Abstract
Learning of most motor skills is constrained in a species-specific manner. However, the proximate mechanisms underlying species-specific learned behaviors remain poorly understood. Songbirds acquire species-specific songs through learning, which is hypothesized to depend on species-specific patterns of gene expression in functionally specialized brain regions for vocal learning and production, called song nuclei. Here, we leveraged two closely related songbird species, zebra finch, owl finch, and their interspecific first-generation (F1) hybrids, to relate transcriptional regulatory divergence between species with the production of species-specific songs. We quantified genome-wide gene expression in both species and compared this with allele-specific expression in F1 hybrids to identify genes whose expression in song nuclei is regulated by species divergence in either cis- or trans-regulation. We found that divergence in transcriptional regulation altered the expression of approximately 10% of total transcribed genes and was linked to differential gene expression between the two species. Furthermore, trans-regulatory changes were more prevalent than cis-regulatory and were associated with synaptic formation and transmission in song nucleus RA, the avian analog of the mammalian laryngeal motor cortex. We identified brain-derived neurotrophic factor (BDNF) as an upstream mediator of trans-regulated genes in RA, with a significant correlation between individual variation in BDNF expression level and species-specific song phenotypes in F1 hybrids. This was supported by the fact that the pharmacological overactivation of BDNF receptors altered the expression of its trans-regulated genes in the RA, thus disrupting the learned song structures of adult zebra finch songs at the acoustic and sequence levels. These results demonstrate functional neurogenetic associations between divergence in region-specific transcriptional regulation and species-specific learned behaviors.
In two closely related songbird species with distinct species-specific songs, divergence in transcriptional regulation (via both cis- and trans-regulatory changes) alters the expression of approximately 10% of the genes transcribed in specific brain regions involved in learned vocal production.
Introduction
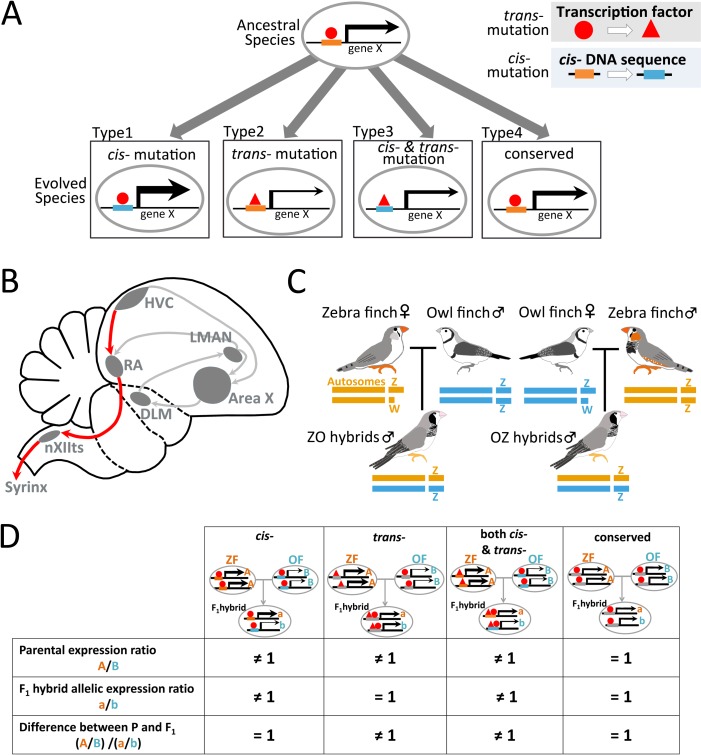
Species-specific behavior plays a role in a variety of inter- and intraspecific interactions, including reproduction and habitat use, in which species differences are thought to be an important factor in species co-occurrence [1–3]. Such species-specific behaviors can arise via species differences in the structure and development of the neural circuits underlying behavior [4–6]. Differences between closely related species are thought to be driven by differential expression and functional changes of orthologous genes in conserved neural circuits, which are often in turn driven by transcriptional regulatory divergence [7–10]. Transcriptional regulatory divergence between species can arise due to species divergence in cis-regulatory elements that affect the transcriptional rate and stability, and/or in trans-regulatory factors that access cis-regulatory elements [11–16] (Fig 1A). However, it remains largely unknown how transcriptional regulatory divergence contributes to the generation of species-specific behavior, especially in the case of learned behavior.
Fig 1. Cis- and/or trans-regulatory changes during species differentiation.
(A) During evolution, cis- and/or trans-regulatory elements change gene expression levels between closely related species. (B) Schematic showing selected song-control regions and connections in the songbird brain. The posterior motor pathway and the anterior cortico-basal ganglia-thalamic circuit (anterior forebrain pathway [AFP]) are represented as red and gray lines, respectively. (C) Genome composition of reciprocal F1 hybrids between zebra finch (ZF) and owl finch (OF). ZO represents F1 hybrid offspring between ZF♀ and OF♂. OZ hybrids are the opposite. Male F1 hybrids share identical sets of auto- and sex chromosomes. (D) Classification of species differences in cis- and/or trans-regulations based on the comparison of the relative gene expression ratio between parental species and the allelic expression ratio in their F1 hybrids. For each gene, “A” and “B” represent gene expression levels in ZF and OF, respectively. “a” and “b” represent gene expression levels from ZF and OF alleles, respectively, in F1 hybrids. “A/B” and “a/b” are the expression ratio between parental species and the allelic expression ratio in F1 hybrids, respectively. Area X, Area X of the striatum; DLM, dorsal lateral nucleus of the medial thalamus; F1, first-generation; HVC, used as a proper name; LMAN, lateral magnocellular nucleus of the anterior nidopallium; nXIIts, tracheosyringeal part of the hypoglossal nucleus; RA, the robust nucleus of the arcopallium.
Songs produced by oscine birds are complex vocal signals acquired through vocal learning [17,18]. Songs are species-specific, and these species differences play an important role in mating interactions and territory defenses within and between species [1,19,20]. In the songbird brain, a conserved neural circuit specialized for vocal learning, called the song system, contributes to song learning and production [18,21,22]. Birdsong is composed of two main traits associated with species specificity: the acoustic elements (syllables) and the temporal pattern (sequence) of song. The production of syllable acoustics and sequence is mainly regulated by the robust nucleus of the arcopallium (RA) and the song nuclei HVC (proper name), respectively, in the vocal motor circuit of the song system (Fig 1B) [22–24]. The importance of these song nuclei in determining species-specific song traits suggests an underlying causative role of species differences in the structure and activity of these regions. Consistent with this, a variety of genes, including transcription factors and neuromodulator receptors, are differentially expressed in these song nuclei between species, even in a laboratory-controlled environment [25–27]. However, a key gap in our knowledge is how species-specific patterns of gene expression in these regions arise via regulatory differences between species.
In this study, we used two closely related songbird species, zebra finch (ZF; Taeniopygia guttata), owl finch (OF; T. bichenovii), and their interspecific first-generation (F1) hybrids, to elucidate how transcriptional regulatory divergence is associated with species-specific song (Fig 1C). These two species diverged about 6.5 million years ago and share overlapping habitats in the north and west of Australia [28,29]. In addition, they produce songs with characteristic species-specific syllable acoustics and sequence. By comparing the gene expression ratio between the two species and the allele-specific expression (ASE) ratio in the F1 hybrids (Fig 1D), we assessed the total number of genes whose expression differs by divergence in cis- versus trans-transcriptional regulation between the two species. On the basis of Gene Ontology (GO) enrichment and the upstream regulatory analyses of transcriptional regulation–altered genes, we identified the candidate key upstream modulators of these differentially regulated genes and examined the functional effects of altered transcriptional regulation in the song nuclei.
Results
Species difference in song phenotypes between ZF and OF
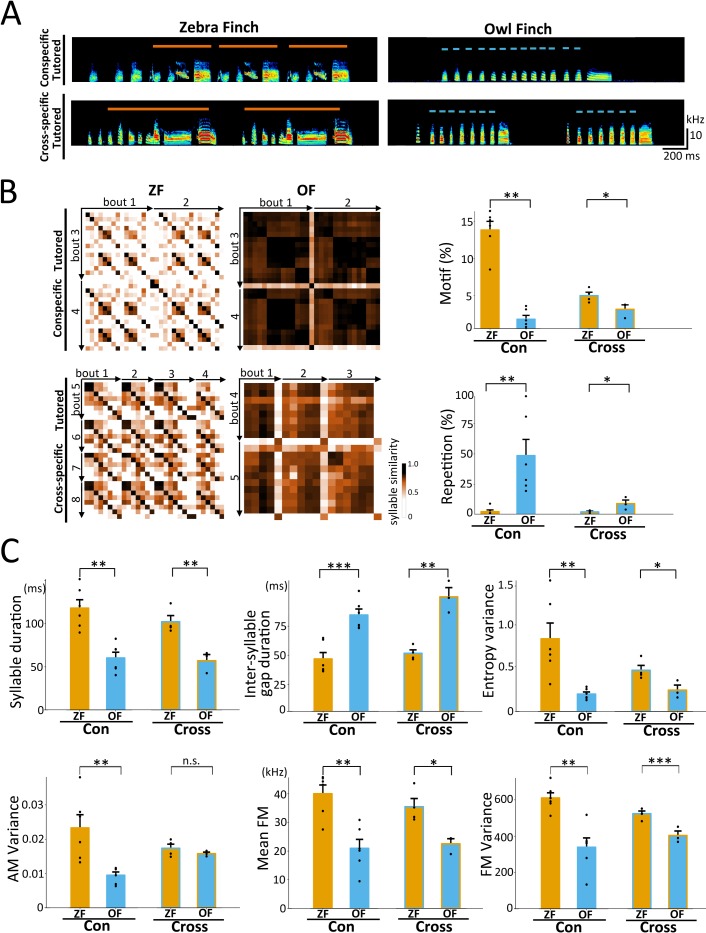
First, we compared the song features of ZF and OF reared with conspecific song tutoring in our breeding colony to confirm whether a laboratory-controlled environment could maintain species-specific song features. We compared the songs of the two species regarding syllable acoustics and sequential features (12 parameters) at the adult stage (Fig 2A) and identified significant differences in six acoustic syllable parameters (i.e., syllable duration, inter-syllable gap duration, entropy variance, amplitude modulation [AM] variance, mean frequency modulation [FM], and FM variance) and in syllable sequence features (motif and repetition transition rates) (n = 6 birds each, p < 0.01, one-way ANOVA) (Fig 2B and 2C and S1 Fig) [30, 31]. We found that the range but not the pattern of each acoustic feature’s distribution overlapped between ZFs and OFs (3,000 syllables from n = 6 birds each and 500 syllables/bird) (S1 Fig), thus suggesting that the species differences in the syllable acoustics were not caused by physical species-specific constraints in the peripheral vocal organs.
Fig 2. Species difference in song structures between ZF and OF.
(A) Typical examples of songs from ZFs and OFs that were reared with conspecific song tutoring and cross-species song tutoring. Orange solid and blue dotted lines represent the motif and repetitive structure of syllables, respectively. (B) Species differences in the syllable sequence of ZF and OF songs. (Left) Syllable similarity matrices for songs produced by ZFs and OFs that were reared with conspecific song tutoring and cross-species song tutoring. (Right) Motif and repetition indices of ZF and OF songs (n = 6 each from conspecific song tutored ZF and OF, n = 4 and 3 from cross-species song tutored ZF and OF, respectively; one-way ANOVA, *p < 0.05, **p < 0.01). Each dot corresponds to an individual bird. (C) Species differences in syllable acoustics (syllable duration, inter-syllable gap duration, entropy variance, AM variance, mean FM, and FM variance) of ZF and OF songs (“Con”: n = 6 each from conspecific song tutored ZF and OF; “Cross”: n = 4 and 3 from cross-species song–tutored ZF and OF, respectively; one-way ANOVA, *p < 0.05, **p < 0.01, ***p < 0.001). Each dot corresponds to an individual bird. Relevant data values are included in S1 Data. AM, amplitude modulation; FM, frequency modulation; OF, owl finch; ZF, zebra finch.
