaffected FXN protein, albeit in low quantities. Although FRDA symptoms typically afflict the nervous system, hypertrophic cardiomyopathy is the predominant cause of death. Our studies were conducted using cardiomyocytes differentiated from induced pluripotent stem cells derived from control individuals, FRDA patients, and isogenic cells corrected by zinc finger nucleases-mediated excision of pathogenic expanded GAA repeats. This correction of the FXN gene removed the primary trigger of the transcription defect, upregulated frataxin expression, reduced pathological lipid accumulation observed in patient cardiomyocytes, and reversed gene expression signatures of FRDA cardiomyocytes. Transcriptome analyses revealed hypertrophy-specific expression signatures unique to FRDA cardiomyocytes, and emphasized similarities between unaffected and ZFN-corrected FRDA cardiomyocytes. Thus, the iPSC-derived FRDA cardiomyocytes exhibit various molecular defects characteristic for cellular models of cardiomyopathy that can be corrected by genome editing of the expanded GAA repeats. These results underscore the utility of genome editing in generating isogenic cellular models of FRDA and the potential of this approach as a future therapy for this disease.
Keywords: Friedreich’s ataxia, GAA repeats, genome editing, isogenic iPSC, cardiomyocytes, lipid metabolism
1. Introduction
Friedreich’s ataxia (FRDA, FA, OMIM229300) is the most prevalent inherited ataxia in humans with a population frequency of 1-2:50,000 and carrier frequency of 1:60 to 1:120 (Campuzano et al., 1996). FRDA is caused by hyperexpansion of the GAA repeats located in the first intron of the FXN gene (Pandolfo, 2006). Unaffected-range alleles contain fewer than 30 triplets and disease-causing expanded alleles can harbor up to 2000 GAA repeats (Campuzano et al., 1996; Montermini et al., 1997). Long GAA tracts inhibit expression of the FXN gene, thus patients homozygous for the GAA expansion have very low frataxin mRNA and protein levels when compared with heterozygous carriers and healthy controls (Campuzano et al., 1996). It has been demonstrated in multiple systems that expanded GAAs induce epigenetic silencing of the FXN gene (Herman et al., 2006; Ku et al., 2010). Frataxin is a small, highly conserved protein expressed in most eukaryotic organisms. Although nuclear-encoded, it functions in mitochondria regulating iron homeostasis primarily as a co-factor in biosynthesis of iron-sulfur clusters (Pastore and Puccio, 2013). Frataxin deficiency results in decreased activity of aconitase and other iron-sulfur-containing proteins as well as an overall deficit in cellular energy production and other metabolic processes relying of iron-sulfur clusters, including DNA synthesis and repair (Bulteau et al., 2004; Martelli et al., 2007).
FRDA is a mitochondrial disorder that affects multiple organ systems. Primary neurodegeneration of the dorsal root ganglia leads to the hallmark clinical findings of progressive ataxia and debilitating scoliosis (Pandolfo, 2009). Other characteristic symptoms of FRDA include discoordination, slurred speech, muscle weakness, and peripheral neuropathy. In addition, optic atrophy, and auditory defects, glucose intolerance and diabetes are observed in some patients (Marmolino, 2011; Parkinson et al., 2013).
Although progressive ataxia is the most prominent clinical symptom of FRDA, hypertrophic cardiomyopathy is the primary cause of death in more than half of patients with Friedreich’s ataxia (Payne and Wagner, 2012). Many other structural heart abnormalities in FRDA cardiomyopathy have been described including fibrosis, accumulation of iron, myocarditis and necrosis of heart fibers (Koeppen et al., 2015). Thus far, modeling cardiac dysfunction in FRDA has been a challenging task. A conditional cardiac knockout of the Fxn gene in mice (MCK Fxn model) demonstrates an overt heart phenotype with an early onset and rapid progression that can be rescued by frataxin overexpression (Perdomini et al., 2014). However, it should be pointed out that the serious cardiac dysfunction in MCK Fxn mice results from a complete lack of Fxn expression and not downregulation of expression as in FRDA. Similarly, inducible, shRNA-mediated knockdown of mouse frataxin resulted in cardiac pathology (Chandran et al., 2017). Other FRDA animal models, including transgenic and knock-in mice, do not exhibit an overt cardiac phenotype. Therefore the ability to differentiate human induced pluripotent stem cells (iPSCs) into cardiomyocytes (Cms) and to generate new model systems directly derived from human cells that closely mimic disease conditions is of critical importance for FRDA.
In this work we demonstrated that Cms differentiated from FRDA iPSCs exhibit a lipid metabolism defect as well as transcriptome changes characteristic for cellular models of hypertrophic cardiomyopathy. We also, for the first time, report significant expression changes in non-coding RNAs of patient-derived Cms. Importantly, both lipid accumulation and the FRDA-specific gene expression signatures can be corrected by ZFN-mediated excision of the expanded GAAs. Correction of a single expanded allele resulted in significant upregulation of FXN mRNA expression and frataxin levels. Current therapeutic approaches are largely based on the reactivation of FXN expression, supplementation of frataxin with gene, protein, or RNA therapy, or amelioration of metabolic impairment and cellular toxicity phenotypes. However, in the later stages of the disease, after cell death becomes apparent and damage irreversible, cell replacement therapy may represent a stronger avenue for treatment. Therefore, results of these studies serve as a proof-of-concept for potential regenerative medicine approaches to treat the cardiac defect in FRDA. The system presented herein may also be an excellent platform to evaluate the therapeutic potential of novel drug candidates targeting cardiomyopathy in FRDA.
2. Methods
2.1. iPS cell culture and cardiac differentiation
The iPS cell lines were derived from fibroblasts obtained from healthy individuals and an FRDA patient with confirmed pathological expansion of GAA triplets. All experiments with human cells were approved by the UAB Institutional Review Board for Human Use (IRB). The FRDA line was obtained from a patient with diagnosed cardiomyopathy. A detailed method of ZFN-based editing of FRDA patient fibroblasts was described previously (Li et al., 2015). Analyses of karyotype, pluripotency and differentiation potential of these cells was described in (Li et al., 2015). Undifferentiated iPSCs were cultured in mTeSR1 medium (STEMCELL Technologies, Vancouver, BC, Canada). Medium was changed daily and cells were passaged every 4-5 days with dispase (STEMCELL Technologies, Vancouver, BC, Canada) as described in (Polak et al., 2016). Cardiac differentiation was performed using a previously published small-molecule protocol based on initial inhibition of GSK3 signaling followed by WNT signaling inhibition (Lian et al., 2012; Lian et al., 2013). Accutase (STEMCELL Technologies, Vancouver, BC, Canada) was used to obtain single iPS cell suspensions. Cells were plated on Geltrex (Life Technologies, Carlsbad, CA) coated wells of 12-well plates (0.4-0.8×106 cells/well) and cultured in standard conditions (mTeSR1, STEMCELL Technologies, Vancouver, BC, Canada) for four days (d-4 – d0). Subsequently, at d0, media was changed to RPMI/B27 without insulin (Life Technologies, Carlsbad, CA) supplemented with 12 μM CHIR99021 (Axon Medchem, Reston, VA) followed by addition (on d3) of the WNT inhibitor IWP2 at a final concentration of 5 μM (Tocris, Minneapolis, MN). Cells were maintained in the RPMI/B27 media starting from d7, with the medium changed every three days. The first spontaneously beating cells typically appeared on d8 or d9. Beating rate (BPM) was measured on d14-d17. Beating areas were identified and marked on the plates. Measurements were conducted by a blinded researcher within the first 5 min after plate removal from the incubator. More than 50 beating areas were analyzed for PCm and CCm cells from several independent differentiation experiments.
2.2. Immunostaining and flow cytometry and cell area measurements
Cardiac markers were evaluated by immunostaining and the efficiency of cardiac differentiation was assessed by flow cytometry. For immunostaining, differentiated cardiomyocytes were treated with TrypLE (Life Technologies, Carlsbad, CA) and seeded on Geltrex-coated coverslips. After 48 h, cells were fixed with 4% paraformaldehyde (PFA, 15 min at 4°C), washed in PBS, and blocked for 1 h in blocking solution (5% goat/donkey serum, 0.5% TritonX-100 in PBS). Primary antibodies to detect cardiac markers (ACTC1 Novus Biologicals, Centennial, CO, EP184E, 1:200; TNNT2 and NKX2.5 Life Technologies, Carlsbad, CA, Human Cardiomyocyte Immunocytochemistry Kit A25973) were diluted in blocking solution and incubated with fixed cells at 4°C overnight. Diluted (1:200) secondary antibody conjugated with Alexa Fluor 555 or Alexa Fluor 488 were added after PBS washing and incubated for 1 h at room temperature (RT). Cells were counterstained with DAPI and imaged on a Zeiss inverted microscope. The TNNT2 and ACTC1 antibodies were also used (at 1:25 dilution) to determine efficiency of cardiac differentiation by flow cytometry. For Cm area analyses, cells were plated on chamber slides and imaged after 48 h. Images were processed by ImageJ as described in (Ronaldson-Bouchard et al., 2018). A total of 60 cells per group derived from two independent differentiation experiments were analyzed by a blinded investigator.