We further performed cross-species song tutoring experiments to examine how genetic and environmental factors contribute to generate species-specific song features of ZF and OF (Fig 2A). Under the cross-species song tutoring condition, juveniles heard only the counter-species songs as tutor songs. By comparing the p-values of song feature differences between conspecific and cross-species song tutoring conditions, we found that song tutoring affected most of the song parameters, including syllable sequence and acoustics (i.e., inter-syllable gap duration, entropy variance, AM variance, mean FM, and FM variance) (Fig 2B and 2C). However, except for AM variance, all song parameters retained species specificity (ZF, n = 4, OF, n = 3; one-way ANOVA, p < 0.05). In line with this result, we performed principal component analysis (PCA) to investigate the song feature distribution of conspecific and cross-species song tutored birds by reducing the dimensionality of the syllable acoustics and sequential features. We observed that clusters were separable by species but not by song tutoring conditions (S1 Fig). As many studies in songbirds reported [32–35], these results also indicate that song learning of these two species is implemented based on species-specific genetic constraint.
Genome-wide transcriptional analysis between ZF, OF, and F1 hybrids
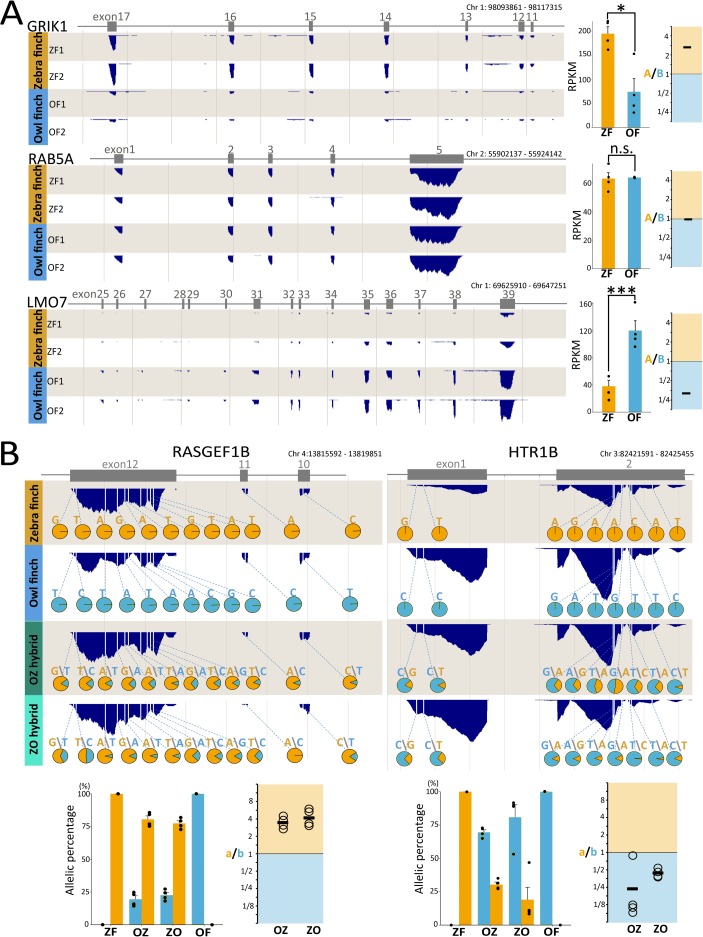
We then conducted a genome-wide transcriptional analysis to elucidate divergence of transcriptional regulation between ZF and OF in their song nuclei. For this purpose, using laser microdissected HVC and RA tissues from ZFs and OFs, we identified 11,501 and 11,487 genes in HVC and RA, respectively, as genes with detectable expression levels in either ZF or OF (reads per kilobase of transcript per million reads mapped [RPKM] ≥ 1). We then calculated the expression ratio between ZF and OF for each gene as “A/B” = RPKM(ZF average)/RPKM(OF average) (n = 4 birds each) (Figs 1D and 3A, S2 and S3 Figs).
Fig 3. Species differences in gene expression between ZF and OF and ASE in F1 hybrids.
(A) Examples of species differences in gene expression between ZF and OF. (Left panels) Expression levels of GRIK1, RAB5A, and LMO7 in song nucleus RA of ZFs and OFs. Gray boxes represent the position of exons for each gene. Dark blue peaks below exons represent read density. (Right panels) Gene expression levels in ZF and OF and the average of the expression ratio between ZF and OF. Each dot represents the RPKM value for individual. Mean ± SEM (n = 4 birds each; one-way ANOVA, *p < 0.05, ***p < 0.001, n.s., not significant). RAB5A is an example with no expression difference between ZF and OF. (B) Examples of ASE in F1 hybrids. (Upper panels) Allelic expression ratios in F1 hybrids at species-specific SNPs (ss-SNPs) of RASGEF1B and HTR1B in song nucleus RA. Dark blue peaks below exons represent read density. White bars in the dark blue–colored peaks represent ss-SNP positions. Pie charts of each ss-SNP represent the percentage of transcribed read numbers from ZF (orange) and OF (blue) alleles. (Bottom panels) The percentage and ratio of parental species-allelic expression of RASGEF1B and HTR1B in OZ and ZO F1 hybrids. Each dot represents average allelic expression ratios of all ss-SNPs in one individual (n = 4 birds each, mean). Orange- and blue-colored bars represent the values from ZF and OF alleles, respectively. Mean ± SEM (n = 4 birds each). Relevant data values are included in S2 Data. ASE, allele-specific expression; Chr, chromosome; F1, first-generation; GRIK1, Glutamate receptor, ionotropic, kainate type 1; HTR1B, 5-hydroxytryptamine receptor 1B; LMO7, LIM domain only protein 7; OF, owl finch; OZ, F1 hybrid offspring between OF♀ and ZF♂; RA, robust nucleus of the arcopallium; RAB5A, Ras-related protein Rab5A; RASGEF1B, Ras-GEF domain-containing family 1B; RPKM, reads per kilobase of transcript per million reads mapped; ZF, zebra finch; ZO, F1 hybrid offspring between ZF♀ and OF♂.
Based on a comparison of whole brain transcriptome between ZF and OF, a total of 2,409,063 SNPs were identified as species-specific SNPs (ss-SNPs) in their transcribed sequences. Using the ss-SNPs for the quantification of ASE ratios in the F1 hybrids, we set a cutoff to extract genes with ≥5 reads at each ss-SNP position and median RPKM ≥ 10 (n = 4 each from ZO and OZ hybrids). Totals of 5,827 and 6,328 genes passed the criteria in HVC and RA, respectively. The ASE ratio of each gene in individual F1 hybrids was calculated as “a/b” = Reads(ZF allele)/Reads(OF allele) (Figs 1D and 3B and S2 Fig). To date, there is no evidence for paternal and maternal genomic imprinting in avian species [36]. In line with this, we identified no genes with a significant paternal or maternal bias in allelic expression in ZO and OZ hybrids. Furthermore, the two reciprocal F1 hybrids (ZO and OZ) have an extremely high correlation in their ASE ratios (Pearson correlation coefficient, r = 0.527, p < 2.2 × 10−16 in HVC; r = 0.550, p < 2.2 × 10−16 in RA) (S4 Fig). Therefore, we treated ZO and OZ hybrids equally when calculating ASE ratios.
Transcriptional regulatory divergence between ZF and OF
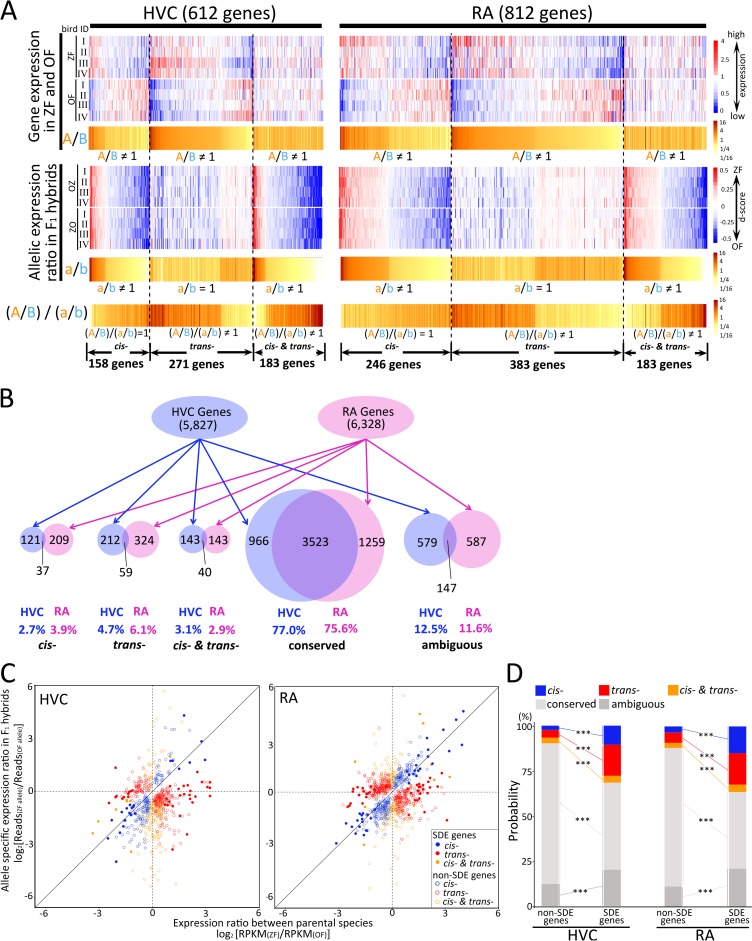
Transcriptional differences, cis- and/or trans-regulation, for each gene can be evaluated using the gene expression ratio between two species and the ASE ratio in the F1 hybrids [14,16,37–39]. ASE in the F1 hybrids reflects cis-dependent differences between the alleles of each parental species, because the two alleles of each gene are exposed to same trans-acting regulatory environment in cells. By comparing the gene expression ratio between parental species and the ASE ratio in F1 hybrids, we determined the following five categories of transcriptional regulatory divergences: (i) “cis-regulation” for genes with significant cis- but not trans-effects (with a/b ≠ 1 and A/B = a/b) as “cis-regulated genes,” (ii) “trans-regulation” for genes with significant trans- but not cis-effects (with a/b = 1 and A/B ≠ a/b) as “trans-regulated genes,” (iii) “both cis- and trans-regulation” for genes with both significant cis- and trans-effects (with a/b ≠ 1 and A/B ≠ a/b) as “both cis- and trans-regulated genes,” (iv) “conserved regulation” for genes with no significant cis- or trans-effects (with a/b = 1 and A/B = a/b) as “conserved genes,” and (v) ambiguous regulation (Figs 1D and 4A and S2 Fig). For this categorization of transcriptional regulatory divergence, we applied a cross-replicate comparison of ASE ratios in the F1 hybrids, through which we could minimize incorrect estimation of cis- and artificial negative correlation in cis- versus trans-comparison (see Materials and methods) [40]. After this procedure, we observed that over 75% and 10% of the examined genes were expressed in both HVC and RA with either “conserved” or “ambiguous” regulation between ZF and OF, respectively (Fig 4B). In contrast, transcriptional regulatory divergence changed the expression of 158 (2.7% of the total 5,827 genes), 271 (4.7%), and 183 (3.1%) genes in HVC categorized as cis-, trans-, and both cis- and trans-regulated genes, respectively. Likewise, in RA, the expression of 246 (3.9% of the total 6,328 genes), 383 (6.1%), and 183 (2.9%) genes was altered by cis-, trans-, and both cis- and trans-regulatory changes between the two species, respectively (Fig 4A and 4B).
Fig 4. Classification of transcriptional regulation divergence between ZF and OF.