2.3. RNA isolation, qRT-PCR and RNAseq analyses
All RNA samples were isolated using a RNeasy Mini Kit (Qiagen, Germantown, MD) followed by treatment with TurboDNase (ThermoFisher Scientific, Grand Island, NY) according to the manufacturer’s recommendations. Quantitative RT-PCR was performed as described (Napierala et al., 2017; Polak et al., 2016). RNAseq data analyses, library details and QC are described in Supplemental Methods and Supplemental Table 1.
2.4. Western Blot
Western blot analyses of frataxin expression were conducted as described earlier (Napierala et al., 2017; Polak et al., 2016). A detailed protocol is provided in Supplemental Methods.
2.5. Lipid droplets evaluation
For cytological analysis of lipid droplets in cardiomyocytes, Oil Red O staining was performed. Cells were fixed with 4% PFA (4°C, 15 min) and washed with PBS. Subsequently, cells were rinsed in 60% isopropanol and stained with 0.3% Oil Red O solution for 15 min. A hematoxylin solution was used to stain nuclei. Also, to quantify lipid droplets, PFA-fixed cardiomyocytes in suspension were stained for 15 min at RT with BODIPY 493/503 (1ug/ml, Life Technologies, Carlsbad, CA) and analyzed using a LSR Fortessa flow cytometer (BD Biosciences, San Jose, CA).
2.6. Amplification of the GAA repeat region and mtDNA copy number assay
To determine the number of GAA repeats in FRDA and ZFN-corrected cells, genomic DNA was extracted using a GenElute™ Mammalian Genomic DNA Miniprep Kit (MilliporeSigma, St. Louis, MO). PCR analyses were carried out exactly as described in (Li et al., 2015; Li et al., 2016). Amplification products were resolved on 1% agarose gels. For mtDNA quantitation, genomic DNA was isolated as above. A qPCR quantitation of mtDNA copy number was performed using primers specific to mtDNA sequence or genomic DNA (Bhalla et al., 2016). Abundance of mtDNA relative to genomic DNA was calculated using the 2−ΔΔCT method (Bhalla et al., 2016).
2.7. Electron microscopy sample preparation and imaging
EM images were collected on a FEI Tecnai T12 120 kV transmission electron microscope (FEI, Hillsboro, OR). Sample preparation is described in Supplemental Methods.
2.8. Statistical analyses
Statistical analyses were conducted using GraphPad Prism 6 software. Statistical significance was determined by two-tailed Student’s t-test, and p<0.05 was considered significant.
3. Results
3.1. Excision of expanded GAA repeats increases FXN expression in FRDA cardiomyocytes
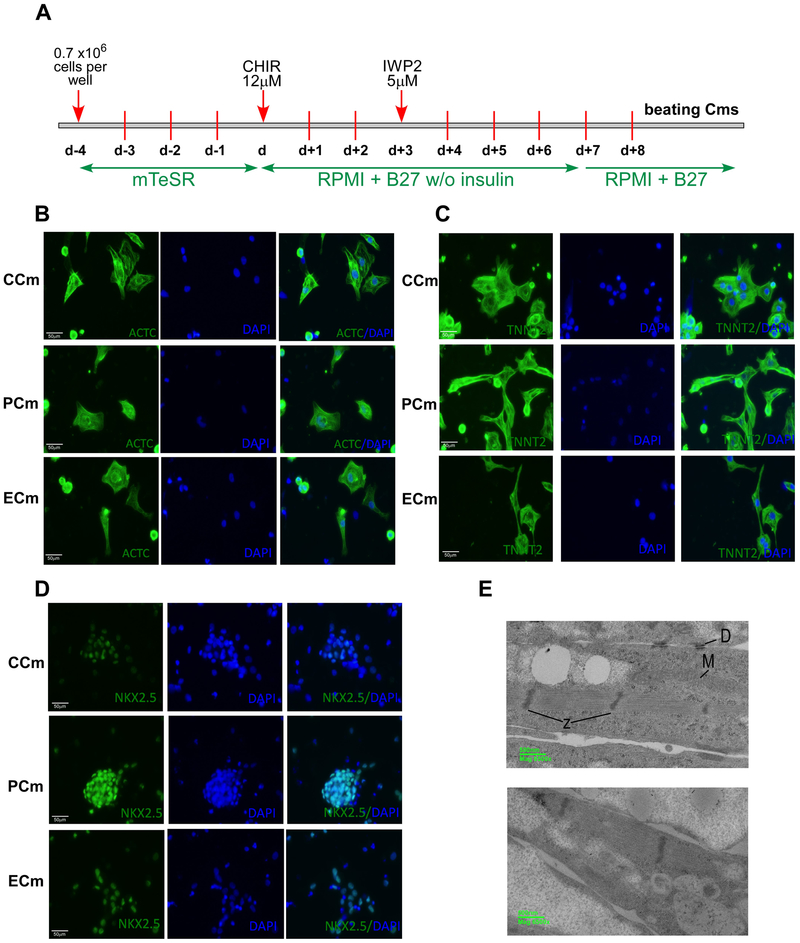
Cardiac pathology is a leading cause of death in FRDA. To determine molecular changes in human cardiac cells caused by frataxin deficiency and reveal molecular and cellular pathways affected in cardiac cells derived from FRDA patients, we differentiated FRDA patient iPSCs along with control iPSCs (derived from unaffected individuals), to beating cardiomyocytes (Cms). Additionally, FRDA iPSCs that underwent zinc finger (ZFN)-mediated editing, which removed the pathogenic expanded repeat tract from a single FXN allele (Li et al., 2015), were differentiated into Cms. We have previously demonstrated that this heterozygous excision of the expanded GAAs from FRDA cells resulted in an increase of FXN expression, aconitase activity and ATP in both iPSCs and iPSC-derived neurons (Li et al., 2015). Thus, the analyses in this study compared features of iPSC-derived Cms from two individual healthy controls (CCms), two Cm lines generated from independent clones of a patient iPSC line (PCms) and two Cm lines differentiated from independent clones of ZFN-edited FRDA iPSCs (ECms). Cardiac differentiation was performed using a previously published protocol based on initial inhibition of GSK3 signaling using CHIR99021 followed by inhibition of WNT signaling with IWP2 (Figure 1A) (Lian et al., 2012; Lian et al., 2013). The first beating areas were typically observed at day 8 and synchronously beating cardiomyocytes covered the entire surface of the well by day 12 of differentiation. We did not observe any gross differences in cardiac differentiation potential between FRDA, control, and ZFN-corrected iPSCs. Immunofluorescence analyses of ACTC1 (Actin, Alpha, Cardiac Muscle 1), TNNT2 (Troponin T2, Cardiac Type) and NKX2.5 (NK2 Homeobox 5) levels revealed robust expression of these cardiac markers in all iPSC-differentiated Cms (Figure 1B-D). Each individual Cm differentiation experiment resulted in a minimum of 80% of cells expressing high levels of TNNT2 and ACTC. Electron microscopy images of Cms at day 15 revealed characteristic features of contractile cells, including appearance of myofilaments with Z-band structures or desmosomes typical for immature Cms (Figure 1E).
Figure 1. Characterization of cardiomyocytes differentiated from healthy (CCm), FRDA (PCm) and ZFN-edited (ECm) iPSCs.
(A) Timeline and major steps of iPSC differentiation into beating cardiomyocytes. See Supplemental Methods for details. (B-D) Expression of cardiac markers analyzed by immunostaining. Nuclei were stained by DAPI and merged images are shown. (B) ACTC1 (Actin, Alpha, Cardiac Muscle 1); (C) TNNT2 (Troponin T2, Cardiac Type); (D) NKX2.5 (NK2 Homeobox 5). (E) Electron microscopy images of Cm ultrastructure (PCm top panel and CCm bottom panel); M - mitochondria, D – desmosomes, and Z – Z-bands.
As the cardiac component of Friedreich’s ataxia manifests predominantly as hypertrophic cardiomyopathy, we analyzed several cellular and molecular phenotypes previously described in stem cell-derived cardiac hypertrophy models (Cohn et al., 2019; Ovchinnikova et al., 2018). No significant differences in the pattern or intensity of F-actin expression was observed between CCm and PCm cells, nor for cardiomyocyte cell area or beating frequency (Supplemental Figure 1A-C). We did not detect differences in oxygen consumption rate (OCR) and total ATP content between PCm and CCm cells, however, quantitative analysis of mitochondrial DNA content revealed a trend towards increased mtDNA copy number in PCms compared to CCm cells (Supplemental Figure 2A-C).