(A) Heatmaps of gene expression in ZFs and OFs, and allelic expression ratios in F1 hybrids for cis-, trans-, and both cis- and trans-regulated genes in song nuclei HVC and RA (blue–red colored). Comparison between species-different gene expression (A/B) and allelic expression ratios in F1 hybrids (a/b) in heatmaps (dark brown–light yellow colored). “A” and “B” represent RPKM(ZF average) and RPKM(OF average), respectively. “a” and “b” represent Reads (ZF allele) and Reads (OF allele), respectively. (B) Gene numbers classified by cis-, trans-, both cis- and trans-, conserved, and ambiguous regulation in HVC and RA. (C) Scatterplots of expression ratios between ZF and OF (x-axis) and allelic expression ratios in F1 hybrids (y-axis) for genes showing differential expression between species. Blue-, red-, and orange-colored spots: cis-, trans-, both cis- and trans-regulated genes, respectively. Filled spots correspond to species-differentially expressed (SDE) genes. (D) Cis- and trans-effects on the expression of species-differentially regulated genes. The percent of cis-, trans-, both cis- and trans-, conserved, and ambiguous transcriptional regulatory genes in the SDE and non-SDE genes (Fisher’s exact test, ***p < 0.001). Relevant data values are included in S3 Data. F1, first-generation; OF, owl finch; RA, robust nucleus of the arcopallium; RPKM, reads per kilobase of transcript per million reads mapped; ZF, zebra finch.
In both HVC and RA, trans-alteration was more prevalent than cis-alteration. These results indicated that the expression of 600–800 genes (approximately 10%–15% of the total expressed genes) in the vocal motor song nuclei was modified by altered transcriptional regulation between the two species. Furthermore, a majority of the genes under conserved regulation were highly expressed in both HVC and RA (3,523 genes of 4,489 [78.5%] and 4,782 [73.7%] genes expressed in HVC and RA, respectively). In contrast, most of the cis- and/or trans-regulated genes were not shared between HVC and RA (Fig 4B), showing a brain region–specific transcriptional regulatory alteration. Although this result was obtained based on a cross-replicate comparison of the ASE ratio using eight F1 hybrids, we confirmed this result through an estimation method using the average of ASE of all F1 hybrids [16,40,41], which showed similar rates of cis- versus trans-regulation divergence (see Materials and methods, S5 Fig).
Cis- and trans-regulatory effects on species-differential expression
We then examined whether the species-differentially expressed (SDE) genes in HVC and RA were affected by the transcriptional regulatory divergence between ZF and OF. Based on the RPKM values of each gene expressed in ZF and OF, 333 and 374 genes showed significantly different expression in HVC and RA, respectively, between the two species (2.9% and 3.3% of the total genes expressed in HVC and RA) (DEseq2 package, p-value corrected by the Benjamini-Hochberg method, p < 0.05; n = 4 each from ZF and OF) (S6 Fig). Totals of 209 and 242 genes of the SDE genes in HVC and RA, respectively, passed the ss-SNPs threshold for calculating the ASE ratio in F1 hybrids. Such SDE genes were significantly enriched with a higher probability of cis-, trans-, and both cis- and trans-regulatory effects compared with those of non-SDE genes, in both HVC and RA (Fisher’s exact test, ***p < 0.001) (Fig 4C and 4D). This shows a significant association of transcriptional regulatory changes with SDE genes in the song nuclei.
A predominant effect on cellular molecular function by trans-regulatory divergence
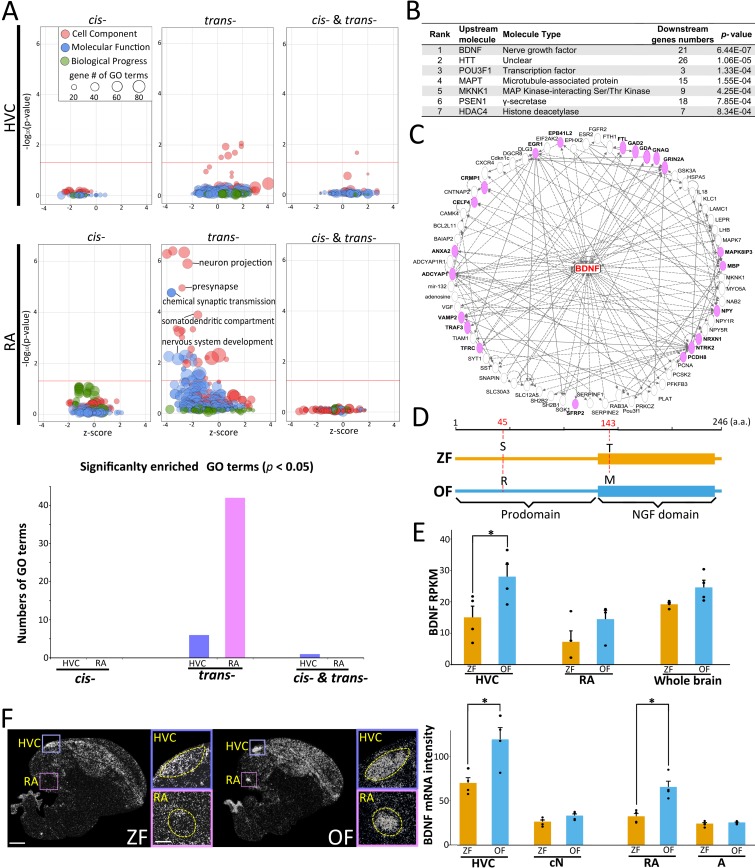
To understand whether transcriptional regulatory divergence has any potential molecular contribution to cellular functions in HVC and RA, we performed GO enrichment analysis using the sets of genes affected by cis-, trans-, and both cis- and trans-regulatory changes. The result showed that more GO categories were enriched for trans-regulated genes compared with the other types of regulatory divergence in both HVC and RA (Fisher’s exact test, p-value corrected by the Benjamini-Hochberg method) (Fig 5A). In particular, we found that GO categories related to neural functions associated with presynapse, chemical synapse transmission, and neuron projection were significantly enriched for RA trans-regulated genes. These results motivated us to focus subsequently on altered trans-regulation in RA.
Fig 5. Predominant effect on cellular molecular function by trans-regulatory divergence.
(A) GO enrichment analysis of the cis-, trans-, and both cis- and trans-regulated genes in HVC and RA. Size of points represents the number of genes assigned to each GO term. Red lines represent the p-value for significant enrichment (Fisher’s exact test adjusted by the Benjamini-Hochberg method, p < 0.05). (B) Top 7 candidate upstream mediators for trans-regulated genes in RA. (C) Gene–gene connections for BDNF downstream genes. Pink-colored genes are trans-regulated genes in RA. Solid and dotted lines represent directed and undirected regulation, respectively, between connected genes. (D) Comparison of BDNF amino acid sequences between ZF and OF. (E) BDNF mRNA expression level in HVC, RA, and whole brain between ZF and OF at the silent condition based on RNA-seq data. (F) BDNF mRNA expression in the HVC, RA, and the surrounding areas (caudal nidopallium [cN] and archopallium [A], respectively) of ZF and OF at the 3-hour undirected singing condition (n = 4 each). White signals: BDNF mRNA. Scale bars, 1 mm (in left panes) and 200 μm (in right panel). Relevant data values are included in S4 Data. a.a., amino acid; BDNF, brain-derived neurotrophic factor; GO, Gene Ontology; NGF, nerve growth factor; OF, owl finch; RA, robust nucleus of the arcopallium; RNA-seq, RNA sequencing; RPKM, reads per kilobase of transcript per million reads mapped; ZF, zebra finch.
To predict the potential regulatory mediators driving species differences in the expression of trans-regulated genes in RA, we performed upstream regulatory analyses using Ingenuity Pathway Analysis (IPA) [42,43]. We found that brain-derived neurotrophic factor (BDNF) was the most significant upstream trans-mediator of genes under trans-regulation in RA, which included genes for neural plasticity and dendritic spine formation (glutamate decarboxylase [GAD] 2, NMDA glutamate receptor [GRIN] 2A, neuropeptide Y [NPY], and collapsin response mediator protein [CRMP] 1) (Fisher’s exact test, p = 6.44 × 10−7) (Fig 5B and 5C and S7 Fig). Amino acid substitution and trans-mediator expression level changes could potentially mediate the trans-regulatory effects to alter downstream gene expression. In line with this prediction of BDNF as a trans-regulatory mediator in RA, we found two amino acid substitutions in BDNF between ZF and OF: Ser45Arg in prodomain and Thr143Met in nerve growth factor (NGF) domain (Fig 5D). Furthermore, BDNF was an SDE gene in HVC between the two species (Student's t test, *p < 0.05) (Fig 5E). In HVC, as an upstream song nucleus connecting to RA, BDNF mRNA is primarily expressed in neurons projecting to RA [44], meaning that HVC could anterogradely secrete BDNF protein to RA via connecting axons as a potential trans-regulation via neural connections. Furthermore, we found differences between species regarding the regulation of the BDNF mRNA expression level in both HVC and RA: OFs had a higher expression level than ZFs at the 3-hour singing condition that induced singing-driven gene expression change, including BDNF (Fig 5F) [44,45]. Therefore, in order to uncover the putative trans-regulatory mechanisms of BDNF and to evaluate its potential impacts in generating species-specific songs, we examined how the amino acid substitution and/or expression level of BDNF relates to song structures.
Correlation between individual variations of the species-biased song phenotypes and the BDNF expression level in F1 hybrids
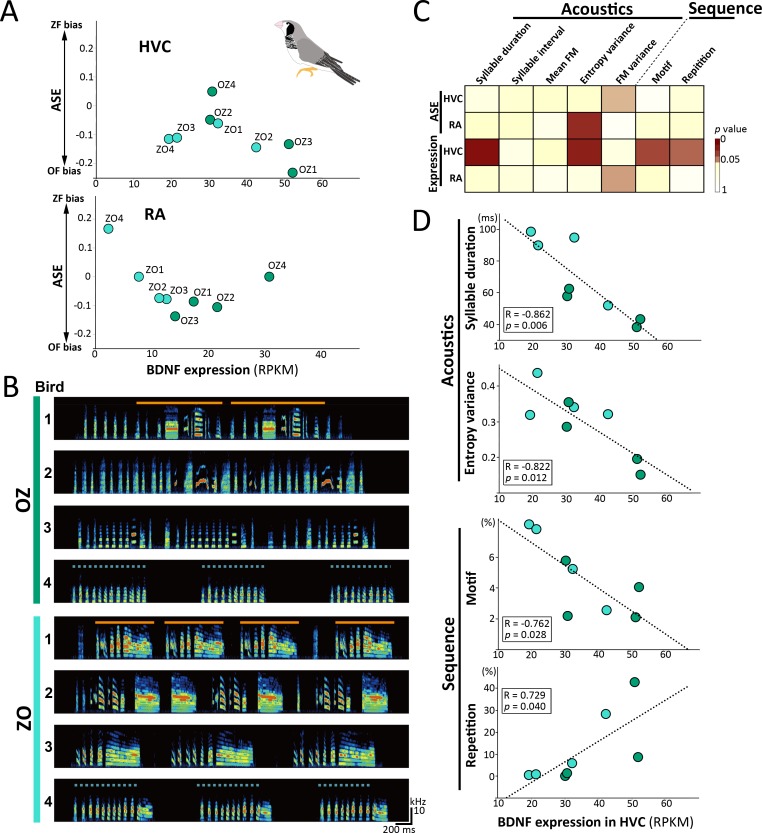
To evaluate the putative trans-regulatory effects mediated by the BDNF amino acid substitution or expression level, we investigated the correlation between song phenotypes and ASE ratio or the expression level of BDNF in F1 hybrids. Considering that neither ZF nor OF are inbred, the interspecies F1 hybrids might present individual variation in the ASE ratio and expression levels of transcribed genes. Consistently, at the transcriptome analysis in F1 hybrids, we realized that F1 hybrids possessed a wide range of individual difference in their ASE ratio and expression level of BDNF mRNA in HVC and RA (Fig 6A), such that each individual F1 hybrid transcribed ZF- and OF-type BDNFs with a unique expression ratio and level. Furthermore, F1 hybrids acquired individually unique songs with a wide range of ZF- and OF-biased features, even though they were reared listening to both ZF and OF songs as models (Fig 6B). We used the same sets of 7 total acoustic and sequential song parameters that showed differences between ZF and OF (5 for acoustic and 2 for sequential parameters) (Fig 2B and 2C). We found only one correlation between the ASE ratio of BDNF in RA and the entropy variance of syllables (r = 0.800, p = 0.017, Pearson correlation) (Fig 6C). In contrast, the expression level of BDNF mRNA in HVC had four significant correlations with acoustic and sequential song parameters in F1 hybrids (acoustics: syllable duration [r = −0.862, p = 0.006] and entropy variance [r = −0.822, p = 0.012]; sequence: motif [r = −0.762, p = 0.028] and repetition [r = 0.729, p = 0.040], Pearson correlation) (Fig 6C and 6D). These correlational analyses in F1 hybrids point to the BDNF mRNA expression level in HVC (instead of the amino acid substitution) being the most likely RA trans-acting mechanism, which induces anterograde secretion of BDNA protein to RA.