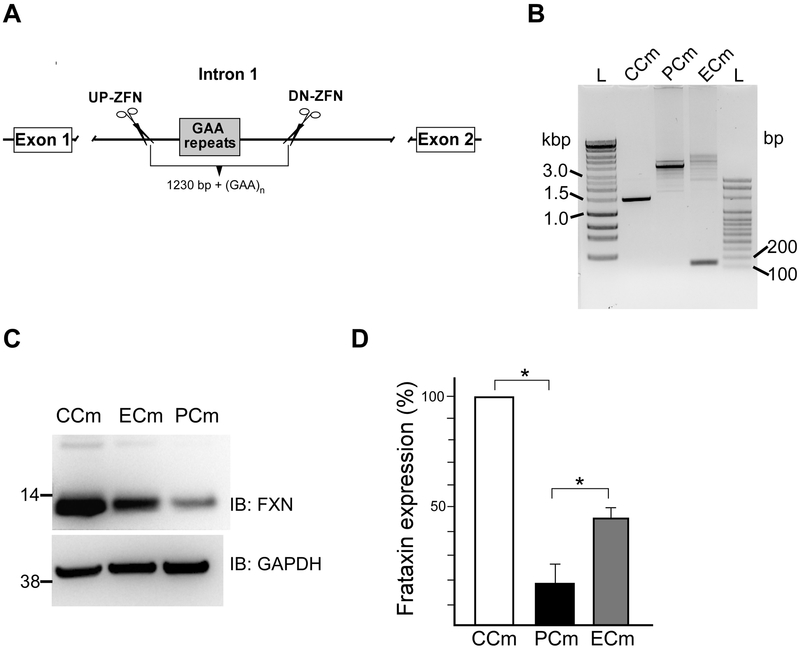
Next, to determine whether differentiation of the iPSCs into Cms affected GAA repeat tract length and frataxin levels, we verified the number of GAAs and FXN expression in Cm cells. Heterozygous excision of one expanded GAA tract using ZFNs resulted in shortening of FXN intron one by ~1.2 kbp of the repeat flanking sequences in addition to the entire expanded GAA repeat region (Figure 2A, Supplemental Figure 3). Amplification of the repeat region in PCm cells showed the presence of only expanded alleles. Substantial heterogeneity of the expanded GAA tract can be noticed as the appearance of multiple PCR bands. This is caused by somatic instability of the expanded GAA repeats occurring during iPSC propagation and/or Cm differentiation (Ku et al., 2010; Polak et al., 2016). Similar somatic instability has been observed in patient tissues (Long et al., 2017). Multiple bands resulting from somatic instability of the GAA tract are detected in ECm cells (containing a single expanded GAA tract), however, a faster migrating product (0.2 kbp) corresponding to the edited allele, which lacks GAA repeats and ~1.2 kbp of the flanking sequences, is also readily amplified (Figure 2B). Control CCm cells contained only alleles with short GAA tracts (Figure 2B).
Figure 2. Editing of the expanded GAA repeats in FRDA cells.
(A) Schematic presentation of the editing strategy to excise intronic expanded GAAs using two ZFNs targeting regions upstream and downstream of the repeats. Details of the approach were described in (Li et al., 2015). (B) Amplification of GAA tracts using PCR. Short GAAs (~1500 bp band) are present in CCm cells while only expanded GAAs (~4000 bp corresponding to ~ 830 GAAs) are present in PCms. The amplified expanded GAA alleles are of similar length. In ECm cells, an expanded GAA allele is amplified as well as short DNA fragment ~180 bp corresponding to the edited allele (GAA repeats are excised along with ~ 1230 bp of flanking sequence). Somatic instability resulting from culturing of the iPSCs prior to cardiac differentiation is responsible for multiple fragments containing various lengths of the expanded GAAs in ECm and PCm cells. (C) Analysis of frataxin levels in CCm, PCm and ECm cells using western blot. (D) Quantitative analysis of frataxin protein expression. Presented data are based on three independent experiments conducted using two CCm lines, two PCm clones and two ECm clones. * indicates p≤0.05.
Repeat expansion is associated with a significant (~90%) decrease of frataxin levels in PCms when compared to the CCm cells, while correction of a single GAA tract increased FXN levels ~3-fold in ECm cells relative to their isogenic counterparts (PCm cells) (Figure 2C, D). Thus, partial restoration of FXN expression in ZFN-edited Cms is comparable to the correction detected in parental ZFN-edited iPSCs (Li et al., 2015).
3.2. Transcriptome profiling reveals a cardiac-specific gene expression pattern in iPSC-differentiated cardiomyocytes
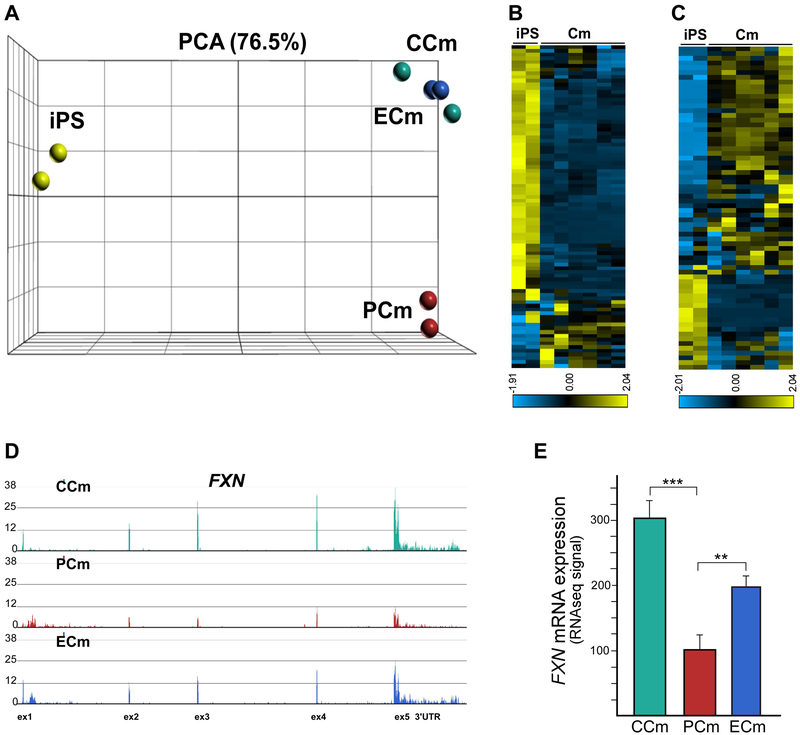
Correction of the expanded GAA tract in FRDA cells presents a unique opportunity to create an isogenic system (e.g. FRDA PCms and carrier equivalent ECms derived from the same patient) that can be used to directly compare patient and control disease-relevant human cells without introducing inter-individual variability. We conducted transcriptome analyses of CCm, PCm, and ECm cells in order to establish an expression signature of FRDA Cms, and to identify pathways associated with the downregulation of FXN expression. To maximize uniformity between samples subjected to RNA sequencing (RNAseq), the purity and homogeneity of all batches of differentiated Cms were determined by flow cytometry using TNNT2 and ACTC1 as markers. Only samples containing ≥ 95% cells positive for both cardiac markers were used in transcriptome sequencing analyses. Poly(A)-enriched mRNA samples were sequenced on an Illumina platform and obtained reads were aligned to the human genome, followed by differential expression analyses using DESeq2 (Materials and Methods and Supplemental Table 1). RNAseq samples obtained from two undifferentiated iPSC lines were included in initial analyses as controls for differentiation. Principle component analyses (PCA) of the transcriptomes obtained from all eight samples demonstrated clear separation of iPSC and cardiac samples (Figure 3A). Significantly, the group of Cm samples was divided into two subpopulations with transcriptomes of PCms separated clearly from the clustered CCms and ECms (Figure 3A). This indicates that FRDA-associated transcriptome changes are the main variance factor, especially considering the fact that PCms and ECms are isogenic lines derived from the same FRDA individual.
Figure 3. Transcriptome analysis of FRDA cardiomyocytes using RNAseq.
(A) Principle component analysis of eight samples based on RNAseq data. In addition to Cm analyses, data from two iPSC lines were included to validate cardiac commitment of Cm cells. (B) A heatmap display representing expression of an 89-gene signature of embryonic stem cells is shown for iPSC and Cm cell lines (Supplemental Table 2, Supplemental Figure 4). (C) Expression of a 79-gene KEGG Cardiac Muscle Contraction signature in iPS and Cm cell lines (Supplemental Table 2, Supplemental Figure 4). (D) Frataxin mRNA expression in CCm, PCm and ECm cells. Normalized RNAseq signal is shown for the FXN locus. Each track represents RNAseq data obtained from two cell lines in two separate RNAseq runs. (E) RNAseq signal quantitation shown as means with a signal range; ** and *** indicate p≤0.01 and p≤0.001, respectively.
To determine whether the Cm cells differentiated from iPSCs lost their pluripotency and acquired a cardiac-specific expression signature, we evaluated the transcription profiles of all six Cm lines and two undifferentiated iPSC lines against an established 89-gene embryonic stem cell signature (Gene Set M4282; Figure 3B and Supplemental Table 2 and Supplemental Figure 4A) and 79-gene KEGG Cardiac Muscle Contraction signature (Gene Set M17673; Figure 3C; Supplemental Table 2 and Supplemental Figure 4B). For both sets, distinct patterns of gene expression were detected for iPSC and Cm cells. Downregulated expression of key pluripotency factors (e.g. OCT4, NANOG, LIN28) was confirmed while cardiac-specific transcripts (e.g. MYH6/7, NPPA) were highly enriched in all Cm cell lines. Lastly, to validate prior qRT-PCR and western blot analyses, we used RNAseq data to determine the level of FXN mRNA in CCm, PCm and ECm cells. As expected, a normalized RNAseq signal detected in CCms was ~3-fold greater than for PCms (Figure 3D, E). Furthermore, excision of a single GAA tract in the ECm cells increased FXN mRNA expression ~2-fold when compared to isogenic PCm cells.