Fig 6. Correlation between individual variation in BDNF expression level and species-biased song structures in F1 hybrids.
(A) Individual variation of BDNF mRNA expression level and ASE ratio between F1 hybrids. (B) Individual variation of learned songs in F1 hybrids that were tutored with ZF and OF songs. Orange solid and blue dotted lines represent the motif and repetitive structure of syllables, respectively. (C) Heatmaps showing the correlation of p-values between the BDNF expression level or ASE ratio and species-biased song phenotypes in F1 hybrids. (D) Correlations between BDNF mRNA expression in HVC and species-biased song structures (syllable duration, entropy variance, motif, and repetition) among F1 hybrid individuals. Relevant data values are included in S5 Data. ASE, allele-specific expression; BDNF, brain-derived neurotrophic factor; F1, first-generation; OF, owl finch; OZ, F1 hybrid offspring between OF♀ and ZF♂; RA, robust nucleus of the arcopallium; RPKM, reads per kilobase of transcript per million reads mapped; ZF, zebra finch; ZO, F1 hybrid offspring between ZF♀ and OF♂.
Alternation of trans-regulated gene expression and obliteration of learned song features by the pharmacological overactivation of BDNF receptors in the RA
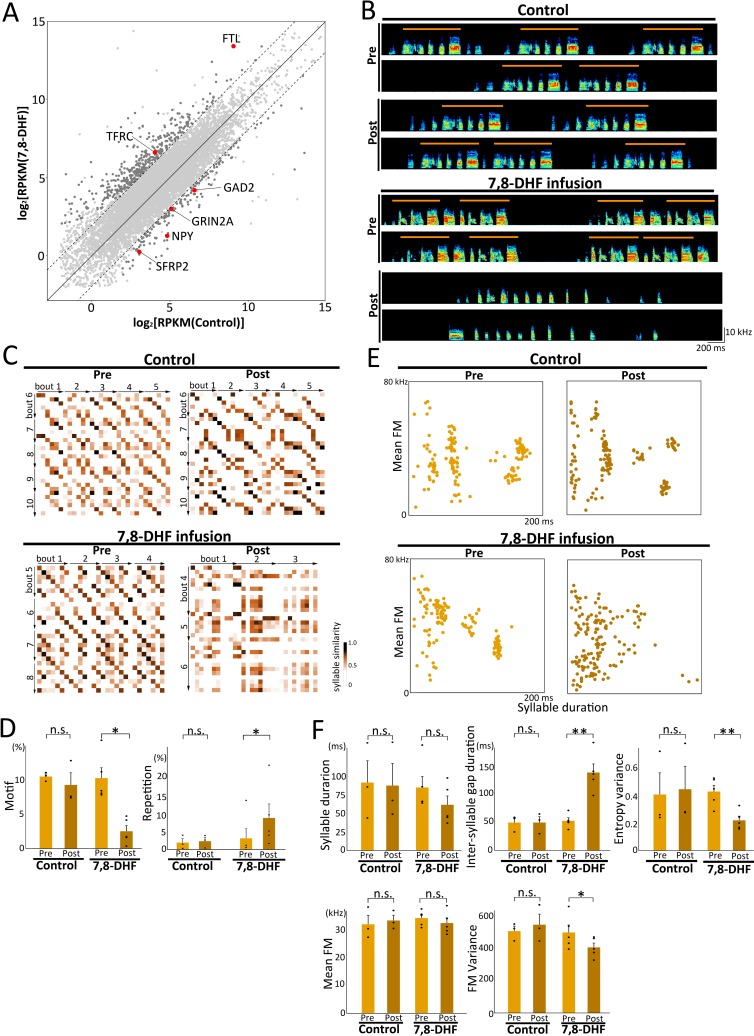
To examine the potential causal links of transcriptional regulation between the BDNF concentration level and the predicted downstream trans-regulated genes, we infused a selective agonist of the BDNF receptor, i.e., tropomyosin receptor kinase B (TrkB), namely 7,8-dihydroxyflavone (7,8-DHF; 10 μg/μL) in vivo, into the RA of adult ZFs by using local retrodialysis (S8 Fig) [46]. Transcriptional analysis to compare control (PBS) and 7,8-DHF–infused birds revealed that 570 genes of the 11,655 genes expressed in the RA were differentially identified, with over 4-fold changes between the two groups (DEseq2, p < 0.05) (Fig 7A). Among the differentially expressed 570 genes, 6 of the 21 putative downstream trans-regulated genes of BDNF (shown in Fig 5C) were found to have an altered expression after the pharmacological activation of the BDNF receptors. This further supports our earlier finding that BDNF could be a potential regulatory mediator of the RA trans-regulated genes.
Fig 7. Obliteration of species specificity of ZF song by BDNF receptor agonist infusion into RA.
(A) Scatterplot indicating RA gene expression in control and 7,8-DHF–infused birds. Dashed lines represent the boundary of the 4-fold expression difference. Darker gray colored dots represent significant differences in expressed genes higher than 4-fold between the control and 7,8-DHF–infused birds. Red colored dots represent downstream trans-regulated genes of BDNF (represented in Fig 5C). (B) Songs before and after infusing BDNF receptor TrkB agonist, 7,8-DHF. Typical examples of songs from control and 7,8-DHF–infused birds. Orange solid lines represent the motif structure of ZF songs. (C) Examples of syllable sequence changes between pre- and post-infusion. Syllable similarity matrices for a pair of songs produced by control and 7,8-DHF–infused birds. (D) Changes in the frequency of motif and repetition in songs at pre- and post-infusion stages (control ZF, n = 3, ZF with 7,8-DHF infusion [7–10 days], n = 5; paired t test, *p < 0.05). Each dot corresponds to individual birds. (E) Examples of syllable acoustic changes between pre- and post-infusion. Scatterplots indicate the distribution of 150 syllables (duration versus mean frequency) from control and 7,8-DHF–infused birds. (F) Changes in syllable acoustics (syllable duration, inter-syllable gap duration, entropy variance, mean FM, and FM variance) of songs at pre- and post-infusion stages (control ZF, n = 3, ZF with 7,8-DFH infusion [7–10 days], n = 5; paired t test, **p < 0.01, *p < 0.05, n.s., not significant). Each dot corresponds to an individual bird. Relevant data values are included in S6 Data. BDNF, brain-derived neurotrophic factor; FM, frequency modulation; RA, robust nucleus of the arcopallium; RPKM, reads per kilobase of transcript per million reads mapped; TrkB, tropomyosin receptor kinase B; ZF, zebra finch; 7,8-DHF, 7,8-dihydroxyflavone.
We also found that song changes following 7,8-DHF infusion, with a lower syllable transition consistency during the early stage (approximately 5 days after drug infusion). In addition, following continuous infusion for up to 2 weeks, adult structured songs gradually became more degraded, leading to the loss of learned song features in adult ZFs (Fig 7B). Although a few of the acoustic parameters (syllable duration and mean FM) maintained the original traits, syllable sequence (i.e., motif and repetitive indexes) and other acoustic parameters (i.e., inter-syllable gap duration, entropy variance, and FM variance) were drastically changed by the infusion of 7,8-DHF (Fig 7C and 7E), thus indicating that a precise amount of BDNF contributes to the maintenance of the learned song structures of ZF.
Discussion
Previous studies have demonstrated monogenic effects on adaptive behavioral phenotypes [7,47–49]. In contrast, the genetic basis of polygenic adaptations has been more challenging to pinpoint. Therefore, elucidating various SDE genes and the transcriptional regulatory divergences could be a promising step towards a better understanding of the contribution of multiple genes to the evolution of behaviors. For these, we examined the distribution of cis- and trans-regulatory divergences underlying the differences in gene expression in specific brain regions associated with the production of learned vocalizations between two closely related songbird species.
A number of studies that used entire organ tissues/body showed that there are more significant changes in cis- than trans-regulation between interspecies/lines of fruit flies [14,50], wasps [51], birds [52], and mouse [37]. In contrast, our study revealed that trans-regulatory changes were more prevalent than cis- in determining gene expression differences in song nuclei between two closely related species (Fig 4B). In addition, biological processes associated with neural functions were more enriched for genes showing trans-regulatory divergence in HVC and RA (Fig 5A). This difference in the effects of cis- or trans-regulations on transcriptional divergence could be caused by different methods of estimation using ASE ratio in the F1 hybrids. However, even when we used an estimation method using the average ASE of F1 hybrids, which has the potential to underestimate trans-regulation [40], we obtained a similar result showing that transcriptional regulatory divergence has occurred primarily in trans-regulation. To examine whether the trans-biased regulatory divergence is specific to song nuclei or not, it will be necessary to perform a similar analysis using samples from non-song nuclei or whole brain in songbirds. Furthermore, we set the cutoff with ≥5 reads at each ss-SNP position and median RPKM ≥ 10 to extract genes that were used for the calculation of the ASE ratio. This cutoff threshold is a stricter setting than other published studies [50,51]. Therefore, if we set a lower threshold to extract the ASE genes, the estimated gene number regulated by different transcriptional divergence would be increased.
In principle, two alleles in the cells of an F1 hybrid are regulated in the same trans-regulatory environment. Therefore, differences in expression of two alleles in F1 hybrids should reflect cis-regulatory divergence between the two parental genomes, generating a consistent ASE ratio among F1 hybrids. However, at a considerable number of genes in the song nuclei, we observed a large variance in ASE ratio among F1 hybrids, which we defined as “ambiguously” regulated genes (S2 Fig). Approximately 10% of the total expressed genes in HVC and RA were categorized as “ambiguous” (Fig 4B). Ambiguous regulation could result from intraspecies genomic variation. Indeed, the experimental ZF and OF have not been genetically selected animals. We found 742,302 and 414,040 polymorphic SNPs in the transcribed sequence from the whole brain of ZFs and OFs, respectively (n = 4 each). Therefore, the individual variability in the ASE ratio between F1 hybrids may be caused by intraspecies polymorphisms, which could in turn be additional trans- and cis-regulatory variants underlying individual difference in gene expression in song nuclei.