3.3. ZFN-mediated excision of the expanded GAA tract corrects the transcription profile of PCms
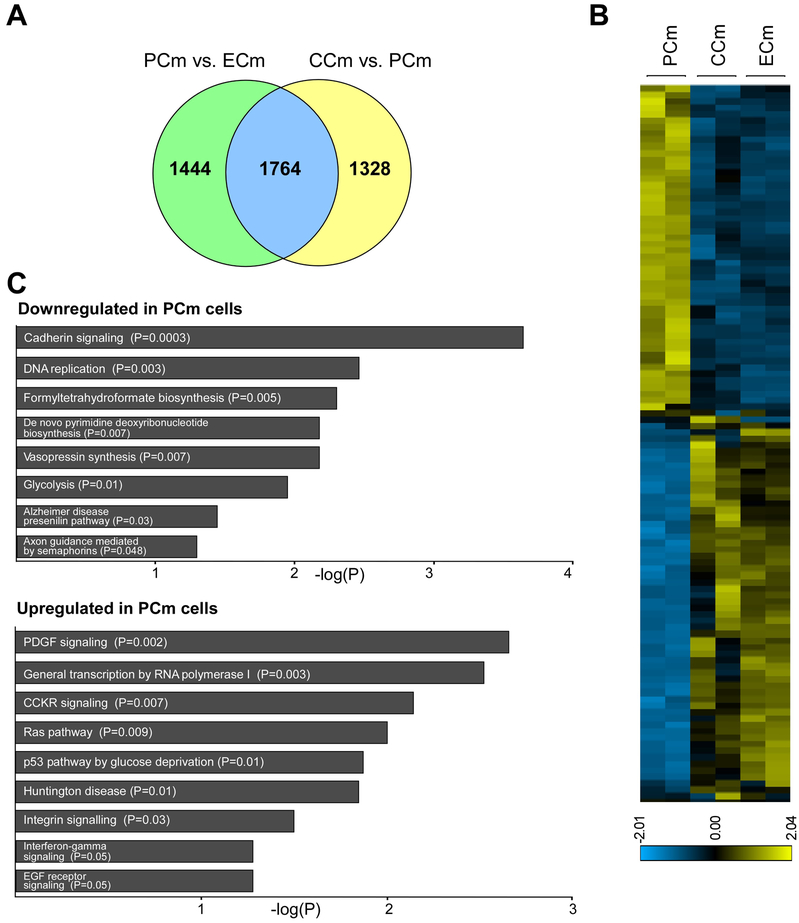
An unbiased PCA separated the Cm cells into two groups: PCms expressing a low level of frataxin or healthy and ZFN-corrected cells (CCms and ECms; Figure 3A). To test if the ZFN-mediated correction of FRDA cells resulted in changes of the PCm transcriptome, we compared differentially expressed genes between PCm and CCm cells as well as PCm and ECm cells (Figure 4A). Using DESeq2, more than 3000 differentially expressed genes were identified between each of these groups (3092 genes for CCm vs. PCm, 3208 genes for PCm vs. ECm; p<0.01; Supplemental Table 3). More than 55% (1764) of the differentially expressed transcripts were common for both compared groups. The transcription profile of ECm cells was similar to CCm cells and strikingly different from their parental, unedited PCm cells (Figure 4B). Out of 1764 differentially expressed genes common for the CCm vs. PCm and PCm vs. ECm groups, 1737 (98.5%) were expressed in ECms at levels similar to CCm cells, indicating rescued expression following GAA excision. Expression of only 27 transcripts was changed in the ECm cells in the opposite direction of the CCm cells (Supplemental Figure 5). In summary, editing of the expanded GAA tract from a single allele was sufficient to correct a significant fraction of transcriptome changes characteristic for FRDA iPSC-derived Cms.
Figure 4. Editing of the expanded GAAs partially corrects FRDA transcriptome.
(A) Venn diagram illustrating the comparison of differentially expressed genes between PCm and ECm cells (p<0.01) with genes differentially expressed between CCm and PCm cells (p<0.01). (B) A heatmap illustrating expression of the 1764 genes differentially expressed in both groups (PCm/ECm and PCm/CCm; Supplemental Table 3). (C) PANTHER pathways analysis of genes downregulated and upregulated in PCm versus CCm cells (p<0.05).
Friedreich’s ataxia cardiomyopathy is hypertrophic, thus we sought to compare FRDA-specific cardiomyocyte transcriptome changes to other established stem cell-derived hypertrophy models (Cohn et al., 2019; Ovchinnikova et al., 2018). To determine global gene expression changes characteristic for FRDA PCm cells, we compared genes downregulated and upregulated in these cells with CCms as annotated by the PANTHER Pathways platform in the Enrichr suite (Figure 4C; Supplemental Table 4). Analyses revealed that PCm downregulated genes were enriched in such pathways as: cadherin signaling, DNA replication, de novo pyrimidine synthesis and glycolysis. Upregulated genes were enriched in PDGF signaling, p53 pathway, integrin and EGF receptor signaling (Figure 4C). Further, we compared the PCm transcriptome with transcription profiles of two previously reported stem cell-derived Cm models of hypertrophy created by either a mutation in a sarcomere gene (MYBPC3; myosin binding protein C, cardiac; (Cohn et al., 2019)) or by mechanical stretching (Supplemental Figure 6 and Supplemental Table 4; (Ovchinnikova et al., 2018)). Analyses of all three datasets identified a signature of 120 genes commonly downregulated in FRDA PCm cells and in the two independent hypertrophy models as compared to CCms (Supplemental Figure 7 and Supplemental Table 4). Interestingly, KEGG pathway analysis of this signature demonstrated changes in the p53 signaling pathway, DNA replication, homologous recombination and Fanconi anemia (Supplemental Figure 7). Recent studies point towards the importance of p53 and proteins involved in DNA metabolism for the development of cardiac hypertrophy (Das et al., 2010; Nakada et al., 2019).
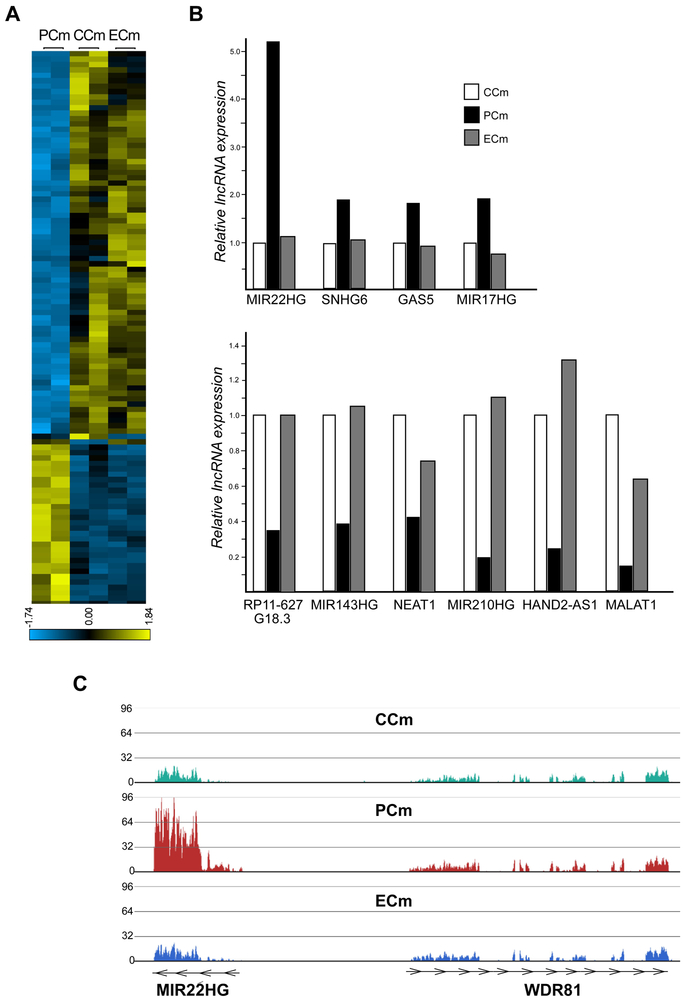
In addition to changes in expression of protein coding genes, a large fraction of long non-coding RNAs (lncRNAs) was differentially expressed between PCm, CCm and ECm cells. LncRNAs are an abundant class of transcripts associated with different physiological and pathological processes (Batista and Chang, 2013). Numerous recent studies indicate the importance of these RNAs in cardiac system development and diseases, including cardiomyopathy and hypertrophy (Han et al., 2014; Hobuss et al., 2019). Overall, more than 12000 non-coding RNAs were identified in our RNAseq datasets, of which 205 were differentially expressed between CCms vs. PCms as well as PCms vs. ECms (p<0.01, Figure 5A, Supplemental Table 5 and Supplemental Figure 8). Similar to the coding transcriptome, excision of the expanded GAAs corrected a significant fraction of aberrantly expressed non-coding RNAs (Figure 5A). In addition, several lncRNAs known to be involved in heart development and cardiovascular diseases were differentially expressed between CCms and PCm cells (Figure 5B). The lncRNA gene MIR22HG was highly upregulated in PCms compared to CCms (5-fold; p<0.00001) and its expression was reduced to control levels in ECms. This lncRNA, reported to be upregulated in myocardial infarction (MI) models, is a host gene for MicroRNA 22 (MIR22) – one of the most important small RNAs regulating hypertrophic response, sarcomere reorganization, and metabolic changes during cardiac remodeling (Huang and Wang, 2014).
Figure 5. Correction of long non-coding RNA expression changes in FRDA Cms by excision of the expanded GAAs.
(A) A heatmap illustrating expression levels of 205 lncRNAs differentially expressed between CCms/PCms and PCms/ECms (p≤0.01; Supplemental Table 5, Supplemental Figure 8). (B) Relative expression values of selected lncRNAs associated with cardiovascular diseases are plotted as bar graphs (p≤0.01 for all CCms/PCms and PCms/ECms comparisons). (C) The normalized RNAseq signal at the MIR22HG locus is depicted for CCm, PCm and ECm cells. The neighboring WDR81 locus is shown for comparison. Each track represents RNAseq data obtained from two cell lines and two separate RNAseq runs.
3.4. Lipid droplet accumulation in FRDA cardiomyocytes
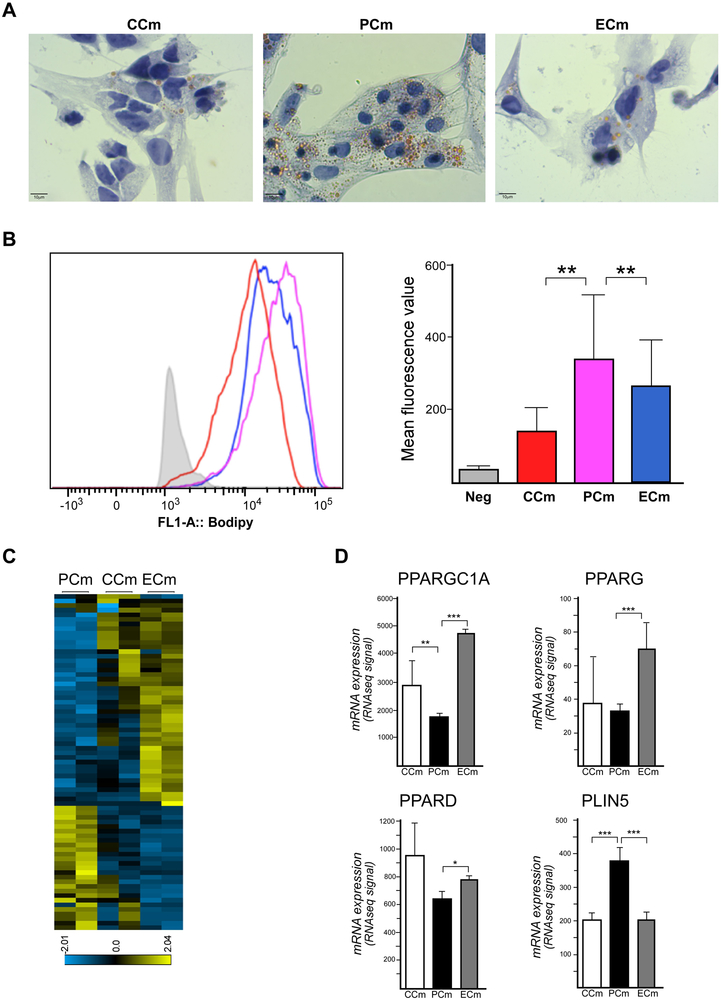
Morphology and immunohistochemistry analyses did not reveal any gross differences between FRDA and control Cms (Figure 1, Supplemental Figures 1 and 2), however, examination of EM images suggested an increase in the amount of vacuoles/lipid droplet structures in PCm compared to CCm cells. Considering prior studies on lipid metabolism in FRDA (reviewed in (Tamarit et al., 2016)), we conducted lipid accumulation analyses in PCm, CCm and ECm cells. Results of Oil Red O staining experiments demonstrated an accumulation of lipid droplets (LDs) in frataxin-deficient PCms relative to CCm cells (Figure 6A). Genetic correction of the expanded GAA repeats decreased the quantity of LDs as revealed by both Oil Red O staining and quantitative flow cytometry analyses of cardiomyocytes stained with a lipophilic dye (BODIPY; Figure 6A, B).
Figure 6. Changes in lipid metabolism in FRDA Cms.
(A) Brightfield microscopy images are shown of CCm, PCm, and ECm cells stained with Oil Red O. (B) Quantitative flow cytometry traces for CCm (shown in red), PCm (purple), and ECm (blue) cells stained with BODIPY are shown as a measure of lipid accumulation. The fluorescence values were recorded and plotted in the bar graph. (C) A heatmap illustrates 109 lipid metabolism signature genes that are differentially expressed between PCm, CCm and ECm cells (p≤0.05; Supplemental Table 2). (D) Expression of selected critical lipid metabolism genes in CCm, PCm, and ECm cells. RNAseq signal quantitation is presented as a mean with a signal range; * p≤0.05, ** p≤0.01 and *** p≤0.001.
To further investigate molecular mechanisms of lipid accumulation in PCm cells, we analyzed changes in the transcriptomes of PCms and isogenic ECm cells. We extracted 498 transcripts of lipid metabolism genes (Supplemental Table 2) from the entire list of ~27000 transcripts identified in our RNAseq analyses. Analyses demonstrated that a large proportion of genes important for lipid metabolism (109 out of 498) was differentially expressed between PCms and ECm cells (Figure 6C). Consistent with previous mouse studies (Marmolino et al., 2010), lower expression of PPARGC1A (Peroxisome Proliferator-Activated Receptor Gamma, Coactivator 1 Alpha) was detected in PCms. The PGC1 alpha protein is a transcriptional coactivator of several genes involved in energy metabolism including PPARG (Peroxisome Proliferator-Activated Receptor Gamma) and PPARD (Peroxisome Proliferator-Activated Receptor Delta) (Liang and Ward, 2006). Importantly, expression of PPARGC1A as well as PPARG and PPARD was increased upon editing of the expanded GAAs (Figure 6D). In the context of LD accumulation observed in PCm cells, we detected increased expression of the PLIN5 (Perilipin 5) gene, which encodes a protein essential for maintaining the balance between lipid storage and utilization in heart cells (Kimmel and Sztalryd, 2014). PLIN5 was significantly upregulated in PCms consistent with the greater quantity of LDs detected in PCms. Again, excision of the GAAs corrected expression of this gene in ECms to levels observed in CCm cells (Figure 6D).
3.5. Cardiac hypertrophy signature of PCms can be corrected
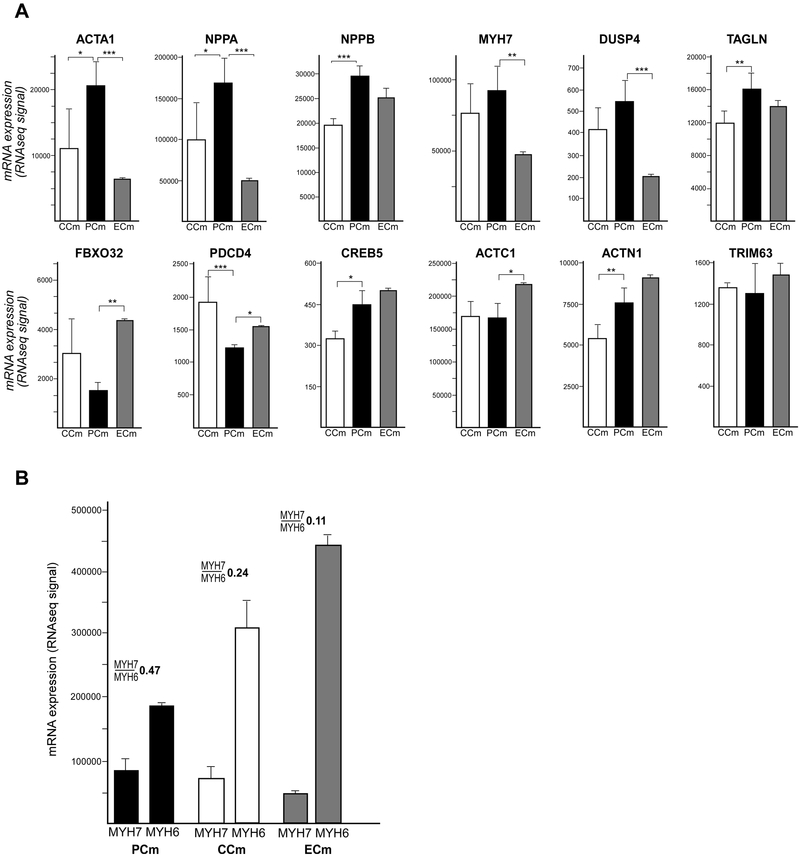
Cardiomyocytes and cardiac organoids differentiated from iPSCs currently represent some of the most suitable model systems to investigate molecular mechanisms of cardiac pathologies in a human cellular context. As the majority of FRDA patients develop progressive hypertrophic cardiomyopathy during the course of their disease, we sought to determine whether expression of hypertrophy-related genes is affected by ZFN-mediated correction of the mutant FXN gene. Recently, Carlson et al. established a hypertrophy signature in immature Cms differentiated from iPSCs that includes 12 differentially expressed genes (nine upregulated and three downregulated) (Carlson et al., 2013). Analysis of this hypertrophy expression signature in PCm, CCm and ECm cells (Figure 7A) revealed that eight out of nine genes highly expressed in the published iPSC-derived Cm hypertrophy model (ACTA1, NPPA, NPPB, MYH7, DUSP4, TAGLN, CREB5 and ACTN1) were also upregulated in PCm cells compared to CCms. Moreover, two out of three genes downregulated in this hypertrophy model (FBXO32 and PDCD4) were also expressed at lower levels in PCm cells. Expression levels of only two genes (ACTC1 and TRIM63) were unchanged irrespective of FRDA status. More importantly, expression of six out of 12 hypertrophy signature genes was corrected (p<0.05) in ECm cells (downregulated: ACTA1, NPPA, MYH7, DUSP4 and upregulated: FBXO32, PDCD4). In the case of NPPB and TAGLN, trends toward correction were detected but did not reach statistical significance (Figure 7A). These results demonstrate that PCm cells exhibit a molecular signature of hypertrophy that can be alleviated by excision of a single expanded GAA tract.