We found that BDNF is one of several potential upstream mediators for trans-regulated genes in RA (Fig 5 and S7 Fig). BDNF transcription, secretion, and actions are directly regulated by neural activity. Secreted BDNF mediates multiple activity-dependent processes, including neuronal differentiation/growth, synapse formation, and plasticity during developmental and adult stages [53–56]. In the song system of songbirds, singing behavior induces BDNF mRNA expression in song nuclei including HVC, suggesting that neural activity-dependent signaling of BDNF regulates neuronal maturation [44,45,57,58]. We had reported that ZFs prevented from singing during the song learning period possess immature dendritic spine density in RA neurons and produced highly unstable song lacking species-specific features when allowed to sing freely, even at the adult stage [59]. Although transient BDNF up-regulation in HVC enhances song learning during the critical period [60], a short-term local injection of BDNF into RA of adult ZFs changed crystallized songs to juvenile-like plastic songs with sequence variability; these changes correlated with an increase in HVC axonal boutons in RA [61]. We further confirmed that the continuous and local infusion of BDNF receptor agonist 7,8-DHF into RA of adult ZFs induced severe song degradation, eliminating both learned acoustic and sequence features. Therefore, we suggest that BDNF mediates the precise synaptic connections and strength of connections allowing HVC to activate populations of RA neurons at specific and precise time points during song rendition. Species differences in BDNF expression in the song nuclei could shape the anatomical and molecular bases for the generation of species-specific learned song structure via the activity-dependent trans-regulation of the downstream genes. Using F1 hybrids, we found that ASE ratios of BDNF were more weakly associated with species-biased song phenotypes compared with BDNF mRNA expression levels. However, this does not rule out a potential trans-regulatory effect mediated by BDNF amino acid substitution on species-specific behaviors. In human, the BDNF polymorphism (Val66Met; rs6265) affects intracellular trafficking and reduces activity-dependent secretion of mature BDNF [62]. The BDNF polymorphism causes altered dendritic spine density, memory formation, and extinction [63,64]. Future application of genome editing technologies would be a powerful tool to elucidate the in vivo contribution of BDNF polymorphisms to species-specific behaviors.
We investigated the divergence between the ZF and OF in terms of gene transcription for the generation of species-specific learned songs (but not for the learning process). Thus, we performed a series of experiments including song comparative analysis, comprehensive RNA sequencing (RNA-seq), and BDNF agonist infusion by using adult birds after the critical period for song learning. However, it is crucial to consider the potential effects of BDNF on the development of neural circuits for species-specific song learning and production during the embryonic and early post-hatching periods. Although we observed that the pharmacological overactivation of BDNF receptors drastically affected song change and led to the loss of learned song structures at both syllable acoustic and sequence levels, we cannot tell whether such song degradation is induced by any species-specific deficiency. Given the wide variety of BDNF cellular functions, the pharmacological experiment was limited by the selective modification of signaling machinery for species-specific song generation. We found that not only predicted downstream trans-regulated genes but also over 550 genes had altered expression levels in the RA, as assessed by comparing control and 7,8-DHF–infused birds. Therefore, future research with more refined experiments for targeted multiple genes, manipulation timing, and cell types will be crucial.
In this study, we investigated the regulatory drivers of species divergence in gene expression in song nuclei in the vocal motor circuit in adults. We suggest these regulatory differences between species could explain a genetic molecular mechanism for the generation and maintenance of the species specificity of learned songs. The anterior forebrain pathway (AFP) is a cortico-basal ganglia-thalamocortical loop, which is a specific pathway for song learning during development and for vocal plasticity maintenance later in life [65–69]. For sensorimotor coordination, AFP generates instructive biased variability and conveys this to the premotor song nuclei RA as a reinforcement signal [46,70]. Currently, we cannot make direct causal links between AFP function and the acquisition of species-specific song patterns. However, lesion of the basal ganglia nucleus, Area X of the striatum (Area X), in the AFP at an early critical period was shown to disrupt motif structure, a sequential trait commonly observed in ZF songs [69]. Furthermore, the expression of transcription factors such as Forkhead box protein P2 (FoxP2) and androgen receptors in Area X shows species-specific patterns [25,26]. These transcription factors could be potential regulators for further species difference due to their regulatory effects on downstream genes, thereby generating species-biased vocal plasticity, which in turn promotes species-specific song learning. Therefore, studying species differences in gene expression in the song nuclei of the AFP through the critical period of song learning would provide vital insight into how species-specific patterns of gene expression underlie species-specific songs.
In conclusion, our results suggest a neurogenetic association between brain region–specific transcriptional divergence and species-specific learned behaviors. Most complex motor skills, such as birdsong and human speech, are acquired through learning and constrained in a species-specific manner [35,71]. Using similar techniques to those developed in the present study on other interspecific hybrids could give additional insights into the existence of more conserved or unique cis-/trans-regulatory changes underlying the evolution of species-specific learned behaviors.
Materials and methods
Ethics statement
All experiments were conducted under the guidelines and approval of the Committee on Animal Experiments of Hokkaido University (Approved No. 18–0053). These guidelines are based on the national regulations for animal welfare in Japan (Law for the Humane Treatment and Management of Animals with partial amendment No. 105, 2011). For brain sampling, the birds were humanely killed by decapitation after injection of an overdose of pentobarbital.
Animals and song tutoring
ZFs (T. guttata) and OFs (T. bichenovii) were obtained from our breeding colony at Hokkaido University and local breeders. Reciprocal F1 hybrids were bred by pairing ZF and OF at our breeding colony. All birds were maintained with food and water available ad libitum under a 13:11-hour light/dark cycle. For song cross-tutoring experiments, ZF chicks were raised by both parents in breeding cages until 10–15 phd, and then the father was removed by 15–25 phd from the cage to prevent male juveniles from listening their father’s song. OF chicks were hand-raised after hatching until they could feed themselves (approximately phd 30–40). After fledging, juveniles were subsequently housed in individual isolation boxes and then individually housed in a sound-attenuating box containing a mirror to reduce social isolation. Cross-species’ tutor songs were played 7 times each in the morning and afternoon at 55–75 decibels from a speaker (SRS-M30, SONY, Tokyo, Japan) passively controlled by Sound Analysis Pro. Similarly, F1 hybrids were song tutored by passively and randomly playing a set of ZF and OF songs with an interval duration of 300–500 ms as the song model.
Song recording and analysis
Songs were recorded using a unidirectional microphone (SM57, Shure) connected to a computer with Sound Analysis Pro (SAP v1.04). For analysis of the acoustic features of songs, 500 syllables were randomly selected from ZF and OF songs (n = 6 birds each). To characterize the syllable that differed between ZF and OF, a total of 10 acoustic features were measured: syllable duration, inter-syllable gap duration, mean pitch, pitch goodness, Wiener entropy, entropy variance, mean AM, AM variance, mean FM, and FM variance [31]. Statistical analysis was performed on these acoustic features between ZF and OF by one-way ANOVA. For the analysis of the sequence feature of songs (motif and repetition rates in a song), a syllable similarity matrix (SSM) analysis was performed following a previously reported method [30] (S1 Fig). This method calculates the contiguous syllables transition frequency of “paired (motif)” and “repetitive” syllables transitions in the songs. To test song structure changes by pharmacological manipulation, we measured both the syllable acoustic and sequential parameters of 150 syllables at pre- and post-time points (7–10 days) after drug infusion. Six acoustic syllable parameters (syllable duration, inter-syllable gap duration, entropy variance, AM variance, mean FM, and FM variance) and sequence features (motif and repetition transition rates) were used for the PCA and 2D view, and this was performed using the prcomp and rgl packages in R, respectively.
Brain tissue sampling and RNA extraction for RNA-seq
For sampling of whole brain tissues, adult male birds were isolated in a soundproof chamber for at least one day before humanely killing (ZF: n = 4, OF: n = 4, adult [>200 phd]). Birds were killed under silent and dark condition in the morning before the lights were turned on. The pallium and pallidum regions were rapidly dissected, frozen on dry ice, and stored at −80°C until RNA extraction. Total RNA was isolated using TRIzol Reagent according to the manufacture’s protocol (Invitrogen) and was treated with RNase-free DNase.
For sampling of HVC and RA tissues by laser capture microdissection (LCM), adult ZF, OF, ZO, and OZ F1 hybrids (n = 4 birds each, >130 phd) and control and 7, 8-DHF–infused ZFs (n = 3 each, >130 phd) were isolated in sound-attenuation boxes and killed under silent and dark condition. Brains were embedded in OCT compound (Sakura Fine Technical) and stored at −80°C until use. Brain sections were cut at a 14-μm thickness in the sagittal plane and mounted onto glass slides with a handmade membrane system for laser microdissection. We confirmed the presence and boundaries of HVC and RA using Nissl staining (LCM Staining kit; Ambion). HVC and RA were microdissected using a laser capture microscope ArcturusXT (Arcturus Bioscience) with the following parameter settings: spot diameter, 100 μm; laser power, 80 mW; and laser duration, 80 ms [72]. The captured tissues were dissolved into RLT buffer (Qiagen) with β-mercaptoethanol, treated with DNase in the column to avoid contamination of genomic DNA, and then stored at −80°C until RNA extraction.
RNA-seq library construction and sequencing
RNA integrity number (RIN) and concentration were measured with Bioanalyzer 2100 (Agilent Technologies) to guarantee the quality of RNA. For RNA-seq of HVC and RA, we performed first-strand cDNA amplification using total RNA (1–2 ng) from HVC and RA under a PCR amplification condition of 14 cycles at 98°C for 10 seconds, 65°C for 15 seconds, and 68°C for 5 minutes, following the Quartz-amplification method [73]. Amplified cDNAs were purified using a PCR purification column (MiniElute PCR Purification Kit; Qiagen) and the concentration was measured using Bioanalyzer 2100 (Agilent Technologies). Non-amplified first-strand cDNAs synthesized using total RNA from the whole brain (telencephalon) and amplified cDNAs using total RNA from HVC and RA tissue were used to construct poly(A) selected paired-end sequencing libraries (TruSeq DNA Sample Prep Kits, Illumina). All libraries were sequenced using the Illumina Hiseq2500 platform for 100-bp paired-end sequencing.
For each telencephalon brain sample, 33.5–47.0 M RNA-seq reads were output from the Illumina Hiseq 2500. Sequencing reads were mapped onto the ZF reference genome obtained from Ensembl (Taeniopygia_guttata taeGut3.2.4.dna.fa) with the Tophat2 program and assembled to predicted transcripts with the Cufflinks program. Through comparison with the previous annotation file using the cuffcompare program, 12,156 transcripts were identified as predicted RNA transcripts expressed in the ZF telencephalon. All RNA-seq data were deposited in the DDBJ Sequence Read Archive (submission numbers DRA005548, DRA002970, and DRA008696).
Identification of ss-SNPs
Adapter sequences of raw data from ZF and OF whole brain NGS results were removed by Trimmonatic. Clean reads from ZF and OF whole brain were mapped to a ZF reference genome obtained from Ensembl (Taeniopygia_guttata.taeGut3.2.4) by TopHat2 to reconstruct pseudo ZF and OF genomes. Mapped reads with longer gaps (>3,000 bp) were removed in the subsequent analysis. ss-SNPs and insert and deletions (indels) between ZF and OF were identified from the mapping result of the whole brain reads. The positions of ss-SNPs and indels were used to reconstruct pseudo genomes of ZF and OF. ss-SNPs were defined as follows: the base variants were same in all individuals of a species, but different from the base found in all individuals of another species. SNPs in individuals of the same species (intraspecies SNPs) were maintained as the same base for both ZF and OF reconstructed genome sequences. MUMmer software was used to identify ss-SNPs using the reconstructed ZF and OF genomes.
Read mapping and quantification of gene expression level
Low-quality reads and adaptor sequences were removed from all HVC and RA raw reads using the Filter FASTQ pipeline (https://cell-innovation.nig.ac.jp) and Flexbar software. Clear reads were mapped to reconstructed ZF genome by TopHat2. Transcript levels were quantified as RPKM value. Cufflinks was used to evaluate the expression levels of each gene by calculating the RPKM of HVC and RA samples of ZF and OF using the improved genome annotation Gene Transfer Format (GTF) file [59]. Based on the RPKM of individual birds (n = 4 each from ZF and OF; n = 3 each from 7,8-DHF–infused and control ZFs), the expression differences of each gene were identified between ZF and OF and between 7,8-DHF and control ZFs as differently expressed genes using the R package DEseq2 (adjusted p-value < 0.05, the Benjamini-Hochberg procedure).