Figure 7. Expression signature of cardiac hypertrophy in PCm cells.
(A) Expression values of a 12-gene cardiac hypertrophy signature (Carlson et al., 2013) are presented as mean with a signal range. Significant differences are indicated by asterisks: * p≤0.05, ** p≤0.01 and *** p≤0.001. (B) Changes in MYH7 and MYH6 expression for PCms compared to CCm and ECm cells along with the calculated MYH7/MYH6 expression ratio are shown by the bar graph.
Transcriptome analyses of the Cms also indicated differences in expression of two genes encoding major cardiac proteins: MYH6 and MYH7 (Myosin Heavy Chain 6 and 7) between PCms, ECms and CCms. Data from human hearts as well as from iPSC-derived genetic models of hypertrophic cardiomyopathy demonstrated an increased MYH7:MYH6 expression ratio in disease tissues or model cells (Mosqueira et al., 2018). RNAseq data from PCms, CCms and ECms are in agreement with published studies (Figure 7B), indicating that the higher MYH7/MYH6 ratio in PCm cells may represent an in vitro biomarker suitable for testing therapeutic approaches in FRDA iPSC-derived cardiomyocytes.
4. Discussion
Although ~95% of Friedreich’s ataxia cases are caused by the same type of mutation, presentation of the disease varies significantly between patients (Marmolino, 2011; Pandolfo, 2009). Differences in age of onset, rate of disease progression as well as penetrance of various symptoms can be only in part attributed to the size of GAA expansions. Clinically, it is difficult to predict the exact course of the disease, moreover, the variability amongst FRDA patients makes this disorder challenging to recapitulate using biological systems because variable presentation is passed onto cellular/animal models.
Genome editing to remove the GAA expansion allows for creation of isogenic or near isogenic pairs of patient and corrected cell lines, and minimizes the influence of individual variability. Access to isogenic models is especially important because experiments on large cohorts of samples is not always feasible, yet studying small groups of samples can lead to inconclusive or false positive/negative findings. What is the best method of creating isogenic cell line models to represent FRDA? Pseudo-isogenic lines generated by si/shRNA knockdown of FXN in control cells or overexpression of frataxin in FRDA cells have been used for the past two decades to study disease mechanisms and test novel therapeutic approaches (Chandran et al., 2017; Lu and Cortopassi, 2007; Palomo et al., 2011; Vannocci et al., 2018). FXN knockdown involves activation of several silencing pathways in order to reduce FXN mRNA levels. On the other hand, ectopic overexpression of frataxin in FRDA cells may increase frataxin beyond physiological levels, leading to potential toxicity and affecting cellular proteostasis (Nabhan et al., 2015; Vannocci et al., 2018). Thus, permanent correction of the mutation by excision of the expanded GAAs (Li et al., 2015) or replacement of long repeats with a shorter tract (Lai et al., 2019) preserves the endogenous locus, promoter and gene copy number, assuring both physiological levels of FXN expression and its appropriate spatial and temporal control.
We previously demonstrated that excision of the GAA repeats and flanking regions bordered by the ZFN-cleavage sites does not affect frataxin expression in control K562 cells (both homozygous and heterozygous excision) and increases expression of FXN in FRDA cells (Li et al., 2015). This data proved that sequences flanking the expanded GAAs are unlikely to be critical regulators of FXN expression. Moreover, the significantly higher levels of FXN mRNA and protein expression in edited cells were maintained following differentiation of the edited iPSCs to neuronal cells and, in this work into cardiomyocytes, indicating that commitment to the specific lineages did not influence frataxin expression.
Although a large fraction of genes differentially expressed between PCm and CCm cells (p<0.01) was corrected by editing of the GAAs, a portion of the transcriptome was not affected by increased FXN expression. This may indicate that regulation of these genes is independent of FXN levels, or could be attributed to the small number of cell lines analyzed, whereby increasing the number of FRDA and edited cell lines analyzed would reduce such background effects (noise). More importantly, correction of a single allele restores FXN expression to the levels of asymptomatic FRDA carriers but not to the levels found in unaffected individuals lacking expanded GAAs on both FXN alleles. Although carriers are asymptomatic, significant expression differences between lymphocyte samples obtained from carriers, unaffected individuals, and FRDA patients have been detected (Coppola et al., 2011). Finally, cells from FRDA patients are functioning under conditions of low frataxin levels starting from the zygote. It is highly likely that many adaptive changes, for example in the epigenome, occur to establish a new intracellular homeostasis under conditions of a reduced frataxin supply. These compensatory changes, especially if imprinted in chromatin, may be difficult to reverse by a rapid increase of FXN expression. Such adaptive modifications may also explain the subset of differentially expressed genes between PCms and ECms that were not revealed when comparing PCms with CCm cells.
Thus far, a limited number of studies reported the generation of Cms from FRDA iPSCs (Crombie et al., 2017; Hick et al., 2013; Lee et al., 2013; Lee et al., 2016). Phenotypic changes, mostly in handling iron overload, calcium metabolism, electrophysiological properties, and mitochondrial abnormalities were identified in FRDA Cms (Hick et al., 2013; Lee et al., 2016). Some of the reported cardiac phenotypes were robust but limited in observation to a single biological replicate. Importantly, Hick et al. quantified contractile properties to assess a potential arrhythmia phenotype of FRDA Cms and detected differences between different clones derived from the same FRDA iPSC line, but no disease-specific differences, thus emphasizing the necessity for analyzing multiple samples, preferably using isogenic lines (Hick et al., 2013). Our comparative analyses of other cellular and molecular markers commonly assessed in iPSC-derived models of cardiac hypertrophy revealed no difference in cell area or beating rate between FRDA PCms and CCms or ECms. In addition, we did not detect differences in cellular bioenergetics (OCR and ATP levels) or cellular ultrastructure. A previous study conducted using a single pair of FRDA and control Cm cell lines demonstrated lower mtDNA content in FRDA Cm cells (Lee et al., 2013). Our data instead indicates a trend towards increased mtDNA copy number in PCms, which is consistent with mtDNA content changes reported in other iPSC-derived Cm hypertrophic cardiomyopathy models (Li et al., 2018). One reason for the general lack of an overt phenotype of FRDA Cms, especially when compared to other cardiac hypertrophy models, is likely related to the metabolic etiology of Friedreich’s ataxia. Robust hypertrophic phenotypes in immature iPSC-derived Cm cells are frequently associated with mutations in genes essential for contractual apparatus development and functions or in models obtained via abrupt chemical or physical cell stimulation. In contrast, FRDA is a relatively slowly progressing disease and cardiac changes are typically demonstrated later in an advanced disease state. Therefore, it is likely that initial metabolic changes, like accumulation of lipid droplets, along with continuous insults caused by low frataxin levels, eventually result in cardiac pathology. Modeling of these gradual changes will likely require maturation of the iPSC-derived Cms, perhaps by aging or adding exogenous mechanical stress during in vitro culture of the Cms. It is also worth mentioning that recent work on iPSC-derived FRDA sensory neurons revealed no phenotypic changes (aconitase activity, oxygen consumption, spare respiratory capacity, ATP production, reactive oxygen species formation, and mitochondrial membrane potential) between FRDA and control cells cultured under typical in vitro conditions (Soragni et al., 2018). Nevertheless, our identification of a ‘correctable’ lipid droplet phenotype and hypertrophy transcriptome signature may be useful in establishing novel cardiac biomarkers, which are necessary for clinical studies focusing on cardiac issues in FRDA.
Our RNAseq analyses revealed significant changes in the lncRNA transcriptome of PCms when compared to CCms and ECms. MIR22HG was found to be the most significantly upregulated lncRNA gene in patient cells (Supplemental Table 5). This lncRNA encodes MIR22, which is a highly conserved small non-coding RNA abundant in cardiac tissues and critical for heart development and functions (Huang and Wang, 2014). Several studies linked aberrant regulation of MIR22 expression with cardiac remodeling (Gurha et al., 2012; Huang et al., 2013). Mice deficient in mir22 fail to properly respond to stress conditions and develop cardiac dilation and fibrosis (Huang et al., 2013). On the other hand, overexpression of MIR22 is sufficient to induce cardiac hypertrophy (Gurha et al., 2013; Jentzsch et al., 2012). Its overexpression also leads to specific changes in the transcriptome, implicating a role of this microRNA in regulation of metabolism (Huang and Wang, 2014). In fact, MIR22 has been demonstrated to directly target PPARGC1A (Gurha et al., 2013). Thus, upregulation of MIR22HG in PCms can lead to increased levels of pro-hypertrophic MIR22 and result in the observed lower levels of PPARGC1A and its downstream targets, ultimately inhibiting lipid catabolism. Impediments in lipid degradation can result in accumulation of lipids in LDs observed in PCms causing lipotoxicity and contributing to cardiomyopathy. Lipid metabolism defects, including accumulation of LDs, have been observed in several FRDA models such as conditional Fxn knock-out mice, glial cells isolated from fhRNAi-1 Drosophila melanogaster, or Fxn shRNA-treated rat neonatal cardiomyocytes (Tamarit et al., 2016).
Increased expression of the PLIN5 transcript, encoding a lipid droplet targeting protein, may be responsible in part for accumulation of LDs in PCm cells. Several studies suggest that PLIN5 is expressed as an adaptive response to protect lipid storage and knockout of this gene results in significantly reduced formation of LDs (Kimmel and Sztalryd, 2014). Aberrant increase of PLIN5 levels can disturb LD homeostasis but also lead to mitochondrial dysfunction and cardiac hypertrophy (Kimmel and Sztalryd, 2014). On the other hand, upregulation of PLIN5 expression may represent a protective response to a decreased level of PPARGC1A. PLIN5 interacts with PGC1 alpha (encoded by PPARGC1A) and promotes its co-activator function, thus promoting efficient fatty acid utilization and protection of mitochondria (Gallardo-Montejano et al., 2016). Further studies of lipid storage, fatty acid catabolism, and mitochondrial functions will be required to define the interplay between these pathways and systems in FRDA Cms.
Our RNAseq data analyses also demonstrated that a hypertrophy signature established previously in iPSC-derived Cms was recapitulated in PCms when compared to CCm cells (Carlson et al., 2013). Importantly, increased expression of NPPA and NPPB (Natriuretic Peptide A and B) genes, recurrently observed in various hypertrophy models (Aggarwal et al., 2014; Carlson et al., 2013), was detected in PCm cells and corrected by editing of the expanded GAAs (Figure 7A). In addition, transcriptome analyses revealed changes in the expression of MYH7 and MYH6 resulting in a higher MYH7:MYH6 expression ratio in PCm cells. An increased ratio between these myosin subunits has been attributed to hypertrophy in human hearts and observed in iPSC-derived hypertrophic cardiomyopathy models (Mosqueira et al., 2018). Again, the MYH7:MYH6 expression ratio is lowered in both CCm and ZFN-corrected ECm cells compared to FRDA PCms.
Considering that classic FRDA typically manifests between the ages of 5 and 15, it is rather unlikely that gross structural and functional differences between FRDA and control Cm cells will be observed in the immature iPSC-derived Cms obtained during in vitro culture. Rather, it is more likely that maturation of iPSC-derived Cms into engineered heart tissue or organoids will emphasize cellular, morphological and developmental problems resulting from decreased frataxin levels. Perhaps application of more sensitive measurements of cellular functions and metabolism using mass spectroscopy or optogenetics will be able to identify additional phenotypic differences between FRDA and isogenic, corrected cells. As cardiac hypertrophy and cardiomyopathy are of the most critical and life-shortening complications of FRDA, development and characterization of appropriate cardiac models is essential to evaluate the efficacy of potential new treatments.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grant R01NS081366 from the National Institute of Neurological Disorders and Stroke (to M.N.) and Friedreich’s Ataxia Research Alliance and FARA Ireland (to M.N. and J.S.N.), Muscular Dystrophy Association (MDA418838).
Footnotes
Declaration of interests
None
References
- Aggarwal P, Turner A, Matter A, Kattman SJ, Stoddard A, Lorier R, Swanson BJ, Arnett DK, and Broeckel U, 2014. RNA expression profiling of human iPSC-derived cardiomyocytes in a cardiac hypertrophy model. PLoS One 9, e108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, and Chang HY, 2013. Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla AD, Khodadadi-Jamayran A, Li Y, Lynch DR, and Napierala M, 2016. Deep sequencing of mitochondrial genomes reveals increased mutation load in Friedreich's ataxia. Annals of clinical and translational neurology 3, 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulteau AL, O'Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, and Szweda LI, 2004. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305, 242–245. [DOI] [PubMed] [Google Scholar]
- Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, De Donato S, Mandel JL, Cocozza S, Koenig M, and Pandolfo M, 1996. Friedreich's ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427. [DOI] [PubMed] [Google Scholar]
- Carlson C, Koonce C, Aoyama N, Einhorn S, Fiene S, Thompson A, Swanson B, Anson B, and Kattman S, 2013. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. J Biomol Screen 18, 1203–1211. [DOI] [PubMed] [Google Scholar]
- Chandran V, Gao K, Swarup V, Versano R, Dong H, Jordan MC, and Geschwind DH, 2017. Inducible and reversible phenotypes in a novel mouse model of Friedreich's Ataxia. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn R, Thakar K, Lowe A, Ladha FA, Pettinato AM, Romano R, Meredith E, Chen YS, Atamanuk K, Huey BD, and Hinson JT, 2019. A Contraction Stress Model of Hypertrophic Cardiomyopathy due to Sarcomere Mutations. Stem Cell Reports 12, 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola G, Burnett R, Perlman S, Versano R, Gao F, Plasterer H, Rai M, Sacca F, Filla A, Lynch DR, Rusche JR, Gottesfeld JM, Pandolfo M, and Geschwind DH, 2011. A gene expression phenotype in lymphocytes from Friedreich ataxia patients. Ann Neurol 70, 790–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombie DE, Curl CL, Raaijmakers AJ, Sivakumaran P, Kulkarni T, Wong RC, Minami I, Evans-Galea MV, Lim SY, Delbridge L, Corben LA, Dottori M, Nakatsuji N, Trounce IA, Hewitt AW, Delatycki MB, Pera MF, and Pebay A, 2017. Friedreich's ataxia induced pluripotent stem cell-derived cardiomyocytes display electrophysiological abnormalities and calcium handling deficiency. Aging (Albany NY) 9, 1440–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Young D, Vasanji A, Gupta S, Sarkar S, and Sen S, 2010. Influence of p53 in the transition of myotrophin-induced cardiac hypertrophy to heart failure. Cardiovasc Res 87, 524–534. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gallardo-Montejano VI, Saxena G, Kusminski CM, Yang C, McAfee JL, Hahner L, Hoch K, Dubinsky W, Narkar VA, and Bickel PE, 2016. Nuclear Perilipin 5 integrates lipid droplet lipolysis with PGC-1alpha/SIRT1-dependent transcriptional regulation of mitochondrial function. Nat Commun 7, 12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurha P, Abreu-Goodger C, Wang T, Ramirez MO, Drumond AL, van Dongen S, Chen Y, Bartonicek N, Enright AJ, Lee B, Kelm RJ Jr., Reddy AK, Taffet GE, Bradley A, Wehrens XH, Entman ML, and Rodriguez A, 2012. Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation 125, 2751–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurha P, Wang T, Larimore AH, Sassi Y, Abreu-Goodger C, Ramirez MO, Reddy AK, Engelhardt S, Taffet GE, Wehrens XH, Entman ML, and Rodriguez A, 2013. microRNA-22 promotes heart failure through coordinate suppression of PPAR/ERR-nuclear hormone receptor transcription. PLoS One 8, e75882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P, Li W, Lin CH, Yang J, Shang C, Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, Xiong Y, Chien H, Zhou B, Ashley E, Bernstein D, Chen PS, Chen HV, Quertermous T, and Chang CP, 2014. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, and Gottesfeld JM, 2006. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat Chem Biol 2, 551–558. [DOI] [PubMed] [Google Scholar]