Allelic expression ratio in F1 hybrids
To distinguish reads of the two alleles in F1 hybrids, the mapping results of HVC and RA of F1 hybrids were used following SNPsplit’s instruction. First, an N-marked genome sequence was constructed by replacing “N” at the ss-SNP position in the ZF pseudo genome. RNA-seq reads of HVC and RA of ZF, OF, and F1 hybrids were mapped to the N-marked genome by TopHat2. ss-SNPs were identified as SNP sites having more than 98% of total reads that were different between ZF and OF alleles. The identified ss-SNPs were reverified by reads from HVC and RA of ZF and OF, to enhance the reliability of ss-SNPs. The mapped HVC and RA reads of F1 hybrids were then separated into ZF or OF allele transcripts based on the ss-SNP information, and the number of reads was counted at each ss-SNP position by SAMtools.
The following thresholds were set for calculating the allelic expression ratio of each gene expressed in HVC and RA of F1 hybrids: (i) existence of at least one ss-SNP, (ii) more than 5 reads at each ss-SNP site, and (iii) a median RPKM of at least 10 for all 16 individuals (including ZF, n = 4; OF, n = 4; ZO, n = 4; OZ, n = 4). The allelic expression ratio was quantified using the d-score [74]:
d-scores of 0 reflect equal expression between the two alleles, whereas d-scores of −0.5 and 0.5 reflect exclusive transcription from OF or ZF alleles, respectively.
Identification of cis- and/or trans-regulatory divergence
The potential of genomic imprinting in F1 hybrids was tested using Spearman’s rank correlation of gene allelic expression ratio between ZO (n = 4) and OZ (n = 4). The difference in the allelic expression ratio of each gene was compared between ZO and OZ hybrids using one-way ANOVA (ZO, n = 4; OZ, n = 4; adjusted p-value by the Benjamini-Hochberg method).
Cis- and/or trans-regulatory divergences were evaluated using a previously reported method [16]. The gene expression ratio between parental species was calculated with the formula X = log2(A/B), where “X” is the gene expression ratio between parental species; “A” and “B” are the average RPKM for ZFs and OFs, respectively (n = 4 each from ZF and OF). The allelic expression ratio of F1 hybrids was calculated as Y = log2(a/b), where “Y” is the allelic expression ratio between two alleles; “a” and “b” are the read counts of ZF and OF alleles in F1 individuals, respectively. Cis- and trans-effects on gene expression divergence were estimated by the scheme described in Fig 1D. In brief, the regulation mechanism of gene expression between ZF and OF was (1) a cis-regulatory difference if X = Y and Y ≠ 0; (2) a trans-regulatory difference if X ≠ Y and Y = 0; (3) both cis- and trans-regulatory differences if X ≠ Y and Y ≠ 0; (4) no cis- and trans-regulatory differences (i.e., conserved) if X = Y and Y = 0. The Student's t test was used to determine the difference between the gene expression ratio in parental species and the allelic expression ratio in F1 hybrids. The SGoF program was employed to correct p-values for multiple testing (adjusted p ≤ 0.05). The previous standard method for estimating regulatory divergence can lead to a negative correlation as an artifact when cis-estimates have any errors [40, 41]. To avoid this bias, first we randomly selected four individual F1 hybrids as a group to estimate cis-effects using their average ASE ratio while the remaining four F1 individual hybrids were used to compare the expression ratio between ZF and OF. For each gene, a total of 70 combinations were constructed by random selection of four of eight F1 hybrid birds (n = 4 each from ZO and OZ). Thus, cis- and/or trans-regulatory identification was done for each gene for each pair of 70 total combinations. During this cross-replicate comparison, some genes were categorized as different transcriptional regulations due to a large variance in ASE ratios among F1 individuals. Therefore, we finally determined which transcriptional divergence made the main regulatory effect on each gene by two steps of statistics following (i) calculation of the difference between four categories (cis-, trans-, both cis- and trans-, and conserved) using the Chi-squared test (with adjusted p-value by FDR < 0.05) and (ii) a comparison of the difference between the first- and second-strongest regulatory effects using a Fisher’s exact-test (adjusted p-value by FDR < 0.05). If genes did not show significance at both tests, such genes were defined as “ambiguous regulatory genes” (S2 Fig).
In addition, we performed analysis of cis- and/or trans-regulatory divergence using a standard method [37,51] and compared these results with those from the above method. The difference of the standard method is that the allelic expression ratios of all eight F1 hybrids (ZO = 4, OZ = 4) were used to estimate cis- and trans-regulatory effects. In brief, the parental expression ratio value X and the allelic expression ratio in F1 hybrid value Y were calculated similarly to our new method. The average values Y of eight F1 hybrid individuals were compared with values X and 0, respectively, to estimate cis- and trans-effects by the scheme described in Fig 1D (Student's t test). The SGoF program was employed to perform multiple testing correction (adjusted p-value ≤ 0.05) (S5 Fig).
Functional analysis of cis- and/or trans-regulated genes
The functions of genes with cis-, trans-, and cis- and trans-regulatory divergences between ZF and OF in HVC and RA were annotated by GO analysis (DAVID Bioinformatics Resources 6.8; https://david.ncifcrf.gov). GO enrichment analysis was performed for each gene group using Fisher’s exact tests (p-value was adjusted by the Benjamini-Hochberg method). As trans-regulated genes in RA were enriched for the most GO terms, an upstream regulatory analysis was performed for RA trans-regulated genes using IPA software.
In situ hybridization
BDNF cDNA fragments used for the synthesis of in situ hybridization probes were cloned from a whole-brain cDNA mixture of a male ZF. Total RNA was transcribed to cDNA using Superscript Reverse Transcriptase (Invitrogen) with oligo dT primers. The cDNAs were amplified by PCR using oligo DNA primers directed to the open reading frame region from the NCBI cDNA database. PCR products were ligated into the pGEM-T Easy plasmid (Promega). The cloned sequences were searched using NCBI BLAST/BLASTX to compare with homologous genes to other species and genome loci identified using BLAT of the UCSC Genome Browser.
Adult male ZFs (n = 4) and OFs (n = 4) were used. Birds were individually housed in sound-attenuating boxes overnight. On the following morning, singing behavior (undirected singing) was recorded for 3 hours after the lights were turned on. After each singing behavior observation session, the birds were euthanized by decapitation. Brains were embedded in OCT compound (Sakura Fine Technical) and stored at −80°C until use. Frozen sections (12-μm thick) were cut in the sagittal plane. Brain sections for a given experiment were simultaneously fixed in 3% paraformaldehyde/1× PBS (pH 7.4), washed in 1× PBS, acetylated, dehydrated in an ascending ethanol series, air-dried, and processed for in situ hybridization with antisense 35S-UTP–labeled riboprobes of genes. To generate the riboprobes, gene inserts in the pGEM-T Easy vector were PCR amplified with plasmid M13 forward and reverse primers and then gel purified. The amplified DNA fragments and SP6 or T7 RNA polymerase were used to transcribe the antisense 35S-riboprobes. A total of 1 × 106 cpm of the 35S-probe was added to a hybridization solution (50% formamide, 10% dextran, 1× Denhardt’s solution, 12 mM EDTA [pH 8.0], 10 mM Tris-HCl [pH 8.0], 300 mM NaCl, 0.5 mg/mL yeast tRNA, and 10 mM dithiothreitol). Hybridization was performed at 65°C for 12–14 hours. The slides were washed in 2× SSPE and 0.1% β-mercaptoethanol at room temperature for 1 hour; 2× SSPE, 50% formamide, and 0.1% β-mercaptoethanol at 65°C for 1 hour; and 0.1× SSPE twice at 65°C for 30 minutes each. Slides were dehydrated in an ascending ethanol series and exposed to X-ray film (Biomax MR, Kodak) for 1–14 days. We carefully attended in order not to overexpose X-ray films to S35-riboprobe hybridized brain sections. The slides were then dipped in an autoradiographic emulsion (NTB2, Kodak), incubated for 1–8 weeks, and processed with D-19 developer (Kodak) and fixer (Kodak). For quantification of mRNA signal, exposed X-ray films of brain images were digitally scanned under a microscope (Leica, Z16 APO) connected to a CCD camera (Leica, DFC490) with Application Suite V3 imaging software (Leica), as previously described [45, 72, 75, 76]. To minimize handling bias for signal detection among experimental groups, we performed in situ hybridization using multiple brain sections at once for each probe and exposed S35-riboprobe hybridized brain sections on the same sheet of X-ray films. The same light settings were used for all images. Photoshop (Adobe Systems) was used to measure the mean pixel intensities in the brain areas of interest from sections after conversion to 256 grayscale images.
Pharmacological manipulation
Custom microdialysis probes were built using a microdialysis membrane (SpectralPor, in vivo microdialysis hollow fiber, O.D. = 216 μm; total weight, <0.035 g) attached to a drug reservoir, based on a previously described method [46]. Probes were bilaterally implanted at positions adjacent to RA using stereotaxic coordinates. Before setting the probe, spontaneous neural activity was measured to verify the location of RA. Microdialysis probes were carefully set outside the RA to avoid physically damaging the RA, because damage to the RA could induce song changes. Following surgery, the reservoir was filled every morning with saline until the bird began to sing consistently and its phonological and syntactical features were confirmed not to be damaged by implantation of probe. To ensure the position of microdialysis probes, tetrodotoxin (TTX; 6–12 μM) was infused into the RA in a hemisphere, and a hemi-RA inactivation-induced song change was confirmed. Saline (n = 3 birds) or 7,8-DHF (10 μg/μL in 0.9% NaCl, pH 7.4–7.6, Santa Cruz; n = 5 birds) was then continuously infused during daytime via the injection of approximately 2.5 uL of solution into the outer reservoir of the microdialysis probes 3 to 4 times daily. The manipulated birds were allowed to move freely in a sound-attenuation chamber, and the song of each individual was recorded over 10 days after initiation of drug infusion. The remaining drug volume and infusion speed were checked by using a transparent polyimide tube as the outer reservoir of the microdialysis probes. Probe positioning was evaluated postmortem by histological staining of tissue sections.
Supporting information
(A) (Upper panels) SSM analysis for the detection of syllable sequential transition patterns. The SSM comprises two steps: First, a correlation matrix including the syllable similarity scores was prepared using the round-robin comparison of all syllables in two songs to maintain the sequential order of the syllables in the songs. These similarity scores in the matrix were binarized at a threshold at 0.595. Second, the occurrence rate of two patterns of binarized “2 row × 2 column” cells in the SSM was calculated as a percentage of the paired (motif) and repetitive-syllable transition types (see the Materials and methods). (Lower panel) Test examples of the SSM method using artificial song models mimicking the songs with motif and repetitive sequences. (B) The similar distribution range of syllable acoustic traits between ZF and OF. Violin plots of the distribution of syllable duration, inter-syllable gap duration, entropy variance, AM variance, mean FM, and FM variance from ZF and OF that were reared with conspecific song tutoring (total 3,000 syllables from n = 6 birds each and 500 syllables/bird). (C) PCA of the song features of ZFs and OFs reared under conspecific and cross-species song tutoring conditions (“Con”: n = 6 each from conspecific song tutored ZF and OF; “Cross”: n = 4 and 3 from cross-species song tutored ZF and OF, respectively). Relevant data values are included in S1 Data for panels B and C. AM, amplitude modulation; FM, frequency modulation; OF, owl finch; PCA, principal component analysis; SSM, syllable similarity matrix; ZF, zebra finch.