- Hick A, Wattenhofer-Donze M, Chintawar S, Tropel P, Simard JP, Vaucamps N, Gall D, Lambot L, Andre C, Reutenauer L, Rai M, Teletin M, Messaddeq N, Schiffmann SN, Viville S, Pearson CE, Pandolfo M, and Puccio H, 2013. Neurons and cardiomyocytes derived from induced pluripotent stem cells as a model for mitochondrial defects in Friedreich's ataxia. Disease models & mechanisms 6, 608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobuss L, Bar C, and Thum T, 2019. Long Non-coding RNAs: At the Heart of Cardiac Dysfunction? Front Physiol 10, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZP, Chen J, Seok HY, Zhang Z, Kataoka M, Hu X, and Wang DZ, 2013. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ Res 112, 1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZP, and Wang DZ, 2014. miR-22 in cardiac remodeling and disease. Trends Cardiovasc Med 24, 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentzsch C, Leierseder S, Loyer X, Flohrschutz I, Sassi Y, Hartmann D, Thum T, Laggerbauer B, and Engelhardt S, 2012. A phenotypic screen to identify hypertrophy-modulating microRNAs in primary cardiomyocytes. J Mol Cell Cardiol 52, 13–20. [DOI] [PubMed] [Google Scholar]
- Kimmel AR, and Sztalryd C, 2014. Perilipin 5, a lipid droplet protein adapted to mitochondrial energy utilization. Curr Opin Lipidol 25, 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppen AH, Ramirez RL, Becker AB, Bjork ST, Levi S, Santambrogio P, Parsons PJ, Kruger PC, Yang KX, Feustel PJ, and Mazurkiewicz JE, 2015. The pathogenesis of cardiomyopathy in Friedreich ataxia. PLoS One 10, e0116396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku S, Soragni E, Campau E, Thomas EA, Altun G, Laurent LC, Loring JF, Napierala M, and Gottesfeld JM, 2010. Friedreich's ataxia induced pluripotent stem cells model intergenerational GAATTC triplet repeat instability. Cell Stem Cell 7, 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JI, Nachun D, Petrosyan L, Throesch B, Campau E, Gao F, Baldwin KK, Coppola G, Gottesfeld JM, and Soragni E, 2019. Transcriptional profiling of isogenic Friedreich ataxia neurons and effect of an HDAC inhibitor on disease signatures. J Biol Chem 294, 1846–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Ho PW, Schick R, Lau YM, Lai WH, Zhou T, Li Y, Ng KM, Ho SL, Esteban MA, Binah O, Tse HF, and Siu CW, 2013. Modeling of Friedreich ataxia-related iron overloading cardiomyopathy using patient-specific-induced pluripotent stem cells. Pflugers Arch. [DOI] [PubMed] [Google Scholar]
- Lee YK, Lau YM, Ng KM, Lai WH, Ho SL, Tse HF, Siu CW, and Ho PW, 2016. Efficient attenuation of Friedreich's ataxia (FRDA) cardiomyopathy by modulation of iron homeostasis-human induced pluripotent stem cell (hiPSC) as a drug screening platform for FRDA. Int J Cardiol 203, 964–971. [DOI] [PubMed] [Google Scholar]
- Li S, Pan H, Tan C, Sun Y, Song Y, Zhang X, Yang W, Wang X, Li D, Dai Y, Ma Q, Xu C, Zhu X, Kang L, Fu Y, Xu X, Shu J, Zhou N, Han F, Qin D, Huang W, Liu Z, and Yan Q, 2018. Mitochondrial Dysfunctions Contribute to Hypertrophic Cardiomyopathy in Patient iPSC-Derived Cardiomyocytes with MT-RNR2 Mutation. Stem Cell Reports 10, 808–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Polak U, Bhalla AD, Rozwadowska N, Butler JS, Lynch DR, Dent SY, and Napierala M, 2015. Excision of Expanded GAA Repeats Alleviates the Molecular Phenotype of Friedreich's Ataxia. Mol Ther 23, 1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Polak U, Clark AD, Bhalla AD, Chen YY, Li J, Farmer J, Seyer L, Lynch D, Butler JS, and Napierala M, 2016. Establishment and Maintenance of Primary Fibroblast Repositories for Rare Diseases-Friedreich's Ataxia Example. Biopreserv Biobank 14, 324–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, and Palecek SP, 2012. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A 109, E1848–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, and Palecek SP, 2013. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc 8, 162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, and Ward WF, 2006. PGC-1alpha: a key regulator of energy metabolism. Adv Physiol Educ 30, 145–151. [DOI] [PubMed] [Google Scholar]
- Long A, Napierala JS, Polak U, Hauser L, Koeppen AH, Lynch DR, and Napierala M, 2017. Somatic instability of the expanded GAA repeats in Friedreich's ataxia. PLoS One 12, e0189990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, and Cortopassi G, 2007. Frataxin knockdown causes loss of cytoplasmic iron-sulfur cluster functions, redox alterations and induction of heme transcripts. Arch Biochem Biophys 457, 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmolino D, 2011. Friedreich's ataxia: past, present and future. Brain Res Rev 67, 311–330. [DOI] [PubMed] [Google Scholar]
- Marmolino D, Manto M, Acquaviva F, Vergara P, Ravella A, Monticelli A, and Pandolfo M, 2010. PGC-1alpha down-regulation affects the antioxidant response in Friedreich's ataxia. PLoS One 5, e10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli A, Wattenhofer-Donze M, Schmucker S, Bouvet S, Reutenauer L, and Puccio H, 2007. Frataxin is essential for extramitochondrial Fe-S cluster proteins in mammalian tissues. Hum Mol Genet 16, 2651–2658. [DOI] [PubMed] [Google Scholar]
- Montermini L, Andermann E, Labuda M, Richter A, Pandolfo M, Cavalcanti F, Pianese L, Iodice L, Farina G, Monticelli A, Turano M, Filla A, De Michele G, and Cocozza S, 1997. The Friedreich ataxia GAA triplet repeat: premutation and normal alleles. Hum Mol Genet 6, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Mosqueira D, Mannhardt I, Bhagwan JR, Lis-Slimak K, Katili P, Scott E, Hassan M, Prondzynski M, Harmer SC, Tinker A, Smith JGW, Carrier L, Williams PM, Gaffney D, Eschenhagen T, Hansen A, and Denning C, 2018. CRISPR/Cas9 editing in human pluripotent stem cell-cardiomyocytes highlights arrhythmias, hypocontractility, and energy depletion as potential therapeutic targets for hypertrophic cardiomyopathy. European heart journal 39, 3879–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Gooch RL, Piatnitski Chekler EL, Pierce B, and Bulawa CE, 2015. Perturbation of cellular proteostasis networks identifies pathways that modulate precursor and intermediate but not mature levels of frataxin. Sci Rep 5, 18251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada Y, Nhi Nguyen NU, Xiao F, Savla JJ, Lam NT, Abdisalaam S, Bhattacharya S, Mukherjee S, Asaithamby A, Gillette TG, Hill JA, and Sadek HA, 2019. DNA Damage Response Mediates Pressure Overload-Induced Cardiomyocyte Hypertrophy. Circulation 139, 1237–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napierala JS, Li Y, Lu Y, Lin K, Hauser LA, Lynch DR, and Napierala M, 2017. Comprehensive analysis of gene expression patterns in Friedreich's ataxia fibroblasts by RNA sequencing reveals altered levels of protein synthesis factors and solute carriers. Disease models & mechanisms 10, 1353–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikova E, Hoes M, Ustyantsev K, Bomer N, de Jong TV, van der Mei H, Berezikov E, and van der Meer P, 2018. Modeling Human Cardiac Hypertrophy in Stem Cell-Derived Cardiomyocytes. Stem Cell Reports 10, 794–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomo GM, Cerrato T, Gargini R, and Diaz-Nido J, 2011. Silencing of frataxin gene expression triggers p53-dependent apoptosis in human neuron-like cells. Hum Mol Genet 20, 2807–2822. [DOI] [PubMed] [Google Scholar]
- Pandolfo M (2006). Friedreich's Ataxia. In Genetic Instabilities and Neurological Diseases, Wells RD, and Ashizawa T, ed. (San Diego, CA: Elsevier-Academic Press; ), pp. 277–296. [Google Scholar]
- Pandolfo M, 2009. Friedreich ataxia: the clinical picture. J Neurol 256 Suppl 1, 3–8. [DOI] [PubMed] [Google Scholar]
- Parkinson MH, Boesch S, Nachbauer W, Mariotti C, and Giunti P, 2013. Clinical features of Friedreich's ataxia: classical and atypical phenotypes. J Neurochem 126 Suppl 1, 103–117. [DOI] [PubMed] [Google Scholar]
- Pastore A, and Puccio H, 2013. Frataxin: a protein in search for a function. J Neurochem 126 Suppl 1, 43–52. [DOI] [PubMed] [Google Scholar]
- Payne RM, and Wagner GR, 2012. Cardiomyopathy in Friedreich ataxia: clinical findings and research. J Child Neurol 27, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdomini M, Belbellaa B, Monassier L, Reutenauer L, Messaddeq N, Cartier N, Crystal RG, Aubourg P, and Puccio H, 2014. Prevention and reversal of severe mitochondrial cardiomyopathy by gene therapy in a mouse model of Friedreich's ataxia. Nat Med 20, 542–547. [DOI] [PubMed] [Google Scholar]
- Polak U, Li Y, Butler JS, and Napierala M, 2016. Alleviating GAA Repeat Induced Transcriptional Silencing of the Friedreich's Ataxia Gene During Somatic Cell Reprogramming. Stem Cells Dev 25, 1788–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, and Vunjak-Novakovic G, 2018. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soragni E, Petrosyan L, Rinkoski TA, Wieben ED, Baratz KH, Fautsch MP, and Gottesfeld JM, 2018. Repeat-Associated Non-ATG (RAN) Translation in Fuchs' Endothelial Corneal Dystrophy. Invest Ophthalmol Vis Sci 59, 1888–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamarit J, Obis E, and Ros J, 2016. Oxidative stress and altered lipid metabolism in Friedreich ataxia. Free Radic Biol Med 100, 138–146. [DOI] [PubMed] [Google Scholar]
- Vannocci T, Notario Manzano R, Beccalli O, Bettegazzi B, Grohovaz F, Cinque G, de Riso A, Quaroni L, Codazzi F, and Pastore A, 2018. Adding a temporal dimension to the study of Friedreich's ataxia: the effect of frataxin overexpression in a human cell model. Disease models & mechanisms 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.