(TIF)
(TIF)
(Left panels) Expression levels of PRKAA1, NR2E1, and CACNA1E in song nucleus HVC of ZFs and OFs. Gray-colored boxes represent the position of exons for each gene. Dark blue peaks below exons represent read density. (Right panels) Gene expression levels in ZF and OF. Each dot represents RPKM value for the individual. Mean ± SEM (n = 4 birds each, one-way ANOVA, *p < 0.05; n.s., not significant). Relevant data values are included in S2 Data. CACNA1E, calcium voltage-gated channel subunit alpha 1E; NR2E1, nuclear receptor subfamily 2 group E member 1; OF, owl finch; PRKAA1, protein kinase AMP-activated catalytic subunit alpha 1; RPKM, reads per kilobase of transcript per million reads mapped; ZF, zebra finch.
(TIF)
Scatterplots of allelic expression ratios of 5,849 and 6,328 genes in HVC and RA, respectively, of OZ and ZO hybrids (Spearman correlation coefficient). Relevant data values are included in S3 Data. F1, first-generation; OF, owl finch; OZ, F1 hybrid offspring between OF♀ and ZF♂; RA, robust nucleus of the arcopallium; ZF, zebra finch; ZO, F1 hybrid offspring between ZF♀ and OF♂.
(TIF)
Relevant data values are included in S3 Data. ASE, allele-specific expression; F1, first-generation; RA, robust nucleus of the arcopallium.
(TIF)
(A) SDE genes in HVC and RA. Orange- and blue-colored spots represent significantly higher expression in ZF or OF, respectively (DEseq2 corrected with the Benjamini-Hochberg method, p < 0.05). (B) Venn diagram representing the number of genes in HVC and RA that are differently expressed between ZF or OF. Relevant data values are included in S3 Data. OF, owl finch; RA, robust nucleus of the arcopallium; SDE, species-differentially expressed; ZF, zebra finch.
(TIF)
Top 7 candidate upstream mediators, including BDNF, HTT, POU3F1, MAPT, MNKK1, PSEN1, and HDAC4. Trans-regulated genes by BDNF in RA are noted in red. Orange- and green-colored genes are trans-regulated genes that are significantly expressed more highly in RA of ZF or OF, respectively. Relevant data values are included in S4 Data. BDNF, brain-derived neurotrophic factor; HDAC4, histone deacetylase 4; HTT, huntingtin; MAPT, microtubule-associated protein tau; MNKK1, MAP kinase-interacting serine/threonine protein kinase 1; OF, owl finch; POU3F1, POU class 3 homeobox 1; PSEN1, presenilin 1; RA, robust nucleus of the arcopallium; ZF, zebra finch.
(TIF)
(Left) Photograph of homemade microdialysis probe. (Right) A ZF with microdialysis probes bilaterally implanted in RA. BDNF, brain-derived neurotrophic factor; RA, robust nucleus of the arcopallium; ZF, zebra finch.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank Keiko Sumida for her excellent bird care and breeding, Drs. C. N. Asogwa and D. Wheatcroft for their comments and discussion, Drs. M. Tanaka and K. Hamaguchi for their instruction for making custom microdialysis probes, and Dr. Y. Suzuki’s laboratory in the Department of Computational Biology, the University of Tokyo, for RNA-seq experiments.
Abbreviations
- AFP
anterior forebrain pathway
- AM
amplitude modulation
- Area X
Area X of the striatum
- ASE
allele-specific expression
- BDNF
brain-derived neurotrophic factor
- CRMP
collapsin response mediator protein
- DLM
dorsal lateral nucleus of the medial thalamus
- FM
frequency modulation
- FoxP2
Forkhead box protein P2
- F1
first-generation
- GAD
glutamate decarboxylase
- GO
Gene Ontology
- GRIK1
Glutamate receptor, ionotropic, kainate type 1
- GRIN
NMDA glutamate receptor
- GTF
Gene Transfer Format
- HTR1B
5-hydroxytryptamine receptor 1B
- indel
insert and deletion
- IPA
Ingenuity Pathway Analysis
- LMAN
lateral magnocellular nucleus of the anterior nidopallium
- LMO7
LIM domain only protein 7
- NGF
nerve growth factor
- NPY
neuropeptide Y
- nXIIts
tracheosyringeal part of the hypoglossal nucleus
- OF
owl finch
- OZ
first-generation hybrid offspring between owl finch female and zebra finch male
- PCA
principal component analysis
- RA
robust nucleus of the arcopallium
- RAB5A
Ras-related protein Rab5A
- RASGEF1B
Ras-GEF domain-containing family 1B
- RIN
RNA integrity number
- RNA-seq
RNA sequencing
- RPKM
reads per kilobase of transcript per million reads mapped
- SDE
species-differentially expressed
- ss-SNP
species-specific SNP
- SSM
syllable similarity matrix
- TrkB
tropomyosin receptor kinase B
- TTX
tetrodotoxin
- ZF
zebra finch
- ZO
first-generation hybrid offspring between zebra finch female and owl finch male
- 7,8-DHF
7,8-dihydroxyflavone
Data Availability
All RNA-seq data were deposited in the DDBJ Sequence Read Archive (submission numbers DRA005548, DRA002970, and DRA008696).
Funding Statement
This work was supported by a Japanese MEXT scholarship and the China Scholarship Council (CSC#201408210091) to HW, MEXT/JSPS KAKENHI Grant Number #4903-JP17H06380, JP17H05932, JP17K19629, and JP18H02520 to KW, and RNA-seq experiments were supported by MEXT KAKENHI 221S0002. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Krebs J, Ashcroft R, Webber M. Song repertoires and territory defence in the great tit. Nature. 1978;271:539 10.1038/271539a0 [DOI] [Google Scholar]
- 2.Searcy WA, Andersson M. Sexual Selection and the Evolution of Song. Annual Review of Ecology and Systematics. 1986;17(1):507–33. 10.1146/annurev.es.17.110186.002451 [DOI] [Google Scholar]
- 3.Tinbergen N. On aims and methods of ethology. Zeitschrift für Tierpsychologie 1963;20:410–33. [Google Scholar]
- 4.Ding Y, Lillvis JL, Cande J, Berman GJ, Arthur BJ, Long X, et al. Neural Evolution of Context-Dependent Fly Song. Current biology: CB. 2019. Epub 2019/03/19. 10.1016/j.cub.2019.02.019 . [DOI] [PubMed] [Google Scholar]
- 5.Seeholzer LF, Seppo M, Stern DL, Ruta V. Evolution of a central neural circuit underlies Drosophila mate preferences. Nature. 2018;559(7715):564–9. 10.1038/s41586-018-0322-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leininger EC, Kelley DB. Evolution of Courtship Songs in Xenopus: Vocal Pattern Generation and Sound Production. Cytogenet Genome Res. 2015;145(3–4):302–14. 10.1159/000433483 . [DOI] [PubMed] [Google Scholar]
- 7.Chandra V, Fetter-Pruneda I, Oxley PR, Ritger AL, McKenzie SK, Libbrecht R, et al. Social regulation of insulin signaling and the evolution of eusociality in ants. Science. 2018;361(6400):398–402. 10.1126/science.aar5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodson JL, Schrock SE, Klatt JD, Kabelik D, Kingsbury MA. Mesotocin and nonapeptide receptors promote estrildid flocking behavior. Science. 2009;325(5942):862–6. Epub 2009/08/15. 10.1126/science.1174929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young LJ, Winslow JT, Nilsen R, Insel TRJBn. Species differences in V₁a receptor gene expression in monogamous and nonmonogamous voles: Behavioral consequences. 1997;111(3):599. [DOI] [PubMed] [Google Scholar]
- 10.Ding Y, Berrocal A, Morita T, Longden KD, Stern DL. Natural courtship song variation caused by an intronic retroelement in an ion channel gene. Nature. 2016;536(7616):329–32. 10.1038/nature19093 . [DOI] [PubMed] [Google Scholar]
- 11.Signor SA, Nuzhdin SV. The Evolution of Gene Expression in cis and trans. Trends in Genetics. 2018;34(7):532–44. 10.1016/j.tig.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prud'homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proceedings of the National Academy of Sciences of the United States of America. 2007;104 Suppl 1:8605–12. 10.1073/pnas.0700488104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wray GA. The evolutionary significance of cis-regulatory mutations. Nature reviews Genetics. 2007;8(3):206–16. 10.1038/nrg2063 . [DOI] [PubMed] [Google Scholar]
- 14.Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85 10.1038/nature02698 https://www.nature.com/articles/nature02698#supplementary-information. [DOI] [PubMed] [Google Scholar]
- 15.McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome research. 2010;20(6):816–25. Epub 2010/04/01. 10.1101/gr.102491.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi X, Ng DW, Zhang C, Comai L, Ye W, Chen ZJ. Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat Commun. 2012;3:950 Epub 2012/07/19. 10.1038/ncomms1954 . [DOI] [PubMed] [Google Scholar]
- 17.Konishi M. The role of auditory feedback in the control of vocalization in the white-crowned saprrow. Zeitschrift fur Tierpsychologie. 1965;22:770–83. [PubMed] [Google Scholar]
- 18.Doupe AJ, Kuhl PK. Birdsong and human speech: common themes and mechanisms. Annu Rev Neurosci. 1999;22:567–631. Epub 1999/04/15. 10.1146/annurev.neuro.22.1.567 . [DOI] [PubMed] [Google Scholar]
- 19.Wheatcroft D, Qvarnstrom A. A blueprint for vocal learning: auditory predispositions from brains to genomes. Biology letters. 2015;11(8). 10.1098/rsbl.2015.0155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nowicki S, Searcy WA. Song function and the evolution of female preferences: why birds sing, why brains matter. Annals of the New York Academy of Sciences. 2004;1016:704–23. Epub 2004/08/18. 10.1196/annals.1298.012 . [DOI] [PubMed] [Google Scholar]
- 21.Jarvis ED. Learned birdsong and the neurobiology of human language. Annals of the New York Academy of Sciences. 2004;1016:749–77. Epub 2004/08/18. 10.1196/annals.1298.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nottebohm F, Stokes TM, Leonard CM. Central control of song in the canary, Serinus canarius. The Journal of comparative neurology. 1976;165(4):457–86. Epub 1976/02/15. 10.1002/cne.901650405 . [DOI] [PubMed] [Google Scholar]
- 23.Hahnloser RH, Kozhevnikov AA, Fee MS. An ultra-sparse code underlies the generation of neural sequences in a songbird. Nature. 2002;419(6902):65–70. Epub 2002/09/06. 10.1038/nature00974 . [DOI] [PubMed] [Google Scholar]
- 24.Sober SJ, Wohlgemuth MJ, Brainard MS. Central contributions to acoustic variation in birdsong. The Journal of neuroscience. 2008;28(41):10370–9. 10.1523/JNEUROSCI.2448-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haesler S, Wada K, Nshdejan A, Morrisey EE, Lints T, Jarvis ED, et al. FoxP2 expression in avian vocal learners and non-learners. The Journal of neuroscience. 2004;24(13):3164–75. Epub 2004/04/02. 10.1523/JNEUROSCI.4369-03.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wada K, Hayase S, Imai R, Mori C, Kobayashi M, Liu WC, et al. Differential androgen receptor expression and DNA methylation state in striatum song nucleus Area X between wild and domesticated songbird strains. The European journal of neuroscience. 2013. 10.1111/ejn.12258 . [DOI] [PubMed] [Google Scholar]
- 27.Asogwa NC, Mori C, Sanchez-Valpuesta M, Hayase S, Wada K. Inter- and intra-specific differences in muscarinic acetylcholine receptor expression in the neural pathways for vocal learning in songbirds. The Journal of comparative neurology. 2018;526(17):2856–69. Epub 2018/09/11. 10.1002/cne.24532 . [DOI] [PubMed] [Google Scholar]
- 28.Hooper DM, Price TD. Rates of karyotypic evolution in Estrildid finches differ between island and continental clades. Evolution. 2015;69(4):890–903. 10.1111/evo.12633 [DOI] [PubMed] [Google Scholar]
- 29.Forshaw JM, Shephard M. Grassfinches in Australia: CSIRO Publishing; 2012. [Google Scholar]
- 30.Imai R, Sawai A, Hayase S, Furukawa H, Asogwa CN, Sanchez M, et al. A quantitative method for analyzing species-specific vocal sequence pattern and its developmental dynamics. Journal of neuroscience methods. 2016. 10.1016/j.jneumeth.2016.06.023 . [DOI] [PubMed] [Google Scholar]
- 31.Tchernichovski O, Nottebohm F, Ho CE, Pesaran B, Mitra PP. A procedure for an automated measurement of song similarity. Animal behaviour. 2000;59(6):1167–76. Epub 2000/07/06. 10.1006/anbe.1999.1416 . [DOI] [PubMed] [Google Scholar]
- 32.Marler P, Peters S. Selective vocal learning in a sparrow. Science. 1977;198(4316):519–21. Epub 1977/11/04. 10.1126/science.198.4316.519 . [DOI] [PubMed] [Google Scholar]
- 33.Eales LA. Song learning in zebra finches: some effects of song model availability on what is learnt and when. Animal behaviour. 1985;33:1293–300. [Google Scholar]
- 34.Clayton NS. The effects of cross-fostering on selective song learning in estrildid finches. Behaviour. 1988;109:163–75. [Google Scholar]
- 35.Gardner TJ, Naef F, Nottebohm F. Freedom and rules: the acquisition and reprogramming of a bird's learned song. Science. 2005;308(5724):1046–9. Epub 2005/05/14. 10.1126/science.1108214 . [DOI] [PubMed] [Google Scholar]
- 36.Frésard L, Leroux S, Servin B, Gourichon D, Dehais P, Cristobal MS, et al. Transcriptome-wide investigation of genomic imprinting in chicken. Nucleic acids research. 2014;42(6):3768–82. 10.1093/nar/gkt1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goncalves A, Leigh-Brown S, Thybert D, Stefflova K, Turro E, Flicek P, et al. Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome research. 2012;22(12):2376–84. Epub 2012/08/25. 10.1101/gr.142281.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tirosh I, Reikhav S, Levy AA, Barkai N. A Yeast Hybrid Provides Insight into the Evolution of Gene Expression Regulation. 2009;324(5927):659–62. 10.1126/science.1169766 [DOI] [PubMed] [Google Scholar]
- 39.Fraser HB. Genome-wide approaches to the study of adaptive gene expression evolution: systematic studies of evolutionary adaptations involving gene expression will allow many fundamental questions in evolutionary biology to be addressed. BioEssays. 2011;33(6):469–77. 10.1002/bies.201000094 . [DOI] [PubMed] [Google Scholar]
- 40.Fraser HB. Improving Estimates of Compensatory cis-trans Regulatory Divergence. Trends in genetics: TIG. 2018. 10.1016/j.tig.2018.09.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Emerson JJ. Inferring Compensatory Evolution of cis- and trans-Regulatory Variation. Trends in genetics: TIG. 2019;35(1):1–3. Epub 2018/12/07. 10.1016/j.tig.2018.11.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ingram M, Wozniak EAL, Duvick L, Yang R, Bergmann P, Carson R, et al. Cerebellar Transcriptome Profiles of ATXN1 Transgenic Mice Reveal SCA1 Disease Progression and Protection Pathways. Neuron. 2016;89(6):1194–207. Epub 2016/03/08. 10.1016/j.neuron.2016.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medugorac I, Graf A, Grohs C, Rothammer S, Zagdsuren Y, Gladyr E, et al. Whole-genome analysis of introgressive hybridization and characterization of the bovine legacy of Mongolian yaks. Nature genetics. 2017;49(3):470–5. Epub 2017/01/31. 10.1038/ng.3775 . [DOI] [PubMed] [Google Scholar]
- 44.Li XC, Jarvis ED, Alvarez-Borda B, Lim DA, Nottebohm F. A relationship between behavior, neurotrophin expression, and new neuron survival. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(15):8584–9. Epub 2000/07/13. 10.1073/pnas.140222497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wada K, Howard JT, McConnell P, Whitney O, Lints T, Rivas MV, et al. A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(41):15212–7. Epub 2006/10/05. 10.1073/pnas.0607098103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andalman AS, Fee MS. A basal ganglia-forebrain circuit in the songbird biases motor output to avoid vocal errors. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(30):12518–23. Epub 2009/07/15. 10.1073/pnas.0903214106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bendesky A, Kwon Y-M, Lassance J-M, Lewarch CL, Yao S, Peterson BK, et al. The genetic basis of parental care evolution in monogamous mice. Nature. 2017. 10.1038/nature22074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Combs PA, Krupp JJ, Khosla NM, Bua D, Petrov DA, Levine JD, et al. Tissue-Specific cis-Regulatory Divergence Implicates eloF in Inhibiting Interspecies Mating in Drosophila. Current biology. 2018;28(24):3969–75 e3. Epub 2018/12/07. 10.1016/j.cub.2018.10.036 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Bono M, Bargmann CI. Natural Variation in a Neuropeptide Y Receptor Homolog Modifies Social Behavior and Food Response in C. elegans. Cell. 1998;94(5):679–89. 10.1016/s0092-8674(00)81609-8 [DOI] [PubMed] [Google Scholar]
- 50.Gomes S, Civetta A. Hybrid male sterility and genome-wide misexpression of male reproductive proteases. Scientific reports. 2015;5:11976 Epub 2015/07/07. 10.1038/srep11976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Werren JH, Clark AG. Allele-Specific Transcriptome and Methylome Analysis Reveals Stable Inheritance and Cis-Regulation of DNA Methylation in Nasonia. PLoS Biol. 2016;14(7):e1002500 Epub 2016/07/06. 10.1371/journal.pbio.1002500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson JH, Balakrishnan CN. Gene Regulatory Evolution During Speciation in a Songbird. G3 (Bethesda). 2016;6(5):1357–64. Epub 2016/03/16. 10.1534/g3.116.027946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park H, Poo MM. Neurotrophin regulation of neural circuit development and function. Nature reviews Neuroscience. 2013;14(1):7–23. Epub 2012/12/21. 10.1038/nrn3379 . [DOI] [PubMed] [Google Scholar]
- 54.Nagappan G, Lu B. Activity-dependent modulation of the BDNF receptor TrkB: mechanisms and implications. Trends in neurosciences. 2005;28(9):464–71. Epub 2005/07/26. 10.1016/j.tins.2005.07.003 . [DOI] [PubMed] [Google Scholar]
- 55.Berardi N, Pizzorusso T, Maffei L. Critical periods during sensory development. Current opinion in neurobiology. 2000;10(1):138–45. 10.1016/s0959-4388(99)00047-1 [DOI] [PubMed] [Google Scholar]
- 56.Schuman EM. Neurotrophin regulation of synaptic transmission. Current opinion in neurobiology. 1999;9(1):105–9. 10.1016/s0959-4388(99)80013-0 [DOI] [PubMed] [Google Scholar]
- 57.Akutagawa E, Konishi M. Transient expression and transport of brain-derived neurotrophic factor in the male zebra finch’s song system during vocal development. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(19):11429–34. 10.1073/pnas.95.19.11429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson F, Hohmann SE, DiStefano PS, Bottjer SW. Neurotrophins suppress apoptosis induced by deafferentation of an avian motor-cortical region. The Journal of neuroscience. 1997;17(6):2101–11. Epub 1997/03/15. 10.1523/JNEUROSCI.17-06-02101.1997 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayase S, Wang H, Ohgushi E, Kobayashi M, Mori C, Horita H, et al. Vocal practice regulates singing activity-dependent genes underlying age-independent vocal learning in songbirds. PLoS Biol. 2018;16(9):e2006537 10.1371/journal.pbio.2006537 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dittrich F, Ter Maat A, Jansen RF, Pieneman A, Hertel M, Frankl-Vilches C, et al. Maximized song learning of juvenile male zebra finches following BDNF expression in the HVC. The European journal of neuroscience. 2013;38(9):3338–44. Epub 2013/08/13. 10.1111/ejn.12329 . [DOI] [PubMed] [Google Scholar]
- 61.Kittelberger JM, Mooney R. Acute injections of brain-derived neurotrophic factor in a vocal premotor nucleus reversibly disrupt adult birdsong stability and trigger syllable deletion. Journal of neurobiology. 2005;62(4):406–24. Epub 2004/11/18. 10.1002/neu.20109 . [DOI] [PubMed] [Google Scholar]
- 62.Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, et al. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. The Journal of neuroscience. 2004;24(18):4401–11. Epub 2004/05/07. 10.1523/JNEUROSCI.0348-04.2004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, et al. The BDNF val66met Polymorphism Affects Activity-Dependent Secretion of BDNF and Human Memory and Hippocampal Function. Cell. 2003;112(2):257–69. 10.1016/s0092-8674(03)00035-7 [DOI] [PubMed] [Google Scholar]
- 64.Giza JI, Kim J, Meyer HC, Anastasia A, Dincheva I, Zheng CI, et al. The BDNF Val66Met Prodomain Disassembles Dendritic Spines Altering Fear Extinction Circuitry and Behavior. Neuron. 2018;99(1):163–78 e6. Epub 2018/06/19. 10.1016/j.neuron.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brainard MS, Doupe AJ. Interruption of a basal ganglia-forebrain circuit prevents plasticity of learned vocalizations. Nature. 2000;404(6779):762–6. Epub 2000/04/28. 10.1038/35008083 . [DOI] [PubMed] [Google Scholar]
- 66.Luo M, Perkel D. Long-range GABAergic projection in a circuit essential for vocal learning. The Journal of comparative neurology. 1999;403(1):68–84. [PubMed] [Google Scholar]
- 67.Reiner A, Perkel DJ, Bruce LL, Butler AB, Csillag A, Kuenzel W, et al. Revised nomenclature for avian telencephalon and some related brainstem nuclei. The Journal of comparative neurology. 2004;473(3):377–414. 10.1002/cne.20118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bottjer SW, Miesner EA, Arnold AP. Forebrain lesions disrupt development but not maintenance of song in passerine birds. Science. 1984;224(4651):901–3. Epub 1984/05/25. 10.1126/science.6719123 . [DOI] [PubMed] [Google Scholar]
- 69.Scharff C, Nottebohm F. A comparative study of the behavioral deficits following lesions of various parts of the zebra finch song system: implications for vocal learning. The Journal of neuroscience. 1991;11(9):2896–913. Epub 1991/09/01. 10.1523/JNEUROSCI.11-09-02896.1991 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Warren TL, Tumer EC, Charlesworth JD, Brainard MS. Mechanisms and time course of vocal learning and consolidation in the adult songbird. J Neurophysiol. 2011;106(4):1806–21. 10.1152/jn.00311.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marler P, Sherman V. Song structure without auditory feedback: emendations of the auditory template hypothesis. The Journal of Neuroscience. 1983;3(3):517–31. 10.1523/JNEUROSCI.03-03-00517.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mori C, Wada K. Audition-Independent Vocal Crystallization Associated with Intrinsic Developmental Gene Expression Dynamics. Journal of Neuroscience. 2015;35(3):878–89. 10.1523/JNEUROSCI.1804-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T, et al. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome biology. 2013;14(4):R31 10.1186/gb-2013-14-4-r31 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eckersley-Maslin MA, Spector DL. Random monoallelic expression: regulating gene expression one allele at a time. Trends in Genetics. 2014. 10.1016/j.tig.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jarvis ED, Nottebohm F. Motor-driven gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(8):4097–102. Epub 1997/04/15. 10.1073/pnas.94.8.4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hayase S, Wada K. Singing activity-driven Arc expression associated with vocal acoustic plasticity in juvenile songbird. The European journal of neuroscience. 2018. Epub 2018/06/24. 10.1111/ejn.14057 . [DOI] [PMC free article] [PubMed] [Google Scholar]